Abstract
BACKGROUND
Sickle cell disease is characterized by hemolytic anemia, pain, and progressive organ damage. A high level of erythrocyte fetal hemoglobin (HbF) comprising α- and γ-globins may ameliorate these manifestations by mitigating sickle hemoglobin polymerization and erythrocyte sickling. BCL11A is a repressor of γ-globin expression and HbF production in adult erythrocytes. Its down-regulation is a promising therapeutic strategy for induction of HbF.
METHODS
We enrolled patients with sickle cell disease in a single-center, open-label pilot study. The investigational therapy involved infusion of autologous CD34+ cells transduced with the BCH-BB694 lentiviral vector, which encodes a short hairpin RNA (shRNA) targeting BCL11A mRNA embedded in a microRNA (shmiR), allowing erythroid lineage–specific knockdown. Patients were assessed for primary end points of engraftment and safety and for hematologic and clinical responses to treatment.
RESULTS
As of October 2020, six patients had been followed for at least 6 months after receiving BCH-BB694 gene therapy; median follow-up was 18 months (range, 7 to 29). All patients had engraftment, and adverse events were consistent with effects of the preparative chemotherapy. All the patients who could be fully evaluated achieved robust and stable HbF induction (percentage HbF/(F+S) at most recent follow-up, 20.4 to 41.3%), with HbF broadly distributed in red cells (F-cells 58.9 to 93.6% of untransfused red cells) and HbF per F-cell of 9.0 to 18.6 pg per cell. Clinical manifestations of sickle cell disease were reduced or absent during the follow-up period.
CONCLUSIONS
This study validates BCL11A inhibition as an effective target for HbF induction and provides preliminary evidence that shmiR-based gene knockdown offers a favorable risk–benefit profile in sickle cell disease. (Funded by the National Institutes of Health; ClinicalTrials.gov number, NCT03282656)
Sickle cell disease is a common inherited disease affecting approximately 100,000 persons in the United States1; worldwide, about 400,000 infants are born with sickle cell disease each year.2 Sickle hemoglobin (HbS) is a variant of normal adult hemoglobin (HbA) that is produced when the β-globin gene (HBB) contains a single E6V missense mutation. On deoxygenation, HbS polymerizes, leading to abnormally shaped red cells and protean downstream clinical sequelae, including painful vaso-occlusive crises, chronic hemolytic anemia, progressive and irreversible organ damage, decreased quality of life, and early death.
The standard curative treatment for sickle cell disease is allogeneic hematopoietic stem-cell transplantation. Matched sibling donor transplantation is curative in more than 90% of patients,3,4 but limitations include a higher risk of complications in older patients,3 a risk of severe graft-versus-host disease (GVHD), and lack of an available matched sibling in approximately 80% of cases.5 Alternative donor allogeneic transplantation is under active investigation. In children, transplantation from an unrelated donor has been associated with higher rates of severe chronic GVHD.6 Haplo-identical transplantation would expand the available donor pool, but it requires depletion of donor T cells either ex vivo or in vivo. Successes have been reported, but the challenges of graft failure and delayed immune reconstitution remain.7,8 For patients with β-hemoglobinopathy who are without a matched sibling donor, transplantation of genetically modified autologous hematopoietic stem cells is an attractive alternative. Clinical trials of lentiviral gene therapy based on the addition of a modified β-globin gene have demonstrated benefit in transfusion-dependent β-thalassemia,9,10 resulting in approval of one product in Europe, and have reported promising — albeit early — results in sickle cell disease.11,12
In utero and during infancy, the abnormal HbS protein is produced at very low levels because the erythrocytes have not yet shifted from expression of the γ-globin gene (HBG), which encodes the developmentally regulated component of HbF, to expression of the HBB gene, a phenomenon known as hemoglobin switching. Thus, infants with sickle cell disease are typically free of clinical symptoms, owing to the potent antisickling properties of HbF combined with the lower levels of HbS.13 In older children and adults, a higher level of HbF is associated with lower disease severity in sickle cell disease,14,15 as has been observed in persons with hereditary persistence of HbF and in those who have a good response to hydroxyurea.15–18 The overall percentage of HbF is important, but equally critical is the level of HbF within each red cell. To exert a beneficial effect, HbF must be distributed broadly enough to protect a high proportion of cells from intracellular HbS polymerization. Prior studies have shown that a 20% HbF level in whole blood is associated with amelioration of sickle cell disease symptoms.18 On an individual cell basis, although less well studied, a level of 10 pg of HbF per F-cell, or approximately one third of the cellular hemoglobin content, is likely to be sufficient to prevent HbS polymerization.19 Because of the prolonged half-life of nonsickling red cells as compared with sickling red cells, the results of allogeneic transplantation suggest that as little as 20% myeloid chimerism in the bone marrow is sufficient to ameliorate sickle cell disease symptoms,4,20–23 a result that is consistent with both mathematical modeling and experimental studies in mice.24
Although hydroxyurea leads to HbF induction in many patients, it is not effective in all patients. The transcription factor BCL11A has been validated as a repressor of HbF levels in model systems.25–27 Inactivation of BCL11A in a transgenic humanized sickle cell mouse model resulted in correction of the hematologic and pathologic defects associated with sickle cell disease.28 The induction of HbF by BCL11A repression is accompanied by coordinated reduction in expression of the HBBS gene, a major advantage of targeting this physiological switch. BCL11A plays key roles outside the erythroid lineage, including a role in the development and function of hematopoietic stem cells and B-lymphocytes,29–31 so clinical translation of a BCL11A-targeting therapy requires lineage specificity to selectively reduce the BCL11A protein in erythrocytes.
We have previously described a lentiviral vector that mediates potent erythroid-specific knockdown of BCL11A through RNA interference (RNAi), using a microRNA (miRNA)-adapted short hairpin RNA (shRNAmiR).31,32 To further optimize the vector titer, this therapeutic shmiR, under transcriptional control of regulatory elements derived from the β-globin locus,33 was inserted in a lentiviral vector backbone previously used in clinical trials,9,11 resulting in the self-inactivating third-generation BCH-BB694 lentiviral vector. In preclinical studies using this vector, lentiviral vector–transduced CD34+ cells derived from donors with sickle cell disease achieved a potent induction of HbF.33 Here we report treatment of patients with sickle cell disease using autologous cells transduced with a shmiR vector, as well as validation of BCL11A inactivation as a strategy for HbF induction.
