Abstract
Syndemics framework describes two or more co-occurring epidemics that synergistically interact with each other and the complex structural social forces that sustain them leading to excess disease burden. The term syndemic was first used to describe the interaction between substance abuse, violence, and AIDS by Merrill Singer. A broader range of syndemic studies has since emerged describing the framework’s applicability to other public health scenarios. With syndemic theory garnering significant attention, the focus is shifting towards developing robust empirical analytical approaches. Unfortunately, the complex nature of the disease-disease interactions nested within several social contexts complicates empirical analyses. In answering the call to analyze syndemics at the population level, we propose the use of spatial epidemiology as an empirical framework for syndemics research.
Spatial epidemiology, which typically relies on geographic information systems (GIS) and statistics, is a discipline that studies spatial variations to understand the geographic landscape and the risk environment within which disease epidemics occur. GIS maps provide visualization aids to investigate the spatial distribution of disease outcomes, the associated social factors, and environmental exposures. Analytical inference, such as estimation of disease risks and identification of spatial disease clusters, can provide a detailed statistical view of spatial distributions of diseases. Spatial and spatiotemporal models can help us to understand, measure, and analyze disease syndemics as well as the social, biological, and structural factors associated with them in space and time.
In this paper, we present a background on syndemics and spatial epidemiological theory and practice. We then present a case study focused on the HIV and HCV syndemic in West Virginia to provide an example of the use of GIS and spatial analytical methods. The concepts described in this paper can be considered to enhance understanding and analysis of other syndemics for which space-time data are available.
Keywords: Spatial epidemiology, Syndemics, Geographic information system (GIS)
1. Introduction
Mainstream biomedicine historically conceptualized disease as an aberration from the generalized state of health with an observable set of symptoms that separated it from other conditions, creating a “bounded entity,” thereby simplifying the process of diagnosis and treatment of the disease (Singer and Clair, 2003). Epidemiologists and public health researchers have played a significant role in suggesting that the approach was too rigid to evaluate the new wave of “social diseases” in the post-industrial world. Recent developments in the field of medical anthropology further challenge this “bounded entity” approach and describe a disease as a physical manifestation of a broader set of issues that include more than an individual’s body (Gaines, 1998; Krieger, 2001). These advances in health research have been partly influenced by the failure of medicine to adequately address the ailments of a modern society, including the diseases of addiction and depression. Broadly termed as a “biocultural approach,” recent efforts examine the interaction of social, cultural, economic, and political environments along with proximal components of health, describing a disease as a result of these complex interactions in an individual (McElroy, 1990; Wiley and Allen, 2009; Singer, 2000). Epidemics, defined as “the occurrence of more cases of a disease than expected in a given area or among a specific group of people over a particular period of time” uses a singular notion of a disease. However, as with many diseases of modern society where many epidemics exist simultaneously and are intertwined with each other, a broader term that describes the dynamic system is needed. The term “syndemic” fits within this biocultural framework, referring to two or more co-occurring epidemics that show synergistic interactions that contribute to the burden of disease (Singer and Clair, 2003; Singer, 2000). Originating from the concepts of co-infection, where health outcomes in individuals were significantly poorer when two or more infections occurred at the same time (e.g., tuberculosis and HIV) (Corbett et al., 2003; Pawlowski et al., 2012), the syndemics framework explains how two or more epidemics interact with each other with an emphasis on the context of the surrounding environment in which these epidemics occur.
The term syndemic was first used to describe the interaction between substance abuse, violence, and AIDS (SAVA) in inner cities by Merrill Singer (2000). Singer observed that substance use was linked with increased risk of HIV transmission and disease progression; domestic violence was associated with substance use and could exacerbate progression of disease when linked with poor health care access, poverty, homelessness and lack of support/stigma from family members. This examination of these three health and social issues, aptly named the SAVA syndemic, illustrates how socioeconomic inequities acted as an accelerant to violence, substance use, and transmission of communicable diseases such as HIV. Following the description of the SAVA syndemic, a broad range of studies emerged that examined the complex interaction between social factors and health conditions. Poor sanitary and working conditions further aided by pathogen-pathogen interaction with acanthamoeba (which causes encephalitis) have resulted in outbreaks of Legionnaire’s disease in areas that create hospitable environments for such interactions such as manufacturing plants and water treatment facilities contributing to a complex pathogen-pathogen and pathogen-environment interaction (Rowbotham, 1980; Herwaldt et al., 1984). Co-infection of HIV and Mycobacterium tuberculosis also emerged in the syndemic literature, with the combined detrimental effects of both infections accelerating deterioration of health (Corbett et al., 2003; Pawlowski et al., 2012). Syndemics theory has also been applied to the domain of non-communicable diseases; violence, immigration, depression, diabetes, and abuse, termed the VIDDA syndemic, highlights the interaction between socio-political processes and diseases such as diabetes and depression (Mendenhall, 2012). The study of substance use, adolescent sexual abuse, violence, internalized homonegativity, and depression among Tanzanian men who have sex with men utilized a syndemic theory framework and has been termed the SAVID syndemic (Adeboye et al., 2017). The application of syndemic theory over the course of almost two decades, within a wide spectrum of multi-epidemic research studies, continues to emphasize the importance of evaluating the interaction and the context of co-occurring epidemics to comprehend what is happening locally, from a public health and clinical perspective, and to inform improved development of intervention and policy frameworks (Singer and Clair, 2003).
With syndemic theory garnering significant attention, the focus has now shifted to the development of robust mixed-methods approaches to evaluate syndemics. The complex nature of the disease-disease interactions within a sophisticated web of underlying social forces makes a robust empirical analysis challenging. Such efforts are necessary to guide policies and interventions aimed at addressing syndemics. However, evidence indicates that empirical approaches have lagged theory in syndemics research. A systematic review concluded that the empirical specification of syndemics in studies of psychosocial health and HIV risk was inconsistent with the definition of the underlying theory (Tsai and Burns, 2015). These claims have largely been made due to the increasing use of the additive forms of predictors in a regression framework, which estimates exposure as a sum of health risk indicators at an individual levelto describe risk for a specific health outcome (Stall et al., 2003, 2015). Critics of such a “sum score approach” in the empirical syndemics literature decry the use of this method, stating that it does not adhere to the theory of syndemics as the sum score approach does not consider the interaction between health risks (Tsai and Burns, 2015; Tsai and Venkataramani, 2016; Tsai, 2018). Proponents of the “sum score approach” argue that their results lend credibility to syndemics theory, and the complex nature of the interaction between epidemics should go beyond adding an interaction term in statistical models, suggesting a need for a robust longitudinal study to examine the emergence of syndemics (Stall et al., 2015).
Before we delve deeper into exploring empirical methodologies, it is crucial to understand the three core concepts underlying a syndemic. The first is the presence of two or more co-occurring epidemics, the second is the interaction between those epidemics that results in synergistic impacts on health outcomes (morbidity, mortality, change in quality of life), and third is the role of the larger socio-economic environment that facilitates the interaction between these epidemics (Tsai et al., 2017). Syndemic studies are broadly operationalized into three different models of co-occurring epidemics: a) mutually causal epidemics: where two or more epidemics are risk factors for each other (e. g.: the Substance Abuse, Violence and AIDS syndemic (Singer, 2000)), b) synergistically interacting epidemics: where the combined burden of each individual epidemic exceeds the sum of two (or more) individual epidemics (e.g.: synergistic effects of violence by partners and substance use (crystal meth) on sexual behaviors (Stoicescu et al., 2019)), and c) serially causal epidemics: in which a series of accumulating health risks are linked together sometimes in a non-additive fashion (e.g., examination of chronic diseases as a results of cumulative exposures over the course of life (Kuh and Shlomo, 2004; Tsai, 2018)). The present challenge is that the three different models have various degrees of agree-ment with the initial concepts put forth by Singer and therefore reach very different conclusions about how each syndemic should be addressed. While there is no consensus on what method should be used to model a syndemic, researchers agree that a nuanced exploration of disease-disease or disease-environment interaction under a given set of socioeconomic conditions is needed (Tsai and Burns, 2015; Stall et al., 2015; Tsai, 2018). It is very likely that any proposed framework will need to be flexible, enabling modifications to be able to study various syndemics.
