Abstract
Background
In the novel coronavirus pandemic, the high infection rate and high mortality have seriously affected people’s health and social order. To better explore the infection mechanism and treatment, the three-dimensional structure of human bronchus has been employed in a better in-depth study on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
Methods
We downloaded a separate microarray from the Integrated Gene Expression System (GEO) on a human bronchial organoids sample to identify differentially expressed genes (DEGS) and analyzed it with R software. After processing with R software, Gene Ontology (GO) and Kyoto PBMCs of Genes and Genomes (KEGG) were analyzed, while a protein–protein interaction (PPI) network was constructed to show the interactions and influence relationships between these differential genes. Finally, the selected highly connected genes, which are called hub genes, were verified in CytoHubba plug-in.
Results
In this study, a total of 966 differentially expressed genes, including 490 upregulated genes and 476 downregulated genes were used. Analysis of GO and KEGG revealed that these differentially expressed genes were significantly enriched in pathways related to immune response and cytokines. We construct protein-protein interaction network and identify 10 hub genes, including IL6, MMP9, IL1B, CXCL8, ICAM1, FGF2, EGF, CXCL10, CCL2, CCL5, CXCL1, and FN1. Finally, with the help of GSE150728, we verified that CXCl1, CXCL8, CXCL10, CCL5, EGF differently expressed before and after SARS-CoV-2 infection in clinical patients.
Conclusions
In this study, we used mRNA expression data from GSE150819 to preliminarily confirm the feasibility of hBO as an in vitro model to further study the pathogenesis and potential treatment of COVID-19. Moreover, based on the mRNA differentiated expression of this model, we found that CXCL8, CXCL10, and EGF are hub genes in the process of SARS-COV-2 infection, and we emphasized their key roles in SARS-CoV-2 infection. And we also suggested that further study of these hub genes may be beneficial to treatment, prognostic prediction of COVID-19.
Keywords: Immune response, Human bronchial organoids, Novel coronavirus infection, Bioinformatics analysis, 3D structure model
Introduction
In late December 2019, Wuhan, China, reported an epidemic situation of viral pneumonia caused by unknown microbial pathogens. Its clinical manifestations are very similar to those of viral pneumonia, including fever, cough, shortness of breath, myalgia, and fatigue [1].
On January 7, the pathogen, which was now named as SARS-CoV-2 [2], of this outbreak was identified by the Chinese Center for Disease Control and Prevention (CDC) from a throat swab sample of a patient [3].
SARS-CoV-2 is a new member of the Coronaviridae family [4]. Similar to other coronaviruses, the main routes of transmissions are respiratory droplets, respiratory secretions, and direct contact [5]. Later, it was reported that SARS-CoV-2 could be isolated from fecal swabs, which explains its rapid spread. The rapid spread and high mortality are responsible for the massive global outbreak followed by the outbreak in China. Based on the situation, the World Health Organization (WHO) declared the outbreak to be a public health emergency of international concern on January 31, 2020 [6]. As of October 15, 2020, there are a total of 38,394,169 confirmed SARS-CoV-2 cases worldwide, including 1,089,047 deaths [7].
Because the novel coronavirus has a huge impact on social order, the global economy and, especially, people’s health, a large number of experiments have been done to study the susceptibility and mechanism of SARS-CoV-2. Many infection models were used to study the reaction and immunity of SARS-CoV-2 infected bodies and these models played an important role in these studies [8–13]. However, all the commonly used models have a limitation. Either they are suboptimal models to represent human bodies, or they are limited in number [10, 11, 14].
In order to better understand the interaction between human body and different pathogens, some gene expression research conducted in vivo or in affected patients has established a three-dimensional structural organ model composed of human stem cells [15–17]. These models have reproduced various cell components and the morphology as well as an arrangement of each kind of cell. They are believed to help in understanding the path mechanism and developing drugs and vaccines [15, 18]. Such an advantaged model was also used to learn about SARS-CoV-2. By now, organoids of different apparatuses have been built and these organoids greatly improved researchers’ knowledge toward SARS-CoV-2. A study of intestinal tract organoids indicated that people get infected by SARS-CoV-2 through the intestinal tract, and studies of nervous system-related organoids may explain SARS-CoV-2-infected patients’ neurologic symptoms [8, 9]. All these studies showed that organoids could be good models to study SARS-CoV-2.
Therefore, we hope to explore the interaction between SARS-COV-2 and human body through the bioinformatics analysis of human bronchial organoids’ mRNA expression data from GSE150819 data set and preliminarily explore whether human bronchial organoids can be used as an in vitro model for the study of SARS-COV-2 infection.
Materials and methods
Gene expression datasets
We searched the GEO (https://www.ncbi.nlm.nih.gov/gds/) database to find relevant datasets for our study. Dataset GSE150819 contains genomic information of human bronchial organoids, which we are interested in. Among all samples in GSE150819, 6 samples are available for our study including 3 samples from SARS-CoV-2 infected organoids and 3 from uninfected organoids, and these 6 samples were all sequenced by GPL24676 Illumina nova seq 6000 (homo sapiens) platform. The sample source is bronchial organoid. The detailed information is listed in Table 1.
Table 1.
Details of the data sources for this study
Identification of DEGs
After the gene expression data were obtained, we analyzed the data using the R package (R Foundation for Statistic Computing). Fold change (FC), P value, and false-positive rate were calculated to define DEGs between SARS-CoV-2 infected organoid samples and uninfected organoid samples. Finally, in the R language program, we selected the DEGs with logFC>1 (upregulated gene) or logFC<−1 (downregulated gene) calculated, while P < 0.05 was considered to be a significant difference and could be used to reduce the false-positive rate.
Functional enrichment analysis of DEGs
The Gene Ontology (GO) database, containing three sections: biological progress (BP), cellular component (CC), molecular function (MF), and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database are the most popular tools to learn about gene functions [19]. Therefore, two databases and the reference value P value < 0.05, which was considered as the significant difference, were used to further study the function of DEGs.
PPI network construction and hub gene analysis
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (https://string-db.org/) is an online database collecting a large amount of protein interaction information. To learn about protein interaction, the STRING online tool was used. Protein interaction is admitted when the combined score was >0.4. We visualized the protein-protein interaction (PPI) network with the usage of Cytoscape software. To study protein interaction further, we used a plug-in to filter to hub genes. And they get the highest degree in the CytoHubba plug-in analysis, which means having the highest degree of connectivity within the PPI network [20].
Validation of gene expression levels
We obtained the PPI network action diagram of differential gene expression, and screened out the central genes through the CytoHubba plug-in. These genes, as key genes, are the genes with the highest degree of connectivity or connecting multiple modules in the PPI network diagram. At the same time, we selected GSE150728 to detect peripheral blood mononuclear cells (PBMCs) and verify the expression of pivotal genes. The expression values of the corresponding central genes were found from the expression matrix and organized into a table, which was then produced into a verification map using the GraphPad Prism8 software.
Results
Identification of DEGs
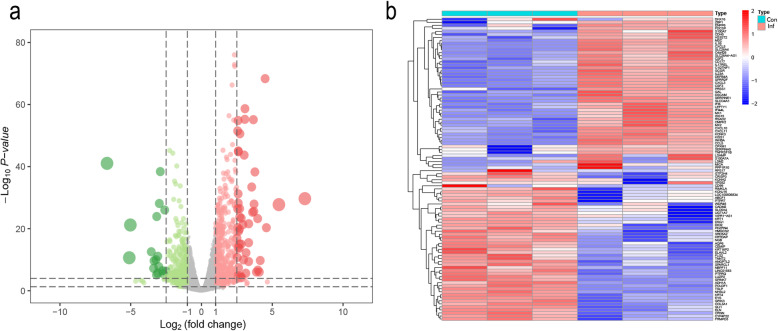
A data set (GSE150819) was obtained from the GEO database and analyzed in the R language. We found 966 DEGs between SARS-CoV-2 infected and uninfected group in total, among which 490 genes were upregulated and 476 genes were downregulated (Table 2). Volcano plot and heatmap of DEGs were also established. A volcanic map was generated to show up (red) and down (green) genes between infection and normal control (Fig. 1a). Afterwards, we used the heatmap package in R to plot a heatmap based on the expression levels of DEGs in GSE150819 (Fig. 1b).
Table 2.
