Abstract
Background
The prevalence of open-angle glaucoma (OAG) varies from 0.5% to 7.0% among populations of diverse ancestry, suggesting the existence of genetic differences. The purposes of this study were to provide insights into genetic causes of OAG, which can result in prevalence and phenotype differences among populations of diverse ancestry for OAG, and to compare allele frequencies of intraocular pressure (IOP) elevation-related SNPs in OAG among Koreans and other ethnic groups.
Methods
We collected the data on a total of 135 OAG-associated single nucleotide polymorphisms (SNPs) from a genome-wide association studies (GWAS) catalog. The population-level allele frequencies of these SNPs were derived based on the 1000 Genomes Project and Korean Reference Genome Database. We used Fisher's exact test to assess whether the effect allele at a given SNP was significantly enriched or depleted.
Results
European, American, and South Asian populations showed similar heatmap patterns, while African, East Asian, and Korean populations had distinct patterns. Korean population presented different profiles compared to other groups; rs1579050 (FMNL2 gene), rs2024211 (CAV2;CAV1), and rs8141433 (GNB1L;TXNRD2 gene), which are known to be associated with IOP variation, were enriched in Americans, Europeans, and Africans, and depleted in Koreans. These can be the candidates for the causative genes of differences in the prevalence of IOP variation in OAG according to ethnic groups.
Conclusions
Differences in allele frequencies associated with IOP related SNPs between Koreans and other ethnicities were observed, which may explain the high prevalence of OAG with normal IOP predominantly in Koreans and East Asians.
Supplementary information
The online version contains supplementary material available at 10.1186/s12920-021-00921-2.
Keywords: Open-angle glaucoma, Allele frequency, Single nucleotide polymorphism, Intraocular pressure, Prevalence, Normal-tension glaucoma
Background
Glaucoma, which is a progressive optic neuropathy characterized by degeneration of the retinal ganglion cells and their axons and a corresponding visual field defect [1], is a leading cause of irreversible blindness worldwide. Prevalence of glaucoma varies among ethnicity and regions, from 0.5% to 8.0% for open-angle glaucoma (OAG) [2–5], and from 0.1% to 2.3% for angle-closure glaucoma [6–8], suggesting the existence of genetic differences among populations of diverse ancestry. As OAG is the predominant form of glaucoma, epidemiologic research regarding diverse ancestry populations has been conducted [2, 5, 9–12]. According to a meta-analysis report [13], the prevalence of OAG was higher in Africans, followed by Europeans and Asians. Still, there were little data on other ethnic groups, such as South and East Asians. A recent study complementing this has shown that the prevalence of OAG was higher in Africans and Latinos, followed by Europeans, South Asians, and East Asians [14]. The relationship between the prevalence and geographical variations might encompass genetic, environmental, and cultural differences.
Evidence for a causal relationship between elevated intraocular pressure (IOP) and OAG is robust, but normal-tension glaucoma (NTG) is considered a part of the spectrum of OAG intrinsically, which is arbitrarily based on the distribution of IOP; OAG with IOP ≤ 21 mmHg is termed as NTG. Specifically, NTG is common in Asians, including Koreans (77.0%) and comprises the majority (52–92%) of cases of OAG in Asian studies [15–18]; the proportions are higher than those in Europeans (30–38%) and African ancestry populations (57.1%) [3, 19, 20]. This difference in the pattern of prevalence of NTG according to populations of diverse ancestry suggested that it may be related to different allele frequencies of glaucoma-related single nucleotide polymorphism (SNP). In addition, first-degree relatives of affected patients are estimated to have as much as 4 to 10 times increased risk compared to the general population [21, 22]. These suggest that genetic aspects should be considered as a cause of glaucoma. Recently, studies based on genome-wide association study (GWAS) of glaucoma with meta-analysis using the results from various cohorts such as International Glaucoma Genetics Consortium (IGGC), Australian and New Zealand Registry of Advanced Glaucoma (ANZRAG), and UK Biobank (UKBB) had revealed novel risk loci and SNPs related with IOP [23–26]. Combining these results using the GWAS catalog (NHGRI-EBI) [27] and utilizing 1000 Genomes Project phase 3 data [28], we could infer the difference in glaucoma-related SNPs according to populations of diverse ancestry. Additionally, it is possible to assess the allele frequency of OAG-associated SNPs and IOP elevation-associated SNPs in OAG among Koreans using 1722 whole-genome sequencing data of healthy Koreans in the Korean Reference Genome Database (KRGDB) [29].
Hence, the purposes of this study were to gain insights into genetic causes of OAG, which can result in prevalence and phenotype differences among populations of diverse ancestry for OAG, and to compare allele frequencies of IOP elevation-related SNPs in OAG among Koreans and other population groups. We also developed the composite genetic risk score for OAG as a whole and OAG with high IOP and tested the correlation between a population-level average of composite risk scores and OAG prevalence.
Methods
This study was approved and monitored by the Institutional Review Board (IRB) of the Veterans Health Service Medical Center, Korea (IRB No. 2019-07-008).
Comparison of OAG-related SNPs in global population and East Asians
According to the International Society of Geographical and Epidemiological Ophthalmology (ISGEO) classification [30], OAG is defined as glaucomatous optic neuropathy in the presence of an open angle and no other ocular abnormality accountable for the neuropathy. We researched the GWAS catalog (NHGRI-EBI, https://www.ebi.ac.uk/gwas/docs/file-downloads, “All associations v1.0.2—with added ontology annotations, GWAS Catalog study accession numbers and genotyping technology”, December 2019) for SNPs that were associated with ‘open-angle glaucoma’-related trait (EFO_0004190). Initially, 146 glaucoma-associated SNPs from the GWAS catalog were collected. Of these, a total of 135 SNPs was used for analysis after removing the repeated ones (Additional files 2, 3: Table S1 and Table S2).
Among SNPs associated with OAG-related trait, we determined OAG risk by examining the sign of beta-coefficient, whether the odds ratio for the effect allele was greater than one, and text description in the primary GWAS reports. The details and advantages of the method have been described elsewhere [31]. In brief, the population-level allele frequencies of these SNPs were derived based on the 1000 Genomes Project phase 3 (n = 2504) and KRGDB (n = 1722). The 1000 Genomes Project surveys genetic variations among 2504 individuals from 26 worldwide populations, which can be grouped into African (AFR), East Asian (EAS), European (EUR), South Asian (SAS), and the American (AMR) based on their geographical locations and ancestries [28]; the data were downloaded from ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/ (last accessed: January 15, 2020). Because the East Asian data in the 1000 Genomes Project did not include data from the Korean population, we compared the data from the five continents with data extracted from KRGDB, which included the whole genome sequencing data for 1722 Korean (KOR) individuals [29]. The data on the population frequency of the SNPs were downloaded from the web-based database (http://152.99.75.168:9090/KRGDB/menuPages/download.jsp/, last accessed: January 15, 2020). For the comparison of the distribution of individual risk alleles of the Korean population, individual genotype results of the 2nd phase of KRGDB (n = 1099) were obtained from the National Human Resource Bank of Korea.
Comparison of SNPs related to IOP elevation in OAG in Global population and East Asians
OAG is defined as an optic nerve state, but IOP is considered a major risk factor. Hence, research for allele frequencies of SNPs influencing IOP may give us insights on the difference in the prevalence of NTG and OAG with high IOP according to different ancestries. The 52 SNPs related to IOP are shown in Table 1, which have been obtained from GWAS catalog data, a study using Genetic Epidemiology Research in Adult Health and Aging (GERA) cohort [32], a large multi-ethnic study for identifying novel loci related to IOP [33], and meta-analysis results of IGGC, ANZRAG, and UKBB [23–26]. The population-level allele frequencies of these SNPs were derived as described above.
Table 1.
Effect allele frequencies (EAFs) of intraocular pressure related single nucleotide polymorphisms in populations of diverse ancestry including Koreans
| SNP ID | Chr | Position | Type | Ref Allele | Alt Allele | Nearby/containing Gene | Global EAF | AMR EAF | AMR log10 P | AFR EAF | AFR log10 P | EAS EAF |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1013278 | chr7 | 117603820 | Intergenic | G | C | CTTNBP2;LSM8 | 0.31 | 0.28 | − 0.735 | 0.33 | 0.738 | 0.1 |
| rs10281637 | chr7 | 116151338 | Intergenic | T | C | CAV2;CAV1 | 0.22 | 0.21 | − 0.177 | 0.4 | 36.671 | 0.0089 |
| rs10483727 | chr14 | 61072875 | Intergenic | T | C | SIX6;SALRNA1 | 0.35 | 0.66 | 51.990 | 0.034 | − 146.666 | 0.21 |
| rs10505100 | chr8 | 108278616 | Intronic | C | A | ANGPT1 | 0.16 | 0.18 | 0.569 | 0.11 | − 5.368 | 0.23 |
| rs10918274 | chr1 | 165714416 | Intronic | T | C | TMCO1 | 0.92 | 0.87 | − 3.857 | 0.92 | 0.000 | 0.99 |
| rs11217878 | chr11 | 120340383 | Intronic | G | A | ARHGEF12 | 0.21 | 0.15 | − 3.286 | 0.25 | 2.601 | 0.24 |
| rs113985657 | chr6 | 597203 | Intronic | C | T | EXOC2 | 0.12 | 0.11 | − 0.249 | 0.087 | − 3.132 | 0.14 |