Among other methods employed to assess syndemics quantitatively, Tsai calls for a multilevel approach to the quantitative synthesis of syndemics research that includes analysis of factors that “go beyond the level of the individual. (Tsai, 2018)” Complex interactions between socioeconomic risk factors can make a populace vulnerable to multiple diseases. As the underlying socioeconomic factors often have unique spatial distributions, understanding how these factors interact in the physical space could be vital to the study of syndemics. Geographic Information Systems (GIS) and their uptake in epidemiologic research have allowed researchers to incorporate spatially-oriented data in assessing the relationship between environment (both social and geographical) and health (Lawson et al., 2016; Lawson, 2013a; Waller and Gotway, 2004). GIS and spatial statistical modeling have been used for a broad range of public health studies ranging from the study of infectious diseases such as cholera (Ali et al., 2002), HIV (Meyers et al., 2014), HCV (Stopka et al., 2017a), and tuberculosis (Vindenes et al., 2018) to non-communicable diseases such as cancer (Schmid and Held, 2004; Brewer, 2006), diabetes (Chen et al., 2014; Geraghty et al., 2010), and chronic obstructive pulmonary diseases (Wang et al., 2015; Nuvolone et al., 2011). It is also increasingly used to inform development, enhancement, and implementation of a variety of public health initiatives ranging from preventative measures (Stopka et al., 2017b) to harm reduction interventions to combat the detrimental effects of substance use and misuse (Davidson et al., 2011). Use of GIS in spatial epidemiology has allowed researchers to examine where diseases are clustered together and how they originate under similar circumstances (Stopka et al., 2017a, 2019a; Meyers et al., 2014). These studies share common features with syndemics research, such as the study of disease clustering and evaluation of the underlying socioeconomic environments that are associated with the emergence of diseases. The possibility of such critical examination of co-occurring epidemics and the underlying environment that fosters such maladies indicates that spatial epidemiology can lay a strong empirical foundation for syndemics research.
However, examples of the utilization of GIS and other spatial methods in syndemics research are limited. Recent studies have aimed to discern the interaction between epidemiology, policy, and the legal milieu that surround disease transmission and overdose risks (Stopka et al., 2019b; Furr-Holden et al., 2016; McLuckie et al., 2019). In other research avenues, Livingston et al. used a mixed-methods approach to study migration, tourism labor, and health outcomes in the Dominican Republic incorporating aspects of GIS and syndemic theory (Livingston et al., 2016). Ramirez et al., described the effect of El Nino on multi-disease risk in Peru, utilizing the term ‘ecosyndemics’ referring to syndemic of infectious diseases mediated by climate change extremes (Ramirez et al., 2018). Recent studies on HIV vulnerability assessments, while not explicitly labeled as syndemics research, have made concerted efforts to incorporate geographical evaluation of underlying social and environmental factors as measures of vulnerability to transmission of diseases related to injection drug use (Van Handel et al., 2016; Rickles et al., 2018). While these studies have made significant strides in employing multilevel approaches (e.g.: hierarchical, nested, and mixed effects models) to study syndemics, future research in this field warrants a robust empirical framework that links syndemic theory to spatial epidemiology. Our objective is to provide an overview of spatial epidemiologic methodologies to examine the complicated relationship between epidemics, thereby providing a new framework for empirical evaluation of syndemics. In the following sections, we provide a brief introduction to spatial epidemiologic methods as applied to syndemics research. We describe spatial epidemiologic data and methods by drawing comparisons with non-spatial data. We then present a case study to elaborate on the applications of spatial epidemiologic methods for syndemics research.
2. A conceptual framework for empirical analysis using spatial epidemiology
2.1. Understanding spatial data
Spatial epidemiology, as previously described, pertains to the study of geographic variations in health conditions with respect to the surrounding risk factors (Ostfeld et al., 2005). The field of study utilizes tools such as GIS to collect, process, and analyze geographically referenced health data to study various diseases and health conditions. GIS-based analysis commonly employs two types of data: 1) vector data (e.g., points, lines, and polygons), and 2) raster data (e.g., grid-based, pixelated). Vector data are commonly used to denote locations of specific events (points), street or road networks (lines), or administrative boundaries (polygons). Raster data are primarily used to visualize data continuously as they change over a surface area (e.g., density maps, heat maps) especially with satellite imagery. The differences between these types of GIS data are integral to their visualization, storage, and use in data analyses.
Health-related data are collected in a variety of different structures and formats. In their raw format, they can take the form of point vectorbased data using an address (e.g., an individual’s residential or work address) or latitudinal and longitudinal measures (e.g., the location where tick bites occur, the location of an opioid overdose), or aggregated polygon level data (e.g., total tick bite counts in a rural town in New England, prevalence of heart disease in Suffolk County, Massachusetts, or HIV infection rates in the state of California). Other demographic, sociocultural, economic, and geographic data can also have a high degree of variation in how they are collected and utilized for research, with varying levels of spatial granularity (e.g., census tract, ZIP Code, municipality, county, state-level). Syndemics research focuses on a multilevel interaction between multiple diseases and/or health conditions (i.e., bio-bio: interaction between acanthamoeba and legionella (Rowbotham, 1980), HIV and HCV coinfection (Singer, 2010)) and the environment within which syndemics thrive. GIS and spatial epidemiology provide tools to facilitate measurement of these outcomes and exposures, as well as generation of visual representations of syndemics in space and time, and they also enable researchers to employ advanced spatiotemporal modeling to analyze and predict syndemics.
2.2. Examining spatial epidemiologic methods
We envision the conceptual framework for syndemics research using spatial epidemiology to similarly span three domains: a) descriptive mapping to assess the burden of disease and exploratory spatial data analysis (ESDA) b) spatial epidemiology to study co-occurrence of intertwined epidemics and c) spatial analyses to assess syndemics.
a). Using descriptive maps to visualize the burden of disease and ESDA:
Various types of maps have been used in epidemiologic research to visualize health data as an explorative approach to identify their geographic patterns. We can broadly classify commonly used maps in health studies into two categories: thematic maps and point vectorbased maps. Thematic maps depict aggregated features within a study area. Examples of thematic maps include choropleth, dot-density, proportional symbols, and graduated symbol maps. Point vectors maps, on the other hand, present address level or geographical point level data (i. e., “pushpins”), which can represent health resources such as hospitals, clinics, health centers or points of economic activities such as shops, workplace or factories, location of residence or sources of health risks such as large industries, or the point source of a specific contaminant. Visual inspection of the incidence or prevalence of two or more separate diseases or health conditions can direct researchers towards the presence of co-occurring epidemics, paving the way for advanced analysis to assess whether a syndemic is present empirically.
ESDA can be considered as a spatial extension of exploratory data analysis (EDA). Like EDA, which is an aggregation of techniques that are used to examine the distribution, moments, and statistically significant aspects of data, ESDA uses a collection of tools to analyze and explore spatial data. These tools can include visual representation or statistical inferences of the distribution of the data. While these techniques do not cross over to correlational analysis or bivariate hypothesis testing, they are widely used to detect spatial patterns, locate outliers and clusters, formulate a hypothesis for subsequent statistical tests, and guide spatial regression models. As in the case of EDA, where the underlying distribution of data is critical to the choice of statistical tests and the formulation of statistical models, ESDA is integral to any spatial modeling and hypothesis testing.
