Abstract
Background
Glutamate and aspartate are preferred nutrients for a variety of microorganisms. In the case for many Pseudomonas spp., utilization of these amino acids is believed to be dependent on a transporter complex comprised of a periplasmic-solute binding protein (AatJ), two permease domains (AatQM) and an ATP-binding component (AatP). Notably, expression of this transporter complex is hypothesized to be regulated at the transcriptional level by the enhancer-binding protein AauR and the alternative sigma factor RpoN. The purpose of the current study was to determine the biological significance of the putative aatJ-aatQMP operon and its regulatory aauR and rpoN genes in the utilization of L-glutamate, L-glutamine, L-aspartate and L-asparagine in Pseudomonas aeruginosa PAO1.
Results
Deletion of the aatJ-aatQMP, aauR or rpoN genes did not affect the growth of P. aeruginosa PAO1 on L-glutamate, L-glutamine, L-aspartate and L-asparagine equally. Instead, only growth on L-glutamate as the sole carbon source was abolished with the deletion of any one of these genes. Interestingly, growth of the aauR mutant on L-glutamate was readily restored via plasmid-based expression of the aatQMP genes, suggesting that it is the function of AatQMP (and not AatJ) that is limiting in the absence of the aauR gene. Subsequent analysis of beta-galactosidase reporters revealed that both aatJ and aatQ were induced in response to L-glutamate, L-glutamine, L-aspartate or L-asparagine in a manner dependent on the aauR and rpoN genes. In addition, both aatJ and aatQ were expressed at reduced levels in the absence of the inducing-amino acids and the regulatory aauR and rpoN genes. The expression of the aatJ-aatQMP genes is, therefore, multifaceted. Lastly, the expression levels of aatJ were significantly higher (> 5 fold) than that of aatQ under all tested conditions.
Conclusions
The primary function of AauR in P. aeruginosa PAO1 is to activate expression of the aatJ-aatQMP genes in response to exogenous acidic amino acids and their amide derivatives. Importantly, it is the AauR-RpoN mediated induction of the aatQMP genes that is the pivotal factor enabling P. aeruginosa PAO1 to effectively utilize or consume L-glutamate as a sole or preferred nutrient.
Keywords: Enhancer-binding protein, Acidic amino acids, Glutamate utilization, AauR, RpoN, Pseudomonas aeruginosa
Background
The assimilation or utilization of dicarboxylates from the environment is wide-spread among soil-dwelling and plant-associated bacteria. For some bacteria, dicarboxylates serve as preferred nutrients for cellular growth whereas for others consumption of these compounds is an important component of their complicated lifestyles. An example of the latter is the establishment of symbiotic relationships between Rhizobia and plants; the exudates from plants are a rich source of C4-dicarobxylates, especially malate [1, 2]. Bacteria belonging to the genus Pseudomonas are some of the best-known examples of the former in which both C4- and C5-dicarboxylates are at the top of the hierarchy for carbon source utilization [3, 4].
In the two instances given above it is the transport of dicarboxylates across the bacterial membrane that is often a major control point or regulated step in their utilization. Regulation of dicarboxylate transport is commonly achieved through a two-component signal transduction system (TCS) comprised of the histidine sensor kinase DctB and its cognate response regulator DctD [1, 5]. In the typical DctBD TCS, extracellular dicarboxylates are expected to bind to the periplasmic input domain of DctB and stimulate the phosphorylation of a conserved histidine residue within its cytoplasmic transmitter domain. The phosphoryl group is subsequently transferred from the phosphohistidine of DctB to a conserved aspartate residue located in the response-regulator domain of DctD. Phosphorylated DctD then activates or upregulates the transcription of genes encoding for dicarboxylate-related transport proteins. In addition to being a response regulator, DctD is part of an unusual family of bacterial transcriptional regulators called enhancer-binding proteins or EBPs [6, 7]. The unusual or unique functionality of EBPs is due to the fact that they are the only known family of transcriptional regulators that activate transcription from the − 12/− 24 promoters recognized by the alternative sigma factor RpoN [8–10]. In other words, transcription from RpoN-controlled − 12/− 24 promoters are dependent on EBPs. This means that genes under the regulation of the DctBD TCS are also potentially governed by RpoN in the bacterial cell.
Interestingly, the genomes of various Pseudomonas spp. encode for three distinct DctBD TCSs wherein each one is specific towards a particular class of dicarboxylates. Two of these systems have been characterized for Pseudomonas aeruginosa PAO1. The classical or prototypical DctBD TCS of P. aeruginosa PAO1 is encoded by the PA5165 – PA5166 genes, and regulates the transport of C4-dicarboxylates, including succinate, fumarate and malate [11]. Specifically, DctD activates transcription of genes encoding for a C4-dicarboxylate-sodium symporter known as DctA and a tripartite ATP-independent periplasmic or TRAP transporter called DctPQM. The second characterized DctBD TCS of P. aeruginosa consists of the sensor kinase MifS (PA5512) and its cognate response regulator MifR (PA5511). The MifSR TCS was originally identified for its role in microcolony formation and named accordingly [12]. It was later demonstrated that the MifSR TCS is required for the utilization of C5-dicarboxylates such as α-ketoglutarate (α-KG) in which MifR activates transcription of the PA5530 gene, encoding for a C5-dicarboxylate-proton symporter [13–15]. The utilization of C4- and C5-dicarboxylates as a sole carbon source in P. aeruginosa PAO1 is dependent on DctD and MifR, respectively [11, 13].
The third and final DctBD TCS occurring in P. aeruginosa PAO1 is that of the acidic-amino acid utilization regulator AauR (PA1335) and its partner sensor histidine kinase AauS (PA1336). The AauSR TCS has only been investigated in Pseudomonas putida KT2440 where it was shown to have an influential effect in the utilization of L-glutamate, L-glutamine and L-aspartate [16, 17]. Notably, expression of an ATP-binding cassette (ABC) transporter complex encoded by the aatJ-aatQMP genes was observed to be upregulated in response to L-glutamate, and the aauR gene was implicated as being required for full expression of this gene cluster in P. putida KT2440 [16]. Subsequent analysis revealed that the periplasmic solute-binding protein AatJ exhibits affinity constants of ~ 0.3 and 1.5 μM for L-glutamate and L-aspartate, respectively, while deletion of the aatJ-aatQMP locus in P. putida KT2440 resulted in drastic growth reductions on L-glutamate, L-aspartate or L-glutamine if present as the sole source of carbon and nitrogen [17]. DNase I footprinting experiments suggest that AauR binds to the consensus sequence TTCGG-N4-CCGAA located in the 5′-regulatory or promoter region of the aatJ-aatQMP operon [17]. Putative AauR-binding sites were also found within the 5′-regulatory regions of gltP (glutamate/aspartate symporter), ppsA (phosphoenolpyruvate synthase), ansB (glutaminase/asparaginase) and dsbC (thiol/disulfide exchange protein), arguing that the AauSR TCS regulates a consortium of genes in response to L-glutamate, L-glutamine and/or L-aspartate in P. putida KT2440 [17].
The 5′-regulatory region of the aatJ-aatQMP genes in P. aeruginosa PAO1 possesses putative binding sites for both AauR and RpoN (Fig. 1). One of the putative AauR-binding sites or motifs upstream of aatJ-aatQMP in P. aeruginosa PAO1 is an exact match to the published consensus sequence for AauR of P. putida KT2440 [17], and it was previously found that RpoN does bind to the aatJ locus in P. aeruginosa PA14 through ChIP seq [18]. Collectively, these findings strongly suggest that the aatJ-aatQMP genes are under the control of AauR and RpoN in P. aeruginosa PAO1. However, the aatJ and aatQMP genes are separated by a 201 bp intergenic region [19]. The presence of such an appreciable intergenic region between the aatJ and aatQMP genes questions the validity of them forming an operon or being necessarily coordinately regulated. Therefore, in order to have a foundational understanding of AauR regulation in P. aeruginosa PAO1, the contributions of both the aatJ and aatQMP loci, as well as the regulatory aauR and rpoN genes, towards the utilization of acidic amino acids and their carboxamide counterparts were investigated.
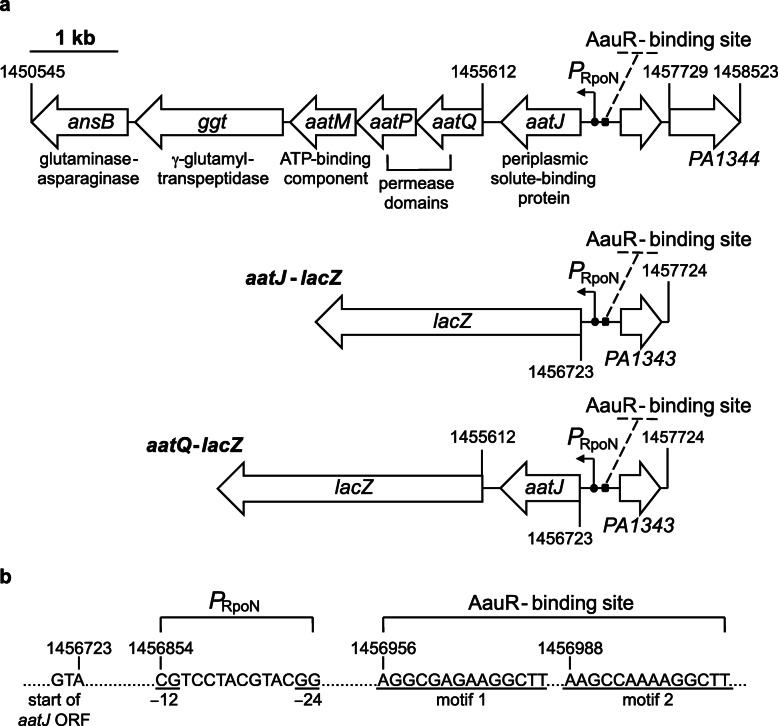
Fig. 1.