| rs11710139 | chr3 | 15005942 | Intergenic | G | A | LINC01214;TSC22D2 | 0.16 | 0.14 | − 0.578 | 0.16 | 0.010 | 0.075 |
| rs12377624 | chr9 | 129373110 | Intergenic | G | C | MVB12B;LMX1B | 0.23 | 0.24 | 0.194 | 0.11 | − 23.178 | 0.13 |
| rs1254276 | chr14 | 60847001 | Intergenic | C | T | LINC02322;C14orf39 | 0.63 | 0.32 | − 51.921 | 0.89 | 82.782 | 0.78 |
| rs12699251 | chr7 | 11679113 | Intronic | A | G | THSD7A | 0.25 | 0.35 | 6.518 | 0.11 | − 29.965 | 0.2 |
| rs1579050 | chr2 | 153364527 | Intronic | A | G | FMNL2 | 0.28 | 0.46 | 19.037 | 0.1 | − 46.833 | 0.026 |
| rs17752199 | chr6 | 51406848 | Intergenic | A | G | TFAP2B;PKHD1 | 0.13 | 0.13 | 0.000 | 0.15 | 1.166 | 0.12 |
| rs1874458 | chr16 | 65080739 | Intronic | G | A | CDH11 | 0.2 | 0.21 | 0.187 | 0.085 | − 24.499 | 0.21 |
| rs2022945 | chr8 | 108251139 | Intergenic | A | G | ABRA;ANGPT1 | 0.84 | 0.83 | − 0.204 | 0.9 | 7.594 | 0.77 |
| rs2024211 | chr7 | 116153025 | Intergenic | A | C | CAV2;CAV1 | 0.21 | 0.2 | − 0.177 | 0.34 | 20.570 | 0.0089 |
| rs2073006 | chr6 | 637465 | Intronic | C | T | EXOC2 | 0.11 | 0.12 | 0.258 | 0.045 | − 13.535 | 0.15 |
| rs2188836 | chr7 | 117635382 | Intergenic | C | T | CTTNBP2;LSM8 | 0.33 | 0.28 | − 1.730 | 0.39 | 4.233 | 0.09 |
| rs2317961 | chr6 | 1533116 | Intergenic | A | G | FOXF2;FOXCUT | 0.63 | 0.59 | − 1.121 | 0.82 | 40.735 | 0.51 |
| rs2472493 | chr9 | 107695848 | Intergenic | G | A | ABCA1;SLC44A1 | 0.61 | 0.65 | 1.095 | 0.69 | 6.970 | 0.49 |
| rs2472496 | chr9 | 107695353 | Intergenic | G | A | ABCA1;SLC44A1 | 0.61 | 0.64 | 0.690 | 0.69 | 6.970 | 0.47 |
| rs2487032 | chr9 | 107703934 | Intergenic | G | A | ABCA1;SLC44A1 | 0.6 | 0.48 | − 7.836 | 0.83 | 58.058 | 0.49 |
| rs2745572 | chr6 | 1548369 | Intergenic | A | G | FOXF2;FOXCUT | 0.36 | 0.38 | 0.395 | 0.15 | − 52.096 | 0.49 |
| rs28500712 | chr4 | 7896213 | Intronic | A | G | AFAP1 | 0.66 | 0.59 | − 3.016 | 0.72 | 4.381 | 0.68 |
| rs28520091 | chr4 | 7846240 | Intronic | C | T | AFAP1 | 0.32 | 0.38 | 2.332 | 0.15 | − 36.294 | 0.29 |
| rs28795989 | chr4 | 7891545 | Intronic | A | G | AFAP1 | 0.29 | 0.32 | 0.743 | 0.14 | − 30.033 | 0.18 |
| rs2935057 | chr6 | 170454915 | Intergenic | A | G | LINC00574;LOC102724511 | 0.8 | 0.82 | 0.502 | 0.74 | − 5.373 | 0.76 |
| rs3013274 | chr6 | 170464367 | Intergenic | G | A | LINC00574;LOC102724511 | 0.61 | 0.58 | − 0.664 | 0.7 | 8.654 | 0.5 |
| rs31918 | chr5 | 14820927 | Intronic | C | T | ANKH | 0.29 | 0.34 | 1.752 | 0.28 | − 0.298 | 0.33 |
| rs327716 | chr7 | 80838977 | Intergenic | A | G | SEMA3C;LOC105369146 | 0.71 | 0.68 | − 0.743 | 0.81 | 12.896 | 0.91 |
| rs33912345 | chr14 | 60976537 | Exonic | C | A | SIX6 | 0.34 | 0.66 | 54.967 | 0.032 | − 143.370 | 0.21 |
| rs3785176 | chr16 | 8896931 | Intronic | A | C | PMM2 | 0.18 | 0.14 | − 1.712 | 0.096 | − 13.535 | 0.28 |
| rs4141671 | chr10 | 60338753 | Intronic | T | C | BICC1 | 0.49 | 0.32 | − 15.716 | 0.55 | 3.871 | 0.48 |
| rs4236601 | chr7 | 116162729 | Intergenic | G | A | CAV2;CAV1 | 0.23 | 0.21 | − 0.434 | 0.4 | 32.449 | 0.0099 |
| rs55892100 | chr7 | 115810676 | Intergenic | A | G | TFEC;TES | 0.54 | 0.43 | − 6.518 | 0.61 | 5.149 | 0.8 |
| rs5756813 | chr22 | 38175477 | Intergenic | G | T | TRIOBP;H1F0 | 0.55 | 0.62 | 2.799 | 0.4 | − 21.169 | 0.71 |
| rs58073046 | chr11 | 120248493 | Intronic | A | G | ARHGEF12 | 0.1 | 0.042 | − 6.316 | 0.013 | − 32.104 | 0.17 |
| rs61394862 | chr5 | 14851094 | Intronic | C | T | ANKH | 0.28 | 0.33 | 1.781 | 0.26 | − 0.777 | 0.32 |
| rs6478746 | chr9 | 129367398 | Intergenic | G | A | MVB12B;LMX1B | 0.8 | 0.85 | 2.421 | 0.73 | − 6.990 | 0.97 |
| rs66602224 | chr8 | 108293718 | Intronic | G | A | ANGPT1 | 0.32 | 0.32 | 0.000 | 0.44 | 14.831 | 0.14 |
| rs6732795 | chr2 | 69411517 | Intronic | A | C | ANTXR1 | 0.66 | 0.49 | − 16.002 | 0.78 | 16.515 | 0.85 |
| rs73174345 | chr3 | 169252883 | Intronic | T | G | MECOM | 0.054 | 0.036 | − 1.095 | 0.13 | 18.287 | 0 |
| rs746491 | chr11 | 86406159 | Intergenic | C | A | ME3;PRSS23 | 0.15 | 0.17 | 0.588 | 0.11 | − 3.715 | 0.14 |
| rs7518099 | chr1 | 165736880 | Intronic | C | T | TMCO1 | 0.92 | 0.87 | − 3.857 | 0.91 | − 0.608 | 0.99 |
| rs7555523 | chr1 | 165718979 | Intronic | C | A | TMCO1 | 0.9 | 0.87 | − 1.399 | 0.85 | − 6.009 | 0.99 |
| rs7924522 | chr11 | 128380742 | Intronic | C | A | ETS1 | 0.75 | 0.71 | − 1.291 | 0.79 | 2.503 | 0.86 |
| rs8141433 | chr22 | 19854006 | Intergenic | A | G | GNB1L;TXNRD2 | 0.28 | 0.16 | − 10.572 | 0.69 | 159.128 | 0.037 |
| rs9284802 | chr3 | 85095766 | Intronic | G | A | CADM2 | 0.32 | 0.33 | 0.177 | 0.13 | − 46.150 | 0.17 |
| rs945686 | chr9 | 129378026 | Intronic | G | C | LMX1B | 0.82 | 0.87 | 2.585 | 0.73 | − 11.619 | 0.99 |
| rs9494457 | chr6 | 136474794 | Intronic | T | A | PDE7B | 0.39 | 0.45 | 2.158 | 0.37 | − 0.696 | 0.33 |
| rs9853115 | chr3 | 186131600 | Intergenic | T | A | DGKG;LINC02052 | 0.56 | 0.47 | − 4.386 | 0.48 | − 6.471 | 0.73 |
| rs9913911 | chr17 | 10031183 | Intronic | A | G | GAS7 | 0.31 | 0.32 | 0.177 | 0.16 | − 28.132 | 0.5 |
| SNP ID | Chr | Position | Type | Ref Allele | Alt Allele | Nearby/containing Gene | EAS log10 P | SAS EAF | SAS log10 P | EUR EAF | EUR log10 P | KOR EAF | KOR log10 P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1013278 | chr7 | 117603820 | Intergenic | G | C | CTTNBP2;LSM8 | − 47.563 | 0.4 | 6.676 | 0.4 | 7.043 | 0.114 | − 102.924 |
| rs10281637 | chr7 | 116151338 | Intergenic | T | C | CAV2;CAV1 | − 80.906 | 0.19 | − 1.271 | 0.26 | 2.100 | 0.007 | − 232.094 |
| rs10483727 | chr14 | 61072875 | Intergenic | T | C | SIX6;SALRNA1 | − 17.809 | 0.42 | 4.030 | 0.6 | 47.063 | 0.271 | − 13.494 |
| rs10505100 | chr8 | 108278616 | Intronic | C | A | ANGPT1 | 6.644 | 0.2 | 2.350 | 0.11 | − 4.159 | 0.283 | 40.539 |
| rs10918274 | chr1 | 165714416 | Intronic | T | C | TMCO1 | 20.362 | 0.96 | 5.017 | 0.86 | − 7.783 | 0.993 | 62.418 |
| rs11217878 | chr11 | 120340383 | Intronic | G | A | ARHGEF12 | 1.395 | 0.23 | 0.659 | 0.16 | − 3.413 | 0.227 | 1.147 |
| rs113985657 | chr6 | 597203 | Intronic | C | T | EXOC2 | 1.036 | 0.12 | 0.000 | 0.14 | 1.014 | 0.208 | 26.399 |
| rs11710139 | chr3 | 150059342 | Intergenic | G | A | LINC01214;TSC22D2 | − 12.606 | 0.16 | 0.000 | 0.23 | 6.403 | 0.096 | − 16.719 |
| rs12377624 | chr9 | 129373110 | Intergenic | G | C | MVB12B;LMX1B | − 12.549 | 0.33 | 9.258 | 0.37 | 18.120 | 0.079 | − 78.578 |
| rs1254276 | chr14 | 60847001 | Intergenic | C | T | LINC02322;C14orf39 | 19.784 | 0.6 | − 0.943 | 0.4 | − 39.746 | 0.727 | 19.784 |
| rs12699251 | chr7 | 11679113 | Intronic | A | G | THSD7A | − 3.064 | 0.25 | 0.000 | 0.4 | 19.691 | 0.229 | − 1.524 |
| rs1579050 | chr2 | 153364527 | Intronic | A | G | FMNL2 | − 89.083 | 0.35 | 4.372 | 0.58 | 69.618 | 0.025 | − 242.856 |
| rs17752199 | chr6 | 51406848 | Intergenic | A | G | TFAP2B;PKHD1 | − 0.364 | 0.12 | − 0.327 | 0.12 | − 0.320 | 0.103 | − 3.773 |
| rs1874458 | chr16 | 65080739 | Intronic | G | A | CDH11 | 0.314 | 0.2 | 0.000 | 0.33 | 17.034 | 0.250 | 7.076 |
| rs2022945 | chr8 | 108251139 | Intergenic | A | G | ABRA;ANGPT1 | − 6.644 | 0.8 | − 2.350 | 0.89 | 4.159 | 0.723 | − 37.209 |
| rs2024211 | chr7 | 116153025 | Intergenic | A | C | CAV2;CAV1 | − 76.273 | 0.19 | − 0.664 | 0.26 | 3.118 | 0.007 | − 218.926 |
| rs2073006 | chr6 | 637465 | Intronic | C | T | EXOC2 | 3.236 | 0.11 | 0.000 | 0.13 | 1.060 | 0.220 | 40.976 |
| rs2188836 | chr7 | 117635382 | Intergenic | C | T | CTTNBP2;LSM8 | − 60.936 | 0.43 | 7.936 | 0.41 | 5.477 | 0.095 | − 150.366 |
| rs2317961 | chr6 | 1533116 | Intergenic | A | G | FOXF2;FOXCUT | − 11.567 | 0.52 | − 8.961 | 0.64 | 0.217 | 0.475 | − 44.116 |
| rs2472493 | chr9 | 107695848 | Intergenic | G | A | ABCA1;SLC44A1 | − 11.416 | 0.65 | 1.567 | 0.58 | − 1.060 | 0.460 | − 40.945 |
| rs2472496 | chr9 | 107695353 | Intergenic | G | A | ABCA1;SLC44A1 | − 15.152 | 0.67 | 3.051 | 0.58 | − 1.060 | 0.428 | − 59.932 |
| rs2487032 | chr9 | 107703934 | Intergenic | G | A | ABCA1;SLC44A1 | − 9.692 | 0.56 | − 1.474 | 0.5 | − 7.924 | 0.476 | − 28.298 |
| rs2745572 | chr6 | 1548369 | Intergenic | A | G | FOXF2;FOXCUT | 13.501 | 0.48 | 10.620 | 0.36 | 0.000 | 0.517 | 45.723 |
| rs28500712 | chr4 | 7896213 | Intronic | A | G | AFAP1 | 0.586 | 0.54 | − 10.735 | 0.72 | 3.416 | 0.709 | 5.547 |
| rs28520091 | chr4 | 7846240 | Intronic | C | T | AFAP1 | − 1.186 | 0.37 | 2.314 | 0.5 | 25.369 | 0.295 | − 1.770 |
| rs28795989 | chr4 | 7891545 | Intronic | A | G | AFAP1 | − 12.655 | 0.26 | − 1.084 | 0.6 | 73.748 | 0.170 | − 36.886 |
| rs2935057 | chr6 | 170454915 | Intergenic | A | G | LINC00574;LOC102724511 | − 2.238 | 0.84 | 2.252 | 0.88 | 8.761 | 0.729 | − 13.364 |
| rs3013274 | chr6 | 170464367 | Intergenic | G | A | LINC00574;LOC102724511 | − 9.753 | 0.68 | 4.063 | 0.57 | − 1.606 | 0.428 | − 59.784 |
| rs31918 | chr5 | 14820927 | Intronic | C | T | ANKH | 1.881 | 0.22 | − 4.799 | 0.29 | 0.000 | 0.334 | 4.776 |
| rs327716 | chr7 | 80838977 | Intergenic | A | G | SEMA3C;LOC105369146 | 45.585 | 0.64 | − 4.367 | 0.43 | − 60.462 | 0.947 | 183.742 |
| rs33912345 | chr14 | 60976537 | Exonic | C | A | SIX6 | − 15.632 | 0.4 | 3.083 | 0.6 | 50.841 | 0.268 | − 11.500 |
| rs3785176 | chr16 | 8896931 | Intronic | A | C | PMM2 | 11.463 | 0.13 | − 3.575 | 0.25 | 6.099 | 0.316 | 45.623 |
| rs4141671 | chr10 | 60338753 | Intronic | T | C | BICC1 | − 0.226 | 0.56 | 3.867 | 0.49 | 0.000 | 0.489 | − 0.037 |
| rs4236601 | chr7 | 116162729 | Intergenic | G | A | CAV2;CAV1 | − 84.231 | 0.19 | − 1.952 | 0.26 | 1.262 | 0.007 | − 244.996 |
| rs55892100 | chr7 | 115810676 | Intergenic | A | G | TFEC;TES | 54.802 | 0.43 | − 8.650 | 0.37 | − 21.708 | 0.835 | 182.195 |
| rs5756813 | chr22 | 38175477 | Intergenic | G | T | TRIOBP;H1F0 | 20.456 | 0.5 | − 2.095 | 0.58 | 0.989 | 0.643 | 16.731 |
| rs58073046 | chr11 | 120248493 | Intronic | A | G | ARHGEF12 | 8.745 | 0.21 | 17.513 | 0.1 | 0.000 | 0.168 | 18.725 |
| rs61394862 | chr5 | 14851094 | Intronic | C | T | ANKH | 1.910 | 0.21 | − 4.971 | 0.28 | 0.000 | 0.333 | 6.636 |