Spatial autocorrelation analysis allows us to look at similarities in values that are near each other. Measures of spatial autocorrelation can be categorized as global indicators of spatial clustering, or local indicators of spatial association (LISA) (Anselin, 1995a). Measures of global spatial autocorrelation assess overall spatial correlation within a dataset on a macro level (e.g., state or county level), and can be measured by the Moran’s I statistic (Cliff and Ord, 1981; Moran, 1948). The construct of Moran’s I is quite similar to a coefficient of correlation; the correlation is measured between the value of a variable and its surrounding features (also known as “spatial lags”). LISA is widely used to assess the significance of clustering on the local level, i.e., to identify where clusters are occurring. Local spatial autocorrelation analysis can help identify hotspots (areas that have higher numbers of events than the expected average number of events), coldspots (areas that have fewer numbers of events than the expected average number of events) and outliers (very high or very low values in relation to all the values in adjacent polygons) that exist spatially. By identifying clusters, we can determine the locations where diseases are showing unusual patterns of incidence, thereby indicating a need for further investigation. Univariate local Moran’s I (Anselin, 1995b), local Geary’s C (Anselin, 1995b), and the Getis-Ord Gi* (Getis and Ord, 2010) statistics are some commonly used indicators for local spatial autocorrelation and clustering.
b). Using spatial epidemiology to study the co-occurrence of intertwined epidemics:
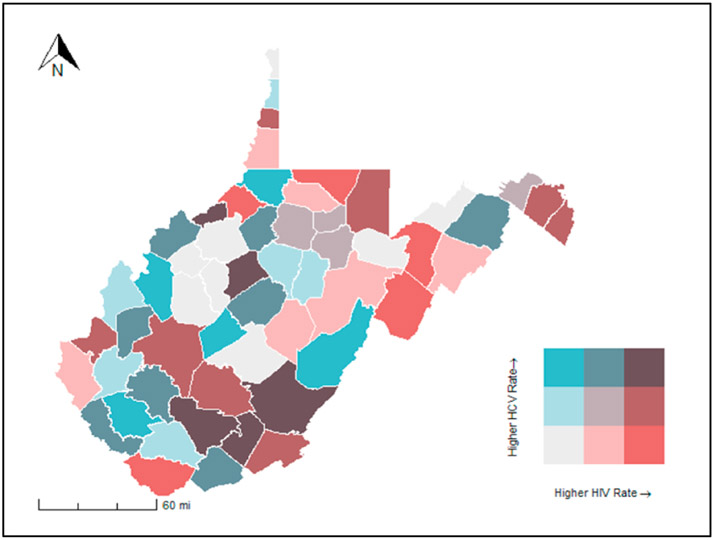
The presence of co-occurring epidemics is one of the core principles of the syndemics theory. Therefore, researchers must be able to assess the spatial relationship between two or more epidemic outcomes. Visually, the relationship between any two features can be observed by overlaying different thematic maps on top of one another. Other visual methods for assessing bivariate relationships include bivariate choropleth maps and bivariate heat maps (Fig. 1). Further, bivariate spatial autocorrelation can be measured using Bivariate Moran’s I and an extension of Geary’s C can be used to measure multivariate spatial autocorrelation (Anselin, 2019; Lee, 2001).
Fig. 1.
HIV and HCV rates in West Virginia by county, 2016–2017. A color matrix of all possible combinations of HIV and HCV rates (high, medium, low) per 100,000 has been used in this figure to display the relationship between these variables.
c). Spatial analysis for understanding syndemics:
After bivariate spatial associations between variables of interest have been explored, we can then focus on constructing a spatial regression model to explain the outcome of interest based on a set of predictors. Consider an ordinary least squares (OLS) model with Xi predictors and outcome Y. One of the assumptions in the OLS model is that the error term is independently and identically distributed. However, it is possible that the error terms in a regression model could be spatially autocorrelated, which would violate the assumption of OLS and affect the estimates of the regression model (Dubin, 1988). A variety of different approaches have been explored to control for the spatial autocorrelation of the error terms, and different models such as spatial lag models and spatial error models have been utilized to account for these effects (Beale et al., 2010).
The spatial lag and spatial error coefficient determine how the spatial distribution of events are related. For example, when the spatial lag coefficient, is positive, it indicates that the value of an outcome in any area is expected to be higher if, on average, neighboring areas also have higher values. On the other hand, when either the spatial lag coefficient or the spatial error coefficient is zero, it means that there are no spatial effects and these spatial models will equate to the simple OLS model. While the technical specifications of these spatial regression models are beyond the scope of this paper, we would like to acknowledge the importance of considering these spatial regression models for syndemics research as we explore the association between spatially co-occurring epidemics. Furthermore, as we explore health outcomes such as disease counts, other methods such as the Poisson regression may need to be employed. Additionally, for over-dispersed count data (when the assumptions of Poisson models are violated), models that integrate a dispersion parameter such as negative binomial regression may need to be used. Hurdle models and Zero-inflated Poisson models have also been developed for situations in which the observed data include a large number of zeros.
The spatial lag and spatial error regression approaches described above belong to the simultaneous autoregressive (SAR) model family which is commonly used in spatial econometrics (LeSage and Pace, 2009). Conditional autoregressive (CAR) models incorporate more flexibility in specifying spatial covariance matrices, which estimate values in an area conditional on the values of the neighbors through a spatial weights matrix (Ver Hoef et al., 2018). Many CAR models (Besag et al., 1991; Besag and Kooperberg, 1995) have been developed using a Bayesian framework implemented via Markov chain Monte Carlo (MCMC), which is computationally expensive. Other computation approaches, such as the integrated nested Laplace approximation (INLA) (Blangiardo and Cameletti, 2015), have been recently proposed as alternative methods for implementation and gained substantial popularity. Government disease surveillance data, which are often in the form of spatially aggregated disease counts, are routinely analyzed using these models. In the univariate case, a spatial CAR model is specified by first assuming the appropriate distribution (Gaussian, Binomial, Poisson) for the data, and then a correlation structure on the spatial random effects (Lee, 2017). Specifically, by letting i = 1, 2, …, I indicate geographic areas and Yi the outcome measure in the ith area, we consider the following spatial model with a given link function g(.),
where α is the intercept, x is the set of covariates with associated coefficients β, and ui the spatial random effects. For normally distributed outcome measures, the link function is the identity function g(E(Yi)) = E(Yi). For count measures, the link function will be the log function.
The spatial effects are constructed using the neighborhood structure, for example, if two areas share the same geographic boundary they are considered as neighbors. This approach is commonly used but other distance-based approaches are also possible. In our case, the conditional distribution of ui on all its neighbors (Leroux et al., 2000) follows
where, ρ is the parameter with values between 0 and 1 for spatial dependency, and τ2 is the variance parameter, mi is the number of area i.
The multivariate conditional autoregressive (MCAR) model has also been developed to analyze multiple disease outcomes jointly, for example, on lung cancer mortality (Richardson et al., 2006). Compared to the routine use of univariate CAR models in disease mapping (Lawson, 2013b), MCAR models are quite underused in practice, particularly in research of co-occurring epidemics. This is likely due to model complexity, and the lack of easy-to-use statistical software. Therefore, we present a case study below where we examine the co-occurrence of HIV and HCV infection under the syndemics framework. We begin with descriptive maps and build up to complex models to demonstrate how co-occurrence can be examined. We used Moran’s I and Geary’s C to examine spatial univariate and bivariate spatial autocorrelation. Then we used the carBayes package to fit the spatial model (Lee, 2017). It is our hope that the step-by-step guidance of the analysis and model development can serve as a foundation for syndemics analyses using a spatial epidemiological framework.
