Abstract
The SARS-CoV-2 spike (S) glycoprotein contains an immunodominant receptor-binding domain (RBD) targeted by most neutralizing antibodies (Abs) in COVID-19 patient plasma. Little is known about neutralizing Abs binding to epitopes outside the RBD and their contribution to protection. Here, we describe 41 human monoclonal Abs (mAbs) derived from memory B cells, which recognize the SARS-CoV-2 S N-terminal domain (NTD) and show that a subset of them neutralize SARS-CoV-2 ultrapotently. We define an antigenic map of the SARS-CoV-2 NTD and identify a supersite (designated site i) recognized by all known NTD-specific neutralizing mAbs. These mAbs inhibit cell-to-cell fusion, activate effector functions, and protect Syrian hamsters from SARS-CoV-2 challenge, albeit selecting escape mutants in some animals. Indeed, several SARS-CoV-2 variants, including the B.1.1.7, B.1.351, and P.1 lineages, harbor frequent mutations within the NTD supersite, suggesting ongoing selective pressure and the importance of NTD-specific neutralizing mAbs for protective immunity and vaccine design.
Keywords: SARS-CoV-2, COVID-19, spike glycoprotein, N-terminal domain, NTD, neutralizing antibody, memory B cells
Graphical abstract
McCallum et al. identify a supersite in the N-terminal domain of SARS-CoV-2 spike protein that is targeted by neutralizing antibodies and exhibits mutation in response to selective pressure.
Introduction
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) at the end of 2019 resulted in the ongoing coronavirus disease 2019 (COVID-19) pandemic, bringing the world to a standstill (Zhou et al., 2020; Zhu et al., 2020). The lack of pre-existing immunity to SARS-CoV-2 combined with its efficient human-to-human transmission has already resulted in more than 109 million infections and over 2.3 million fatalities as of February 2021. Although vaccines are being developed and deployed at an unprecedented pace, the timeline for large-scale manufacturing and distribution to a large enough population for achieving community protection remains uncertain. As a result, prophylactic and/or therapeutic anti-viral drugs are expected to play a role in controlling COVID-19 disease and the ongoing pandemic. Such drugs may be helpful for unvaccinated individuals or those who respond poorly to vaccination, as well as upon waning of immunity and emergence of antigenically distinct variants or strains.
SARS-CoV-2 infects host cells through attachment of the viral transmembrane spike (S) glycoprotein to angiotensin-converting enzyme 2 (ACE2) followed by fusion of the viral and host membranes (Letko et al., 2020; Walls et al., 2020b). SARS-CoV-2 S also engages cell-surface heparan-sulfates (Clausen et al., 2020), neuropilin-1 (Cantuti-Castelvetri et al., 2020; Daly et al., 2020) and L-SIGN/DC-SIGN (Chiodo et al., 2020; Gao et al., 2020; Soh et al., 2020; Thépaut et al., 2020), which were proposed to serve as co-receptors, auxiliary receptors, or adsorption factors. SARS-CoV-2 S is the main target of neutralizing antibodies (Abs) in infected individuals and the focus of the many nucleic acid, vectored, and protein subunit vaccines currently deployed or in development (Corbett et al., 2020a, 2020b; Erasmus et al., 2020; Hassan et al., 2020; Keech et al., 2020; Mercado et al., 2020; Walls et al., 2020a). Besides blocking ACE2 attachment (Piccoli et al., 2020; Tortorici et al., 2020), some neutralizing Abs are alternatively expected to interfere with heparan-sulfate, neuropilin-1, or L-SIGN/DC-SIGN interactions.
The SARS-CoV-2 S protein comprises an N-terminal S1 subunit responsible for virus-receptor binding and a C-terminal S2 subunit that promotes virus-cell membrane fusion (Walls et al., 2020b; Wrapp et al., 2020). The S1 subunit comprises an N-terminal domain (NTD) and a receptor-binding domain (RBD), also known as domain A and B, respectively (Tortorici and Veesler, 2019). Abs targeting the RBD account for ∼90% of the neutralizing activity in COVID-19 convalescent sera (Greaney et al., 2021; Piccoli et al., 2020), and numerous monoclonal antibodies (mAbs) recognizing this domain have been isolated and characterized (Barnes et al., 2020a, 2020b; Baum et al., 2020b; Brouwer et al., 2020; Hansen et al., 2020; Ju et al., 2020; Piccoli et al., 2020; Pinto et al., 2020; Tortorici et al., 2020; Wang et al., 2020; Wu et al., 2020). Several RBD-specific mAbs capable of protecting small animals and non-human primates from SARS-CoV-2 challenge are able to neutralize viral infection by targeting multiple distinct antigenic sites (Baum et al., 2020a; Hansen et al., 2020; Jones et al., 2020; Pinto et al., 2020; Rogers et al., 2020; Tortorici et al., 2020; Zost et al., 2020). A subset of these mAbs is currently being evaluated in clinical trials or have recently received emergency use authorization from the FDA.
