Abstract
There is increasing interest among cancer researchers in the study of Piwi-interacting RNAs (piRNAs), a group of small RNAs important for maintaining genome stability in the germline. Aberrant expression of piRNAs in cancer could imply an involvement of these regulatory RNAs in neoplastic transformation. On top of that, it could enable early cancer diagnosis based on RNA analysis in liquid biopsies, as piRNAs are not expected to widely circulate in the bloodstream of healthy individuals. Indeed, it has recently been shown that serum piR-54265 allows for excellent discrimination between colorectal cancer patients and healthy controls. However, we have also shown that most somatic piRNAs reported to date in mammals are actually fragments of other noncoding RNAs. Herein, we show that reports positioning piR-54265 as a noninvasive biomarker for colorectal cancer were actually measuring variations in the levels of a full-length (72 nt) small nucleolar RNA in serum. This should place a cautionary note for future research in somatic and cancer-specific piRNAs. We deeply encourage this line of research but discuss proper ways to identify somatic piRNAs without the interference of erroneous entries contained in piRNA databases. We also introduce the concept of miscellaneous-piRNAs (m-piRNAs) to distinguish between canonical piRNAs and other small RNAs circumstantially associated with PIWI proteins in somatic cells.
Keywords: PIWI proteins, Piwi-interacting RNAs, cancer piRNAs, somatic piRNAs, piRNA databases
INTRODUCTION
Aberrant expression of Piwi-interacting RNAs (piRNAs) could be exploited for early cancer diagnosis based on minimally invasive liquid biopsies. In this regard, it has recently been shown that serum piR-54,265 could be used as a biomarker for early detection and clinical surveillance of colorectal cancer (CRC) (Mai et al. 2020). However, we have analyzed the sequence of piR-54265 and realized it corresponds to a 5′ fragment derived from the C/D box small nucleolar RNA SNORD57. Moreover, the analytical method used to validate the 29 nt piR-54265 as a biomarker of CRC is not specific for this fragment, and actually measures the levels of the full-length (72 nt) SNORD57.
From a biomarker point of view, this distinction could be deemed irrelevant as long as this specific sequence enables accurate discrimination between cancer patients and normal donors. Nevertheless, it is conceptually different to detect a cancer-specific transcript than to observe variations in the levels of an RNA that is expressed in virtually all cells of the body, including nucleated blood cells.
This example illustrates the problems of using piRNA databases, known to contain erroneous entries (Tosar et al. 2018), to identify piRNAs in somatic tissues and infer the roles and biomarker potential of PIWI-associated RNAs in cancer.
We consider as well that a further molecular and biochemical redefinition of PIWI-associated RNAs is indispensable to continue with a deeper understanding of the potential role of piRNAs in somatic contexts. In 2006 several groups characterized the small RNAs coimmunoprecipitating with PIWI-clade Argonaute proteins in mouse testes (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006) or in the Drosophila germline (Vagin et al. 2006). These papers coined the term “piRNA” for “Piwi-interacting RNAs” (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006) but immediately moved forward and showed the clustering of these sequences in the genome and their connection to selfish genetic elements. In subsequent years, the molecular machinery responsible for piRNA biogenesis, processing, and function was characterized (for review, see Czech and Hannon 2016). Thus, from a biological perspective, piRNAs are a group of PIWI-bound sequences of small size (24–32 nt) that are derived from piRNA clusters codified in the genome. Alternatively, they can be generated by PIWI-mediated cleavage of antisense transcripts, and typically bear a 2′-O-methyl mark at their 3′ end upon maturation. Albeit not exclusively, they are functionally linked to the silencing of transposable elements (TE) and tend to be expressed in the germline, where uncontrolled expression of TEs can affect genetic inheritance. However, from a semantic point of view, the term “Piwi-interacting RNA” is inclusive of any RNA interacting with a PIWI protein in any given cellular context. This leads to what we will call the “definition problem,” which summed up to the widespread use of noncurated piRNA databases and “black-box” bioinformatics pipelines, explains the erratic path toward an understanding of somatic piRNAs in cancer and biofluids.
