Abstract
Long noncoding RNAs (lncRNAs) have been widely recognized to play an important role in a variety of diseases. Abnormal regulation of lncRNA GATA3-antisense RNA 1 (AS1) occurs in several cancers, but whether it is involved in the progression of pancreatic cancer (PC) remains unknown. The present study aimed to investigate the biological effects of GATA3-AS1 in PC and to explore the underlying molecular mechanisms. Upregulation of GATA3-AS1 was revealed in PC tissues and cell lines. Knockdown of GATA3-AS1 in PANC-1 or AsPC-1 cells markedly reduced cell viability, cell proliferation, and cell invasion abilities, while cell apoptosis was increased. In addition, GATA3-AS1 knockdown suppressed the stemness of PANC-1 and AsPC-1 cells by decreasing the spheroid formation ability. A tumor xenograft in vivo assay demonstrated that GATA3-AS1 knockdown inhibited tumorigenicity of AsPC-1 cells. Furthermore, the microRNA (miR)-30b-5p downregulation and GATA3-AS1 upregulation were revealed in PC tissues and cell lines. Negative correlations were present between GATA3-AS1 and miR-30b-5p and between miR-30b-5p and testis-expressed protein 10 (Tex10) in the PC tissues, while GATA3-AS1 and Tex10 were positively correlated. GATA3-AS1 was then revealed to act as a competing endogenous RNA (ceRNA) for miR-30b-5p in regulating Tex10 expression. Moreover, the miR-30b-5p-Tex10 axis was confirmed to be involved in the regulation of biological effects of GATA3-AS1, including cell viability, cell proliferation, cell invasion, cell apoptosis, and cell stemness, as well as Wnt1/β-catenin signaling. Collectively, these data indicated that the GATA3-AS1-miR-30b-5p-Tex10 axis modulates tumorigenesis in PC, which may be associated with the Wnt/β-catenin signaling pathway.
Keywords: long non-coding RNA GATA3-antisense RNA 1, microRNA-30b-5p, testis-expressed protein 10, tumorigenesis, pancreatic cancer
Introduction
Pancreatic cancer (PC) is an aggressive malignancy and was predicted to be the second leading cause of cancer-related deaths in 2020 (1,2). Pancreatic ductal adenocarcinoma is the most frequent histological subtype. Despite advances in cancer treatment in recent decades, PC has continued to have poor clinical outcomes due to its late presentation with nonspecific symptoms and early metastatic nature (3). The overall 5-year survival rate for PC is 8%, which is the lowest compared with all other solid tumors (4). Given the high mortality, novel therapies to maximize life length and quality of life of patients with PC are urgently required. Thus, a thorough investigation of the mechanisms underlying the progression of PC is warranted.
Long noncoding RNAs (lncRNAs) are a class of noncoding transcripts longer than 200 nucleotides that have been recognized to play important roles in various physiological and pathological processes, such as cell proliferation and apoptosis, cell growth and senescence, and immune activation or inactivation (5–7). One well-defined mechanism by which lncRNAs act is by competition with endogenous RNAs (ceRNAs) to sponge microRNAs (miRNAs), thus affecting their regulatory function (8). miRNAs are a class of noncoding transcripts of approximately 22 nucleotides, which are widely known to participate in posttranscriptional regulation of gene expression via binding to the 3′-untranslated regions (3′-UTRs) of target messenger RNAs (mRNAs) and thus are deeply involved in a variety of biological activities (9,10). The sophisticated regulatory network involving lncRNA, miRNA, and mRNA is attracting increasing research attention worldwide.
LncRNA GATA3-antisense RNA 1 (GATA3-AS1) is a divergent lncRNA neighboring GATA3. GATA3-AS1 has been reported to share a promoter region with GATA3, and it is necessary for efficient transcription of GATA3 (11). One study demonstrated that GATA3 was markedly overexpressed in PC samples (12). Moreover, GATA3-AS1 was recently recognized as an oncogene in hepatocellular carcinoma (HCC) (13). Therefore, based on existing research, it was hypothesized that GATA3-AS1 is dysregulated and acts as a modulating molecular mechanism in the development of PC. The aim of the present study was to characterize the dysregulation and the biological roles of GATA3-AS1 in PC and to clarify the possible miRNA-mRNA regulatory axis involved.
Materials and methods
Patient samples
Cancer tissues and adjacent noncancerous tissues (normal tissues) were collected from 30 patients with PC at the First Affiliated Hospital of Xi'an Jiaotong University who underwent curative resection for PC between August 2017 and October 2019. Among them, 17 were males, and 13 were females. Their ages were between 35 and 71 years, and the mean age was 56 years. None of the subjects had received any biotherapy or chemotherapy before recruitment to this study. Informed consent was obtained from all participants, and the study protocol was approved by the Ethics Committee of the First Affiliated Hospital of Xi'an Jiaotong University.
Cell culture
The human immortalized pancreatic duct epithelial cell line HPDE6-C7 and PC cell lines BxPC-3, Capan-2, SW1990, PANC-1, and AsPC-1 were obtained from American Type Culture Collection and cultured in Dulbecco's modified Eagle's medium (DMEM) or RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) and 100 U/ml penicillin/streptomycin (all from Gibco; Thermo Fisher Scientific, Inc.), in a humidified atmosphere with 5% CO2 at 37°C.
Cell transfection
Small interfering RNAs (siRNAs) specifically targeting GATA3-AS1 (sequence: 5′-UCUCCGCGCGUCAAUCGA-3′), as well as the corresponding negative control (NC; sequence: 5′-CUACACCGUAUUCUACUACUA-3′), miR-30b-5p mimic (sequence: 5′-UGUAAACAUCCUACACUCAGCU-3′) and mimic-NC (sequence: 5′-UUCUCCGAACGUGUCACGUTT-3′) were obtained from Shanghai GenePharma Co., Ltd. pcDNA3.1-GATA3-AS1 and pcDNA3.1-Tex10 plasmids were constructed by Shanghai Sangon Biotech Co., Ltd. In a 24-well plate, cell transfection with siRNAs or si-NC (30 pmol/well), mimics or mimic-NC (30 pmol/well), or plasmids (0.5 µg/well) was performed using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions at room temperature. Unless otherwise specified, subsequent experimentation was performed after 48 h of transfection. An untransfected (normal cultured cells) group was identified as the control group.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from cancer tissues and cell lines using Trizol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) in accordance with the manufacturer's instructions. Complementary DNA (cDNA) was reverse transcribed using SuperScript First Strand cDNA System (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's instructions. The levels of GATA3-AS1 and mRNA expression of Tex10 were assessed using SYBR Premix Ex Taq kit (Takara Biotechnology Co., Ltd.) following the manufacturer's instructions. GAPDH was used as an internal control. The levels of miR-30b-5p were assessed using TaqMan microRNA assays (Applied Biosystems; Thermo Fisher Scientific, Inc.) following the manufacturer's instructions. U6 was used as an internal control. qPCR was performed on an Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) under the following thermocycling conditions: Initial denaturation at 95°C for 5 min followed by 40 cycles at 95°C for 15 sec (denaturation), 58°C for 30 sec (annealing) and 72°C for 30 sec (elongation), and final extension at 72°C for 5 min. The relative expression was calculated using the 2−ΔΔCq method (14). The primer sequences are presented in Table I.