Upregulated genes and downregulated genes that meet the screening criteria
| DEGs | Gene terms |
|---|---|
| Upregulated | KPNA7,RAB32,TMEM132A,CD7,A1BG-AS1,CDH2,ANGPTL4,BAMBI,ULBP1,DDX58,CYP8B1,CSTB,PNMA2,NEFM,SOCS1,NCF2,LINC00152,SOD2,THBS1,CYR61,HCG4B,IFITM2,COL6A2,TAGLN,TGFBR3L,TINAGL1,RASGRP3,TNFAIP2,ST6GALNAC5,GOLT1A,NALCN,TMIE,ABCA12,LRRC3,HMGA2,RELT,MTNR1A,CAPN14,ELOVL3,CEACAM19,GRAP,DGKI,NLRC4,BST2,DDIT4L,GBP4,HGD,ISG20,ZBED2,FEZ1,TMEM92,PRSS30P,FTH1,C10orf55,SIRPB2,MTHFD2,ARL4C,SUSD2,ITGB2,THY1,FUOM,RAB3B,KIF12,CBS,CPXM2,IFITM3,GAS6,LOC644936,MMD,LCP1,CTGF,FNDC4,EGOT,NRIP3,GFI1,WNT11,ARL9,ZC3H12A,MOGAT2,RCN3,IL2RG,MPV17L,TMEM106A,TMEM163,ADM2,LINC01215,FAP,SLC2A6,KRT7,HSD17B14,GREB1L,UCHL1,ALOXE3,NCF1,S100A12,RAC2,CYGB,ITGA5,NDP,PTAFR,NWD2,RFTN1,FAM155B,ALOX15B,REN,BATF2,PNMAL2,CPNE4,MEOX1,IRF7,ZNF876P,SLC34A2,HSH2D,GLDC,TNFSF14,SLC26A9,HDAC9,NKILA,RASGRF2,BIRC3,EGF,TUBB3,DPYSL4,DDX60,SLC8A2,FSTL3,ADAMTS1,SERPIND1,GSTM3,NFKBIA,TREM1,ADAM8,SCG5,SLC15A3,PARM1,DDO,RHEBL1,TSPAN2,CHD5,LHX5,FBLN7,PDCD1LG2,DUSP5,UCA1,C3,ARSF,BICC1,HIST1H1E,XKR9,PDZK1IP1,SAMD14,BCL2L15,IL24,GRIP2,FZD9,1-Dec,FGF2,MT1L,NOTCH4,IL7,KYNU,RBM24,TMPRSS2,TNIP1,THEMIS2,RENBP,FAM181B,VNN3,LOC101927865,LOC101927571,EDN1,RASD2,USP18,TNFAIP3,CYB5R2,CPA6,MAPK8IP2,SLC39A8,TYMP,DYSF,IER3,XAF1,CSF1R,OAS3,CAMK1G,CD83,IL15,LOC100130476,TRIM36,HKDC1,TAGLN3,RHCG,PDGFRL,PTP4A3,CILP2,ITPKA,LOC100505622,FABP6,PRRX2,CD200,BVES,TRIB3,KLHDC7B,IFI35,CST6,LOC102724279,RNU6-2,LAPTM5,SLC13A5,MFI2,BMP2,KRTAP2-3,KLK8,CCRL2,DHRS2,CD79A,SRGN,KLK9,PTPRH,ZFP42,CXCL6,ZG16B,WFDC5,HSD17B2,HBEGF,MUC3A,SEMA7A,PTGS2,PI3,MAFF,ADAMTS4,MEI1,GRB14,KMO,TMEM171,C6orf223,LCN2,HLA-B,KCNG1,LOC102467080,ATP6V0D2,IFI44,RPSAP52,GAS6-AS2,SERPINB7,OAS2,GPR176,TFPI2,CD70,GGT1,OTOP3,SHISA2,FN1,CTSE,DUOX2,LEMD1,NTM,KCCAT211,ANXA6,MIR146A,SH3RF3,RNASE1,PRSS22,LOC101929705,NODAL,LAMC2,IFIT2,ADAM19,INSC,C3orf70,IGFN1,DAAM2,CXCL1,NOG,HERC6,BPGM,HHIP,FAM169B,NEURL3,AMTN,IL6,PIWIL2,NCF1C,TBXAS1,FPR1,AANAT,PLAUR,CFB,TAF7L,BAG6,CDA,ASPHD1,LGALS2,TRABD2A,ICAM1,CCL2,ESM1,VNN1,4-Mar,KLK6,SLC7A7,HLA-F,NT5E,IL7R,POU2F2,POPDC3,C10orf10,SLC5A1,NOXO1,SELL,SSC4D,ZNF114,STC2,ART3,ALDOB,SAA2,PLAU,IL36G,BEST1,SPRR2D,FAM205A,PARVB,MUC13,SH2D2A,EPSTI1,LIF,TNFRSF9,NES,SAA4,ADRA1B,AWAT2,RASD1,ANXA2P3,ADAMTS9,MMP7,XDH,IL4I1,RAB42,PSG9,LAMP3,ANKRD1,HSD11B1,RND1,TNIP3,NOX1,ADAMTS6,UBD,IL1R2,MPZ,WNT7A,SV2B,IL32,MYCT1,MT2A,BCL2A1,DIO3,IDO2,TRPV2,NTNG1,LOC102723649,CXCL9,SNAI1,SYNGR3,IDO1,OLFM1,KLK5,KCNN3,RNASE7,C15orf48,SPRR2C,APOBEC3A,ROBO4,HEPHL1,SYNDIG1,HMGN2P46,SAA1,CUX2,IFITM1,SLC5A8,B4GALNT2,LINC00880,COL22A1,GFPT2,L1CAM,GDF15,CXCL8,MSC-AS1,IL19,PLA2G4C,TFF1,IFIT3,TRHDE,CXCL2,ATRNL1,CHAC1,G0S2,GBP5,CLLU1OS,SELP,C8orf4,MMP8,ZNF385C,MYH16,FOLR3,PDE4B,MRGPRX3,MMP9,SLC7A2,C19orf38,GRIN2A,MCHR1,PITX3,IFIT1,IL12RB1,COL13A1,IL17C,NCF1B,IL1A,CEACAM7,SMOC1,CCL20,SERPINA3,CD69,IFI27,ALPL,EPS8L3,OASL,LOC100126784,BEAN1,AMZ1,SPRR2A,SLC5A5,PDGFB,SPRR2E,HERC5,EBI3,CDH19,CYP27A1,DUOXA2,LOC101929427,SLC26A4-AS1,SERPINE1,MX1,ISG15,CXCL3,INHBA,HS3ST2,C4orf26,DSCAM,MSC,CD177,KISS1,SLCO4A1,C1QTNF1,CPXM1,MICA,IL17REL,LSAMP,IFI6,LEFTY1,IFI44L,SERPINA5,CXCL10,CXCL5,SPRR2F,S100A7A,GAL,KCNK3,VPS52,PPP1R10,GCGR,S100A7,CCL5,IL1B,IL23A,CMPK2,SLC26A4,MX2,CXCL11,DEFB4A,TNFRSF1B,RSAD2,ZBP1,ENPP6,CDH5,CSF3,CSF2,LSM2,FDCSP,PRSS1 |
| Downregulated | KRT1,DIO2,DSG1,GTF2H4,RPS17,WDR46,DHX16,GRIK3,ABCF1,LUZP2,CYP4F22,UGT1A7,SPINK7,PDZRN4,ANGPTL2,KCNJ16,COL3A1,KRT4,LOC100506834,SRD5A2,CADM2,KCNH2,CRNN,AQP6,HMGCS2,GLI1,VIPR1-AS1,LINC01583,SPARCL1,SLC6A4,KRTDAP,PTPRT,FRMPD3,PTPRQ,TSLP,NHSL2,ADH1A,CEMIP,ELN,FLG2,KRT16P2,POU3F1,NBPF11,TNNT2,ELAVL2,EYS,NGB,RIMKLA,CRISP3,CD99,PLXDC1,EPB41L3,HMCN1,RPTN,APCDD1,AJAP1,IGFBP5,MAP7D2,SDR9C7,SNORD59B,PPP1R42,FAT3,PSAPL1,PRKCB,SLURP1,STEAP3-AS1,KRT15,EVPLL,BGN,ANPEP,POSTN,MPPED2,SMIM17,SLC10A5,EPHA3,SHISA9,C1orf168,SLC1A3,ASXL3,KRT13,RBM20,ANXA10,AADAC,FGF14-AS2,SBSN,MANEA-AS1,CACNA1D,PPP1R16B,RERG,IGFL1,KRT10,LOC101927023,CCDC136,DCLK1,ELFN1,CCDC73,AR,RTN4RL1,LOC730101,HOXA2,HYDIN2,IGFL2,TNNI2,HTR2B,GATA3-AS1,GLI2,SYNPO2,GRM4,HLF,FAM153A,KANTR,SLC24A3,GPR20,MSI1,DAPL1,FAM46C,CXCL14,PCDH18,CLDN8,GPC3,MESP1,CCDC3,DNAH6,SLC47A2,DNASE1L3,COL21A1,TPTE2P1,LINC01121,CYP4Z2P,RTCA-AS1,UMODL1,ANKH,P2RY1,GSTA2,RDH12,DMRT3,ZNF157,CHRNE,CNR1,LY6H,KRT16P3,EPGN,MIR4523,FABP4,TMPRSS11B,GUSBP9,LINC01116,EPHA7,C14orf132,MCM10,H19,FOXP2,APOBEC2,TP73,SMAD5-AS1,MIR4653,SPOCK2,SAMD5,SOSTDC1,MTUS2,TAS2R14,TMEM200A,SNX31,BEND5,DNAJC22,CLIC5,CYP2C8,IL2RB,C2orf73,SYT13,NKAPP1,CILP,TGM3,LOC200772,ARHGEF33,OR7E47P,FAM25A,COLCA1,MAP2K6,TMEM220-AS1,KRT6C,NUDT10,CXCL13,PAK7,PDE6A,EPPIN,HOPX,MFAP4,RNF222,CYP4B1,POU2AF1,SCN2B