The limited immunogenicity of the SARS-CoV-2 NTD in COVID-19 patients (Piccoli et al., 2020; Rogers et al., 2020) has been hypothesized to result from its extensive N-linked glycan shielding (Walls et al., 2020b; Watanabe et al., 2020). However, recent studies have reported the isolation of NTD-targeted mAbs and their ability to neutralize SARS-CoV-2 infection in vitro, suggesting they could be useful for COVID-19 prophylaxis or treatment (Chi et al., 2020; Liu et al., 2020a). Although the NTD has been proposed to interact with auxiliary receptors in cell types that do not express ACE2 (e.g., DC-SIGN/L-SIGN), its role and the mechanism of action of NTD-targeted neutralizing mAbs remain unknown (Soh et al., 2020). Understanding the immunogenicity of different S domains and the function of mAbs targeting them, including the NTD, is critical to a full understanding of immunity during the pandemic.
Here, we analyze Ab responses in three COVID-19 convalescent individuals and describe 41 NTD-specific human mAbs. Integrating cryoelectron microscopy (cryo-EM), binding assays, and Ab escape mutant analysis, we define a SARS-CoV-2 NTD antigenic map and identify a supersite (designated site i) recognized by potent neutralizing mAbs. We show that these mAbs exhibit neutralization activities on par with best-in-class RBD-specific mAbs and efficiently activate Fc-mediated effector functions. We also identify immunologically important variation of the SARS-CoV-2 NTD suggesting that the S glycoprotein is under selective pressure from the host humoral immune response. Finally, we provide proof-of-principle that prophylactic administration of a highly potent NTD mAb confers protection of Syrian hamsters against SARS-CoV-2 challenge albeit selecting for escape mutants in some animals. Overall, these findings highlight the importance of NTD-specific neutralizing mAbs for protective immunity, virus evolution, and vaccine design.
Results
NTD-specific mAbs with potent neutralizing activity
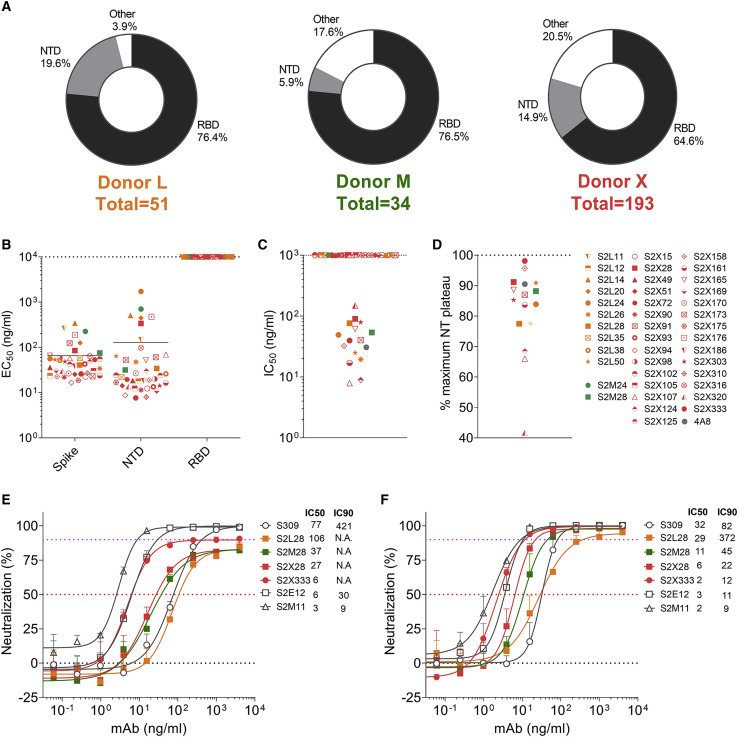
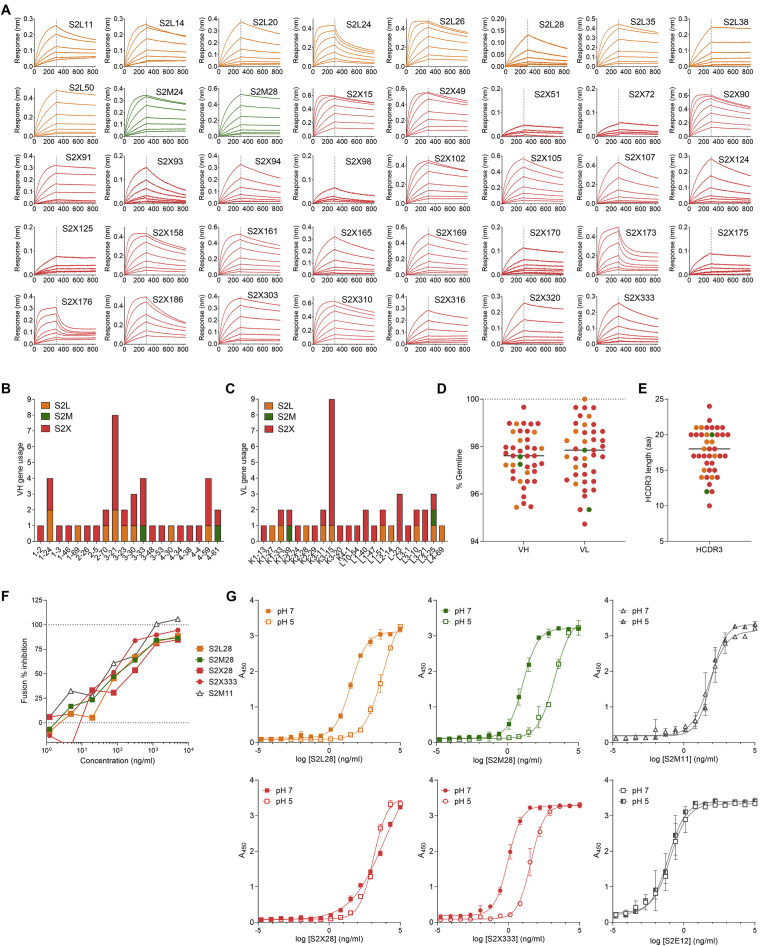
To discover mAbs targeting diverse SARS-CoV-2 epitopes, we sorted immunoglobulin (Ig)G+ memory B cells from peripheral blood mononuclear cells (PBMCs) of three COVID-19 convalescent individuals (L, M, X) using biotinylated prefusion SARS-CoV-2 S as a bait. The percentage of SARS-CoV-2 S-reactive IgG+ memory B cells ranged between 1.1%–1.3%. A total of 278 mAbs were isolated and recombinantly produced as human IgG1 (Figure 1 A). Characterization by ELISA showed that most mAbs isolated from the three donors recognize the RBD (65%–77%), with a smaller fraction targeting the NTD (6%–20%). The remaining mAbs (4%–20%) are expected to bind to either the