| rs6478746 | chr9 | 129367398 | Intergenic | G | A | MVB12B;LMX1B | 50.342 | 0.78 | − 0.668 | 0.7 | − 10.654 | 0.967 | 127.390 |
| rs66602224 | chr8 | 108293718 | Intronic | G | A | ANGPT1 | − 32.704 | 0.28 | − 1.639 | 0.4 | 5.548 | 0.204 | − 31.901 |
| rs6732795 | chr2 | 69411517 | Intronic | A | C | ANTXR1 | 34.845 | 0.68 | 0.540 | 0.4 | − 50.841 | 0.900 | 152.394 |
| rs73174345 | chr3 | 169252883 | Intronic | T | G | MECOM | − 21.410 | 0.014 | − 7.882 | 0.059 | 0.230 | 0.000 | − 59.854 |
| rs746491 | chr11 | 86406159 | Intergenic | C | A | ME3;PRSS23 | − 0.338 | 0.13 | − 0.815 | 0.21 | 5.097 | 0.193 | 6.653 |
| rs7518099 | chr1 | 165736880 | Intronic | C | T | TMCO1 | 20.362 | 0.96 | 5.017 | 0.86 | − 7.783 | 0.993 | 63.362 |
| rs7555523 | chr1 | 165718979 | Intronic | C | A | TMCO1 | 27.970 | 0.96 | 9.258 | 0.86 | − 3.391 | 0.993 | 85.262 |
| rs7924522 | chr11 | 128380742 | Intronic | C | A | ETS1 | 14.150 | 0.75 | 0.000 | 0.63 | − 13.128 | 0.907 | 78.316 |
| rs8141433 | chr22 | 19854006 | Intergenic | A | G | GNB1L;TXNRD2 | − 78.658 | 0.22 | − 3.655 | 0.15 | − 18.042 | 0.040 | − 200.793 |
| rs9284802 | chr3 | 85095766 | Intronic | G | A | CADM2 | − 22.155 | 0.41 | 6.585 | 0.63 | 72.241 | 0.176 | − 49.830 |
| rs945686 | chr9 | 129378026 | Intronic | G | C | LMX1B | 61.221 | 0.79 | − 1.348 | 0.76 | − 4.495 | 0.987 | 156.676 |
| rs9494457 | chr6 | 136474794 | Intronic | T | A | PDE7B | − 3.323 | 0.44 | 2.158 | 0.37 | − 0.570 | 0.282 | − 23.733 |
| rs9853115 | chr3 | 186131600 | Intergenic | T | A | DGKG;LINC02052 | 23.247 | 0.57 | 0.193 | 0.53 | − 1.014 | 0.701 | 38.881 |
| rs9913911 | chr17 | 10031183 | Intronic | A | G | GAS7 | 28.572 | 0.27 | − 1.662 | 0.37 | 3.416 | 0.554 | 109.722 |
Chr: chromosome EAF: effect allele frequency ref allele: reference allele alt allele: alterative allele AMR: Americans AFR: Africans, EAS: East Asians, SAS: South Asians, EUR: Europeans, KOR: Koreans, P-value: adjusted Fischer’s test, statistical significance was set at P < 0.05 and |log10 P|> 1.301
Composite genetic risk score calculation using SNPs related to OAG and OAG with high IOP
To compare the composite genetic risk of OAG, we adopted the equation suggested by Mao et al. [31] The composite genetic risk score is calculated using the following formula:
where “I” refers to the number of OAG-related SNPs, and “Xi” refers to copies of risk alleles (Xi ∈ {0,1,2}) at the ith SNP. In one extreme case, if a person has two copies of risk alleles at each OAG-related SNP, then the person’s risk score will become 1. On the other hand, if a person has no copy of risk alleles at each OAG-related SNP, then the person’s risk score will become 0. A person with a composite score of 1 has the highest possible genetic risk for OAG, while a person with a score of 0 has the lowest possible genetic risk. If copies of effect alleles (0/1/2) are randomly assigned to each SNP, the expected value of the risk score will be 0.5. SNPs with a frequency difference of more than 10% between the total (n = 1722) and the 2nd phase (n = 1099) data of KRGDB were excluded from the genetic score calculation. We used the average of composite genetic risk scores for the populations for correlation with the country-wise OAG prevalence data. In addition, the composite genetic risk score was calculated using IOP elevation-related SNPs for OAG with high IOP. The prevalence of OAG with high IOP was calculated as (1—the NTG proportion of OAG [12]) × the total OAG prevalence. The correlation analysis with the composite genetic risk score and the prevalence of OAG with high IOP was performed.
Data analyses
We used the Kruskal–Wallis rank-sum test to assess OAG related SNP frequencies according to populations of diverse ancestry and Fisher's exact test to assess whether the effect allele at a given SNP is significantly enriched or depleted compared to the global population frequency in the 1000 Genomes Project database, and the P values were first log10-transformed. In the heatmap generated to visualize allele enrichment or depletion patterns in different populations, red and purple colors meant higher and lower frequencies than the global average, respectively. If the effect allele of an SNP is enriched in a population, then the negative of log10 of the enrichment P-value (a positive number) was used to represent the SNP associated with that population in a heatmap. On the other hand, if the allele of an SNP is depleted in a population, the value of log10 of the depletion P-value (a negative number) was used to represent the SNP for that population in the heatmap. Statistical analyses were performed using R software version 3.6.0 (R Foundation, Vienna, Austria). Statistical significance was set at P < 0.05 and |log10 P|> 1.301.
Results
Patterns of OAG risk alleles among populations
A total of 135 OAG-related SNPs was obtained from 24 GWAS studies. Among these, 15 studies were performed in Europeans, 13 in East Asians, 4 in South Asians, 7 in Africans, and 2 in Americans (9 studies were performed in mixed ethnic populations). Clearly, populations except Europeans were understudied. However, there was no significant difference in the SNP frequency among these populations with the Kruskal–Wallis rank-sum test (Fig. 1). This result suggests that many SNPs found in Europeans are also applicable to other populations. Following collecting the OAG-associated SNPs, we obtained their effect allele frequencies (EAFs) in each of the continental groups and Koreans based on genotype information from the 1000 Genomes Project and KRGDB (Additional file 2: Table 1). A heatmap showed how significantly the effect allele was enriched or depleted across the Korean and continental groups (Additional files 1: Figure S1) with a log scale among 135 OAG-related SNPs. For Koreans, 63 OAG-related SNPs were significantly enriched, 63 SNPs were depleted, and 9 SNPs were similar to global EAF. The heatmap visualized the proportion of effect alleles in each continental group compared to the global average. There were certain patterns of risk allele frequencies depending on continental groups. The hierarchical clustering tree showed the differences among the populations; Europeans, Americans, and South Asians were in one cluster, and Africans, East Asians, and Koreans were in another cluster. In addition, we compared the EAFs of East Asians and Koreans (Additional file 3: Table S2, Additional file 1: Figure S2). Although the EAF was not much different among East Asians, 25 SNPs were enriched, 26 SNPs were depleted, and 84 SNPs were similar to those in Koreans. Moreover, the heatmap clearly showed that the main pattern of the allele frequency in East Asians in the 1000 Genomes Project was very similar to that in Koreans, while few alleles showed the opposite pattern in allele frequency between East Asians and Koreans.
Fig. 1.
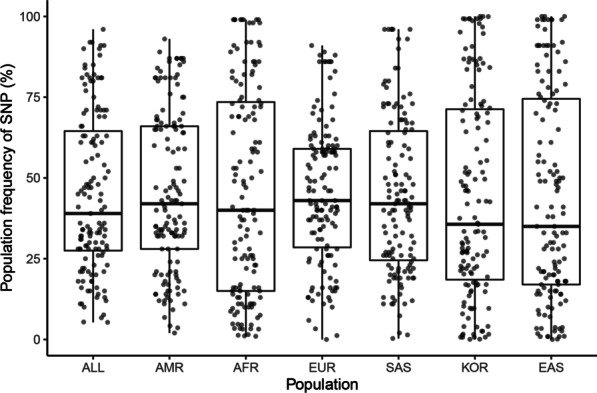
Comparison of frequency of open-angle glaucoma (OAG)-related single nucleotide polymorphisms (SNP) according to populations of diverse ancestry. This figure shows no difference in OAG-related SNP frequency between populations with the Kruskal–Wallis rank-sum test (P = 0.8893). AMR: American, EUR: Europeans, SAS: South Asians, AFR: Africans, EAS: East Asians, KOR: Koreans
Patterns of IOP elevation-related OAG risk alleles among populations
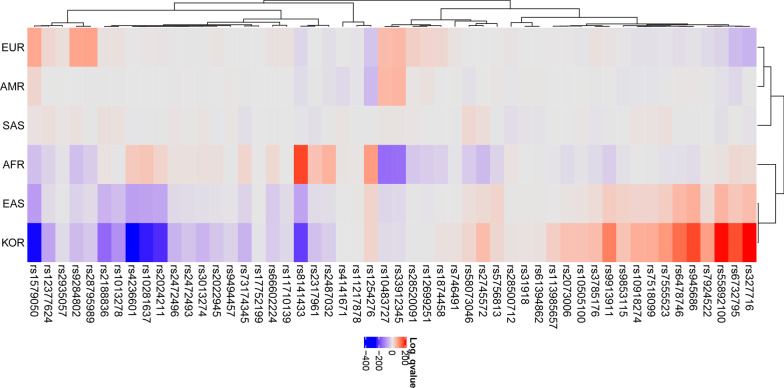
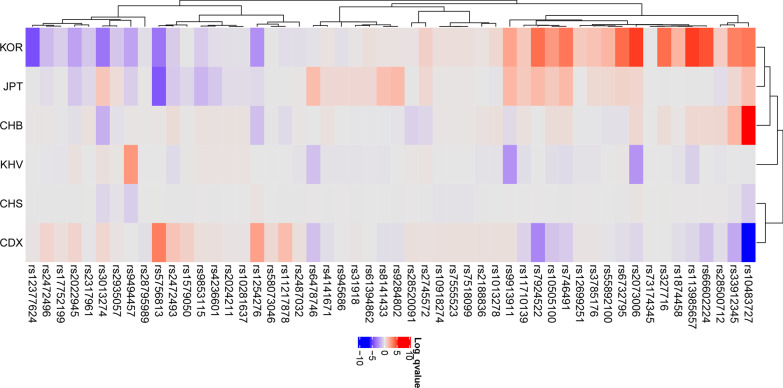
Fifty-two IOP-related SNP traits were selected from 135 OAG-related SNPs (Table 1), and a heatmap showing how significantly the EAF was enriched or depleted across the Koreans and continental groups (Fig. 2) with a log scale among 52 IOP-related SNPs was generated. For Koreans, 23 OAG-related SNPs were significantly enriched, 27 SNPs depleted, and 2 SNPs were similar to global EAF. For example, rs2024211, located in the Intergenic of CAV2; CAV1 that is expressed in the trabecular meshwork cells cultured from OAG eyes and encoding Caveolins [34], has T/C alleles in which the C allele was tested in European populations to identify the risk of OAG with high IOP [25]. The C allele frequencies were 26%, 36%, and 0.7% in European, African, and Korean populations, respectively. Further, rs1579050, located in FMNL2 gene belongs to the formin-related family of proteins that acts as a downstream effector of CDC42 (Rho family member) [33], has A/G alleles in which the G allele frequencies were 58%, 10%, and 2.5% in European, African, and Korean populations. The rs8141433, located in the Intergenic of GNB1L; TXNRD2, TXNRD2 gene encodes a mitochondrial protein required for redox homeostasis [35], revealed the G allele frequencies of 15%, 69%, and 4% in European, African, and Korean populations. A hierarchical clustering tree showed the differences among the populations; Europeans, Americans, and South Asians were in one cluster, and Africans, East Asians, and Koreans were in another cluster. These allele frequencies may be related to a difference in the NTG proportion of OAG among the populations. In addition, a heatmap showed how significantly the EAF was enriched or depleted across the Koreans and East Asians (Table 2, Fig. 3). For Koreans, rs12377624, LMX1B gene had G/C alleles in which the C allele frequencies were 14%, 12.5%, and 7.9% in Chinese, Japanese, and Koreans, respectively, whereas, rs2073006, EXOC2 gene had C/T alleles in which the T allele frequencies were 11.8%, 8.1%, and 22.0% in Chinese, Vietnamese, and Koreans, respectively. However, the EAF was not much different among East Asians; for Koreans, 13 SNPs were enriched, 7 SNPs were depleted, and 32 SNPs were similar to those in Global East Asians. A hierarchical clustering tree showed the differences among East Asians, KOR, Japanese, and Han Chinese in Beijing in one cluster, and Southern Han Chinese and Chinese Dai in Xishuangbanna in another cluster.
