Abstract
Helicobacter pylori, a gastric pathogen associated with a broad range of stomach diseases, has a high tendency to become resistant to antibiotics. One of the most important factors related to therapeutic failures is its ability to change from a spiral to a coccoid form. Therefore, the main aim of our original article was to determine the influence of myricetin, a natural compound with an antivirulence action, on the morphological transformation of H. pylori and check the potential of myricetin to increase the activity of antibiotics against this pathogen. We observed that sub-minimal inhibitory concentrations (sub-MICs) of this compound have the ability to slow down the process of transformation into coccoid forms and reduce biofilm formation of this bacterium. Using checkerboard assays, we noticed that the exposure of H. pylori to sub-MICs of myricetin enabled a 4–16-fold reduction in MICs of all classically used antibiotics (amoxicillin, clarithromycin, tetracycline, metronidazole, and levofloxacin). Additionally, RT-qPCR studies of genes related to the H. pylori morphogenesis showed a decrease in their expression during exposure to myricetin. This inhibitory effect was more strongly seen for genes involved in the muropeptide monomers shortening (csd3, csd6, csd4, and amiA), suggesting their significant participation in the spiral-to-coccoid transition. To our knowledge, this is the first research showing the ability of any compound to synergistically interact with all five antibiotics against H. pylori and the first one showing the capacity of a natural substance to interfere with the morphological transition of H. pylori from spiral to coccoid forms.
Keywords: Helicobacter pylori, coccoid forms, morphological transformation, myricetin, checkerboard assay, synergism, biofilm
1. Introduction
The use of antibiotics in human treatment is considered to be one of the greatest medical achievements of the 20th century [1]. Nevertheless, about 100 years after their widespread introduction, the applicability of antibiotics has decreased drastically. This phenomenon seems to be strongly influenced by the dynamic spreading of antibiotic-resistant microorganisms, especially multidrug-resistant and pandrug-resistant strains. Currently, the annual mortality of patients caused by antibiotic-resistant microorganisms is estimated at about 700,000, while statistical analyses indicate that in 2050 this number may rise to a dramatic 10 million deaths per year [2]. For this reason, the World Health Organization is trying to highlight the seriousness of the above situation and encourage scientists for the intensification of research on new therapies against antibiotic-resistant pathogens [3].
Resistance to antibiotics is a multifactorial phenomenon with a tendency of quick spreading in society [4]. Thus, fighting this requires a variety of strategies, including not only improving the management of currently available antibiotics, but also the application of alternative methods targeting pathogens. Among them, there is increasing interest in antivirulence therapies, i.e., the use of substances that interfere with the factor(s) being crucial for the pathogenicity of a given microorganism [4,5]. The compounds that hinder the production of pathogens’ virulence factors may deprive them of their ability to develop infections or defend themselves against the host, making them easier to eradicate.
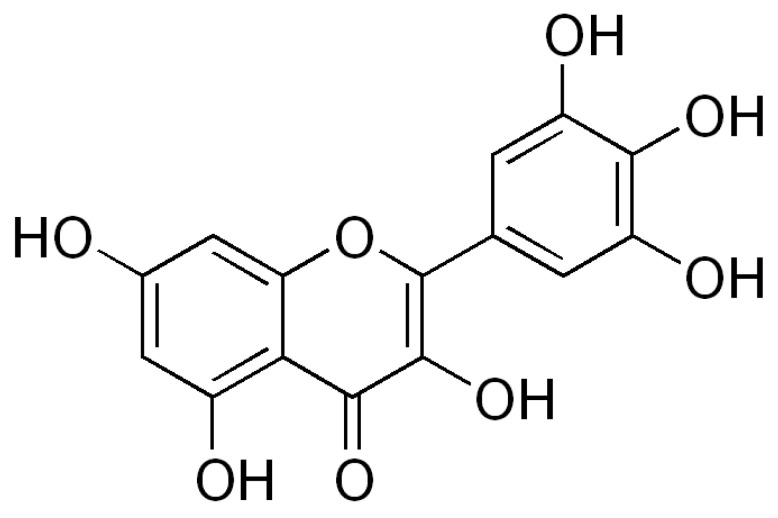
Within the natural substances with promising antivirulence properties, special attention is now paid to polyphenols, secondary plant metabolites characterized by multiples of phenol units [6,7]. This diverse group of compounds includes myricetin (MYR; 3,5,7,3′,4′,5′-hexahydroxyflavone, Figure 1), where, in recent years, a particularly high ability to inhibit a number of virulence factors has been demonstrated [8]. MYR was originally isolated from the bark of the Myrica tree, which gave the compound its name [9,10]. In its pure form MYR appears as yellow crystals, a property with which its initial application as a biopigment was related to [10]. Over the years, the usefulness of this compound in the fight against allergies, inflammations, hypertension, and diabetes has been recognized [8]. Additionally, a very promising antibacterial activity has been observed [8,9,10]. However, the antibacterial features of MYR are not related to the disturbance of microbial growth, but are rather associated with a capacity to decrease pathogenicity of microbes, including their cytotoxic action (production of lytic enzymes or toxins), eukaryotic cell colonization (adhesion and invasion), and defense mechanisms (e.g., biofilm formation) [11,12,13,14,15]. Therefore, MYR appears to be an ideal candidate for use in antivirulence therapies for difficult-to-treat pathogens.
Figure 1.
The chemical structure of myricetin (MYR; 3,5,7,3′,4′,5′-hexahydroxyflavone).
Helicobacter pylori is a Gram-negative bacterium that colonizes the gastric mucosa [16,17,18]. Infections most often occur during early childhood and, in the absence of the appropriate treatment, are life-long. The gastric environment is an unfavorable niche and for this reason H. pylori has developed various adaptations enabling the effective colonization of this organ. Strains that produce multiple virulence factors are believed to have a greater potential to persistently infect the stomach [19]. The classically mentioned virulence determinants include urease [20], vacuolating toxin (VacA) [21], cytotoxic oncoprotein (CagA) [22], and numerous adhesins [23]. However, more and more often attention is paid to the morphological transformation of this bacterium as one of the key properties conditioning the survival of H. pylori [24]. The capacity to change from the spiral/rod-shaped form to the coccoid form is associated with a number of physiological changes in cells of this bacterium, which are accompanied by an increased ability to avoid detection from the host’s immune system and a significant reduction in antibiotic sensitivity [24,25]. It has even been indicated that the presence of coccoid H. pylori forms is an independent risk factor related to patients’ therapeutic failures [26,27] and that coccoid forms of this bacterium are able to produce virulence factors, as well [28]. Therefore, it seems that finding a substance capable of interfering with the process of morphological transformation of H. pylori may turn out to be very useful for increasing the degree of eradication of this pathogen [24,25].
Based on the above deduction, the aim of this study was to determine the influence of MYR on the morphological transformation of H. pylori and to determine the potential of MYR to increase the activity of classically used antibiotics against this pathogen.
2. Results
2.1. Antibacterial Activity of MYR
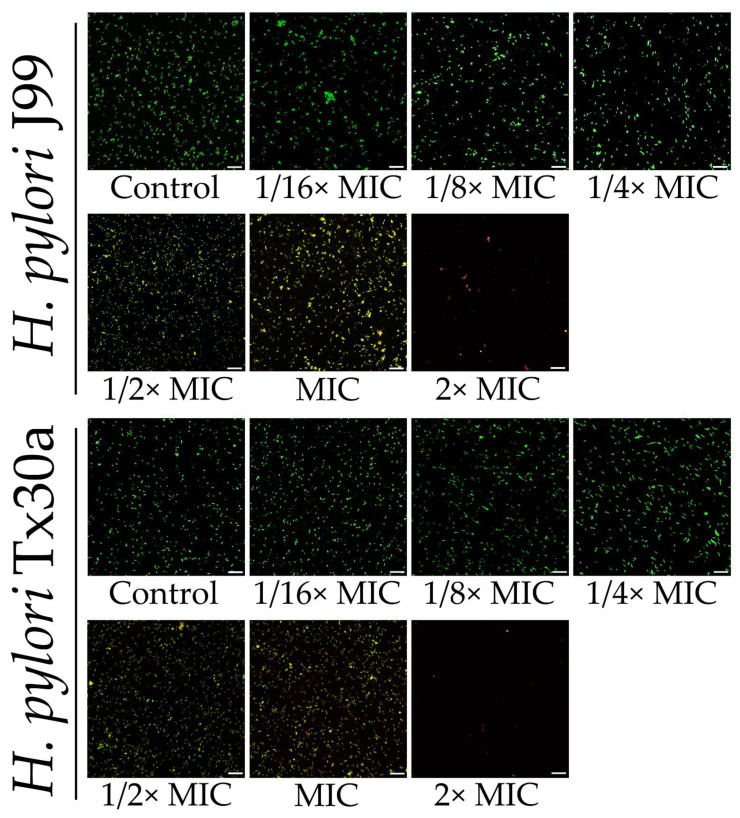
In the first stage of our research, we decided to determine the antibacterial activity of MYR against H. pylori. We observed that minimal inhibitory concentrations (MICs) and minimal bactericidal concentrations (MBCs) were convergent and counted for 160 and 320 µg/mL, respectively, for both tested strains (H. pylori J99 and Tx30a) (Table 1). As we were interested in establishing the antivirulence potential of MYR and not affecting the growth of H. pylori, we additionally conducted a fluorescence analysis of LIVE/DEAD-stained cells exposed to different concentrations of this compound. We observed that sub-minimal inhibitory concentrations (sub-MICs) of MYR up to 1/4× MIC had a marginal effect on the green/red fluorescence ratio of bacterial cells, but some antibacterial effect was noticeable starting from 1/2× MIC (Figure 2). Therefore, in further analyses we decided to focus on MYR in a range of 1/256×–1/4× MIC when checking its modulatory effect on the morphological transformation of H. pylori.