,MAP6,SEMA3G,PIK3C2G,LOC101929378,ECM2,MBNL1-AS1,LOC101927318,KIT,IL12RB2,SCN9A,PDE3B,KCNH4,SLAMF8,FOXI1,GOLGA8T,C1orf101,LINC01206,MIR27B,CLSTN2,SESN3,HERC2P3,SEMA5A,CLEC18A,ADGRL3,EDN2,FAIM2,CH25H,NLGN4X,ZNF233,DUSP9,DLK2,CTD-2201I18.1,UGT2A1,CDCA7,MIR6753,NEIL3,KNDC1,ERICH3,SEPP1,SOD3,ADGRF5,LINC01094,LOC100507283,MIR4517,GOLGA6L3,KIAA2022,TINCR,LOC100506127,HS3ST6,FOXN1,LOC100286922,LINC01132,ITGA10,PAQR5,RGAG4,GPR63,CPA4,TGFBR3,SPON2,PNPLA1,LOC441178,ATP8B4,EML6,CYP7B1,WSCD2,ACVR2B-AS1,AQP5,RGS22,LOC100129203,SKIDA1,PI16,LOC105373383,ENDOU,UGT2B28,METTL7A,SPTSSB,FKBP5,TET1,FETUB,ARHGAP28,CKAP2L,SNORD31,MPL,E2F2,SALRNA1,BMS1P5,PDGFD,ANXA2R,SNHG21,MIR99AHG,LOC100130093,EXO1,CARD18,UNC5C,BCL11B,DNAH9,CDHR3,C2orf71,CD36,SYT8,IGSF11,OPCML,INO80B,PKMYT1,SPOCK3,USP6,ADH7,MIR200B,LOC100128076,SLC25A34,HRASLS,DMRT2,SEPT5-GP1BB,CENPA,CYP4F3,LOC101929567,ECT2L,BAALC,CYP4F29P,LMO2,LEF1,DEPTOR,HSD17B13,FAM216B,ARHGAP11B,ALS2CR12,ADAD2,AGBL2,CARNS1,KIF11,F5,RGMA,PDE7B,CYP2C9,PALM,UGT2B17,FIBIN,COL5A3,KLHL31,VIPR1,CCDC62,HEY2,SUSD5,LOC100130417,LINC00683,SNHG19,MKI67,IL5RA,GAS1,ADAMTS5,MIR23B,TUB,MROH9,DEPDC7,OR7E91P,JMJD1C-AS1,TDRD1,AQP2,TEN1-CDK3,MIR186,COMP,ADAM23,LGI3,TOP2A,AADACP1,NOS2,SAXO2,ZRANB2-AS1,HEPACAM2,C1orf87,PCSK1,WNT3A,FLRT1,LMNB1,SEMA6D,HECW2,TREX2,LOC100507462,FOXD2,MKNK1-AS1,NWD1,PTGS1,TRPC6,GSTA1,GATA3,RGS4,IL6R,HSPB3,ARL14,FAM19A2,NDC80,CCR7,ACSS3,SPAG8,ITM2A,ZNF853,LOC100131655,GPR78,IL33,CYP4F12,EPHA4,BMP7,IGSF10,CFAP74,DTL,LOC339260,CCNE2,FAM131B,FGFR1,KCNJ5,AQP1,KIF26A,CHGB,TNNT3,C5,CUBN,NFIA,TFF3,PROC,ARHGEF26,TDRD6,CEP55,MBNL3,IL16,LOC100507006,KCNQ5,ZNF367,SNORA63,NPNT,CHRNA7,LINC01410,INHBB,KLLN,PLA2G4F,NCAPG,EGFL6,LGALS4,COL2A1,FZD1,MERTK,DNAAF1,LINC00664,FAM20A,DACT2,KC6,ENTPD8,UCP3,CASC5,EPHA10,RAET1E,ZNF608,AMH,LOC100129917,MYO3B,FAM181A-AS1,KCNAB3,VWA3B,CFAP43,SUSD4,WDR76,GATM,IP6K3,TCF4,FAM65B,LINC01481,DNAH3,KIF18B,POLQ,DLGAP5,STAG3L1,ZFHX4,ABLIM2,ABCA9,LHX6,ITGB7,LOC101928020,PDK4,CYP2W1,C6orf118,FILIP1,LOC100130705,LRGUK,PRUNE2,DTHD1 |
Fig. 1.
Heat map (a) and volcano plot (b) of the differentially expressed genes (DEGs). a Heat map of DEGs. The abscissa axis represents sample types and the ordinate axis represents gene names. b Volcanic plot analysis of upregulated genes and downregulated genes in infected and control groups. In total, 966 DEGs, including 490 upregulated and 476 downregulated DEGs, were identified in the GSE150819 when comparing the infected group and control group. Red indicates log2FC>1 while green indicates log2FC<−1, which means different gene expression between two groups (red for upregulated DEGs and green for downregulated DEGs). And gray indicates no difference
Functional enrichment analysis of DEGs
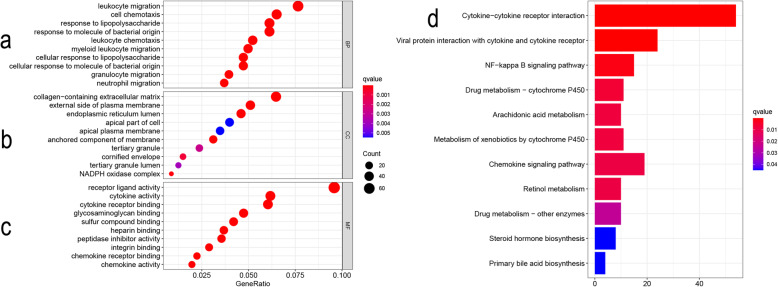
We analyzed the enrichment of DEGs, with two set databases GO and KEGG used. The first 10 biological processes of DEGs enrichment are shown in Fig. 2, including leukocyte migration, cell chemotaxis, and response to lipopolysaccharide (Fig. 2a). With regard to cellular components, the results showed that the collagen containing extra-cellular matrix was enriched, and the importance of extracellular matrix collagen components and plasma membrane in the progression of infection in the COVID-19 was gradually recognized (Fig. 2b). In addition, regarding the classification of molecular functions, DEGs focuses on the participation in the following functions: receptor–ligand activity, cytokine activity, and cytokine receptor binding (Fig. 2c). Importantly, KEGG analysis showed that the overlapping DEGs were mainly enriched in the cytokine receptor interaction and the viral protein interaction with cytokine and cytokine receptor (Fig. 2d). We described the difference in gene expression in the above pathway between the SARS-CoV-2 infected group and the uninfected group, respectively (Tables 3 and 4).
Fig. 2.
Bubble diagram results of GO analysis and histogram results of KEGG pathway analysis. Bubble diagram: The abscissa axis represents gene ratio while the ordinate axis represents term names. The size of a single bubble represents the degree of enrichment; the color variety represents different q value (those with red color are considered to be of significance). Histogram: The abscissa axis represents counts; the ordinate axis represents KEGG pathways; color represents the same meaning as the bubble diagram. a Top 10 enriched GO terms in biological process (BP); b top 10 enriched GO terms in cellular component (CC); c top 10 enriched GO terms in the molecular function (MF); d top 11 of enriched KEGG pathways
Table 3.
