Summary
The phosphoinositide PI(3,5)P2, generated exclusively by the PIKfyve lipid kinase complex, is key for lysosomal biology. Here we explore how PI(3,5)P2 levels within cells are regulated. We find the PIKfyve complex comprises 5 copies of the scaffolding protein Vac14, and one copy each of the lipid kinase PIKfyve, generating PI(3,5)P2 from PI3P, and the lipid phosphatase Fig4, reversing the reaction. Fig4 is active as a lipid phosphatase in the ternary complex, whereas PIKfyve within the complex cannot access membrane-incorporated phosphoinositides due to steric constraints. We find further that the phosphoinositide-directed activities of both PIKfyve and Fig4 are regulated by protein-directed activities within the complex. PIKfyve autophosphorylation represses its lipid kinase activity and stimulates Fig4 lipid phosphatase activity. Further, Fig4 is also a protein phosphatase acting on PIKfyve to stimulate its lipid kinase activity, explaining why catalytically active Fig4 is required for maximal PI(3,5)P2 production by PIKfyve in vivo.
Graphical Abstract
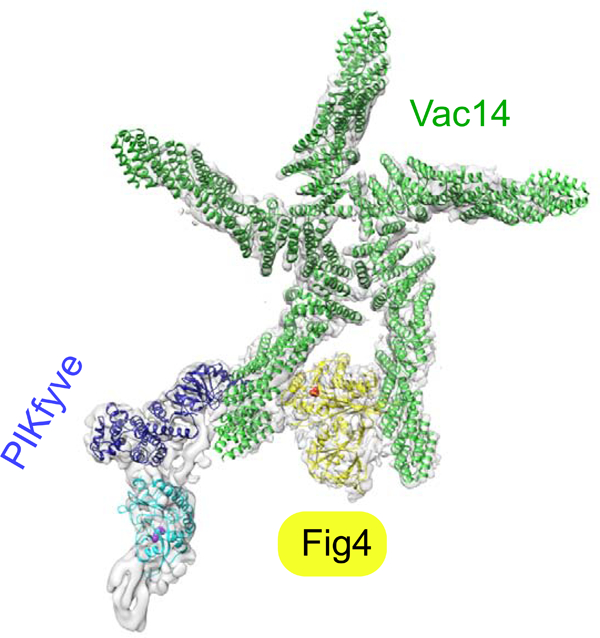
Eukaryotic cells have seven species of phosphoinositide lipids differentially enriched at different compartments. Although these lipids are present only in minute quantities, they play major roles in organelle dynamics and signaling. Two phosphoinositides critical for lysosome biology are PI3P and its derivative PI(3,5)P2, which is generated by the still poorly characterized PIKfyve complex (Hasegawa et al., 2017; Jin et al., 2016). PIKfyve is conserved in all eukaryotes and is their only source of PI(3,5)P2. Highlighting the importance of this complex in cell physiology, mutations in its subunits result in disabling neurodegenerative diseases such as forms of Charcot-Marie Tooth (CMT) disease (Chow et al., 2007) and Amyotropic Lateral Sclerosis (ALS) (Chow et al., 2009). PIKfyve inhibition has been proposed as an anti-viral treatment (Hulseberg et al., 2019; Nelson et al., 2017), and a selective inhibitor of PIKfyve activity, apilimod, is being evaluated as a treatment for non-Hodgkins lymphoma (Gayle et al., 2017).
In addition to the lipid kinase PIKfyve itself (also called Fab1), the complex includes a lipid phosphatase in the Sac family, Fig4 (also called Sac3), and a scaffolding protein Vac14 (also called ArPIKfyve) (Fig. 1A). While PIKfyve transfers a phosphate group from ATP to convert PI3P to PI(3,5)P2, Fig4 dephosphorylates PI(3,5)P2 in the reverse reaction. How these activities are coordinated within the complex to avoid futile cycles of ATP hydrolysis has been enigmatic. Further mysterious is the observation that PI(3,5)P2 levels in cells deficient in Fig4 activity, as for example in patients with CMT, are reduced rather than upregulated (Chow et al., 2007; Shisheva et al., 2019), as might have been expected, and that catalytically active Fig4 is required for PI(3,5)P2 upregulation (Duex et al., 2006a; Duex et al., 2006b; Strunk et al., 2020).
Figure 1. Assembly and gross architecture of the human PIKfyve complex by negative stain EM analysis.
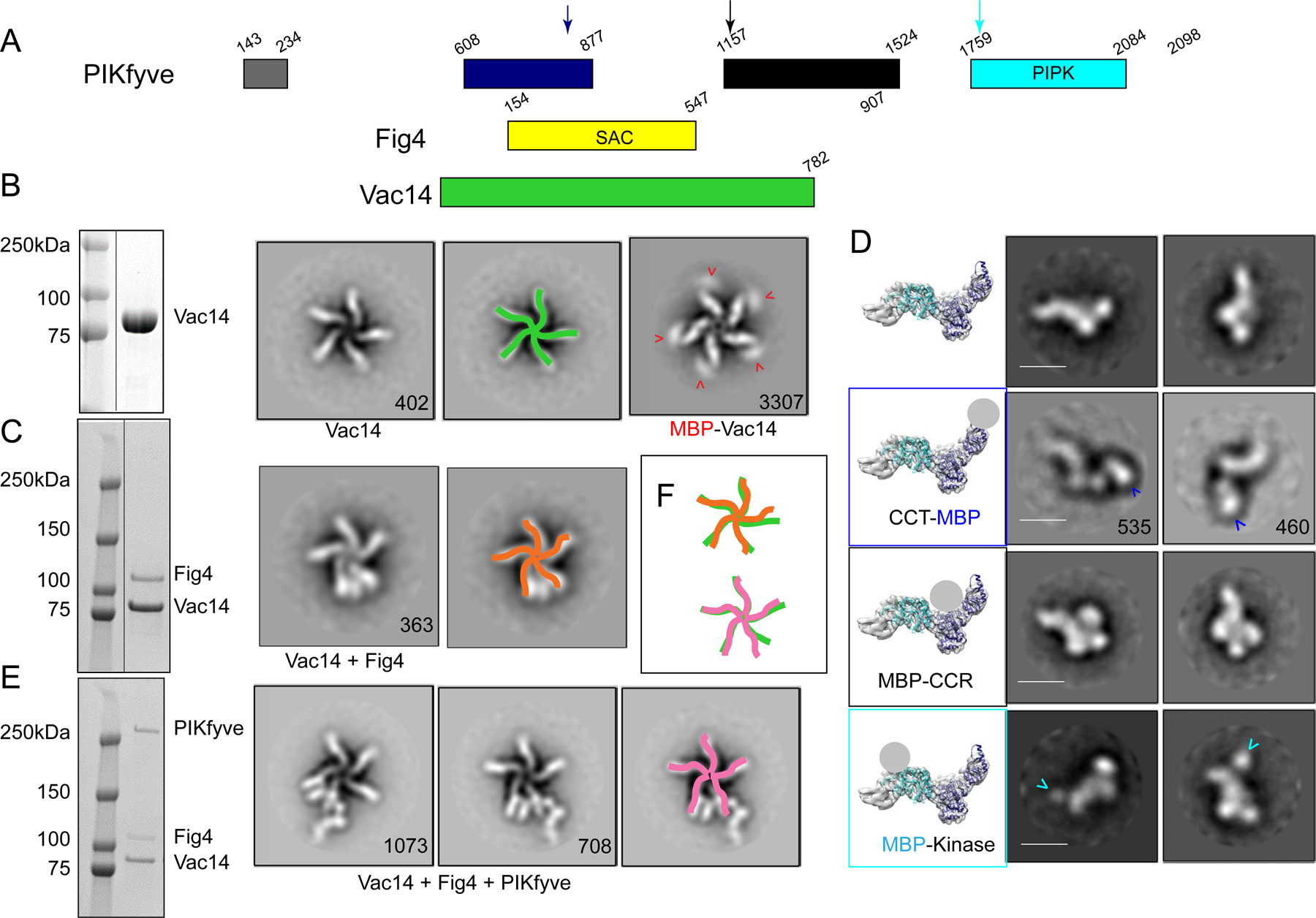
(A) Domain organization of PIKfyve, Fig4, and Vac14. (B) The Vac14 sample used for negative staining EM is pure per SDS-PAGE (line indicates lanes not shown). 2D class averages of Vac14 or MBP-Vac14 show that it pentamerizes. Numbers of particles for each class average are indicated. Maltose binding protein (MBP) fused to the Vac14 N-terminus is at the tip of the Vac14 “leg”. (C) The Fig4/Vac14 sample used for negative staining EM is pure per SDS-PAGE (line indicates lanes not shown). 2D class averages of Fig4/Vac14 show Fig4 between two Vac14 “legs”. (D) Negative stain EM analysis of PIKfyve or MBP+PIKfyve fusions. MBP was inserted into the PIKfyve sequence as indicated by arrows in 1A. The left-most column shows PIKfyve density from the cryo-EM maps (Fig. 2) with docked models of the CCT and kinase modules; a grey ball indicates the location of MBP in the class averages. The other columns show 2D class averages of PIKfyve by itself or with MBP-insertions. (E) The PIKfyve/Fig4/Vac14 sample used for negative staining and cryo-EM is pure per SDS-PAGE. 2D class averages of PIKfyve/Fig4/Vac14 show PIKfyve close to Fig4 at the tip of the Vac14 legs. (F) Schematic overlay of the Vac14 pentamers in Vac14, Fig4/Vac14, and PIKfyve/Fig4/Vac14, showing that the 5-fold symmetry of the Vac14 pentamer is distorted in Fig4/Vac14 and PIKfyve/Fig4/Vac14 complexes.