S2 subunit or the C-D domains within the S1 subunit (Figure 1A). The low proportion of NTD-specific mAbs isolated from these donors is in line with the previously observed limited NTD immunogenicity in SARS-CoV-2-exposed individuals (Piccoli et al., 2020; Rogers et al., 2020). Overall, we identified 41 mAbs recognizing the SARS-CoV2 NTD with EC50s ranging between 7.6–698 ng/mL and nanomolar binding affinities, as evaluated using ELISA and biolayer interferometry, respectively (Figures 1B and S1 A; Table S1). These NTD-specific mAbs use a large repertoire of V genes, with an over-representation of IGHV3-21 and IGK3-15 genes (Figures S1B and S1C; Table S1), and harbor few somatic hypermutations (VH and VL are 97.57% and 97.54% identical to V germline genes, respectively; Figure S1D; Table S1), as previously described for many SARS-CoV-2 neutralizing mAbs binding to the RBD (Piccoli et al., 2020; Seydoux et al., 2020). CDRH3 lengths range between 10 and 24 amino acid residues (Figure S1E). Collectively, these data indicate that the Ab response to the SARS-CoV-2 NTD is polyclonal.
Figure 1.
Discovery of potent NTD-specific SARS-CoV-2 neutralizing mAbs from three convalescent individuals
(A) Pie charts showing the frequency of mAbs (cloned from IgG+ memory B cells) recognizing the SARS-CoV-2 NTD, RBD, or other S regions for patients L, M, and X.
(B) Binding of the 41 isolated NTD mAbs to immobilized SARS-CoV-2 S, NTD, or RBD analyzed by ELISA.
(C and D) Neutralization potencies (IC50, C) and maximal neutralization plateau (NT, D) of 15 NTD-specific neutralizing mAbs against SARS-CoV-2 S MLV pseudotyped virus. Data are from one out of two independent experiments performed.
(E and F) Dose-dependent neutralization of selected NTD- and RBD-specific mAbs against authentic SARS-CoV-2-Nluc assessed 6 h (at MOI 0.1) (E) or 24 h (at MOI of 0.01) (F) after infection. One independent experiment out of at least two is shown. Error bars indicate standard deviation of triplicates.
Figure S1.
Characteristics of SARS-CoV-2 NTD mAbs related to Figure 1
(A) Biolayer interferometry binding kinetic analysis of the SARS-CoV-2 NTD to immobilized mAbs.
(B and C) V gene usage for the heavy (B) and light (C) chains of the NTD mAbs.
(D) Nucleotide sequence identity of the mAbs isolated relative to the respective V germline genes.
(E) HCDR3 amino acid length for individual mAbs.
(F) Cell-to-cell fusion inhibition assay with Vero E6 cells transfected with SARS-CoV-2 S and incubated with varying concentrations of S2L28, S2M28, S2X28, S2X333 or the RBD-specific mAb S2M11.
(G) Binding of NTD- and RBD-specific mAbs to immobilized SARS-CoV-2 S at pH7 and pH5 as analyzed by ELISA. One independent experiment out of two is shown.