Table I.
Primer sequences.
| Gene | Primer sequences (5′-3′) |
|---|---|
| GATA3-AS1 | F: AAGTTGAGCGGGGTATGT |
| R: TTTCTGGCCTTTGGTGTC | |
| miR-30b-5p | F: ACACTCCAGCTGGGTGTAAACATCCTACAC |
| R: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAGCTGAGT | |
| Tex10 | F: TTGCAACTTGCTCATCTTGG |
| R: AGAGTCTGCAGGGAGAACCA | |
| GAPDH | F: AGAACGGGAAGCTTGTCATC |
| R: CATCGCCCCACTTGATTTTG | |
| U6 | F: CTCGCTTCGGCAGCACA |
| R: AACGCTTCACGAATTTGCGT |
GATA3-AS1, GATA3-antisense RNA 1; miR-30b-5p, microRNA-30b-5p; Tex10, testis-expressed protein 10; F, forward; R; reverse.
Cell viability assay
Cell viability was assessed using Cell Counting Kit (CCK)-8 (Beyotime Institute of Biotechnology) at 0, 24, 48, 72 and 96 h post transfection according to the manufacturer's instructions. Briefly, 10 µl of CCK-8 solution was added to each well (4×104 cells) of 96-well plates, and the cells were incubated with CCK-8 solution for 4 h at 37°C. The absorbance value was measured at 450 nm using a microplate reader (Thermo Fisher Scientific, Inc.).
Colony formation assay
Cells (1×106) were seeded into 6-well plates and cultured in DMEM or RPMI-1640 medium with 10% FBS at 37°C. After 2 weeks, the cells were fixed using 4% paraformaldehyde for 20 min, and then stained using 1% crystal violet dye for 20 min at room temperature, and then the number of colonies was counted.
Cell apoptosis assay
Cell apoptosis was assessed by flow cytometry 48 h post transfection using an Annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit (Beyotime Institute of Biotechnology) according to the manufacturer's instructions. Briefly, 1×106 cells were seeded into 6-well plates and 48 h post-transfection, the cells were collected, washed, and resuspended in binding buffer. Then, the cells were incubated with 5 µl of Annexin V-FITC and 10 µl of propidium iodide at room temperature in the dark for 20 min. Cell apoptosis was assessed using a FACSCalibur (BD Biosciences) within 1 h. FlowJo software Ver.10 (Tree Star, Inc.) was used for analysis.
Transwell assay
The invasion assay was performed using 24-well Transwell chambers with 8-µm pores (Corning, Inc.); the chambers were precoated with Matrigel for 30 min at 37°C. Cells were harvested at 24 h post transfection and then 200 µl of cell suspension in serum-free medium (4×104 cells) was seeded in the upper chamber of Transwell plates, while 600 µl of medium containing 20% FBS was added to the lower chamber. After 48 h of incubation (72 h post transfection), cells remaining in the upper chamber were removed by cotton swabs, while the cells on the filter surface were fixed using 4% paraformaldehyde for 20 min and then stained using 0.1% crystal violet dye for 20 min at room temperature, and then counted in five random fields under an optical inverted microscope (magnification, ×200).
Spheroid formation assay
Cells (1×103) were seeded in Ultra Low Attachment 24-well plates (Corning, Inc.) and cultured in serum-free DMEM/F12 medium (Invitrogen; Thermo Fisher Scientific, Inc.), supplemented with 2% B27 (Gibco; Thermo Fisher Scientific, Inc.), 20 ng/ml epidermal growth factor, and 10 ng/ml basic fibroblast growth factor (PeproTech, Inc.) containing 100 U/ml penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.). Seven days later, the formed spheroids were counted under an optical inverted microscope (magnification, ×200).
Dual-luciferase reporter assay
The starBase v2.0 (http://starbase.sysu.edu.cn/) was used in the bioinformatics analysis to predict the binding sites between GATA3-AS1 and miR-30b-5p, and between miR-30b-5p and Tex10 (15). The sequences of GATA3-AS1 wild-type (wt, chr10:8,092,655-8,092,876) and Tex10 wt (chr9:103,064,259-103,064,483), as well as their corresponding mutant sequences were synthesized and inserted into the pGL3 vectors by Shanghai Sangon Biotech Co., Ltd., and named as pGL3-GATA3-AS1 wt, pGL3-GATA3-AS1 mut, pGL3-Tex10 wt, and pGL3-Tex10 mut plasmids, respectively. Cells (4×104) were co-transfected with indicated pGL3 plasmids and miR-30b-5p mimic or mimic NC, along with pRL-TK plasmids (Promega Corporation), using Lipofectamine 2000 according to the manufacturer's instructions. The luciferase activity was monitored 48 h post transfection using Dual-Glo Luciferase assay (Promega Corporation) according to the manufacturer's instructions. The relative firefly luciferase activity was normalized to Renilla luciferase activity.
Western blotting
Total protein was extracted from PC tissues and cell lines using RIPA buffer (Beyotime Institute of Biotechnology) according to the manufacturer's instructions. The BCA method was used for protein quantification. Equal amounts (30 µg) of proteins were separated by 10% SDS-PAGE and transferred onto nitrocellulose membranes (EMD Millipore), followed by blocking with 5% nonfat milk for 1 h at room temperature. Then, the membranes were incubated with anti-Tex10 (1:1,000; cat. no. 720257; Invitrogen; Thermo Fisher Scientific, Inc.), anti-Wnt1 (1:500; ab15251; Abcam) or anti-β-catenin (1:500; ab16051; Abcam) at 4°C overnight and then incubated with horseradish-peroxidase-conjugated secondary antibody (1:10,000; cat. no. BA1055; Boster Biological Technology) at room temperature for 1 h. The protein bands were visualized using an enhanced chemiluminescence detection kit (Amersham Biosciences; Cytiva). GAPDH was used as an internal control. The density of the bands was quantified by ImageJ 1.52a (National Institutes of Health).
Lentiviral vectors and animal experiments
Lentiviral vectors encoding short hairpin (sh)-GATA3-AS1 or shRNA NC were constructed and packaged by Shanghai GenePharma Co., Ltd. Briefly, the 2nd generation system was used. Lentiviral plasmid (20 µg), and two packaging components (15 µg or 5 µg, respectively) were co-transfected into 293T cells using Lipofectamine 2000 according to the manufacturer's protocol. At 8 h post-transfection, the medium was replaced with medium containing 10% serum, and the cells were cultured for an additional 48-72 h. Subsequently, the supernatant containing lentiviral particles was harvested and concentrated by ultracentrifugation to obtain a high-titer lentivirus concentration. Then the virus solution was added to the cells at the multiplicity of infection (MOI) of 40 for 72 h. Finally, after 5-7 days of purinomycin screening, the stable cells were collected and passaged at least three times for subsequent animal experiments.