Genetic organization of the aatJ-aatQMP genes of P. aeruginosa PAO1. a The uptake of acidic amino acids is believed to mediated through an ABC transporter complex encoded by the aatJ-aatQMP cluster. Positioned 131–144 bp upstream of the aatJ ORF is a putative −12/−24 promoter (PRpoN), which is the promoter specifically recognized by the sigma factor RpoN. Located upstream of the − 12/− 24 promoter are two sequences or motifs resembling the consensus DNA-binding site for the EBP AauR of P. putida KT2440. The presence of these predicted sites is a strong indicator that both AauR and RpoN are involved in the regulation of aatJ-aatQMP genes, and therefore, the transport of acidic amino acids. To investigate this regulation, two β-galactosidase (LacZ) reporters were constructed and are illustrated: aatJ-lacZ and aatQ-lacZ. Although not the subject of the current study, the ansB and ggt genes potentially form an operon with aatJ-aatQMP, and collectively, are coordinately regulated in response to acidic amino acids and their amide derivatives in P. aeruginosa PAO1. b Close-up view of the putative − 12/− 24 promoter (PRpoN) and AauR-binding site (motifs 1 and 2) located upstream of the aatJ ORF in P. aeruginosa PAO1
Results
The aauR and rpoN genes are required for optimal growth on L-glutamate and L-glutamine as sole carbon sources
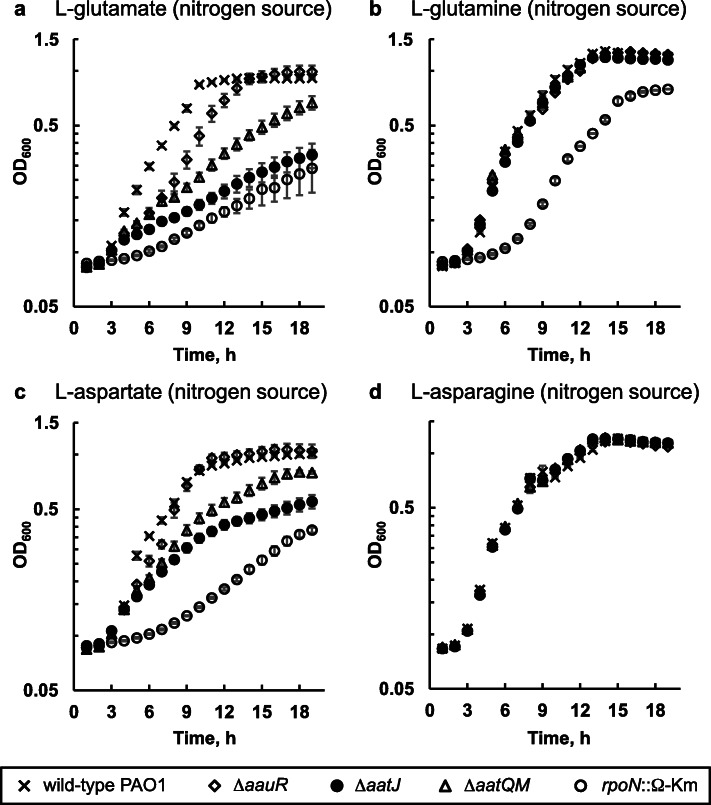
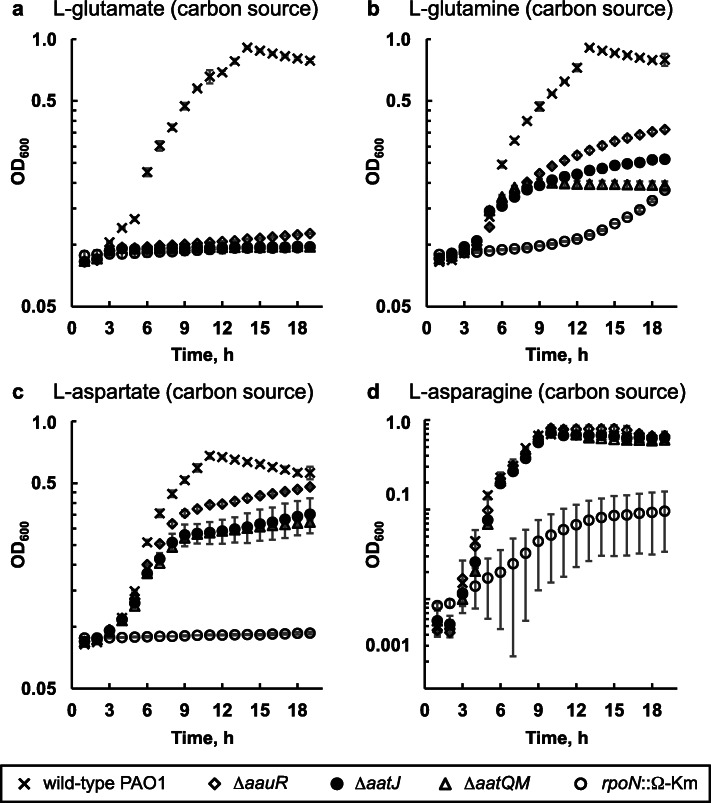
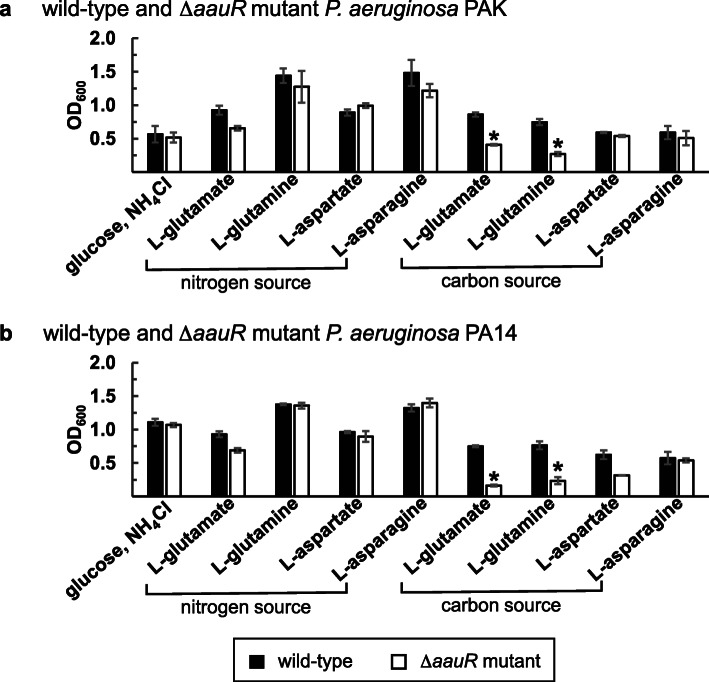
The aauR (PA1335), aatJ (PA1342) and aatQM (PA1341 – PA1340) genes were individually deleted from the genome of P. aeruginosa PAO1. The resulting markerless ΔaauR, ΔaatJ, and ΔaatQM mutants, in addition to an rpoN::Ω-Km mutant and wild-type P. aeruginosa PAO1, were then grown in minimal media in which L-glutamate, L-glutamine, L-aspartate or L-asparagine served as the sole source of nitrogen or carbon. As shown in Fig. 2, the growth of the ΔaauR mutant did not differ significantly from wild-type P. aeruginosa PAO1 when each amino acid served as the sole source of nitrogen. However, growth deficiencies were observed for the ΔaauR mutant when either L-glutamate or L-glutamine served as the sole carbon source (Fig. 3). After 18 h of growth on L-glutamate as the sole carbon source, wild-type P. aeruginosa PAO1 reached a cell density (OD600) of ~ 1.0 while the ΔaauR mutant yielded an OD600 of ~ 0.1 (Fig. 3a). The ΔaauR mutant fared better on L-glutamine as a carbon source, reaching an OD600 of ~ 0.3, which was 3-fold lower than that observed for wild-type P. aeruginosa PAO1 (Fig. 3b). The final cell densities for wild-type P. aeruginosa PAO1 and the ΔaauR mutant on L-aspartate as the sole carbon source were similar (OD600 of ~ 0.6) (Fig. 3c), and the growth curves for both of these strains were identical to one another when L-asparagine was the carbon source (Fig. 3d). These results suggest that the aauR gene of P. aeruginosa PAO1 is not essential for the utilization of acidic amino acids and their amide derivatives in general, but instead, is specific towards the utilization of L-glutamate and L-glutamine as sole or preferred carbon sources. Comparable results were observed for ΔaauR mutants of P. aeruginosa PA14 and PAK, both of which displayed significant growth reductions (> 2-fold) on L-glutamate or L-glutamine as the sole carbon source (Fig. 4).
Fig. 2.
Growth of ΔaauR, rpoN::Ω-Km, ΔaatJ and ΔaatQM mutants of P. aeruginosa PAO1 on L-glutamate (a), L-glutamine (b), L-aspartate (c) and L-asparagine (d) as nitrogen sources. Bacteria were grown at 37 °C in minimal media supplemented with 20 mM glucose and 10 mM of indicated amino acid as the sole nitrogen source. Data points represent mean values (n = 3) ± SD
Fig. 3.
Growth of ΔaauR, rpoN::Ω-Km, ΔaatJ and ΔaatQM mutants of P. aeruginosa PAO1 on L-glutamate (a), L-glutamine (b), L-aspartate (c) and L-asparagine (d) as carbon sources. Bacteria were grown at 37 °C in minimal media supplemented with 10 mM NH4Cl and 20 mM of indicated amino acid as the sole carbon source. Data points represent mean values (n = 3) ± SD
Fig. 4.
Growth of ΔaauR mutants of P. aeruginosa strains PAK (a) and PA14 (b) on acidic amino acids and their amide derivatives as nitrogen or carbon sources. Bacteria were grown at 37 °C for 24 h in minimal media supplemented with 20 mM carbon source and 10 mM nitrogen source as indicated. Data points represent mean values (n = 3) ± SD. ANOVA was performed with a Dunnett’s post hoc test (α value, 0.05) to identify significant differences (P < 0.0001), which are indicated with asterisks
Growth of the rpoN::Ω-Km mutant was significantly decreased compared to that of wild-type P. aeruginosa PAO1 under all conditions tested except when L-asparagine served as the sole nitrogen source (Fig. 2d). In the presence of L-glutamate (Fig. 2a) and L-aspartate (Fig. 2c) as nitrogen sources, the rpoN::Ω-Km mutant grew to a final OD600 of ~ 0.3. The rpoN::Ω-Km mutant displayed an extended lag phase on L-glutamine as a nitrogen source (Fig. 2b), but its final OD600 was 0.8, which was only slightly below the value of ~ 1.2 observed for wild-type P. aeruginosa PAO1 and the ΔaauR mutant. When the four amino acids were used as the sole carbon sources, the growth of the rpoN::Ω-Km mutant became even more limited. No growth was observed for the rpoN::Ω-Km mutant when L-glutamate (Fig. 3a) or L-aspartate (Fig. 3c) was the carbon source. In the presence of L-glutamine (Fig. 3b) or L-asparagine (Fig. 3d) as the carbon source, the cell densities of the rpoN::Ω-Km mutant were 5-fold lower than that of wild-type P. aeruginosa PAO1. These results support earlier findings demonstrating the essentiality of the rpoN gene in the utilization of these amino acids as carbon sources in P. aeruginosa PA14 and P. putida [20, 21].
The aatJ and aatQM genes are essential for optimal growth on L-glutamate, L-glutamine and L-aspartate as sole carbon sources
In the presence of L-glutamate as the sole nitrogen source, the final cell densities of the ΔaatJ and ΔaatQM mutants were ~ 0.3 and 0.6, respectively, both of which were significantly lower than the OD600 of ~ 1.0 observed for the ΔaauR mutant and wild-type P. aeruginosa PAO1 (Fig. 2a). In contrast, the ΔaatJ and ΔaatQM mutants displayed wild-type growth when L-glutamine was the only available nitrogen source (Fig. 2b). Hindered growth was observed for the ΔaatJ and ΔaatQM mutants when L-aspartate was the sole nitrogen source (Fig. 2c), but both mutants grew similar to wild-type P. aeruginosa PAO1 in the presence of L-asparagine as the nitrogen source (Fig. 2d). Interestingly, deletions of the aatJ and aatQM genes had negative effects on the utilization of L-glutamate and L-aspartate as nitrogen sources, which was not observed when the regulatory aauR gene was deleted.
There were substantial growth deficiencies observed for the ΔaatJ and ΔaatQM mutants in the presence of L-glutamate, L-glutamine and L-aspartate as carbon sources (Fig. 3). Both mutants did not grow on L-glutamate as a carbon source (Fig. 3a), and both reached a final OD600 of ~ 0.2 on L-glutamine as the sole carbon source (Fig. 3b) compared to ~ 0.9 for wild-type P. aeruginosa PAO1. When L-aspartate was the carbon source, the ΔaatJ and ΔaatQM mutants each yielded an OD600 of ~ 0.3 (Fig. 3c), which was about 2-fold lower than that of wild-type P. aeruginosa PAO1. The ΔaatJ and ΔaatQM mutants did, however, display wild-type growth on L-asparagine as the sole carbon source (Fig. 3d).
Elevated concentrations of L-glutamate are present in the spent medium of the ΔaauR mutant compared to wild-type P. aeruginosa PAO1
Deletion of the aauR gene reduced the growth of P. aeruginosa PAO1 on L-glutamate as the sole source of carbon but not nitrogen. This suggested that the uptake or assimilation of L-glutamate was not completely abolished in the ΔaauR mutant. To gain further insight into this matter, the depletion of L-glutamate, L-glutamine, L-aspartate and L-asparagine from the growth medium was measured for both the ΔaauR mutant and wild-type P. aeruginosa PAO1. Cells were grown in glucose-minimal media that was supplemented with one of these four amino acids as the sole nitrogen source at an initial concentration of 20 mM, and the concentration of these four amino acids in the cell-free spent medium were quantified by HPLC at 6.0 and 9.0 h post inoculation.