Table 1.
Antibacterial activity of myricetin against tested Helicobacter pylori strains.
| H. Pylori Strains | Collection Number | Myricetin Activity [µg/mL] | |
|---|---|---|---|
| MIC | MBC | ||
| J99 | ATCC 700824 | 160 ± 23.1 | 320 ± 53.3 |
| Tx30a | ATCC 51932 | 160 ± 26.7 | 320 ± 46.2 |
MIC: Minimal inhibitory concentration; MBC: Minimal bactericidal concentration. Results are presented as means ± standard deviations from three independent biological experiments with three repetitions each.
Figure 2.
Representative fluorescence microscopy images of reference H. pylori J99 and Tx30a strains exposed to a gradient concentration of myricetin. The minimal inhibitory concentration (MIC) in both strains counts for 160 µg/mL, while minimal bactericidal concentration (MBC) is equal to 2× MIC and counts for 320 µg/mL. Bacterial cells were stained with a LIVE/DEAD kit. The scale bar shows 20 µm.
2.2. Inhibitory Effect of MYR against the Spiral-to-Coccoid Transformation
When assessing the ability of MYR to inhibit the transformation of H. pylori into coccoid forms, we applied serum starving conditions, a well-known factor stimulating this process [29,30]. The two reference H. pylori strains used in this study provide a good model to study this type of interaction. The highly virulent H. pylori J99 has a high potential for the spiral-to-coccoid transition, contrary to the H. pylori Tx30a, a non-pathogenic strain with the low transformative potential [31].
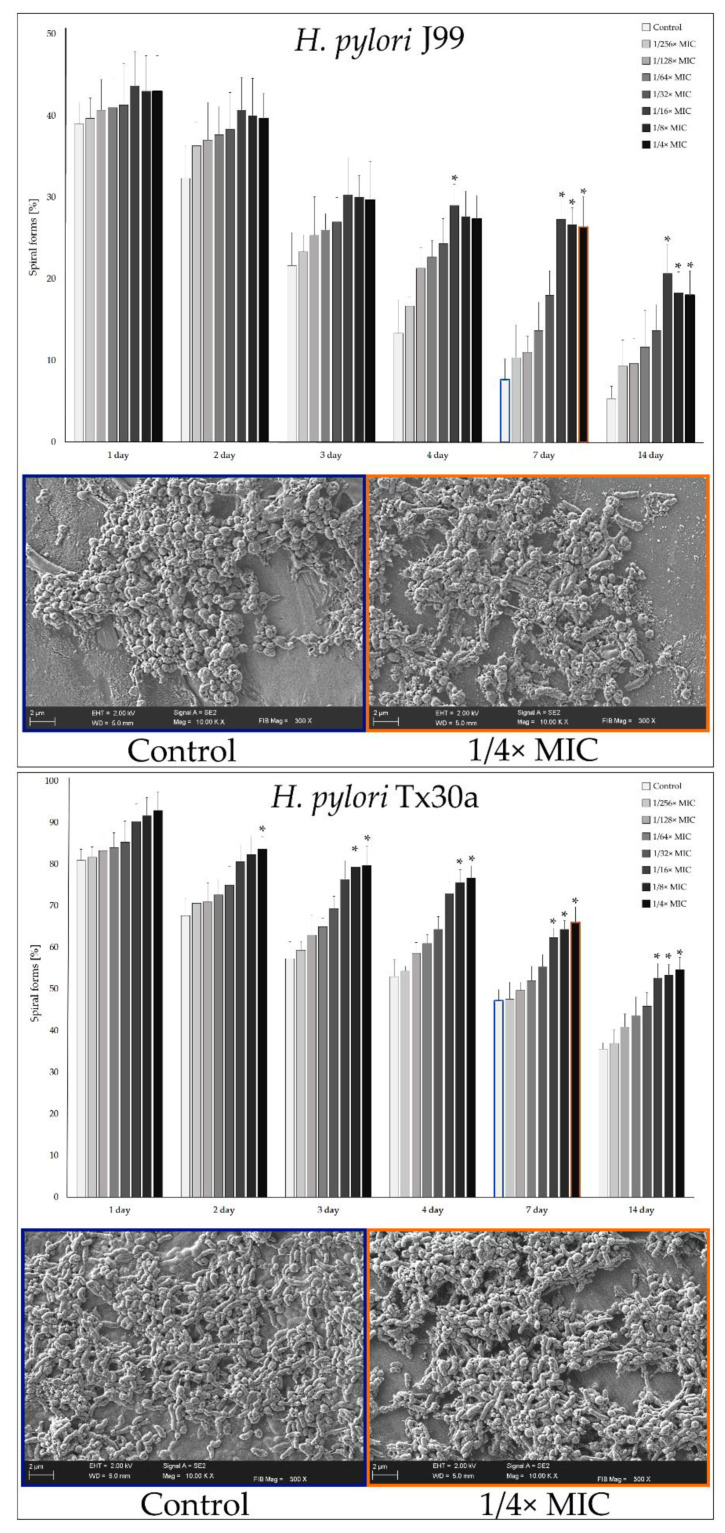
We observed that the effect of slowing down the transformation of H. pylori is dependent on both the concentration and the duration of exposure to MYR. The H. pylori J99 tended to reduce the number of spiral forms very quickly in the conditions we used (a decrease from 90% to 39%–43% in the first day) (Figure 3). The 7- and 14-day exposure of this strain to 1/16×–1/4× MIC of MYR allowed us to maintain about 3–4-times higher amount of spiral forms than in the control samples. The H. pylori Tx30a, in line with previous assumptions, lowered the number of spiral forms much slower than H. pylori J99 (from 98% to 81%–93% in the first day) (Figure 3). Here, differences between control samples and these exposed to sub-MICs of MYR were statistically significant from the 2nd day of culture, while the difference was most noticeable on the 7th and 14th day of incubation (approx. 1.5-times higher number of spiral forms than in the control). Our observations, showing a significantly higher number of spiral forms in samples treated with 1/4× MIC of MYR for 1 week, were additionally confirmed by scanning electron microscopy (SEM) (Figure 3). In conclusion, we showed that MYR at concentrations between 1/16× MIC and 1/4× MIC has a strong potential for modulating the speed of transformation into coccoids by H. pylori.
Figure 3.
The concentration- and time-dependent activity of myricetin in inhibiting the transition of reference H. pylori J99 and Tx30 strains from spiral to coccoid forms during serum starving conditions. The MIC of myricetin for both strains is equal to 160 µg/mL. The initial number of spiral forms (a 0-h time point) was 90% and 98% for H. pylori J99 and Tx30a, respectively. Results are presented as means ± standard deviations from three independent biological experiments. * Indicates statistically significant difference (p ≤ 0.05).
2.3. Inhibitory Effect of MYR against Biofilm Formation
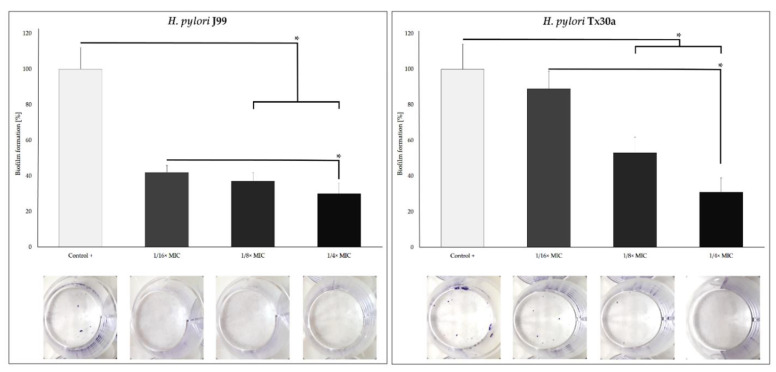
Keeping in mind that biofilm formation is another important feature that may facilitate antibiotic tolerance of H. pylori [32], we decided to use a crystal violet staining method to determine the ability of H. pylori to form this structure during the presence of 1/16×–1/4× MIC of MYR. We found that a 3-day incubation of this pathogen with sub-MICs of MYR caused the concentration-dependent inhibitory effect on biofilm development (Figure 4). Exposure to 1/8× and 1/4× MIC resulted in a 50% and 70% reduction in the biofilm formation of H. pylori Tx30a and 60% and 70% decrease for H. pylori J99, respectively.
Figure 4.
Anti-biofilm activity of myricetin against reference H. pylori J99 and Tx30a strains measured by a crystal violet staining method. The MIC of myricetin for both strains is equal to 160 µg/mL. Results are presented as means ± standard deviations from three independent biological experiments with three repetitions each. * Indicates statistically significant difference (p ≤ 0.05).
2.4. Synergistic Activity of MYR with Antibiotics
In further stages of our research, we focused on the analysis of the potential of sub-MICs of MYR (1/16×–1/4× MIC, concentrations being the strongest spiral-to-coccoid inhibitors) to enhance the antibacterial activity of five clinically used antibiotics (amoxicillin, clarithromycin, tetracycline, metronidazole, and levofloxacin) against H. pylori.