METHODS
STUDY DESIGN AND INVESTIGATIONAL THERAPY
The primary aim of this pilot study is to assess the safety and feasibility of gene therapy with the BCH-BB694 BCL11A shmiR-encoding lentiviral vector in patients with severe sickle cell disease. The study protocol (available with the full text of this article at NEJM.org) was approved by the requisite regulatory and human-protection committees and was registered at ClinicalTrials.gov. Patients were enrolled after providing written informed consent or, if the patient was younger than 18 years old, after a guardian provided consent. Patients 12 to 17 years old provided assent. Eligibility was restricted to patients who had a genotype of HbSS, HbS/β0, HbSD, or HbSO and had clinically severe sickle cell disease, defined, as in many allogeneic transplantation sickle cell disease studies, by two or more episodes of acute chest syndrome during the preceding 2 years, three or more severe pain events during the preceding 2 years, recurrent priapism, red-cell alloimmunization in patients requiring transfusions, or an indication for regular transfusions for primary or secondary stroke prophylaxis. Patients were eligible for enrollment if they met these severity criteria despite receiving hydroxyurea therapy. Patients with an HLA-matched sibling who could serve as donor for transplantation were excluded from the study.
As of October 2020, a total of 11 patients had provided informed consent: one did not meet eligibility criteria, one had complications of underlying disease after consent and withdrew from the study, two had less than 6 months follow-up, and one had not yet been infused, leaving six patients with at least 6 months of follow-up (see Table S1 in the Supplementary Appendix, available at NEJM.org).
At least 3 months before infusion of gene-modified cells, ongoing hydroxyurea therapy was stopped and a protocol-prescribed transfusion regimen was initiated to reduce the level of HbS and thereby improve the safety of mobilization of hematopoietic stem cells and procedures requiring anesthesia. Apheresis for manufacture of the BCH-BB694 BCL11A shmiR drug product was performed after mobilization with 240 μg per kilogram of plerixafor, with a targeted collection for cell manufacturing of at least 4 million CD34+ cells per kilogram. An additional 2 million CD34+ cells per kilogram were collected by apheresis and stored, without manipulation, for rescue in the event of engraftment failure. CD34+ cells were selected with the use of a cell-isolation platform (CliniMACS), transduced with the BCH-BB694 BCL11A shmiR lentiviral vector (Fig. S1), and cryopreserved. Cell transduction was performed at the Dana–Farber Cell Manufacturing Core Facility as described elsewhere,33 in accordance with Good Manufacturing Practice conditions and with the use of uniform and validated standard operating procedures. Patients received fully myeloablative conditioning with intravenous busulfan for 4 consecutive days, with pharmacokinetic dose adjustment to target a daily area under the curve of approximately 5500 μM per minute. The transduced CD34+ cells were infused at least 24 hours after the last dose of busulfan. Patients are followed for 2 years before being offered enrollment in a 13-year long-term follow-up study.
CLINICAL, IMAGING, AND LABORATORY ASSESSMENTS
Patients were assessed regularly for adverse events (according to the Common Terminology Criteria for Adverse Events, version 4) and clinical and laboratory markers of sickle cell disease. The primary end point was neutrophil engraftment, as measured by an absolute neutrophil count of at least 0.5×109 per liter for 3 sequential days during the 7 weeks after infusion. Secondary end points included the presence and function of the transgene in various cell populations, as well as laboratory and clinical features of sickle cell disease. All statements about before-and-after comparisons are qualitative, are based on inspection of the data, and do not imply statistical significance.
Vector copy number was determined with the use of a quantitative polymerase-chain-reaction (qPCR) assay performed on the bulk manufactured cell product after in vitro erythroid differentiation and on whole blood, blood mononuclear cells, and hematopoietic lineage-isolated peripheral blood and bone marrow cells. To determine the percentage of progenitor cells within the product that were transduced, clonogenic hematopoietic progenitors were plated in methylcellulose for 12 to 14 days, followed by qPCR assay of individual colonies to detect the integrated lentiviral vector. Peripheral blood samples were assessed for hemoglobin by routine complete blood counts, HbF by high-performance liquid chromatography, and percentage of F-cells by flow cytometry. At each study visit, clinical data were collected, including any reports of vaso-occlusive pain or other sickle-related events and transfusion frequency.
STUDY OVERSIGHT
The study was performed under an approved Food and Drug Administration investigational new drug application and was reviewed at regularly scheduled intervals by an independent data and safety monitoring board assigned by the National Heart, Lung, and Blood Institute. Data reported were current as of October 2020.
RESULTS
PATIENTS AND TREATMENT
Six patients (7 to 25 years of age at enrollment) received the investigational gene therapy between February 2018 and March 2020, and the patients had a median follow-up of 18 months (range, 7 to 29) after infusion. The severe manifestations of sickle cell disease that qualified the patients for enrollment included history of stroke (3 patients), frequent vaso-occlusive events (2), and frequent episodes of priapism (1) (Table 1). Patient 3 had previously diagnosed and surgically repaired moyamoya neurovascular disease. Because of a potential higher risk of subsequent stroke, a predetermined transfusion regimen was described in the consent process and was implemented after infusion of gene-modified cells in order to maintain a maximum HbS level no greater than the patient’s baseline level before gene therapy. Patients 2 and 7, who were undergoing regular transfusion, had a history of stroke more than 10 years before enrollment but had no evidence of neurovascular disease on magnetic resonance angiography images. Transfusions were not continued after engraftment in these two patients, once suitable induction of HbF was observed.
Table 1.