Most piRNAs described in human colorectal cancer are ncRNA fragments
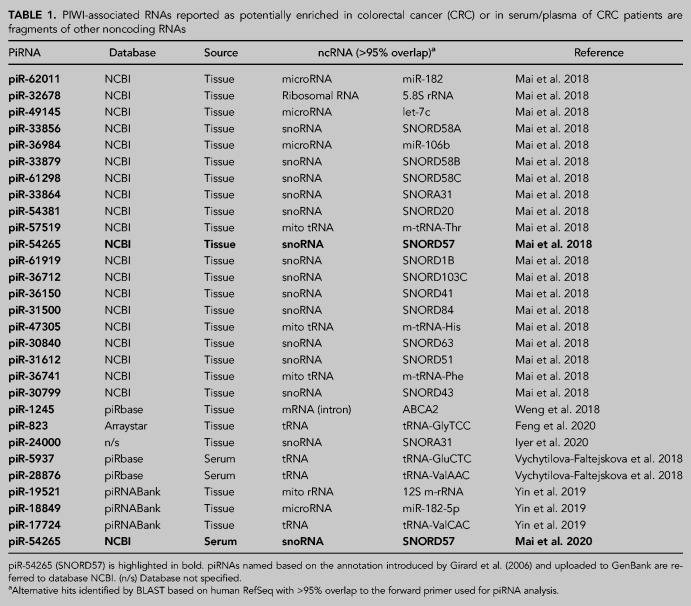
In 2018 we surveyed several reports describing piRNAs in nongonadal mammalian tissues and realized that virtually all putative somatic piRNAs could be classified as fragments of tRNAs, rRNAs, YRNAs, snoRNAs, or miRNAs (Tosar et al. 2018). This is remarkable, given that <1% of sequences present in piRNA databases belonged to this ambiguous category. Since then, several reports have described the presence of piRNAs in cancer, and particularly in CRC or in plasma/serum from CRC patients (Mai et al. 2018; Vychytilova-Faltejskova et al. 2018; Weng et al. 2018; Yin et al. 2019; Feng et al. 2020; Iyer et al. 2020; Mai et al. 2020). Strikingly, all of these CRC-specific piRNAs are mature miRNAs or ncRNA fragments (Table 1), consistent with our previous findings (Tosar et al. 2018).
TABLE 1.
PIWI-associated RNAs reported as potentially enriched in colorectal cancer (CRC) or in serum/plasma of CRC patients are fragments of other noncoding RNAs
Analytical assays for serum piR-54265 also detect full-length SNORD57
Among the list of CRC-specific piRNAs identified by analysis of TCGA (The Cancer Genome Atlas) data sets (Table 1), only piR-54265 could be validated by RT-qPCR in an independent cohort of paired CRC and nontumor samples (Mai et al. 2018). Furthermore, levels of this piRNA in cancer tissue correlated with clinical outcome of the patients. It was further showed that piR-54265 is oncogenic in CRC and associated with PIWIL2 by coimmunoprecipitation. Targeting of piR-54265 with an antisense oligonucleotide reduced tumor growth in mouse xenografts. However, it should be noted that the length of piR-54265 (29 nt) is not consistent with the modal length of piRNAs bound to PIWIL2 (26 nt) (Williams et al. 2015)
In a later study, the authors analyzed piR-54265 levels in the sera of 725 patients with CRC, 1303 patients with other digestive cancers, 192 patients with benign colorectal tumors and 209 healthy controls (Mai et al. 2020). Their RT-digital PCR assay showed elevated levels of piR-54265 in the sera of CRC patients vs controls (P = 1 × 10−67; Student t-test) or compared to sera from patients with other cancers of the gastrointestinal tract or benign colorectal tumors. Serum piR-54265 levels also correlated with clinical stage in CRC patients, decreased after surgery, and were again elevated in the case of tumor relapse. Furthermore, serum piR-54265 was a better indicator of tumor relapse than other biomarkers currently used in the clinical setting, including carcinoembryonic antigen, carbohydrate antigens 125 and 19-9 and methylated SEPTIN9.