The in vitro neutralization activity of the NTD-specific mAbs was subsequently evaluated using a SARS-CoV-2 S pseudotyped murine leukemia virus (MLV) system (Millet and Whittaker, 2016; Walls et al., 2020b). Out of 41 mAbs, 9 are potent neutralizers (IC50 < 50 ng/mL) and 6 are moderate neutralizers (IC50 of 50–150 ng/mL) (Figure 1C). The remaining 25 mAbs were non-neutralizing. Most of the mAbs plateaued around 80%–90% maximum neutralization in this assay (Figure 1D). Evaluation of the neutralization potency of a subset of NTD-specific mAbs measured 6 h post-infection of Vero E6 cells infected with authentic SARS-CoV-2 virus confirmed that these mAbs did not completely block viral entry and instead plateaued at 80%–90% neutralization, as opposed to the RBD-specific mAbs S309, S2E12, and S2M11 that achieved 100% neutralization (Figure 1E) (Pinto et al., 2020; Tortorici et al., 2020). When the activity was measured at 24 h post-infection, however, all mAbs tested achieved 95%–100% neutralization with a marked enhancement of neutralization potency (Figure 1F). For instance, S2X333 neutralized SARS-CoV-2 with an IC50 of 2 ng/mL and an IC90 of 12 ng/mL, on par with the best-in-class ultrapotent RBD-targeting mAbs S2E12 and S2M11 (Figure 1F). The time-dependent difference of the results may reflect inhibition of extensive viral spread during the 24 h assay as opposed to the sole inhibition of viral entry measured after 6 h.
Previous studies established that SARS-CoV-2 infection of Vero E6 cells proceeds through cathepsin-activated endosomal fusion, as opposed to TMPRSS2-dependent entry, which is supposed to occur at the level of the plasma membrane and to be the most relevant route of lung cells infection (Hoffmann et al., 2020a, 2020b, 2020c). Although S2L28, S2M28, S2X28, and S2X333 efficiently blocked cell-cell membrane fusion of Vero E6 cells transiently transfected with full-length wild-type SARS-CoV-2 S (Figure S1F), binding of S2L28, S2M28, and S2X333 to SARS-CoV-2 S was dampened by 2 orders of magnitude at endosomal pH (pH5) compared to neutral pH (pH7) (Figure S1G). As cell-cell membrane fusion bypasses the endosomal compartment, S2L28, S2M28, S2X28, and S2X333 efficiently blocked fusion of Vero E6 cells transiently transfected with full-length wild-type SARS-CoV-2 S (Figure S1F). Therefore, partial neutralization may have been a result of reduced blocking of the endosomal entry route at 6 h post-infection, whereas cell-cell spread of the virus after 24 h was efficiently blocked.
NTD-specific neutralizing mAbs delineate an antigenic supersite
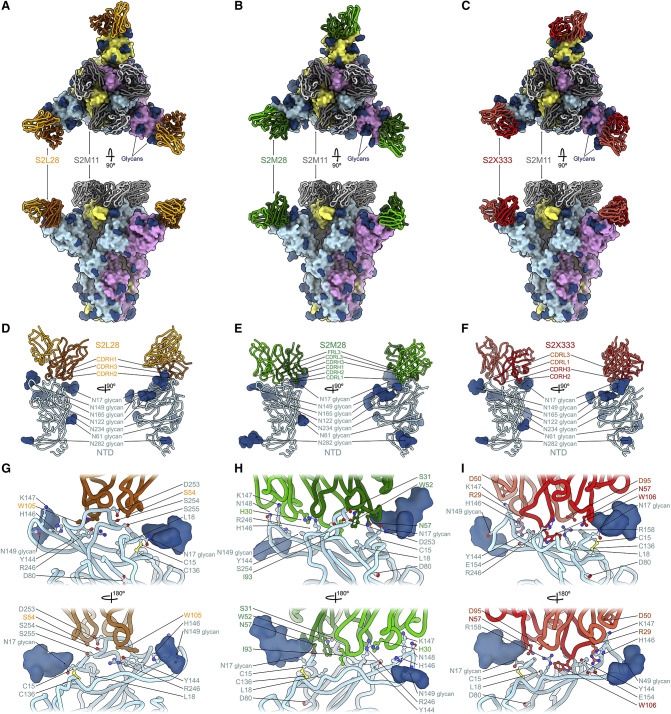