Baseline Characteristics of the Treated Patients and the Drug Product.*
| Patient Number | Age in Yr, Sex | Genotype | Severe Symptoms | Receiving Transfusion, Hydroxyurea | CD34+ Cell Dose | VCN of Product | CD34+ Cells Transduced | Follow-up |
|---|---|---|---|---|---|---|---|---|
| cells per kg | copies per diploid genome | % | mo | |||||
| 2 | 20, M | βS/βS | Previous stroke | Yes, No | 5.1 | 3.3 | 95.8 | 29 |
| 3 | 25, M | βS/βS | Previous stroke | Yes, No | 6.7 | 5.0 | 98.6 | 19 |
| 4 | 24, M | βS/βS | Priapism | No, Yes | 5.2 | 3.3 | 95.5 | 20 |
| 6 | 16, F | βS/β0 | Vaso-occlusive event, ACS | No, Yes | 8.3 | 6.9 | 100.0 | 16 |
| 7 | 12, F | βS/βS | Previous stroke | Yes, No | 6.1 | 2.8 | 93.1 | 12 |
| 8 | 7, M | βS/βS | Vaso-occlusive event | No, Yes | 4.9 | 1.8 | 62.0 | 7 |
Baseline laboratory values represent the most recent samples collected before cell infusion for which all relevant data are available. ACS denotes acute chest syndrome, and VCN vector copy number.
CD34+ cell products were the result of one mobilization cycle consisting of plerixafor and apheresis on 1 or 2 sequential days, yielding a median of 9.3 million CD34+ cells per kilogram (range, 7.8 million to 11.4 million) (Table S2). The unmanipulated back-up collection required additional apheresis of 1 to 2 days. One patient, Patient 4, was discovered to have restarted hydroxyurea inadvertently before and during the first round of mobilization and collection. After cessation of hydroxyurea, an additional collection of back-up cells was performed. The transduction protocol yielded a vector copy number in the manufactured cell product of 1.8 to 6.9 copies per diploid genome (Table 1). The bulk vector copy number appeared to correlate with the percentage of cells transduced, as measured by qPCR of individual CD34-derived progenitor cells in the drug product (Fig. S2). Pharmacokinetic analysis of plasma busulfan was performed after the first three doses, followed by dose adjustments for a combined total target dose of approximately 20,740 to 23,180 μM per minute per liter (approximately 85 to 95 mg × hour per liter) (Table S3).
Patients were infused with the BCH-BB694 BCL11A shmiR drug product, which contained 4.9 million to 8.3 million CD34+ cells per kilogram of body weight (Table 1). After the infusion, all patients achieved neutrophil engraftment (median, 22 days [range, 18 to 26]) and platelet engraftment (median, 33 days [range, 26 to 62]). One patient, Patient 8, received a short course of filgrastim to treat a postinfusion respiratory infection.
SAFETY OUTCOMES
No adverse events of grade 3 or higher were associated with mobilization, collection, or infusion in the patients described here. Patient 1, whose product infusion was delayed owing to underlying medical complications and whose data are not reported here, had a serious adverse event. With an indwelling central venous catheter in place, and after the patient was exposed to estrogen for fertility preservation, a right atrial thrombus and pulmonary emboli developed. Most adverse effects occurred before neutrophil engraftment and were effects known to be associated with central venous access devices or myeloablative chemotherapy. Patient 8 received a new diagnosis of type 1 diabetes mellitus 2 weeks after infusion, while the patient had a severe respiratory infection. The presence of autoantibodies suggests that the patient had an underlying predisposition to type 1 diabetes mellitus that was unmasked by intercurrent illness or conditioning. All nonlaboratory adverse events of grade 3 or higher are listed in Table S4. After the patients were discharged from the hospital where transplantation was performed, three serious adverse events occurred: hospital admission for less than 24 hours for fever and influenza infection (1 patient), recurrent priapism leading to two hospital admissions approximately 4 months after gene therapy (1 patient), and leg pain (atypical of sickle-related pain, osteomyelitis ruled out, and no intervention required) leading to a readmission for less than 24 hours shortly after discharge (1 patient).
LABORATORY OUTCOMES
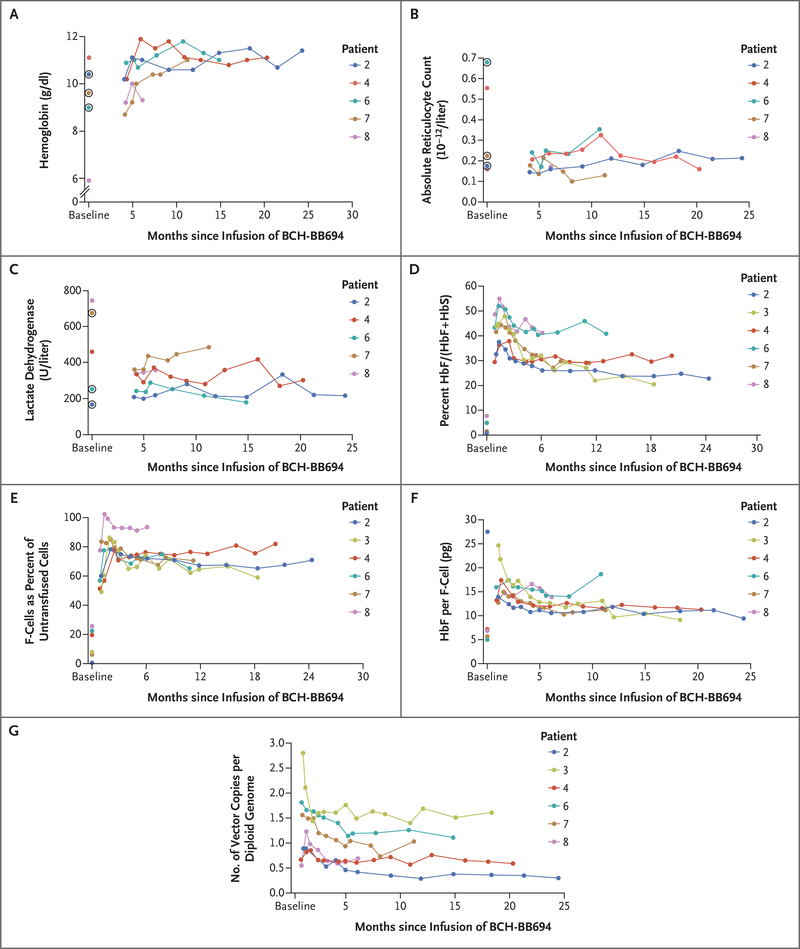
After infusion with the BCH-BB694 BCL11A shmiR drug product, the HbF fraction of total Hb increased and then remained stable in all patients. Hemoglobin increased to a new baseline of 9.3 to 11.4 g per deciliter in the five untransfused patients and was stable over time (Table 2 and Fig. 1A). Hemolysis continued in all patients but was reduced from baseline, with the absolute reticulocyte count falling from 175,000–680,000 per microliter to 160,000–355,000 per microliter (Fig. 1B) and with lactate dehydrogenase falling from 167–726 U per liter to 217–485 U per liter (Fig. 1C). Patient 3, who continued regular pre–gene therapy transfusions, required less frequent transfusions to maintain HbS suppression at his pre–gene therapy level (Table S5). At the most recent study visit as of October 2020 (6 to 24 months after infusion), the median percentage of HbF as measured by high-performance liquid chromatography in the five untransfused patients was 30.4% (range, 21.6 to 40.0). In all six patients, the induction of HbF was uniformly high, with a median HbF/(HbF+HbS) of 30.5% (range, 20.4 to 41.3). The level of total HbF appeared to be stable during follow-up (Fig. 1D). The induction of HbF was distributed broadly across erythrocytes. At the most recent study visit as of October 2020, the median percentage of F-cells among untransfused red cells was 70.8% (range, 58.9 to 93.6) which represented a robust increase from baseline (median, 14% [range, 0.7 to 25.5]) (Fig. 1E). The average HbF per F-cell was calculated and was uniformly high, with concomitant reduction in the cellular concentration of HbS. In patients 3, 4, 6, 7, and 8, HbF per F-cell increased from 5.0–7.1 pg per cell to an average of 12.7 pg per cell (range, 9.0 to 18.6) (Table 2 and Fig. 1F). One patient, Patient 2, had only 0.3% F-cells at baseline, resulting in an outlying calculated HbF per F-cell of 27.4 pg per cell. At 24 months of follow-up, Patient 2 had 71% F-cells, with 9.4 pg HbF per F-cell (Table 2).