Fig. 2.
The heatmap generated using intraocular pressure elevation in open-angle glaucoma-related single nucleotide polymorphisms in the global population. The heatmap shows how significantly the effect alleles are enriched or depleted in each population. Each row shows SNPs, and each column shows populations of diverse ancestry. Red color means effect allele is enriched, whereas purple color means effect allele is depleted (log10 P > 1.301 indicated enrichment, log10 P < − 1.301 indicated depletion). A hierarchical clustering tree shows the differences among continents; EUR, AMR, and SAS are in one cluster, and AFR, EAS, and KOR are in another cluster. AMR: American, EUR: Europeans, SAS: South Asians, AFR: Africans, EAS: East Asians, KOR: Koreans
Table 2.
Effect allele frequencies (EAFs) of intraocular pressure related single nucleotide polymorphisms in East Asian groups including Koreans
| SNP ID | Chr | Position | Type | Ref Allele | Alt Allele | Nearby/containing Gene | Global East Asian EAF | CHS EAF | CHS log10 P | CDX EAF | CDX log10 P | KHV EAF | KHV log10 P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1013278 | chr7 | 117603820 | Intergenic | G | C | CTTNBP2;LSM8 | 0.1 | 0.105 | 0.000 | 0.118 | 0.162 | 0.076 | − 0.079 |
| rs10281637 | chr7 | 116151338 | Intergenic | T | C | CAV2;CAV1 | 0.0089 | 0.005 | 0.000 | 0.011 | 0.061 | 0.015 | 0.079 |
| rs10483727 | chr14 | 61072875 | Intergenic | T | C | SIX6;SALRNA1 | 0.21 | 0.167 | − 0.381 | 0.113 | − 5.663 | 0.202 | − 0.034 |
| rs10505100 | chr8 | 108278616 | Intronic | C | A | ANGPT1 | 0.23 | 0.233 | 0.000 | 0.156 | − 0.852 | 0.182 | − 0.142 |
| rs10918274 | chr1 | 165714416 | Intronic | T | C | TMCO1 | 0.99 | 0.976 | − 0.046 | 1.000 | 0.175 | 0.990 | 0.000 |
| rs11217878 | chr11 | 120340383 | Intronic | G | A | ARHGEF12 | 0.24 | 0.224 | 0.000 | 0.339 | 1.234 | 0.227 | 0.000 |
| rs113985657 | chr6 | 597203 | Intronic | C | T | EXOC2 | 0.14 | 0.167 | 0.000 | 0.097 | − 0.467 | 0.086 | − 0.448 |
| rs11710139 | chr3 | 150059342 | Intergenic | G | A | LINC01214;TSC22D2 | 0.075 | 0.071 | 0.000 | 0.038 | − 0.586 | 0.045 | − 0.137 |
| rs12377624 | chr9 | 129373110 | Intergenic | G | C | MVB12B;LMX1B | 0.13 | 0.114 | 0.000 | 0.140 | 0.048 | 0.131 | 0.000 |
| rs1254276 | chr14 | 60847001 | Intergenic | C | T | LINC02322;C14orf39 | 0.78 | 0.833 | 0.126 | 0.887 | 1.936 | 0.778 | 0.000 |
| rs12699251 | chr7 | 11679113 | Intronic | A | G | THSD7A | 0.2 | 0.224 | 0.000 | 0.215 | 0.061 | 0.167 | − 0.079 |
| rs1579050 | chr2 | 153364527 | Intronic | A | G | FMNL2 | 0.026 | 0.010 | 0.000 | 0.054 | 0.658 | 0.030 | 0.034 |
| rs17752199 | chr6 | 51406848 | Intergenic | A | G | TFAP2B;PKHD1 | 0.12 | 0.105 | 0.000 | 0.145 | 0.280 | 0.106 | − 0.054 |
| rs1874458 | chr16 | 65080739 | Intronic | G | A | CDH11 | 0.21 | 0.229 | 0.000 | 0.177 | − 0.186 | 0.182 | − 0.079 |
| rs2022945 | chr8 | 108251139 | Intergenic | A | G | ABRA;ANGPT1 | 0.77 | 0.762 | 0.000 | 0.828 | 0.586 | 0.813 | 0.096 |
| rs2024211 | chr7 | 116153025 | Intergenic | A | C | CAV2;CAV1 | 0.0089 | 0.005 | 0.000 | 0.011 | 0.061 | 0.015 | 0.079 |
| rs2073006 | chr6 | 637465 | Intronic | C | T | EXOC2 | 0.15 | 0.181 | 0.046 | 0.118 | − 0.447 | 0.081 | − 2.070 |
| rs2188836 | chr7 | 117635382 | Intergenic | C | T | CTTNBP2;LSM8 | 0.09 | 0.076 | 0.000 | 0.113 | 0.182 | 0.066 | − 0.079 |
| rs2317961 | chr6 | 1533116 | Intergenic | A | G | FOXF2;FOXCUT | 0.51 | 0.519 | 0.000 | 0.500 | − 0.023 | 0.495 | − 0.009 |
| rs2472493 | chr9 | 107695848 | Intergenic | G | A | ABCA1;SLC44A1 | 0.49 | 0.471 | 0.000 | 0.543 | 0.945 | 0.455 | − 0.170 |
| rs2472496 | chr9 | 107695353 | Intergenic | G | A | ABCA1;SLC44A1 | 0.47 | 0.457 | 0.000 | 0.538 | 0.544 | 0.449 | − 0.034 |
| rs2487032 | chr9 | 107703934 | Intergenic | G | A | ABCA1;SLC44A1 | 0.49 | 0.467 | 0.000 | 0.532 | 0.198 | 0.449 | − 0.079 |
| rs2745572 | chr6 | 1548369 | Intergenic | A | G | FOXF2;FOXCUT | 0.49 | 0.481 | 0.000 | 0.505 | 0.137 | 0.510 | 0.079 |
| rs28500712 | chr4 | 7896213 | Intronic | A | G | AFAP1 | 0.68 | 0.695 | 0.000 | 0.640 | − 0.198 | 0.657 | − 0.041 |
| rs28520091 | chr4 | 7846240 | Intronic | C | T | AFAP1 | 0.29 | 0.329 | 0.000 | 0.328 | 0.199 | 0.313 | 0.041 |
| rs28795989 | chr4 | 7891545 | Intronic | A | G | AFAP1 | 0.18 | 0.181 | 0.000 | 0.156 | − 0.155 | 0.187 | 0.000 |
| rs2935057 | chr6 | 170454915 | Intergenic | A | G | LINC00574;LOC102724511 | 0.76 | 0.700 | − 0.155 | 0.774 | 0.054 | 0.783 | 0.054 |
| rs3013274 | chr6 | 170464367 | Intergenic | G | A | LINC00574;LOC102724511 | 0.5 | 0.419 | − 0.286 | 0.565 | 0.497 | 0.556 | 0.142 |
| rs31918 | chr5 | 14820927 | Intronic | C | T | ANKH | 0.33 | 0.319 | 0.000 | 0.344 | 0.046 | 0.298 | − 0.079 |
| rs327716 | chr7 | 80838977 | Intergenic | A | G | SEMA3C;LOC105369146 | 0.91 | 0.919 | 0.000 | 0.909 | 0.000 | 0.909 | 0.000 |
| rs33912345 | chr14 | 60976537 | Exonic | C | A | SIX6 | 0.21 | 0.167 | − 0.046 | 0.113 | − 1.659 | 0.212 | 0.000 |
| rs3785176 | chr16 | 8896931 | Intronic | A | C | PMM2 | 0.28 | 0.271 | 0.000 | 0.296 | 0.061 | 0.237 | − 0.079 |
| rs4141671 | chr10 | 60338753 | Intronic | T | C | BICC1 | 0.48 | 0.443 | 0.000 | 0.441 | − 0.182 | 0.439 | − 0.079 |
| rs4236601 | chr7 | 116162729 | Intergenic | G | A | CAV2;CAV1 | 0.0099 | 0.005 | 0.000 | 0.016 | 0.172 | 0.015 | 0.079 |
| rs55892100 | chr7 | 115810676 | Intergenic | A | G | TFEC;TES | 0.8 | 0.805 | 0.000 | 0.785 | − 0.061 | 0.803 | 0.000 |
| rs5756813 | chr22 | 38175477 | Intergenic | G | T | TRIOBP;H1F0 | 0.71 | 0.724 | 0.000 | 0.849 | 2.955 | 0.717 | 0.000 |
| rs58073046 | chr11 | 120248493 | Intronic | A | G | ARHGEF12 | 0.17 | 0.167 | 0.000 | 0.215 | 0.447 | 0.167 | 0.000 |
| rs61394862 | chr5 | 14851094 | Intronic | C | T | ANKH | 0.32 | 0.319 | 0.000 | 0.323 | 0.000 | 0.298 | − 0.041 |
| rs6478746 | chr9 | 129367398 | Intergenic | G | A | MVB12B;LMX1B | 0.97 | 0.976 | 0.000 | 0.946 | − 0.945 | 0.944 | − 0.944 |
| rs66602224 | chr8 | 108293718 | Intronic | G | A | ANGPT1 | 0.14 | 0.167 | 0.000 | 0.075 | − 0.945 | 0.131 | 0.000 |
| rs6732795 | chr2 | 69411517 | Intronic | A | C | ANTXR1 | 0.85 | 0.819 | 0.000 | 0.823 | − 0.175 | 0.869 | 0.040 |
| rs73174345 | chr3 | 169252883 | Intronic | T | G | MECOM | 0 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| rs746491 | chr11 | 86406159 | Intergenic | C | A | ME3;PRSS23 | 0.14 | 0.129 | 0.000 | 0.086 | − 0.715 | 0.096 | − 0.170 |
| rs7518099 | chr1 | 165736880 | Intronic | C | T | TMCO1 | 0.99 | 0.976 | − 0.046 | 1.000 | 0.175 | 0.990 | 0.000 |
| rs7555523 | chr1 | 165718979 | Intronic | C | A | TMCO1 | 0.99 | 0.976 | − 0.046 | 1.000 | 0.175 | 0.990 | 0.000 |
| rs7924522 | chr11 | 128380742 | Intronic | C | A | ETS1 | 0.86 | 0.876 | 0.000 | 0.742 | − 2.617 | 0.859 | 0.000 |
| rs8141433 | chr22 | 19854006 | Intergenic | A | G | GNB1L;TXNRD2 | 0.037 | 0.033 | 0.000 | 0.022 | − 0.175 | 0.020 | − 0.079 |
| rs9284802 | chr3 | 85095766 | Intronic | G | A | CADM2 | 0.17 | 0.152 | 0.000 | 0.134 | − 0.215 | 0.172 | 0.000 |
| rs945686 | chr9 | 129378026 | Intronic | G | C | LMX1B | 0.99 | 0.995 | 0.000 | 0.989 | 0.000 | 0.980 | − 0.079 |