A total of 16 BALB/c nude mice (male; aged 4-6 weeks) were purchased from the Chengdu DOSSY Experimental Animals Co., Ltd. All the mice were housed under 20-25°C in a humidified room (humidity, 40-55%) with a 12-h light-dark cycle and free access to food and water. The PANC-1 cells or AsPC-1 cells infected with lentivirus-shRNA NC or lentivirus-sh-GATA3-AS1 were subcutaneously injected into the axilla of nude mice, respectively (n=4 per group). Animal health was monitored every day throughout the study, and the mice were to be euthanized whenever they exhibited persistent lethargy and loss of appetite. However, no mice exhibited the aforementioned humane endpoints and the 16 tumor-bearing mice all survived throughout the present study. Every 3 days, the tumor size was monitored and the maximum tumor size was ≤1 cm3. Two weeks later, the mice were euthanized by cervical dislocation. Both, no fluctuation in the chest cavity (respiratory arrest) and no breathing and heartbeat sound (cardiac arrest) confirmed the sacrifice of the mice. Then the tumors were isolated and weighed. The animal study was approved by the Animal Care and Use Committee of the First Affiliated Hospital of Xi'an Jiaotong University.
Statistical analysis
Statistical analysis was carried out with GraphPad Prism 8.0 (GraphPad Software, Inc). All data are expressed as the mean ± standard deviation of at least three independent experiments. The differences were analyzed by paired t-test between two groups, and one-way analysis of variance followed by Bonferroni's post hoc test among more than two groups. Correlation of molecular expression was analyzed by linear regression. P<0.05 was considered to indicate a statistically significant difference.
Results
LncRNA GATA3-AS1 expression is upregulated in human PC tissues and cell lines
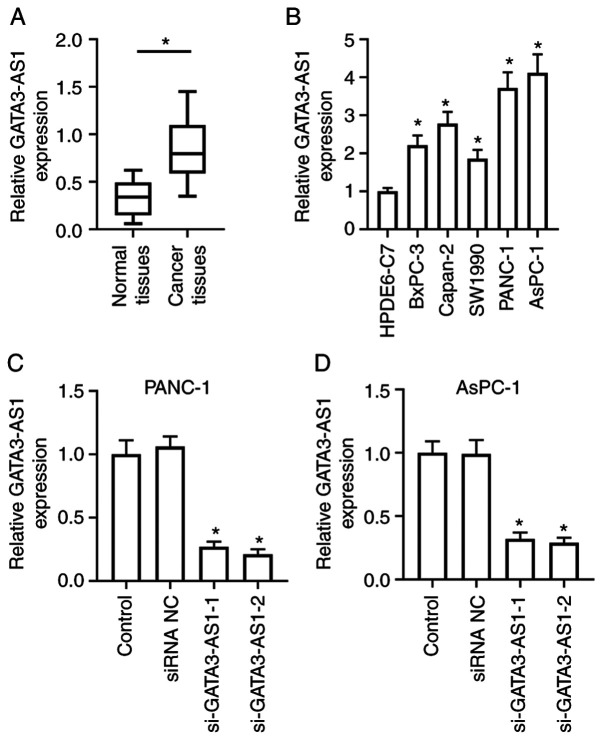
The expression level of lncRNA GATA3-AS1 was first assessed in PC tissues and cell lines. As revealed in Fig. 1A, the expression of GATA3-AS1 in the cancer tissues from patients with PC was significantly upregulated, compared with the normal tissues. As revealed in Fig. 1B, the expression of GATA3-AS1 in the PC cell lines BxPC-3, Capan-2, SW1990, PANC-1, and AsPC-1 was also significantly upregulated, compared with the normal human pancreatic duct epithelial cell line HPDE6-C7. To assess the biological effects of GATA3-AS1 upregulation, its expression was knocked down in PANC-1 and AsPC-1 cells by transfecting them with siRNAs. As revealed in Fig. 1C and D, compared with the siRNA NC group, the interference efficiency exhibited no obvious difference between si-GATA3-AS1-1 and si-GATA3-AS1-2, and both decreased the expression of GATA3-AS1 in PANC-1 and AsPC-1 cells. In the following experiments, si-GATA3-AS1-2 was selected to interfere with GATA3-AS1 expression.
Figure 1.
LncRNA GATA3-AS1 expression is upregulated in human PC tissues and cell lines. (A) The expression of GATA3-AS1 in 30 pairs of PC tissues and matched normal tissues was assessed by qPCR. *P<0.05, compared with the normal tissues. (B) The expression of GATA3-AS1 in normal human pancreatic duct epithelial cell line HPDE6-C7, and in PC cell lines BxPC-3, Capan-2, SW1990, PANC-1, and AsPC-1 was assessed by qPCR. *P<0.05, compared with the HPDE6-C7 group. (C and D) Two GATA3-AS1 siRNAs (si-GATA3-AS1-1 and si-GATA3-AS1-2) and negative control siRNA (siRNA NC) were transfected into PANC-1 or AsPC-1 cells, respectively. Then, the expression of GATA3-AS1 was detected by qPCR. *P<0.05, compared with the siRNA NC group. LncRNA, long noncoding RNA; GATA3-AS1, GATA3-antisense RNA 1; PC, pancreatic cancer; qPCR, quantitative polymerase chain reaction; siRNA or si-, small interfering RNA; NC, negative control.
Knockdown of GATA3-AS1 affects the cell biology and stem cell-like characteristics of PC cells
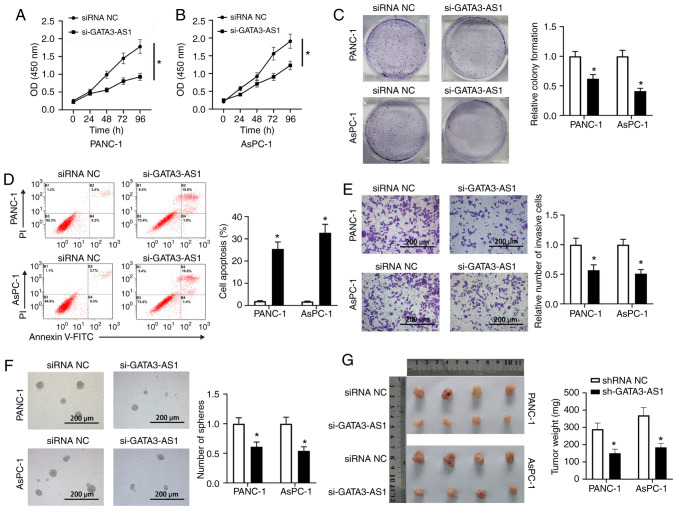
As revealed in Fig. 2A and B, the CCK-8 assay revealed that the OD (450 nm) in the si-GATA3-AS1 group was significantly decreased compared with that in the siRNA NC group, indicating that the cell viability of both PANC-1 and AsPC-1 cells was reduced by the knockdown of GATA3-AS1. Furthermore, compared with the siRNA NC group, the si-GATA3-AS1 group exhibited decreased proliferation ability (Fig. 2C), increased cell apoptosis (Fig. 2D), and reduced invasion ability (Fig. 2E) in both PANC-1 and AsPC-1 cells. In addition, the spheroid formation ability in the si-GATA3-AS1 group was also significantly reduced compared with the siRNA NC group (Fig. 2F). Moreover, the in vivo tumorigenicity experiment revealed that the tumor volumes and weights of nude mice in the sh-GATA3-AS1 group were significantly decreased compared with the nude mice in the shRNA NC group (Fig. 2G).