Table 2.
Hematologic Data.*
| Patient Number | Months since Infusion | Hb | Hct | MCV | MCHC | HbF‡ | F-Cells§ | HbF per F-Cell, Baseline, Most Recent¶ | ARC (Normal, 43–85) | LDH (Normal, 100–210) |
|---|---|---|---|---|---|---|---|---|---|---|
| g/dl | % | fl | g/dl | % | % | pg | × 10−9/liter | U per liter | ||
| 2 | 24 | 11.4 | 32.5 | 87.8 | 35.1 | 22.7 | 71.0 | 27.4,¶ 9.4 | 215 | 217 |
| 3† | 18 | 9.5 | 28.9 | 86.3 | 32.9 | 20.4 | 58.9 | 5.6, 9.0 | 355 | 369 |
| 4 | 21 | 11.1 | 31.2 | 84.8 | 35.6 | 31.9 | 81.9 | 7.1, 11.2 | 160 | 301 |
| 6 | 15 | 11.0 | 32.5 | 76.0 | 33.8 | 38.8∥ | 65.3 | 5.0, 18.6∥ | 244 | 180 |
| 7 | 12 | 11.0 | 31.4 | 81.1 | 35.0 | 29.0 | 70.6 | 5.6, 11.1 | 130 | 485 |
| 8 | 6 | 9.3 | 25.5 | 88.9 | 36.5 | 41.3 | 93.6 | 6.8, 13.8 | 170 | 359 |
Data represent the most recent values available before the July 2020 data snapshot. ARC denotes absolute reticulocyte count, Hb hemoglobin, HbF fetal hemoglobin, Hct hematocrit, LDH lactate dehydrogenase, MCHC mean corpuscular hemoglobin concentration, and MCV mean corpuscular volume.
As of October 2020, Patient 3 has continued transfusions. At the most recent pretransfusion time point, HbA was 46.3%, HbS 39.0%, and HbF 12.1%.
HbF values are presented as HbF/(HbF + HbS).
F cells are reported as a percentage of untransfused red cells.
The average HbF per F-cell was calculated with the following formula: (MCH × HbF) per% F-cells. The HbF per F-cell calculation at baseline in Patient 2 was influenced by a very low baseline percentage of F-cells of 0.3%.
The most recent HbF was at 12 months, and the most recent HbF per F-cell calculation was at 10 months.
Figure 1. Fetal Hemoglobin Induction and Gene Marking after Gene Therapy.
Panel A shows hemoglobin (Hb), Panel B absolute reticulocyte count (ARC), and Panel C lactate dehydrogenase over time after infusion. In Panels A, B, and C, circled dots indicate the presence of transfused red cells at the time of the baseline sample. In these three panels, Patient 3 is not shown owing to reinitiation of regular red-cell transfusions. Panel D shows HbF as a percent of HbF + HbS at various times after infusion. Panel E shows the percent of F-cells (circulating untransfused red cells that express HbF) at various times after infusion. In Panels A through F, the baseline values shown indicate the values before gene therapy, which are the latest samples collected before cell infusion for which relevant data are available. Patients 2, 3, and 7 at the baseline time point were receiving regular transfusions; for Patient 4, the baseline time point is 2 months after hydroxyurea was discontinued and after 3 months of pre–gene therapy exchange transfusions; for Patient 6, the baseline time point is 2 weeks after hydroxyurea was discontinued and 1 month after a transfusion given for clinical indications; and Patient 8 at the baseline time point was receiving hydroxyurea before starting pre–gene therapy transfusions. Panel G shows the whole-blood vector copy number in peripheral blood for each of the six patients at various times after infusion.
Since BCL11A expression is required for normal B-cell development, immunoglobulins and CD19+ and CD3+ levels were evaluated at baseline and at 6, 12, and 24 months after infusion. The results indicate normal development and function of the B-cell compartment and suggest an absence of toxicity consistent with erythroid-specific BCL11A suppression (Table S6).
The vector copy number in peripheral whole blood ranged from 0.42 to 1.49 copies per diploid genome at 6 months after infusion and remained generally stable at subsequent time points (Fig. 1G). We specifically examined the stability of gene marking within the CD19+ B-cell lineage because B-cell precursors inappropriately expressing the transgene would be selected against over time. Between the early engraftment period of 3 to 6 months, vector copy number in CD19+ cells was stable in all six patients. Gene marking within the peripheral blood erythroid lineage, which is the intended cellular target, was 0.23 to 1.99 copies per diploid genome at the latest time point. Bone marrow CD34+, CD15+ myeloid compartment, and erythroid precursors defined as glycophorin A+, CD71+ cells at 6 months showed similar vector copy numbers (Table 3).