At 6.0 h post inoculation, L-glutamate decreased from an initial concentration of 20 mM to 8.4 ± 0.2 and 17.8 ± 0.1 mM in the spent medium of wild-type P. aeruginosa PAO1 and the ΔaauR mutant, respectively (Fig. 5a). Three hours later at 9.0 h post inoculation, L-glutamate was not detected in the spent medium from wild-type cells but was still present at a concentration of 12.2 ± 0.6 mM for the ΔaauR mutant. The depletion of L-glutamine as the sole nitrogen source was also noticeably different between the two strains (Fig. 5b). At 6.0 h post inoculation, L-glutamine was not detected in the spent medium of wild-type cells but L-glutamate was found to be present at a concentration of 11.0 ± 0.4 mM. Neither L-glutamine nor L-glutamate was observed in the spent medium from wild-type P. aeruginosa PAO1 at 9.0 h post inoculation. In sharp contrast, L-glutamine (but not L-glutamate) was detected at a concentration of 10.8 ± 0.3 mM in the spent medium from the ΔaauR mutant at 6.0 h post inoculation. Subsequently, at 9.0 h post inoculation, L-glutamine was no longer detected in the spent medium from the ΔaauR mutant while L-glutamate was detected at a concentration of 14.1 ± 0.6 mM.
Fig. 5.
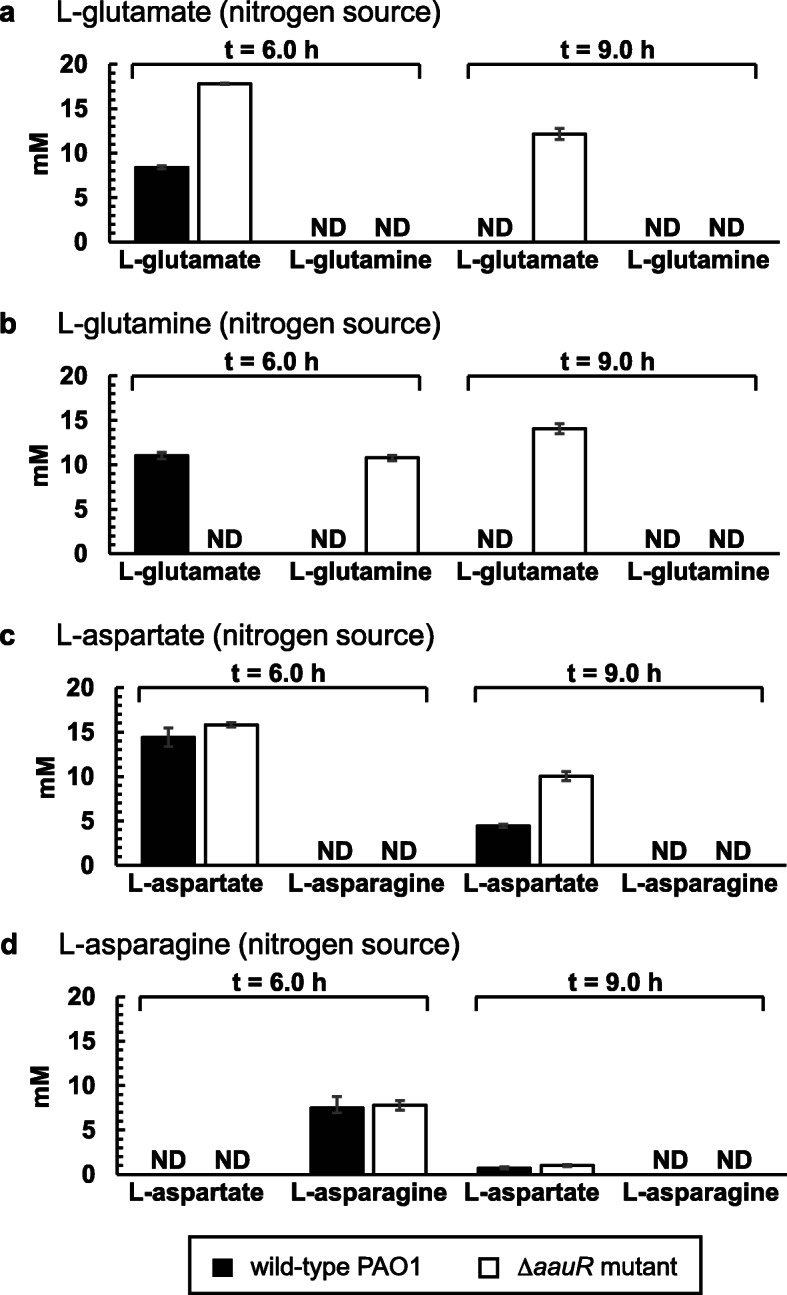
HPLC-measured concentration of L-glutamate (a), L-glutamine (b), L-aspartate (c) and L-asparagine (d) in the spent medium of wild-type P. aeruginosa PAO1 and the ΔaauR mutant at 6.0 and 9.0 h post inoculation. Data points represent mean values ± SD obtained from triplicate bacterial samples grown at 37 °C in glucose-minimal media supplemented with an initial concentration of 20 mM of indicated amino acid. ND denotes not detected
When L-aspartate served as the sole nitrogen source, its concentration in the spent medium was 14.4 ± 1.1 and 15.8 ± 0.2 for wild-type P. aeruginosa PAO1 and the ΔaauR mutant, respectively, at 6.0 h post inoculation (Fig. 5c). The extracellular concentration of L-aspartate decreased to 4.4 ± 0.2 for wild-type P. aeruginosa PAO1 at 9.0 h post inoculation whereas for the ΔaauR mutant it was 2-fold higher, yielding a concentration of 10.0 ± 0.5 mM. The depletion of L-asparagine was similar between the ΔaauR mutant and wild-type P. aeruginosa PAO1 (Fig. 5d). For both strains at 6.0 h post inoculation, extracellular L-asparagine was at a concentration of ~ 8.0 mM and no extracellular L-aspartate was detected. At 9.0 h post inoculation, L-asparagine was completely absent from the spent medium from both strains while L-aspartate was detected at concentrations of ~ 1.0 mM.
Plasmid-based expression of aatQMP or aatJ-aatQMP in the ΔaauR mutant restores its growth to wild-type levels when L-glutamate or L-glutamine served as the sole carbon source
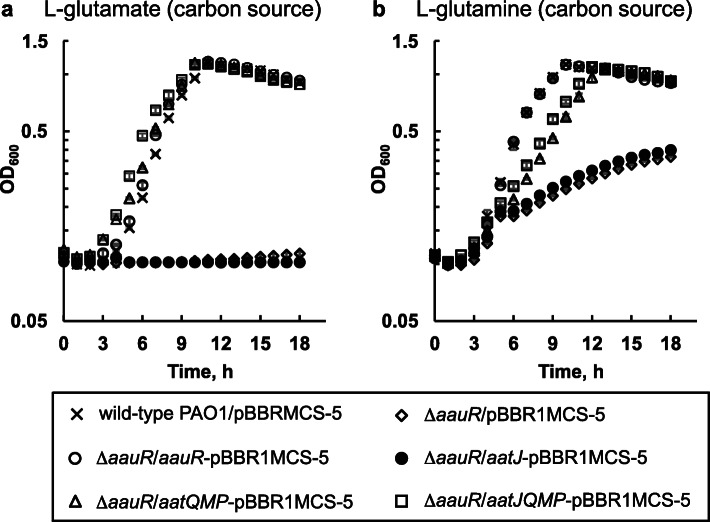
The findings from the aforementioned experiments indicated that the growth deficiencies observed for the ΔaauR mutant might be the result of insufficiencies in L-glutamate uptake or transport associated with deregulation of the aatJ-aatQMP locus. As a first step to address this possibility, the aatJ, aatQMP, and the aatJ-aatQMP genes were individually cloned under the lac promoter of the expression plasmid pBBR1MCS-5, and the resulting constructs were introduced into the ΔaauR mutant. Recombinant strains were subsequently grown in minimal media supplemented with 30 μg mL− 1 gentamicin and 20 mM L-glutamate or L-glutamine as the sole carbon source. The ΔaauR mutant harboring empty pBBR1MCS-5 and an aauR-pBBR1MCS-5 derivative were included in the analysis.
The ΔaauR mutant transformed with aauR-, aatQMP-, or aatJ-aatQMP-pBBR1MCS-5 displayed wild-type levels of growth in the presence of either L-glutamate (Fig. 6a) or L-glutamine (Fig. 6b) as the sole carbon source. In contrast, no growth (OD600 < 0.1) was observed for the ΔaauR mutant harboring aatJ-pBBR1MCS-5 on either amino acid. It is interesting to note that plasmid-based expression of the aatQMP genes was sufficient to rescue the L-glutamate and L-glutamine related growth deficiencies of the ΔaauR mutant. This suggests that the expression of the aatQMP genes, and not aatJ nor the entire aatJ-aatQMP cluster, are the main limiting factor for utilization of L-glutamate and L-glutamine in the absence of the regulatory aauR gene.
Fig. 6.
Genetic-complementation experiments for the ΔaauR mutant grown on L-glutamate (a) and L-glutamine (b) as carbon sources. The ΔaauR mutant was transformed with expression plasmids encoding for aauR (aauR-pBBR1MCS-5), aatJ (aatJ-pBBR1MCS-5), aatQMP (aatQMP-pBBR1MCS-5) or aatJQMP (aatJQMP-pBBR1MCS-5). Both wild-type P. aeruginosa PAO1 and the ΔaauR mutant harboring empty plasmid (pBBR1MCS-5) were included as controls. Bacteria were grown at 37 °C in minimal media supplemented with 30 μg mL− 1 gentamicin, 10 mM NH4Cl and 20 mM of indicated amino acid as the sole carbon source. Data points represent mean values (n = 3) ± SD
The aauR and rpoN genes are essential for the induction of aatJ-lacZ and aatQ-lacZ in response to acidic amino acids and their amide derivatives
Based on the findings of AauR regulation in P. putida KT2440 [16], it was expected that expression of the aatJ-aatQMP genes in P. aeruginosa PAO1 would be regulated to some degree by the availability of L-glutamate in a mechanism consisting of the EBP AauR and the sigma factor RpoN. It was uncertain, however, as to whether the expression of the aatJ-aatQMP genes would be completely dependent on AauR and RpoN, or if these regulatory proteins are only necessary for the upregulation or induction of this locus in response to L-glutamate. To evaluate the expression of the aatJ-aatQMP genes, two plasmid-based LacZ reporters were constructed: aatJ-lacZ and aatQ-lacZ. As shown in Fig. 1, both aatJ-lacZ and aatQ-lacZ have the same ~ 1000 bp 5′-regulatory region immediately upstream of the aatJ ORF. The aatQ-lacZ also possesses the aatJ ORF and the 201 bp intergenic region between the aatJ and aatQ ORFs.