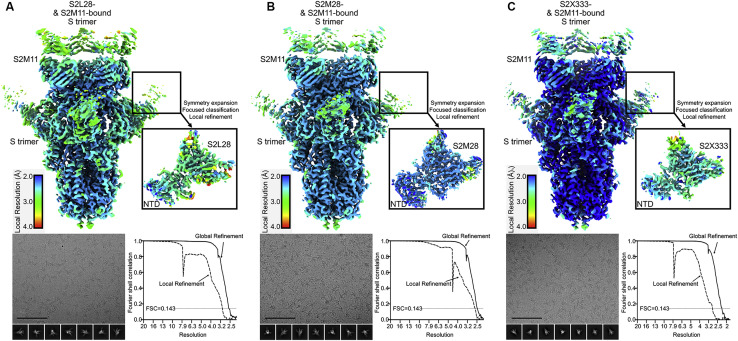
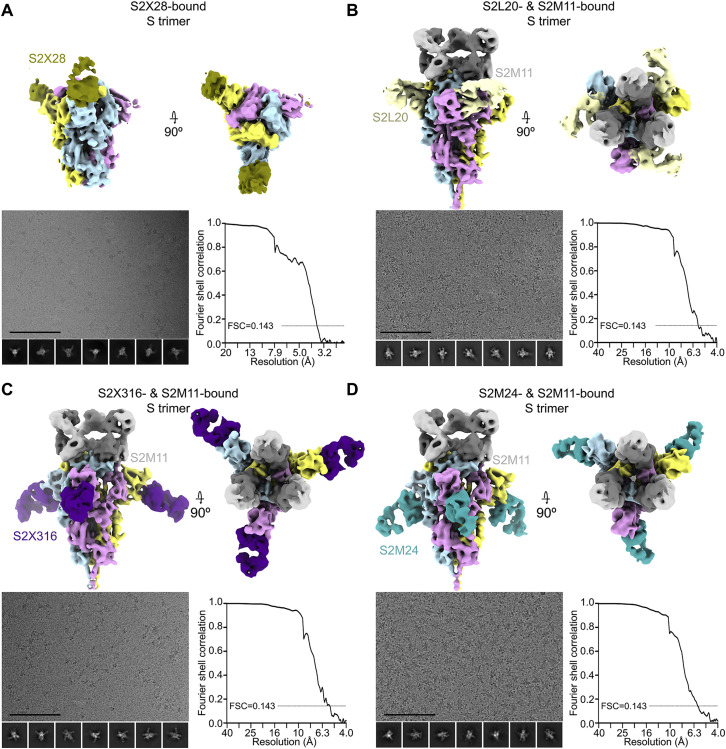
To elucidate the mechanism of potent SARS-CoV-2 neutralization by NTD mAbs, we carried out single-particle cryo-EM analysis of the SARS-CoV-2 S ectodomain trimer bound to one NTD-specific mAb from each donor—S2L28, S2M28 or S2X333—in combination with the RBD-specific mAb S2M11. S2M11 was used as it locks the RBDs in the closed state by recognizing a quaternary epitope spanning two adjacent RBDs, thus enabling the use of 3-fold symmetry during reconstruction (Tortorici et al., 2020). 3D classification of the particle images belonging to each dataset revealed the presence of homogeneous ternary complexes with three S2M11 Fabs bound to the RBDs and three bound NTD Fabs radiating from the trimer periphery. We determined reconstructions at 2.6 Å, 2.5 Å, and 2.2 Å for the S2L28/S2M11/S, S2M28/S2M11/S, and S2X333/S2M11/S complexes (Figures 2 A–2I and S2 A–S2C; Table S2). We subsequently used local refinement to account for the pronounced conformational dynamics of S2L28, S2M28, and S2X333 and obtained reconstructions at 2.6–3.0 Å resolution for the region comprising the Fab variable domains and their bound epitope in the NTD (Figures 2A–2I and S2A–S2C; Table S2). In parallel, we determined a crystal structure of the SARS-CoV-2 NTD in complex with the S2M28 Fab at 3.0 Å resolution revealing several additional ordered loops and N-linked glycans (Figures 2E–2H; Table S3).
Figure 2.
SARS-CoV-2 NTD neutralizing mAbs target the same antigenic supersite
(A–C) Ribbon diagrams in two orthogonal orientations of the SARS-CoV-2 S ectodomain trimer (surface) bound to the RBD-specific S2M11 Fab (gray) and to the NTD-targeted Fab S2L28 (A), S2M28 (B), or S2X333 (C).
(D–F) S2L28 (D), S2M28 (E), and SX333 (F) binding pose relative to the NTD.
(G–I) Zoomed-in views showing selected interactions of S2L28 (G), S2M28 (H), or S2X333 (I) with the NTD. SARS-CoV-2 S protomers are colored pink, cyan, and gold whereas N-linked glycans are rendered as dark blue surfaces.
See also Figure S2 and Tables S2–S4.
Figure S2.
Cryo-EM data processing of SARS-CoV-2 S bound to S2L28, S2M28, or S2X333, related to Figure 2
(Top) Unsharpened maps colored by local resolution calculated using cryoSPARC for the S trimer bound to S2M11 Fab and either S2L28 (A), S2M28 (B), or S2X333 Fab (C), as well as the locally refined reconstruction of NTD-bound Fab (inset). (Bottom) Representative electron micrograph and class averages (bottom left of each panel) are shown for SARS-CoV-2 S in complex with the indicated Fabs embedded in vitreous ice (Scale bar: 100 nm).