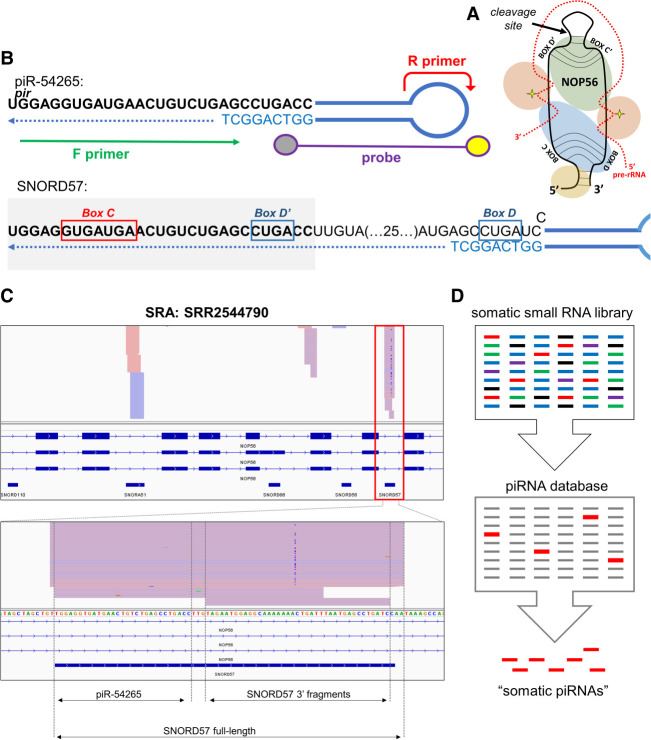
Despite these extremely promising results, we were concerned about piR-54265 showing a 100% overlap with the C/D box snoRNA SNORD57 (NCBI RefSeq: NR_002738.1), which is coded inside an intron of the NOP56 gene (Table 1). Of note, NOP56 is one of the proteins which bind C/D box snoRNAs to make a ribonucleoprotein particle (RNP) involved in pre-rRNA processing in the nucleolus (Fig. 1A).
FIGURE 1.
The analytical method described in Mai et al. (2020) is not specific for piR-54265. (A) Schematic diagram of a C/D box snoRNA (black), with its associated proteins and pre-rRNA (red). (B) Schematic representation of the stem–loop RT-ddPCR strategy designed in Mai et al. (2020) for the analysis of piR-54265 (top), also amplifying full-length SNORD57 (bottom). (C) Genome browser coverage tracks of TGIRT-seq reads obtained in plasma from a healthy human subject. (Bottom panel) Read coverage in the SNORD57 gene, showing no support for circulating piR-54265. (D) Illustration of the problem of identifying piRNAs based on mapping small RNA-seq data to piRNA databases, which contain entries that correspond to ncRNA fragments (shown in red) as well as bona fide piRNAs (shown in gray).
The SNORD57 transcript is composed of 72 nt, and the sequence annotated as piR-54265 corresponds to its first 29 nt. In theory, a 29-nt 5′ fragment of SNORD57 could be recruited by a PIWI-clade Argonaute protein and become Piwi-interacting RNA. However, a careful analysis of the reverse transcription-droplet digital PCR (RT-ddPCR) assay designed in (Mai et al. 2020) shows that this method is not specific for piR-54265/SNORD57 5′ fragments, as it is also predicted to amplify the full-length SNORD57 snoRNA (Fig. 1B).
Low abundancy of piR-54265 in serum compared to SNORD57
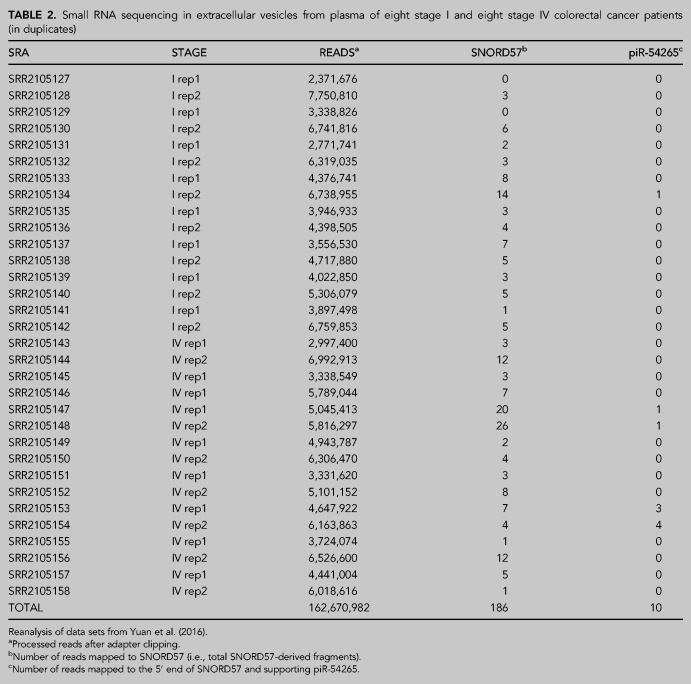
Is serum piR-54265 a biomarker of CRC (Mai et al. 2020) or could the aforementioned results be explained by variations in the levels of circulating SNORD57? To answer this question, we resorted to the extracellular RNA atlas (exrna-atlas.org) to identify small RNA sequencing studies performed in plasma or serum of CRC patients (Murillo et al. 2019). We identified one study containing a large data set of small RNAs in extracellular vesicles (EVs) purified from blood plasma of CRC patients representing different stages of the disease (Yuan et al. 2016). Remarkably, reanalysis of representative data sets suggests that the levels of piR-54265 are negligible in plasma of CRC patients, at least inside EVs (Table 2). However, a low but consistent number of reads corresponding to the 3′ half of SNORD57 was evident.