| rs9494457 | chr6 | 136474794 | Intronic | T | A | PDE7B | 0.33 | 0.262 | − 0.381 | 0.328 | 0.000 | 0.439 | 2.084 |
| rs9853115 | chr3 | 186131600 | Intergenic | T | A | DGKG;LINC02052 | 0.73 | 0.752 | 0.000 | 0.753 | 0.169 | 0.753 | 0.079 |
| rs9913911 | chr17 | 10031183 | Intronic | A | G | GAS7 | 0.5 | 0.519 | 0.000 | 0.522 | 0.175 | 0.414 | − 2.084 |
| SNP ID | Chr | Position | Type | Ref Allele | Alt Allele | Nearby/containing Gene | CHB EAF | CHB log10 P | JPT EAF | JPT log10 P | KOR EAF | KOR log10 P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1013278 | chr7 | 117603820 | Intergenic | G | C | CTTNBP2;LSM8 | 0.126 | 0.216 | 0.087 | − 0.081 | 0.114 | 0.413 |
| rs10281637 | chr7 | 116151338 | Intergenic | T | C | CAV2;CAV1 | 0.015 | 0.133 | 0.000 | − 0.232 | 0.007 | − 0.169 |
| rs10483727 | chr14 | 61072875 | Intergenic | T | C | SIX6;SALRNA1 | 0.316 | 4.865 | 0.260 | 1.092 | 0.271 | 3.218 |
| rs10505100 | chr8 | 108278616 | Intronic | C | A | ANGPT1 | 0.272 | 0.285 | 0.303 | 0.819 | 0.283 | 2.435 |
| rs10918274 | chr1 | 165714416 | Intronic | T | C | TMCO1 | 0.995 | 0.059 | 0.995 | 0.047 | 0.993 | 0.229 |
| rs11217878 | chr11 | 120340383 | Intronic | G | A | ARHGEF12 | 0.189 | − 0.328 | 0.231 | − 0.023 | 0.227 | − 0.229 |
| rs113985657 | chr6 | 597203 | Intronic | C | T | EXOC2 | 0.189 | 0.430 | 0.144 | 0.004 | 0.208 | 4.434 |
| rs11710139 | chr3 | 150059342 | Intergenic | G | A | LINC01214;TSC22D2 | 0.097 | 0.195 | 0.120 | 0.779 | 0.096 | 0.939 |
| rs12377624 | chr9 | 129373110 | Intergenic | G | C | MVB12B;LMX1B | 0.146 | 0.078 | 0.125 | − 0.004 | 0.079 | − 4.387 |
| rs1254276 | chr14 | 60847001 | Intergenic | C | T | LINC02322;C14orf39 | 0.689 | − 1.099 | 0.745 | − 0.295 | 0.727 | − 2.493 |
| rs12699251 | chr7 | 11679113 | Intronic | A | G | THSD7A | 0.184 | − 0.059 | 0.207 | 0.023 | 0.229 | 0.876 |
| rs1579050 | chr2 | 153364527 | Intronic | A | G | FMNL2 | 0.029 | 0.038 | 0.010 | − 0.343 | 0.025 | − 0.017 |
| rs17752199 | chr6 | 51406848 | Intergenic | A | G | TFAP2B;PKHD1 | 0.112 | − 0.059 | 0.115 | − 0.023 | 0.103 | − 0.624 |
| rs1874458 | chr16 | 65080739 | Intronic | G | A | CDH11 | 0.228 | 0.078 | 0.212 | 0.000 | 0.250 | 1.588 |
| rs2022945 | chr8 | 108251139 | Intergenic | A | G | ABRA;ANGPT1 | 0.733 | − 0.195 | 0.702 | − 0.779 | 0.723 | − 2.027 |
| rs2024211 | chr7 | 116153025 | Intergenic | A | C | CAV2;CAV1 | 0.015 | 0.133 | 0.000 | − 0.232 | 0.007 | − 0.169 |
| rs2073006 | chr6 | 637465 | Intronic | C | T | EXOC2 | 0.189 | 0.430 | 0.183 | 0.440 | 0.220 | 4.434 |
| rs2188836 | chr7 | 117635382 | Intergenic | C | T | CTTNBP2;LSM8 | 0.107 | 0.133 | 0.091 | 0.000 | 0.095 | 0.098 |
| rs2317961 | chr6 | 1533116 | Intergenic | A | G | FOXF2;FOXCUT | 0.549 | 0.195 | 0.466 | − 0.313 | 0.475 | − 0.904 |
| rs2472493 | chr9 | 107695848 | Intergenic | G | A | ABCA1;SLC44A1 | 0.519 | 0.285 | 0.447 | − 0.685 | 0.460 | − 0.711 |
| rs2472496 | chr9 | 107695353 | Intergenic | G | A | ABCA1;SLC44A1 | 0.485 | 0.059 | 0.438 | − 0.203 | 0.428 | − 1.306 |
| rs2487032 | chr9 | 107703934 | Intergenic | G | A | ABCA1;SLC44A1 | 0.510 | 0.059 | 0.505 | 0.047 | 0.476 | − 0.209 |
| rs2745572 | chr6 | 1548369 | Intergenic | A | G | FOXF2;FOXCUT | 0.451 | − 0.430 | 0.519 | 0.346 | 0.517 | 0.624 |
| rs28500712 | chr4 | 7896213 | Intronic | A | G | AFAP1 | 0.752 | 0.489 | 0.649 | − 0.191 | 0.709 | 0.767 |
| rs28520091 | chr4 | 7846240 | Intronic | C | T | AFAP1 | 0.214 | − 0.598 | 0.279 | − 0.025 | 0.295 | 0.076 |
| rs28795989 | chr4 | 7891545 | Intronic | A | G | AFAP1 | 0.184 | 0.035 | 0.188 | 0.035 | 0.170 | − 0.196 |
| rs2935057 | chr6 | 170454915 | Intergenic | A | G | LINC00574;LOC102724511 | 0.733 | − 0.133 | 0.793 | 0.236 | 0.729 | − 0.913 |
| rs3013274 | chr6 | 170464367 | Intergenic | G | A | LINC00574;LOC102724511 | 0.379 | − 1.458 | 0.587 | 0.867 | 0.428 | − 3.235 |
| rs31918 | chr5 | 14820927 | Intronic | C | T | ANKH | 0.316 | − 0.051 | 0.375 | 0.339 | 0.334 | 0.054 |
| rs327716 | chr7 | 80838977 | Intergenic | A | G | SEMA3C;LOC105369146 | 0.942 | 0.285 | 0.904 | − 0.025 | 0.947 | 3.495 |
| rs33912345 | chr14 | 60976537 | Exonic | C | A | SIX6 | 0.311 | 1.376 | 0.255 | 0.351 | 0.268 | 2.976 |
| rs3785176 | chr16 | 8896931 | Intronic | A | C | PMM2 | 0.267 | − 0.052 | 0.322 | 0.329 | 0.316 | 1.128 |
| rs4141671 | chr10 | 60338753 | Intronic | T | C | BICC1 | 0.519 | 0.195 | 0.529 | 0.339 | 0.489 | 0.113 |
| rs4236601 | chr7 | 116162729 | Intergenic | G | A | CAV2;CAV1 | 0.015 | 0.133 | 0.000 | − 0.779 | 0.007 | − 0.229 |
| rs55892100 | chr7 | 115810676 | Intergenic | A | G | TFEC;TES | 0.772 | − 0.133 | 0.841 | 0.346 | 0.835 | 1.552 |
| rs5756813 | chr22 | 38175477 | Intergenic | G | T | TRIOBP;H1F0 | 0.728 | 0.059 | 0.529 | − 4.121 | 0.643 | − 3.218 |
| rs58073046 | chr11 | 120248493 | Intronic | A | G | ARHGEF12 | 0.155 | − 0.059 | 0.159 | − 0.035 | 0.168 | − 0.023 |
| rs61394862 | chr5 | 14851094 | Intronic | C | T | ANKH | 0.301 | − 0.059 | 0.365 | 0.339 | 0.333 | 0.196 |
| rs6478746 | chr9 | 129367398 | Intergenic | G | A | MVB12B;LMX1B | 0.971 | 0.000 | 0.990 | 1.200 | 0.967 | − 0.106 |
| rs66602224 | chr8 | 108293718 | Intronic | G | A | ANGPT1 | 0.189 | 0.430 | 0.144 | 0.004 | 0.204 | 4.208 |
| rs6732795 | chr2 | 69411517 | Intronic | A | C | ANTXR1 | 0.825 | − 0.133 | 0.894 | 0.489 | 0.900 | 3.633 |
| rs73174345 | chr3 | 169252883 | Intronic | T | G | MECOM | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| rs746491 | chr11 | 86406159 | Intergenic | C | A | ME3;PRSS23 | 0.180 | 0.285 | 0.221 | 1.329 | 0.193 | 3.194 |
| rs7518099 | chr1 | 165736880 | Intronic | C | T | TMCO1 | 0.995 | 0.059 | 0.995 | 0.047 | 0.993 | 0.229 |
| rs7555523 | chr1 | 165718979 | Intronic | C | A | TMCO1 | 0.995 | 0.059 | 0.995 | 0.047 | 0.993 | 0.229 |
| rs7924522 | chr11 | 128380742 | Intronic | C | A | ETS1 | 0.859 | 0.000 | 0.933 | 1.329 | 0.907 | 3.495 |
| rs8141433 | chr22 | 19854006 | Intergenic | A | G | GNB1L;TXNRD2 | 0.029 | − 0.059 | 0.077 | 1.092 | 0.040 | 0.113 |
| rs9284802 | chr3 | 85095766 | Intronic | G | A | CADM2 | 0.150 | − 0.099 | 0.255 | 1.296 | 0.176 | 0.106 |
| rs945686 | chr9 | 129378026 | Intronic | G | C | LMX1B | 0.990 | 0.000 | 1.000 | 0.339 | 0.987 | − 0.169 |
| rs9494457 | chr6 | 136474794 | Intronic | T | A | PDE7B | 0.340 | 0.051 | 0.284 | − 0.530 | 0.282 | − 1.983 |
| rs9853115 | chr3 | 186131600 | Intergenic | T | A | DGKG;LINC02052 | 0.757 | 0.195 | 0.654 | − 1.329 | 0.701 | − 0.768 |
| rs9913911 | chr17 | 10031183 | Intronic | A | G | GAS7 | 0.466 | − 0.285 | 0.572 | 1.376 | 0.554 | 2.055 |
Chr: chromosome, EAF: effect allele frequency, ref allele: reference allele, alt allele: alterative allele, CHS: Southern Han Chinese, China, CDX: Chinese Dai in Xishuangbanna, China, KHV: Kinh in Ho Chi Minh City, Vietnam, CHB: Han Chinese in Beijing, China, JPT: Japanese in Tokyo in 1000 genome project, KOR: Korean Reference Genome data base, P-value: adjusted Fischer’s test, statistical significance was set at P < 0.05 and |log10 P|> 1.301
Fig. 3.