Here we combine negative stain- and cryo-EM studies elucidating the architecture of the human PIKfyve complex at medium to low resolution with a biochemical analysis to obtain insights as to how the antagonistic activities within the complex are regulated. Our structural studies show that Vac14 pentamerizes into a star-shaped structure, which can bind a single copy each of PIKfyve and Fig4. We found that Fig4 is active as a lipid phosphatase in the intact complex including PIKfyve, whereas the ability of PIKfyve to generate PI(3,5)P2 in membranes is suppressed in the complex as compared to by itself. Our structural data indicate that complex formation conformationally restrains the kinase domain to prevent its access to membranes containing the PI3P substrate. We further report that in addition to its activity on lipid, Fig4 is a serine phosphatase that acts on PIKfyve to increase its lipid kinase activity. While PIKfyve within the complex cannot access PI3P, it may nevertheless be primed by Fig4 for maximal activity following a conformational change, such as complex dissociation, in response to a still unknown stimulus. This model would explain why the ablation of Fig4 activity depresses PI(3,5)P2 production in cells.
Results and Discussion
Vac14 forms a pentameric scaffold through which PIKfyve and Fig4 interact.
As a first step in understanding the regulation of the PIKfyve complex, we examined its architecture and that of its subcomplexes by negative stain electron microscopy. The protein assemblies were produced in mammalian cells (Expi293F) by transfecting these with DNAs coding for one or combinations of PIKfyve components. Fig4 was N-terminally 3xFLAG-tagged to capture Fig4/Vac14 complex, PIKfyve was N-terminally 3xFLAG-tagged to capture PIKfyve alone or ternary complex, and Vac14 alone was purified via an N-terminal 2xStrepII tag. Vac14, predicted to comprise almost entirely alpha helical HEAT repeats, forms a rod that oligomerizes into a pentameric star-like structure (Fig 1B). As C-terminal deletions abrogate Vac14 oligomerization (Ikonomov et al., 2009), Vac14 self association at the center of the star-shaped assembly must be via its C-terminal portions. The N- terminus of Vac14 is at the other end of each Vac14 “leg”, as identified by density corresponding to maltose binding protein (MBP) in 2D class averages of an MBP- Vac14 construct (Fig 1B). (We could not find density corresponding to MBP in 2D class averages of a Vac14-MBP construct (distinct from the MBP-Vac14 construct).) We next produced Fig4/Vac14, unexpectedly finding that only one copy of Fig4 associates with each Vac14 pentamer. The density corresponding to Fig4 appears between the N-terminal tips of two of the Vac14 “legs” (Fig 1C). PIKfyve alone is elongated and kinked at one end (Fig 1D, top row). In the ternary complex, there is density for a single copy of PIKfyve appended to one of the Vac14 legs that flanks Fig4, extending away from the Vac14 pentamer (Fig 1E). We observed only a single stoichiometry, a pentamer of Vac14 associated with one copy each of the phosphatase and kinase, irrespective of the ratios of Fig4:Vac14 or PIKfyve:Fig4:Vac14 DNAs used in transfection. Addition of independently expressed and purified Fig4 and PIKfyve constructs did not change this stoichiometry. The five-fold symmetry of the Vac14 pentamer is disrupted in both the binary and ternary complexes, apparent as a different spacing between Vac14 legs interacting with Fig4 and PIKfyve (Fig. 1F).
The Fig4 and PIKfyve active sites cannot access membranes simultaneously in the complex.
A ternary complex comprising PIKfyveS2053E, which harbors a point- mutation in the kinase activation loop (see characterization below), along with wild- type versions of Fig4 and Vac14 was analyzed further by cryo-EM, yielding a map at a final nominal resolution of 5.25 Å (FSC=0.143 criterion). Due to the extended shape and high flexibility of the complex, it was not possible to obtain a single map with well-ordered density for all three proteins. Rather, a reconstruction of the complete complex was synthesized from three independent maps obtained from the same micrograph dataset, centered on different portions of the complex, which were calculated separately (as described in Methods; Fig. S1, S2) and superimposed based on their overlaps, to form a composite reconstruction (Fig. 2). In the reconstruction, the Vac14 pentamer is cup-shaped, measuring 240 Å across and 110 Å deep, with both Fig4 and PIKfyve occupying sites near the “rim” of the cup as defined by the Vac14 N termini. Each Vac14 “leg” was modeled as a series of alpha helices, although their connectivity could not be established at the resolution of the reconstruction. The Sac homology module of Fig4 comprises an N-terminal regulatory and the upstream catalytic domain and was modeled based on a crystal structure of the same module in Sac1 (PDB 3lwt). It could be confidently docked into density between opposite surfaces of two neighboring copies of Vac14 (Fig. 2), corresponding to the highest resolution region in our maps. The Fig4 active site is oriented to face the top of the Vac14 cup. The catalytic domain of the Fig4 Sac- homology module primarily mediates the interactions with both neighboring Vac14’s. This interaction slightly twists one of the Vac14 copies, likely modifying a surface on its opposite face to facilitate PIKfyve binding, consistent with PIKfyve’s reported inability to bind Fig4 or Vac14 alone (Ikonomov et al., 2009). We did not observe density that could be attributed to portions of Fig4 downstream of its catalytic domain, although Fig4 C-terminal regions interact with Vac14 (Ikonomov et al., 2009; Strunk et al., 2020).
Figure 2. Cryo-EM reconstruction of the human PIKfyve complex at medium- low resolution.