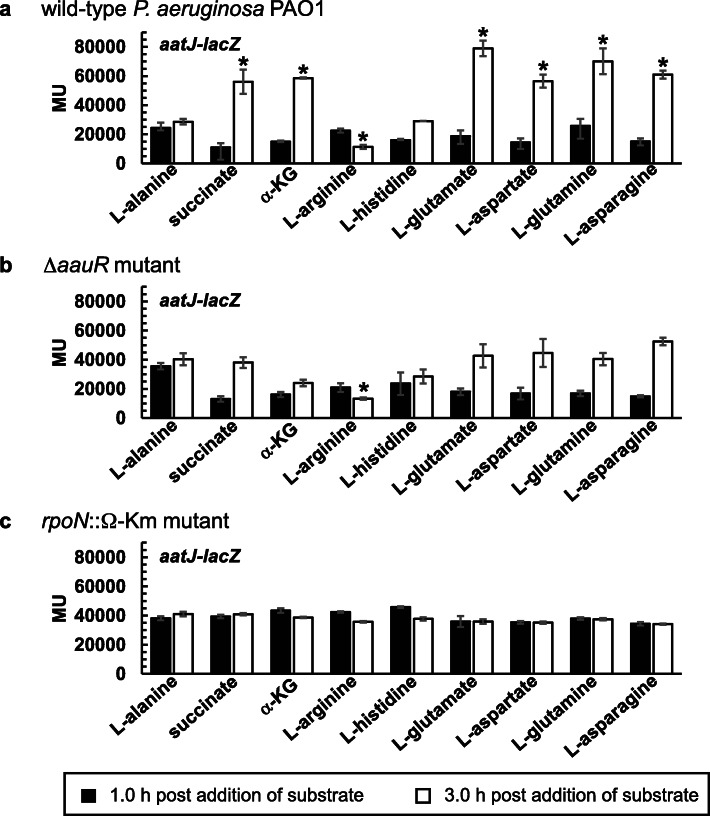
The aatJ-lacZ and aatQ-lacZ reporters were introduced into the ΔaauR and rpoN::Ω-Km mutants, as well as wild-type P. aeruginosa PAO1. Recombinant strains were grown in L-alanine-minimal media to an OD600 of 0.2, and subsequently challenged with the addition of 20 mM of succinate, α-KG, L-arginine, L-histidine, L-glutamate, L-aspartate, L-glutamine or L-asparagine. Because L-alanine was reported to have minimal influence in catabolite repression and global-gene regulation in Pseudomonas [22, 23], it was an ideal choice for establishing a baseline level of expression for the aatJ-lacZ and aatQ-lacZ reporters while simultaneously serving as a permissible or growth-sustaining nutrient for the ΔaauR and rpoN::Ω-Km mutants. Representative C4- and C5-dicarboxylates were included as substrates in the analysis due to the fact that AauR is a predicted member of the DctD-family of EBPs. In addition, L-glutamate is an intermediate in the catabolism of L-arginine and L-histidine [24, 25], so both of these amino acids were included as substrates in these assays. Because a lag phase of ~ 3.0 h was observed for wild-type P. aeruginosa PAO1 when grown on acidic amino acids and their amide derivatives, LacZ activity was measured near the beginning and end of this phase, i.e., 1.0 and 3.0 h post addition of substrate. LacZ activity is reported in Miller Units (MU).
The expression levels for aatJ-lacZ in response to L-alanine as the sole carbon source were ~ 25,000 MU in wild-type P. aeruginosa PAO1 at both the 1.0 and 3.0 h timepoints (Fig. 7a). Expression of aatJ-lacZ in response to L-alanine was only slightly higher for the ΔaauR (Fig. 7b) and rpoN::Ω-Km (Fig. 7c) mutants, yielding LacZ activities of ~ 35,000 MU for both timepoints. The expression levels for aatJ-lacZ in the sole presence of L-alanine were not statistically different among the three strains, and thus, served as the baseline for all subsequent comparisons.. When challenged with L-glutamate, L-aspartate, L-glutamine or L-asparagine, the expression levels of aatJ-lacZ increased by > 2-fold in wild-type P. aeruginosa PAO1 at 3.0 h post addition (Fig. 7a) whereas no such induction was observed for the ΔaauR (Fig. 7b) or rpoN::Ω-Km mutant (Fig. 7c). The expression levels of aatJ-lacZ did not significantly increase at 1.0 h post addition for any tested substrate or strain.
Fig. 7.
LacZ activity of aatJ-lacZ in wild-type P. aeruginosa PAO1 (a), the ΔaauR mutant (b) and the rpoN::Ω-Km mutant (c). Bacteria were grown in alanine-minimal media to an OD600 of ~ 0.2 and then challenged with 20 mM of indicated substrate. LacZ activity was subsequently measured at 1.0 and 3.0 h post addition of substrate and is reported in Miller Units (MU). Data points represent mean values (n = 3) ± SD. For each strain or mutant, an ANOVA was performed with a Dunnett’s post hoc test (α value, 0.05) to identify significant changes in the expression of aatJ-lacZ relative to the alanine-treated control. Significant differences (P < 0.0001) are indicated with asterisks
Expression of aatJ-lacZ was significantly influenced by the presence of succinate, α-KG and L-arginine in wild-type P. aeruginosa PAO1 (Fig. 7a). At 3.0 h post addition of succinate or α-KG, expression of aatJ-lacZ increased by 2-fold (~ 55,000 MU) in wild-type P. aeruginosa PAO1. The presence of L-arginine actually repressed the expression of aatJ-lacZ, yielding LacZ activities of ~ 15,000 MU for wild-type P. aeruginosa PAO1 at the 3.0 timepoint. Similar repression of aatJ-lacZ by L-arginine was observed for the ΔaauR mutant (Fig. 7b). The changes or fluctuations in the expression of aatJ-lacZ caused by succinate, α-KG or L-arginine were not observed in the rpoN::Ω-Km mutant (Fig. 7c). Lastly, the presence of L-histidine did not significantly affect the expression of aatJ-lacZ in any of the three investigated strains.
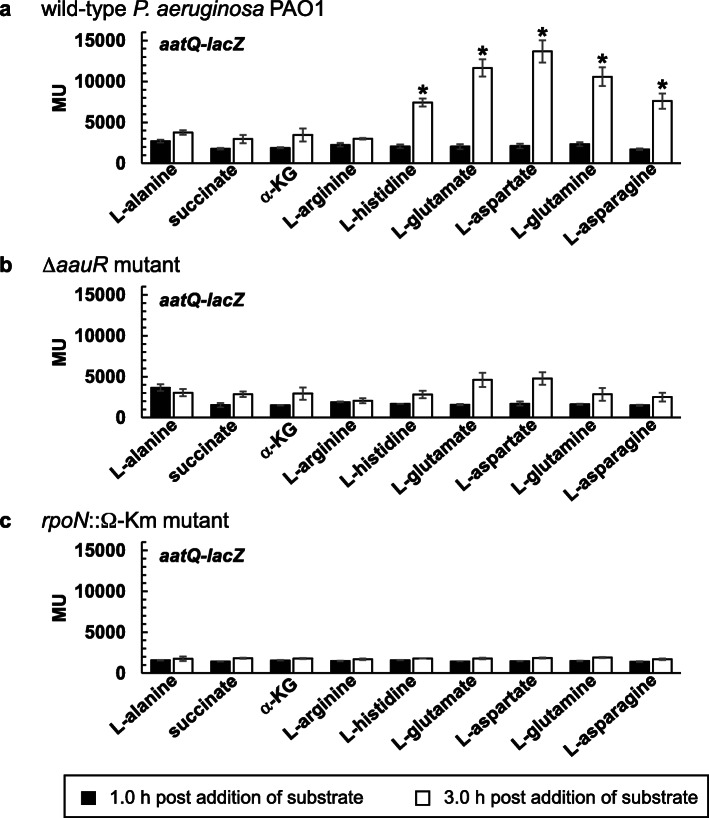
The expression levels for aatQ-lacZ were significantly lower than that of aatJ-lacZ for all tested substrates in all three strains (Fig. 8). In wild-type P. aeruginosa PAO1 (Fig. 8a), the expression levels for aatQ-lacZ in response to L-alanine as the sole carbon source generated LacZ activities of ~ 3500 MU for both the 1.0 and 3.0 h timepoints. This value increased by > 2-fold at 3.0 h post addition of L-glutamate, L-aspartate, L-glutamine, L-asparagine, or unexpectedly, L-histidine. Unlike that of aatJ-lacZ, the addition of succinate, α-KG or L-arginine had no significant effect on the expression of aatQ-lacZ in wild-type P. aeruginosa PAO1. Expression of aatQ-lacZ in the ΔaauR (Fig. 8b) and rpoN::Ω-Km (Fig. 8c) mutants was not significantly affected by the addition of any substrate. The average LacZ activities for aatQ-lacZ in these mutants ranged from 2000 to 4500 MU.
Fig. 8.
LacZ activity of aatQ-lacZ in wild-type P. aeruginosa PAO1 (a), the ΔaauR mutant (b) and the rpoN::Ω-Km mutant (c). Bacteria were grown in alanine-minimal media to an OD600 of ~ 0.2 and then challenged with 20 mM of indicated substrate. LacZ activity was subsequently measured at 1.0 and 3.0 h post addition of substrate and is reported in Miller Units (MU). Data points represent mean values (n = 3) ± SD. For each strain or mutant, an ANOVA was performed with a Dunnett’s post hoc test (α value, 0.05) to identify significant changes in the expression of aatQ-lacZ relative to the alanine-treated control. Significant differences (P < 0.0001) are indicated with asterisks
Discussion
The transporter AatJQMP has a fundamental role in the utilization of L-glutamate and L-glutamine in P. aeruginosa PAO1
The genome of P. aeruginosa PAO1 encodes for three putative glutamate transporters: the ABC-transporter complex AatJQMP (PA1339 – PA13342), the sodium-glutamate symporter GltS (PA3176), and the proton-glutamate symporter GltP (PA5479). Deletion of the aatJ or aatQM genes in P. aeruginosa PAO1 significantly reduced the growth of this bacterium on L-glutamate as either the sole source of nitrogen or carbon (Figs. 2a and 3a); although as a sole carbon source, growth was more severely hindered. Similar growth deficiencies on L-glutamate have been observed for aatQMP-transposon mutants of P. aeruginosa PAK [26]. It would appear that the importance of AatJQMP in L-glutamate utilization is not restricted to PAO1, but is likely common among other various strains and isolates of P. aeruginosa. The roles of GltS, GltP and other dicarboxylate-transport proteins in L-glutamate utilization have yet to be resolved, but the presence of these transporters would explain the lingering or residual growth observed for the ΔaatJ and ΔaatQM mutants of P. aeruginosa PAO1 on L-glutamate. Lastly, it should be mentioned that transposon insertions of the aatJ, aatQ, and aatP genes were previously reported to abolish the growth of P. aeruginosa PAO1 on D-glutamate as the sole carbon and nitrogen source [27]. This suggest that the transporter AatJQMP is not stereospecific, and thus, plays a central role in the utilization of both L- and D-isomers of glutamate in P. aeruginosa PAO1.
The aatJ-aatQMP genes were required for optimal growth of P. aeruginosa PAO1 on L-glutamine as the sole source of carbon but not nitrogen (Figs. 2b and 3b). The PA5073 – PA5076 operon of P. aeruginosa PAO1 encodes for a putative glutamine ABC-transporter complex, but a previous analysis of transposon mutants indicates that this locus is not required for growth on any of the twenty common amino acids [26]. Nonetheless, it is still plausible that L-glutamine enters the cell through PA5073 – PA5076 and then undergoes transamination and/or deamidation to yield L-glutamate. Another route for assimilation of L-glutamine involves deamidation in the periplasm catalyzed by the glutaminase-asparaginase AnsB (PA1337) [28] with the subsequent transport of the L-glutamate product via AatJQMP, GltS, GltP and/or additional dicarboxylate-transport proteins. Data from the HPLC analysis (Fig. 5b) supports the existence of this extracellular metabolic conversion. Notably, because the periplasmic deamidation of L-glutamine liberates ammonia, a readily consumable nitrogen source, the bacterium does not have a strict requirement for the uptake of L-glutamine directly or the L-glutamate product to fulfill its nitrogen demands. This would account for the unperturbed growth observed for the ΔaatJ and ΔaatQM mutants of P. aeruginosa PAO1 on L-glutamine as a source of nitrogen but not carbon (Figs. 2b and 3b); the latter of which requires the L-glutamate product as a precursor for cellular biosynthesis. A similar result was observed for P. aeruginosa PAK in which a transposon mutation of the aatM gene eliminated its growth on L-glutamine as a source of carbon but not nitrogen [26].