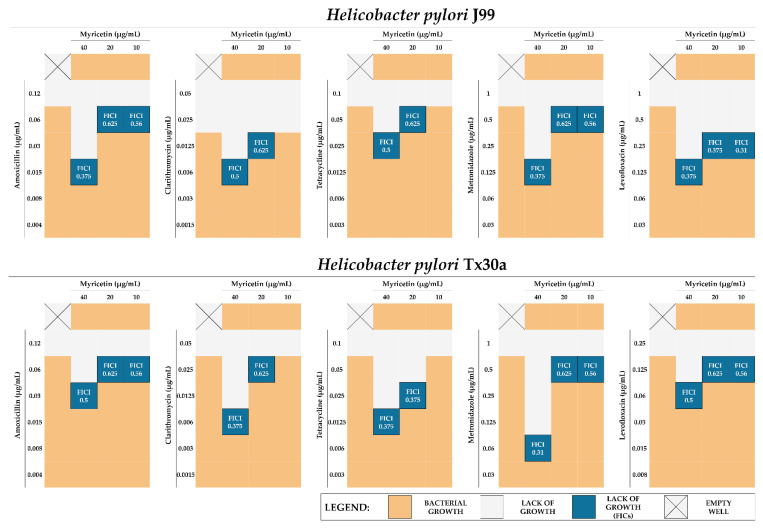
We discovered that 1/4× MIC of MYR is able to lower the MICs of all tested antibiotics by 4–16 times, which allows us to conclude its potential to synergistically enhance the antibiotics’ action (fractional inhibitory concentration indexes, FICI = 0.31–0.5) (Figure 5). The 1/8× MIC of MYR also increased the antimicrobial activity of all tested antibiotics, although in this case most of the interactions were additive (for most of them FICI = 0.625) (Figure 5). The 1/16× MIC of MYR additively increased the activity only of bactericidal antibiotics (amoxicillin, metronidazole, and levofloxacin; FICI = 0.56), but not bacteriostatic ones (clarithromycin and tetracycline, FICI = 2.0) (Figure 5).
Figure 5.
Graphical representation of the results of checkerboard assays testing synergistic interactions between myricetin and antibiotics against reference H. pylori J99 and Tx30a strains. The MIC of myricetin for both strains is equal to 160 µg/mL. The MICs for amoxicillin, tetracycline, and metronidazole for both strains count for 0.12, 0.1, and 1 µg/mL, respectively. The MICs for clarithromycin and levofloxacin are equal to 0.025 and 1 µg/mL for H. pylori J99, and 0.05 and 0.25 µg/mL for H. pylori Tx30a. The blue field marks the wells of the titration plates, in which no bacterial growth and a positive interaction between the tested compounds was observed. The interactions were interpreted based on the calculation of the fractional inhibitory concentration index (FICI), in which ≤0.5 and >0.5 but ≤1 was considered as synergism and additivity, respectively.
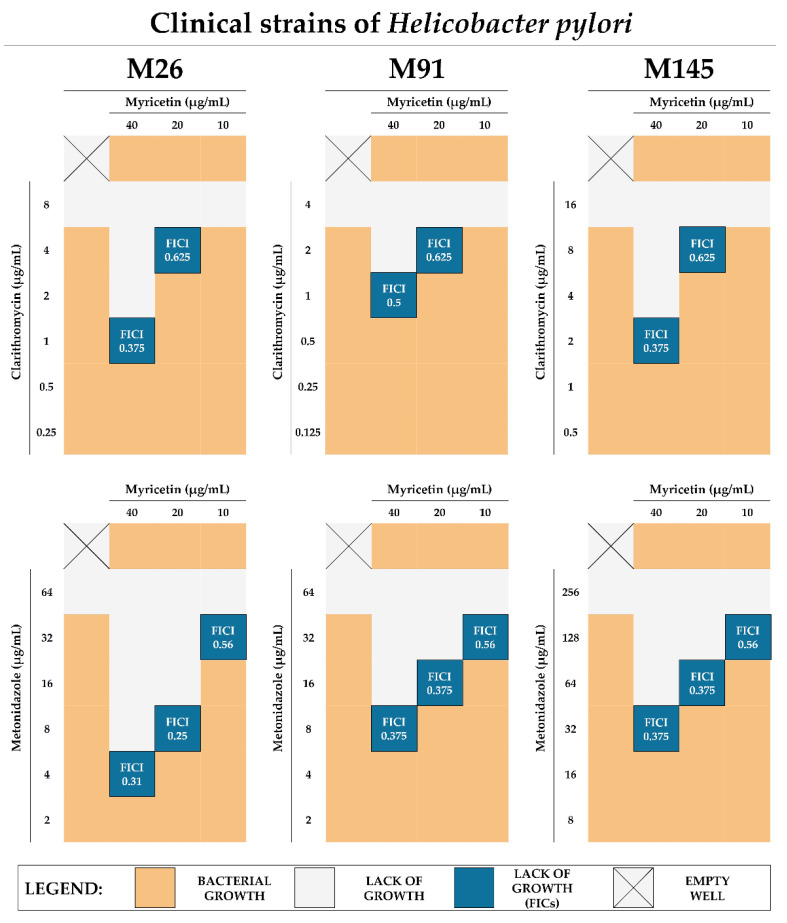
Dual resistance to clarithromycin and metronidazole of H. pylori is considered to be the most important resistance profile determining therapeutic failures of this pathogen [33,34], therefore we extended our research with three clinical H. pylori strains with the above-mentioned resistance. Checkerboard assays determining the sub-MICs of MYR with clarithromycin or metronidazole confirmed our previously described observations, indicating synergism of 1/4× MIC of MYR with these antibiotics (FICI = 0.31–0.5) and suggesting that resistance to antibiotics of H. pylori is not adversely affecting the antibiotic-boosting effect of MYR (Figure 6).
Figure 6.
Graphical representation of the results of checkerboard assays testing synergistic interactions between myricetin and two selected antibiotics (clarithromycin and metronidazole) against clinical, double-resistant H. pylori strains (clarithromycin- and metronidazole-resistant). The MIC of myricetin for all strains is equal to 160 µg/mL. The MICs for clarithromycin count for 8, 4, and 16 µg/mL for H. pylori M26, M91, and M145, respectively. The MICs for metronidazole are equal to 64, 64, and 256 µg/mL for H. pylori M26, M91, and M145, respectively. The blue field marks the wells of the titration plates, in which no bacterial growth and a positive interaction between the tested compounds was observed. The interactions were interpreted based on the calculation of the FICI, in which ≤0.5 and >0.5 but ≤1 was considered as synergism and additivity, respectively.
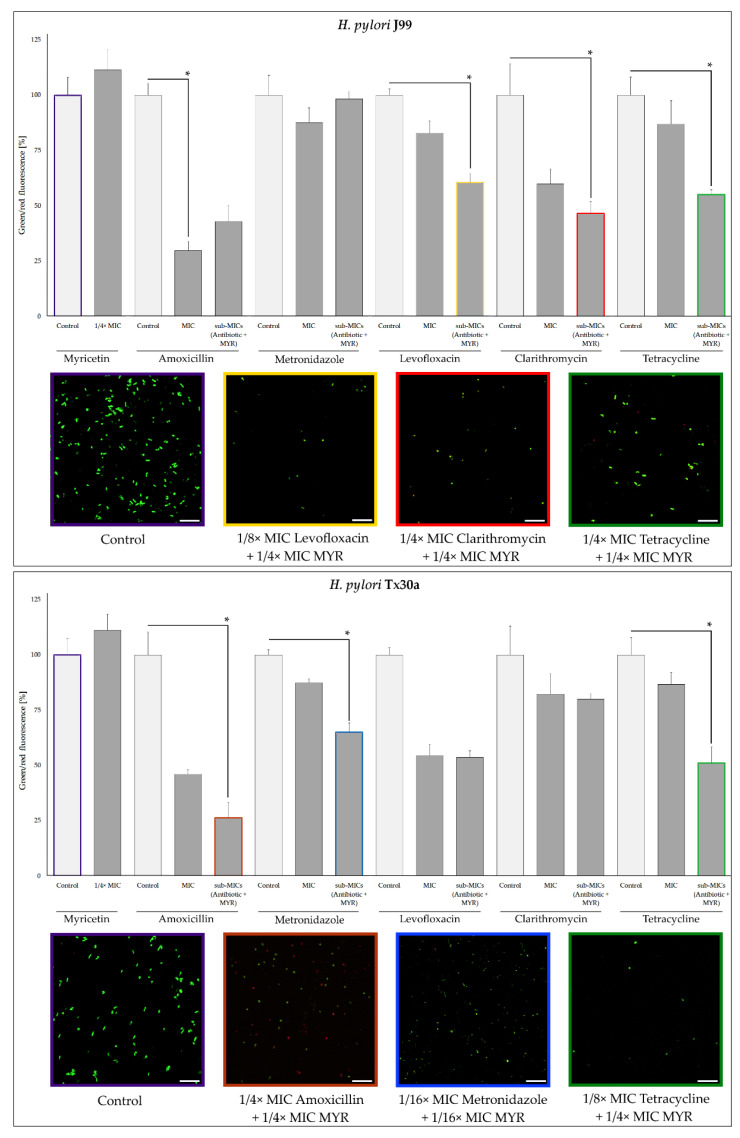
To broaden the information on the synergism of MYR with antibiotics, we performed the fluorescence analysis of H. pylori J99 and Tx30a cells treated with MICs of antibiotics alone or in the combination with MYR (fractional inhibitory concentrations (FICs), being 1/4× MIC of MYR + sub-MICs of the selected antibiotic) (Figure 7). We observed that the green/red fluorescence ratio of bacterial cells was reduced in most samples compared to the controls. Among the 7/10 of biological samples in which we noticed a significant decrease in cell viability, six of them constituted samples of two combined substances (sub-MICs of both MYR + antibiotic) (Figure 7). These results show that MYR at 1/4× MIC is not only able to reduce the MIC values of all tested antibiotics while maintaining their antimicrobial activity, but even intensify this antibacterial activity against H. pylori.