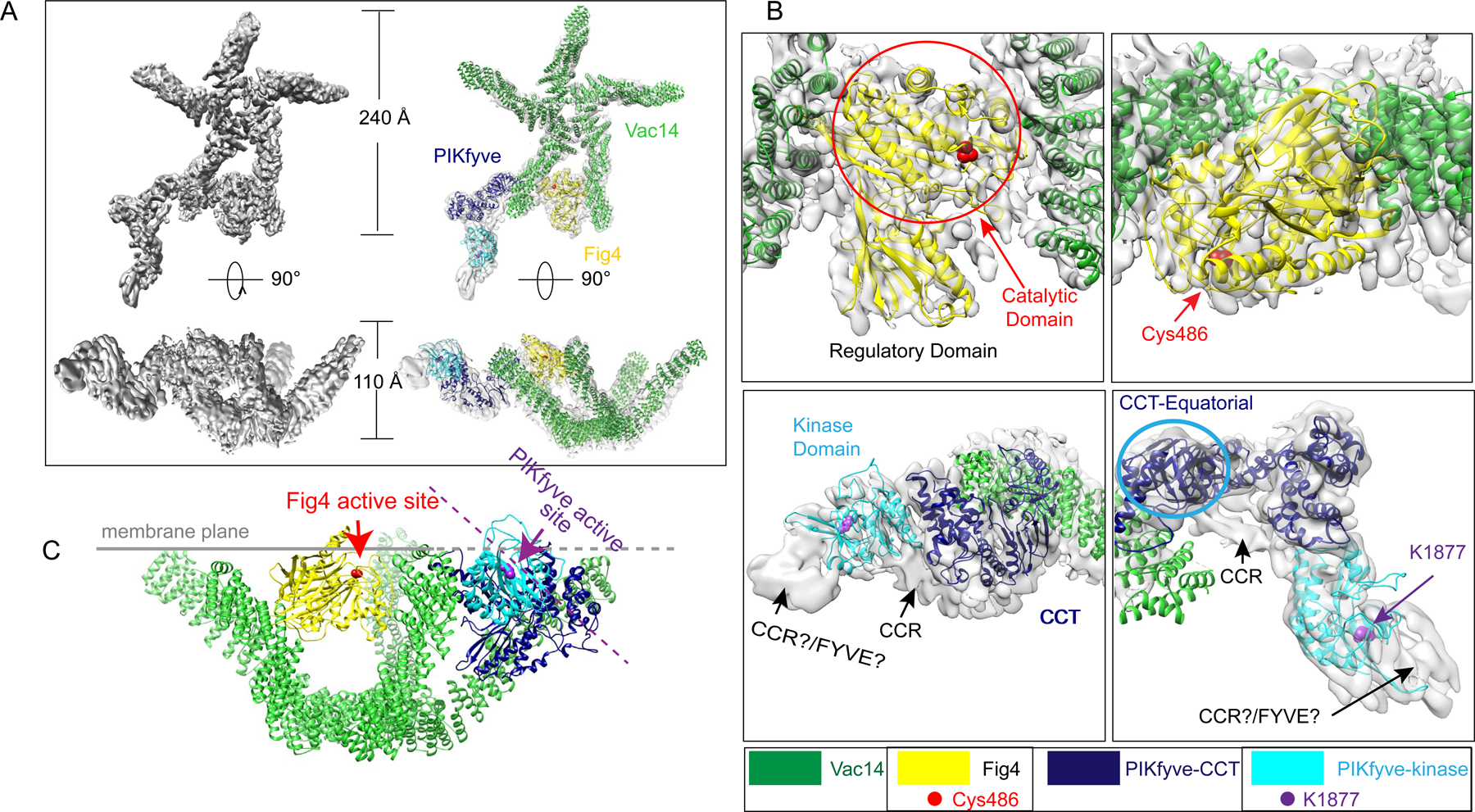
(A) The composite reconstruction alone and with model. The left panels show the maps only; the right panels also show docked protein models colored according to panel 1A. (B) Enlarged views showing the fit of the Fig4 Sac homology module (top panels) and the PIKfyve CCT and kinase modules (bottom panels) to the map. Two different views for Fig4 and PIKfyve are related by 90° rotations. (C) Model for PIKfyve complex interacting with membrane. The Fig4 active site is oriented to face the membrane, whereas the PIKfyve active site is twisted away from the membrane by ~45° and so cannot access membrane incorporated phosphoinositides.
PIKfyve comprises four structured regions, including a FYVE domain and CCR, CCT and kinase modules, separated by long unstructured sequences (Fig 1A). We generated homology models for the kinase and CCT modules of PIKfyve using threading software (RaptorX and I-Tasser (Kallberg et al., 2012; Yang et al., 2015), PDB 4tz7 and 3pn9). The CCT module was manually fitted into PIKfyve density closest to Vac14 (Fig. 2B). The CCT module consists of three domains, termed axial, intermediate and equatorial in the chaperonin TCP1/TRiC where this module is also present, with the so-called equatorial domain abutting Vac14. For the best fit into the map, the axial/intermediate domains of PIKfyve were reoriented with respect to the equatorial domain as compared to in other contexts (PDBIDs 1a6d, 5gw5, 3pn9). Consistent with our assignment, the PIKFyve CCT-domain is required for the formation of the ternary complex (Botelho et al., 2008; Ikonomov et al., 2009). The kinase domain was docked into the map adjacent to the CCT-domain, with the kinase active site twisted by ~45° away from the top of the Vac14 “cup” (Fig. 2B,C). As the resolution in this part of the map was low (local resolutions are ~8–10 Å, Fig. S2D), locations of both the CCT and kinase domains were independently confirmed by negative stain EM of MBP-labeled PIKfyve alone (Fig. 1D). In the cryo-EM maps, there is additional unassigned density in PIKfyve between the CCT and kinase domains as well as adjacent to the kinase domain N-terminus. An MBP-label inserted N-terminally to the CCR domain in PIKfyve is positioned between the CCT and kinase domains in negative stain micrographs (Fig 1D), suggesting that the density between the CCT and kinase modules in the cryo-EM maps corresponds to an N-terminal portion of the CCR domain. The remaining unassigned density could correspond to C-terminal portions of the CCR region and/or portions or the FYVE domain. The fold of the CCR module is unknown.
The PIKfyve complex is recruited to endo-lysosomal membranes via interactions between PI3P and the FYVE domain of PIKfyve (Hasegawa et al., 2017; Jin et al., 2016). Given the location of PIKfyve close to one of the Vac14 legs, it is plausible that the complex associates with membranes via the “rim” of the Vac14 cup (Fig 2C). If the complex is bound in this fashion, the Fig4 active site is oriented for optimal access to its lipid substrate while the PIKfyve active site is rotated away from the membrane (Fig 2C) and so is sub-optimally positioned for contact with substrate. The different orientations of their active site pockets in the complex indicates that either Fig4 or PIKfyve, but not both, can access membrane bound phosphoinositides at one time. This suggests a structural switching mechanism that controls the relative access of Fig4 and PIKfyve to substrate, and thus, their activities.
PI3P/PI35P2 metabolism by individual PIKfyve components is modulated within the intact complex.
To better understand how lipid phosphorylation and dephosphorylation activities are coordinated within the complex, we next compared these activities in subcomplexes, containing either PIKfyve or Fig4/Vac14, and intact complex. To make the intact complex for these experiments, we co-expressed Fig4 and Vac14, purified them together, and then mixed the Fig4/Vac14 subcomplex with PIKfyve that had been expressed and purified separately. We were unable to make Fig4 alone as it is proteolytically degraded in the absence of Vac14, likely because its C-terminal portions are unstructured.
We monitored PIKfyve lipid kinase activity on soluble diC6-PI3P (C6-C6-BODIPY-FL- PI3P) as well as PI3P incorporated in liposomes, with different outcomes. We followed diC6-PI3P conversion to diC6-PI(3,5)P2 by thin layer chromatography, finding that PIKfyve alone and PIKfyve/Fig4/Vac14 phosphorylated diC6-PI3P with similar efficiencies (Fig. 3A). Although Fig4 in the intact complex dephosphorylates soluble PI(3,5)P2 (see below), neither the presence of Fig4 or a catalytically inactivated version (C486S) significantly affected PI(3,5)P2 generation. This could be because the PI3P phosphorylation reaction is more efficient. In contrast, when we used a radiometric assay to monitor PI(3,5)P2 production from liposome incorporated PI3P and 32P-ATP, we found that only PIKfyve alone but not PIKfyve/Fig4/Vac14 was active (Fig. 3B). This supports the notion that the kinase domain cannot access PI3P in the membrane because of a conformational restraint imposed within the complex, as suggested by the structure.
Figure 3. Complex formation affects the enzymatic activities of PIKfyve and Fig4 with respect to phosphoinositide lipids.