The transporter AatJQMP affects the utilization of L-aspartate but not L-asparagine in P. aeruginosa PAO1
Growth was observed for both the ΔaatJ and ΔaatQM mutants on L-aspartate as a nitrogen or carbon source, but the final cell densities for these mutants were 2-fold lower than that of wild-type P. aeruginosa PAO1 (Figs. 2c and 3c). This indicates that the transporter AatJQMP is required for optimal utilization of L-aspartate as a preferred or sole nutrient. In an earlier study, data from LacZ assays and microarray analysis clearly demonstrated that exogenous L-aspartate induced expression of the dctA and dctPQM genes in P. aeruginosa PAO1 [29]. These previous results suggest that the C4-dicarboxylate transporters DctA and DctPQM might facilitate the uptake of L-aspartate, and therefore, would give reason as to why the ΔaatJ and ΔaatQM mutations had a less deleterious effect in the utilization of L-aspartate compared to that of L-glutamate, a C5-dicarboxylate. For comparison purposes, neither L-aspartate nor L-glutamate induced expression of the C5-dicarboxylate transporter gene PA5530 [13, 29].
The utilization of L-asparagine is not dependent on AatJQMP in P. aeruginosa PAO1. Analogous to that of L-glutamine, one might expect that exogenous L-asparagine is deaminated in the periplasm through the actions of AnsB, and the resulting L-aspartate transported intracellularly via AatJQMP, DctA and DctPQM. However, results from our HPLC analysis indicated that very little L-aspartate (< 1.0 mM) accumulated in the extracellular milieu of P. aeruginosa PAO1 when fed an initial concentration of 20 mM L-asparagine as the sole nitrogen source (Fig. 5d). This is in sharp contrast to the > 10 mM L-glutamate that amassed in the spent medium when cells were given a starting concentration of 20 mM L-glutamine as the sole nitrogen source (Fig. 5b). In a previous study, exogenous L-asparagine was shown to induce expression of two transporter-gene lacZ fusions, PA5530-lacZ and PA2252-lacZ [29]. The PA2252 gene encodes for a putative sodium-amino acid symporter and potentially forms an operon with the ansA gene, encoding for a cytoplasmic glutaminase-asparaginase. The direct transport of L-asparagine through PA5530 and/or PA2252 followed by AnsA-catalyzed deamidation would account for the non-essential nature of the aatJ-aatQMP genes in the utilization this amino acid in P. aeruginosa PAO1.
AauR and RpoN are essential for the induction of aatJ-aatQMP in response to acidic amino acids and their amide derivatives P. aeruginosa PAO1
The results from the LacZ assays for aatJ-lacZ and aatQ-lacZ revealed several key factors surrounding the expression and regulation of the aatJ-aatQMP genes in P. aeruginosa PAO1. First, expression of both aatJ-lacZ and aatQ-lacZ were induced or upregulated in the presence of exogenous L-glutamate (Figs. 7a and 8a), suggesting that the expression of the aatJ-aatQMP genes are coordinately upregulated in response to this amino acid. Such coordinated upregulation is not limited to L-glutamate, because the presence of L-aspartate, L-glutamine and L-asparagine also induced expression of aatJ-lacZ and aatQ-lacZ (Figs. 7a and 8a). Second, the upregulation of aatJ-lacZ and aatQ-lacZ in response to these four amino acids was completely dependent on the aauR and rpoN genes. This is consistent with the EBP AauR and the sigma factor RpoN being directly involved in the activation of the aatJ-aatQMP genes in P. aeruginosa.
Third, both aatJ-lacZ and aatQ-lacZ were still expressed in the absence of (i) the aauR and rpoN genes, and (ii) exogenous L-glutamate, L-aspartate, L-glutamine or L-asparagine. These expression levels, however, were less than that of their induced states. The same appears to be true for P. putida KT2440 in which deletion of the aauR gene did not eliminate expression of an aatJ-lacZ reporter, but instead, reduced its expression by ~ 50% in response to L-glutamate [16]. Fourth and last, the expression levels of aatJ-lacZ were significantly greater (> 5-fold) than that of aatQ-lacZ under both inducing and non-inducing conditions. Collectively, these findings indicate that although the aatJ-aatQMP genes in P. aeruginosa PAO1 are coordinately upregulated by AauR-RpoN in response to certain amino acids, they are also expressed independently of these two regulatory proteins, including a potential mechanism in which the aatJ gene is expressed separately or distinctly from that of aatQMP.
The multivariate expression surrounding the aatJ-aatQMP genes of P. aeruginosa PAO1 is evident in the response of aatJ-lacZ and aatQ-lacZ to various substrates (Figs. 7 and 8). The presence of L-arginine repressed expression of aatJ-lacZ but not aatQ-lacZ whereas exogenous L-histidine induced expression of the latter and not the former. An earlier study did report that the transcript levels of aatJ in P. aeruginosa PAO1 were ~ 2-fold higher for cells grown in L-glutamate compared to L-arginine as the sole nitrogen source [27]. However, the analysis of the ArgR regulon of P. aeruginosa PAO1 revealed that the transcript levels for aatJ were not significantly different between cells grown in L-glutamate versus a mixture of L-glutamate and L-arginine [24]. Therefore, it would appear that L-arginine represses the expression of aatJ when present as the sole nutrient, i.e., in the absence of L-glutamate. ArgR was reported to repress genes associated with glutamate metabolism [24], so it possible that aatJ is also a target of negative control by this regulator. In the presence of both L-arginine and L-glutamate, expression of aatJ might be a competition between repression and activation via ArgR and AauR, respectively. Curiously, the presence of dicarboxylates such as succinate and α-KG, which are preferred carbon sources in Pseudomonas [3, 4], caused ~ 2-fold increase in the expression levels of aatJ-lacZ and aatQ-lacZ. An earlier transcriptomic study did identify the aatJ gene as a potential regulatory target of catabolite repression in P. aeruginosa PAO1 [30], and results from ChIP seq showed that the sigma factor FecI binds to the aatJ locus in P. aeruginosa PA14 [18] . Further investigation is needed to determine how such control points and regulatory factors contribute to the expression of the aatJ-aatQMP genes in P. aeruginosa.
The EBP AauR plays a pivotal role in the utilization of L-glutamate and L-glutamine as sole or preferred nutrients
Two significant growth deficiencies were observed for the ΔaauR mutant of P. aeruginosa PAO1. The ΔaauR mutant did not grow on L-glutamate as the sole carbon source (Fig. 3a), and it displayed a ~ 2-fold growth reduction on L-glutamine as the sole carbon source (Fig. 3b). The results of the genetic-complementation experiments (Fig. 6) and LacZ assays (Figs. 7b and 8b) suggest that these growth deficiencies were due to inadequate expression of the aatQMP genes. Namely, plasmid-based expression of the aatQMP genes restored the growth of the ΔaauR mutant to wild-type levels, and expression of aatQ-lacZ was downregulated by > 2-fold in the absence of the aauR gene. In comparison, plasmid-based expression of aatJ did not significantly impact or improve the growth of the ΔaauR mutant, and even though expression of aatJ-lacZ was not induced in the ΔaauR mutant, the non-induced or basal expression levels were still substantial and greater than that of aatQ-lacZ. It is AauR-mediated upregulation of the aatQMP genes, and not aatJ, which is a decisive factor in the utilization of L-glutamate and L-glutamine as sole or preferred nutrients in P. aeruginosa PAO1. Unlike the ΔaatJ and ΔaatQM mutants, the growth of the ΔaauR mutant did not differ significantly from wild-type P. aeruginosa PAO1 when L-glutamate served as the only nitrogen source (Fig. 2a) or L-aspartate served as the sole nitrogen or carbon source (Figs. 2c and 3c). The upregulation of the aatJ-aatQMP genes via AauR-RpoN is non-essential under such conditions.
The sigma factor RpoN is necessary for the utilization of acidic amino acids and their amide derivatives
The rpoN::Ω-Km mutant of P. aeruginosa PAO1 exhibited growth deficiencies on both L-glutamate and L-glutamine when either amino acid served as the sole source of carbon or nitrogen (Figs. 2 and 3). Based on the results of the LacZ assays (Figs. 7c and 8c), the observed growth deficiencies of the rpoN::Ω-Km mutant were likely caused to some degree by the downregulation of the aatJ-aatQMP genes. In addition, the rpoN::Ω-Km mutant displayed growth deficiencies on L-aspartate as either a nitrogen or carbon source (Figs. 2c and 3c). As noted earlier, the dicarboxylate-transport proteins DctA and DctPQM have been implicated in the transport of L-aspartate [29], and the expression of the corresponding dctA and dctPQM genes are dependent on RpoN [11]. The downregulation or reduced expression of the aatJ-aatQMP, dctA and dctPQM genes is expected to have a negative effect in the utilization of L-aspartate as observed for the rpoN::Ω-Km mutant. The possible role or function of RpoN in the utilization of L-asparagine is less clear. While RpoN was required for growth on L-asparagine as the sole carbon source (Fig. 3d), it was unnecessary when this amino acid was the only available nitrogen source (Fig. 2d). One potential contributing factor is the transporter PA5530, whose expression is positively regulated by both RpoN and L-asparagine [13, 29]. In contrast, the PA2252-ansA genes, which have also been associated with L-asparagine utilization [29], are neither predicted nor experimentally-proven targets of RpoN regulation in P. aeruginosa.
The growth deficiencies observed for the rpoN::Ω-Km mutant can be attributed to some extent to the downregulation of specific target genes, but it is plausible that pleiotropic effects were also an underlying factor. The RpoN regulon of P. aeruginosa is extensive - second only to that of the housekeeping sigma factor RpoD [18]. The transcription of > 500 genes are affected by RpoN [18], which regulates a consortium of diverse cellular processes such as motility [31], quorum sensing [32, 33], transport of dicarboxylates [11, 13], catabolite repression and nitrogen assimilation [34, 35]. Consequently, the deregulation of core metabolic and assimilatory pathways is expected to hinder or limit the overall growth of the rpoN::Ω-Km mutant. The expression patterns of aatJ-lacZ (Fig. 7c) and aatQ-lacZ (Fig. 8c) in the rpoN::Ω-Km mutant is also suggestive of such deregulation. For example, exogenous L-arginine repressed the expression of aatJ-lacZ in the ΔaauR mutant and wild-type P. aeruginosa PAO1, whereas in the rpoN::Ω-Km mutant, it had no effect. Furthermore, both aatJ-lacZ and aatQ-lacZ were unresponsive in the rpoN::Ω-Km mutant.
The scope and magnitude of AauR regulation in P. aeruginosa PAO1
The last 38 amino acid residues of the C-terminus of AauR form a putative FIS-type helix-turn-helix (HTH) motif. This FIS-type HTH or DNA-binding domain is 79% identical between the AauR proteins of P. aeruginosa PAO1 and P. putida KT2440, suggesting that these two EBPs recognize similar if not identical DNA-binding sites. Located 131–144 and 233–278 bp upstream of the aatJ ORF in P. aeruginosa PAO1 are sequences resembling the unique − 12/− 24 promoter recognized by RpoN and the consensus DNA-binding site for AauR, respectively (Fig. 1). Apart from aatJ, no other genes in P. aeruginosa PAO1 possess sequences matching the consensus DNA-recognition site of AauR in their 5′-regulatory regions. Biochemical characterization of AauR from P. aeruginosa PAO1 should reveal the exact DNA-binding site for this regulator and will be invaluable in generating a more definitive answer as to the number and range of genes that are directly governed by this EBP.