3. Syringe mediated syndemics: The West Virginia HIV-HCV syndemic case study
3.1. Background
We build on the earlier work of Bulled and Singer on “Syringe-Mediated Syndemics” (Bulled and Singer, 2011), assessing how infectious diseases such as HIV and HCV proliferate in the midst of the opioid crisis and a mix of socioeconomic factors that co-occur in local communities. In the following sections, we first provide a brief overview of syringe mediated syndemics. Next, we discuss transmission of infectious diseases with an emphasis on the recent resurgence of HIV and HCV infections in injection drug-using communities. Finally, we present an analytical framework using thematic mapping building up to advanced spatial modeling of syndemic social, environmental, and biological disease interactions.
Despite our understanding of the relationship between syringe sharing and transmission of infectious diseases, the recent resurgence of HIV within drug injecting communities (Conrad et al., 2015; Peters et al., 2016; Cranston et al., 2019) reminds us that systemic co-occurring risks and public health service deficiencies can lead to increases in the incidence of infectious diseases in communities at risk. In 2017, 35,000 individuals were diagnosed with HIV, and the CDC estimated that approximately 1 in 11 new HIV infections occurred in injection drug users (Centers for Disease Contr, 2017). HIV outbreaks have become increasingly prevalent among people who inject drugs (PWID) in recent years. Large spikes in HIV infections have been correlated with injection drug use (IDU) in Scott County, Indiana (Conrad et al., 2015; Peters et al., 2016), Cabell County, West Virginia (Evans et al., 2018; Bradley et al., 2019), and the Merrimack Valley of Massachusetts (Cranston et al., 2019). Similarly, rates of acute HCV infection have steadily climbed over the past decade doubling from 0.3 per 100,000 in 2004 to 0.7 cases per 100,000 in 2014 (Zibbell et al., 2018). In comparison, the rate of acute HCV infection among PWID increased fourfold over the same period (Zibbell et al., 2018). Syringe sharing has been identified as one of the principal causes of transmission of these diseases (Degenhardt et al., 2017). Historically, policies designed to curtail drug use such as regulating syringe sales led to negative consequences such as needle sharing and increased risk of infection. Complicating factors tied to syringe access and possession, ranging from stigma to policing incidents, have shaped injection drug use behavior, contributing to risky drug injection practices (Cooper et al., 2009, 2012; Miller et al., 2008). The recent resurgence of HIV infection among PWID in Indiana and Massachusetts, along with an increase in the daily frequency of injection drug use attributed to fentanyl (in Massachusetts) and oxymorphone (in Indiana) portray a rapidly changing terrain, and point to deficits in local prevention efforts to curtail syringe-mediated risks (Peters et al., 2016; Cranston et al., 2019).
A spatio-temporal cluster of incident HIV infections was reported among three West Virginia counties in 2017 (Evans et al., 2018; Bradley et al., 2019). Contact tracing identified co-infections with syphilis, hepatitis B, and HCV in as many as 50% of cases (Evans et al., 2018). Sexual or injection drug using contacts among cases implicated a further 12 counties as high risk. Geo-targeted HIV screening media campaigns and interviews with patients and providers were initiated to increase preventive measures and identify barriers to testing. Cited most frequently among the barriers to disease testing in rural locations were stigma, transportation, and poor health literacy (Evans et al., 2018).
Findings from the 2017 surge in HIV cases in West Virginia support the use of syndemics theory to study the co-occurrence of infectious diseases among persons who misuse drugs in underserved areas. Vulnerability analyses conducted at the state and national level have incorporated social and biological variables as predictors of HIV vulnerability using acute HCV as an indicator of injection drug use and high risk for HIV infection (Van Handel et al., 2016). While these studies do not explicitly incorporate advanced GIS or spatial epidemiological approaches, they do demonstrate the potential application of thematic GIS mapping and a regression-based approach to assess HIV transmission vulnerability among at-risk populations.
3.2. Proposed framework
Step one of our proposed framework to incorporate GIS and spatial epidemiology into infectious disease syndemics analyses is to identify individual associations between salient variables through basic mapping or exploratory spatial data analyses (ESDA). While integral to a public health response, visualizing individual variables does not provide as comprehensive an understanding as is needed to gain a broader appreciation for the complex underlying factors driving co-occurrence of diseases across geographic space and time. Here we present a thorough application of our proposed conceptual model, utilizing West Virginia county-level data. A detailed description of each variable included in our case study is presented in Table 1.
Table 1.
County-Level variables for the West Virginia HIV and HCV Syndemic.
| Variable | Definition | Source |
|---|---|---|
| HCV | Acute HCV rate per 100,000 in 2017 | West Virginia Department of Health and Human Resources |
| HIV | Reported HIV cases per 100,000 in 2016. | AIDSVu86 |
| OD | Opioid overdose death rate per 100,000 population in 2017 | CDC WONDER87 |
| INCOME | Log of median per capita income in 2017. | American Community Survey, US Census Bureau88 |
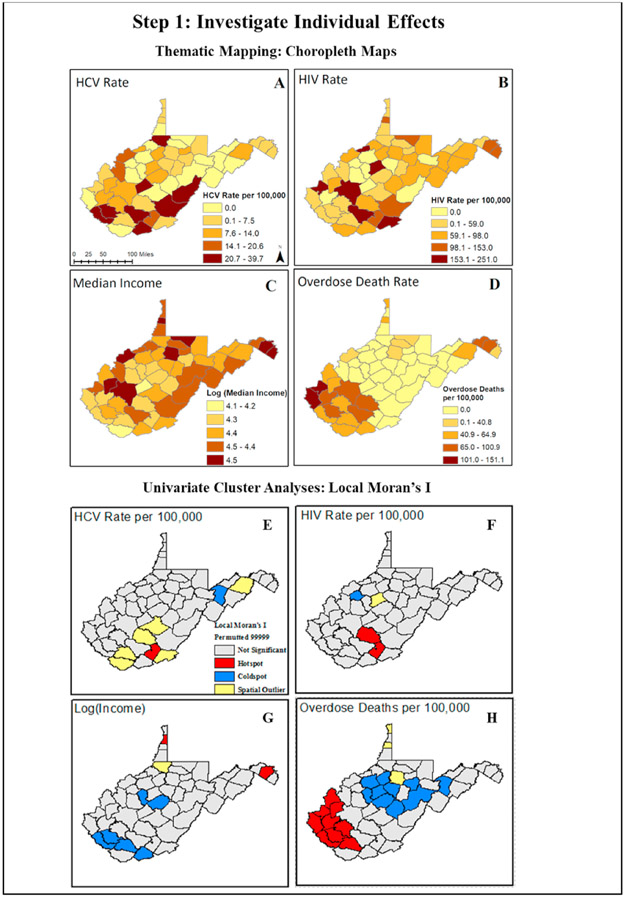
We begin by displaying unique spatial trends in each variable using thematic risk mapping (Fig. 2, Maps A-D). Here we observe county-level variation in spatial patterns for HCV, HIV, and overdose death rates per 100,000, as well as median income. We note that higher rates of HCV, HIV, and opioid-involved deaths are displayed in the southern and eastern regions of WV (indicated by darker color shades). Unique geographic trends among individual variables are further quantified using univariate spatial statistics, like univariate Local Moran’s I and local indicators of spatial autocorrelation. Here we incorporate a queen’s contiguity neighborhood matrix to characterize the influence of surrounding areas based upon the median values of the variable of interest (Burt et al., 2009). When using areal data in spatial statistics, it is most often necessary to define neighbors whose spatial influence is specified by a spatial weight (W). These weights can be specified using boundaries of nearby objects or distance. A queen’s contiguity method takes into consideration the effect of bordering counties (boundaries include both sides and vertices). We conducted a permutation test to evaluate the statistical significance of the spatial autocorrelation. In our case, for each variable we used 99,999 random permutations of the observed values for the given spatial weight matrix and computed the Moran’s I. The pseudo P-value was then established by comparing the observed Moran’s I to those from the permutation (Hendricks and Mark-Carew, 2017). Hotspots (indicated in red) appear in southern and eastern counties and reflect patterns observed in the univariate descriptive maps, presenting statistically significant clusters of counties with elevated values. Coldspots (blue counties) and spatial outliers (yellow counties) are also identified in the analysis (Fig. 2 Maps E-H). While hotspots are often the subject of interest in epidemiological studies, identifying areas that are coldspots and spatial outliers are also useful in gaining a broader understanding of factors that could drive the occurrence of diseases. Considering the 2017 surge in HIV cases, it would also be useful to know the number of harm reduction programs (e. g., syringe services programs) in each county and whether counties regarded as coldspots had a higher number of harm reduction and public health services.