The heatmap generated using intraocular pressure elevation in open-angle glaucoma-related single nucleotide polymorphisms in the East Asian population. The heatmap shows how significantly the effect alleles are enriched or depleted in each population. Each row shows SNPs, and each column shows populations of diverse ancestry. Red color means effect allele is enriched, whereas purple color means effect allele is depleted (log10 P > 1.301 indicated enrichment, log10 P < − 1.301 indicated depletion). A hierarchical clustering tree shows the differences among East Asians, KOR, JPT, and CHB in one cluster, and CHS, and CDX in another cluster. CDX: Chinese Dai in Xishuangbanna, CHB: Han Chinese in Beijing, China, CHS: Southern Han Chinese, China, JPT: Japanese in Tokyo, Japan, KOR: Korean in Republic Korea, KHV: Kinh in Ho Chi Minh City, Vietnam
Composite genetic risk scores calculated using SNPs related to OAG as whole and OAG with high IOP
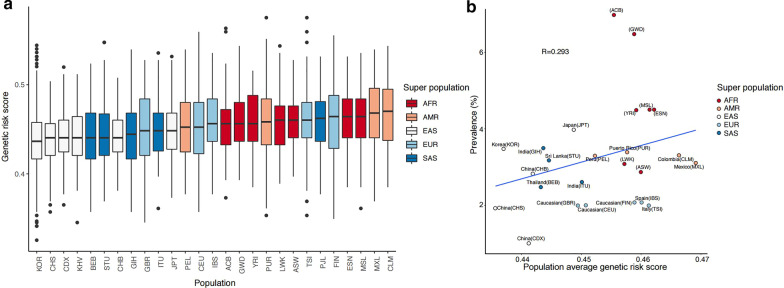
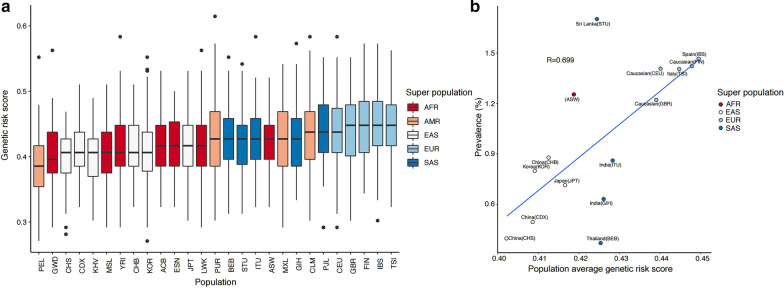
We calculated the composite genetic risk scores based on copies of effect alleles at OAG-associated SNPs, with the assumption that allelic associations from a significant majority of GWAS-identified variants can be replicated in non-European populations [36]. The genetic risk score of OAG was highest in Africans, followed by Europeans, South Asians, and East Asians (Fig. 4). The prevalence of OAG was correlated with the population average genetic risk score (R = 0.293, Fig. 4). In addition, the genetic risk score of OAG with high IOP was highest in Europeans, followed by South Asians, Africans, and East Asians (Fig. 5). The prevalence of OAG with elevated IOP was positively correlated with the population average genetic risk score (R = 0.699, Fig. 5).
Fig. 4.
Correlation plots of the prevalence of open-angle glaucoma and genetic risk score using related single nucleotide polymorphisms. a The genetic risk score of OAG according to populations of different ancestry was highest in Africans, followed by Europeans, South Asians, and East Asians. b The prevalence of OAG was correlated with the population average genetic risk score (R = 0.293). AMR: American, EUR: Europeans, SAS: South Asians, AFR: Africans, EAS: East Asians. ACB: African Caribbean in Barbados, ASW: African Ancestry in Southwest US, BEB: Bengali in Bangladesh, CDX: Chinese Dai in Xishuangbanna, CEU: Utah residents with Northern and Western European ancestry, CHB: Han Chinese in Beijing, China, CHS: Southern Han Chinese, China, CLM: Colombian in Medellin, Colombia, ESN: Esan in Nigeria, FIN: Finnish in Finland, GBR: British in England and Scotland, GIH: Gujarati Indian in Houston, TX, GWD: Gambian in Western Division, The Gambia, IBS: Iberian populations in Spain, ITU: Indian Telugu in the UK, JPT: Japanese in Tokyo, Japan, KOR: Korean in Republic Korea, KHV: Kinh in Ho Chi Minh City, Vietnam, LWK: Luhya in Webuye, Kenya, MSL: Mende in Sierra Leone, MXL: Mexican Ancestry in Los Angeles, California, PEL: Peruvian in Lima, Peru, PJL: Punjabi in Lahore, Pakistan, PUR: Puerto Rican in Puerto Rico, STU: Sri Lankan Tamil in the UK, TSI: Toscani in Italy, YRI: Yoruba in Ibadan, Nigeria
Fig. 5.
Correlation plots of the prevalence of open-angle glaucoma with intraocular pressure and genetic risk score using related single nucleotide polymorphisms. a The genetic risk score of OAG with high IOP according to populations of different ancestry was highest in Europeans, followed by South Asians, Africans, and East Asians. b The prevalence of OAG with elevated IOP was positively correlated with the population average genetic risk score (R = 0.699). The prevalence of open-angle glaucoma with normal intraocular pressure (normal-tension glaucoma) in AMR was excluded from the analysis since there were not enough research results. AMR: American, EUR: Europeans, SAS: South Asians, AFR: Africans, EAS: East Asians. ACB: African Caribbean in Barbados, ASW: African Ancestry in Southwest US, BEB: Bengali in Bangladesh, CDX: Chinese Dai in Xishuangbanna, CEU: Utah residents with Northern and Western European ancestry, CHB: Han Chinese in Beijing, China, CHS: Southern Han Chinese, China, CLM: Colombian in Medellin, Colombia, ESN: Esan in Nigeria, FIN: Finnish in Finland, GBR: British in England and Scotland, GIH: Gujarati Indian in Houston, TX, GWD: Gambian in Western Division, The Gambia, IBS: Iberian populations in Spain, ITU: Indian Telugu in the UK, JPT: Japanese in Tokyo, Japan, KOR: Korean in Republic Korea, KHV: Kinh in Ho Chi Minh City, Vietnam, LWK: Luhya in Webuye, Kenya, MSL: Mende in Sierra Leone, MXL: Mexican Ancestry in Los Angeles, California, PEL: Peruvian in Lima, Peru, PJL: Punjabi in Lahore, Pakistan, PUR: Puerto Rican in Puerto Rico, STU: Sri Lankan Tamil in the UK, TSI: Toscani in Italy, YRI: Yoruba in Ibadan, Nigeria
Discussion
Genetic factors are likely to play an essential role in the development of OAG [37]. Well-designed glaucoma cohort studies [23–26] had demonstrated the relationships of genetic loci with OAG or IOP elevation in OAG [25, 32, 33]. Although there is a Genome Aggregation Database (gnomAD) with more numbers, the authors thought that 1000 Genomes Projects would fit the design of this study, which being representative genome data and well-designed public data of whole-genome sequencing data from various populations around the world. Also, data of 1000 Genomes Projects was used for the gene target prediction model in the previous large POAG/IOP study [25]. Combining the results of these studies and the Korean whole-genome data, our study had identified the differences in allele frequencies of SNPs related to OAG or IOP elevation in OAG for worldwide populations as well as the relationship between the composite genetic risk scores and the prevalence in OAG or OAG with high IOP according to populations of diverse ancestry. The present study highlighted that the genetic risk of OAG with high IOP was present in Europeans, Americans, South Asians, Africans, and East Asians, in that order and showed a positive correlation with actual prevalence.
Our findings demonstrated that the prevalence of OAG differed among populations of diverse ancestry and was positively correlated with genetic factors. These results are consistent with those of the previous study by Kapetanakis et al., which demonstrated the global variations and time trends in the prevalence of OAG [14]. In their study, among people over 80 years of age, Latinos (AMR) had the highest OAG prevalence, followed by blacks (AFR), whites (EUR), South Asians, and East Asians, and this distribution was consistent with our genetic risk score results. For people with an age of 40 years, the population prevalence of OAG was highest in Africans (4.5%), followed by Americans (3.5%), Europeans (2.1%), South Asians (2.0%), and East Asians (1.8%).
The advantage of our study is that we used the data of IOP-related SNPs in OAG, derived from a study using GERA cohort [32], a large multi-ethnic study for identifying novel loci related to IOP [33], and meta-analysis results of IGGC, ANZRAG, and UKBB [23–26]. Although NTG is considered as a type of OAG with an IOP ≤ 21 mmHg, controversy exists regarding whether NTG should be regarded as a disease within the spectrum of OAG or a distinctive disease. The focus of this study was not on the pathogenesis of NTG but on the SNPs known to be related to IOP in OAG and the prevalence of OAG with high IOP. In countries other than Asia, there are limited data on NTG proportion [12], but our study showed that the IOP-dependent mechanism in OAG could be explained according to populations of diverse ancestry. On the aspect of allele frequency, the IOP-dependent mechanism was highest in Europeans, followed by South Asians, Americans, Africans, and finally, East Asians. These results suggested that the IOP-dependent mechanism was less prominent in Koreans and Japanese.
Our results suggested that IOP-related SNPs for African ancestry might have been less enrolled, as SNPs were different because most of SNPs were derived from Europeans studies. A recent study on the association of genetic variants with OAG with individuals with African ancestry reported that rs59892895T > C risk allele was the appreciable frequency in Africans, but not in Europeans or Asians [38]. As these alleles were not included in our study, then other results would be expected, considering the prevalence of OAG in African ancestry. In addition, further studies on glaucoma cohorts of Asians and Americans are necessary to identify the differences in genetic variants with respect to those associated with African ancestry.
A recent study on multi-trait analysis of glaucoma used polygenic prediction for glaucoma progression in early manifest glaucoma cases and surgical intervention in advanced glaucoma cases, which could facilitate the development of a personalized approach for treatment [39]. In addition, another study showed that the association of polygenic risk score with IOP improved the prediction of OAG [40]. Our findings also showed a trend in genetic risk and glaucoma prevalence. These results demonstrated the importance of IOP as well as genetic factors in glaucoma development and progression. The particular contributions of the genes to the pathogenesis of OAG, however, remain to be elucidated. Moreover, the major heritable component of OAG is still unexplained. Further studies are necessary for glaucoma specialists to discover genetic variants to explain the identified associations and investigate any gene–gene or gene-environment interactions.
This study may insight the prevalence difference of OAG and NTG according to region and country. In European descent, most glaucoma is known to be POAG with high IOP, and POAG with normal IOP is known to be the minority. However, in East Asians, especially in Japan and Korea, most of the OAG is NTG. Since glaucoma treatment is a management of lowering IOP and other risk factors, understanding the genetic difference in SNP frequencies with IOP would provide sufficient insights regarding the pathogenesis of glaucoma for clinical ophthalmologists and glaucoma specialists. A major strength of our study was the inclusion of the large Korean whole-genome data (n = 1722) to reflect the allele frequency of SNPs related to OAG and OAG with high IOP. Additionally, we did not systematically organize the new glaucoma cohort and analyze the effects; instead, we compared the 1000 Genomes Project data with OAG-related SNP data from the GWAS catalog. However, there are a few limitations to this study. First, the GWAS catalog contains data for which the risk allele is not clearly defined in the minor allele frequency (MAF). However, we did not exclude these in our study because the majority of MAFs are likely to be risk alleles, so removing all of the undefined alleles would result in inaccurate subgroup analysis. Further study is needed for the data curation of 32 undefined SNPs. To solve this problem, risk allele curation is necessary for the GWAS catalog, based on further results of large population studies using glaucoma cohorts. Second, the statistical significance of EAF in Koreans was high and should be interpreted with caution since the Fisher’s test can decrease the P-value as the number of subjects increases, even with the same odds ratio values as the Korean reference genome number was 1722, which was very high, assuming that the genome number of 26 populations belonging to the 1000 Genomes Project was about 100, ranging from 61 to 113. Third, we used the composite risk score that did not include the effect size weights, as the weighted-odd ratios vary according to the ethnic group even for the same SNP, and there are inaccuracies due to insufficient study data on OAG-related SNPs in the Africans and Asians population. In the future, the polygenic risk score with the effect size weighted odd ratio will be a more desirable study. Fourth, our study analyzed what is currently known; it is not a study that shows new genetic loci or pathway analysis using cell culture and animal study. Additionally, our findings should be interpreted in consideration of the fact that the penetrance is variable even when causative SNPs are present in specific individuals. Nevertheless, it would be meaningful to understand genetic research from the point of view of a clinical glaucoma specialist.