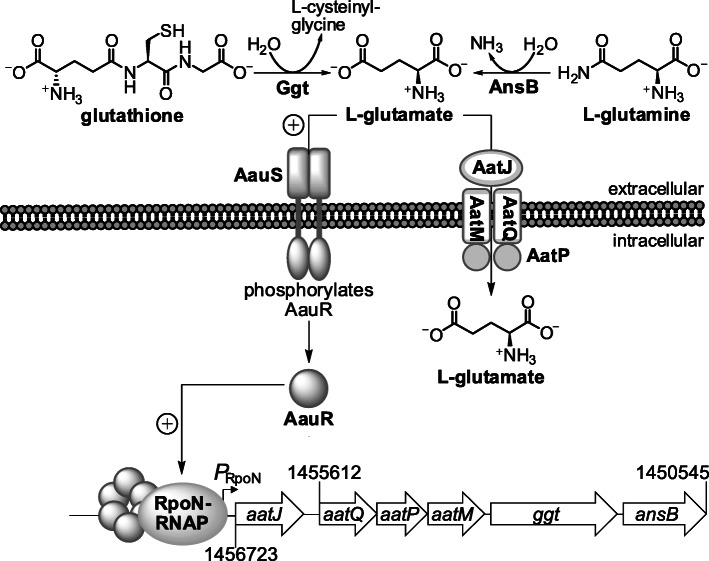
While the aatJ-aatQMP genes were the focus of the current study, it is probable that AauR and RpoN regulate the expression of two other genes associated with this locus (Figs. 1 and 9). Namely, the aatJ-aatQMP genes might form an operon with genes encoding for the periplasmic glutaminase-asparaginase AnsB and a periplasmic gamma-glutamyltranspeptidase (Ggt) [19]. As mentioned earlier, the utilization of L-glutamine is thought to involve AnsB, which catalyzes the deamidation of this amino acid in the periplasm to liberate L-glutamate [28]. Deletion of the aauR did hinder the growth of P. aeruginosa PAO1 on L-glutamine as the sole carbon source, but the total disappearance or depletion of exogenously fed L-glutamine was only delayed and not abolished in the ΔaauR mutant, arguing that the deamidation of L-glutamine is not strictly dependent on AauR in P. aeruginosa PAO1. Interestingly, growth on L-asparagine but not L-glutamine was significantly reduced in an ΔansA ΔansB double mutant of P. aeruginosa PAO1 [29]. This suggests that additional glutaminases apart from the cytoplasmic AnsA and periplasmic AnsB are involved in the deamidation and metabolism of L-glutamine in P. aeruginosa. The enzyme Ggt catalyzes the transfer of γ-glutamyl groups from donor molecules, most notably glutathione, to target or acceptor substrates consisting of amino acids, peptides or even water. Although Ggt has been implicated in the metabolism of glutathione and cysteine in some bacteria [36], little is known on the biological importance of this enzyme in Pseudomonas, including any potential role it may have in glutamate-related metabolism.
Fig. 9.
The AauSR TCS in the utilization of L-glutamate and L-glutamine in P. aeruginosa PAO1. Extracellular L-glutamate is sensed by the histidine kinase AauS, which subsequently phosphorylates its cognate partner EBP AauR. Phosphorylated AauR (shown as a hexamer [6]) interacts with the RpoN-RNA polymerase complex and mediates the transcriptional activation of the aatJ-aatQMP genes from the − 12/− 24 promoter (PRpoN). The ansB and ggt genes are potentially transcribed with aatJ-aatQMP, but experimental verification is needed. The periplasmic glutaminase-asparaginase AnsB catalyzes the deamidation of L-glutamine to yield L-glutamate whereas the metabolic role of Ggt is unknown but might involve the liberation of L-glutamate from the hydrolysis of γ-glutamyl compounds such as glutathione
The sensor histidine kinase AauS is hypothesized to be essential for optimal utilization of L-glutamate and L-glutamine in P. aeruginosa
The sensor histidine kinase AauS and the EBP AauR form a putative DctBD-type TCS. The transcriptional activity of AauR is, therefore, believed to be controlled through phosphorylation catalyzed by AauS. Because AauS controls the transcriptional activity of AauR, it is reasonable to predict that optimal utilization of L-glutamate and L-glutamine in P. aeruginosa will be dependent on AauS. Indeed, an earlier study reported that the growth phenotypes of aauS and aauR mutants of P. putida KT2440 were similar to one another [16]. Thus, comparable results are expected in P. aeruginosa.
The results of the LacZ assays strongly suggest that the AauSR TCS regulates the expression of the aatJ-aatQMP genes in response to L-glutamate, L-aspartate, L-glutamine and L-asparagine. The question then becomes: what is the substrate specificity of AauS? Does AauS recognize each amino acid, or are only the acidic amino acids capable of binding to and stimulating the histidine kinase activity of AauS? In the latter scenario, the periplasmic deamidation of L-glutamine and L-asparagine would generate the requisite acidic amino acid substrates for AauS. Another potential and unexpected substrate of AauS is that of L-histidine, which was found to induce expression of aatQ-lacZ in a manner dependent on AauR. Intracellular L-glutamate is an intermediate in the catabolism of L-histidine, and studies involving P. putida and Rhizobium leguminosarum have indicated that L-glutamate efflux may occur under certain conditions [17, 37, 38]. Perhaps the AauSR TCS of P. aeruginosa facilitates the recapture of effluxed or escaped L-glutamate during L-histidine catabolism. Lastly, given that the transporter AatJQMP is required for growth on both D- and L-glutamate in P. aeruginosa, it would suggest that AauS is also not stereospecific, i.e., either isomer of glutamate is sufficient for stimulating AauS activity. Current efforts are underway to determine the substrate specificity and biological significance of AauS in P. aeruginosa PAO1.
Conclusions
The results of this study provide some much-needed insight into the biological function and significance of the EBP AauR in P. aeruginosa (Fig. 9). In response to extracellular acidic amino acids and their amide derivatives, AauR and the sigma factor RpoN upregulate the expression of the aatJ-aatQMP genes, encoding for an ABC-transporter complex that mediates the uptake of L-glutamate and L-aspartate. Specifically, it is the AauR-RpoN mediated upregulation of the aatQMP genes (and not aatJ) that is a crucial, growth-limiting step in the utilization of L-glutamate and L-glutamine as sole or preferred nutrients in P. aeruginosa PAO1. The upregulation of aatJ-aatQMP via AauR-RpoN was not essential for the utilization of L-aspartate or L-asparagine, suggesting that this particular control mechanism is more influential or important towards the catabolism of the L-glutamate/L-glutamine pair.
Methods
Bacteria and bacteriological media
Bacteria used in the study are given in Table 1. Bacteria were grown in Lennox broth (LB) or minimal media (22 mM KH2PO4, 42 mM Na2HPO4, 8.6 mM NaCl, 2.0 mM MgSO4, 5.0 μM FeSO4, pH 7.0), which was supplemented with carbon and nitrogen sources as indicated per experiment described below. Solid bacteriological media was prepared with the addition of BD Difco™ agar at 15 g L− 1. Plasmids and chromosomal-genetic markers were selected with the following antibiotics at the indicated final concentrations in μg mL− 1: carbenicillin (200 for P. aeruginosa, 100 for E. coli); gentamicin (30 for P. aeruginosa, 25 for E coli; kanamycin (50 for E. coli).
Table 1.
Bacteria used in the current study
| Strains | Relevant characteristics | Source |
|---|---|---|
| Pseudomonas aeruginosa | ||
| PAO1 | wild type | [32] |
| ∆aauR PAO1 | ∆aauR derivative of PAO1 | This study |
| ∆aatQM PAO1 | ∆aatQM derivative of PAO1 | This study |
| ∆aatJ PAO1 | ∆aatJ derivative of PAO1 | This study |
| rpoN:Ω-Km PAO1 | ΔrpoN derivative of PAO1 | [32] |
| PA14 | wild type | [39] |
| ∆aauR PA14 | ∆aauR derivative of PA14 | This study |
| PAK | wild type | [40] |
| ∆aauR PAK | ∆aauR derivative of PAK | This study |
| Escherichia coli MG1655 | F− λ− ilvG− rfb-50 rph-1 | [41] |
| Escherichia coli Top10a | F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ (ara-leu)7697 galE15 galK16 rpsL(Strr) endA1 λ− | Invitrogen |
aUsed for cloning purposes and plasmid maintenance
Molecular biology methods
Plasmids and oligonucleotides used in the study are given in Tables 2 and 3, respectively. Molecular biology enzymes (restriction endonucleases, DNA polymerases and ligases) were purchased from New England BioLabs (Ipswich, Massachusetts, USA) whereas the purification of nucleic acids was completed using commercially available kits from Promega (Madison, Wisconsin, USA). Cloned DNA was sequenced to confirm its identity.
Table 2.
Plasmids used in the current study
| Plasmids | Relevant characteristicsa | Source |
|---|---|---|
| pCR-Blunt | Cloning plasmid; Kmr | Invitrogen |
| pBBR1MCS-5 | Broad-host-range plasmid; Gmr | [42] |
| pDONR221 | Gateway cloning plasmid; Kmr | Invitrogen |
| pEX18ApGW | Gene-deletion plasmid for P. aeruginosa; Cbr | [43] |
| ∆Plac-pBBR1MCS-5 | pBBR1MCS-5 minus lac promoter; Gmr | [13] |
| pJMS02 | aauR in pCR-Blunt; Kmr | This study |
| pJMS03 | aauR in pBBR1MCS-5; Gmr | This study |
| pJMS04 | aatQMP in pCR-Blunt; Kmr | This study |
| pJMS05 | aatQMP in pBBR1MCS-5; Gmr | This study |
| pJMS07 | aatJ-lacZ in pCR-Blunt; Kmr | This study |
| pJMS08 | aatQM-lacZ in pCR-Blunt; Kmr | This study |
| pJMS09 | aatJ-lacZ in ∆Plac-pBBR1MCS-5; Gmr | This study |
| pJMS10 | aatQM-lacZ in ∆Plac-pBBR1MCS-5; Gmr | This study |
| pJMS18 | aatJQMP in pCR-Blunt; Kmr | This study |
| pJMS19 | aatJQMP in pBBR1MCS-5; Gmr | This study |
| pJMS20 | aatJ in pCR-Blunt; Kmr | This study |
| pJMS21 | aatJ in pBBR1MCS-5; Gmr | This study |
| pJMS298 | aauR::FRT-aacC1-FRT in pDONR221; Kmr Gmr | This study |
| pJMS299 | aauR::FRT-aacC1-FRT in pEX18ApGW; Cbr Gmr | This study |
| pJMS300 | aatJ::FRT-aacC1-FRT in pDONR221; Kmr Gmr | This study |
| pJMS301 | aatJ::FRT-aacC1-FRT in pEX18ApGW; Cbr Gmr | This study |
| pJMS302 | aatQM::FRT-aacC1-FRT in pDONR221; Kmr Gmr | This study |
| pJMS303 | aatQM::FRT-aacC1-FRT in pEX18ApGW; Cbr Gmr | This study |
aNote that Cbr, Gmr and Kmr denote resistance to carbenicillin, gentamicin and kanamycin, respectively
Table 3.