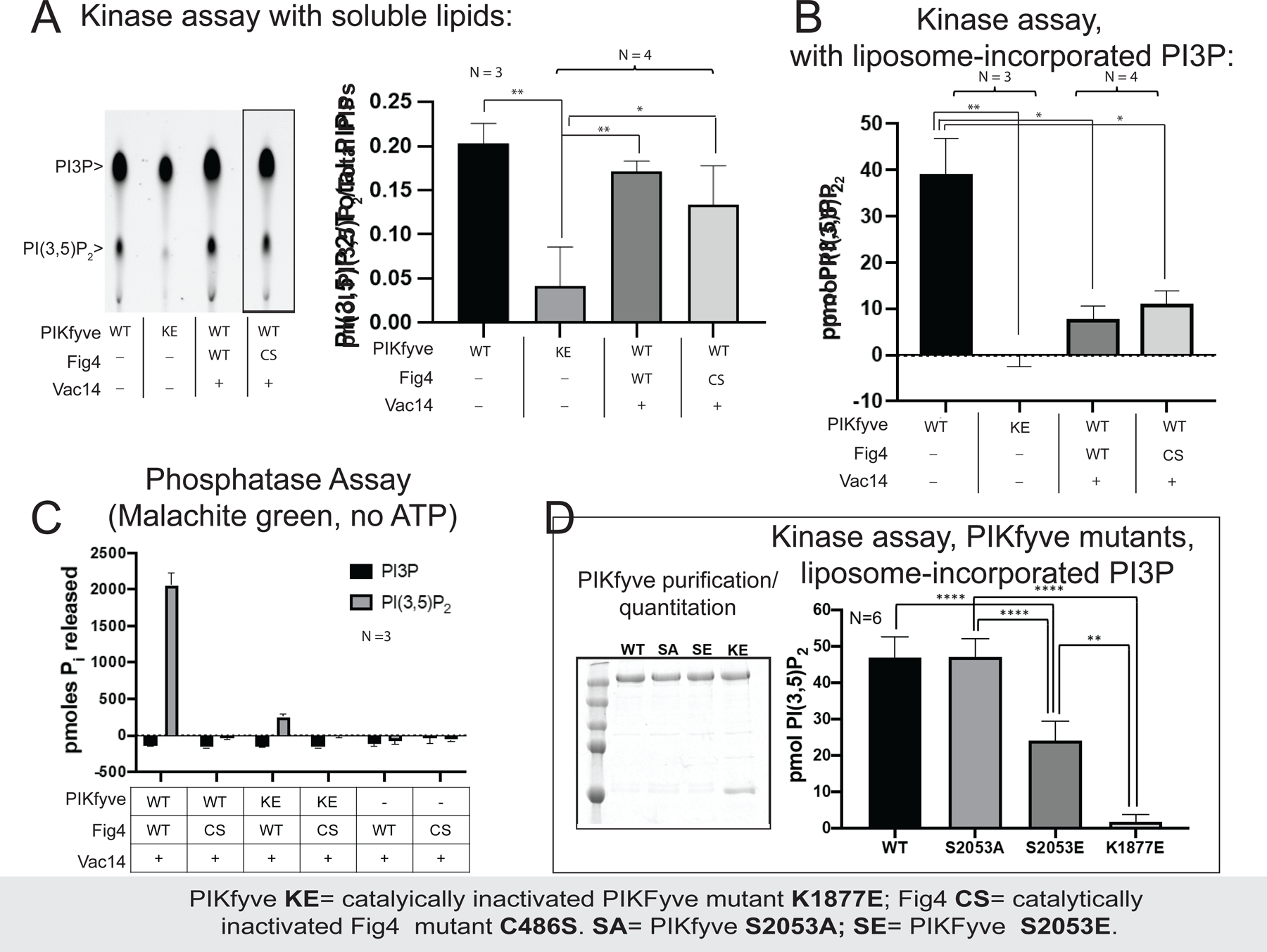
(A) PIKfyve alone and in the PIKfyve/Fig4/Vac14 complex phosphorylate soluble C6-C6-BODIPY-FL-PI3P with similar efficiency, independent of whether the lipid phosphatase Fig4 is catalytically active (WT) or not (C486S). The reaction was monitored by thin layer chromatography; representative image shown at left (lanes rearranged, as indicated by line, to match graph). (B) PIKfyve alone phosphorylates liposome incorporated PI3P in a radiometric assay more efficiently that the PIKfyve/Fig4/Vac14 complex. The PIKfyve active site may be sterically constrained in the complex and so unable to access membrane-incorporated PI3P. (C) In a malachite green assay monitoring dephosphorylation of soluble di-C8 PI(3,5)P2, robust Fig4 lipid phosphatase activity was observed only for Fig4 in a complex with WT PIKfyve. Complexes comprising a catalytically inactivated PIKfyve mutant (K1877E) were significantly less active; binary complex lacking PIKfyve was the least active. (D) PIKfyve lipid kinase activity is inhibited when Ser2053 in the kinase activation loop is mutated to glutamate to mimic its phosphorylation but not when it is changed to alanine. Experiments were carried out at least 3 times, as indicated. Error bars are standard deviations, and significance was determined by Welch’s t-test.
We next compared the lipid phosphatase activities of Fig4/Vac14 and PIKfyve/Fig4/Vac14 on diC8-PI(3,5)P2 using the malachite green assay, which measures the levels of orthophosphate released upon dephosphorylation. Robust Fig4 lipid phosphatase activity was observed only in the intact complex, including PIKfyve (Fig. 3C). The conformational changes in Fig4/Vac14 induced when PIKfyve is added may relieve an inhibition, perhaps by unblocking or rearranging the Fig4 active site. They may also correctly orient the Fig4 active site with respect to the membrane, but such a reorientation would not be necessary for activity with respect to the soluble diC8-lipids as used in the assay.
The protein kinase and phosphatase activities of PIKfyve and Fig4 regulate phosphoinositide metabolism.
PIKfyve has been well established as a ser kinase that autophosphorylates itself to inhibit its lipid kinase activity (Sbrissa et al., 2000). We found, unexpectedly, that PIKfyve protein kinase activity is also required for the lipid phosphatase activity of Fig4 within the complex, for only wild-type PIKfyve but not a catalytically inactivated version (K1877E) stimulates lipid hydrolysis by Fig4 in the malachite green assays (Fig. 3C). Because PIKfyve and Fig4/Vac14 were prepared separately and then mixed in the absence of ATP, it is improbable that Fig4 stimulation is due to its phosphorylation by PIKfyve. A likely explanation is that it is PIKfyve autophosphorylation that stimulates Fig4 activity.
To determine which sites in PIKfyve are autophosphorylated and to obtain insights as to how PIKfyve autophosphorylation might affect the activities of the complex with respect to phosphoinositides, we prepared two versions of PIKfyve/Fig4/Vac14, one containing wild-type and another the catalytically inactivated PIKfyve (K1877E). The purified reconstituted complexes were given the opportunity to auto-phosphorylate in the presence of ATP, Mg2+, and Mn2+. We then analyzed the PIKfyve phosphosites by mass spectrometry (MS/MS), finding serines that were phosphorylated only in the wild-type construct (S23, S48, S1522, S1669, S1969, and S2053) (Fig. 4). One of these sites (S2053) was of particular interest as it is located in the activation loop (residues 2036–2069) near the kinase active site, where it might play a role in inhibiting kinase activity. Consistent with such a role, we found a ~2-fold reduction in lipid kinase activity when we mutated this serine to glutamate, mimicking phosphorylation, versus no reduction when the serine was changed to alanine (Fig. 3D). This reduction in activity is similar to that reported before for an autophosphorylated versus a dephosphorylated form of the kinase (Sbrissa et al., 2000). We speculate that PIKfyve autophosphorylation at one or more of the six phosphosites we identified also affects Fig4 lipid phosphatase activity, although this remains to be investigated.
Figure 4. PIKfyve autophosphorylates itself, and Fig4 is also a protein phosphatase that dephosphorylates PIKfyve.