The collection of significantly enricher GO pathways (top 10 according to P value)
| Ontology | ID | Description | Count | P value | Gene ID |
|---|---|---|---|---|---|
| BP | GO:0060326 | Cell chemotaxis | 51 | 1.77E−17 | IL1B/CCL20/CXCL8/CXCL5/SERPINE1/CXCL2/CXCL3/CXCL10/DEFB4A/CXCL1/IL23A/PDGFB/IL36G/SAA2/SAA1/CXCL11/RAC2/GAS6/CXCL6/HBEGF/CCL5/EDN1/ADAM8/SEMA5A/CXCL14/ITGB2/S100A7/THBS1/CXCL13/DYSF/IL6R/PDE4B/CH25H/IL16/KIT/EDN2/CXCL9/S100A12/CCL2/SAA4/TNFSF14/C5/FGFR1/LEF1/FGF2/CCRL2/PDGFD/CCR7/IL6/CYP7B1/SLAMF8 |
| BP | GO:0030595 | Leukocyte chemotaxis | 41 | 1.21E−15 | IL1B/CCL20/CXCL8/CXCL5/SERPINE1/CXCL2/CXCL3/CXCL10/CXCL1/IL23A/PDGFB/IL36G/SAA1/CXCL11/RAC2/GAS6/CXCL6/CCL5/EDN1/ADAM8/CXCL14/ITGB2/S100A7/THBS1/CXCL13/DYSF/IL6R/PDE4B/CH25H/IL16/KIT/EDN2/CXCL9/S100A12/CCL2/TNFSF14/C5/CCR7/IL6/CYP7B1/SLAMF8 |
| BP | GO:0097529 | Myeloid leukocyte migration | 39 | 3.68E−15 | IL1B/CCL20/CXCL8/CXCL5/SERPINE1/CXCL2/CXCL3/CXCL10/CXCL1/IL23A/PDGFB/IL36G/SAA1/CXCL11/RAC2/CXCL6/CCL5/EDN1/ADAM8/CD177/ITGB2/S100A7/THBS1/CXCL13/DYSF/IL6R/PDE4B/KIT/EDN2/CXCL9/S100A12/CCL2/C5/CD99/PDGFD/CCR7/IL6/CD200/SLAMF8 |
| BP | GO:1990266 | Neutrophil migration | 29 | 6.55E−15 | IL1B/CCL20/CXCL8/CXCL5/CXCL2/CXCL3/CXCL10/CXCL1/IL23A/IL36G/SAA1/CXCL11/RAC2/CXCL6/CCL5/EDN1/ADAM8/CD177/ITGB2/CXCL13/DYSF/PDE4B/EDN2/CXCL9/S100A12/CCL2/CD99/CCR7/SLAMF8 |
| BP | GO:0097530 | Granulocyte migration | 31 | 2.10E−14 | IL1B/CCL20/CXCL8/CXCL5/CXCL2/CXCL3/CXCL10/CXCL1/IL23A/IL36G/SAA1/CXCL11/RAC2/CXCL6/CCL5/EDN1/ADAM8/CD177/ITGB2/S100A7/THBS1/CXCL13/DYSF/PDE4B/EDN2/CXCL9/S100A12/CCL2/CD99/CCR7/SLAMF8 |
| BP | GO:0032496 | Response to lipopolysaccharide | 48 | 4.74E−14 | IL1B/CSF3/CXCL8/CXCL5/SERPINE1/CXCL2/CXCL3/CXCL10/ALPL/CXCL1/IDO1/TNFAIP3/IL36G/CXCL11/CXCL6/ICAM1/NFKBIA/CMPK2/PTGS2/SPON2/LCN2/ZC3H12A/CCL5/PTAFR/CSF2/GGT1/TNIP3/EDN1/ANKRD1/S100A7/CXCL13/PDCD1LG2/NOS2/PDE4B/KMO/CD36/TNFRSF1B/IL12RB2/CXCL9/CCL2/SELP/MIR146A/GFI1/CCR7/IL6/REN/CNR1/IL24 |
| BP | GO:0071222 | Cellular response to lipopolysaccharide | 37 | 4.80E−14 | IL1B/CSF3/CXCL8/CXCL5/SERPINE1/CXCL2/CXCL3/CXCL10/CXCL1/TNFAIP3/IL36G/CXCL11/CXCL6/ICAM1/NFKBIA/CMPK2/SPON2/LCN2/ZC3H12A/CCL5/PTAFR/CSF2/TNIP3/ANKRD1/CXCL13/PDCD1LG2/NOS2/PDE4B/KMO/CD36/TNFRSF1B/CXCL9/CCL2/MIR146A/GFI1/IL6/IL24 |
| BP | GO:0071219 | Cellular response to molecule of bacterial origin | 37 | 1.42E−13 | IL1B/CSF3/CXCL8/CXCL5/SERPINE1/CXCL2/CXCL3/CXCL10/CXCL1/TNFAIP3/IL36G/CXCL11/CXCL6/ICAM1/NFKBIA/CMPK2/SPON2/LCN2/ZC3H12A/CCL5/PTAFR/CSF2/TNIP3/ANKRD1/CXCL13/PDCD1LG2/NOS2/PDE4B/KMO/CD36/TNFRSF1B/CXCL9/CCL2/MIR146A/GFI1/IL6/IL24 |
| BP | GO:0050900 | Leukocyte migration | 60 | 1.91E−13 | IL1B/CCL20/CXCL8/CXCL5/SERPINE1/CXCL2/CXCL3/CXCL10/CXCL1/IL23A/PDGFB/IL36G/L1CAM/SAA1/CXCL11/RAC2/ITGA5/GAS6/CXCL6/ICAM1/CCL5/PTAFR/EDN1/SLC7A7/ADAM8/CD177/CXCL14/GATA3/FN1/IL33/ITGB2/S100A7/SELL/THBS1/CXCL13/DYSF/IL6R/PDE4B/CH25H/IL16/KIT/EDN2/CXCL9/ITGB7/S100A12/CCL2/MERTK/TNFSF14/THY1/C5/GRB14/TREM1/SELP/CD99/PDGFD/CCR7/IL6/CD200/CYP7B1/SLAMF8 |
| BP | GO:0002237 | Response to molecule of bacterial origin | 48 | 2.08E−13 | IL1B/CSF3/CXCL8/CXCL5/SERPINE1/CXCL2/CXCL3/CXCL10/ALPL/CXCL1/IDO1/TNFAIP3/IL36G/CXCL11/CXCL6/ICAM1/NFKBIA/CMPK2/PTGS2/SPON2/LCN2/ZC3H12A/CCL5/PTAFR/CSF2/GGT1/TNIP3/EDN1/ANKRD1/S100A7/CXCL13/PDCD1LG2/NOS2/PDE4B/KMO/CD36/TNFRSF1B/IL12RB2/CXCL9/CCL2/SELP/MIR146A/GFI1/CCR7/IL6/REN/CNR1/IL24 |
| CC | GO:0062023 | Collagen-containing extracellular matrix | 52 | 2.39E−13 | SERPINE1/ADAM19/HMCN1/SERPINA3/KRT1/GDF15/PDGFB/PRSS1/L1CAM/COL3A1/MMP9/ICAM1/SEMA7A/CSTB/LAMC2/GPC3/NPNT/SMOC1/TINAGL1/ANXA6/FN1/ELN/BMP7/COL21A1/ANGPTL2/EYS/COMP/ADAMTS1/EGFL6/SERPINA5/S100A7/CDH2/POSTN/THBS1/IL7/AMTN/CILP/COL5A3/COL13A1/ADAMTS5/BGN/COL6A2/SPARCL1/ADAMTS9/MMP8/MFAP4/SOD3/ADAMTS4/NDP/ECM2/LGALS4/COL2A1 |
| CC | GO:0005788 | Endoplasmic reticulum lumen | 37 | 4.67E−09 | WNT7A/PLAUR/COL22A1/IL23A/PDGFB/IGFBP5/COL3A1/GAS6/STC2/C3/PTGS2/FSTL3/TMEM132A/EBI3/GPC3/EDN1/FN1/COL21A1/PROC/CDH2/THBS1/AMTN/COL5A3/COL13A1/RCN3/ADAMTS5/COL6A2/SPARCL1/F5/ARSF/WNT3A/CYP2W1/PDGFD/CHGB/SERPIND1/IL6/COL2A1 |
| CC | GO:0031225 | Anchored component of membrane | 25 | 2.69E−08 | CEACAM7/ALPL/PLAUR/NT5E/PRSS22/SEMA7A/NTM/VNN1/GPC3/CD177/VNN3/RGMA/OPCML/BST2/ENPP6/FOLR3/NTNG1/GAS1/THY1/RAB3B/LSAMP/ART3/ULBP1/RTN4RL1/LY6H |
| CC | GO:0009897 | External side of plasma membrane | 41 | 3.80E−08 | CXCL10/ITGA5/ICAM1/SEMA7A/EBI3/HLA-B/SSC4D/CD83/IL12RB1/SERPINA5/ITGB2/KCNJ5/CDH5/THBS1/IL6R/RAET1E/IL7R/HLA-F/PDCD1LG2/TNFRSF9/CD69/CD36/KIT/TGFBR3/IL12RB2/CXCL9/FOLR3/ANPEP/MICA/IL2RG/IL2RB/THY1/CUBN/SELP/IL5RA/ULBP1/RTN4RL1/CCRL2/CD79A/UMODL1/CCR7 |
| CC | GO:0043020 | NADPH oxidase complex | 7 | 8.82E−07 | DUOX2/NCF2/NOXO1/NOX1/NCF1B/NCF1/NCF1C |
| CC | GO:0001533 | Cornified envelope | 12 | 1.07E−05 | SPRR2F/SPRR2D/KRT1/KRT10/SPRR2A/CST6/DSG1/PI3/SPRR2E/FLG2/RPTN/EVPLL |
| CC | GO:0070820 | Tertiary granule | 19 | 4.14E−05 | CXCL1/PLAU/MMP9/CSTB/DSG1/PTAFR/CDA/ADAM8/CD177/FTH1/METTL7A/ITGB2/FLG2/FOLR3/FRMPD3/CRISP3/MMP8/ATP8B4/FPR1 |
| CC | GO:1904724 | Tertiary granule lumen | 10 | 6.64E−05 | CXCL1/MMP9/CSTB/CDA/FTH1/METTL7A/FLG2/FOLR3/CRISP3/MMP8 |
| CC | GO:0045177 | Apical part of cell | 32 | 0.000111784 | CEACAM7/SLC26A4/DUOXA2/SLC5A1/RHCG/DUOX2/KISS1/DSG1/PTPRH/AQP5/SLC34A2/FN1/P2RY1/AJAP1/SLC26A9/CDH2/SLC5A8/MUC13/BST2/CLIC5/IL6R/CYP4F12/AQP6/CD36/FAP/THY1/ATP6V0D2/CUBN/AQP2/ADGRF5/AQP1/REN |