Fig. 2.
Investigating the spatial distribution of outcome and covariate data using thematic maps and a combination of exploratory spatial data analyses (ESDA) to visualize spatial trends in individual variables.
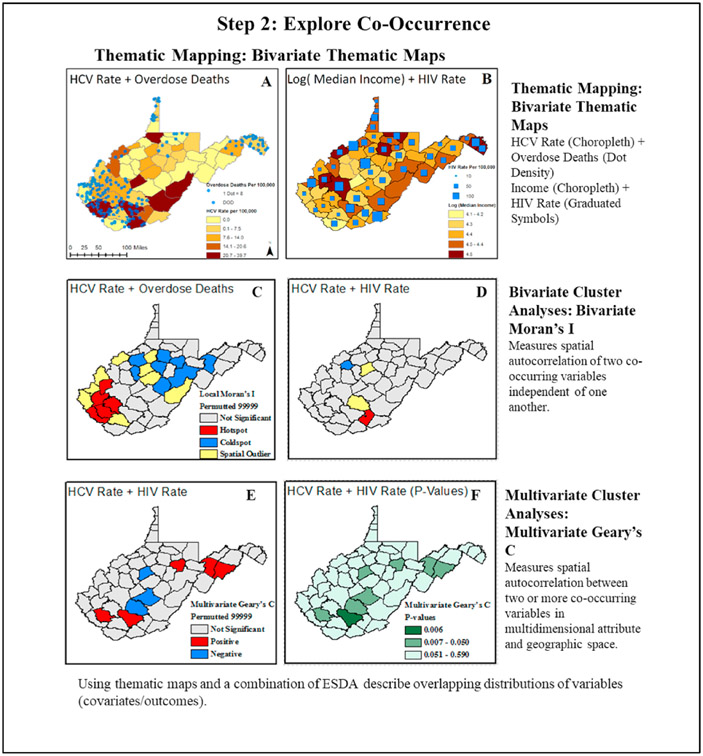
Next, we explore the co-occurrence of disease and potential social and biological drivers through bivariate and multivariate approaches. In this case study, we highlight the co-occurrence of the HCV and opioid overdose death rates per 100,000, as well as median income and the HIV rate per 100,000 in bivariate thematic maps utilizing a combination of thematic and point vector mapping. In the first display, darker shades of color indicate that higher HCV rates align with higher rates of opioid-involved deaths depicted by a dot density map layer (Fig. 3 Map A). In the second display, darker shades of color indicate that counties with higher income do not appear to align with larger points on the maps that depict a higher frequency of HIV infections (Fig. 3 Map B). Bivariate trends in the data are further quantified using ESDA approaches, such as the bivariate Local Moran’s I and multivariate Geary’s C. The focus of these analyses shifts to examine the co-occurrence of HCV, HIV, and opioid overdose deaths. We also incorporate the same color scheme and queen’s contiguity weight from the univariate Moran’s I into the bivariate and multivariate analyses for consistency. In the bivariate Local Moran’s I and local indicators of spatial autocorrelation (LISA) maps, we identify hotspots, coldspots, and spatial outliers based on independent values for HIV, HCV, and opioid overdose death rates (Lee, 2001). In both displays, we see hotpots indicating a higher frequency of HCV and opioid-involved deaths (Fig. 3 Maps C-D) and HCV and HIV in southern West Virginia. Slightly different patterns are observed between HCV and HIV in the multivariate Geary’s C. Here, we identify counties which cluster positively or negatively, and are able to capture clustering in relation to attributes (disease outcomes and/or other socioeconomic variables) and geographic space (Fig. 3 Maps E-F) (Anselin, 2019). Going one step further and incorporating the multivariate Geary’s C provides a necessary first impression as to how frequency of HCV and HIV interact and cluster together.
Fig. 3.
Exploring co-occurrence and bivariate associations between hepatitis C virus (HCV) rates, HIV rates, and opioid overdose rates, in West Virginia counties, 2016–2017.
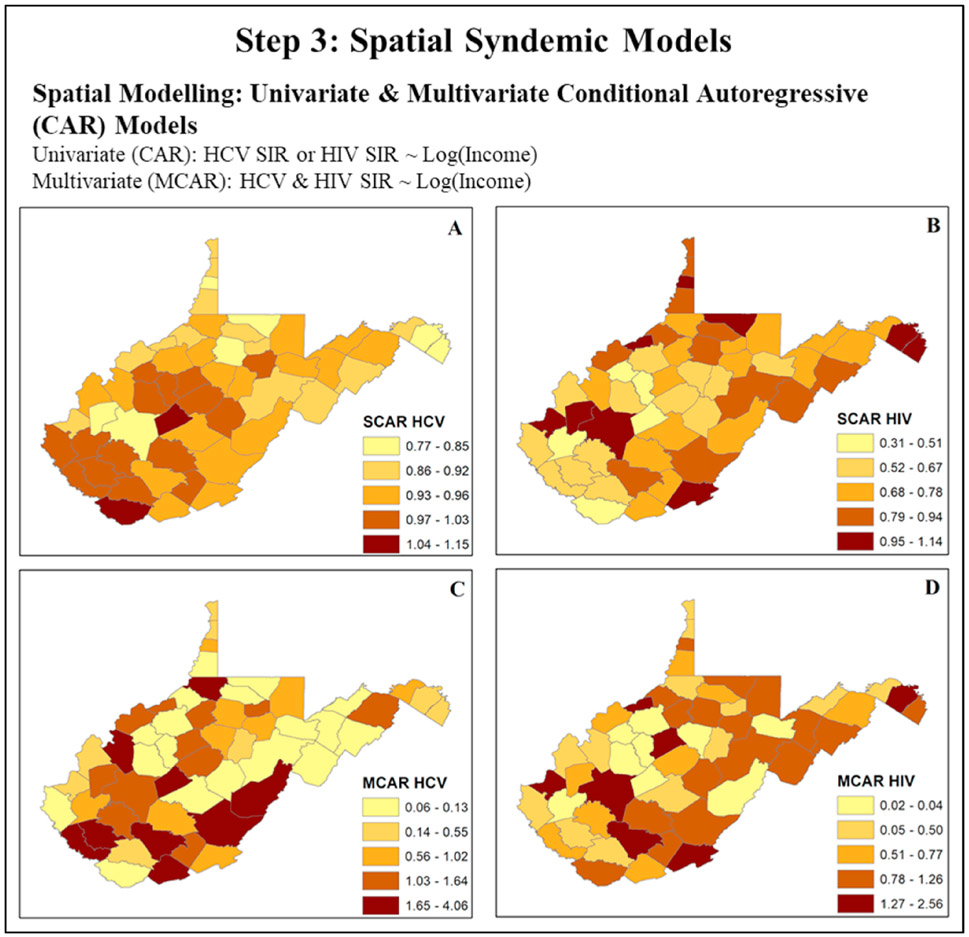
In the final step of our case study, we use spatial modeling techniques to identify geographic variations in co-occurring diseases, adjusting for social and biological drivers. To further illustrate differences between univariate vs. multivariate approaches for syndemics research, we display county-level maps for the predicted standardized incidence ratio (SIR, ratio of observed number of cases to expected number of cases) for HCV and HIV, adjusting for the effect of income using conditional autoregressive (CAR) models. Maps are presented in three sets: 1) simple CAR (Fig. 4 Maps A-B) and 2) multivariate CAR (MCAR) model fitted SIR (Fig. 4 Maps C-D). At first glance, the most apparent difference between methods is the use of spatial smoothing. Smoothers applied during spatial modeling stabilize predicted rates and imposed “shrinking to the mean” by borrowing strength from neighboring counties based upon the designated spatial weight (W). As a result, zero values observed in some counties in the raw data, are not seen later in the model fitted results. Similarly, extreme values in the raw SIR county-level map are weighted towards the average value of the neighbors specified through the spatial weight. This effect is most evident and probably can been considered as overshrinkage in the univariate CAR model, where all predicted SIR values for counties are contained within at most three mapped categories. While useful in ascertaining smoothed patterns of disease, adjusting for spatial variation in covariates, univariate models do not accommodate the complex multidimensional structure of syndemics. Based on previous studies that incorporated vulnerability analyses, we recognize that acute HCV is correlated with HIV, thus making it a valid indicator of HIV risk (McLuckie et al., 2019; Van Handel et al., 2016; Rickles et al., 2018). Additionally, we understand that the co-occurrence of these infections in space and time are influenced by social and biological variables unique to the local geographic space, such as the number of harm reduction programs and local drug availability in an area (Peters et al., 2016). In Scott County, Indiana, for instance, the local HIV and HCV epidemics were driven by the use of Opana®, a pre-scription opioid that fostered multiple injections in one sitting, thus increasing risk for syringe-mediated disease transmission (Broz et al., 2018). In the Merrimack Valley of Massachusetts, the introduction of fentanyl within the local drug supply, with its shorter biological half-life, appears to have contributed to more frequent injection practices, again increasing risk for injection-mediated HIV and HCV transmission (Cranston et al., 2019). Thus, the co-occurrence of these correlated diseases must be jointly modeled using multivariate approaches to gain a broader understanding of drivers for the HIV/HCV syndemic in underserved areas (Gelfand and Vounatsou, 2003). The results of the MCAR model are presented separately for HIV and HCV. Similar to our univariate approach, spatial smoothing is included through the designation of a spatial weight. However, with MCAR analyses, we consider joint differences in the spatial distribution of spatially correlated outcomes (HIV-HCV). Additionally, we can compare the coefficients for median income across outcomes without assuming that the occurrence of HIV and HCV are independent of one another. Predicted SIRs of HIV and HCV from our multivariate analysis highlight a higher degree of spatial variability in spatial trends of HIV and HCV than are inferred from the univariate analysis alone. The MCAR model seems to most closely reflect patterns in the crudely estimated SIR for both diseases after adjusting for county-level median income.