Oligonucleotides used in the current study
| Oligonucleotides | Sequence | Purposea |
|---|---|---|
| BL342.r | gcagttatttttgacaccagaccaactggta | E. coli lacZ |
| JS05.r | tgaatccgtaatcatggtcatgttggggtttcccctcgg | aatQ-lacZ |
| JS06.f | ccgaggggaaaccccaacatgaccatgattacggattcactgg | E. coli lacZ (aatQ overlap) |
| JS42.f | ccgaggggaaaccccaacatgaccatgattacggattcactgg | E. coli lacZ (aatJ overlap) |
| JS43.r | tgaatccgtaatcatggtcatgaatttcttcctcgacttgttttg | aatJ-lacZ |
| JS35P.f | cgtctcctgtcgagtgacgaacc | aatJ-lacZ and aatQ-lacZ |
| JS50.f | gcatctagaccgccagtgtctgcaacc | aatQMP genes |
| JS51.r | gcagagctctcagtgcgggaggatcttgg | aatQMP genes |
| JS391K.f | aggaacttcaagatccccaattcgggaaatggtcaacgaagtgctcg | aatQM DnF-Gm |
| JS391K.r | tacaagaaagctgggtgatcttggcgaggaactgc | aatQM DnR-GWR |
| JS401K.f | tacaaaaaagcaggctggacttcagtggaatcgttcc | aatQM UpF-GWL |
| JS401K.r | tcagagcgcttttgaagctaattcgcgatgttcgacagcaacttgc | aatQM UpR-Gm |
| JS351.f | tacaaaaaagcaggctggaggagctggaggacatc | aauR UpF-GWL |
| JS351.r | tcagagcgcttttgaagctaattcggatgatccgttgcgccagg | aauR UpR-Gm |
| JS352.f | aggaacttcaagatccccaattcggaacacttcctccagcagtc | aauR DnF-Gm |
| JS352.r | tacaagaaagctgggtgcaggccgtacttcttcac | aauR DnR-GWR |
| JS100.f | tacaaaaaagcaggctccggatttcataagctggc | aatJ UpF-GWL |
| JS100.r | tcagagcgcttttgaagctaattcgggtaggagaagggaatcgaagc | aatJ UpR-Gm |
| JS101.f | aggaacttcaagatccccaattcggcgaggtcaacaagatctacg | aatJ DnF-Gm |
| JS101.r | tacaagaaagctgggtcctcggttcaatcggttgc | aatJ DnR-GWR |
| JS111.f | gcactcgaggccggggaaaaccgcgaaacaccc | aatJ gene |
| JS111.r | gcatctagattacatctgctcggcggccttgtcgg | aatJ gene |
| JS117.f | gcaggatccccggggaaaaccgcgaaacaccctgtccc | aatJQMP genes |
| JS117.r | gcatctagatcagtgcgggaggatcttggcgaggaactgc | aatJQMP genes |
| JS118.f | gcatctagagcatgtcttgctcggctgccagcaggc | aauR gene |
| JS118.r | gcagagctccccggcctattgcaggccgtacttcttcaccttgtcg | aauR gene |
aNote that oligonucleotides used in the cloning of gene-deletion plasmids were designated as DnF-Gm, DnR-GWR, UpF-GWL or UpR-Gm in accordance with the original methods publication [43]
Cloning of expression vectors for the aatJQMP and aauR genes
The aauR, aatJ, aatQMP, and aatJQMP genes were PCR-amplified from genomic DNA of P. aeruginosa PAO1 with the primers JS118.f-JS118.r, JS111.f-JS111.r, JS50.f-JS51.r, and JS117.f-JS117.r, respectively. The desired PCR products were gel purified and cloned into pCR-Blunt (Invitrogen) according to the manufacturer’s instructions. The aauR, aatJ, aatQMP, and aatJQMP genes were subsequently subcloned into the XbaI-SacI, XhoI-XbaI, XbaI-SacI, and KpnI-XbaI sites of pBBR1MCS-5 [42] to yield the plasmids pJMS03, pJMS21, pJMS05, and pJMS19, respectively.
Cloning of aatJ-lacZ and aatQ-lacZ reporters
The 5′ regulatory regions of the aatJ and aatQ genes were PCR-amplified from genomic DNA of P. aeruginosa PAO1 with the primers JS35P.f-JS43.r and JS35P.f-JS05.r, respectively, while the lacZ ORF was PCR-amplified from genomic DNA of E. coli MG1655 with the primers JS42.f-BL342.r (aatJ overlap) or JS06.f-BL342.r (aatQ overlap). The PCR-amplified 5′ regulatory regions of aatJ and aatQ were then fused to their cognate E. coli lacZ ORFs via fusion PCR [13], and the resulting aatJ-lacZ and aatQ-lacZ fusions were gel purified and subsequently cloned into pCR-Blunt (Invitrogen) to yield pJMS07 and pJMS08, respectively. The aatJ-lacZ and aatQ-lacZ fusions were then excised from pJMS07 and pJMS08 through double digestion with XbaI-SpeI, and the liberated fragments were separately cloned into the XbaI site of the promoter-less plasmid ΔPlac-pBBR1MCS-5 [13] to give pJMS09 and pJMS10, respectively.
Construction of gene-deletion mutants of P. aeruginosa
The aauR, aatJ, and aatQM genes were deleted from the genome of P. aeruginosa PAO1 using established methods [43]. Briefly, the chromosomal mutations aauR::FRT-aacC1-FRT, aatJ::FRT-aacC1-FRT, and aatQM::FRT-aacC1-FRT were generated by electroporating P. aeruginosa PAO1 with the gene-deletion plasmids pJMS299, pJMS301, and pJMS303, respectively. Following selection on LB-agar supplemented with 30 μg mL− 1 gentamicin and verification of double-crossover mutations through PCR, the gentamicin-selection markers (aacC1) were removed via FLP-mediated excision to yield the unmarked ΔaauR, ΔaatJ, and ΔaatQM mutants. The ΔaauR mutants of P. aeruginosa PA14 and PAK were constructed in similar fashion.
Cell-culture experiments
Analysis was performed in triplicate for each strain and/or condition. Individual colonies of wild-type P. aeruginosa and its isogenic mutants were each inoculated into 2.0 mL of LB (in a 16 × 125 mm culture tube) and subsequently grown at 37 °C at 200 rpm for 18 h. The LB-grown cells were collected via centrifugation and then washed two times with equal volumes of carbon- and nitrogen-free minimal media. The wash steps ensured the removal of residual carbon and nitrogen sources. Each washed cell pellet was ultimately suspended in an equal volume of carbon- and nitrogen-free minimal media, and the resulting cell suspensions were used at 1% (vol/vol), i.e., 2.0 μL, for the inoculation of 0.2 mL of test medium housed in a 96-well flat-bottom polystyrene plate (Falcon). Test media used for nitrogen-source analysis consisted of minimal media supplemented with 20 mM glucose and 10 mM NH4Cl, L-glutamate, L-aspartate, L-glutamine or L-asparagine. Test media used for carbon-source analysis consisted of minimal media supplemented with 10 mM NH4Cl and 20 mM L-glutamate, L-aspartate, L-glutamine or L-asparagine. Inoculated plates were incubated in a BioTek Synergy™ H4 microplate reader set at 37 °C with medium shaking for 18 h. The absorbances or optical densities at 600 nm (OD600) were measured at 1.0 h intervals.
The ΔaauR mutant of P. aeruginosa PAO1 was electroporated with expression plasmids for aauR (pJMS03), aatJ (pJMS21), aatQMP (pJMS05) and aatJQMP (pJMS19). For controls, both wild-type P. aeruginosa PAO1 and the ΔaauR were transformed with empty pBBR1MCS-5. The resulting recombinant strains were grown in minimal media supplemented with 10 mM NH4Cl and either 20 mM L-glutamate or L-glutamine. Experiments were done exactly as described above except that growth media was supplemented with 30 μg mL− 1 gentamicin for plasmid selection.
HPLC analysis of extracellular amino acids
Analysis was performed in triplicate for each strain and/or condition. Wild-type P. aeruginosa PAO1 and the ∆aauR mutant were grown in 10 mL of minimal media (in a 50 mL conical centrifuge tube) supplemented with 20 mM glucose and 20 mM L-glutamate, L-glutamine, L-aspartate or L-asparagine. Cultures were grown at 37 °C and 200 rpm. At 6.0 and 9.0 h post inoculation, 0.5 mL of culture was withdrawn, and cells were removed by centrifugation and subsequent passage of the supernatant through a 0.2 μm syringe filter. The resulting cell-free samples were then treated with o-phthalaldehyde to derivative the amino acids, which were subsequently separated through a reverse-phase Agilent Zorbax Eclipse amino acid analysis column (75 mm by 4.6-mm ID, 3.5-μm particle size) maintained at a temperature of 40 °C [44]. Separation involved the mobile phases A (40 mM Na2HPO3, pH 7.8) and B (45% acetonitrile:45% methanol:10% H2O) in a gradient program consisting of 2.0 min of 100% phase A, a 12.0 min linear gradient to 50% phase B, a 10 min linear gradient back to 100% phase A, and 4.0 min of 100% phase A at a flow rate of 1 mL min− 1 [44]. The dynamic range for the detection and quantification of derivatized amino acids was 45–450 pmol.
LacZ assays
Analysis was performed in triplicate for each strain and/or condition. Wild-type P. aeruginosa PAO1, the ∆aauR and the rpoN::Ω-Km mutants were each electroporated with the aatJ-lacZ (pJMS09) and aatQ-lacZ (pJMS10) reporter plasmids. Following selection on LB agar supplemented with 30 μg mL-1 gentamicin, individual colonies were directly inoculated into minimal media supplemented with 20 mM L-alanine, 10 mM NH4Cl and 30 μg mL− 1 gentamicin. Bacteria were grown at 37 °C to an OD600 of ~ 0.2 and subsequently challenged with the addition of a final concentration of 20 mM of substrate: L-alanine, succinate, α-ketoglutarate, L-arginine, L-histidine, L-glutamate, L-aspartate, L-glutamine and L-asparagine. LacZ activity was measured at 1.0 and 3.0 h post addition of substrate using a microplate reader protocol as previously described [45]. Briefly, in a 96-well flat-bottom polystyrene plate (Falcon), 5.0 μL of cell culture was mixed with 45 μL of permeabilization solution (100 mM Na2HPO4, 80 μg mL− 1 CTAB, 40 μg mL− 1 deoxycholate, 2.0 mM MgSO4, 5.4 μL mL− 1 β-mercaptoethanol). The mixture was incubated for 30 min at 30 °C. Subsequently, 225 μL of substrate solution (100 mM Na phosphate, pH 7.0, 20 mM KCl, 1.0 mg mL− 1 o-nitrophenyl-β-D-galactoside, 2.7 μL mL− 1 β-mercaptoethanol) was added to the permeabilized-cell mixture, and the resulting reaction was incubated at 30 °C with shaking. The absorbance at 420 nm was measured every 1.5 min. Analysis of variance (ANOVA) was done using Dunnett’s post hoc test (α value, 0.05) to identify significant differences (P < 0.0001) in LacZ activities.
Acknowledgements
We would like to thank the Support of Scholarly Activities (SOSA) and Mentored Undergraduate Summer Experience (MUSE) programs at The College of New Jersey for their involvement in student participation in this project.
Abbreviations
- ABC
ATP-binding cassette
- EBP
Enhancer-binding protein
- TCS
Two-component signal transduction system
- ND
Not detected.
Authors’ contributions
BRL, JMS, ZS and CTM designed the overall study and prepared the manuscript. BRL carried out the β-galactosidase assays and genetic-complementation experiments. JMS performed all the molecular biology experiments and construction of bacterial mutants. JMS and SS carried out the cell-culture experiments. JMS and RAS conducted the HPLC analysis. Author order was determined based on contribution to the study. The author(s) read and approved the final manuscript.
Funding
This project was funded by the National Institute of Allergy and Infectious Diseases (Award R15GM104880–03).