| CC | GO:0016324 | Apical plasma membrane | 28 | 0.000117339 | CEACAM7/SLC26A4/SLC5A1/RHCG/DUOX2/KISS1/DSG1/PTPRH/AQP5/SLC34A2/FN1/P2RY1/AJAP1/SLC26A9/CDH2/SLC5A8/MUC13/BST2/CLIC5/IL6R/CYP4F12/AQP6/CD36/THY1/ATP6V0D2/CUBN/AQP2/AQP1 |
| MF | GO:0048018 | Receptor ligand activity | 73 | 2.69E−21 | IL1B/IL1A/CCL20/CSF3/CXCL8/CXCL5/CXCL2/TSLP/CXCL3/INHBA/CXCL10/LIF/DEFB4A/CXCL1/WNT7A/GDF15/IL23A/PDGFB/IL36G/SAA2/TYMP/SAA1/CXCL11/GAS6/CXCL6/SEMA7A/STC2/BMP2/HBEGF/CCL5/IL32/EBI3/CSF2/EDN1/SEMA5A/CXCL14/IL19/BMP7/IL33/ADM2/IL17C/CXCL13/IL6R/EGF/TFF1/IL15/IL7/IL16/SEMA6D/EDN2/GAL/CXCL9/CCL2/EPGN/CD70/SAA4/SLURP1/LEFTY1/TNFSF14/SEMA3G/INHBB/C5/FGF2/WNT3A/PDGFD/CHGB/EPHA7/NDP/IL6/ENDOU/AMH/IL24/NODAL |
| MF | GO:0005125 | Cytokine activity | 47 | 3.18E−20 | IL1B/IL1A/CCL20/CSF3/CXCL8/CXCL5/CXCL2/TSLP/CXCL3/INHBA/CXCL10/LIF/CXCL1/WNT7A/GDF15/IL23A/IL36G/CXCL11/CXCL6/BMP2/CCL5/IL32/EBI3/CSF2/EDN1/CXCL14/IL19/BMP7/IL33/IL17C/CXCL13/IL15/IL7/IL16/CXCL9/CCL2/CD70/SLURP1/LEFTY1/TNFSF14/INHBB/C5/FGF2/NDP/IL6/IL24/NODAL |
| MF | GO:0005126 | Cytokine receptor binding | 46 | 8.30E−15 | IL1B/IL1A/CCL20/CSF3/CXCL8/CXCL5/CXCL2/TSLP/CXCL3/INHBA/CXCL10/LIF/DEFB4A/CXCL1/GDF15/IL23A/IL36G/CXCL11/ITGA5/CXCL6/BMP2/CCL5/EBI3/CSF2/CXCL14/GATA3/BMP7/IL12RB1/CDH5/BAMBI/CXCL13/IL6R/IL15/IL7/TGFBR3/CXCL9/CCL2/CD70/LEFTY1/TNFSF14/INHBB/C5/CCRL2/IL6/AMH/NODAL |
| MF | GO:0005539 | Glycosaminoglycan binding | 36 | 1.43E−11 | CXCL10/MMP7/SAA1/CXCL11/CXCL6/HBEGF/LAMC2/RNASE7/CEMIP/SEMA5A/SPOCK3/ANXA6/FN1/BMP7/COMP/ADAMTS1/SERPINA5/SELL/POSTN/THBS1/CXCL13/COL5A3/COL13A1/TGFBR3/ADAMTS5/BGN/FGFR1/SPOCK2/SELP/FGF2/RTN4RL1/FBLN7/SOD3/ECM2/SERPIND1/SUSD5 |
| MF | GO:0008201 | Heparin binding | 28 | 7.83E−10 | CXCL10/MMP7/SAA1/CXCL11/CXCL6/HBEGF/LAMC2/FN1/BMP7/COMP/ADAMTS1/SERPINA5/SELL/POSTN/THBS1/CXCL13/COL5A3/COL13A1/TGFBR3/ADAMTS5/FGFR1/SELP/FGF2/RTN4RL1/FBLN7/SOD3/ECM2/SERPIND1 |
| MF | GO:0008009 | Chemokine activity | 15 | 1.11E−09 | CCL20/CXCL8/CXCL5/CXCL2/CXCL3/CXCL10/CXCL1/CXCL11/CXCL6/CCL5/CXCL14/CXCL13/CXCL9/CCL2/C5 |
| MF | GO:0042379 | Chemokine receptor binding | 17 | 1.71E−09 | CCL20/CXCL8/CXCL5/CXCL2/CXCL3/CXCL10/DEFB4A/CXCL1/CXCL11/CXCL6/CCL5/CXCL14/CXCL13/CXCL9/CCL2/C5/CCRL2 |
| MF | GO:0030414 | Peptidase inhibitor activity | 27 | 1.83E−08 | SERPINE1/SERPINA3/GAS6/CST6/C3/SPINK7/CSTB/PI3/TFPI2/GPC3/SPOCK3/SERPINB7/BIRC3/WFDC5/SERPINA5/BST2/FETUB/TNFSF14/RENBP/C5/LEF1/SPOCK2/UMODL1/CARD18/SERPIND1/PI16/EPPIN |
| MF | GO:1901681 | Sulfur compound binding | 32 | 3.47E−08 | CXCL10/MMP7/SAA1/CXCL11/CXCL6/HBEGF/LAMC2/SEMA5A/ANXA6/GSTM3/FN1/BMP7/COMP/ADAMTS1/SERPINA5/SELL/POSTN/CBS/THBS1/CXCL13/COL5A3/COL13A1/TGFBR3/ADAMTS5/FGFR1/SELP/FGF2/RTN4RL1/FBLN7/SOD3/ECM2/SERPIND1 |
| MF | GO:0005178 | Integrin binding | 22 | 4.57E−08 | IL1B/ISG15/COL3A1/ITGA5/ICAM1/SEMA7A/NPNT/CD177/FN1/COMP/EGFL6/ITGB2/THBS1/ITGB7/ADAMTS5/FAP/THY1/LCP1/ESM1/FGF2/ECM2/ADAM23 |
Table 4.
Eleven most significantly enriched KEGG pathways
| ID | Description | Count | P value | P. adjust | Gene ID |
|---|---|---|---|---|---|
| hsa04060 | Cytokine-cytokine receptor interaction | 54 | 3.59E−20 | 3.41E−18 | IL1B/IL1A/CCL20/CSF3/CXCL8/CXCL5/CXCL2/TSLP/CXCL3/INHBA/CXCL10/LIF/CXCL1/GDF15/IL23A/IL36G/CXCL11/CXCL6/BMP2/CCL5/IL32/CSF2/CXCL14/IL19/BMP7/IL33/RELT/IL12RB1/IL17C/CXCL13/IL6R/IL7R/TNFRSF9/IL15/IL7/IL16/TNFRSF1B/IL12RB2/CXCL9/CSF1R/CCL2/CD70/IL2RG/IL2RB/TNFSF14/INHBB/IL1R2/IL5RA/CCR7/IL6/AMH/IL24/NODAL/MPL |
| hsa04061 | Viral protein interaction with cytokine and cytokine receptor | 24 | 6.71E−12 | 3.19E−10 | CCL20/CXCL8/CXCL5/CXCL2/CXCL3/CXCL10/CXCL1/CXCL11/CXCL6/CCL5/CXCL14/IL19/CXCL13/IL6R/TNFRSF1B/CXCL9/CSF1R/CCL2/IL2RG/IL2RB/TNFSF14/CCR7/IL6/IL24 |
| hsa04064 | NF-kappa B signaling pathway | 15 | 5.89E−05 | 0.00186373 | IL1B/CXCL8/CXCL2/CXCL3/CXCL1/PLAU/TNFAIP3/BCL2A1/ICAM1/NFKBIA/PTGS2/BIRC3/PRKCB/DDX58/TNFSF14 |
| hsa00982 | Drug metabolism—cytochrome P450 | 11 | 0.000306597 | 0.007281679 | ADH7/GSTM3/GSTA2/GSTA1/CYP2C9/UGT1A7/ADH1A/UGT2A1/CYP2C8/UGT2B28/UGT2B17 |
| hsa00590 | Arachidonic acid metabolism | 10 | 0.000471477 | 0.008885837 | PTGS2/GGT1/ALOX15B/PLA2G4C/CYP4F3/PLA2G4F/PTGS1/TBXAS1/CYP2C9/CYP2C8 |
| hsa00980 | Metabolism of xenobiotics by cytochrome P450 | 11 | 0.000561211 | 0.008885837 | ADH7/GSTM3/GSTA2/GSTA1/CYP2C9/UGT1A7/ADH1A/HSD11B1/UGT2A1/UGT2B28/UGT2B17 |
| hsa04062 | Chemokine signaling pathway | 19 | 0.000840976 | 0.010436507 | CCL20/CXCL8/CXCL5/CXCL2/CXCL3/CXCL10/CXCL1/CXCL11/RAC2/CXCL6/NFKBIA/CCL5/CXCL14/PRKCB/CXCL13/CXCL9/CCL2/NCF1/CCR7 |
| hsa00830 | Retinol metabolism | 10 | 0.000878864 | 0.010436507 | ADH7/AWAT2/CYP2C9/UGT1A7/RDH12/ADH1A/UGT2A1/CYP2C8/UGT2B28/UGT2B17 |
| hsa00983 | Drug metabolism—other enzymes | 10 | 0.002812844 | 0.029691134 | XDH/TYMP/CDA/GSTM3/GSTA2/GSTA1/UGT1A7/UGT2A1/UGT2B28/UGT2B17 |
| hsa00140 | Steroid hormone biosynthesis | 8 | 0.005895127 | 0.053494911 | HSD17B2/SRD5A2/UGT1A7/HSD11B1/UGT2A1/UGT2B28/UGT2B17/CYP7B1 |
| hsa00120 | Primary bile acid biosynthesis | 4 | 0.006194148 | 0.053494911 | CYP27A1/CH25H/CYP7B1/CYP8B1 |
PPI network construction and hub gene analysis
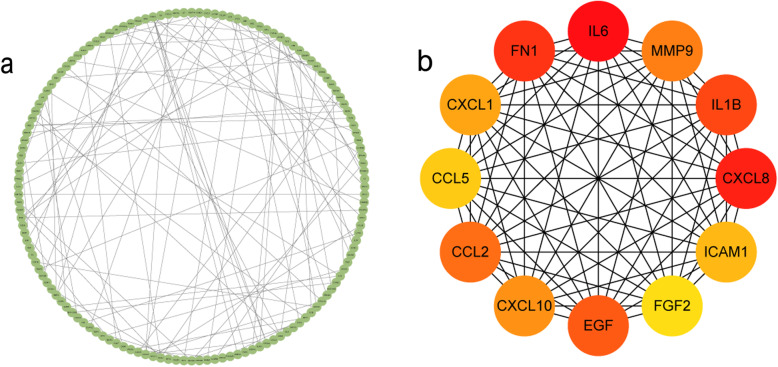
We finally established the PPI network (Fig. 3a) through the STRING online tool and Cytoscape [21]. The network provides a clear and direct expression of complex interactions between gene node, and many domain analyses are based on network analysis. There are 140 nodes and 105 edges in the PPI network. We calculated the degree with the help of the CytoHubba plug-in. By doing so, we found out 12 hub genes and showed their interaction (Fig. 3b).