Fig. 4.
Spatial syndemics models using advanced spatial epidemiological approaches to estimate the associations between biological/social factors and the occurrence of HIV and HCV in West Virginia counties, 2016–2017. (Abbreviations: SIR: Standardized incidence ratio, CAR: Conditional autoregressive, MCAR: Multivariate Conditional autoregressive, HIV: Human immune-deficiency Virus, HCV: Hepatitis C Virus).
It should be noted that models presented in our case study contain only a years’ worth of data and that spatio-temporal extensions are possible to examine the effect of time in addition to spatial autocorrelation. Additionally, models are kept simple to illustrate the use of these methods, but more covariates can be added to represent other biological (e.g., effect of half-life of synthetic opioids) or social phenomena (e.g., homelessness, social network composition) which potentially influence co-occurrence of these infectious diseases. Bayesian spatiotemporal models, described earlier, also serve as good candidates for analytical tools in disease mapping, association studies, and cluster detection.
4. Discussion
The theory of syndemics focuses on a broad set of conditions that exist both on a population and individual level that create an environment where multiple epidemics can co-exist and exacerbate their adverse effects on human health. However, most studies that focus of syndemics are largely based on individual level outcomes (Tsai and Burns, 2015). In answering the call to utilize a population level approach to syndemics research, we elaborated on how spatial epidemiologic methods could be applied to study the co-occurrence of multiple diseases and the environmental context surrounding these diseases. Sound epidemiological inquiries, which consider broader socio-cultural, and economic contexts, supported by accurate spatiotemporal data can be used to accurately guide public health research. As illustrated throughout the case study, descriptive illustrations can be used to inform community members and public health officials alike about the risk landscape, and spatial epidemiological models can allow researchers and health policy experts to monitor and forecast health outcomes across local geographies. The spatial modeling techniques described earlier can be used to examine the correlation and interaction between two co-occurring epidemics while accounting for other population level characteristics within the study region. In addition, these models can be further improved by the addition of time-varying measures. This would be critical in understanding how these epidemics evolve over time, which could further our understanding of these complex relationships between social, biological, and structural factors, as well as individual decisions, that lead to the emergence of syndemics.
While there are significant advantages of using spatial epidemiologic methods to study syndemics, we should clarify that these empirical methods should go in tandem with qualitative and traditional quantitative studies to overcome some of the limitations of ecological analysis. Issues regarding loss of information when aggregating data into a discrete unit of analysis (e.g., census tract), sampling biases, cartographic confounding, modifiable areal unit problem, and ecological fallacies should be carefully considered while utilizing spatial epidemiologic methods. The spatial framework itself draws the researcher away from the individual as a unit of analysis. This, while useful for examining clustering of diseases and the socio-political environment affecting the diseases and the outcome, often limits the ability to study bio-bio interactions that happen at an individual or even cellular levels. Furthermore, as models describing a specific phenomenon get complex, study findings and interpretation can be driven mostly by available data and methods utilized, which could lead to biased results. However, a strong theoretical framework backed by evidence from qualitative and quantitative studies can help minimize biases. The evidence and the methodology presented in this paper demonstrates how spatial epidemiology can lend a strong supporting empirical framework to syndemics theory and research. Application of these methods can enable researchers, public health experts and policymakers to effectively leverage local spatially-oriented data to understand and address the micro and macro-level factors that contribute to a syndemic.
Supplementary Material
Acknowledgements
BH’s effort was supported by the National Institute Of General Medical Sciences, 2U54GM104942–02. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Declaration of competing interest
None.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.socscimed.2020.113352.