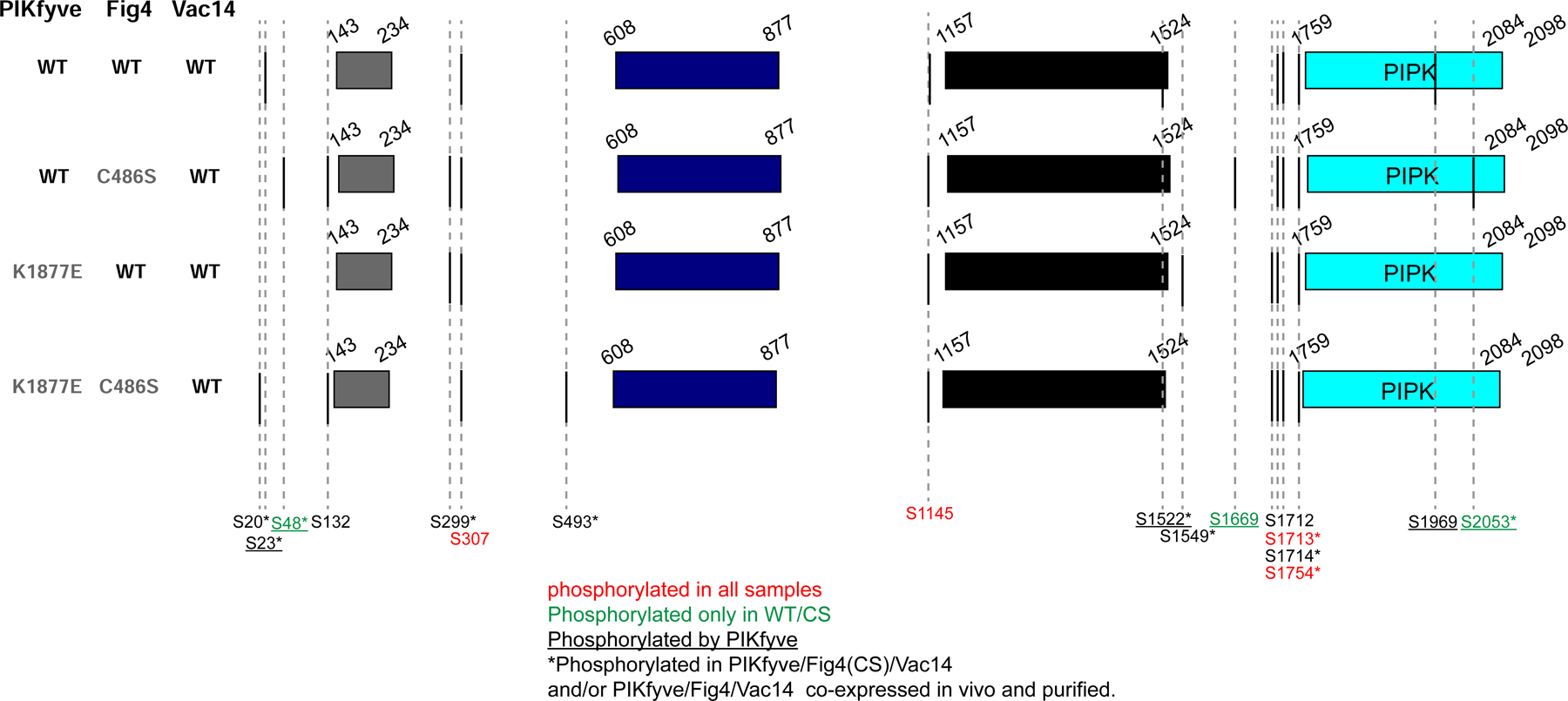
To reconstitute ternary complexes, WT PIKfyve or a catalytically inactivated mutant (K1877E) were mixed in equimolar amounts with Fig4 (WT or C486S)/Vac14 complexes. After incubation with ATP to allow phosphorylation, samples were resolved by SDS-PAGE. Bands for PIKfyve excised from the gels was analyzed by MS/MS to identify phosphosites. MS/MS analysis was also used to identify PIKfyve phosphosites in complexes produced by co-expressing all three proteins, with either WT or catalytically inactivated Fig4 (C486S). Importantly, Ser2053 in the kinase activation loop is phosphorylated in the presence of WT PIKfyve but not K1877E mutant and is dephosphorylated in complexes containing WT Fig4 but not C486S mutant. This suggests a model where Fig4 activates PIKfyve by dephosphorylating PIKfyve’s activation loop Ser2053, explaining why maximal PI(3,5)P2 production in vivo requires catalytically active Fig4.
The observations (1) that PIKfyve is both a lipid and a protein kinase whose autophosphorylation affects both lipid kinase and phosphatase activities of the PIKfyve complex (Sbrissa et al., 2000)(and above) and (2) that Fig4 catalytic activity is required for PIKfyve upregulation in vivo (Duex et al., 2006a; Duex et al., 2006b; Strunk et al., 2020) prompted us to explore the possibility that Fig4 might affect PIKfyve activity as a protein phosphatase. To determine whether Fig4 might act on PIKfyve, we prepared two versions of the PIKfyve/Fig4/Vac14 complex, as described before, one containing wild-type and the other a catalytically inactivated Fig4 mutant (C486S). We again analyzed PIKfyve phosphosites in these complexes by mass spectrometry after allowing for autophosphorylation in the presence of ATP, Mg2+, and Mn2+, finding that three of the sites phosphorylated by PIKfyve are dephosphorylated by Fig4 (S48, S1669, S2053). These sites include Ser2053 in the PIKfyve activation loop, whose phosphorylation state affects lipid kinase activity (Fig 3D). As another approach, we also overexpressed and purified the two versions of the complex in mammalian cells (Expi293F) and similarly analyzed PIKfyve phosphosites, with similar results (S48 and S2053 are differentially phosphorylated). These results strongly support the idea that in addition to acting as PI(3,5)P2 phosphatase, Fig4 is a serine phosphatase that regulates PIKfyve lipid kinase activity, explaining why an active form of Fig4 is required for normal levels of PI(3,5)P2 in vivo (Duex et al., 2006a; Duex et al., 2006b; Strunk et al., 2020). PTEN, the phosphatase that dephosphorylates PI(3,4,5)P3 during PI3K/Akt signaling, also is active on protein substrates (Myers et al., 1997), setting a precedent for phosphatases with dual specificity on lipids and proteins.
Although relatively long, the PIKfyve activation loop cannot reach the Fig4 active site if PIKfyve and Fig4 are placed as in the cryo-EM structure. It is plausible, though, that the kinase and phosphatase active sites can move closer together as the complex flexes, or that there are other conformations that allow for closer proximity, or that PIKfyve and Fig4 from different complexes interact in trans, or that Fig4 can act on PIKfyve either as the complex assembles or disassembles. Conformational changes within the complex will be subject to further investigation.
Concluding thoughts.
The unusual 1:1:5 stoichiometry in the PIKfyve/Fig4/Vac14 complex is intriguing. From our structural studies, a key role of Vac14 pentamerization is as a scaffold that coordinates Fig4 and PIKfyve activities to avoid futile cycles of ATP hydrolysis. The Vac14 scaffold helps to optimally orient Fig4 in the complex with respect to the membrane for its lipid-targeted catalytic activities whereas PIKfyve is constrained and unable to access membrane incorporated PI3P. Still, there may be additional roles for Vac14 pentamerization. For example, the Vac14 pentamer could help define membrane microzones that are enriched in either PI3P or PI(3,5)P2, and these roles remain to be investigated.
Our studies have also uncovered an exquisite interplay between the lipid-directed activities of PIKfyve and Fig4 and their protein-directed activities that affect phosphoinositide metabolism. We have found that PIKfyve autophosphorylation downregulates its lipid kinase activities (as also reported in (Sbrissa et al., 2000)) while simultaneously upregulating Fig4 activity with respect to phosphoinositide lipids. We have also discovered a role for Fig4 as a protein phosphatase required for PIKfyve activation. Likely, PIKfyve and Fig4 will additionally be regulated by protein kinases and phosphatases extrinsic to the complex, and this remains to be explored.
Limitations.
While these studies represent a major advance in understanding the regulation within the PIKfyve complex and lysosomal phosphoinositide homeostasis, we acknowledge limitations in the scope and interpretation of our data. First, the moderate resolution of our cryo-EM reconstruction precludes finer-grained analysis of the PIKfyve complex inter-subunit binding interfaces and the positioning and folds of some elements, such as the PIKfyve activation loop, CCR and FYVE domains, and the Fig4 C-terminus. Second, we have so far visualized the PIKfyve complex in one conformation, although our studies suggest that additional states exist. These considerations limit the structure-based conclusions we can draw about the activation and inactivation mechanisms of PIKfyve and Fig4. Further, we have examined PIKfyve and Fig4 activities only in the context of the purified complex. The PIKfyve complex is a well-known regulatory target of other signaling pathways whose influence is not considered here. Indeed, our mass spectrometry experiments (in Fig. 4) revealed several phosphorylated sites on PIKfyve unattributable to its auto-kinase activity. Additionally, Vac14 has been reported to interact with a host of proteins, including Rabs, RabGEFs, RabGAPs, BAR domain proteins, scaffolding proteins, and others (Currinn et al., 2016; Ikonomov et al., 2015; Jin et al., 2016; Malia et al., 2018; Mayer et al., 2018; Schulze et al., 2014), which could modulate and be modulated by its interaction with the membrane and with PIKfyve and Fig4. These interactions likely play a role in spatio-temporal regulation of PIKfyve and Fig4 enzymatic activities, as well as signaling between the PIKfyve complex and regulators and effectors. The influence of outside factors on the activities within the PIKfyve complex offers clear avenues for follow-up work.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact (karin.reinisch@yale.edu).
Materials Availability
Plasmids generated in this study will be made available upon request made to the Lead Contact.
Data and Code Availability
Cryo-EM maps and atomic models generated for this study are available at the EM Data Bank (EMD-22631, 22647, 22634) and the Protein Data Bank (7K1W, 7K2V, 7K1Y).
Experimental Model and Subject Details
All proteins were produced by transfection of Expi293F cell suspension culture (Gibco Thermo Fisher) in Expi293 Expression Medium (Gibco Thermo Fisher) at 37°C, in the presence of 8% CO2, according to manufacturer instructions.
METHOD DETAILS
Plasmid construction.