Table 3.
Vector Copy Number of Peripheral Blood and Bone Marrow (Copies per Diploid Genome).
| Source and Time Point | Patient 2 | Patient 3 | Patient 4 | Patient 6 | Patient 7 | Patient 8 |
|---|---|---|---|---|---|---|
| Peripheral blood | ||||||
| 1.5 mo | ||||||
| Whole blood | 0.89 | 2.11 | 0.82 | 1.66 | 1.49 | 1.23 |
| CD19+ | 0.003 | 0.02 | 0.03 | 0.03 | 0.06 | 0.03 |
| GlyA+, CD71+* | 0.47 | 2.39 | 1.31 | 1.02 | 1.38 | 1.07 |
| 3 mo | ||||||
| Whole blood | 0.53 | 1.62 | 0.65 | 1.51 | 1.14 | 0.64 |
| CD19+ | 0.3 | 1.2 | 0.48 | 1.11 | 0.68 | 0.55 |
| GlyA+, CD71+ | 0.33 | 2.79 | 1.03 | 1.86 | 0.88 | 0.90 |
| 6 mo | ||||||
| Whole blood | 0.42 | 1.49 | 0.61 | 1.19 | 1.04 | 0.69 |
| CD19+ | 0.5 | 1.21 | 0.5 | 1.13 | 0.66 | 0.64 |
| GlyA+, CD71+ | 0.2 | 2.85 | 0.96 | 1.35 | 0.76 | 0.68 |
| Most recent time point† | ||||||
| Whole blood | 0.30 | 1.61 | 0.63 | 1.11 | 1.03 | 0.69‡ |
| CD19+ | 0.43 | 1.37 | 0.61 | 1.11 | 0.78 | 0.64‡ |
| GlyA+, CD71+ | 0.23 | 1.99 | 0.77 | 1.95 | 1.39 | 0.68‡ |
| Bone marrow | ||||||
| 6 mo | ||||||
| CD34+ | 0.53 | 1.52 | 0.60 | 1.20 | 0.86 | 0.59 |
| CD15+ | 0.65 | 1.59 | 0.79 | 1.65 | 1.25 | 0.77 |
| GlyA+, CD71+ | 0.63 | 1.62 | 0.80 | 1.42 | 1.34 | 0.73 |
CD19+ describes B-lymphoid cells, and GlyA+, CD71+ describes erythroid precursors.
Data are the most recent values available with vector copy number lineage sorting before the October 2020 data snapshot (range, 6 to 24 months).
Month 6 was the most recent time point for Patient 8.
CLINICAL OUTCOMES
No patient has had a vaso-occlusive crisis, acute chest syndrome, or stroke since the gene therapy infusion (Table 4). Patient 4 (whose infusion had been performed 20 months ago as of October 2020) had required frequent hospitalizations for priapism before gene therapy. Between 4 and 8 months after gene therapy, he had several severe episodes of priapism. He has not had a priapism-related emergency department visit or hospitalization since month 8, although intermittent episodes of priapism have recurred. Patient 6 had left hip avascular necrosis before study enrollment. At 9 months after gene therapy, symptoms consistent with avascular necrosis of the right hip had developed in this patient.
Table 4.
Clinical Events before and after Infusion.*
| Patient Number | Months since Infusion | No. of Transfusions (Annualized) | No. of Severe Sickle Cell Clinical Events† | |||
|---|---|---|---|---|---|---|
| Prestudy | After Engraftment | Prestudy | After Gene Therapy (<5 mo) | After Gene Therapy (≥5 mo) | ||
| 2 | 29 | 12.5 | 0 | 0 | 0 | 0 |
| 3 | 19 | 10.5 | 5.7 | 0 | 0 | 0 |
| 4 | 20 | 2 | 0 | 13 | 5 | 1‡ |
| 6 | 16 | 3 | 0 | 6 | 0 | 0 |
| 7 | 12 | 11 | 0 | 0 | 0 | 0 |
| 8 | 7 | 1 | 0 | 3 | 0 | 0 |
Prestudy events represent the 2 years preceding study enrollment. Events that occurred after gene therapy represent the time from engraftment to the most recent follow-up.
Events include vaso-occlusive crisis pain requiring an emergency department visit or admission for opioid treatment, admission for acute chest syndrome, and priapism events requiring an emergency department visit or admission for a procedural intervention.
The most recent severe event was an emergency department visit that occurred 8 months after gene therapy.
Three patients (Patients 2, 3, and 7) were receiving regular exchange transfusion regimens for secondary stroke prophylaxis before gene therapy. Patients 2 and 7 have not received a red-cell transfusion since engraftment.
DISCUSSION
We report findings in six patients with severe sickle cell disease treated with therapy directed against BCL11A, in this case through infusion with BCH-BB694 BCL11A shmiR transduced CD34+ cells, an approach that reverses hemoglobin switching in order to increase intracellular HbF concentration and concomitantly reduce HbS. All six had reached more than 6 months of follow-up as of October 2020 and had achieved robust induction of HbF, with a favorable safety profile for peripheral autologous stem-cell mobilization and collection, conditioning, and transplantation.
In transduced cells, BCL11A is suppressed by an shRNA embedded in a microRNA architecture that harnesses the endogenous cellular machinery to produce a simultaneous increase in HbF protein and decrease in HbS protein. Altering the balance of intracellular globin production is particularly advantageous, because even a small decrease in HbS concentration results in a large increase in the time to polymer formation, the main determinant of the sickling process in red cells.34,35 The HbF per F-cell for the reported patients appears to be stable over time and across patients, and the percentage of red cells that are F-cells is consistently high, indicating broad distribution of HbF in transduced cells. The HbF levels achieved in this study are similar to or exceed those seen in patients with nondeletional hereditary persistence of HbF,36 who have a milder clinical course,37,38 and above a threshold level of 20% HbF that is associated with significant amelioration of clinical symptoms.18 BCH-BB694 BCL11A shmiR–transduced cells demonstrated high levels of HbF per F-cell regardless of the in vivo vector copy numbers, which suggests that both potent effects of the vector and a maximum induction of HbF are possible at low vector copy numbers.