S2L28, S2M28, and S2X333 contact the N-terminal region (residues 14–20, NTD N terminus), a β-hairpin formed by residues 140–158 (supersite β-hairpin), and a loop spanning residues 245–264 (supersite loop). These three regions collectively form an antigenic supersite on the pinnacle of the NTD on the side distal to the viral membrane (Figures 2A–2I). The epitopes targeted by S2L28, S2M28, and S2X333 are overlapping and flanked by the oligosaccharides at position N17 and N149, which are located at opposite sides of the antigenic supersite (Figures 2D–2I). As a stunning example of convergent binding, each mAb uses hydrophobic residues at the tip of the HCDR3 loop to contact the NTD supersite near residue R246. For S2L28, S2M28, and S2X333, this hydrophobic residue is W105, I93, and W106, respectively (Figures 2G–2I).
Although these Fabs recognize overlapping epitopes within this antigenic supersite, the detailed nature of the interactions formed with the NTD varies. In particular, S2L28 primarily binds to residues in the NTD supersite loop including residue D253 (Figure 2G), while S2X333 interacts primarily with the N-terminal region including residue N17 and the supersite β-hairpin including residues E156 and R158 (Figure 2I). S2M28 interacts extensively with all three supersite regions, including residues 146–148 in the supersite β-hairpin, D253 in the supersite loop, and N17 in the N-terminal region (Figure 2H). As a result of these distinct interactions, whereas the S2L28-bound NTD adopts an identical conformation to that observed in the apo S structures (Walls et al., 2020b; Wrapp et al., 2020; Wrobel et al., 2020b), S2M28 and S2X333 induce remodeling of the NTD supersite loop and β-hairpin, respectively (Figures 2G–2I).
Cryo-EM maps of S2L28-, S2M28-, S2X28-, and S2X333-bound SARS-CoV-2 S delineate an antigenic supersite recognized by potent NTD-specific neutralizing mAbs isolated from COVID-19 individuals (Figures 2A–2I and S3 A). Likewise, the previously described mAb 4A8 (Chi et al., 2020) as well as the recently described FC05 (Zhang et al., 2020) and CM25 (Voss et al., 2020), bind to the same NTD antigenic supersite to neutralize SARS-CoV-2. Collectively, these results show that NTD-specific neutralizing mAbs have adopted a convergent mechanism of viral inhibition through the use of multiple V genes (Table S1), which points to a key role of the NTD supersite in viral neutralization and putatively also infection.
Figure S3.
Cryo-EM data processing of SARS-CoV-2 S bound to S2X28, S2L20, S2X316, or S2M24, related to Figures 2 and 3
(A) 6 Å low-pass filtered map of the SARS-CoV-2 2P DS S trimer (McCallum et al., 2020) bound to S2X28 (gold).
(B–D) Sharpened maps of the S trimer bound to S2M11 Fabs and S2L20 (B, gray), S2X316 (C, purple), or S2M24 Fabs (D, sky blue). Representative electron micrographs and class averages are shown at the bottom left of each panel (Scale bar: 100 nm). The corresponding Fourier shell correlation curves (bottom right of each panel) are shown with the 0.143 cutoff indicated by horizontal dashed lines.
Definition of a SARS-CoV-2 NTD antigenic map
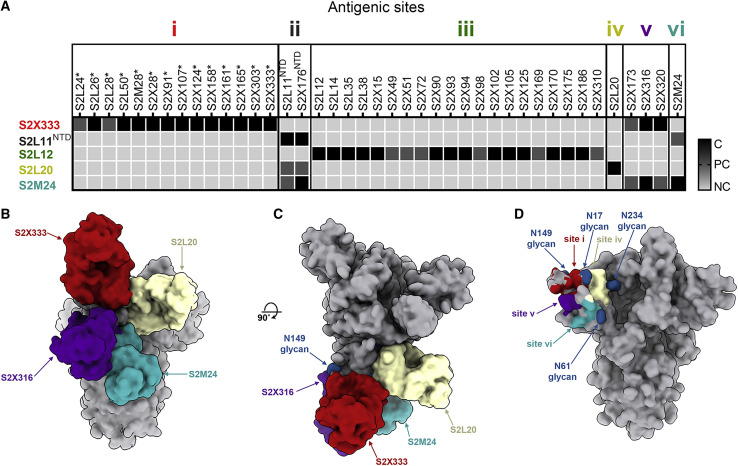
To understand the markedly different neutralization potencies across the 41 NTD-specific mAbs identified, we carried out competition biolayer interferometry binding assays using recombinant SARS-CoV-2 S ectodomain trimer or isolated NTD. The data indicated that the mAbs recognize six distinct antigenic sites, which we designated i, ii, iii, iv, v, and vi. Most mAbs clustered within antigenic sites i and iii whereas sites ii, iv, v, and vi each accounted for only one or a small number of mAbs from our panel (Figure 3 A). All potently neutralizing mAbs competed for binding to the NTD site i, indicating they recognize overlapping epitopes within the structurally identified antigenic supersite (Figures 1B–1E, 2, and 3A).
Figure 3.
The SARS-CoV-2 NTD comprises multiple antigenic sites
(A) Epitope binning of the 41 NTD-specific mAbs isolated led to the identification of six antigenic sites based on competition binding assays using biolayer interferometry. The competition assays were performed either using the S ectodomain trimer or the NTD (NTD). NC, no competition; PC, partial competition; C, competition; ∗, potently neutralizing mAbs. One independent experiment out of two is shown.