Constructs of wildtype Vac14, Fig4, and PIKfyve were PCR amplified from plasmids gifted by the laboratory of L. Weisman and cloned into indicated linearized plasmids using Gibson Assembly (NEB) (Tables S1, S2). Plasmids were linearized via BamH1 and Not1 restriction enzymes (NEB). StrepII- tagged Vac14 and His-tagged Fig4 were cloned into pcDNA3.1 vector. 3xFLAG-Fig4 and -PIKfyve were cloned into pCMV-10 vector. The sequence for MBP was inserted into the vectors for PIKfyve or Vac14, as indicated in Fig. 1A, using Gibson Assembly (NEB).
Protein expression and purification.
For EM specimens, constructs encoding 3xFLAG-PIKfyve(S2053E), His8-Fig4, and 2xStrepII-Vac14 were co-transfected into Expi293F cells according to manufacturer instructions. Proteins were expressed for 48 hours after transfection, then harvested by centrifugation and flash-frozen for storage or immediately used for protein purification. Cells were lysed by 3 cycles of freezing and thawing in buffer containing 100 mM HEPES, 700 mM NaCl, 0.5 mM TCEP, 1 mM ATP, 1 mM MgCl2, and 1x EDTA-free protease inhibitor (Roche), pH 7.8. Insoluble debris was removed by centrifugation at 15000 x g for 30 minutes. The supernatant was incubated with anti-FLAG M2 beads (Sigma) for 2 hours, then washed with 3 x 10 bed volumes of buffer and eluted in 3 x 0.5 bed volume of buffer containing 0.25 mg/ml 3xFLAG peptide (APExBio). Elutions were immediately analyzed by negative-stain electron microscopy to assess sample quality and concentration. Grids were frozen immediately for cryoEM data collection.
For enzymatic assays and in vitro sample preparation for mass spectrometry, constructs encoding 3xFLAG-tagged versions of PIKfyve (WT, S2053E, S2053A, and K1877E) were transfected into Expi293F cells according to manufacturer instructions and expressed for 48 hours. For formation of ternary complexes, 3xFLAG-Fig4 and 2xStrepII-Vac14 or 3xFLAG-Fig4(C486S) and 2xStrepII-Vac14 were co-transfected into Expi cells for 48 hours. Cell pellets were resuspended and lysed by freeze-thaw in buffer containing 100 mM HEPES, 500 mM NaCl, 0.5 mM TCEP, 1x EDTA-free protease inhibitor (Roche), pH 7.8. After centrifugation to remove insoluble material, supernatants were incubated with anti-FLAG M2 beads, washed with 3 x 10 bed volumes of buffer, then incubated overnight in 10 bed volumes buffer with 1 mM ATP and 1 mM MgCl2 added. Beads were then washed again with 3 x 10 bed volumes of buffer, then eluted in 3 x 0.5 bed volume of buffer containing 0.25 mg/ml 3xFLAG peptide.
Specimen preparation and imaging by negative stain EM.
Indicated protein samples were diluted to 50nM–100nM. 4ul of sample was loaded onto glow discharged copper grids coated with 10nm of carbon film. Samples were negatively stained with 2% of uranyl acetate and visualized on a FEI Tecnai T12 electron microscope operating at voltage of 120kV with a nominal magnification of 52000x (2.14 Å/pixel at specimen level). Images were further analyzed via RELION 3.0.4 (Zivanov et al., 2018) to obtain 2D class averages.
Cryo-EM specimen preparation and data collection
(Table 1). Purified PIKfyve complex samples were supplemented with 0.02% ß-octylglucoside immediately before freezing and applied to Quantifoil Cu 300 mesh R1.2/1.3 grids glow discharged in residual air for 30 seconds at 20 mA. Grids were then blotted and plunge-frozen in liquid ethane cooled by liquid nitrogen using a Vitrobot Mark IV plunge-freezing robot (FEI) (Fig. S1B).
Table 1.
Data Collection and Processing Statistics
| Map1, Vac14-centered | Map2, PIKfyve-centered | Map3, Fig4-centered | |
|---|---|---|---|
| Magnification | 130000x | 130000x | 130000x |
| Voltage (kV) | 300 | 300 | 300 |
| Electron exposure (e−/Å2) | 64 | 64 | 64 |
| Defocus range (µm) | −1.5 to −3.5 | −1.5 to −3.5 | −1.5 to −3.5 |
| Pixel size (Å) | 1.05 | 1.05 | 1.05 |
| Symmetry imposed | C1 | C1 | C1 |
| Initial particle images (no.) | 649270 | 614008 | 614008 |
| Final particle images (no.) | 53279 | 19998 | 19998 |
| Map resolution (Å) | 5.25 | 6.6 | 5.1 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
1913 movies were collected in super-resolution mode on an FEI Titan Krios at 300 kV equipped with a Gatan K2 Summit direct electron detector, using a nominal magnification of 130000x in super-resolution mode with a magnified pixel size of 1.05 Å on the specimen level (counting mode). Movies were dose-fractionated into 40 frames of 0.2 seconds each at a dose rate of 1.6 e-/Å2/frame for a total dose of 64 e-/Å2. All micrographs were collected with the stage tilted at a 30º angle to overcome a preferred particle orientation bias.
Cryo-EM data processing
(Fig. S1 and S2). Motion correction for 1913 micrographs was performed in MotionCor2 (Zheng et al., 2017) with a binning factor of 2 and dividing micrographs into 5x5 patches. Global CTF calculation was performed with CTFFIND4.1 (Rohou and Grigorieff, 2015). 954 particles were manually picked and subjected to 2D reference-free classification in Relion 3.0.4 (Zivanov et al., 2018). Classes showing good particle definition were chosen as references for automated particle picking in Relion, yielding a dataset of 649,270 particles. Multiple rounds of 2D classification were used to remove ice contamination and bad particles, leaving 218356 particles representing 21 2D classes. These particles were used to generate an initial model using the stochastic gradient descent algorithm as implemented in Relion. The resulting reference model was used for 3D classification and refinement in Relion. After Bayesian polishing, followed by further 3D classification, refinement, and post-processing, a final model centered on the Vac14 pentamer, comprising 53279 particles with a resolution of 5.25 Å (FSC=0.143 criterion), was obtained.
The density of PIKfyve in the 5.25 Å map was diffuse and incomplete. To obtain a more complete map of the region around PIKfyve, particle subtraction was used in Relion to remove partial density for Vac14 and Fig4 from the particles, after which 2D classification was performed on the subtracted particles, yielding improved alignment of the kinase region. Particle selection from these 2D classes was used to re-center the classes on the center of mass of the remaining density, closer to the kinase, and these were used as references to re-autopick the dataset. The re-picked particles were subjected to 2D classification, initial modeling, 3D classification and refinement. This process of subtraction, re-centering, autopicking, and re-processing was repeated twice more before a set of 614008 particles, centered close to the kinase, which showed improved density after modeling, was obtained. These were subjected to 2D classification with circular masking to remove ice and bad particles, which yielded a good dataset of 39125 particles. Particles from the best-defined 2D classes representing a range of different views were used for initial modeling by stochastic gradient descent. This model was used for 3D classification and refinement. After polishing and re-refinement, a model comprising 19998 particles was used for multibody refinement to independently refine the kinase and the region around Fig4 comprising two copies of Vac14, yielding maps of these regions with nominal resolutions of 6.6 and 5.1 Å according to the FSC=0.143 criterion, respectively, after post-processing (Fig. S2B,C). Each of the reconstructions was generated by processing particles from only one autopick job, so as to avoid any chance of particle duplication after re-centering on the kinase. Local resolution was estimated using the algorithm implemented in RELION 3.0.4, and much of the map for the kinase domain is estimated ~8–10 Å resolution by this means (Fig. S2D). We attribute the discrepancy in part to the inherent difficulty of measuring local resolution in cryoEM maps, which can vary between algorithms. Importantly, this map exhibits well-defined tubular densities that closely match the helical arrangements of the CCT and kinase modules, and allowed confident fitting of these modules, confirmed further by analysis of MBP-tagged constructs by negative-stain EM.
Homology models of Fig4 Sac homology and PIKfyve kinase modules were obtained using RaptorX (Kallberg et al., 2012) against coordinates from PDB entries 3lwt and 4tz7, respectively. The PIKfyve CCT domain was modeled using I-TASSER (Yang et al., 2015) against coordinates from PDB entry 3p9e. For Fig4, the model was rigid- body fitted into the 5.1 Å cryoEM map, allowing for slight reorientation of the regulatory and catalytic domains, then manually adjusted in Coot (Emsley et al., 2010). The PIKfyve kinase domain was rigid-body fitted into congruent density in the map of the kinase, then manually adjusted in Coot. The PIKfyve CCT was split into separate models for the equatorial domain and the remainder of the module, which were rigid-body fitted independently to allow for the domains to rotate relative to one another. Vac14 helices were modeled de novo as poly-alanine into the 5.25 Å map using Coot, but their connectivity could not be ascertained from the density, nor sequence assigned.