Availability of data and materials
All data generated or analyzed during this study are included in the submitted manuscript.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yurgel SN, Kahn ML. Dicarboxylate transport by rhizobia. FEMS Microbiol Rev. 2004;28(4):489–501. doi: 10.1016/j.femsre.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Udvardi M, Poole PS. Transport and metabolism in legume-rhizobia symbioses. Annu Rev Plant Biol. 2013;64(1):781–805. doi: 10.1146/annurev-arplant-050312-120235. [DOI] [PubMed] [Google Scholar]
- 3.Clarke PH, Meadow PM. Evidence for the occurrence of permeases for tricarboxylic acid cycle intermediates in Pseudomonas aeruginosa. Microbiology. 1959;20:144–155. doi: 10.1099/00221287-20-1-144. [DOI] [PubMed] [Google Scholar]
- 4.Tiwari NP, Campbell JJ. Enzymatic control of the metabolic activity of Pseudomonas aeruginosa grown in glucose or succinate medium. Biochim Biophys Acta. 1969;192(3):395–401. doi: 10.1016/0304-4165(69)90388-2. [DOI] [PubMed] [Google Scholar]
- 5.Janausch IG, Zientz ET, Q. H., Kröger a, Unden G. C4-dicarboxylate carriers and sensors in bacteria. Biochim Biophys Acta. 2002;1553(1-2):39–56. doi: 10.1016/S0005-2728(01)00233-X. [DOI] [PubMed] [Google Scholar]
- 6.Bush M, Dixon R. The role of bacterial enhancer binding proteins as specialized activators of sigma54-dependent transcription. Microbiol Mol Biol Rev. 2012;76(3):497–529. doi: 10.1128/MMBR.00006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Studholme DJ, Buck M. The biology of enhancer-dependent transcriptional regulation in bacteria: insights from genome sequences. FEMS Microbiol Lett. 2000;186(1):1–9. doi: 10.1111/j.1574-6968.2000.tb09074.x. [DOI] [PubMed] [Google Scholar]
- 8.Morett E, Segovia L. The sigma 54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;175(19):6067–6074. doi: 10.1128/JB.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buck M, Gallegos MT, Studholme DJ, Guo Y, Gralla JD. The bacterial enhancer-dependent sigma(54) (sigma(N)) transcription factor. J Bacteriol. 2000;182(15):4129–4136. doi: 10.1128/JB.182.15.4129-4136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schumacher J, Joly N, Rappas M, Zhang X, Buck M. Structures and organisation of AAA+ enhancer binding proteins in transcriptional activation. J Struct Biol. 2006;156(1):190–199. doi: 10.1016/j.jsb.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Valentini M, Storelli N, Lapouge K. Identification of C4-dicarboxylate transport systems in Pseudomonas aeruginosa PAO1. J Bacteriol. 2011;193(17):4307–4316. doi: 10.1128/JB.05074-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petrova OE, Sauer K. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 2009;5(11):e1000668. doi: 10.1371/journal.ppat.1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lundgren BR, Villegas-Peñaranda LR, Harris JR, Mottern AM, Dunn DM, Boddy CN, et al. Genetic analysis of the assimilation of C5-dicarboxylic acids in Pseudomonas aeruginosa PAO1. J Bacteriol. 2014;196(14):2543–2551. doi: 10.1128/JB.01615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tatke G, Kumari H, Silva-Herzog E, Ramirez L, Mathee K. Pseudomonas aeruginosa MifS-MifR two-component system is specific for α-ketoglutarate utilization. PLoS One. 2015;10(6):e0129629. doi: 10.1371/journal.pone.0129629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarwar Z, Wang MX, Lundgren BR, Nomura CT. MifS, a DctB family histidine kinase, is a specific regulator of α-ketoglutarate response in Pseudomonas aeruginosa PAO1. Microbiology. 2020;166(9):867–879. doi: 10.1099/mic.0.000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sonawane AM, Singh B, Rohm KH. The AauR-AauS two-component system regulates uptake and metabolism of acidic amino acids in Pseudomonas putida. Appl Environ Microbiol. 2006;72(10):6569–6577. doi: 10.1128/AEM.00830-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh B, Rohm KH. Characterization of a Pseudomonas putida ABC transporter (AatJMQP) required for acidic amino acid uptake: biochemical properties and regulation by the Aau two-component system. Microbiology. 2008;154(3):797–809. doi: 10.1099/mic.0.2007/013185-0. [DOI] [PubMed] [Google Scholar]
- 18.Schulz S, Eckweiler D, Bielecka A, Nicolai T, Franke R, Dotsch A, et al. Elucidation of sigma factor-associated networks in Pseudomonas aeruginosa reveals a modular architecture with limited and function-specific crosstalk. PLoS Pathog. 2015;11(3):e1004744. doi: 10.1371/journal.ppat.1004744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FS. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016;44(D1):D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Totten PA, Lara JC, Lory S. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J Bacteriol. 1990;172(1):389–396. doi: 10.1128/JB.172.1.389-396.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Köhler T, Harayama S, Ramos JL, Timmis KN. Involvement of Pseudomonas putida RpoN sigma factor in regulation of various metabolic functions. J Bacteriol. 1989;171(8):4326–4333. doi: 10.1128/JB.171.8.4326-4333.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreno R, Martínez-Gomariz M, Yuste L, Gil C, Rojo F. The Pseudomonas putida Crc global regulator controls the hierarchical assimilation of amino acids in a complete medium: evidence from proteomic and genomic analyses. Proteomics. 2009;9(11):2910–2928. doi: 10.1002/pmic.200800918. [DOI] [PubMed] [Google Scholar]
- 23.La Rosa R, Behrends V, Williams HD, Bundy JG, Rojo F. Influence of the Crc regulator on the hierarchical use of carbon sources from a complete medium in Pseudomonas. Environ Microbiol. 2016;18(3):807–818. doi: 10.1111/1462-2920.13126. [DOI] [PubMed] [Google Scholar]
- 24.Lu CD, Yang Z, Li W. Transcriptome analysis of the ArgR regulon in Pseudomonas aeruginosa. J Bacteriol. 2004;186(12):3855–3861. doi: 10.1128/JB.186.12.3855-3861.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bender RA. Regulation of the histidine utilization (hut) system in bacteria. Microbiol Mol Biol Rev. 2012;76(3):565–584. doi: 10.1128/MMBR.00014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson DA, Tetu SG, Phillippy KH, Chen J, Ren Q, Paulsen IT. High-throughput phenotypic characterization of Pseudomonas aeruginosa membrane transport genes. PLoS Genet. 2008;4(10):e1000211. doi: 10.1371/journal.pgen.1000211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He W, Li G, Yang CK, Lu CD. Functional characterization of the dguRABC locus for D-Glu and D-Gln utilization in Pseudomonas aeruginosa PAO1. Microbiology. 2014;160(10):2331–2340. doi: 10.1099/mic.0.081141-0. [DOI] [PubMed] [Google Scholar]
- 28.Sonawane AM, Klöppner U, Derst C, Röhm KH. Utilization of acidic amino acids and their amides by pseudomonads: role of periplasmic glutaminase-asparaginase. Arch Microbiol. 2003;179(3):151–159. doi: 10.1007/s00203-002-0511-6. [DOI] [PubMed] [Google Scholar]
- 29.Li G, Lu CD. Molecular characterization and regulation of operons for asparagine and aspartate uptake and utilization in Pseudomonas aeruginosa. Microbiology. 2018;164(2):205–216. doi: 10.1099/mic.0.000594. [DOI] [PubMed] [Google Scholar]
- 30.Sonnleitner E, Valentini M, Wenner N, Haichar F, Haas D, Lapouge K. Novel targets of the CbrAB/Crc carbon catabolite control system revealed by transcript abundance in Pseudomonas aeruginosa. PLoS One. 2012;7(10):e44637. doi: 10.1371/journal.pone.0044637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dasgupta N, Wolfgang MC, Goodman AL, Arora SK, Jyot J, Lory S, Ramphal R. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol. 2003;50(3):809–824. doi: 10.1046/j.1365-2958.2003.03740.x. [DOI] [PubMed] [Google Scholar]
- 32.Heurlier K, Denervaud V, Pessi G, Reimmann C, Haas D. Negative control of quorum sensing by RpoN (sigma54) in Pseudomonas aeruginosa PAO1. J Bacteriol. 2003;185(7):2227–2235. doi: 10.1128/JB.185.7.2227-2235.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shao X, Zhang X, Zhang Y, Zhu M, Yang P, Yuan J, et al. RpoN-dependent direct regulation of quorum sensing and the type VI secretion system in Pseudomonas aeruginosa PAO1. J Bacteriol. 2018;200:e00205–e00218. doi: 10.1128/JB.00205-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishijyo T, Haas D, Itoh Y. The CbrA-CbrB two-component regulatory system controls the utilization of multiple carbon and nitrogen sources in Pseudomonas aeruginosa. Mol Microbiol. 2001;40(4):917–931. doi: 10.1046/j.1365-2958.2001.02435.x. [DOI] [PubMed] [Google Scholar]
- 35.Li W, Lu CD. Regulation of carbon and nitrogen utilization by CbrAB and NtrBC two-component systems in Pseudomonas aeruginosa. J Bacteriol. 2007;189(15):5413–5420. doi: 10.1128/JB.00432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki H, Fukuyama K, Kumagai H. Bacterial γ-glutamyltranspeptidases, physiological function, structure, catalytic mechanism and application. Proc Jpn Acad Ser B Phys Biol Sci. 2020;96(9):440–469. doi: 10.2183/pjab.96.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walshaw DL, Poole PS. The general L-amino acid permease of Rhizobium leguminosarum is an ABC uptake system that also influences efflux of solutes. Mol Microbiol. 1996;21(6):1239–1252. doi: 10.1046/j.1365-2958.1996.00078.x. [DOI] [PubMed] [Google Scholar]
- 38.Walshaw DL, Wilkinson AJ, Mundy M, Smith M, Poole PS. Regulation of the TCA cycle and the general amino acid permease by overflow metabolism in Rhizobium leguminosarum. Microbiology. 1997;143(7):2209–2221. doi: 10.1099/00221287-143-7-2209. [DOI] [PubMed] [Google Scholar]
- 39.Price-Whelan A, Dietrich LE, Newman DK. Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189(17):6372–6381. doi: 10.1128/JB.00505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sadovskaya I, Vinogradov E, Li J, Hachani A, Kowalska K, Filloux A. High-level antibiotic resistance in Pseudomonas aeruginosa biofilm: the ndvB gene is involved in the production of highly glycerol-phosphorylated beta-(1->3)-glucans, which bind aminoglycosides. Glycobiology. 2010;20(7):895–904. doi: 10.1093/glycob/cwq047. [DOI] [PubMed] [Google Scholar]
- 41.Kang Y, Durfee T, Glasner JD, Qiu Y, Frisch D, Winterberg KM, Blattner FR. Systematic mutagenesis of the Escherichia coli genome. J Bacteriol. 2004;186(15):4921–4930. doi: 10.1128/JB.186.15.4921-4930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, 2nd, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166(1):175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 43.Choi KH, Schweizer HP. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 2005;5(1):30. doi: 10.1186/1471-2180-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lundgren BR, Thornton W, Dornan MH, Villegas-Peñaranda LR, Boddy CN, Nomura CT. Gene PA2449 is essential for glycine metabolism and pyocyanin biosynthesis in Pseudomonas aeruginosa PAO1. J Bacteriol. 2013;195(9):2087–2100. doi: 10.1128/JB.02205-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lundgren BR, Sarwar Z, Feldman KS, Shoytush JM, Nomura CT. SfnR2 regulates dimethyl sulfide-related utilization in Pseudomonas aeruginosa PAO1. J Bacteriol. 2019;201:e00606–e00618. doi: 10.1128/JB.00606-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in the submitted manuscript.