Figure 7.
Graphical representation of changes in cell viability of reference H. pylori J99 and Tx30a strains when exposed to MICs of antibiotics or a combination of sub-minimal concentrations (sub-MICs) of the selected antibiotic with 1/4× MIC of myricetin (MYR). Fluorescence in the “control” and “1/4× MIC” samples collectively labeled as “myricetin” is the mean of all other experiments, being their joint element. Bacterial cells were stained with a LIVE/DEAD kit. The scale bar shows 20 µm. Results are presented as means ± standard deviations from three independent, biological experiments. * Indicates statistically significant difference (p ≤ 0.05).
2.5. Modulatory Effect of MYR on Genes Related to Morphogenesis
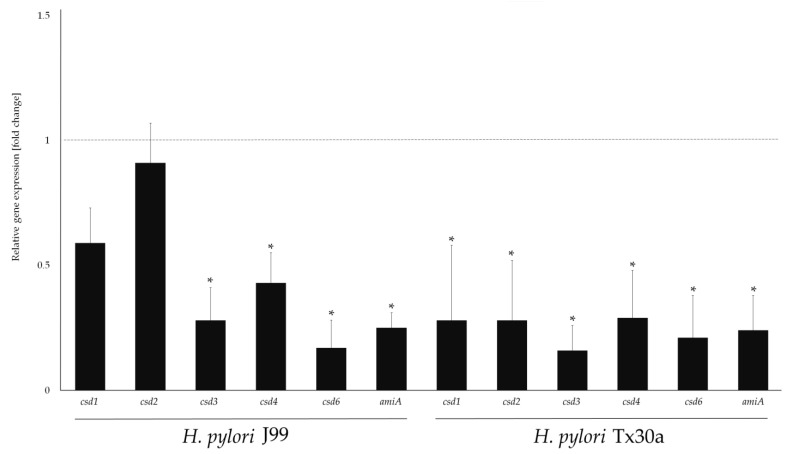
The last stage of our research was to determine the effect of a 3-day exposure of H. pylori on 1/4× MIC of MYR, the concentration that most effectively potentiated the action of antibiotics, on the expression of genes related to the morphogenesis of this pathogen. In this case, we selected genetical determinants involved in the peptidoglycan modeling, i.e., cleaving of cross-linking bridges of muropeptide dimers (csd1, csd2, and csd3) and shortening of muropeptide monomers (csd3, csd6, csd4, and amiA) [25]. In particular, the activity of genes associated with the second peptidoglycan rearrangement pathway seemed important to us as it has been suggested that the trimming of penta-, tetra-, and tripeptides to dipeptides is a molecular marker associated with the formation of spherical H. pylori forms [35,36,37].
The results of these analysis were similar for both H. pylori strains. Exposure to MYR caused a reduction in the expression of all tested genes with the inhibition most strongly observed for csd3 (3.6- and 6.2-fold for H. pylori J99 and Tx30a, respectively), csd6 (5.8- and 4.8-fold for H. pylori J99 and Tx30a, respectively), and amiA (about 4-fold for both H. pylori strains) (Figure 8). For csd4, we also observed some inhibitory effect (2.3- and 3.5-fold for H. pylori J99 and Tx30a, respectively), but not as strong as for the genes mentioned before (Figure 8). Despite the large convergence of the results obtained for both H. pylori strains, we observed some differences in the expression of csd1 and csd2. Exposure of H. pylori J99 to 1/4× MIC of MYR had a marginal effect on the expression of these genes (less than a 2-fold change), while in H. pylori Tx30a this effect was significant (about a 3.5-fold reduction in their expression) (Figure 8).
Figure 8.
Modulatory activity of 1/4× MIC of myricetin (40 µg/mL) on the expression of the morphogenesis-associated genes of reference H. pylori J99 and Tx30a strains. Expression of the tested genes was related to the expression of the house-keeping gene—ureA, which presented sufficient stability during these studies (the difference between the expression in the control and experimental samples was less than 1). The dashed line indicates no change in gene expression. A 2-fold change in expression was considered statistically significant (*). Results are presented as means ± standard deviations from three independent, biological experiments.
3. Discussion
The main goal of pathogenic microorganisms is to achieve a preferred niche in the host, replicate and avoid destruction, and transfer to the next organism [38]. To obtain this, pathogens have developed the ability to produce many virulence determinants, some of which have been known and characterized widely for many years (toxins, lytic enzymes, siderophores, envelopes, etc.), while the importance of other, more sophisticated strategies has been recently described [38,39]. Bacterial morphology affects the capacity to survive stressful environmental conditions, colonize specific niches within hosts or escape the immune system detection [40,41]. Therefore, scientists speculate to consider the microbial shape and the possibility of its transformation as an important pathogenicity strategy. High morphological heterogeneity and the ability to change from spiral to coccoid forms is one of the key features of H. pylori associated with antibiotics tolerance and therapeutic failures [24,25]. For this reason, in this original paper, we attempted to inhibit this process in order to increase the sensitivity of H. pylori to classically used antibiotics.
In recent years, very promising antivirulence properties have been reported for MYR. Although this substance possesses some antibacterial action, its activity is described as relatively low. The MIC values of MYR reported by others are in the range of 62.5–256 µg/mL [13,42,43,44,45] and are in line with those obtained by us (MIC = 160 µg/mL) (Table 1). Moreover, it was observed that sub-MICs of this compound do not disturb the growth of microorganisms but may inhibit the production of many virulence factors, including sortase A of Gram-positive bacteria [46]; adhesins, proteases, and collagenases of Porphyromonas gingivalis [13]; suilysin of Streptococcus suis [14]; and invasiveness of Salmonella Typhimurium [15]. Research groups focusing on Staphylococcus aureus have also reported very promising results [11,12,42,47], demonstrating the capacity of MYR to disrupt adhesion, biofilm formation, and synthesis of hemolysin α and staphyloxanthin.
In this study, we were focused on determining the ability of MYR to interfere with the transformation of H. pylori into coccoid forms. We observed a concentration- and time-dependent slowing in the transition of H. pylori into spherical forms when treated with this compound (Figure 3). For example, an 1-week incubation under starving conditions resulted in 2–4 times (H. pylori J99) and 1.5 times (H. pylori Tx30a) higher amount of spiral forms during exposure to 1/16×–1/4× MIC of MYR than in control samples. These observations were additionally confirmed by scanning electron microscopy. Ideally, exposure of H. pylori to MYR would allow a complete and long-term inhibition of the spiral-to-coccoid transformation. Compared to spiral forms, coccoids have a lower level of metabolism, a tendency to self-aggregate and form biofilms, and a higher efflux pumps expression, all of which generate their antibiotic tolerance [24,32]. As demonstrated by Obonyo et al. (2012) [48] and Faghri et al. (2014) [49], preformed coccoid H. pylori forms require much higher concentrations of antimicrobial substances to be destroyed than spiral forms (even if these can transform to spherical forms over time). It seems that spiral forms accumulate sufficiently high concentrations of antimicrobial compounds in their interior that the subsequent conversion to coccoid forms no longer has a protective function and is rather an expression of cell death. Thus, on the basis of the above considerations and the deduction presented in a review by Krzyżek et al. (2020) [24], we conclude that the slowing down of the morphological transition might be sufficient to break down the coccoid-related antibiotic tolerance of H. pylori.
For MYR, apart from the affinity to diminish the virulence of various pathogens, the ability to enhance the activity of antimicrobial substances has also been demonstrated [43,44,47]. In our research, we showed very promising properties of MYR as an enhancer of antibiotics against H. pylori. We demonstrated the synergistic effect of 1/4× MIC of MYR in combination with all five tested antibiotics (amoxicillin, clarithromycin, tetracycline, metronidazole, and levofloxacin), allowing us to decrease 4–16 times the MIC values of these antibiotics (Figure 5 and Figure 6). In addition, observations made by fluorescence microscopy and LIVE/DEAD staining confirmed the results of the checkerboard assays (Figure 7). Based on our review published in 2020 [50], being a holistic revision of in vitro studies on H. pylori synergistic treatments, we can conclude that MYR is the first described compound capable of synergistically interacting with all five antibiotics against H. pylori. The enhancement of antibiotics’ activity was not related to the bacterial resistance profile as convergent results were obtained for both the two reference H. pylori strains (J99 and Tx30a) (Figure 5) and the three clinical, double-resistant H. pylori strains (M26, M91, and M145) (Figure 6). The targets of MYR in microbial cells are replication and transcription, which results from the inhibition of DNA and RNA polymerases [51,52,53]. It seems that the blockage of transcription may condition the decreased expression of genes related to the microbial stress response, making pathogens more sensitive to the action of antimicrobial substances [54]. For many representatives of polyphenols, the ability to interact and disrupt the function of bacterial cell membrane and/or cell wall has been demonstrated [55], and although this type of activity has not been demonstrated for MYR so far, we cannot conclusively state that this type of phenomenon did not occur in this case.