(B and C) Composite model of the SARS-CoV-2 S trimer viewed along two orthogonal orientations with four bound Fab fragments representative of antigenic site i (S2X333), site iv (S2L20), site v (S2X316), and site vi (S2M24).
(D) Footprints of the antigenic sites identified structurally are shown along with neighboring N-linked glycans (blue spheres).
To evaluate the presence of site i-specific mAbs in previously infected or vaccinated individuals, we carried out competition ELISA using S2X28. Plasma from infected individuals and convalescent subjects that received a first dose of mRNA vaccine (convalescent-Vx1), but not naive-vaccinated individuals (Vx1), efficiently competed with S2X28 for binding to the antigenic supersite (site i) (Table S5). These results confirm that in some individuals, antigenic site i-specific Abs can be elicited as a result of SARS-CoV-2 infection and are possibly boosted upon vaccination despite the dense glycosylation of the NTD.
To delineate the different NTD antigenic sites, we mapped the epitopes targeted by representative mAbs through determination of cryo-EM structures of S2M24, S2X316, or S2L20 in complex with SARS-CoV-2 S, together with S2M11 to expedite structural determination at 6–8 Å resolution (Figure S3; Table S4). S2L20 (site iv) binds to an epitope flanked by glycans at positions N17, N61, and N234 located within the NTD side, which faces the RBD belonging to the same protomer (Figures 3B–3D). S2M24 (site vi) recognizes the viral membrane proximal side of the NTD, which is devoid of glycans except for the presence of the N61 oligosaccharide on the edge of the epitope (Figures 3B–3D). S2M24 recognizes a similar antigenic site as the one described for the polyclonal Fabs COV57 isolated directly from COVID-19 patient sera (Barnes et al., 2020b). S2X316 (site v) interacts with an epitope residing nearby glycans N74 and N149 at the peripheral-most part of the NTD, located in between antigenic sites i and vi (Figures 3B–3D). As all NTD-specific mAbs with neutralizing activity target the structurally identified antigenic supersite site (site i), our data points to the existence of a site of vulnerability that is harnessed by the immune system of multiple individuals.