As is true with other autologous gene therapy approaches for hemoglobinopathies, patients who have completed this treatment have a partial correction of disease, based on laboratory measures. Bernaudin et al. recently reported that chimerism of less than 50% in allogeneic transplantation is associated with persistent laboratory evidence of hemolysis4; however, multiple studies have demonstrated that mixed-chimerism patients are free of other sickle cell disease-related symptoms as long as chimerism exceeds approximately 20%.4,20–22 This is because patients with even a relatively small proportion of donor engrafted cells are capable of sustaining much higher percentages of donor-derived erythrocytes owing to the prolongation of the red-cell lifespan in the peripheral blood.23 In this regard, the post–autologous therapy cellular phenotype is distinct from that of post-allogeneic transplantation. In allogeneic transplantation, there are distinct populations of autologous (HbSS) and donor (HbAA or HbAS) cells, whereas the post–gene therapy patients in this study most likely retain a small population of cells containing only HbS from nonablated autologous cells or nontransduced infused cells and a population of F-cells that contain a mixture of HbS and antisickling HbF. It will be important to assess whether the population of residual HbS-containing cells and associated hemolysis contribute to any ongoing microvascular damage.
Unlike earlier trials of gene therapy in which enhancer-containing γ retroviruses caused leukemia in some patients owing to insertional mutagenesis, the current generation of lentiviral vectors has not so far been associated with insertional mutagenesis, with up to 12 years of follow-up.39 However myeloablative busulfan conditioning also carries a small risk of secondary cancer, and in one patient with sickle cell disease who underwent gene therapy in another trial, myelodysplastic syndrome and subsequent acute myeloid leukemia developed that were unrelated to the lentiviral vector.40 In this trial and other autologous genetic treatment studies, it will be important to evaluate the risk of post-alkylator myelodysplasia and to determine whether factors such as busulfan exposure affect this risk.
The initial results of this trial provide validation that BCL11A, a gene discovered to be involved in globin developmental regulation through population-based genomewide association studies,25–27 can be targeted to lead to successful HbF induction in humans. The field of autologous gene therapies for hemoglobinopathies is advancing rapidly, including lentiviral trials of gene addition in which the nonsickling hemoglobin is formed from an exogenous γ-globin or modified β-globin gene. In the approach described here, targeting the physiologic hemoglobin switch allows transcription of the endogenous γ-globin gene. This approach is also a direct intent of the current gene editing trials targeting BCL11A. The lineage-specific reduction in this trial predicts that clustered regularly interspaced short palindromic repeats (CRISPR)–mediated disruption of erythroid-specific enhancer in the BCL11A gene41,42 will also be beneficial if a sufficient percentage of hematopoietic stem cells can be edited at clinical scale and if the edits lead to adequate reduction of BCL11A expression in each edited cell.43
This study reports in humans the use of a short hairpin RNA embedded within an endogenous microRNA scaffold, termed a shmiR vector, to alter genetic expression, rather than reliance on the addition of a protein coding sequence. This development has potential implications for other diseases that could benefit from down-regulation of gene expression rather than addition of a gene. On the basis of current HbF levels, percentage of F-cells, and HbF per F-cell, we predict that the patients in this study will have protection from sickling to prevent or significantly ameliorate both acute and chronic complications of sickle cell disease. Additional follow-up will clarify the long-term effects.
Supplementary Material
Acknowledgments
Supported by a grant (5R01HL137848) from the National Heart, Lung, and Blood Institute, National Institutes of Health. The GMP vector was produced and furnished by Bluebird Bio as part of a licensing agreement between Boston Children’s Hospital and Bluebird Bio, and as part of the agreement, Bluebird Bio prereviewed an earlier version of the manuscript.
Dr. Esrick reports receiving grant support, paid to Boston Children’s Hospital, and steering committee fees from bluebird bio, and grant support, paid to Boston Children’s Hospital, from Sangamo Therapeutics; Dr. Achebe, receiving advisory board fees from Fulcrum Therapeutics, Global Blood Therapeutics, and Pharmacosmos; Dr. Shaw, receiving consulting fees from Orchard Therapeutics; Dr. O. Negre, being employed by and owning shares in bluebird bio; Dr. Ritz, receiving advisory board fees from Aleta Biotherapeutics, Celgene, Falcon Therapeutics, LifeVaultBio, Rheos Medicines, Talaris Therapeutics, and TScan Therapeutics, grant support from Amgen, Equillium, and Kite, fees for serving on a data and safety monitoring board from AvroBio, and consulting fees from Infinity Pharmaceuticals; Dr. London, receiving fees for serving on a data and safety monitoring board from Jubilant Draximage and Merck; Dr. Heeney, receiving consulting fees, advisory board fees, and clinical trial support from AstraZeneca and Novartis, fees for serving on a data and safety monitoring board from CRISPR Therapeutics and Vertex Pharmaceuticals, advisory board fees and clinical trial support from Cyclerion and Global Blood Therapeutics, advisory board fees from Forma Therapeutics, and consulting fees and clinical trial support from Micelle Biopharma (Sancilio & Co), and Pfizer; Dr. Manis, receiving advisory board fees from Sanofi US Services; Dr. Williams, receiving advisory board fees from Beam Therapeutics and Geneception, grant support and advisory board fees from bluebird bio and Orchard Therapeutics, consulting fees from Emerging Therapy Solutions, serving as an advisor for Novartis, serving as cofounder of Alerion Biosciences, and serving as site investigator for a trial sponsored by Sangamo. No other potential conflict of interest relevant to this article was reported.
We thank current and previous employees of Bluebird Bio for helpful discussions; Mursal Hassan and Pauleen Faynberg for administrative assistance; Pei-Chi Kao for assistance with statistics and graphics; Stephanie Patriarca for clinical research coordination assistance; Ellen Proeung and the BCH Therapeutic Apheresis Unit for transfusion-related patient care; BCH Hematology and HSCT providers for inpatient and post-transplantation clinical care; the hematology teams at Massachusetts General Hospital, Hasbro Children’s Hospital, Phoenix Children’s Hospital, and Wyckoff Heights Medical Center for patient care collaboration; and David G. Nathan, Stuart Orkin, Vijay Sankaran, Franklin Bunn, and Orah Platt for critical review of an earlier version of the manuscript and for helpful discussions.