TABLE 2.
Small RNA sequencing in extracellular vesicles from plasma of eight stage I and eight stage IV colorectal cancer patients (in duplicates)
To analyze the presence of full-length SNORD57, we reanalyzed a data set produced by TGIRT-seq in normal human plasma (Qin et al. 2016). TGIRT-seq uses a thermostable group II intron RT and has been shown to be an effective sequencing technique to retrieve highly structured transcripts such as full-length tRNAs. It does not show the bias toward ncRNA fragments observed with conventional small RNA sequencing (Shurtleff et al. 2017) and gives extracellular RNA profiles more consistent with northern blot results (Tosar et al. 2020). Interestingly, TGIRT-seq supported the presence of full-length SNORD57 in human plasma (and other snoRNAs encoded inside the NOP56 gene, such as SNORA51) and of small RNA fragments derived from its 3′ end. No reads supported piR-54265 in the analyzed data set (Fig. 1C).
Because piRNAs are 2′-O-methylated (2′-O-Me) at their 3′ end (Horwich et al. 2007; Kirino and Mourelatos 2007a,b; Ohara et al. 2007; Saito et al. 2007), it could be argued that this modification was preventing piR-54265 detection in plasma using TGIRT-seq. Indeed, the template switching reaction characteristic of group II intron RT enzymes is inhibited by the presence of 2′-O-Me groups in the RNA (Lentzsch et al. 2019). However, it is possible to overcome this problem by performing the RT reaction under lower salt conditions (Lentzsch et al. 2019), as it was used in a more recent TGIRT-seq based RNA profiling of human plasma (Yao et al. 2020). Therefore, we reanalyzed representative data sets of this new study (SRA: SRR12047399; SRR12047420) and confirmed detection of full-length SNORD57 and the absence of reads supporting piR-54265. In summary, we conclude that SNORD57 rather than piR-54265 should be considered and further validated as a serum/plasma biomarker of CRC.
Are piRNAs expressed in somatic cancer cells?
Aberrant expression of PIWI proteins has been extensively documented in human cancer cells (Genzor et al. 2019; Li et al. 2020; Shi et al. 2020). Additionally, many reports have described the expression of piRNAs in cancer (Martinez et al. 2015; Yin et al. 2017, 2019; Mai et al. 2018; Vychytilova-Faltejskova et al. 2018; Weng et al. 2018; Tan et al. 2019; Iyer et al. 2020) or circulating in human body fluids (Bahn et al. 2015; Yang et al. 2015; Freedman et al. 2016; Yuan et al. 2016; Umu et al. 2018; Vychytilova-Faltejskova et al. 2018; Mai et al. 2020). However, the vast majority of these cancer or blood-derived piRNAs are fragments of other noncoding RNAs and probably correspond to false positives due to the use of noncurated piRNA databases (Table 1; Tosar et al. 2018).
Other authors have gone beyond simply mapping small RNA-seq data to piRNA databases and performed functional interrogation of candidate cancer-specific piRNAs. For instance, piR-651 has been identified by microarrays and then showed by RT-qPCR to be significantly up-regulated in gastric cancer (Cheng et al. 2011). Strikingly, the inhibition of piR-651 caused cell growth arrest in cancer cells, suggesting piRNAs could also act as therapeutic targets. However, analysis of piR-651 shows it is a fragment of the 28S ribosomal RNA, explaining the observed growth arrest phenotype. This example highlights the importance of understanding ambiguities in piRNA annotation.
Nongonadal somatic piRNAs are well documented in arthropods (Lewis et al. 2018) and mollusks (Jehn et al. 2018). We have recently performed de novo identification of piRNA clusters in Gastrotricha (Fromm et al. 2019), although the relative contribution of germline and somatic cells in this data is uncertain. As we have shown here and in a previous report (Tosar et al. 2018), most nongonadal somatic piRNAs described in mammals have ambiguous annotations. This leads to the question whether somatic piRNAs in mammals, and cancer piRNAs in particular, are real or an artefact due to improper data analysis.