Fig. 3.
Protein–protein interactions (PPI) network, module analysis, and hub gene identification. The most significant modules were screened out by using the CytoHubba plug-in. Nodes represent gene; lines represent interactions between gene-encoded protein. a Construction of PPI networks. Direct molecular interaction network between novel coronavirus differently regulated genes supported by experiments, encoded protein; b PPI network of the top 12 hub genes (the redder the color, the more important it is)
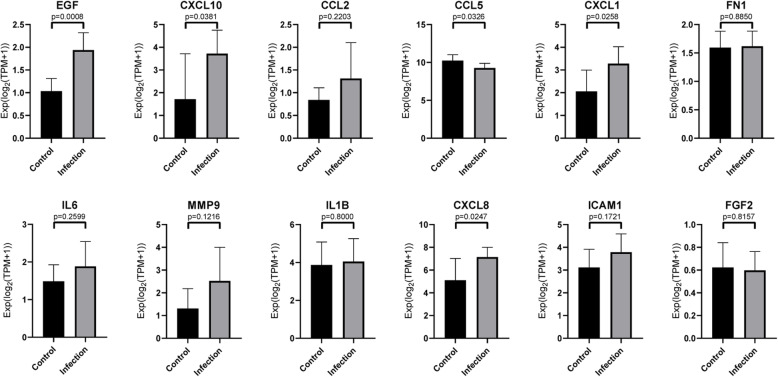
Electronic validation of hub gene by GSE150728
GSE150728, performed on peripheral blood mononuclear cells (PBMCs), was selected to validate the expression of hub gene. In GSE150728 database, CXCL1, EGF, CXCL10, CXCL8, and CCL5 were differently expressed before and after SARS-CoV-2 infection. Among these genes, CCL5 was the only gene, whose levels decreased. Anyhow the electronic validation support the finding that CXCL1, EGF, CXCL10, and CXCL8 were significantly expressed in SARS-CoV-2 infected bronchial organoid when compared to normal bronchial organoid (Fig. 4)
Fig. 4.
Validation of hub gene expression level. Genes are the top 12 selected from DEGs and PPI networks co-regulated by control and infected samples
Discussion
Pneumonia caused by an infection in novel coronavirus has become a popular public health crisis in the world [22]. Its early diagnosis and treatment are crucial for later disease progression. In this paper, the related molecular and infectious mechanisms of the human bronchus-like tube after its infection with novel coronavirus were discussed from the perspective of bioinformatics analysis.
In this study, we analyzed six samples from the GSE150819GEO database and found 966 differential genes, of which 490 were upregulated and 476 were downregulated, resulting in the establishment of a volcanic map. In addition, we performed GO and KEGG analyses on the dataset. Among them, biological processes are highly enriched in molecules with leukocyte migration, cell chemotaxis, response to lipopolysaccharide, and bacterial origin, which may be related to an inflammatory reaction [23–25]. In the inflammatory response, cytokines secreted by endothelial cells react with leukocyte ligands to express adhesion molecules, chemokines, and chemoattractants. Chemokines bind to glycosaminoglycans on the surface of epithelial cells, triggering leukocyte recruitment and inducing integrin activation. Second, leukocytes also enter the basement membrane and migrate to the mesenchyme, causing significant enrichment and expression in the collagen in the extracellular matrix, the outside of the plasma membrane, and the endoplasmic reticulum lumen of the cell components, which is directly related to the mechanism of pneumonia caused by the novel coronavirus [26]. In the KEGG analysis, the enrichment of cytokine–cytokine receptor interactions and chemokine signaling pathways correspond to the enrichment of cellular components and molecular functions. The high aggregation of the interaction of viral proteins with cytokines and cytokine receptors suggested that viral proteins might be involved in related processes such as inflammation factor storm and inflammation factor regulation. The expression of endoplasmic reticulum collagen in cellular components also suggested that viral proteins might affect the transport process of endoplasmic reticulum. As a pathway to promote cell function, NF−kappa B signaling pathways activation may be related to the synthesis of viral proteins. In summary, the potential host-virus protein interactions may provide us with new ideas and strategies for drug screening and the development of drug-related research in SARS-CoV-2 [27]. At the same time, based on the PPI network diagram, we used the CytoHubba plug-in to select 12 central genes with high connectivity, all of which occupied the core nodes of the network, suggesting that these central genes might play a pivotal role in the process of infection in the novel coronavirus [20].
There are several highly interacting genes in the PPI network, such as CXCL1, EGF, CXCL10, CXCL8, CCL5, and what calls for special attention is that CCL5 levels decreased in peripheral blood mononuclear cells (PBMCs), whose data is form GSE150728, the dataset we used for electronic validation of hub gene. However, CCL5 was repeatedly reported to be increased in patients infected by SARS-CoV-2 [28], which supported the result we get from hBO. The unique inflammatory chemokine CCL5 (RANTES) induced at a later phase of inflammation guides white blood cells to migrate into inflammatory lesions during varied pathological processes and maintains local immune cells [29]. It has been demonstrated that CCL5 functions through activating the pathways including transcription factor 3 signaling transducers and activators, nuclear factor-κb, and MAPK pathways across three cell surface receptors named CCR1, CCR3, and CCR5. In addition, CCL5 can directly activate M1 (pro-inflammatory) polarization and prevent M2 (anti-inflammatory) polarization [30]. The lack of homeostatic CCL5 expression will largely influence the activated state of lung TRM (tissue-resident memory) T cell and natural killer cell components [31].
The chemokine CXCL1 plays a significant role in the immune response, mediating its function by combining with the CXCR2 receptor and GAG [32]. It highly regulates the transport, collection, and activation of neutrophilic granulocyte [33]. What is more, it activates the release of proteases as well as reactive oxygen species (ROS), killing the microorganisms at tissue [32]. But it is also associated with damage in numerous tissues, including the lung [34].
Data from previous coronavirus prevalence, including severe acute respiratory syndrome as well as the Middle East respiratory, and current data from the COVID-19 pandemic indicated that there could be pulmonary fibrosis after SARS-CoV-2 infection [35]. EGF may play an important role in pulmonary fibrosis after SARS-CoV-2 infection. Studies of SARS-CoV and SARS showed that abnormal expressions of EGFR are vital in the pathogenesis of fibrosis during coronavirus infection [36, 37]. An animal study also reported EGFR potential role in enhancing disease when infected by SARS-CoV [37]. So, it is of great significance to study EGF function in SARS-CoV-2 infected body, which may help prevent pulmonary fibrosis and learn further about the pathogenesis of SARS-CoV-2 induced lung injury.