With RT-qPCR, we inspect the capacity of MYR to interfere with the expression of genes responsible for the H. pylori morphogenesis. Among the most important genetic determinants involved in H. pylori peptidoglycan rearrangement, there are two groups of genes related to this process [25]. The first group is responsible for cleaving cross-linking bridges of muropeptide dimers (csd1, csd2, and csd3) and the second is involved in muropeptide monomers shortening (csd3, csd6, csd4, and amiA). The csd3 is associated with cutting pentapeptides into tetrapeptides, which are then used by csd6 to produce tripeptides, which in turn are processed by csd4 to create dipeptides [25]. The amiA, on the other hand, is associated with the coordination of functioning of these genes [36]. In the present study, we observed that MYR seems to have a stronger influence on the second stage of H. pylori murein modification (trimming of muropeptide monomers), with an approximately 4-fold reduction in the expression of these genes (Figure 8). This is consistent with the observations showing the importance of dipeptides accumulation in coccoid H. pylori forms [35,36,37], and the participation of csd4 and amiA as determinants of this morphological transformation in both H. pylori [36,56] and Campylobacter jejuni [57]. Taking into account our results, we suspect that MYR may be responsible for the disturbance of the csd3 and csd6 activity (contributing to the decrease of substrates for the csd4 functioning), as well as the inhibition of amiA, a gene coordinating the work of the aforementioned determinants. The interruption of morphogenesis may also affect other virulence traits, such as biofilm formation, as previously shown for C. jejuni [58]. Although the effect of MYR on the formation of H. pylori biofilm was not the main goal of our current original work, we undertook a preliminary verification on the influence of sub-MICs of MYR on the initial stages of this structure development. We observed that 1/4× MIC of MYR reduced the amount of H. pylori biofilm by 70% (Figure 4). These results encourage us to undertake further research on the modulating effects of MYR on H. pylori biofilms in the future.
An issue worth considering in the context of H. pylori therapy is the delivery method and the stability of substances in the gastric environment. MYR is a slightly acidic compound [59,60], which may positively influence its use in the therapy of H. pylori. In laboratory conditions simulating the stomach environment, it was shown that MYR had a very high level of stability with a t1/2 at pH = 2 of approx. 10 h [59]. The solubility was also enhanced by an acidic environment, with an increase in this parameter by 46- to 9-fold at pH between 1 and 3. Although MYR is found in many vegetables, fruits, nuts, and herbs [8], it is highly doubtful that these concentrations would be sufficient to achieve the desired therapeutic effect against H. pylori. However, it was shown that the use of carriers for MYR significantly improves its bioavailability [61,62,63,64]. One of the strategies may be the use of biocellulose, the utilization of which has been shown by our research group in ex vivo experiments in burning wounds/bones infections [65,66] and preliminary in vitro tests on H. pylori [67]. The applicability of this type of delivery system in H. pylori therapy with MYR will be determined by us in the future.
4. Materials and Methods
4.1. H. pylori Strains
Two reference H. pylori strains, J99 (ATCC 700824) and Tx30a (ATCC 51932), were used in all steps of the research. Additionally, three clinical strains of H. pylori (M26, M91, and M145), having dual antibiotic resistance against clarithromycin and metronidazole, were also utilized during the checkerboard assays. These strains come from the collection of microorganisms of the Wrocław Medical University in Poland and were isolated during previous studies [34].
All H. pylori strains were stored frozen at −80 °C until needed [68,69]. In order to revive them, they were sown on Columbia agars (Difco, Lublin, Poland) with 10% hemolyzed horse blood and incubated for 3 days at 37 °C under microaerophilic conditions (Genbox microaer kits, BioMerieux, Marcy I’Etoile, France) and shaking speed of 100 rpm (MaxQ 6000, Thermo Fisher, Waltham, MA, USA).
4.2. Assessment of the Bactericidal Activity of MYR
The determination of MICs of MYR was carried out using flat-bottom, ventilated 12-well microtiter plates (Bionovo, Legnica, Poland) [68,69]. MYR (Sigma-Aldrich, St. Louis, MO, USA) was dissolved in dimethyl sulfoxide (DMSO; Chempur, Piekary Śląskie, Poland) and a series of dilutions were made in Brain Heart Infusion broth (BHI; Oxoid, Dardilly, France) with 7% foetal calf serum (Gibco, Paisley, Scotland, UK) to obtain a gradient MYR range of 640–0.625 µg/mL and the final concentration of DMSO lower than 1% (v/v). Each well contained 1 mL of culture broth, the desired concentration of MYR, and 107 CFU/mL of H. pylori.
The MIC was taken to be the lowest concentration in which no microbial growth was observed visually after a 3-day incubation (microaerophilic, 37 °C, 100 rpm shaking). The MBC value was determined by taking 10 µL of samples from each well of the titration plate and spotting them on Columbia agars with 10% hemolyzed horse blood (3 days of microaerophilic incubation at 37 °C). Both tests determining MICs and MBCs were performed using three biological assays with three technical repetitions each.
Independently, the assessment of the bacterial viability was performed using fluorescence microscopy (see Section “4.5. Assessment of Cell Viability”).
4.3. Assessment of an Inhibitory Effect of MYR on Morphological Transformation
The slowdown in morphological transformation into coccoid forms of H. pylori was investigated with exposure of these bacteria to various sub-MICs of MYR (40–0.625 µg/mL corresponding to 1/4×–1/256× MIC) at specific time points, i.e., after 1, 2, 3, 4, 7, and 14 days of incubation. Each well of 12-well titration plates contained 2 mL of BHI (the lack of serum served as a stress factor stimulating morphological transition) and 107 CFU/mL of H. pylori. The well containing bacteria without MYR and well without bacteria consisted of positive and negative controls, respectively. The plates were incubated for 3 days under classical conditions (microaerophilic, 37 °C, 100 rpm shaking).
At each time point, 50 µL of culture was used to make Gram-stained preparations [31]. From each slide, 100 bacterial cells were counted from three independent biological tests (n = 300). The exception was the beginning of the experiment (a 0-h time point), in which a total of 800 cells obtained from all samples were counted and presented as the mean initial number of spiral forms. The bacteria were observed under an Olympus BX50 microscope (Olympus Optical, Tokyo, Japan).
In order to confirm the observations from light microscopy, control samples (untreated with MYR) and those exposed to 1/4× MIC of MYR, both coming from a 7-day incubation, were additionally submitted for a SEM analysis [31,68,69]. Such samples were fixed by adding a 2.5% solution of glutaraldehyde (Sigma-Aldrich) with a 1-day incubation at 4 °C. These samples were then centrifuged several times (600× g for 5 min) and washed in a 0.1 M cacodyl buffer (Sigma-Aldrich). After this step, the bacteria were passed through an ethanol series (50%–99.9%) and sputtered with gold. Observations were made using an Auriga 60 electron microscope (Zeiss, Oberkochen, Germany).
4.4. Assessment of the Interaction of MYR with Antibiotics
The potential existence of a positive interaction in the antibacterial activity of MYR with the classically used antibiotics, i.e., amoxicillin, clarithromycin, tetracycline, metronidazole, and levofloxacin (all from Sigma-Aldrich), was performed using the checkerboard assay. The research methodology was similar to that used by us previously [68,69] with minor modifications. Three selected concentrations of MYR (1/4×, 1/8×, and 1/16× MIC) and a range of antibiotic concentrations were applied during the checkerboard assays. Bacteria in the amount of 107 CFU/mL were incubated in 1 mL of BHI with 7% serum (3 days of microaerophilic incubation at 37 °C, 100 rpm shaking). Since MYR alone in the concentrations used in these assays did not inhibit the growth of H. pylori, reading of MICs of control wells (a single compound used) was limited to antibiotics only.
The interactions were interpreted based on the calculation of FICI, in which ≤0.5; >0.5 but ≤1, >1 was considered as synergism, additivity, and neutral interaction, respectively [68,69]. In addition, the viability of bacterial cells derived from the four wells, corresponding to the control, 1/4× MIC of MYR, MIC of antibiotics, as well as the FIC of both compounds, was determined (for analysis, see Section “4.5. Assessment of Cell Viability”).
4.5. Assessment of Cell Viability
The viability of H. pylori was examined by staining bacterial cells with the LIVE/DEAD Kit (Thermo Fisher, Waltham, MA, USA) and analyzing them by fluorescence microscopy (Olympus BX51, Olympus Optical, Tokyo, Japan) [67]. The staining procedure was performed according to the manufacturer’s recommendations. In brief, 0.1 mL of the sample was centrifuged at 10,000× g for 15 min, the supernatant was collected, the resulting bacterial pellet was resuspended in 1 mL of a phosphate buffer solution (PBS; Sigma-Aldrich), and then centrifuged again at the same speed. The supernatant was taken from the sample and the bacteria were suspended in 0.2 mL of a 0.85% NaCl solution with 0.6 µL of SYTO9 and propidium iodide (1:1 ratio), followed by incubation for 15 min in the dark. Then, 10 µL of samples was spotted on microscope slides and capped with coverslips. The green (viable cells) and red (degenerated cells) fluorescence were counted for 10 regions of interest from three independent bioassays (n = 30). The intensity of these two signals was counted separately from the same regions and then reported as the ratio of green to red fluorescence. The fluorescence analysis was performed with the ImageJ program.
4.6. Assessment of Biofilm Formation
Determination of the influence of selected sub-MICs of MYR (1/4×, 1/8×, and 1/16× MIC) on the formation of H. pylori biofilm was performed by a crystal violet method [70]. For this purpose, bacteria were incubated for 3 days under classical conditions (microaerophilic, 37 °C, 50 rpm shaking) in 1 mL of BHI broth with 2% foetal calf serum (this concentration stimulates both biofilm formation and ensures multiplication of H. pylori). After this period, the bacterial suspension was collected, the wells were washed twice with 1 mL PBS, dried, and stained with 1 mL of 0.1% crystal violet (Sigma-Aldrich) for 15 min. Then, the dye was removed, the wells were washed twice with PBS, dried, and flushed with 1 mL of 96% ethanol (Stanlab, Lublin, Poland) to dissolve the crystal violet adsorbed in the H. pylori biofilms. The 200 µL of these solutions was transferred to a 96-well microtiter plate (Bionovo, Legnica, Poland). Absorbance was measured at OD590 using an Asys UVM 340 microplate reader (Biochrom Ltd., Cambridge, UK). In each case, the absorbance of the negative control (wells without bacterial biofilms, being a pure culture medium) was subtracted from the absorbance of the remaining samples. The tests were performed three times in three technical repetitions.