The easier way to answer this question is to ask whether there are reports describing mammalian somatic piRNAs that cannot be classified as anything else than a piRNA. We might have missed other examples, but there are at least two very recent reports that would match this criterion. These includes the description of piR-30473 (GenBank: DQ570262) in large B-cell lymphoma (Han et al. 2020) and of piR-31470 (GenBank: DQ571358) in prostate cancer (Zhang et al. 2020). The former is codified in a single genomic locus in the immediacy of many other annotated piRNAs, in what corresponds to a prototypical piRNA cluster located in chromosome 18. The latter maps in multiple genomic loci because its sequence is derived from an ERV1 LTR retrotransposon.
A related question is whether fragments of other noncoding RNA families could be classified as piRNAs under certain circumstances. A recent preprint describes coexpression of the murine PIWI gene Mili (PiwiL2) and piRNAs in neural progenitor cells from the adult mouse brain (aNPCs) (Gasperini et al. 2020). Although this work describes five “piRNA clusters” composed of fragments of tRNAs, rRNAs, and snoRNAs, it is remarkable that the expression of these ncRNA fragments was affected by the knockdown of MILI. This suggests that MILI could stabilize some of these ncRNA fragments in aNPCs, in agreement with RNAs sequenced by immunoprecipitation of HIWI2 (human Piwi-like protein 4) in human cancer cells (Keam et al. 2014). However, others have found that RNA fragments copurifying with PIWIL1 (Piwi-like protein 1) in human colon cancer cells are indistinguishable from those found in background pull-downs (Genzor et al. 2019), suggesting piRNA expression is not reactivated in cancer cells. Consistent with these results, Li et al. (2020) labeled total RNAs with [γ-32P]-ATP and showed coimmunoprecpitation of ≈29 nt RNAs with PIWIL1 in testis, but not in PIWIL1-expressing pancreatic cancer cells. Overall, the involvement of PIWI proteins in cancer pathogenesis is well supported (Genzor et al. 2019; Li et al. 2020; Shi et al. 2020), and these reports do not show concurrent expression of canonical piRNAs in cancer, nor robust expression of other genes in the piRNA pathway (Genzor et al. 2019). However, it could be possible that aberrantly expressed PIWI proteins could recruit some ncRNA fragments that are abundant in the cytoplasm, in the absence of their canonical germline-specific piRNA cofactors. This would be favored by the lack of strong sequence constrains that is characteristic of these RNA-binding proteins (Stein et al. 2019). For example, human Argonaute-2 expressed in insect cells co-purifies with virtually any small RNA of 20 nt found in the host cell (Elkayam et al. 2012; Schirle and MacRae 2012). Many of these copurifying RNAs are not classified as microRNAs, but this classification would change if microRNAs were defined as “Ago-interacting RNAs.”
The problem with a context-dependent definition of “Piwi-interacting RNAs” is that it is incompatible with standard bioinformatics practices in small RNA-seq data analysis. If any small RNA could be a piRNA under certain circumstances, piRNA databases would be fed with miscellaneous entries and piRNAs will appear to be expressed everywhere. This is actually happening nowadays, as we tried to show. For instance, assume we sequence small RNAs in a hypothetical cell line where all four human PIWI genes have been silenced and hence there can be no bona fide piRNAs. If we then map sequencing reads against a noncurated piRNA database which contains contaminating entries, we will retrieve a collection of sequences that will be erroneously classified as piRNAs (Fig. 1D). Alternatively, defining piRNAs based on their biogenesis makes sense from the perspective of evolutionary biology and allows unambiguous genome annotation, but excludes some “Piwi-interacting RNAs” that could be relevant in cancer or in specific somatic cells.
In summary, the identification of somatic nongonadal piRNAs in mammals could be highly relevant to understand putative roles of PIWI proteins in cancer. Additionally, cancer piRNAs released into the bloodstream could constitute very promising biomarkers for the era of liquid biopsies. However, these putative piRNAs cannot be identified by simply mapping small RNA-seq data to piRNA databases. Doing so will produce inconsistent outputs that jeopardize our understanding of piRNA biology. As a rule of thumb, whenever working with a piRNA that might be of diagnostic use, we urge researchers to map back the sequence to the corresponding transcriptome and identify any alternative annotations of said sequence.