Figure 2.
Knockdown of GATA3-AS1 affects the cell biology and stem cell-like characteristics of PC cells. (A and B) Cell Counting Kit-8 assay at 0, 24, 48, 72 and 96 h post transfection. (C) Cell proliferation of PANC-1 and AsPC-1 cells was assessed by colony formation assay. (D) Cell apoptosis of PANC-1 and AsPC-1 cells was assessed by flow cytometry. (E) Cell invasion of PANC-1 and AsPC-1 cells was assessed by Transwell assay. (F) The stemness of PANC-1 and AsPC-1 cells was assessed by spheroid formation assay. *P<0.05, compared with the siRNA NC group. (G) The PANC-1 or AsPC-1 cells infected with lentivirus-shRNA NC or lentivirus-sh-GATA3-AS1 were subcutaneously injected into the axilla of nude mice, respectively. Two weeks later, the primary tumors were weighed. *P<0.05, compared with the shRNA NC group. GATA3-AS1, GATA3-antisense RNA 1; PC, pancreatic cancer; siRNA or si-, small interfering RNA; shRNA or sh-, short hairpin RNA; NC, negative control.
GATA3-AS1 acts as a ceRNA of miR-30b-5p
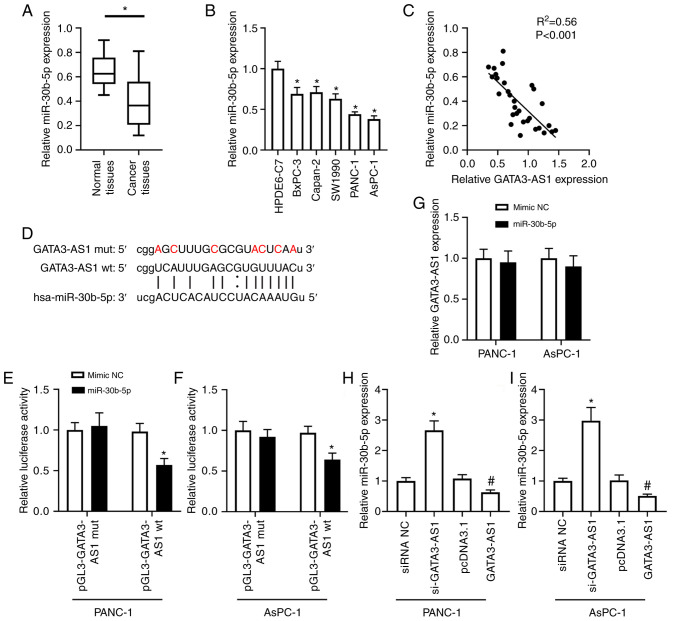
The bioinformatics analysis revealed that GATA3-AS1 contained a miR-30b-5p binding site (Fig. 3D). The expression of miR-30b-5p in the PC tissues and cell lines was then assessed. As revealed in Fig. 3A, the expression of miR-30b-5p in the cancer tissues from patients with PC was significantly downregulated, compared with the normal tissues. In addition, as revealed in Fig. 3B, compared with the HPDE6-C7 cells, the expression of GATA3-AS1 in BxPC-3, Capan-2, SW1990, PANC-1, and AsPC-1 cell lines was also significantly downregulated. A negative correlation was revealed between the expression of GATA3-AS1 and miR-30b-5p in the PC tissues (Fig. 3C). Furthermore, the dual-luciferase reporter assay revealed that, in comparison with the mimic-NC group, miR-30b-5p overexpression significantly reduced the firefly luciferase activity of the pGL3-GATA3-AS1 wt plasmids, but it had no effect on the firefly luciferase activity of pGL3-GATA3-AS1 mut plasmids in either PANC-1 or AsPC-1 cells (Fig. 3E and F). Moreover, as revealed in Fig. 3G, compared with the mimic NC group, the expression of GATA3-AS1 had no obvious difference when miR-30b-5p was overexpressed in both PANC-1 and AsPC-1 cells. In addition, compared with the siRNA NC group, the expression of miR-30b-5p was significantly upregulated in the si-GATA3-AS1 group; compared with the pcDNA 3.1 group, the expression of miR-30b-5p was significantly downregulated in the GATA3-AS1 overexpression group (Fig. 3H and I).
Figure 3.
GATA3-AS1 acts as a ceRNA of miR-30b-5p. (A) The expression of miR-30b-5p in 30 pairs of PC tissues and matched normal tissues was assessed by qPCR. *P<0.05, compared with the normal tissues. (B) The expression of miR-30b-5p in HPDE6-C7, BxPC-3, Capan-2, SW1990, PANC-1, and AsPC-1 cell lines was assessed by qPCR. *P<0.05, compared with the HPDE6-C7 group. (C) The negative correlation between the expression of GATA3-AS1 and miR-30b-5p in PC tissues. (D) The wild-type and mutant sequences of the binding site between GATA3-AS1 and miR-30b-5p. (E and F) The target relationship between GATA3-AS1 and miR-30b-5p was confirmed by dual-luciferase reporter assay in PANC-1 and AsPC-1 cells. *P<0.05, compared with the mimic-NC group. (G) PANC-1 and AsPC-1 cells were transfected with mimic NC or miR-30b-5p mimic. Then, the expression of GATA3-AS1 was assessed by qPCR. (H and I) PANC-1 and AsPC-1 cells were transfected with si-GATA3-AS1 or pcDNA 3.1-GATA3-AS1 plasmids to knockdown or overexpress GATA3-AS1. Then, the expression of miR-30b-5p was assessed by qPCR. *P<0.05, compared with the siRNA NC group; #P<0.05, compared with the pcDNA 3.1 group. GATA3-AS1, GATA3-antisense RNA 1; ceRNA; competing endogenous RNA; miR-30b-5p, microRNA-30-5p; PC, pancreatic cancer; qPCR, quantitative polymerase chain reaction; siRNA or si-, small interfering RNA; NC, negative control; wt, wild-type; mut, mutant.