References
- Adeboye A, Ross M, Wilkerson M, Springer A, Ahaneku H, Yusuf R, 2017. Syndemic production of HIV infection among Tanzanian MSM. J Health Educ Res Dev 5 (3), 1000231. [Google Scholar]
- Ali M, Emch M, Donnay JP, Yunus M, Sack RB, 2002. Identifying environmental risk factors for endemic cholera: a raster GIS approach. Health Place 8 (3), 201–210. [DOI] [PubMed] [Google Scholar]
- Anselin L, 1995a. Local indicators of spatial association - LISA. Geogr. Anal 27 (2), 93–115. [Google Scholar]
- Anselin L, 1995b. Local indicators of spatial association—LISA. Geogr. Anal 27 (2), 93–115. [Google Scholar]
- Anselin L, 2019. A local indicator of multivariate spatial association: extending Geary’s C. Geogr. Anal 51 (2), 133–150. [Google Scholar]
- Beale CM, Lennon JJ, Yearsley JM, Brewer MJ, Elston DA, 2010. Regression analysis of spatial data. Ecol. Lett 13 (2), 246–264. [DOI] [PubMed] [Google Scholar]
- Besag J, Kooperberg C, 1995. On conditional and intrinsic autoregressions. Biometrika 82 (4), 733–746. [Google Scholar]
- Besag J, York J, Mollié A, 1991. Bayesian image restoration, with two applications in spatial statistics. Ann. Inst. Stat. Math 43 (1), 1–20. [Google Scholar]
- Blangiardo M, Cameletti M, 2015. Spatial and Spatio-Temporal Bayesian Models with R-INLA. John Wiley & Sons. [Google Scholar]
- Bradley H, Hogan V, Agnew-Brune C, et al. , 2019. Increased HIV diagnoses in West Virginia counties highly vulnerable to rapid HIV dissemination through injection drug use: a cautionary tale. Ann. Epidemiol 34, 12–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer CA, 2006. Basic mapping principles for visualizing cancer data using Geographic Information Systems (GIS). Am. J. Prev. Med 30 (2 Suppl. l), S25–S36. [DOI] [PubMed] [Google Scholar]
- Broz D, Zibbell J, Foote C, et al. , 2018. Multiple injections per injection episode: high-risk injection practice among people who injected pills during the 2015 HIV outbreak in Indiana. Int. J. Drug Pol 52, 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulled N, Singer M, 2011. Syringe-mediated syndemics. AIDS Behav. 15 (7), 1539–1545. [DOI] [PubMed] [Google Scholar]
- Burt JE, Barber GM, Rigby DL, 2009. Elementary Statistics for Geographers. Guilford Press. [Google Scholar]
- Centers for Disease Control and Prevention, 2017. HIV Surveillance Report, 2018. [Google Scholar]
- Chen C, Wakefield J, Lumely T, 2014. The use of sampling weights in Bayesian hierarchical models for small area estimation. Spat Spatiotemporal Epidemiol 11, 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliff AD, Ord JK, 1981. Spatial Processes: Models and Applications. [Google Scholar]
- Conrad C, Bradley HM, Broz D, et al. , 2015. Community outbreak of HIV infection linked to injection drug use of oxymorphone–Indiana. MMWR Morb Mortal Wkly Rep. 2015 64 (16), 443–444. [PMC free article] [PubMed] [Google Scholar]
- Cooper HL, Bossak B, Tempalski B, Des Jarlais DC, Friedman SR, 2009. Geographic approaches to quantifying the risk environment: drug-related law enforcement and access to syringe exchange programmes. Int. J. Drug Pol 20 (3), 217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper HL, Des Jarlais DC, Tempalski B, Bossak BH, Ross Z, Friedman SR, 2012. Drug-related arrest rates and spatial access to syringe exchange programs in New York City health districts: combined effects on the risk of injection-related infections among injectors. Health Place 18 (2), 218–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett EL, Watt CJ, Walker N, et al. , 2003. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch. Intern. Med 163 (9), 1009–1021. [DOI] [PubMed] [Google Scholar]
- Cranston K, Alpren C, John B, et al. , 2019. Notes from the field: HIV diagnoses among persons who inject drugs - northeastern Massachusetts, 2015-2018. MMWR Morb. Mortal. Wkly. Rep 68 (10), 253–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson PJ, Scholar S, Howe M, 2011. A GIS-based methodology for improving needle exchange service delivery. Int. J. Drug Pol 22 (2), 140–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt L, Peacock A, Colledge S, et al. , 2017. Global prevalence of injecting drug use and sociodemographic characteristics and prevalence of HIV, HBV, and HCV in people who inject drugs: a multistage systematic review. Lancet Glob Health 5 (12), e1192–e1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin RA, 1988. Estimation of regression coefficients in the presence of spatially autocorrelated error terms. Rev. Econ. Stat 466–474. [Google Scholar]
- Evans ME, Labuda SM, Hogan V, et al. , 2018. Notes from the field: HIV infection investigation in a rural area - West Virginia, 2017. MMWR Morb. Mortal. Wkly. Rep 67 (8), 257–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furr-Holden DM, Milam AJ, Nesoff ED, et al. , 2016. Triangulating syndemic services and drug treatment policy: improving drug treatment portal locations in baltimore city. Prog Community Health Partnersh 10 (2), 319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaines AD, 1998. From margin to center: from medical anthropology to cultural studies of science. JSTOR. [Google Scholar]
- Gelfand AE, Vounatsou P, 2003. Proper multivariate conditional autoregressive models for spatial data analysis. Biostatistics 4 (1), 11–25. [DOI] [PubMed] [Google Scholar]
- Geraghty EM, Balsbaugh T, Nuovo J, Tandon S, 2010. Using Geographic Information Systems (GIS) to assess outcome disparities in patients with type 2 diabetes and hyperlipidemia. J. Am. Board Fam. Med 23 (1), 88–96. [DOI] [PubMed] [Google Scholar]
- Getis A, Ord JK, 2010. The analysis of spatial association by use of distance statistics. Perspectives on Spatial Data Analysis. Springer, pp. 127–145. [Google Scholar]
- Hendricks B, Mark-Carew M, 2017. Using exploratory data analysis to identify and predict patterns of human Lyme disease case clustering within a multistate region, 2010–2014. Spatial Spatio-temporal Epidemiol 20, 35–43. [DOI] [PubMed] [Google Scholar]
- Herwaldt LA, Gorman GW, McGrath T, et al. , 1984. A new Legionella species, Legionella feeleii species nova, causes Pontiac fever in an automobile plant. Ann. Intern. Med 100 (3), 333–338. [DOI] [PubMed] [Google Scholar]
- Krieger N, 2001. Theories for social epidemiology in the 21st century: an ecosocial perspective. Int. J. Epidemiol 30 (4), 668–677. [DOI] [PubMed] [Google Scholar]
- Kuh D, Shlomo YB, 2004. A Life Course Approach to Chronic Disease Epidemiology. Oxford University Press. [Google Scholar]
- Lawson AB, 2013a. Statistical Methods in Spatial Epidemiology. John Wiley & Sons. [Google Scholar]
- Lawson AB, 2013b. Bayesian Disease Mapping: Hierarchical Modeling in Spatial Epidemiology. Chapman and Hall/CRC. [Google Scholar]
- Lawson AB, Banerjee S, Haining RP, Ugarte MD, 2016. Handbook of Spatial Epidemiology. CRC Press. [Google Scholar]
- Lee S-I, 2001. Developing a bivariate spatial association measure: an integration of Pearson’s r and Moran’s I. J. Geogr. Syst 3 (4), 369–385. [Google Scholar]
- Lee D, 2017. CARBayes Version 4.6: an R Package for Spatial Areal Unit Modelling with Conditional Autoregressive Priors. University of Glasgow, Glasgow. [Google Scholar]
- Leroux BG, Lei X, Breslow N, 2000. Estimation of disease rates in small areas: a new mixed model for spatial dependence. Statistical Models in Epidemiology, the Environment, and Clinical Trials. Springer, pp. 179–191. [Google Scholar]
- LeSage JP, Pace RK, 2009. Introduction to Spatial Econometrics. CRC Press, Boca Raton. [Google Scholar]