Liposome preparation.
For PIKfyve kinase enzymatic assays, substrate liposomes were prepared by mixing lipid stocks dissolved in chloroform in glass test tubes in the following molar ratio: 75% DOPC (Avanti Polar Lipids), 20% liver PE (Avanti Polar Lipids), 5% PI3P diC16 (Echelon Biosciences). Lipid mixtures were evaporated to dryness under a nitrogen stream, then dried further under vacuum for 30 minutes. Dried lipid films were rehydrated under 1 ml buffer (100 mM HEPES pH 7.8, 500 mM NaCl, 0.5 mM TCEP, 0.75 M sucrose) at 85°C for 30 minutes, then vortexed for 30 seconds to generate crude liposomes. Liposomes were subjected to 7 freeze-thaw cycles to eliminate multi-lamellar structures, then mixed with 2 volumes sucrose-free buffer and pelleted for 40 minutes at 16,000 x g. The resulting liposome pellet was resuspended in sucrose-free buffer and pelleted again for 30 minutes. The second pellet was resuspended to 5 mM total lipid concentration in sucrose-free buffer and used immediately.
Lipid kinase enzymatic assays—on liposome incorporated PI3P.
Purified PIKfyve and Fig4/Vac14 samples were analyzed by SDS-PAGE and Coomassie staining, then quantitated by band densitometry against BSA standards. For complexes, PIKfyve WT or K1877E samples were mixed in equimolar amounts with Fig4/Vac14 complexes. All protein samples were diluted to a 100 nM final stock concentration (based on kinase concentration). Shortly before preparing reactions, liposomes were swollen by pelleting and resuspending in sucrose-free buffer containing 150 mM NaCl. Reaction mixtures were prepared by combining protein (10 nM final concentration), sucrose-free buffer, a 10x ATP/Mg2+/Mn2+ solution (1 mM ATP, 20 mM MgCl2, and 5 mM MnCl2 final concentration), and 32P-ATP (2.5 uCi per reaction), then adding substrate liposomes to a final concentration of 2.5 mM total lipid. Reactions were incubated at room temperature for one hour, then removed to tubes containing EDTA (10 mM final) and proteinase K (0.1 mg/ml final) and incubated for 15 minutes at room temperature. Liposomes were then pelleted by centrifugation for 15 minutes at 16,000 x g. Pellets were resuspended in 100 μl buffer, then transferred to scintillation vials for measurement of 32P radioactivity in a scintillation counter.
Lipid kinase enzymatic assay –on soluble PI3P lipids.
For the assays on soluble diC6-PI3P, the protein samples were diluted to 40nM final stock concentration (based on PIKfyve quantitation). Shortly before preparing reactions, green fluorophore labeled C6-C6-BODIPY-FL-PI3P (Echelon Biosciences) was dissolved in water to make a 50μM stock solution. Reaction mixtures (20μl total volume) were prepared by combining PIKfyve or the PIKfyve complex (10 nM final concentration) with diC6-PI3P (20 μM final concentration) in buffer (100 mM HEPES, pH 7.8, 150 mM NaCl, 0.5 mM TCEP) supplemented with 1 mM ATP, 2 mM MgCl2, and 0.5 mM MnCl2. The reactions were incubated at room temperature for 1.5 hours and terminated by introducing ten volumes of Chloroform:Methanol (1:2). The samples were dried by SpeedVac (ThermoFisher) at 35℃ for two hours, then dissolved in chloroform and separated by thin layer chromatography as in (Taylor and Dixon, 2001). The phosphoinositide spots were visualized under ultraviolet light and quantitated fluorometrically, using ImageJ.
Lipid phosphatase enzymatic assays.
Purified PIKfyve was mixed with purified Fig4/Vac14 complexes to achieve an equimolar ratio of PIKfyve to Fig4, with a final concentration of 100 nM ternary complex. For Fig4/Vac14 only assays, Fig4/Vac14 complexes were directly diluted to 100 nM. Reactions were carried out in a 50 ul volume containing 50 nM protein complex, 50 uM diC8 PI3P or PI(3,5)P2 (Echelon Biosciences), and 2 mM MgCl2 for 1 hour at 37°C. Released phosphate was quantitated using a malachite green colorimetric assay in 96-well plate format according to the manufacturer’s instructions (Cayman Chemical #30412051), with measurements performed on a BioTek Synergy H1M microplate reader.
PIKfyve phosphosite analysis by mass spectrometry.
For in vitro phosphorylation/dephosphorylation analysis, purified PIKfyve (WT or K1877E) and Fig4 (WT and C486S)/Vac14 complex samples were quantitated by band densitometry against BSA standards after SDS-PAGE and Coomassie staining. PIKfyve WT or K1877E samples were mixed in equimolar amounts with Fig4 (WT or C486S)/Vac14 complexes to give four different combinations. Samples were incubated in the presence of 1 mM ATP, 2 mM MgCl2, and 0.5 mM MnCl2 for 1 hour at room temperature. After incubation, samples were resolved by SDS-PAGE and stained with Coomassie Brilliant Blue. PIKfyve bands were excised from the gels after extensive washing in deionized water, and submitted to the University of Alabama-Birmingham Proteomics core facility for tryptic digestion and phosphopeptide identification by MS/MS. Samples of PIKfyve/Fig4/Vac14 and PIKfyve/Fig4(C486S)/Vac14 complexes produced by co-transfection in Expi293F cells were affinity purified (in the presence of phosphatase inhibitor and in the absence of ATP) and submitted for phosphosite analysis at the University of Alabama-Birmingham Proteomics core facility.
Quantification and Statistical Analysis.
All lipid kinase and lipid phosphatase assays were carried out at least three times, as indicated (in Fig. 3 and legend), with the indicated standard deviations. Statistical comparisons between each group of samples were calculated using Welch two- tailed unpaired t test. Results were indicated in the following manner: * for P < 0.05, ** for P < 0.01, *** for P < 0.001, where P < 0.05 is considered as significantly different.
Supplementary Material
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| cOmplete™, EDTA-free Protease Inhibitor Cocktail | Roche Applied Science | Cat # 11873580001 |
| 3X (DYKDDDDK) Peptide | APExBio | Cat # A6001 |
| Critical Commercial Assays | ||
| Gibson Assembly® Master Mix | New England BioLabs | Cat # E2611S |
| ANTI-FLAG® M2 Affinity Gel | Sigma Aldrich | Cat # A2220 |
| Octyl-beta-Glucoside Detergent | Thermo Fisher | Cat # 28310 |
| 18:1 (Δ9-Cis) PC (DOPC) | Avanti Polar Lipids | Cat # 850375 |
| 16:0–18:1 PE | Avanti Polar Lipids | Cat # 850757 |
| PI3P diC16 | Echelon Biosciences | Cat # P-3016 |
| PI(3,5)P2 diC16 | Echelon Biosciences | Cat # P-3516 |
| PI3P diC8 | Echelon Biosciences | Cat # P-3008 |
| PI(3,5)P2 diC8 | Echelon Biosciences | Cat # P-3508 |
| C6-C6-BODIPY-FL-PI3P | Echelon Biosciences | Cat # C-03F6 |
| C6-C6-BODIPY-FL-PI(3,5)P2 | Echelon Biosciences | Cat # C-35F6 |
| Malachite Green Phosphate Assay Kit | Cayman Chemical | Cat # 10009325 |
| Deposited Data | ||
| PIKfyve complex centered on Vac14 - map1 | This paper | EMD: 22634 |
| Vac14 backbone models | This paper | PDB: 7K1Y |
| PIKfyve complex centered on PIKfyve - map2 | This paper | EMD: 22647 |
| PIKfyve Kinase domain, CCT domain models | This paper | PDB: 7K2V |
| PIKfyve complex centered on Fig4 – map3 | This paper | EMD: 22631 |
| Fig4 Sac homology domain model | This paper | PDB: 7K1W |
| Experimental Models: Cell Lines | ||
| Expi293F™ Cells | Gibco™ Thermo Fisher | Cat # A14527 |
| Oligonucleotides | ||
| Primers for Recombinant DNA, see Table S2 | This paper | N/A |
| Recombinant DNA | ||
| Construct of wildtype Vac14, Fig4, PIKfyve | L. Weisman | N/A |
| Plasmid: pcDNA3.1 – 2xStrepII-tagged Vac14 | This paper | N/A |
| Plasmid: pcDNA3.1 – 2xStrepII-tagged Vac14 - MBP | This paper | N/A |
| Plasmid: pcDNA3.1 - His-tagged Fig4 -WT | This paper | N/A |
| Plasmid: pcDNA3.1 - His-tagged Fig4 -CS | This paper | N/A |
| Plasmid: pCMV-10 – 3xFLAG-Fig4 - WT | This paper | N/A |
| Plasmid: pCMV-10 – 3xFLAG-Fig4 - CS | This paper | N/A |
| Plasmid: pCMV-10 – 3xFLAG-PIKfyve - WT | This paper | N/A |
| Plasmid: pCMV-10 – 3xFLAG-PIKfyve – S2053E | This paper | N/A |
| Plasmid: pCMV-10 – 3xFLAG-PIKfyve – S2053A | This paper | N/A |
| Plasmid: pCMV-10 – 3xFLAG-PIKfyve – K1877E | This paper | N/A |
| Plasmid: pCMV-10 – 3xFLAG-PIKfyve – 722-MBP-723 | This paper | N/A |
| Plasmid: pCMV-10 – 3xFLAG-PIKfyve – 1192-MBP-1193 | This paper | N/A |
| Plasmid: pCMV-10 – 3xFLAG-PIKfyve – 1806-MBP-1807 | This paper | N/A |
| Software and Algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| RELION 3.0.4 | Zivanov et al., 2018 | DOI: 10.7554/eLife.42166 |
| RaptorX | Kallberg et al., 2012 | http://raptorx.uchicago.edu/ |
| I-TASSER | Yang et al., 2015 | https://zhanglab.ccmb.med.umich.edu/I-TASSER/ |
| Other | ||
| Quantifoil®, Orthogonal Array of 1.2µm Diameter Holes - 1.3µm Separation, mounted on a 300M Cu grid | TED PELLA, INC | Cat # 658–300-Cu |
Acknowledgements.