Footnotes
Contributor Information
Erica B. Esrick, Dana–Farber/Boston Children’s Cancer and Blood Disorders Center, Harvard Medical School, Boston
Leslie E. Lehmann, Dana–Farber/Boston Children’s Cancer and Blood Disorders Center, Harvard Medical School, Boston
Alessandra Biffi, Dana–Farber/Boston Children’s Cancer and Blood Disorders Center, Harvard Medical School, Boston Harvard Stem Cell Institute, Harvard Medical School, Boston; Gene Therapy Program, Dana–Farber/Boston Children’s Cancer and Blood Disorders Center, Boston.
Maureen Achebe, Division of Hematology, Brigham and Women’s Hospital, Harvard Medical School, Boston
Christian Brendel, Dana–Farber/Boston Children’s Cancer and Blood Disorders Center, Harvard Medical School, Boston Harvard Stem Cell Institute, Harvard Medical School, Boston.
Marioara F. Ciuculescu, Dana–Farber/Boston Children’s Cancer and Blood Disorders Center, Harvard Medical School, Boston Gene Therapy Program, Dana–Farber/Boston Children’s Cancer and Blood Disorders Center, Boston.
Heather Daley, Connell and O’Reilly Families Cell Manipulation Core Facility, Dana–Farber Cancer Institute, Boston
Brenda MacKinnon, Dana–Farber/Boston Children’s Cancer and Blood Disorders Center, Harvard Medical School, Boston Gene Therapy Program, Dana–Farber/Boston Children’s Cancer and Blood Disorders Center, Boston.
Emily Morris, Gene Therapy Program, Dana–Farber/Boston Children’s Cancer and Blood Disorders Center, Boston
Amy Federico, Gene Therapy Program, Dana–Farber/Boston Children’s Cancer and Blood Disorders Center, Boston
Daniela Abriss, TransLab, Boston Children’s Hospital, Boston
Kari Boardman, Dana–Farber/Boston Children’s Cancer and Blood Disorders Center, Harvard Medical School, Boston
Radia Khelladi, Connell and O’Reilly Families Cell Manipulation Core Facility, Dana–Farber Cancer Institute, Boston
Kit Shaw, Connell and O’Reilly Families Cell Manipulation Core Facility, Dana–Farber Cancer Institute, Boston
Helene Negre, Connell and O’Reilly Families Cell Manipulation Core Facility, Dana–Farber Cancer Institute, Boston
Olivier Negre, Bluebird Bio, Cambridge, MA
Sarah Nikiforow, Connell and O’Reilly Families Cell Manipulation Core Facility, Dana–Farber Cancer Institute, Boston
Jerome Ritz, Connell and O’Reilly Families Cell Manipulation Core Facility, Dana–Farber Cancer Institute, Boston
Sung-Yun Pai, Dana–Farber/Boston Children’s Cancer and Blood Disorders Center, Harvard Medical School, Boston the Gene Therapy Program, Dana–Farber/Boston Children’s Cancer and Blood Disorders Center, Boston.
Wendy B. London, Dana–Farber/Boston Children’s Cancer and Blood Disorders Center, Harvard Medical School, Boston
Colleen Dansereau, Dana–Farber/Boston Children’s Cancer and Blood Disorders Center, Harvard Medical School, Boston Gene Therapy Program, Dana–Farber/Boston Children’s Cancer and Blood Disorders Center, Boston.
Matthew M. Heeney, Dana–Farber/Boston Children’s Cancer and Blood Disorders Center, Harvard Medical School, Boston
Myriam Armant, TransLab, Boston Children’s Hospital, Boston
John P Manis, Department of Laboratory Medicine, Boston Children’s Hospital, Harvard Medical School, Boston
David A. Williams, Dana–Farber/Boston Children’s Cancer and Blood Disorders Center, Harvard Medical School, Boston Gene Therapy Program, Dana–Farber/Boston Children’s Cancer and Blood Disorders Center, Boston.
REFERENCES
- 1.Heeney MM, Ware RE. Hydroxyurea for children with sickle cell disease. Hematol Oncol Clin North Am 2010;24:199–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Piel FB, Patil AP, Howes RE, et al. Global epidemiology of sickle haemoglobin in neonates: a contemporary geostatistical model-based map and population estimates. Lancet 2013;381:142–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gluckman E, Cappelli B, Bernaudin F, et al. Sickle cell disease: an international survey of results of HLA-identical sibling hematopoietic stem cell transplantation. Blood 2017;129:1548–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernaudin F, Dalle J-H, Bories D, et al. Long-term event-free survival, chimerism and fertility outcomes in 234 patients with sickle-cell anemia younger than 30 years after myeloablative conditioning and matched-sibling transplantation in France. Haematologica 2020;105:91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krishnamurti L, Abel S, Maiers M, Flesch S. Availability of unrelated donors for hematopoietic stem cell transplantation for hemoglobinopathies. Bone Marrow Transplant 2003;31:547–50. [DOI] [PubMed] [Google Scholar]
- 6.Shenoy S, Eapen M, Panepinto JA, et al. A trial of unrelated donor marrow transplantation for children with severe sickle cell disease. Blood 2016;128:2561–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leonard A, Tisdale J, Abraham A. Curative options for sickle cell disease: haploidentical stem cell transplantation or gene therapy? Br J Haematol 2020;189:408–23. [DOI] [PubMed] [Google Scholar]
- 8.Bolaños-Meade J, Cooke KR, Gamper CJ, et al. Effect of increased dose of total body irradiation on graft failure associated with HLA-haploidentical transplantation in patients with severe haemoglobinopathies: a prospective clinical trial. Lancet Haematol 2019;6(4):e183–e193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavazzana-Calvo M, Payen E, Negre O, et al. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature 2010;467: 318–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson AA, Walters MC, Kwiatkowski J, et al. Gene therapy in patients with transfusion-dependent β-thalassemia. N Engl J Med 2018;378:1479–93. [DOI] [PubMed] [Google Scholar]
- 11.Ribeil J-A, Hacein-Bey-Abina S, Payen E, et al. Gene therapy in a patient with sickle cell disease. N Engl J Med 2017;376: 848–55. [DOI] [PubMed] [Google Scholar]
- 12.Kanter JWTJ, Mapara MY, Kwiatkowski JL, et al. Resolution of sickle cell disease manifestations in patients treated with lentiglobin gene therapy: updated results from the phase 1/2 Hgb-206 Group C Study. Blood 2019;134:Suppl 1:990. [Google Scholar]