- Livingston K, Padilla M, Scott D, Colon-Burgos JF, Reyes AM, Varas-Diaz N, 2016. Methods of mapping ethnographic data on migration, tourism labor, and health risk in the Dominican Republic. Fla. Geogr 47. [PMC free article] [PubMed] [Google Scholar]
- McElroy A, 1990. Biocultural models in studies of human health and adaptation. Med. Anthropol. Q 4 (3), 243–265. [Google Scholar]
- McLuckie C, Pho MT, Ellis K, et al. , 2019. Identifying areas with disproportionate local health department services relative to opioid overdose, HIV and hepatitis C diagnosis rates: a study of rural Illinois. Int. J. Environ. Res. Publ. Health 16 (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall E, 2012. The VIDDA Syndemic: Distress and Diabetes in Social and Cultural Context. [Google Scholar]
- Meyers DJ, Hood ME, Stopka TJ, 2014. HIV and hepatitis C mortality in Massachusetts, 2002-2011: spatial cluster and trend analysis of HIV and HCV using multiple cause of death. PloS One 9 (12), e114822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CL, Firestone M, Ramos R, et al. , 2008. Injecting drug users’ experiences of policing practices in two Mexican-U.S. border cities: public health perspectives. Int. J. Drug Pol 19 (4), 324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran PAP, 1948. The interpretation of statistical maps. J. Roy. Stat. Soc. B 10 (2), 243–251. [Google Scholar]
- Nuvolone D, Della Maggiore R, Maio S, et al. , 2011. Geographical information system and environmental epidemiology: a cross-sectional spatial analysis of the effects of traffic-related air pollution on population respiratory health. Environ. Health 10, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostfeld RS, Glass GE, Keesing F, 2005. Spatial epidemiology: an emerging (or reemerging) discipline. Trends Ecol. Evol 20 (6), 328–336. [DOI] [PubMed] [Google Scholar]
- Pawlowski A, Jansson M, Sköld M, Rottenberg ME, Källenius G, 2012. Tuberculosis and HIV co-infection. PLoS Pathog. 8 (2), e1002464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters PJ, Pontones P, Hoover KW, et al. , 2016. HIV infection linked to injection use of oxymorphone in Indiana, 2014-2015. N. Engl. J. Med 375 (3), 229–239. [DOI] [PubMed] [Google Scholar]
- Ramirez IJ, Lee J, Grady SC, 2018. Mapping multi-disease risk during El niño: an ecosyndemic approach. Int. J. Environ. Res. Publ. Health 15 (12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S, Abelian JJ, Best N, 2006. Bayesian spatio-temporal analysis of joint patterns of male and female lung cancer risks in Yorkshire (UK). Stat. Methods Med. Res 15 (4), 385–407. [DOI] [PubMed] [Google Scholar]
- Rickies M, Rebeiro PF, Sizemore L, et al. , 2018. Tennessee’s in-state vulnerability assessment for a "rapid dissemination of human immunodeficiency virus or hepatitis C virus infection" event utilizing data about the opioid epidemic. Clin. Infect. Dis 66 (11), 1722–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowbotham TJ, 1980. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol 33 (12), 1179–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid V, Held L, 2004. Bayesian extrapolation of space-time trends in cancer registry data. Biometrics 60 (4), 1034–1042. [DOI] [PubMed] [Google Scholar]
- Singer M, 2000. A dose of drugs, a touch of violence, a case of AIDS: conceptualizing the SAVA syndemic. Free Inq. Creativ. Sociol 28 (1), 13–24. [Google Scholar]
- Singer M, 2010. Pathogen-pathogen interaction: a syndemic model of complex biosocial processes in disease. Virulence 1 (1), 10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M, Clair S, 2003. Syndemics and public health: reconceptualizing disease in bio-social context. Med. Anthropol. Q 17 (4), 423–441. [DOI] [PubMed] [Google Scholar]
- Stall R, Mills TC, Williamson J, et al. , 2003. Association of co-occurring psychosocial health problems and increased vulnerability to HIV/AIDS among urban men who have sex with men. Am. J. Publ. Health 93 (6), 939–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stall R, Coulter RW, Friedman MR, Plankey MW, 2015. Commentary on "Syndemics of psychosocial problems and HIV risk: a systematic review of empirical tests of the disease interaction concept" by A. Tsai and B. Burns. Soc. Sci. Med 145, 129–131. [DOI] [PubMed] [Google Scholar]
- Stoicescu C, Ameilia R, Irwanto, Praptoraharjo I, Mahanani M, 2019. Syndemic and synergistic effects of intimate partner violence, crystal methamphetamine, and depression on HIV sexual risk behaviors among women who inject drugs in Indonesia. J. Urban Health 96 (3), 477–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopka TJ, Amaravadi H, Kaplan AR, et al. , 2019a. Opioid overdose deaths and potentially inappropriate opioid prescribing practices (PIP): a spatial epidemiological study. Int. J. Drug Pol 68, 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopka TJ, Jacque E, Kelso P, et al. , 2019b. The opioid epidemic in rural northern New England: an approach to epidemiologic, policy, and legal surveillance. Prev. Med 105740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopka TJ, Goulart MA, Meyers DJ, et al. , 2017a. Identifying and characterizing hepatitis C virus hotspots in Massachusetts: a spatial epidemiological approach. BMC Infect. Dis 17 (1), 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopka TJ, Donahue A, Hutcheson M, Green TC, 2017b. Nonprescription naloxone and syringe sales in the midst of opioid overdose and hepatitis C virus epidemics: Massachusetts, 2015. J. Am. Pharmaceut. Assoc 57 (2S), S34–S44, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai AC, 2018. Syndemics: a theory in search of data or data in search of a theory? Soc. Sci. Med 206, 117–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai AC, Burns BF, 2015. Syndemics of psychosocial problems and HIV risk: a systematic review of empirical tests of the disease interaction concept. Soc. Sci. Med 139, 26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai AC, Venkataramani AS, 2016. Syndemics and health disparities: a methodological note. AIDS Behav. 20 (2), 423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai AC, Mendenhall E, Trostle JA, Kawachi I, 2017. Co-occurring epidemics, syndemics, and population health. Lancet 389 (10072), 978–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Handel MM, Rose CE, Hallisey EJ, et al. , 2016. County-level vulnerability assessment for rapid dissemination of HIV or HCV infections among persons who inject drugs, United States. J. Acquir. Immune Defic. Syndr 73 (3), 323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ver Hoef JM, Hanks EM, Hooten MB, 2018. On the relationship between conditional (CAR) and simultaneous (SAR) autoregressive models. Spat Stat 25, 68–85. [Google Scholar]
- Vindenes T, Jordan MR, Tibbs A, Stopka TJ, Johnson D, Cochran J, 2018. A genotypic and spatial epidemiologic analysis of Massachusetts’ Mycobacterium tuberculosis cases from 2012 to 2015. Tuberculosis 112, 20–26. [DOI] [PubMed] [Google Scholar]
- Waller LA, Gotway CA, 2004. Applied Spatial Statistics for Public Health Data, vol. 368. John Wiley & Sons. [Google Scholar]
- Wang W, Ying Y, Wu Q, Zhang H, Ma D, Xiao W, 2015. A GIS-based spatial correlation analysis for ambient air pollution and AECOPD hospitalizations in Jinan, China. Respir. Med 109 (3), 372–378. [DOI] [PubMed] [Google Scholar]
- Wiley AS, Allen JS, 2009. Medical Anthropology: A Biocultural Approach. Oxford University Press, Oxford. [Google Scholar]
- Zibbell JE, Asher AK, Patel RC, et al. , 2018. Increases in acute hepatitis C virus infection related to a growing opioid epidemic and associated injection drug use, United States, 2004 to 2014. Am. J. Publ. Health 108 (2), 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.