miR-30b-5p directly targets the 3′-UTR of Tex10 to suppress its expression
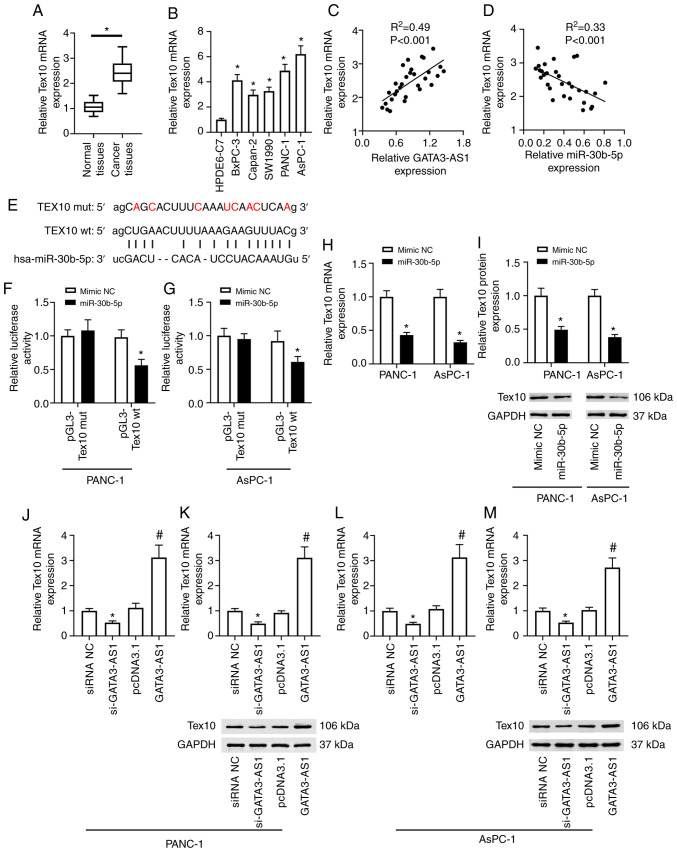
The bioinformatics analysis revealed that Tex10 also contained a miR-30b-5p binding site (Fig. 4E). The expression of Tex10 was then assessed in the PC tissues and cell lines. As revealed in Fig. 4A, compared with the normal tissues, the mRNA expression of Tex10 in the cancer tissues from patients with PC was significantly upregulated. In addition, as revealed in Fig. 4B, compared with the HPDE6-C7 cells, the mRNA expression of Tex10 in BxPC-3, Capan-2, SW1990, PANC-1, and AsPC-1 cell lines was also significantly upregulated. A positive correlation was revealed between the expression of GATA3-AS1 and Tex10 (Fig. 4C), and a negative correlation between the expression of miR-30b-5p and Tex10 (Fig. 4D) in the PC tissues. Furthermore, the dual-luciferase reporter assay revealed that compared with the mimic-NC group, miR-30b-5p overexpression significantly reduced the firefly luciferase activity of the pGL3-Tex10 wt plasmids, but it had no effect on the firefly luciferase activity of pGL3-Tex10 mut plasmids in either PANC-1 or AsPC-1 cells (Fig. 4F and G). Moreover, as revealed in Fig. 4H and I, compared with the mimic NC group, the mRNA and protein expression of Tex10 in the miR-30b-5p overexpression group were markedly downregulated in both PANC-1 and AsPC-1 cells. In addition, compared with the siRNA NC group, the mRNA and protein expression of Tex10 were significantly downregulated in the si-GATA3-AS1 group; compared with the pcDNA 3.1 group, the expression of Tex10 was significantly upregulated in the GATA3-AS1 overexpression group in the PANC-1 cells (Fig. 4J and K). Similar results were observed in the AsPC-1 cells (Fig. 4L and M).
Figure 4.
miR-30b-5p directly targets the 3′-UTR of Tex10 to suppress its expression. (A) The mRNA expression of Tex10 in 30 pairs of PC tissues and matched normal tissues was assessed by qPCR. *P<0.05, compared with the normal tissues. (B) The mRNA expression of Tex10 in HPDE6-C7, BxPC-3, Capan-2, SW1990, PANC-1, and AsPC-1 cell lines was assessed by qPCR. *P<0.05, compared with the HPDE6-C7 group. (C) The positive correlation between the expression of GATA3-AS1 and Tex10 in PC tissues. (D) The negative correlation between the expression of miR-30b-5p and Tex10 in PC tissues. (E) The wild-type and mutant sequences of the binding site between miR-30b-5p and Tex10. (F and G) The target relationship between miR-30b-5p and Tex10 was confirmed by dual-luciferase reporter assay in PANC-1 and AsPC-1 cells. *P<0.05, compared with the mimic-NC group. (H and I) PANC-1 and AsPC-1 cells were transfected with mimic NC or miR-30b-5p mimic. The mRNA and protein expression levels of Tex10 were assessed by qPCR and western blotting. *P<0.05, compared with the mimic-NC group. PANC-1 and AsPC-1 cells were transfected with si-GATA3-AS1 or pcDNA 3.1-GATA3-AS1 plasmids. (J and K) The mRNA and protein expression levels of Tex10 in PANC-1 cells were assessed by qPCR and western blotting. (L and M) The mRNA and protein expression levels of Tex10 in AsPC-1 cells were assessed by qPCR and western blotting. *P<0.05, compared with the siRNA NC group, #P<0.05, compared with the pcDNA 3.1 group. miR-30b-5p, microRNA-30-5p; Tex10, testis-expressed protein 10; PC, pancreatic cancer; qPCR, quantitative polymerase chain reaction; GATA3-AS1, GATA3-antisense RNA 1; siRNA or si-, small interfering RNA; NC, negative control; wt, wild-type; mut, mutant.
miR-30b-5p-Tex10 axis partially mediates the effects of GATA3-AS1
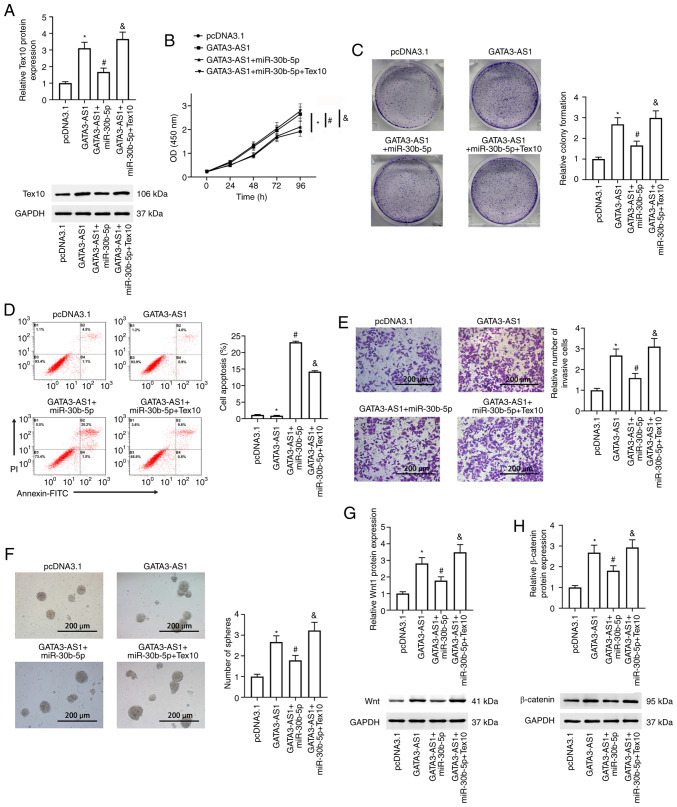
To determine whether the miR-30b-5p-Tex10 axis was involved in the regulation of GATA3-AS1 bioactivities, pcDNA 3.1 empty plasmids, pcDNA 3.1-GATA3-AS1 plasmids, a combination of pcDNA 3.1-GATA3-AS1 plasmids and miR-30b-5p mimic, or a combination of pcDNA 3.1-GATA3-AS1 plasmids, miR-30b-5p mimic and pcDNA3.1-Tex10 plasmids were transfected into AsPC-1 cells. As revealed in Fig. 5A, compared with the GATA3-AS1 group, the upregulated protein expression of Tex10 was significantly suppressed in the GATA3-AS1 + miR-30b-5p group. However, this was reversed and it was upregulated again in the GATA3-AS1 + miR-30b-5p + Tex10 group. The cell viability, cell proliferation, cell apoptosis and invasion, and stem cell characteristics were further assessed. Compared with the pcDNA 3.1 group, increased cell viability and proliferation, invasion, and spheroid formation abilities were observed in the GATA3-AS1 group; all of these were suppressed in the GATA3-AS1 + miR-30b-5p group. Furthermore, compared with the GATA3-AS1 + miR-30b-5p group, the cell viability and proliferation, invasion, and spheroid formation abilities once more exhibited a rising trend, and cell apoptosis exhibited a decreasing trend in the GATA3-AS1 + miR-30b-5p + Tex10 group (Fig. 5B-F). In addition, compared with the pcDNA 3.1 group, the protein expression levels of Wnt1 and β-catenin were upregulated in the GATA3-AS1 group; compared with the GATA3-AS1 group, the protein expression levels of Wnt1 and β-catenin were downregulated in the GATA3-AS1 + miR-30b-5p group but upregulated in the GATA3-AS1 + miR-30b-5p + Tex10 group (Fig. 5G and H).