The piRNA community should agree on minimal experimental standards to claim the presence of piRNAs in mammalian somatic cells. Without closing the door to the possibility of PIWI proteins recruiting alternative small RNAs in the absence of canonical germline-specific piRNAs. And in such a case, what are these opportunistic PIWI-associated RNAs to be called? We would like to suggest the name of “miscellaneous piRNAs” (m-piRNAs). The most important aspect of this definition is that m-piRNAs should not be assigned a number (e.g., piR-1234) nor should they appear in piRNA databases. Their identity as a piRNA would be purely circumstantial, and they will probably always have alternative annotations. More importantly, pull-down and PIWI silencing assays should be essential to claim the presence of m-piRNAs, as their sole identification by RNA-seq would be considered irrelevant. The main advantage of this disengagement between piRNAs and m-piRNAs is that it secures the biological coherence of the germline piRNA silencing pathway while not limiting research on somatic piRNAs, especially in cancer.
ACKNOWLEDGMENTS
Funding for this work was received from Agencia Nacional de Investigación e Innovación, (ANII, Uruguay; FCE_3_2018_1_148745 and FSS_X_2018_1_149070) and from the National Institutes of Health, USA (UG3CA241694, supported by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director). J.P.T. and A.C. are researchers and receive general funding from PEDECIBA, ANII and Universidad de la República, Uruguay.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078444.120.
REFERENCES
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207. 10.1038/nature04916 [DOI] [PubMed] [Google Scholar]
- Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, Wong DTW, Xiao X. 2015. The landscape of microRNA, piwi-interacting RNA, and circular RNA in human saliva. Clin Chem 61: 221–230. 10.1373/clinchem.2014.230433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Guo JM, Xiao BX, Miao Y, Jiang Z, Zhou H, Li QN. 2011. PiRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin Chim Acta 412: 1621–1625. 10.1016/j.cca.2011.05.015 [DOI] [PubMed] [Google Scholar]
- Czech B, Hannon GJ. 2016. One loop to rule them all: the Ping-Pong cycle and piRNA-guided silencing. Trends Biochem Sci 41: 324–337. 10.1016/j.tibs.2015.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L. 2012. The structure of human argonaute-2 in complex with miR-20a. Cell 150: 100–110. 10.1016/j.cell.2012.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Yang M, Wei Q, Song F, Zhang Y, Wang X, Liu B, Li J. 2020. Novel evidence for oncogenic piRNA-823 as a promising prognostic biomarker and a potential therapeutic target in colorectal cancer. J Cell Mol Med 24: 9028–9040. 10.1111/jcmm.15537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman JE, Gerstein M, Mick E, Rozowsky J, Levy D, Kitchen R, Das S, Shah R, Danielson K, Beaulieu L, et al. 2016. Diverse human extracellular RNAs are widely detected in human plasma. Nat Commun 7: 11106. 10.1038/ncomms11106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm B, Tosar JP, Aguilera F, Friedländer MR, Bachmann L, Hejnol A. 2019. Evolutionary implications of the microRNA- and piRNA complement of Lepidodermella squamata (Gastrotricha). Noncoding RNA 5: 19. 10.3390/ncrna5010019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperini C, Pelizzoli R, Lo Van A, Mangoni D, Cossu RM, Pascarella G, Bianchini P, Bielefeld P, Scarpato M, Pons-Espinal M, et al. 2020. The piRNA pathway sustains adult neurogenesis by repressing protein synthesis. bioRxiv 10.1101/2020.09.15.297739 [DOI] [Google Scholar]
- Genzor P, Cordts SC, Bokil N V, Haase AD. 2019. Aberrant expression of select piRNA-pathway genes does not reactivate piRNA silencing in cancer cells. Proc Natl Acad Sci 166: 11111–11112. 10.1073/pnas.1904498116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202. 10.1038/nature04917 [DOI] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H. 2006. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev 20: 1709–1714. 10.1101/gad.1434406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Fan G, Song S, Jiang Y, Qian C, Zhang W, Su Q, Xue X, Zhuang W, Li B. 2020. piRNA-30473 contributes to tumorigenesis and poor prognosis by regulating m6A RNA methylation in DLBCL. Blood 10.1182/blood.2019003764 [DOI] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. 2007. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol 17: 1265–1272. 10.1016/j.cub.2007.06.030 [DOI] [PubMed] [Google Scholar]