- 13.Bunn HF. Pathogenesis and treatment of sickle cell disease. N Engl J Med 1997; 337:762–9. [DOI] [PubMed] [Google Scholar]
- 14.Platt OS, Thorington BD, Brambilla DJ, et al. Pain in sickle cell disease: rates and risk factors. N Engl J Med 1991;325: 11–6. [DOI] [PubMed] [Google Scholar]
- 15.Platt OS, Brambilla DJ, Rosse WF, et al. Mortality in sickle cell disease: life expectancy and risk factors for early death. N Engl J Med 1994;330:1639–44. [DOI] [PubMed] [Google Scholar]
- 16.Perrine RP, Brown MJ, Clegg JB, Weatherall DJ, May A. Benign sickle-cell anaemia. Lancet 1972;2:1163–7. [DOI] [PubMed] [Google Scholar]
- 17.Charache S, Terrin ML, Moore RD, et al. Effect of hydroxyurea on the frequency of painful crises in sickle cell anemia. N Engl J Med 1995;332:1317–22. [DOI] [PubMed] [Google Scholar]
- 18.Powars DR, Weiss JN, Chan LS, Schroeder WA. Is there a threshold level of fetal hemoglobin that ameliorates morbidity in sickle cell anemia? Blood 1984; 63:921–6. [PubMed] [Google Scholar]
- 19.Steinberg MH, Chui DHK, Dover GJ, Sebastiani P, Alsultan A. Fetal hemoglobin in sickle cell anemia: a glass half full? Blood 2014;123:481–5. [DOI] [PubMed] [Google Scholar]
- 20.Hsieh MM, Wu CJ, Tisdale JF. In mixed hematopoietic chimerism, the donor red cells win. Haematologica 2011;96: 13–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fitzhugh CD, Cordes S, Taylor T, et al. At least 20% donor myeloid chimerism is necessary to reverse the sickle phenotype after allogeneic HST. Blood 2017;130: 1946–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abraham A, Hsieh M, Eapen M, et al. Relationship between mixed donor-recipient chimerism and disease recurrence after hematopoietic cell transplantation for sickle cell disease. Biol Blood Marrow Transplant 2017;23:2178–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andreani M, Testi M, Gaziev J, et al. Quantitatively different red cell/nucleated cell chimerism in patients with long-term, persistent hematopoietic mixed chimerism after bone marrow transplantation for thalassemia major or sickle cell disease. Haematologica 2011;96:128–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altrock PM, Brendel C, Renella R, Orkin SH, Williams DA, Michor F. Mathematical modeling of erythrocyte chimerism informs genetic intervention strategies for sickle cell disease. Am J Hematol 2016;91:931–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sankaran VG, Menne TF, Xu J, et al. Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science 2008; 322:1839–42. [DOI] [PubMed] [Google Scholar]
- 26.Menzel S, Garner C, Gut I, et al. A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat Genet 2007;39: 1197–9. [DOI] [PubMed] [Google Scholar]
- 27.Uda M, Galanello R, Sanna S, et al. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc Natl Acad Sci U S A 2008;105:1620–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu J, Peng C, Sankaran VG, et al. Correction of sickle cell disease in adult mice by interference with fetal hemoglobin silencing. Science 2011;334:993–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu P, Keller JR, Ortiz M, et al. Bcl11a is essential for normal lymphoid development. Nat Immunol 2003;4:525–32. [DOI] [PubMed] [Google Scholar]
- 30.Tsang JCH, Yu Y, Burke S, et al. Single-cell transcriptomic reconstruction reveals cell cycle and multi-lineage differentiation defects in Bcl11a-deficient hematopoietic stem cells. Genome Biol 2015;16:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brendel C, Guda S, Renella R, et al. Lineage-specific BCL11A knockdown circumvents toxicities and reverses sickle phenotype. J Clin Invest 2016;126:3868–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guda S, Brendel C, Renella R, et al. miRNA-Embedded shRNAs for lineage-specific BCL11A knockdown and hemoglobin F Induction. Mol Ther 2015;23: 1465–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brendel C, Negre O, Rothe M, et al. Preclinical evaluation of a novel lentiviral vector driving lineage-specific BCL11A knockdown for sickle cell gene therapy. Mol Ther Methods Clin Dev 2020;17:589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eaton WA, Hofrichter J, Ross PD. Editorial: delay time of gelation: a possible determinant of clinical severity in sickle cell disease. Blood 1976;47:621–7. [PubMed] [Google Scholar]
- 35.Hofrichter J, Ross PD, Eaton WA. Kinetics and mechanism of deoxyhemoglobin S gelation: a new approach to understanding sickle cell disease. Proc Natl Acad Sci U S A 1974;71:4864–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wienert B, Martyn GE, Funnell APW, Quinlan KGR, Crossley M. Wake-up sleepy gene: reactivating fetal globin for β-hemoglobinopathies. Trends Genet 2018;34: 927–40. [DOI] [PubMed] [Google Scholar]
- 37.Jacob GF, Raper AB. Hereditary persistence of foetal haemoglobin production, and its interaction with the sickle-cell trait. Br J Haematol 1958;4:138–49. [DOI] [PubMed] [Google Scholar]
- 38.Weatherall DJ. Phenotype-genotype relationships in monogenic disease: lessons from the thalassaemias. Nat Rev Genet 2001;2:245–55. [DOI] [PubMed] [Google Scholar]
- 39.Cavazzana M, Bushman FD, Miccio A, André-Schmutz I, Six E. Gene therapy targeting haematopoietic stem cells for inherited diseases: progress and challenges. Nat Rev Drug Discov 2019;18:447–62. [DOI] [PubMed] [Google Scholar]
- 40.Hsieh MM, Bonner M, Pierciey FJ, et al. Myelodysplastic syndrome unrelated to lentiviral vector in a patient treated with gene therapy for sickle cell disease. Blood Adv 2020;4:2058–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bauer DE, Kamran SC, Lessard S, et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science 2013;342:253–7.24115442 [Google Scholar]
- 42.Canver MC, Smith EC, Sher F, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature 2015;527:192–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frangoul H, Altshuler D, Cappellini MD, et al. CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. N Engl J Med 2021;384:252–60. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.