Figure 5.
miR-30b-5p-Tex10 axis partially mediates the effects of GATA3-AS1. AsPC-1 cells were transfected with pcDNA 3.1 empty plasmids, pcDNA 3.1-GATA3-AS1 plasmids, a combination of pcDNA 3.1-GATA3-AS1 plasmids and miR-30b-5p mimic, or a combination of pcDNA 3.1-GATA3-AS1 plasmids, miR-30b-5p mimic, and pcDNA3.1-Tex10 plasmids. (A) The protein expression of Tex10 was assessed by western blotting. (B) Cell viability was assessed by Cell Counting Kit-8 assay at 0, 24, 48, 72 and 96 h post transfection. (C) Cell proliferation was assessed by colony formation assay. (D) Cell apoptosis was assessed by flow cytometry. (E) Cell invasion was assessed by Transwell assay. (F) The stemness was assessed by spheroid formation assay. The protein expression of (G) Wnt1 and (H) β-catenin was assessed by western blotting. *P<0.05, compared with the pcDNA 3.1 group; #P<0.05, compared with the GATA3-AS1 group; &P<0.05, compared with the GATA3-AS1 + miR-30b-5p group. miR-30b-5p, microRNA-30-5p; Tex10, testis-expressed protein 10; GATA3-AS1, GATA3-antisense RNA 1.
Discussion
During the past few decades, it has become widely recognized that the noncoding portion of the mammalian transcriptome is more complex than the protein-coding genes. Among these ncRNAs, 18% of lncRNAs have been revealed to be associated with the progression of cancers, compared with 9% of protein-coding genes (16,17), indicating that lncRNAs could have a major role in modulating carcinogenesis. Some well-defined lncRNAs have been recognized as oncogenes or suppressors in various cancers (7), such as H19 (18,19), HOTAIR (20,21), MALAT1 (22,23), UCA1 (24,25), and MEG3 (26,27). In the present study, it was demonstrated that GATA3-AS1 may function as an oncogene in the progression of PC.
GATA3-AS1 is a newly discovered antisense lncRNA, which was first identified from human CD4+ T-cell subsets (28). Subsequently, Zhu et al (29) revealed that GATA3-AS1 was overexpressed in human bladder cancer tissues. Recently, Luo et al (13) revealed that GATA3-AS1 was significantly upregulated in HCC tissues. In the present study, GATA3-AS1 expression in the PC tissues and cell lines was significantly upregulated, indicating that the aberrant expression may contribute to the progression of PC, similar to its oncogenic roles in HCC. To better understand the biological roles of GATA3-AS1 in PC, in vitro loss-of-function experiments were performed. Knockdown of GATA3-AS1 in PANC-1 and AsPC-1 cells obviously inhibited cell viability, proliferation, and invasion and increased cell apoptosis. Furthermore, knockdown of GATA3-AS1 in PANC-1 and AsPC-1 cells resulted in reduced stemness, which was characterized by suppressed spheroid formation ability. Moreover, decreased volumes and weights of primary tumors by GATA3-AS1 knockdown were revealed in the in vivo tumorigenicity experiment. Collectively, these data demonstrated that GATA3-AS1 also acts as an oncogene to accelerate the progression of PC.
Accumulating evidence has demonstrated that lncRNAs act as endogenous ceRNAs for specific miRNAs and thus regulate gene expression. For instance, Gong et al (30) have revealed that UCA1 interacts with miR-203, resulting in the release of miR-203-targeted transcripts ZEB2, and thus promotes metastatic gastric cancer. Xu et al (31) have shown that MALAT1 facilitates colorectal cancer cell proliferation, invasion, and migration by sponging the miR-145-SOX9 regulatory axis. Chang et al (32) have shown that HOTAIR regulates CCND1 and CCND2 expression by sponging miR-206 to stimulate the proliferation and invasion of ovarian cancer cells. Ding et al (33) have shown that the H19-miR-29b-3p-PGRN axis promotes epithelial-mesenchymal transition of colorectal cancer cells. In short, the previous studies collectively indicated that the lncRNA-miRNA-mRNA axis is an important regulatory mechanism in cancer progression. In order to gain insights into the underlying regulatory mechanism of GATA3-AS1, noteworthy miR-30b-5p, which has a binding site with GATA3-AS1, was predicted by bioinformatics analysis. Past studies have demonstrated that the miR-30 family plays a crucial regulatory role in the pathogenesis of different diseases, which indicates it may be a promising regulator (34,35). miR-30b was reported to be downregulated in PC, serving as a novel therapeutic agent for the treatment of PC (36,37). In the present study, consistent with the aforementioned result, miR-30b expression was significantly decreased in PC tissues and cell lines. Furthermore, a negative correlation between the expression of GATA3-AS1 and miR-30b-5p in the PC tissues was revealed, implying that GATA3-AS1 could directly bind to miR-30b-5p to sponge it. This implication was confirmed by dual-luciferase reporter assay. These data indicated that GATA3-AS1 acts as a ceRNA of miR-30b-5p in PC cells.