- Iyer DN, Wan TMH, Man JHW, Sin RWY, Li X, Lo OSH, Foo DCC, Pang RWC, Law WL, Ng L. 2020. Small RNA profiling of piRNAs in colorectal cancer identifies consistent overexpression of piR-24000 that correlates clinically with an aggressive disease phenotype. Cancers (Basel) 12: 188. 10.3390/cancers12010188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehn J, Gebert D, Pipilescu F, Stern S, Kiefer JST, Hewel C, Rosenkranz D. 2018. PIWI genes and piRNAs are ubiquitously expressed in mollusks and show patterns of lineage-specific adaptation. Commun Biol 1: 137. 10.1038/s42003-018-0141-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keam SP, Young PE, McCorkindale AL, Dang THY, Clancy JL, Humphreys DT, Preiss T, Hutvagner G, Martin DIK, Cropley JE, et al. 2014. The human Piwi protein Hiwi2 associates with tRNA-derived piRNAs in somatic cells. Nucleic Acids Res 42: 8984–8995. 10.1093/nar/gku620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z. 2007a. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat Struct Mol Biol 14: 347–348. 10.1038/nsmb1218 [DOI] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z. 2007b. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA 13: 1397–1401. 10.1261/rna.659307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentzsch AM, Yao J, Russell R, Lambowitz AM. 2019. Template-switching mechanism of a group II intron-encoded reverse transcriptase and its implications for biological function and RNA-Seq. J Biol Chem 294: 19764–19784. 10.1074/jbc.RA119.011337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SH, Quarles KA, Yang Y, Tanguy M, Frézal L, Smith SA, Sharma PP, Cordaux R, Gilbert C, Giraud I, et al. 2018. Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nat Ecol Evol 2: 174–181. 10.1038/s41559-017-0403-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Yuan P, Rao M, Jin CH, Tang W, Rong YF, Hu YP, Zhang F, Wei T, Yin Q, et al. 2020. piRNA-independent function of PIWIL1 as a co-activator for anaphase promoting complex/cyclosome to drive pancreatic cancer metastasis. Nat Cell Biol 22: 425–438. 10.1038/s41556-020-0486-z [DOI] [PubMed] [Google Scholar]
- Mai D, Ding P, Tan L, Zhang J, Pan Z, Bai R, Li C, Li M, Zhou Y, Tan W, et al. 2018. PIWI-interacting RNA-54265 is oncogenic and a potential therapeutic target in colorectal adenocarcinoma. Theranostics 8: 5213–5230. 10.7150/thno.28001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai D, Zheng Y, Guo H, Ding P, Bai R, Li M, Ye Y, Zhang J, Huang X, Liu D, et al. 2020. Serum piRNA-54265 is a new biomarker for early detection and clinical surveillance of human colorectal cancer. Theranostics 10: 8468–8478. 10.7150/thno.46241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez VD, Vucic EA, Thu KL, Hubaux R, Enfield KSS, Pikor LA, Becker-Santos DD, Brown CJ, Lam S, Lam WL. 2015. Unique somatic and malignant expression patterns implicate PIWI-interacting RNAs in cancer-type specific biology. Sci Rep 5: 10423. 10.1038/srep10423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murillo OD, Thistlethwaite W, Rozowsky J, Subramanian SL, Lucero R, Shah N, Jackson AR, Srinivasan S, Chung A, Laurent CD, et al. 2019. exRNA atlas analysis reveals distinct extracellular RNA Cargo types and their carriers present across human biofluids. Cell 177: 463–477.e15. 10.1016/j.cell.2019.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K, Suzuki T. 2007. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat Struct Mol Biol 14: 349–350. 10.1038/nsmb1220 [DOI] [PubMed] [Google Scholar]