4.7. Assessment of Expression of Genes Encoding Morphogenesis
The determination of the expression of genes related to the morphogenesis of MYR-exposed H. pylori was performed according to the procedures described by Fernandes et al. (2017) [71] with minor modifications.
Bacteria in 107 CFU/mL were grown in 12-well microtiter plates containing 1 mL BHI with 7% foetal calf serum (with 1/4× MIC of MYR or without this compound (serving as a control)) and incubated for 3 days under standard conditions (microaerophilic, 37 °C, 100 rpm shaking). After this time, the microorganisms were transferred to microcentrifuge tubes (Bionovo), centrifuged at 10,000 rpm (Eppendorf 5424R, Hamburg, Germany), the supernatant was collected, and the samples were followed to the steps provided by the manufacturer of the MINI RNA Kits (A&A Biotechnology, Gdynia, Poland). The purity and concentration of the obtained RNA was determined spectrometrically using a NanoDrop (DenoviX DS-11, Wilmington, USA) and the samples were stored at −80 °C until needed.
The TranScriba Kit (A&A Biotechnology) was applied to transcribe the obtained RNA into cDNA. The synthesis was performed in a two-step reaction with random hexamers, using incubation at 65 °C for 5 min in a T100 Thermal Cycler (Bio-Rad, CA, USA). Then, a reaction buffer, RNase inhibitor, dNTP mix, and reverse transcriptase were all added and incubated (25 °C for 5 min, then 42 °C for 60 min). The obtained cDNA was stored at −80 °C until the analysis was done.
The qPCR reaction was performed using the RT PCR SYBR A Kit (A&A Biotechnology) and primers of the tested (csd1, csd2, csd3, csd4, csd6, and amiA) and reference genes (ureA, gyrB, and glmM), obtained from Genomed (Warsaw, Poland) (Table 2). All primer sequences were taken from Fernandes et al. (2017) [71] and used in the present study. The exception was the csd6 gene (not present in that study), in which the primers were created by comparing the csd6 sequences of different H. pylori strains, obtained from the NCBI database, and then designed using the Primer3 v.4.1 program (https://bioinfo.ut.ee/primer3/ (accessed on 10 February 2021)). The procedure was performed using a CFX Connect Real-Time PCR Detection System (Bio-Rad, CA, USA) and the listed reaction parameters, i.e., initial denaturation (98 °C for 2 min), followed by 39 cycles: denaturation (98 °C for 30 s), annealing (51–57 °C for 10 s; depending on primers in Table 2), and elongation (65 °C for 5 s). In line with the previous study, the reference gene with sufficient stability during these studies was ureA (the difference between the expression in the control and experimental samples was <1), which served to compare the obtained expression of the tested genes. The other two reference genes, glmM and gyrB, underwent too high changes in the expression in experimental settings compared to controls (more than 3) and were therefore not used in our research. The negative controls were wells with primers but without cDNA. Gene expression was determined by calculating 2−∆∆Ct and a 2-fold change in the expression was considered as statistically significant. The tests were performed in three biological repetitions.
Table 2.
Primer sequences used during the quantitative RT-PCR (RT-qPCR).
| Gene | Primer Sequence (5’ to 3’) | Annealing Temperature | |
|---|---|---|---|
| csd1 | Forward | TCGCATACACAGGGGTGTTA | 54 °C |
| Reverse | TGCGCCTTATCCCTAATGAC | ||
| csd2 | Forward | CCTTTCTTTGGTGGGTTTGA | 54 °C |
| Reverse | GCTCTTTATTGTGGGGCAAA | ||
| csd3 | Forward | CGCTCATTCAAGCCCTTATC | 57 °C |
| Reverse | GCTAAAAGGGGGTCATTGGT | ||
| csd4 | Forward | TTAAACCCACCAGGCTCATC | 54 °C |
| Reverse | GGCTTGTGTTCTTGGGTGTT | ||
| csd6 | Forward | GCAGAAATTAGAGCGCTTGG | 51 °C |
| Reverse | GCCCTTGGTGTTCAATTCAT | ||
| amiA | Forward | ATACGGTTTGCTTTGGATGC | 54 °C |
| Reverse | GTCCGCAAAAATTACCCTGA | ||
| ureA | Forward | TTTCACGCTAACGGCTTTTT | 54 °C |
| Reverse | AACCGGATGATGTGATGGAT | ||
| glmM | Forward | CAACCGCTTGAGAAGAAAGG | 54 °C |
| Reverse | CCAACCAATTAAGCCAGGAA | ||
| gyrB | Forward | CGTCGCTTTGGATCATTTTT | 55 °C |
| Reverse | AATGGCGTGCCACTTTTAAC | ||
4.8. Statistical Analysis
The statistical significance of data within groups and between groups was determined by the Kruskal-Wallis test with a Bonferroni correction. For all tests, RStudio and a significance level of α = 0.05 were used.
5. Conclusions
In this original study, we demonstrated the potential of MYR to slow down the morphological transformation of H. pylori from spiral/rod-shaped forms to coccoid forms, the latter being classically involved in antibiotic tolerance and therapeutic failures. This inhibitory mechanism is most likely associated with the decreased expression of genes involved in the muropeptide monomers shortening (csd3, csd6, csd4, and amiA). Additionally, we have determined the ability of MYR to reduce the biofilm formation of H. pylori and obtain synergism with all five tested antibiotics (amoxicillin, clarithromycin, tetracycline, metronidazole, and levofloxacin) against this pathogen.
Author Contributions
Conceptualization, P.K.; methodology, P.K.; validation, P.K.; microbiological research and antibacterial activity testing, P.K.; light microscopy images obtaining and analysis, P.K.; fluorescence microscopy images obtaining, E.P.; fluorescence microscopy images analysis, P.K.; electron microscopy images obtaining, P.M.; genetic research, P.K. and M.K.; statistical analysis, P.M.; data curation, P.K.; writing—original draft preparation, P.K.; writing—review and editing, P.K., E.P., M.K., A.W., and G.G.; visualization, P.K.; supervision, P.K.; funding acquisition, P.K. All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported by the Wroclaw Medical University grant number STM.A130.20.002. The funders had no role in the study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wang C.H., Hsieh Y.H., Powers Z.M., Kao C.Y. Defeating Antibiotic-Resistant Bacteria: Exploring Alternative Therapies for a Post-Antibiotic Era. Int. J. Mol. Sci. 2020;21:1061. doi: 10.3390/ijms21031061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tagliabue A., Rappuoli R. Changing Priorities in Vaccinology: Antibiotic Resistance Moving to the Top. Front. Immunol. 2018;9:1068. doi: 10.3389/fimmu.2018.01068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D.L., Pulcini C., Kahlmeter G., Kluytmans J., Carmeli Y., et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 4.Van Giau V., An S.S.A., Hulme J. Recent Advances in the Treatment of Pathogenic Infections using Antibiotics and Nano-Drug Delivery Vehicles. Drug Des. Devel. Ther. 2019;13:327–343. doi: 10.2147/DDDT.S190577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buroni S., Chiarelli L.R. Antivirulence Compounds: A Future Direction to Overcome Antibiotic Resistance? Future Microbiol. 2020;15:299–301. doi: 10.2217/fmb-2019-0294. [DOI] [PubMed] [Google Scholar]
- 6.Cory H., Passarelli S., Szeto J., Tamez M., Mattei J. The Role of Polyphenols in Human Health and Food Systems: A Mini-Review. Front. Nutr. 2018;5:87. doi: 10.3389/fnut.2018.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silva L.N., Zimmer K.R., Macedo A.J., Trentin D.S. Plant Natural Products Targeting Bacterial Virulence Factors. Chem. Rev. 2016;116:9162–9236. doi: 10.1021/acs.chemrev.6b00184. [DOI] [PubMed] [Google Scholar]
- 8.Taheri Y., Suleria H.A.R., Martins N., Sytar O., Beyatli A., Yeskaliyeva B., Seitimova G., Salehi B., Semwal P., Painuli S., et al. Myricetin Bioactive Effects: Moving from Preclinical Evidence to Potential Clinical Applications. BMC Complement. Med. Ther. 2020;20:241. doi: 10.1186/s12906-020-03033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Semwal D.K., Semwal R.B., Combrinck S., Viljoen A. Myricetin: A Dietary Molecule with Diverse Biological Activities. Nutrients. 2016;8:90. doi: 10.3390/nu8020090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song X., Tan L., Wang M., Ren C., Guo C., Yang B., Ren Y., Cao Z., Li Y., Pei J. Myricetin: A Review of the Most Recent Research. Biomed. Pharmacother. 2021;134:111017. doi: 10.1016/j.biopha.2020.111017. [DOI] [PubMed] [Google Scholar]