Notable Tex10, which shares a binding site with miR-30b-5p, was further predicted by bioinformatics analysis. Tex10, a component of the five friends of methylated Chtop and Rix complexes, is deeply involved in transcriptional regulation and ribosome biogenesis, as well as the cell cycle (38,39). Ding et al (40) identified Tex10 as an evolutionarily conserved key pluripotency factor that is required for embryonic stem cell self-renewal and pluripotency. Recently, Xiang et al (41,42) demonstrated that Tex10 is upregulated and plays a potent carcinogenic role in both HCC and esophageal squamous cell carcinoma. In the present study, similar to the aforementioned result, Tex10 was significantly upregulated in PC tissues and cell lines. The direct target relationship between miR-30b-5p and Tex10 was confirmed by dual-luciferase reporter assay. Furthermore, a positive correlation between the expression of GATA3-AS1 and Tex10 and a negative correlation between the expression of miR-30b-5p and Tex10 was revealed in PC tissues. In addition, GATA3-AS1 knockdown upregulated the expression of miR-30b-5p, but downregulated the expression of Tex10, whereas GATA3-AS1 overexpression had the opposite effects. Collectively, these data indicated that GATA3-AS1 acts as a ceRNA of miR-30b-5p, leading to the release of miR-30b-5p-targeted transcript Tex10 in PC cells.
The function of the GATA3-AS1/miR-30b-5p/Tex10 axis involved in PC was further addressed. The increased cell viability and proliferation, cell invasion, and stemness induced by GATA3-AS1 overexpression were all suppressed by the co-overexpression of GATA3-AS1 and miR-30b-5p; whereas the cell survival revealed an increasing trend with co-overexpression of GATA3-AS1, miR-30b-5p, and Tex10. These data indicated that the biological roles of GATA3-AS1 in PC are at least in part mediated by the miR-30b-5p-Tex10 axis. The aberrant activation of Wnt/β-catenin signaling has been reported to contribute to carcinogenesis and tumor progression of several cancers, including PC (43,44). Furthermore, the effects of miR-30b-5p and Tex10 have been revealed to be associated with Wnt/β-catenin signaling (42,45). Therefore, it was further assessed whether Wnt/β-catenin signaling participates in the regulation of the GATA3-AS1/miR-30b-5p/Tex10 axis. It was revealed that the upregulated expression of Wnt1 and β-catenin induced by GATA3-AS1 overexpression was suppressed by the co-overexpression of GATA3-AS1 and miR-30b-5p, but was upregulated once more by the co-overexpression of GATA3-AS1, miR-30b-5p, and Tex10. These results indicated that the GATA3-AS1/miR-30b-5p/Tex10 axis modulated Wnt/β-catenin signaling in PC cells.
In conclusion, the present findings demonstrated a significant increase of GATA3-AS1 expression in both PC tissues and cell lines. In vitro loss- or gain-of-function experiments revealed that the GATA3-AS1-miR-30b-5p-Tex10 axis modulated cell proliferation, invasion, apoptosis, and stemness in PC, which may be associated with the Wnt/β-catenin signaling pathway. The present study may provide some novel insights into identifying an informative biomarker and treatment targets for PC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
YL and LL conceived and designed the study. YL and GX performed the experiments, and analyzed the data. YL wrote the paper. YL, GX and LL reviewed and edited the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All patients were informed before their inclusion; written consent of the patients was obtained. All experimental protocols were approved by the Ethics Committee of The First Affiliated Hospital of Xi'an Jiaotong University (Xi'an, China). Animal protocols were approved by the Animal Care and Use Committee of the First Affiliated Hospital of Xi'an Jiaotong University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing of interests.
References
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 2.Henriksen A, Dyhl-Polk A, Chen I, Nielsen D. Checkpoint inhibitors in pancreatic cancer. Cancer Treat Rev. 2019;78:17–30. doi: 10.1016/j.ctrv.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 5.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. Adv Exp Med Biol. 2017;1008:1–46. doi: 10.1007/978-981-10-5203-3_1. [DOI] [PubMed] [Google Scholar]
- 7.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Z, Wang P, Zhang L, Huang C, Gao J, Li Y, Yang B. Identification of key genes and long noncoding RNA-associated competing endogenous RNA (ceRNA) networks in early-onset preeclampsia. Biomed Res Int. 2020;2020:1673486. doi: 10.1155/2020/1673486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vishnoi A, Rani S. MiRNA Biogenesis and regulation of diseases: An overview. Methods Mol Biol. 2017;1509:1–10. doi: 10.1007/978-1-4939-6524-3_1. [DOI] [PubMed] [Google Scholar]
- 10.Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141:1202–1207. doi: 10.1016/j.jaci.2017.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbons HR, Shaginurova G, Kim LC, Chapman N, Spurlock CF, III, Aune TM. Divergent lncRNA GATA3-AS1 regulates GATA3 transcription in T-Helper 2 cells. Front Immunol. 2018;9:2512. doi: 10.3389/fimmu.2018.02512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gulbinas A, Berberat PO, Dambrauskas Z, Giese T, Giese N, Autschbach F, Kleeff J, Meuer S, Büchler MW, Friess H. Aberrant gata-3 expression in human pancreatic cancer. J Histochem Cytochem. 2006;54:161–169. doi: 10.1369/jhc.5A6626.2005. [DOI] [PubMed] [Google Scholar]
- 13.Luo X, Zhou N, Wang L, Zeng Q, Tang H. Long noncoding RNA GATA3-AS1 promotes cell proliferation and metastasis in hepatocellular carcinoma by suppression of PTEN, CDKN1A, and TP53. Can J Gastroenterol Hepatol. 2019;2019:1389653. doi: 10.1155/2019/1389653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42((Database issue)):D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khachane AN, Harrison PM. Mining mammalian transcript data for functional long non-coding RNAs. PLoS One. 2010;5:e10316. doi: 10.1371/journal.pone.0010316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei W, Liu Y, Lu Y, Yang B, Tang L. LncRNA XIST promotes pancreatic cancer proliferation through miR-133a/EGFR. J Cell Biochem. 2017;118:3349–3358. doi: 10.1002/jcb.25988. [DOI] [PubMed] [Google Scholar]
- 18.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren J, Fu J, Ma T, Yan B, Gao R, An Z, Wang D. LncRNA H19-elevated LIN28B promotes lung cancer progression through sequestering miR-196b. Cell Cycle. 2018;17:1372–1380. doi: 10.1080/15384101.2018.1482137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang Q, Hann SS. HOTAIR: An oncogenic long non-coding RNA in human cancer. Cell Physiol Biochem. 2018;47:893–913. doi: 10.1159/000490131. [DOI] [PubMed] [Google Scholar]