Conclusions
Our study showed substantial population differentiation in allele frequencies in both of OAG-related SNPs and IOP-related SNPs in OAG. From the allele frequency of these SNPs, we calculated the composite risk scores for OAG and OAG with high IOP for 26 ethnic groups in the 1000 Genomes Project and Koreans. In addition, the prevalence of OAG and OAG with high IOP correlated with genetic risk scores. We observed differences in allele frequencies associated with SNPs related to IOP in OAG between Koreans and other populations of diverse ancestry, which may explain the high prevalence of OAG with normal IOP predominantly in Koreans and East Asians.
Supplementary Information
Additional file 1. Figure S1. A heatmap generated using open-angle glaucoma-related single nucleotide polymorphisms in the global population and the East Asian population. Figure S2. A Heatmap generated using open-angle glaucoma-related single nucleotide polymorphisms in the East Asian population
Additional file 2. Table S1. Effect allele frequencies (EAFs) of open-angle glaucoma related single nucleotide polymorphisms in populations of diverse ancestry including Koreans.
Additional file 3. Table S2. Effect allele frequencies (EAFs) of open-angle glaucoma-related single nucleotide polymorphisms in East Asian groups, including Koreans.
Acknowledgements
This study was conducted with bio resources from the National Biobank of Korea, the Center for Disease Control and Prevention, Republic of Korea (KBN-2019-053).
Abbreviations
- AFR
African
- AMR
American
- ANZRAG
Australian and New Zealand Registry of Advanced Glaucoma
- EAFs
Effect allele frequencies
- EAS
East Asian
- EUR
European
- GWAS
Genome-wide association studies
- IGGC
International Glaucoma Genetics Consortium
- IOP
Intraocular pressure
- ISGEO
International Society of Geographical and Epidemiological Ophthalmology
- KOR
Korean
- KRGDB
Korean Reference Genome Database
- MAF
Minor allele frequency
- NTG
Normal-tension glaucoma
- OAG
Open-angle glaucoma
- SAS
South Asian
- SNP
Single nucleotide polymorphism
- UKBB
UK Biobank
Authors' contributions
HTS, and JHS participated in the study concept and design. HTS, BWY, and JHS contributed to acquisition, analysis, or interpretation of data. HTS carried out statistical analysis, administrative, technical, or material support. HTS, BWY, and JHS wrote and revise the manuscript. HTS and JHS did study supervision. All authors read and approved the final manuscript.
Funding
This study was supported by a Veterans Health Service Medical Center Research Grant (grant number: VHSMC19022). The sponsor or funding organization had no role in the design or conduct of this research.
Availability of data and materials
The raw datasets generated and analyzed during the current study are not publicly available since any data providing the whole-genome sequencing data is considered to be personal property by the Korea Bioethics law. However, the raw whole-genome sequencing data for research are available at the reasonable request under the permission of the National Biobank of Korea contact at [http://nih.go.kr/biobank/cmm/main/mainPage.do?/] and e-mail [biobank@korea.kr]. The allele frequency of Korea reference genome data base (KRGDB) is available [http://152.99.75.168:9090/KRGDBDN/dnKRGinput.jsp], files required are all three of ‘the totally merged sets’ of common variants, rare variants, and indels. The 1000genomes data is available, all the files from the following folder were downloaded, [ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/] (last accessed: January 15, 2020). The genome-wide association study (GWAS) catalog data is available in the (NHGRI-EBI, [https://www.ebi.ac.uk/gwas/docs/file-downloads], “All associations v1.0.2—with added ontology annotations, GWAS Catalog study accession numbers and genotyping technology”, December 2019).
Declarations
Ethics approval and consent to participate
This study was approved and monitored by the Institutional Review Board (IRB) of the Veterans Health Service Medical Center, Korea (IRB No. 2019-07-008) and was conducted in accordance with the Declaration of Helsinki. HTS approved administrative permission after the committee review of the National Biobank of Korea (KBN-2019–053) to access the raw data of Korea Reference Genome Data Base under Korea Bioethics law.
Informed consent for this study was waived by the Institutional Review Board (IRB) of the Veterans Health Service Medical Center, Korea (IRB No. 2019-07-008), because the National Biobank of Korea had obtained informed consent to participate for general research and de-identified data granted by National Biobank of Korea by Korea Bioethics law.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hyun-Tae Shin and Byung Woo Yoon contributed equally to this work
References
- 1.Quigley HA. Glaucoma Lancet. 2011;377:1367–1377. doi: 10.1016/S0140-6736(10)61423-7. [DOI] [PubMed] [Google Scholar]
- 2.Tielsch JM, Sommer A, Katz J, Royall RM, Quigley HA, Javitt J. Racial variations in the prevalence of primary open-angle glaucoma. Baltimore Eye Survey JAMA. 1991;266:369–374. [PubMed] [Google Scholar]
- 3.Klein BE, Klein R, Sponsel WE, Franke T, Cantor LB, Martone J, Menage MJ. Prevalence of glaucoma. The Beaver Dam Eye Study Ophthalmology. 1992;99:1499–1504. doi: 10.1016/s0161-6420(92)31774-9. [DOI] [PubMed] [Google Scholar]
- 4.Wensor MD, McCarty CA, Stanislavsky YL, Livingston PM, Taylor HR. The prevalence of glaucoma in the Melbourne Visual Impairment Project. Ophthalmology. 1998;105:733–739. doi: 10.1016/S0161-6420(98)94031-3. [DOI] [PubMed] [Google Scholar]
- 5.Bonomi L, Marchini G, Marraffa M, Bernardi P, De Franco I, Perfetti S, Varotto A. Epidemiology of angle-closure glaucoma: prevalence, clinical types, and association with peripheral anterior chamber depth in the Egna-Neumarket Glaucoma Study. Ophthalmology. 2000;107:998–1003. doi: 10.1016/s0161-6420(00)00022-1. [DOI] [PubMed] [Google Scholar]
- 6.Quigley HA, West SK, Rodriguez J, Munoz B, Klein R, Snyder R. The prevalence of glaucoma in a population-based study of Hispanic subjects: Proyecto VER. Arch Ophthalmol. 2001;119:1819–1826. doi: 10.1001/archopht.119.12.1819. [DOI] [PubMed] [Google Scholar]
- 7.Day AC, Baio G, Gazzard G, Bunce C, Azuara-Blanco A, Munoz B, Friedman DS, Foster PJ. The prevalence of primary angle closure glaucoma in European derived populations: a systematic review. Br J Ophthalmol. 2012;96:1162–1167. doi: 10.1136/bjophthalmol-2011-301189. [DOI] [PubMed] [Google Scholar]
- 8.Foster PJ, Oen FT, Machin D, Ng TP, Devereux JG, Johnson GJ, Khaw PT, Seah SK. The prevalence of glaucoma in Chinese residents of Singapore: a cross-sectional population survey of the Tanjong Pagar district. Arch Ophthalmol. 2000;118:1105–1111. doi: 10.1001/archopht.118.8.1105. [DOI] [PubMed] [Google Scholar]
- 9.Stein JD, Kim DS, Niziol LM, Talwar N, Nan B, Musch DC, Richards JE. Differences in rates of glaucoma among Asian Americans and other racial groups, and among various Asian ethnic groups. Ophthalmology. 2011;118:1031–1037. doi: 10.1016/j.ophtha.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mwanza JC, Tulenko SE, Barton K, Herndon LW, Mathenge E, Hall A, Kim HY, Hay-Smith G, Budenz DL. Eight-year incidence of open-angle glaucoma in the tema eye survey. Ophthalmology. 2019;126:372–380. doi: 10.1016/j.ophtha.2018.10.016. [DOI] [PubMed] [Google Scholar]
- 11.Budenz DL, Barton K, Whiteside-de Vos J, Schiffman J, Bandi J, Nolan W, Herndon L, Kim H, Hay-Smith G, Tielsch JM, Tema Eye Survey Study G Prevalence of glaucoma in an urban West African population: the Tema Eye Survey. JAMA Ophthalmol. 2013;131:651–658. doi: 10.1001/jamaophthalmol.2013.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim KE, Park KH. Update on the prevalence, etiology, diagnosis, and monitoring of normal-tension glaucoma. Asia Pac J Ophthalmol (Phila) 2016;5:23–31. doi: 10.1097/APO.0000000000000177. [DOI] [PubMed] [Google Scholar]
- 13.Rudnicka AR, Mt-Isa S, Owen CG, Cook DG, Ashby D. Variations in primary open-angle glaucoma prevalence by age, gender, and race: a Bayesian meta-analysis. Invest Ophthalmol Vis Sci. 2006;47:4254–4261. doi: 10.1167/iovs.06-0299. [DOI] [PubMed] [Google Scholar]
- 14.Kapetanakis VV, Chan MP, Foster PJ, Cook DG, Owen CG, Rudnicka AR. Global variations and time trends in the prevalence of primary open angle glaucoma (POAG): a systematic review and meta-analysis. Br J Ophthalmol. 2016;100:86–93. doi: 10.1136/bjophthalmol-2015-307223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamamoto S, Sawaguchi S, Iwase A, Yamamoto T, Abe H, Tomita G, Tomidokoro A, Araie M. Primary open-angle glaucoma in a population associated with high prevalence of primary angle-closure glaucoma: the Kumejima Study. Ophthalmology. 2014;121:1558–1565. doi: 10.1016/j.ophtha.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Iwase A, Suzuki Y, Araie M, Yamamoto T, Abe H, Shirato S, Kuwayama Y, Mishima HK, Shimizu H, Tomita G, Inoue Y, Kitazawa Y, Tajimi Study Group JGS The prevalence of primary open-angle glaucoma in Japanese: the Tajimi Study. Ophthalmology. 2004;111:1641–1648. doi: 10.1016/j.ophtha.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 17.Song W, Shan L, Cheng F, Fan P, Zhang L, Qu W, Zhang Q, Yuan H. Prevalence of glaucoma in a rural northern china adult population: a population-based survey in kailu county, inner mongolia. Ophthalmology. 2011;118:1982–1988. doi: 10.1016/j.ophtha.2011.02.050. [DOI] [PubMed] [Google Scholar]
- 18.Kim CS, Seong GJ, Lee NH, Song KC, Namil Study Group KGS Prevalence of primary open-angle glaucoma in central South Korea the Namil study. Ophthalmology. 2011;118:1024–1030. doi: 10.1016/j.ophtha.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 19.Sommer A, Tielsch JM, Katz J, Quigley HA, Gottsch JD, Javitt JC, Martone JF, Royall RM, Witt KA, Ezrine S. Racial differences in the cause-specific prevalence of blindness in east Baltimore. N Engl J Med. 1991;325:1412–1417. doi: 10.1056/NEJM199111143252004. [DOI] [PubMed] [Google Scholar]