- 11.Wang T., Zhang P., Lv H., Deng X., Wang J. A Natural Dietary Flavone Myricetin as an α-Hemolysin Inhibitor for Controlling Staphylococcus aureus Infection. Front. Cell. Infect. Microbiol. 2020;10:330. doi: 10.3389/fcimb.2020.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silva L.N., Da Hora G.C.A., Soares T.A., Bojer M.S., Ingmer H., Macedo A.J., Trentin D.S. Myricetin Protects Galleria mellonella against Staphylococcus aureus Infection and Inhibits Multiple Virulence Factors. Sci. Rep. 2017;7:2823. doi: 10.1038/s41598-017-02712-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grenier D., Chen H., Lagha A.B., Fournier-Larente J., Morin M.P. Dual Action of Myricetin on Porphyromonas gingivalis and the Inflammatory Response of Host Cells: A Promising Therapeutic Molecule for Periodontal Diseases. PLoS ONE. 2015;10:e0131758. doi: 10.1371/journal.pone.0131758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li G., Wang G., Si X., Zhang X., Liu W., Li L., Wang J. Inhibition of Suilysin Activity and Inflammation by Myricetin Attenuates Streptococcus suis Virulence. Life Sci. 2019;223:62–68. doi: 10.1016/j.lfs.2019.03.024. [DOI] [PubMed] [Google Scholar]
- 15.Lv Q., Lv Y., Dou X., Wassy S.L., Jia G., Wei L., Yu Q., Deng X., Zhang C., Wang J. Myricetin Inhibits the Type III Secretion System of Salmonella enterica serovar Typhimurium by Downregulating the Salmonella Pathogenic Island I Gene Regulatory Pathway. Microb. Pathog. 2021;150:104695. doi: 10.1016/j.micpath.2020.104695. [DOI] [PubMed] [Google Scholar]
- 16.Rhee K.H., Park J.S., Cho M.J. Helicobacter pylori: Bacterial Strategy for Incipient Stage and Persistent Colonization in Human Gastric Niches. Yonsei Med. J. 2014;55:1453–1466. doi: 10.3349/ymj.2014.55.6.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jonaitis L., Pellicano R., Kupcinskas L. Helicobacter pylori and Nonmalignant Upper Gastrointestinal Diseases. Helicobacter. 2018;23:e12522. doi: 10.1111/hel.12522. [DOI] [PubMed] [Google Scholar]
- 18.Ribaldone D.G., Simondi D., Petrini E., Astegiano M., Durazzo M. Non-Invasive Biomarkers for Gastric Cancer Diagnosis: Ready for Prime Time? Minerva Biotecnol. 2019;31:3–10. doi: 10.23736/S1120-4826.18.02463-1. [DOI] [Google Scholar]
- 19.Sgouras D.N., Trang T.T.H., Yamaoka Y. Pathogenesis of Helicobacter pylori Infection. Helicobacter. 2015;20:8–16. doi: 10.1111/hel.12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olivera-Severo D., Uberti A.F., Marques M.S., Pinto M.T., Gomez-Lazaro M., Figueiredo C., Leite M., Carlini C.R. A New Role for Helicobacter pylori Urease: Contributions to Angiogenesis. Front. Microbiol. 2017;8:1883. doi: 10.3389/fmicb.2017.01883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ansari S., Yamaoka Y. Role of Vacuolating Cytotoxin A in Helicobacter pylori Infection and Its Impact on Gastric Pathogenesis. Expert Rev. Anti. Infect. Ther. 2020;18:987–996. doi: 10.1080/14787210.2020.1782739. [DOI] [PubMed] [Google Scholar]
- 22.Ansari S., Yamaoka Y. Helicobacter pylori Virulence Factor Cytotoxin-Associated Gene A (CagA)-Mediated Gastric Pathogenicity. Int. J. Mol. Sci. 2020;21:7430. doi: 10.3390/ijms21197430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu C., Soyfoo D.M., Wu Y., Xu S. Virulence of Helicobacter pylori Outer Membrane Proteins: An Updated Review. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:1821–1830. doi: 10.1007/s10096-020-03948-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krzyżek P., Grande R. Transformation of Helicobacter pylori into Coccoid Forms as a Challenge for Research Determining Activity of Antimicrobial Substances. Pathogens. 2020;9:184. doi: 10.3390/pathogens9030184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krzyżek P., Gościniak G. Morphology of Helicobacter pylori as a Result of Peptidoglycan and Cytoskeleton Rearrangements. Prz. Gastroenterol. 2018;13:182–195. doi: 10.5114/pg.2018.78284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Figura N., Moretti E., Vaglio L., Langone F., Vernillo R., Vindigni C., Giordano N. Factors Modulating the Outcome of Treatment for the Eradication of Helicobacter pylori Infection. New Microbiol. 2012;35:335–340. [PubMed] [Google Scholar]
- 27.Kadkhodaei S., Siavoshi F., Akbari Noghabi K. Mucoid and Coccoid Helicobacter pylori with Fast Growth and Antibiotic Resistance. Helicobacter. 2020;25:e12678. doi: 10.1111/hel.12678. [DOI] [PubMed] [Google Scholar]
- 28.Sisto F., Brenciaglia M.I., Scaltrito M.M., Dubini F. Helicobacter pylori: ureA, cagA and vacA Expression during Conversion to the Coccoid Form. Int. J. Antimicrob. Agents. 2000;15:277–282. doi: 10.1016/S0924-8579(00)00188-6. [DOI] [PubMed] [Google Scholar]
- 29.Mizoguchi H., Fujioka T., Kishi K., Nishizono A., Kodama R., Nasu M. Diversity in Protein Synthesis and Viability of Helicobacter pylori Coccoid Forms in Response to Various Stimuli. Infect. Immun. 1998;66:5555–5560. doi: 10.1128/IAI.66.11.5555-5560.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shao C., Sun Y., Wang N., Yu H., Zhou Y., Chen C., Jia J. Changes of Proteome Components of Helicobacter pylori Biofilms Induced by Serum Starvation. Mol. Med. Rep. 2013;8:1761–1766. doi: 10.3892/mmr.2013.1712. [DOI] [PubMed] [Google Scholar]
- 31.Krzyżek P., Biernat M.M., Gościniak G. Intensive Formation of Coccoid Forms as a Feature Strongly Associated with Highly Pathogenic Helicobacter pylori Strains. Folia Microbiol. 2019;64:273–281. doi: 10.1007/s12223-018-0665-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krzyżek P., Grande R., Migdał P., Paluch E., Gościniak G. Biofilm Formation as a Complex Result of Virulence and Adaptive Responses of Helicobacter pylori. Pathogens. 2020;9:1062. doi: 10.3390/pathogens9121062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang J.Y., Kim C., Kwon Y.H., Lee J.E., Jeon S.W., Nam S.Y., Seo A.N., Han M.-H., Park J.H. Dual Clarithromycin and Metronidazole Resistance is the Main Cause of Failure in Ultimate Helicobacter pylori Eradication. Dig. Dis. 2021 doi: 10.1159/000514278. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 34.Krzyżek P., Pawełka D., Iwańczak B., Kempiński R., Leśniakowski K., Mégraud F., Łaczmański Ł., Biernat M., Gościniak G. High Primary Antibiotic Resistance of Helicobacter pylori Strains Isolated from Pediatric and Adult Patients in Poland during 2016–2018. Antibiotics. 2020;9:228. doi: 10.3390/antibiotics9050228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costa K., Bacher G., Allmaier G., Dominguez-Bello M.G., Engstrand L., Falk P., de Pedro M.A., García-del Portillo F. The Morphological Transition of Helicobacter pylori Cells from Spiral to Coccoid is Preceded by a Substantial Modification of the Cell Wall. J. Bacteriol. 1999;181:3710–3715. doi: 10.1128/JB.181.12.3710-3715.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaput C., Ecobichon C., Pouradier N., Rousselle J.-C., Namane A., Boneca I.G. Role of the N-acetylmuramoyl-L-alanyl Amidase, AmiA, of Helicobacter pylori in Peptidoglycan Metabolism, Daughter Cell Separation, and Virulence. Microb. Drug Resist. 2016;22:477–486. doi: 10.1089/mdr.2016.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.El Mortaji L., Tejada-Arranz A., Rifflet A., Boneca I.G., Pehau-Arnaudet G., Radicella J.P., Marsin S., De Reuse H. A Peptide of a Type I Toxin-Antitoxin System Induces Helicobacter pylori Morphological Transformation from Spiral Shape to Coccoids. Proc. Natl. Acad. Sci. USA. 2020;117:31398–31409. doi: 10.1073/pnas.2016195117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webb S.A.R., Kahler C.M. Bench-to-Bedside Review: Bacterial Virulence and Subversion of Host Defences. Crit. Care. 2008;12:234. doi: 10.1186/cc7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leitão J.H. Microbial Virulence Factors. Int. J. Mol. Sci. 2020;21:5320. doi: 10.3390/ijms21155320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang D.C., Blair K.M., Salama N.R. Staying in Shape: The Impact of Cell Shape on Bacterial Survival in Diverse Environments. Microbiol. Mol. Biol. Rev. 2016;80:187–203. doi: 10.1128/MMBR.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Teeseling M.C.F., de Pedro M.A., Cava F. Determinants of Bacterial Morphology: From Fundamentals to Possibilities for Antimicrobial Targeting. Front. Microbiol. 2017;8:1264. doi: 10.3389/fmicb.2017.01264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lopes L.A.A., dos Santos Rodrigues J.B., Magnani M., de Souza E.L., de Siqueira-Júnior J.P. Inhibitory Effects of Flavonoids on Biofilm Formation by Staphylococcus aureus that Overexpresses Efflux Protein Genes. Microb. Pathog. 2017;107:193–197. doi: 10.1016/j.micpath.2017.03.033. [DOI] [PubMed] [Google Scholar]