- 21.Chang YT, Lin TP, Tang JT, Campbell M, Luo YL, Lu SY, Yang CP, Cheng TY, Chang CH, Liu TT, et al. HOTAIR is a REST-regulated lncRNA that promotes neuroendocrine differentiation in castration resistant prostate cancer. Cancer Lett. 2018;433:43–52. doi: 10.1016/j.canlet.2018.06.029. [DOI] [PubMed] [Google Scholar]
- 22.Kim J, Piao HL, Kim BJ, Yao F, Han Z, Wang Y, Xiao Z, Siverly AN, Lawhon SE, Ton BN, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet. 2018;50:1705–1715. doi: 10.1038/s41588-018-0252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S, Mei Z, Hu HB, Zhang X. The lncRNA MALAT1 contributes to non-small cell lung cancer development via modulating miR-124/STAT3 axis. J Cell Physiol. 2018;233:6679–6688. doi: 10.1002/jcp.26325. [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Guan Z, He K, Qian J, Cao J, Teng L. LncRNA UCA1 in anti-cancer drug resistance. Oncotarget. 2017;8:64638–64650. doi: 10.18632/oncotarget.18344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao F, Wang Q, Wu Q. The prognostic value and mechanisms of lncRNA UCA1 in human cancer. Cancer Manag Res. 2019;11:7685–7696. doi: 10.2147/CMAR.S200436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uroda T, Anastasakou E, Rossi A, Teulon JM, Pellequer JL, Annibale P, Pessey O, Inga A, Chillón I, Marcia M. Conserved pseudoknots in lncRNA MEG3 are essential for stimulation of the p53 pathway. Mol Cell. 2019;75:982–995.e9. doi: 10.1016/j.molcel.2019.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang W, Xie Y, Chen F, Liu X, Zhong LL, Wang HQ, Li QC. LncRNA MEG3 acts a biomarker and regulates cell functions by targeting ADAR1 in colorectal cancer. World J Gastroenterol. 2019;25:3972–3984. doi: 10.3748/wjg.v25.i29.3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Nestor CE, Zhao S, Lentini A, Bohle B, Benson M, Wang H. Profiling of human CD4+ T-cell subsets identifies the TH2-specific noncoding RNA GATA3-AS1. J Allergy Clin Immunol. 2013;132:1005–1008. doi: 10.1016/j.jaci.2013.05.033. [DOI] [PubMed] [Google Scholar]
- 29.Zhu YP, Bian XJ, Ye DW, Yao XD, Zhang SL, Dai B, Zhang HL, Shen YJ. Long noncoding RNA expression signatures of bladder cancer revealed by microarray. Oncol Lett. 2014;7:1197–1202. doi: 10.3892/ol.2014.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong P, Qiao F, Wu H, Cui H, Li Y, Zheng Y, Zhou M, Fan H. LncRNA UCA1 promotes tumor metastasis by inducing miR-203/ZEB2 axis in gastric cancer. Cell Death Dis. 2018;9:1158. doi: 10.1038/s41419-018-1170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Y, Zhang X, Hu X, Zhou W, Zhang P, Zhang J, Yang S, Liu Y. The effects of lncRNA MALAT1 on proliferation, invasion and migration in colorectal cancer through regulating SOX9. Mol Med. 2018;24:52. doi: 10.1186/s10020-018-0050-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang L, Guo R, Yuan Z, Shi H, Zhang D. LncRNA HOTAIR Regulates CCND1 and CCND2 expression by sponging miR-206 in ovarian cancer. Cell Physiol Biochem. 2018;49:1289–1303. doi: 10.1159/000493408. [DOI] [PubMed] [Google Scholar]
- 33.Ding D, Li C, Zhao T, Li D, Yang L, Zhang B. LncRNA H19/miR-29b-3p/PGRN axis promoted epithelial-mesenchymal transition of colorectal cancer cells by acting on wnt signaling. Mol Cells. 2018;41:423–435. doi: 10.14348/molcells.2018.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao L, Liu S, Hu L, Jia L, Wang H, Guo M, Chen C, Liu Y, Xu L. MiR-30 family: A promising regulator in development and disease. Biomed Res Int. 2018;2018:9623412. doi: 10.1155/2018/9623412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang SJ, Yang SY, Wang DD, Chen X, Shen HY, Zhang XH, Zhong SL, Tang JH, Zhao JH. The miR-30 family: Versatile players in breast cancer. Tumour Biol. 2017;39:1010428317692204. doi: 10.1177/1010428317692204. [DOI] [PubMed] [Google Scholar]
- 36.Xiong Y, Wang Y, Wang L, Huang Y, Xu Y, Xu L, Guo Y, Lu J, Li X, Zhu M, Qian H. MicroRNA-30b targets Snail to impede epithelial-mesenchymal transition in pancreatic cancer stem cells. J Cancer. 2018;9:2147–2159. doi: 10.7150/jca.25006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Azmi AS, Li Y, Aboukameel A, Muqbil I, Philip PA, Mohammad RM. DNA-methylation-caused downregulation of miR-30 contributes to the high expression of XPO1 and the aggressive growth of tumors in pancreatic ductal adenocarcinoma. Cancers (Basel) 2019;11:1101. doi: 10.3390/cancers11081101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fanis P, Gillemans N, Aghajanirefah A, Pourfarzad F, Demmers J, Esteghamat F, Vadlamudi RK, Grosveld F, Philipsen S, van Dijk TB. Five friends of methylated chromatin target of protein-arginine-methyltransferase[prmt]-1 (chtop), a complex linking arginine methylation to desumoylation. Mol Cell Proteomics. 2012;11:1263–1273. doi: 10.1074/mcp.M112.017194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castle CD, Cassimere EK, Denicourt C. LAS1L interacts with the mammalian Rix1 complex to regulate ribosome biogenesis. Mol Biol Cell. 2012;23:716–728. doi: 10.1091/mbc.e11-06-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding J, Huang X, Shao N, Zhou H, Lee DF, Faiola F, Fidalgo M, Guallar D, Saunders A, Shliaha PV, et al. Tex10 coordinates epigenetic control of super-enhancer activity in pluripotency and reprogramming. Cell Stem Cell. 2015;16:653–668. doi: 10.1016/j.stem.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiang X, Deng L, Xiong R, Xiao D, Chen Z, Yang F, Liu K, Feng G. Tex10 is upregulated and promotes cancer stem cell properties and chemoresistance in hepatocellular carcinoma. Cell Cycle. 2018;17:1310–1318. doi: 10.1080/15384101.2018.1480208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiang X, Xiong R, Yu C, Deng L, Bie J, Xiao D, Chen Z, Zhou Y, Li X, Liu K, Feng G. Tex10 promotes stemness and EMT phenotypes in esophageal squamous cell carcinoma via the Wnt/β-catenin pathway. Oncol Rep. 2019;42:2600–2610. doi: 10.3892/or.2019.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shang S, Hua F, Hu ZW. The regulation of beta-catenin activity and function in cancer: Therapeutic opportunities. Oncotarget. 2017;8:33972–33989. doi: 10.18632/oncotarget.15687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ram Makena M, Gatla H, Verlekar D, Sukhavasi S, K Pandey M, C Pramanik K. Wnt/β-Catenin signaling: The culprit in pancreatic carcinogenesis and therapeutic resistance. Int J Mol Sci. 2019;20:4242. doi: 10.3390/ijms20174242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang S, Miao D, Wang M, Lv J, Wang Y, Tong J. MiR-30-5p suppresses cell chemoresistance and stemness in colorectal cancer through USP22/Wnt/β-catenin signaling axis. J Cell Mol Med. 2019;23:630–640. doi: 10.1111/jcmm.13968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the present study are available from the corresponding author upon reasonable request.