- 20.Rotchford AP, Johnson GJ. Glaucoma in Zulus: a population-based cross-sectional survey in a rural district in South Africa. Arch Ophthalmol. 2002;120:471–478. doi: 10.1001/archopht.120.4.471. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Harmon J, Zabrieskie N, Chen Y, Grob S, Williams B, Lee C, Kasuga D, Shaw PX, Buehler J, Wang N, Zhang K. Using the Utah Population Database to assess familial risk of primary open angle glaucoma. Vision Res. 2010;50:2391–2395. doi: 10.1016/j.visres.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 22.Sung VC, Koppens JM, Vernon SA, Pawson P, Rubinstein M, King AJ, Tattersall CL. Longitudinal glaucoma screening for siblings of patients with primary open angle glaucoma: the Nottingham Family Glaucoma Screening Study. Br J Ophthalmol. 2006;90:59–63. doi: 10.1136/bjo.2005.072751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ozel AB, Moroi SE, Reed DM, Nika M, Schmidt CM, Akbari S, Scott K, Rozsa F, Pawar H, Musch DC, Lichter PR, Gaasterland D, Branham K, Gilbert J, Garnai SJ, Chen W, Othman M, Heckenlively J, Swaroop A, Abecasis G, Friedman DS, Zack D, Ashley-Koch A, Ulmer M, Kang JH, Consortium N, Liu Y, Yaspan BL, Haines J, Allingham RR, Hauser MA, Pasquale L, Wiggs J, Richards JE, Li JZ Genome-wide association study and meta-analysis of intraocular pressure. Hum Genet. 2014;133:41–57. doi: 10.1007/s00439-013-1349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiggs JL, Hauser MA, Abdrabou W, Allingham RR, Budenz DL, Delbono E, Friedman DS, Kang JH, Gaasterland D, Gaasterland T, Lee RK, Lichter PR, Loomis S, Liu Y, McCarty C, Medeiros FA, Moroi SE, Olson LM, Realini A, Richards JE, Rozsa FW, Schuman JS, Singh K, Stein JD, Vollrath D, Weinreb RN, Wollstein G, Yaspan BL, Yoneyama S, Zack D, Zhang K, Pericak-Vance M, Pasquale LR, Haines JL. The NEIGHBOR consortium primary open-angle glaucoma genome-wide association study: rationale, study design, and clinical variables. J Glaucoma. 2013;22:517–525. doi: 10.1097/IJG.0b013e31824d4fd8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacGregor S, Ong JS, An J, Han X, Zhou T, Siggs OM, Law MH, Souzeau E, Sharma S, Lynn DJ, Beesley J, Sheldrick B, Mills RA, Landers J, Ruddle JB, Graham SL, Healey PR, White AJR, Casson RJ, Best S, Grigg JR, Goldberg I, Powell JE, Whiteman DC, Radford-Smith GL, Martin NG, Montgomery GW, Burdon KP, Mackey DA, Gharahkhani P, Craig JE, Hewitt AW. Genome-wide association study of intraocular pressure uncovers new pathways to glaucoma. Nat Genet. 2018;50:1067–1071. doi: 10.1038/s41588-018-0176-y. [DOI] [PubMed] [Google Scholar]
- 26.Gao XR, Huang H, Nannini DR, Fan F, Kim H. Genome-wide association analyses identify new loci influencing intraocular pressure. Hum Mol Genet. 2018;27:2205–2213. doi: 10.1093/hmg/ddy111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J, Weber JA, Jho S, Jang J, Jun J, Cho YS, Kim HM, Kim H, Kim Y, Chung O, Kim CG, Lee H, Kim BC, Han K, Koh I, Chae KS, Lee S, Edwards JS, Bhak J. KoVariome: Korean National Standard Reference Variome database of whole genomes with comprehensive SNV, indel, CNV, and SV analyses. Sci Rep. 2018;8:5677. doi: 10.1038/s41598-018-23837-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foster PJ, Buhrmann R, Quigley HA, Johnson GJ. The definition and classification of glaucoma in prevalence surveys. Br J Ophthalmol. 2002;86:238–242. doi: 10.1136/bjo.86.2.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mao L, Fang Y, Campbell M, Southerland WM. Population differentiation in allele frequencies of obesity-associated SNPs. BMC Genomics. 2017;18:861. doi: 10.1186/s12864-017-4262-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choquet H, Paylakhi S, Kneeland SC, Thai KK, Hoffmann TJ, Yin J, Kvale MN, Banda Y, Tolman NG, Williams PA, Schaefer C, Melles RB, Risch N, John SWM, Nair KS, Jorgenson E. A multiethnic genome-wide association study of primary open-angle glaucoma identifies novel risk loci. Nat Commun. 2018;9:2278. doi: 10.1038/s41467-018-04555-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choquet H, Thai KK, Yin J, Hoffmann TJ, Kvale MN, Banda Y, Schaefer C, Risch N, Nair KS, Melles R, Jorgenson E. A large multi-ethnic genome-wide association study identifies novel genetic loci for intraocular pressure. Nat Commun. 2017;8:2108. doi: 10.1038/s41467-017-01913-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen F, Klein AP, Klein BE, Lee KE, Truitt B, Klein R, Iyengar SK, Duggal P. Exome array analysis identifies CAV1/CAV2 as a susceptibility locus for intraocular pressure. Invest Ophthalmol Vis Sci. 2014;56:544–551. doi: 10.1167/iovs.14-15204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bailey JN, Loomis SJ, Kang JH, Allingham RR, Gharahkhani P, Khor CC, Burdon KP, Aschard H, Chasman DI, Igo RP, Jr., Hysi PG, Glastonbury CA, Ashley-Koch A, Brilliant M, Brown AA, Budenz DL, Buil A, Cheng CY, Choi H, Christen WG, Curhan G, De Vivo I, Fingert JH, Foster PJ, Fuchs C, Gaasterland D, Gaasterland T, Hewitt AW, Hu F, Hunter DJ, Khawaja AP, Lee RK, Li Z, Lichter PR, Mackey DA, McGuffin P, Mitchell P, Moroi SE, Perera SA, Pepper KW, Qi Q, Realini T, Richards JE, Ridker PM, Rimm E, Ritch R, Ritchie M, Schuman JS, Scott WK, Singh K, Sit AJ, Song YE, Tamimi RM, Topouzis F, Viswanathan AC, Verma SS, Vollrath D, Wang JJ, Weisschuh N, Wissinger B, Wollstein G, Wong TY, Yaspan BL, Zack DJ, Zhang K, Study EN, Consortium A, Weinreb RN, Pericak-Vance MA, Small K, Hammond CJ, Aung T, Liu Y, Vithana EN, MacGregor S, Craig JE, Kraft P, Howell G, Hauser MA, Pasquale LR, Haines JL, Wiggs JL Genome-wide association analysis identifies TXNRD2, ATXN2 and FOXC1 as susceptibility loci for primary open-angle glaucoma. Nat Genet. 2016;48:189–194. doi: 10.1038/ng.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carlson CS, Matise TC, North KE, Haiman CA, Fesinmeyer MD, Buyske S, Schumacher FR, Peters U, Franceschini N, Ritchie MD, Duggan DJ, Spencer KL, Dumitrescu L, Eaton CB, Thomas F, Young A, Carty C, Heiss G, Le Marchand L, Crawford DC, Hindorff LA, Kooperberg CL, Consortium P Generalization and dilution of association results from European GWAS in populations of non-European ancestry: the PAGE study. PLoS Biol. 2013;11:e1001661. doi: 10.1371/journal.pbio.1001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abu-Amero K, Kondkar AA, Chalam KV. An updated review on the genetics of primary open angle glaucoma. Int J Mol Sci. 2015;16:28886–28911. doi: 10.3390/ijms161226135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Genetics of Glaucoma in People of African Descent C, Hauser MA, Allingham RR, Aung T, Van Der Heide CJ, Taylor KD, Rotter JI, Wang SJ, Bonnemaijer PWM, Williams SE, Abdullahi SM, Abu-Amero KK, Anderson MG, Akafo S, Alhassan MB, Asimadu I, Ayyagari R, Bakayoko S, Nyamsi PB, Bowden DW, Bromley WC, Budenz DL, Carmichael TR, Challa P, Chen YI, Chuka-Okosa CM, Cooke Bailey JN, Costa VP, Cruz DA, DuBiner H, Ervin JF, Feldman RM, Flamme-Wiese M, Gaasterland DE, Garnai SJ, Girkin CA, Guirou N, Guo X, Haines JL, Hammond CJ, Herndon L, Hoffmann TJ, Hulette CM, Hydara A, Igo RP, Jr., Jorgenson E, Kabwe J, Kilangalanga NJ, Kizor-Akaraiwe N, Kuchtey RW, Lamari H, Li Z, Liebmann JM, Liu Y, Loos RJF, Melo MB, Moroi SE, Msosa JM, Mullins RF, Nadkarni G, Napo A, Ng MCY, Nunes HF, Obeng-Nyarkoh E, Okeke A, Okeke S, Olaniyi O, Olawoye O, Oliveira MB, Pasquale LR, Perez-Grossmann RA, Pericak-Vance MA, Qin X, Ramsay M, Resnikoff S, Richards JE, Schimiti RB, Sim KS, Sponsel WE, Svidnicki PV, Thiadens A, Uche NJ, van Duijn CM, de Vasconcellos JPC, Wiggs JL, Zangwill LM, Risch N, Milea D, Ashaye A, Klaver CCW, Weinreb RN, Ashley Koch AE, Fingert JH, Khor CC Association of genetic variants with primary open-angle glaucoma among individuals with African ancestry. JAMA. 2019;322:1682–1691. doi: 10.1001/jama.2019.16161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Craig JE, Han X, Qassim A, Hassall M, Cooke Bailey JN, Kinzy TG, Khawaja AP, An J, Marshall H, Gharahkhani P, Igo RP, Jr, Graham SL, Healey PR, Ong JS, Zhou T, Siggs O, Law MH, Souzeau E, Ridge B, Hysi PG, Burdon KP, Mills RA, Landers J, Ruddle JB, Agar A, Galanopoulos A, White AJR, Willoughby CE, Andrew NH, Best S, Vincent AL, Goldberg I, Radford-Smith G, Martin NG, Montgomery GW, Vitart V, Hoehn R, Wojciechowski R, Jonas JB, Aung T, Pasquale LR, Cree AJ, Sivaprasad S, Vallabh NA, consortium N, Eye UKB, Vision C, Viswanathan AC, Pasutto F, Haines JL, Klaver CCW, van Duijn CM, Casson RJ, Foster PJ, Khaw PT, Hammond CJ, Mackey DA, Mitchell P, Lotery AJ, Wiggs JL, Hewitt AW, MacGregor S, Multitrait analysis of glaucoma identifies new risk loci and enables polygenic prediction of disease susceptibility and progression. Nat Genet. 2020 doi: 10.1038/s41588-019-0556-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao XR, Huang H, Kim H. Polygenic Risk Score Is Associated With Intraocular Pressure and Improves Glaucoma Prediction in the UK Biobank Cohort. Transl Vis Sci Technol. 2019;8:10. doi: 10.1167/tvst.8.2.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Figure S1. A heatmap generated using open-angle glaucoma-related single nucleotide polymorphisms in the global population and the East Asian population. Figure S2. A Heatmap generated using open-angle glaucoma-related single nucleotide polymorphisms in the East Asian population
Additional file 2. Table S1. Effect allele frequencies (EAFs) of open-angle glaucoma related single nucleotide polymorphisms in populations of diverse ancestry including Koreans.
Additional file 3. Table S2. Effect allele frequencies (EAFs) of open-angle glaucoma-related single nucleotide polymorphisms in East Asian groups, including Koreans.
Data Availability Statement
The raw datasets generated and analyzed during the current study are not publicly available since any data providing the whole-genome sequencing data is considered to be personal property by the Korea Bioethics law. However, the raw whole-genome sequencing data for research are available at the reasonable request under the permission of the National Biobank of Korea contact at [http://nih.go.kr/biobank/cmm/main/mainPage.do?/] and e-mail [biobank@korea.kr]. The allele frequency of Korea reference genome data base (KRGDB) is available [http://152.99.75.168:9090/KRGDBDN/dnKRGinput.jsp], files required are all three of ‘the totally merged sets’ of common variants, rare variants, and indels. The 1000genomes data is available, all the files from the following folder were downloaded, [ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/] (last accessed: January 15, 2020). The genome-wide association study (GWAS) catalog data is available in the (NHGRI-EBI, [https://www.ebi.ac.uk/gwas/docs/file-downloads], “All associations v1.0.2—with added ontology annotations, GWAS Catalog study accession numbers and genotyping technology”, December 2019).