CXCL10, an interferon-inducible protein, increases greatly in SARS-CoV-2 infected patients [38, 39]. Abnormal increase of CXCL10 abnormal is also incurred in SARS infected patients and indicates the worse clinical outcome [40, 41]. A study conducted on mice showed that CXCL10 may be involved in fulminant lung injury which can be modified when CXCL10 was neutralized [42]. Back to SARS-CoV-2, serum concentrations of CXCL10, combining with GM-CSF and SARS-CoV-2 viral load are associated with day-28 mortality [43]. All these indicated that CXCL10 can be a potential indicator and a therapeutic target for SARS-CoV-2.
In our study, increased expressions of CXCL10 and CXCL8 are observed, which also seem to characterize COVID-19. CXCL8 (IL-8) is a member of the chemokine family, and its receptors are CXCR1/2 [41]. The combination of CXCL8 and CXCR1 or CXCR2 will lead to lung injury [22, 44]. Once blocked, the injury reduces [22, 44]. Hence, the study also indicates that CXR1/2 receptor stimulation has a positive effect on pulmonary recovery [41]. In this case, understanding its role in COVID-19 may contribute to clinical drug development. For patients suffering from ARDS, both CXCL8 and anti-CXCL8 levels increase which enhanced the survival of neutrophils [44]. CXCL8 plays a major role in the initial control of respiratory tract infection due to its chemotactic activity for neutrophils and monocytes [45]. And CXCL8 is considered a potential prognostic bio-marker for ARDS clinical course, which is a significant syndrome of SARS-CoV-2 [41].
In addition, this study has its limitations. The small sample size increases the error of the analysis results. If the analysis can be performed based on a large sample size, it may be possible to study the relationship between each central gene and the role of the pathway more fully so that the accuracy is better.
In conclusion, by studying the mRNA expression of human bronchial organoids, we have found that a series of DEGs (CXCL1, CCL5, EGF, CXCL10, CXCL8), which were also reported in SARS-CoV-2 infected patients, in vitro and in vivo models, plays an important role in the COVID-19 and are helpful for diagnosis and treatment. Our findings can better study the molecular mechanism of infection.
Conclusion
In summary, using a series of biological analyses, we have obtained a new understanding and description of human bronchial organoids infected with novel coronavirus, and through the analysis and verification of differential genes, we have identified that CXCL1, CCL5, EGF, CXCL10, and CXCL8 play a key role in the infection process in novel coronavirus and a series of pathways related to cytokine expression, leukocyte migration, and cytokine-cytokine receptor interaction have been significantly changed. These findings will help study the infection mechanism and therapeutic control in novel coronavirus. In order for these biomarkers and targets to become a powerful tool for clinically diagnosing and studying the infection mechanism, more samples should be used to further study the expression differences of these genes. What is more, the pathways and genes mentioned above also play an important role in patients, in vitro and in vivo models infected with SARS-COV-2, and they may be a key link in the pathogenesis of COVID-19. Therefore, we preliminarily concluded that the bronchial organ model can be used as an in vitro model for the study of SARS-COV-2.
Acknowledgements
Not applicable.
Editorial policies
Not applicable.
Authors’ contributions
Xu-Guang Guo provided the original idea and constructed the article outline. Ke-Ying Fang, Wen-Chao Ca, Tian-Ao Xie, Jie Lv and Jia-Xin Chen, Xun-Jie Cao, Zhong-Wei Li, Shu-Ting Deng were responsible for data analysis, table making, and manuscript constructing. All the authors read and approved the final version of the manuscript.
Funding
Not applicable.
Availability of data and materials
The GSE150819 datasets used during the current study are available from GEO database (https://www.ncbi.nlm.nih.gov/gds/).
Declarations
Consent for publication
All of the authors have read and approved the content, and agree to submit the whole article in your journal.
Competing interests
All the authors declare that there are no competing interests associated with the manuscript.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ke-Ying Fang, Wen-Chao Cao and Tian-Ao Xie contributed equally to this work.
References
- 1.Zhou J, Li C, Liu X, Chiu MC, Zhao X, Wang D, Wei Y, Lee A, Zhang AJ, Chu H, Cai JP, Yip CCY, Chan IHY, Wong KKY, Tsang OTY, Chan KH, Chan JFW, To KKW, Chen H, Yuen KY. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat Med. 2020;26(7):1077–1083. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- 2.Xu X, Yu C, Qu J, Zhang L, Jiang S, Huang D, Chen B, Zhang Z, Guan W, Ling Z, Jiang R, Hu T, Ding Y, Lin L, Gan Q, Luo L, Tang X, Liu J. Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2. Eur J Nucl Med Mol Imaging. 2020;47(5):1275–1280. doi: 10.1007/s00259-020-04735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J', Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. The Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh A, Singh RS, Sarma P, Batra G, Joshi R, Kaur H, Sharma AR, Prakash A, Medhi B. A comprehensive review of animal models for coronaviruses: SARS-CoV-2, SARS-CoV, and MERS-CoV. Virol Sin. 2020;35(3):290–304. doi: 10.1007/s12250-020-00252-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rakib A, Sami SA, Mimi NJ, Chowdhury MM, Eva TA, Nainu F, Paul A, Shahriar A, Tareq AM, Emon NU, Chakraborty S, Shil S, Mily SJ, Ben Hadda T, Almalki FA, Emran TB. Immunoinformatics-guided design of an epitope-based vaccine against severe acute respiratory syndrome coronavirus 2 spike glycoprotein. Comput Biol Med. 2020;124:103967. doi: 10.1016/j.compbiomed.2020.103967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng W, Zong W, Wang F, Ju S. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): a review. Mol Cancer. 2020;19(1):100. doi: 10.1186/s12943-020-01218-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO. 2020/10/15; Available from: https://covid19.who.int/.
- 8.Desole G, et al. Modelling neurotropic flavivirus infection in human induced pluripotent stem cell-derived systems. Int J Mol Sci. 2019;20:21. doi: 10.3390/ijms20215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacob F, et al. Human pluripotent stem cell-derived neural cells and brain organoids reveal SARS-CoV-2 neurotropism. bioRxiv. 2020;27(6):937–950.e9. 10.1016/j.stem.2020.09.016. [DOI] [PMC free article] [PubMed]
- 10.Shan C, Yao YF, Yang XL, Zhou YW, Gao G, Peng Y, Yang L, Hu X, Xiong J, Jiang RD, Zhang HJ, Gao XX, Peng C, Min J, Chen Y, Si HR, Wu J, Zhou P, Wang YY, Wei HP, Pang W, Hu ZF, Lv LB, Zheng YT, Shi ZL, Yuan ZM. Infection with novel coronavirus (SARS-CoV-2) causes pneumonia in Rhesus macaques. Cell Res. 2020;30(8):670–677. doi: 10.1038/s41422-020-0364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun SH, Chen Q, Gu HJ, Yang G, Wang YX, Huang XY, Liu SS, Zhang NN, Li XF, Xiong R, Guo Y, Deng YQ, Huang WJ, Liu Q, Liu QM, Shen YL, Zhou Y, Yang X, Zhao TY, Fan CF, Zhou YS, Qin CF, Wang YC. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe. 2020;28(1):124–133. doi: 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imai M, Iwatsuki-Horimoto K, Hatta M, Loeber S, Halfmann PJ, Nakajima N, Watanabe T, Ujie M, Takahashi K, Ito M, Yamada S, Fan S, Chiba S, Kuroda M, Guan L, Takada K, Armbrust T, Balogh A, Furusawa Y, Okuda M, Ueki H, Yasuhara A, Sakai-Tagawa Y, Lopes TJS, Kiso M, Yamayoshi S, Kinoshita N, Ohmagari N, Hattori SI, Takeda M, Mitsuya H, Krammer F, Suzuki T, Kawaoka Y. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci U S A. 2020;117(28):16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Israelow B, Song E, Mao T, Lu P, Meir A, Liu F, Alfajaro MM, Wei J, Dong H, Homer RJ, Ring A, Wilen CB, Iwasaki A. Mouse model of SARSCoV-2 reveals inflammatory role of type I interferon signaling. bioRxiv [Preprint]. 2020:2020.05.27.118893. 10.1101/2020.05.27.118893. [DOI] [PMC free article] [PubMed]