- Qin Y, Yao J, Wu DC, Nottingham RM, Mohr S, Hunicke-Smith S, Lambowitz AM. 2016. High-throughput sequencing of human plasma RNA by using thermostable group II intron reverse transcriptases. RNA 22: 111–128. 10.1261/rna.054809.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC. 2007. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev 21: 1603–1608. 10.1101/gad.1563607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle NT, MacRae IJ. 2012. The crystal structure of human argonaute2. Science 336: 1037–1040. 10.1126/science.1221551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Yang Z-Z, Liu S, Yang F, Lin H. 2020. PIWIL1 promotes gastric cancer via a piRNA-independent mechanism. Proc Natl Acad Sci 117: 22390–22401. 10.1073/pnas.2008724117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff MJ, Yao J, Qin Y, Nottingham RM, Temoche-Diaz MM, Schekman R, Lambowitz AM. 2017. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc Natl Acad Sci 114: E8987–E8995. 10.1073/pnas.1712108114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein CB, Genzor P, Mitra S, Elchert AR, Ipsaro JJ, Benner L, Sobti S, Su Y, Hammell M, Joshua-Tor L, et al. 2019. Decoding the 5′ nucleotide bias of PIWI-interacting RNAs. Nat Commun 10: 828. 10.1038/s41467-019-08803-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Mai D, Zhang B, Jiang X, Zhang J, Bai R, Ye Y, Li M, Pan L, Su J, et al. 2019. PIWI-interacting RNA-36712 restrains breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA. Mol Cancer 18: 9. 10.1186/s12943-019-0940-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosar JP, Rovira C, Cayota A. 2018. Non-coding RNA fragments account for the majority of annotated piRNAs expressed in somatic non-gonadal tissues. Commun Biol 1: 2. 10.1038/s42003-017-0001-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosar JP, Segovia M, Gámbaro F, Akiyama Y, Fagúndez P, Costa B, Possi T, Hill M, Ivanov P, Cayota A. 2020. Fragmentation of extracellular ribosomes and tRNAs shapes the extracellular RNAome. Nucleic Acids Res 48: 12874–12888. 10.1093/nar/gkaa674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umu SU, Langseth H, Bucher-Johannessen C, Fromm B, Keller A, Meese E, Lauritzen M, Leithaug M, Lyle R, Rounge TB. 2018. A comprehensive profile of circulating RNAs in human serum. RNA Biol 15: 242–250. 10.1080/15476286.2017.1403003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324. 10.1126/science.1129333 [DOI] [PubMed] [Google Scholar]
- Vychytilova-Faltejskova P, Stitkovcova K, Radova L, Sachlova M, Kosarova Z, Slaba K, Kala Z, Svoboda M, Kiss I, Vyzula R, et al. 2018. Circulating PIWI-interacting RNAs piR-5937 and piR-28876 are promising diagnostic biomarkers of colon cancer. Cancer Epidemiol Biomarkers Prev 27: 1019–1028. 10.1158/1055-9965.EPI-18-0318 [DOI] [PubMed] [Google Scholar]
- Weng W, Liu N, Toiyama Y, Kusunoki M, Nagasaka T, Fujiwara T, Wei Q, Qin H, Lin H, Ma Y, et al. 2018. Novel evidence for a PIWI-interacting RNA (piRNA) as an oncogenic mediator of disease progression, and a potential prognostic biomarker in colorectal cancer. Mol Cancer 17: 16. 10.1186/s12943-018-0767-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams Z, Morozov P, Mihailovic A, Lin C, Puvvula PK, Juranek S, Rosenwaks Z, Tuschl T. 2015. Discovery and characterization of piRNAs in the human fetal ovary. Cell Rep 13: 854–863. 10.1016/j.celrep.2015.09.030 [DOI] [PubMed] [Google Scholar]
- Yang X, Cheng Y, Lu Q, Wei J, Yang H, Gu M. 2015. Detection of stably expressed piRNAs in human blood. Int J Clin Exp Med 8: 13353–13358. [PMC free article] [PubMed] [Google Scholar]
- Yao J, Wu DC, Nottingham RM, Lambowitz AM. 2020. Identification of protein-protected mRNA fragments and structured excised intron RNAs in human plasma by TGIRT-seq peak calling. Elife 9: 60743. 10.7554/eLife.60743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Jiang XY, Qi W, Ji CG, Xie XL, Zhang DX, Cui ZJ, Wang CK, Bai Y, Wang J, et al. 2017. piR-823 contributes to colorectal tumorigenesis by enhancing the transcriptional activity of HSF1. Cancer Sci 108: 1746–1756. 10.1111/cas.13300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Qi W, Ji CG, Zhang DX, Xie XL, Ding Q, Jiang XY, Han J, Jiang HQ. 2019. Small RNA sequencing revealed aberrant piRNA expression profiles in colorectal cancer. Oncol Rep 42: 263–272. 10.3892/or.2019.7158 [DOI] [PubMed] [Google Scholar]
- Yuan T, Huang X, Woodcock M, Du M, Dittmar R, Wang Y, Tsai S, Kohli M, Boardman L, Patel T, et al. 2016. Plasma extracellular RNA profiles in healthy and cancer patients. Sci Rep 6: 19413. 10.1038/srep19413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Meng X, Pan C, Han R, Qu F, Gan W, Xiang Z, Han X, Li D. 2020. piR-31470 epigenetically suppresses the expression of glutathione S-transferase pi 1 in prostate cancer via DNA methylation. Cell Signal 67: 109501. 10.1016/j.cellsig.2019.109501 [DOI] [PubMed] [Google Scholar]