This work was supported by the NIH (R01 GM114068 and R35 GM131715 to KMR and R01 NS099340 to LSW), a Ruth L. Kirchstein NRSA to NK, a Yale China Scholarship Council Award to PL. We are grateful to P. De Camilli and T. Melia for their comments and suggestions for the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflict of interest.
References
- Botelho RJ, Efe JA, Teis D, and Emr SD (2008). Assembly of a Fab1 phosphoinositide kinase signaling complex requires the Fig4 phosphoinositide phosphatase. Mol Biol Cell 19, 4273–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CY, Landers JE, Bergren SK, Sapp PC, Grant AE, Jones JM, Everett L, Lenk GM, McKenna-Yasek DM, Weisman LS, et al. (2009). Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. Am J Hum Genet 84, 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CY, Zhang Y, Dowling JJ, Jin N, Adamska M, Shiga K, Szigeti K, Shy ME, Li J, Zhang X, et al. (2007). Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature 448, 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currinn H, Guscott B, Balklava Z, Rothnie A, and Wassmer T (2016). APP controls the formation of PI(3,5)P(2) vesicles through its binding of the PIKfyve complex. Cell Mol Life Sci 73, 393–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duex JE, Nau JJ, Kauffman EJ, and Weisman LS (2006a). Phosphoinositide 5- phosphatase Fig 4p is required for both acute rise and subsequent fall in stress-induced phosphatidylinositol 3,5-bisphosphate levels. Eukaryot Cell 5, 723–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duex JE, Tang F, and Weisman LS (2006b). The Vac14p-Fig4p complex acts independently of Vac7p and couples PI3,5P2 synthesis and turnover. J Cell Biol 172, 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayle S, Landrette S, Beeharry N, Conrad C, Hernandez M, Beckett P, Ferguson SM, Mandelkern T, Zheng M, Xu T, et al. (2017). Identification of apilimod as a first-in-class PIKfyve kinase inhibitor for treatment of B-cell non-Hodgkin lymphoma. Blood 129, 1768–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa J, Strunk BS, and Weisman LS (2017). PI5P and PI(3,5)P2: Minor, but Essential Phosphoinositides. Cell Struct Funct 42, 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulseberg CE, Feneant L, Szymanska-de Wijs KM, Kessler NP, Nelson EA, Shoemaker CJ, Schmaljohn CS, Polyak SJ, and White JM (2019). Arbidol and Other Low-Molecular-Weight Drugs That Inhibit Lassa and Ebola Viruses. J Virol 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomov OC, Sbrissa D, Compton LM, Kumar R, Tisdale EJ, Chen X, and Shisheva A (2015). The Protein Complex of Neurodegeneration-related Phosphoinositide Phosphatase Sac3 and ArPIKfyve Binds the Lewy Body-associated Synphilin-1, Preventing Its Aggregation. J Biol Chem 290, 28515–28529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomov OC, Sbrissa D, Fenner H, and Shisheva A (2009). PIKfyve-ArPIKfyve- Sac3 core complex: contact sites and their consequence for Sac3 phosphatase activity and endocytic membrane homeostasis. J Biol Chem 284, 35794–35806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin N, Lang MJ, and Weisman LS (2016). Phosphatidylinositol 3,5-bisphosphate: regulation of cellular events in space and time. Biochem Soc Trans 44, 177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, and Xu J (2012). Template-based protein structure modeling using the RaptorX web server. Nat Protoc 7, 1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malia PC, Numrich J, Nishimura T, Gonzalez Montoro A, Stefan CJ, and Ungermann C (2018). Control of vacuole membrane homeostasis by a resident PI-3,5- kinase inhibitor. Proc Natl Acad Sci U S A 115, 4684–4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer L, Jasztal M, Pardo M, Aguera de Haro S, Collins J, Bariana TK, Smethurst PA, Grassi L, Petersen R, Nurden P, et al. (2018). Nbeal2 interacts with Dock7, Sec16a, and Vac14. Blood 131, 1000–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers MP, Stolarov JP, Eng C, Li J, Wang SI, Wigler MH, Parsons R, and Tonks NK (1997). P-TEN, the tumor suppressor from human chromosome 10q23, is a dual-specificity phosphatase. Proc Natl Acad Sci U S A 94, 9052–9057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson EA, Dyall J, Hoenen T, Barnes AB, Zhou H, Liang JY, Michelotti J, Dewey WH, DeWald LE, Bennett RS, et al. (2017). The phosphatidylinositol-3- phosphate 5-kinase inhibitor apilimod blocks filoviral entry and infection. PLoS Negl Trop Dis 11, e0005540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A, and Grigorieff N (2015). CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbrissa D, Ikonomov OC, and Shisheva A (2000). PIKfyve lipid kinase is a protein kinase: downregulation of 5’-phosphoinositide product formation by autophosphorylation. Biochemistry 39, 15980–15989. [DOI] [PubMed] [Google Scholar]
- Schulze U, Vollenbroker B, Braun DA, Van Le T, Granado D, Kremerskothen J, Franzel B, Klosowski R, Barth J, Fufezan C, et al. (2014). The Vac14-interaction network is linked to regulators of the endolysosomal and autophagic pathway. Mol Cell Proteomics 13, 1397–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shisheva A, Sbrissa D, Hu B, and Li J (2019). Severe Consequences of SAC3/FIG4 Phosphatase Deficiency to Phosphoinositides in Patients with Charcot-Marie-Tooth Disease Type-4J. Mol Neurobiol 56, 8656–8667. [DOI] [PubMed] [Google Scholar]
- Strunk BS, Steinfeld N, Lee S, Jin N, Munoz-Rivera C, Meeks G, Thomas A, Akemann C, Mapp AK, MacGurn JA, et al. (2020). Roles for a lipid phosphatase in the activation of its opposing lipid kinase. Mol Biol Cell, mbcE18090556. [DOI] [PMC free article] [PubMed]
- Taylor GS, and Dixon JE (2001). An assay for phosphoinositide phosphatases utilizing fluorescent substrates. Anal Biochem 295, 122–126. [DOI] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, and Zhang Y (2015). The I-TASSER Suite: protein structure and function prediction. Nat Methods 12, 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo- electron microscopy. Nat Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM maps and atomic models generated for this study are available at the EM Data Bank (EMD-22631, 22647, 22634) and the Protein Data Bank (7K1W, 7K2V, 7K1Y).