- 43.Lin R.D., Chin Y.P., Lee M.H. Antimicrobial Activity of Antibiotics in Combination with Natural Flavonoids against Clinical Extended-Spectrum β-lactamase (ESBL)-Producing Klebsiella pneumoniae. Phyther. Res. 2005;19:612–617. doi: 10.1002/ptr.1695. [DOI] [PubMed] [Google Scholar]
- 44.Lechner D., Gibbons S., Bucar F. Modulation of Isoniazid Susceptibility by Flavonoids in Mycobacterium. Phytochem. Lett. 2008;1:71–75. doi: 10.1016/j.phytol.2008.01.002. [DOI] [Google Scholar]
- 45.Brown A.R., Ettefagh K.A., Todd D., Cole P.S., Egan J.M., Foil D.H., Graf T.N., Schindler B.D., Kaatz G.W., Cech N.B. A Mass Spectrometry-Based Assay for Improved Quantitative Measurements of Efflux Pump Inhibition. PLoS ONE. 2015;10:e0124814. doi: 10.1371/journal.pone.0124814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nitulescu G., Nicorescu I.M., Olaru O.T., Ungurianu A., Mihai D.P., Zanfirescu A., Nitulescu G.M., Margina D. Molecular Docking and Screening Studies of New Natural Sortase A Inhibitors. Int. J. Mol. Sci. 2017;18:2217. doi: 10.3390/ijms18102217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pinto H.B., Brust F.R., Macedo A.J., Trentin D.S. The Antivirulence Compound Myricetin Possesses Remarkable Synergistic Effect with Antibacterials upon Multidrug Resistant Staphylococcus aureus. Microb. Pathog. 2020;149:104571. doi: 10.1016/j.micpath.2020.104571. [DOI] [PubMed] [Google Scholar]
- 48.Obonyo M., Zhang L., Thamphiwatana S., Pornpattananangkul D., Fu V., Zhang L. Antibacterial Activities of Liposomal Linolenic Acids against Antibiotic-Resistant Helicobacter pylori. Mol. Pharm. 2012;9:2677–2685. doi: 10.1021/mp300243w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Faghri J., Poursina F., Moghim S., Zarkesh Esfahani H., Nasr Esfahani B., Fazeli H., Mirzaei N., Jamshidian A., Ghasemian Safaei H. Morphological and Bactericidal Effects of Different Antibiotics on Helicobacter pylori. Jundishapur J. Microbiol. 2014;7:e8704. doi: 10.5812/jjm.8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krzyżek P., Paluch E., Gościniak G. Synergistic Therapies as a Promising Option for the Treatment of Antibiotic-Resistant Helicobacter pylori. Antibiotics. 2020;9:658. doi: 10.3390/antibiotics9100658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Griep M.A., Blood S., Larson M.A., Koepsell S.A., Hinrichs S.H. Myricetin Inhibits Escherichia coli DnaB Helicase but Not Primase. Bioorg. Med. Chem. 2007;15:7203–7208. doi: 10.1016/j.bmc.2007.07.057. [DOI] [PubMed] [Google Scholar]
- 52.Lin H.H., Huang C.Y. Characterization of Flavonol Inhibition of DnaB Helicase: Real-Time Monitoring, Structural Modeling, and Proposed Mechanism. J. Biomed. Biotechnol. 2012;2012:735368. doi: 10.1155/2012/735368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shinozuka K., Kikuchi Y., Nishino C., Mori A., Tawata S. Inhibitory Effect of Flavonoids on DNA-Dependent DNA and RNA Polymerases. Experientia. 1988;44:882–885. doi: 10.1007/BF01941188. [DOI] [PubMed] [Google Scholar]
- 54.Gottesman S. Trouble is Coming: Signaling Pathways that Regulate General Stress Responses in Bacteria. J. Biol. Chem. 2019;294:11685–11700. doi: 10.1074/jbc.REV119.005593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Górniak I., Bartoszewski R., Króliczewski J. Comprehensive Review of Antimicrobial Activities of Plant Flavonoids. Phytochem. Rev. 2019;18:241–272. doi: 10.1007/s11101-018-9591-z. [DOI] [Google Scholar]
- 56.Chaput C., Ecobichon C., Cayet N., Girardin S.E., Werts C., Guadagnini S., Prévost M.-C., Mengin-Lecreulx D., Labigne A., Boneca I.G. Role of AmiA in the Morphological Transition of Helicobacter pylori and in Immune Escape. PLoS Pathog. 2006;2:e97. doi: 10.1371/journal.ppat.0020097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frirdich E., Biboy J., Pryjma M., Lee J., Huynh S., Parker C.T., Girardin S.E., Vollmer W., Gaynor E.C. The Campylobacter jejuni Helical to Coccoid Transition Involves Changes to Peptidoglycan and the Ability to Elicit an Immune Response. Mol. Microbiol. 2019;112:280–301. doi: 10.1111/mmi.14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frirdich E., Biboy J., Adams C., Lee J., Ellermeier J., Gielda L.D., DiRita V.J., Girardin S.E., Vollmer W., Gaynor E.C. Peptidoglycan-Modifying Enzyme Pgp1 Is Required for Helical Cell Shape and Pathogenicity Traits in Campylobacter jejuni. PLoS Pathog. 2012;8:e1002602. doi: 10.1371/annotation/aac1c73d-b014-4cee-8dbc-eb8472de211d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yao Y., Lin G., Xie Y., Ma P., Li G., Meng Q., Wu T. Preformulation Studies of Myricetin: A Natural Antioxidant Flavonoid. Pharmazie. 2014;69:19–26. [PubMed] [Google Scholar]
- 60.Xiang D., Wang C.G., Wang W.Q., Shi C.Y., Xiong W., Wang M.D., Fang J.G. Gastrointestinal Stability of Dihydromyricetin, Myricetin, and Myricitrin: An In Vitro Investigation. Int. J. Food Sci. Nutr. 2017;68:704–711. doi: 10.1080/09637486.2016.1276518. [DOI] [PubMed] [Google Scholar]
- 61.Guo H., Chen Y.F., Tang Y., Qian J.Q. Method for Enhancing Bioavailability of Myricetin Based on Self-Assembly of Casein–Myricetin Nanomicelles. IET Nanobiotechnol. 2020;14:239–244. doi: 10.1049/iet-nbt.2018.5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yao Y., Xie Y., Hong C., Li G., Shen H., Ji G. Development of a Myricetin/Hydroxypropyl-β-Cyclodextrin Inclusion Complex: Preparation, Characterization, and Evaluation. Carbohydr. Polym. 2014;110:329–337. doi: 10.1016/j.carbpol.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 63.Guo R.X., Fu X., Chen J., Zhou L., Chen G. Preparation and Characterization of Microemulsions of Myricetin for Improving Its Antiproliferative and Antioxidative Activities and Oral Bioavailability. J. Agric. Food Chem. 2016;64:6286–6294. doi: 10.1021/acs.jafc.6b02184. [DOI] [PubMed] [Google Scholar]
- 64.Qian J., Meng H., Xin L., Xia M., Shen H., Li G., Xie Y. Self-Nanoemulsifying Drug Delivery Systems of Myricetin: Formulation Development, Characterization, and In Vitro and In Vivo Evaluation. Colloids Surf. B Biointerfaces. 2017;160:101–109. doi: 10.1016/j.colsurfb.2017.09.020. [DOI] [PubMed] [Google Scholar]
- 65.Junka A., Bartoszewicz M., Dziadas M., Szymczyk P., Dydak K., Żywicka A., Owczarek A., Bil-Lula I., Czajkowska J., Fijałkowski K. Application of Bacterial Cellulose Experimental Dressings Saturated with Gentamycin for Management of Bone Biofilm In Vitro and Ex Vivo. J. Biomed. Mater. Res. Part B: Appl. Biomater. 2020;108:30–37. doi: 10.1002/jbm.b.34362. [DOI] [PubMed] [Google Scholar]
- 66.Ciecholewska-Juśko D., Żywicka A., Junka A., Drozd R., Sobolewski P., Migdał P., Kowalska U., Toporkiewicz M., Fijałkowski K. Superabsorbent Crosslinked Bacterial Cellulose Biomaterials for Chronic Wound Dressings. Carbohydr. Polym. 2021;253:117247. doi: 10.1016/j.carbpol.2020.117247. [DOI] [PubMed] [Google Scholar]
- 67.Krzyżek P., Gościniak G., Fijałkowski K., Migdał P., Dziadas M., Owczarek A., Czajkowska J., Aniołek O., Junka A. Potential of Bacterial Cellulose Chemisorbed with Anti-Metabolites, 3-Bromopyruvate or Sertraline, to Fight against Helicobacter pylori Lawn Biofilm. Int. J. Mol. Sci. 2020;21:9507. doi: 10.3390/ijms21249507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krzyżek P., Franiczek R., Krzyżanowska B., Łaczmański Ł., Migdał P., Gościniak G. In Vitro Activity of 3-Bromopyruvate, an Anti-Cancer Compound, Against Antibiotic-Susceptible and Antibiotic-Resistant Helicobacter pylori Strains. Cancers. 2019;11:229. doi: 10.3390/cancers11020229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krzyżek P., Franiczek R., Krzyżanowska B., Łaczmański Ł., Migdał P., Gościniak G. In Vitro Activity of Sertraline, an Antidepressant, Against Antibiotic-Susceptible and Antibiotic-Resistant Helicobacter pylori Strains. Pathogens. 2019;8:228. doi: 10.3390/pathogens8040228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hathroubi S., Zerebinski J., Ottemann K.M. Helicobacter pylori Biofilm Involves a Multigene Stress-Biased Response, Including a Structural Role for Flagella. MBio. 2018;9:e01973-18. doi: 10.1128/mBio.01973-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fernandes R.M., Silva H., Oliveira R., Almeida C., Azevedo N.F., Vieira M.J. Morphological Transition of Helicobacter pylori Adapted to Water. Future Microbiol. 2017;12:1167–1179. doi: 10.2217/fmb-2016-0174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data is contained within the article.