- 14.Kim YI, Kim SG, Kim SM, Kim EH, Park SJ, Yu KM, Chang JH, Kim EJ, Lee S, Casel MAB, Um J, Song MS, Jeong HW, Lai VD, Kim Y, Chin BS, Park JS, Chung KH, Foo SS, Poo H, Mo IP, Lee OJ, Webby RJ, Jung JU, Choi YK. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe. 2020;27(5):704–709. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou J, Li C, Sachs N, Chiu MC, Wong BHY, Chu H, Poon VKM, Wang D, Zhao X, Wen L, Song W, Yuan S, Wong KKY, Chan JFW, To KKW, Chen H, Clevers H, Yuen KY. Differentiated human airway organoids to assess infectivity of emerging influenza virus. Proc Natl Acad Sci U S A. 2018;115(26):6822–6827. doi: 10.1073/pnas.1806308115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porotto M, et al. Authentic modeling of human respiratory virus infection in human pluripotent stem cell-derived lung organoids. mBio. 2019;10(3):e00723. doi: 10.1128/mBio.00723-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strange DP, Zarandi NP, Trivedi G, Atala A, Bishop CE, Sadri-Ardekani H, Verma S. Human testicular organoid system as a novel tool to study Zika virus pathogenesis. Emerg Microbes Infect. 2018;7(1):82. doi: 10.1038/s41426-018-0080-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monteil V, Kwon H, Prado P, Hagelkrüys A, Wimmer RA, Stahl M, Leopoldi A, Garreta E, Hurtado del Pozo C, Prosper F, Romero JP, Wirnsberger G, Zhang H, Slutsky AS, Conder R, Montserrat N, Mirazimi A, Penninger JM. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181(4):905–913. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Huan W, Wu J, Zou S, Qu L. IGFBP6 is downregulated in unstable carotid atherosclerotic plaques according to an integrated bioinformatics analysis and experimental verification. J Atheroscler Thromb. 2020;27(10):1068–1085. doi: 10.5551/jat.52993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li N, Zhou JN. Screening and validating the immune-related gene expression signatures in peripheral blood mononuclear cells of nonischaemic cardiomyopathy. Math Biosci Eng. 2020;17(3):2330–2347. doi: 10.3934/mbe.2020124. [DOI] [PubMed] [Google Scholar]
- 21.Chen X, Wang J, Peng X, Liu K, Zhang C, Zeng X, Lai Y. Comprehensive analysis of biomarkers for prostate cancer based on weighted gene co-expression network analysis. Medicine (Baltimore) 2020;99(14):e19628. doi: 10.1097/MD.0000000000019628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jayne JG, Bensman TJ, Schaal JB, Park AYJ, Kimura E, Tran D, Selsted ME, Beringer PM. Rhesus theta-Defensin-1 attenuates endotoxin-induced acute lung injury by inhibiting proinflammatory cytokines and neutrophil recruitment. Am J Respir Cell Mol Biol. 2018;58(3):310–319. doi: 10.1165/rcmb.2016-0428OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petruk G, Puthia M, Petrlova J, Samsudin F, Strömdahl AC, Cerps S, Uller L, Kjellström S, Bond PJ, Schmidtchen A. SARS-CoV-2 Spike protein binds to bacterial lipopolysaccharide and boosts proinflammatory activity. J Mol Cell Biol. 2020:mjaa067. 10.1093/jmcb/mjaa067. [DOI] [PMC free article] [PubMed]
- 24.Shaath H, Alajez NM. Computational and transcriptome analyses revealed preferential induction of chemotaxis and lipid synthesis by SARS-CoV-2. Biology (Basel) 2020;9(9):260. doi: 10.3390/biology9090260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alon R, Sportiello M, Kozlovski S, Kumar A, Reilly EC, Zarbock A, Garbi N, Topham DJ. Leukocyte trafficking to the lungs and beyond: lessons from influenza for COVID-19. Nat Rev Immunol. 2021;21(1):49–64. 10.1038/s41577-020-00470-2. [DOI] [PMC free article] [PubMed]
- 26.Bradley BT, Maioli H, Johnston R, Chaudhry I, Fink SL, Xu H, Najafian B, Deutsch G, Lacy JM, Williams T, Yarid N, Marshall DA. Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: a case series. Lancet. 2020;396(10247):320–332. doi: 10.1016/S0140-6736(20)31305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin Y, Qin X. Comprehensive analysis of transcriptome data for identifying biomarkers and therapeutic targets in head and neck squamous cell carcinoma. Ann Transl Med. 2020;8(6):282. doi: 10.21037/atm.2020.03.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montalvo Villalba MC, Valdés Ramírez O, Muné Jiménez M, Arencibia Garcia A, Martinez Alfonso J, González Baéz G, Roque Arrieta R, Rosell Simón D, Alvárez Gainza D, Sierra Vázquez B, Resik Aguirre S, Guzmán Tirado MG. Interferon gamma, TGF-beta1 and RANTES expression in upper airway samples from SARS-CoV-2 infected patients. Clin Immunol. 2020;220:108576. doi: 10.1016/j.clim.2020.108576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marques RE, Guabiraba R, Russo RC, Teixeira MM. Targeting CCL5 in inflammation. Expert Opin Ther Targets. 2013;17(12):1439–1460. doi: 10.1517/14728222.2013.837886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M, Sun X, Zhao J, Xia L, Li J, Xu M, Wang B, Guo H, Yu C, Gao Y, Wu H, Kong X, Xia Q. CCL5 deficiency promotes liver repair by improving inflammation resolution and liver regeneration through M2 macrophage polarization. Cell Mol Immunol. 2020;17(7):753–764. doi: 10.1038/s41423-019-0279-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seo W, Shimizu K, Kojo S, Okeke A, Kohwi-Shigematsu T, Fujii SI, Taniuchi I. Runx-mediated regulation of CCL5 via antagonizing two enhancers influences immune cell function and anti-tumor immunity. Nat Commun. 2020;11(1):1562. doi: 10.1038/s41467-020-15375-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawant KV, Poluri KM, Dutta AK, Sepuru KM, Troshkina A, Garofalo RP, Rajarathnam K. Chemokine CXCL1 mediated neutrophil recruitment: role of glycosaminoglycan interactions. Sci Rep. 2016;6(1):33123. doi: 10.1038/srep33123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajarathnam K, Schnoor M, Richardson RM, Rajagopal S. How do chemokines navigate neutrophils to the target site: dissecting the structural mechanisms and signaling pathways. Cell Signal. 2019;54:69–80. doi: 10.1016/j.cellsig.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sawant KV, Xu R, Cox R, Hawkins H, Sbrana E, Kolli D, Garofalo RP, Rajarathnam K. Chemokine CXCL1-mediated neutrophil trafficking in the lung: role of CXCR2 activation. J Innate Immun. 2015;7(6):647–658. doi: 10.1159/000430914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.George PM, Wells AU, Jenkins RG. Pulmonary fibrosis and COVID-19: the potential role for antifibrotic therapy. Lancet Respir Med. 2020;8(8):807–815. doi: 10.1016/S2213-2600(20)30225-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lechowicz K, et al. COVID-19: the potential treatment of pulmonary fibrosis associated with SARS-CoV-2 infection. J Clin Med. 2020;9(6):1917. doi: 10.3390/jcm9061917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coleman CM, Venkataraman T, Liu YV, Glenn GM, Smith GE, Flyer DC, Frieman MB. MERS-CoV spike nanoparticles protect mice from MERS-CoV infection. Vaccine. 2017;35(12):1586–1589. doi: 10.1016/j.vaccine.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sims JT, Krishnan V, Chang CY, Engle SM, Casalini G, Rodgers GH, Bivi N, Nickoloff BJ, Konrad RJ, de Bono S, Higgs RE, Benschop RJ, Ottaviani S, Cardoso A, Nirula A, Corbellino M, Stebbing J. Characterization of the cytokine storm reflects hyperinflammatory endothelial dysfunction in COVID-19. J Allergy Clin Immunol. 2021;147(1):107–11. 10.1016/j.jaci.2020.08.031. [DOI] [PMC free article] [PubMed]
- 39.Mamber SW, et al. Can unconventional immunomodulatory agents help alleviate COVID-19 symptoms and severity? mSphere. 2020;5(3):e00288. doi: 10.1128/mSphere.00288-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oliviero A, de Castro F, Coperchini F, Chiovato L, Rotondi M. COVID-19 Pulmonary and Olfactory Dysfunctions: Is the Chemokine CXCL10 the Common Denominator? Neuroscientist. 2020:1073858420939033. 10.1177/1073858420939033. [DOI] [PubMed]
- 41.Coperchini F, Chiovato L, Croce L, Magri F, Rotondi M. The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lang S, Li L, Wang X, Sun J, Xue X, Xiao Y, Zhang M, Ao T, Wang J. CXCL10/IP-10 neutralization can ameliorate lipopolysaccharide-induced acute respiratory distress syndrome in rats. PLoS One. 2017;12(1):e0169100. doi: 10.1371/journal.pone.0169100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hue S, Beldi-Ferchiou A, Bendib I, Surenaud M, Fourati S, Frapard T, Rivoal S, Razazi K, Carteaux G, Delfau-Larue MH, Mekontso-Dessap A, Audureau E, de Prost N. Uncontrolled innate and impaired adaptive immune responses in patients with COVID-19 acute respiratory distress syndrome. Am J Respir Crit Care Med. 2020;202(11):1509–1519. doi: 10.1164/rccm.202005-1885OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allen TC, Kurdowska A. Interleukin 8 and acute lung injury. Arch Pathol Lab Med. 2014;138(2):266–269. doi: 10.5858/arpa.2013-0182-RA. [DOI] [PubMed] [Google Scholar]
- 45.Metzemaekers M, Vandendriessche S, Berghmans N, Gouwy M, Proost P. Truncation of CXCL8 to CXCL8(9-77) enhances actin polymerization and in vivo migration of neutrophils. J Leukoc Biol. 2020;107(6):1167–1173. doi: 10.1002/JLB.3AB0220-470R. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The GSE150819 datasets used during the current study are available from GEO database (https://www.ncbi.nlm.nih.gov/gds/).