NTD mAbs are affected by the genetic diversity of circulating SARS-CoV-2 isolates and sarbecoviruses
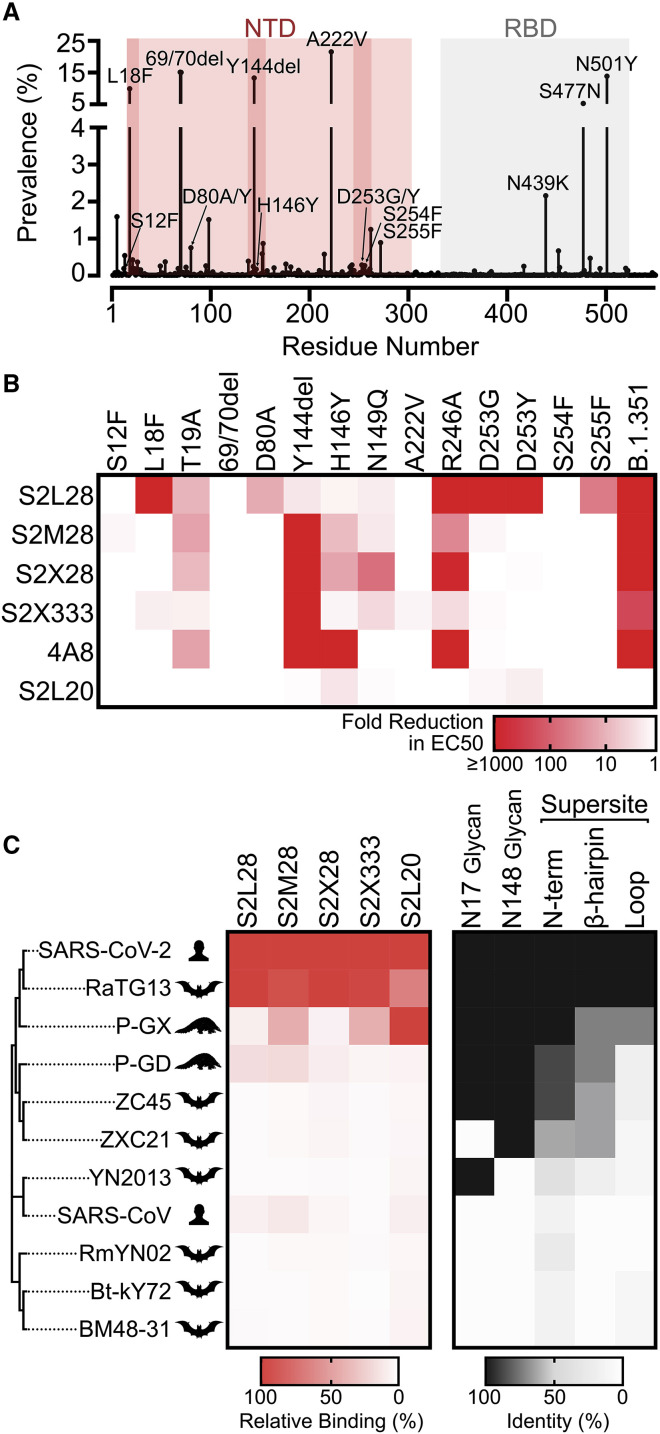
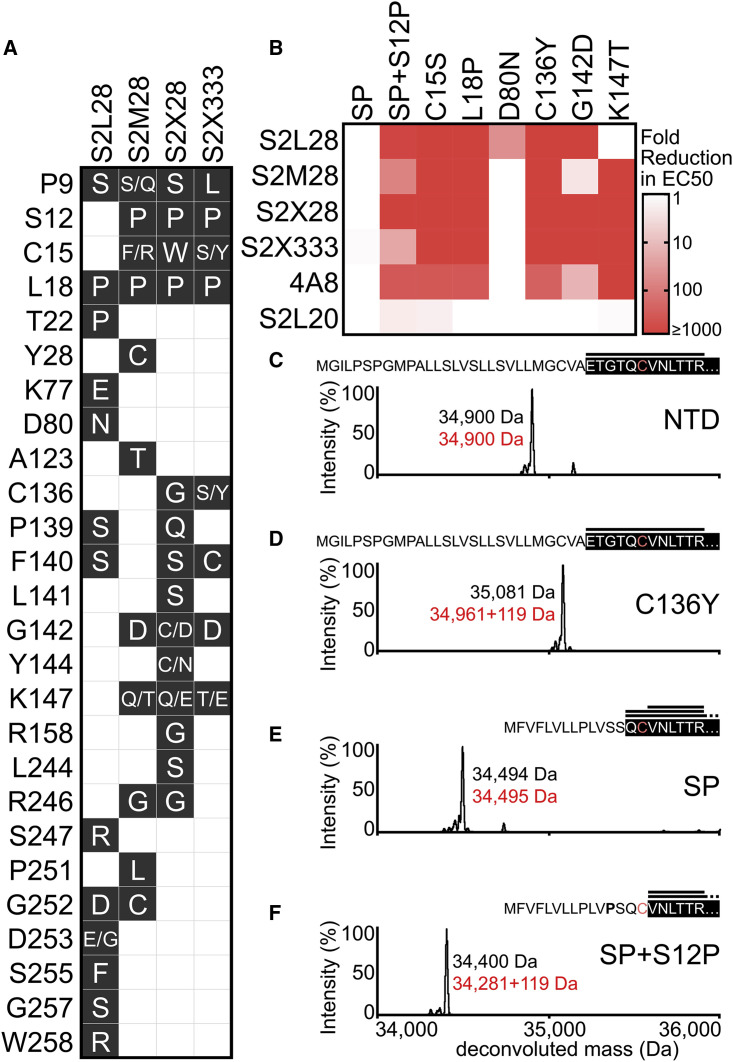
Analysis of the 508,771 SARS-CoV-2 genome sequences deposited in GISAID as of February 12, 2021 reveals a larger number of prevalent mutations and deletions in the NTD relative to other regions of the S glycoprotein (Figure 4 A). We therefore set out to evaluate the impact on NTD-specific mAb binding of several mutations identified within and outside the NTD antigenic supersite along with a few mutants designed based on the structural data. The recombinant mutant NTDs tested include S12F, L18F, T19A (removes the N15 glycan), a 69/70 deletion, D80A, a Y144 deletion, H146Y, N149Q (removes the N149 glycan), A222V, R246A, D253G/Y, S254F, S255F, and the B1.351 NTD (comprising the L18F, D80A, D215G, 242–244 deletion, and R246I mutations).
Figure 4.
NTD mAbs are affected by the genetic diversity of circulating SARS-CoV-2 variants and sarbecoviruses
(A) Prevalence of SARS-CoV-2 S variants among circulating isolates as of February 12, 2021 (508,771 sequences). The NTD and RBD are highlighted with red and gray backgrounds, respectively. The NTD supersite residues are highlighted by darker red background shading.
(B) S2L28, S2M28, S2X28, S2X333, 4A8, and S2L20 binding to recombinant SARS-CoV-2 NTD variants analyzed by ELISA and displayed as a heatmap. 69/70del, deletion of residues 69/70; Y144del, deletion of residue Y144. EC50s were calculated as mean of duplicates from two technical replicates.
(C) Heatmap of NTD mAb binding to a panel of sarbecovirus S glycoproteins expressed at the surface of ExpiCHO cells analyzed by flow cytometry. Binding is expressed as mean fluorescence intensities relative to SARS-CoV-2 S binding for each mAb. One independent experiment out of two is shown. A coronavirus NTD cladogram is shown on the left. The conservation of relevant glycans and NTD supersite regions is also shown (right).
All neutralizing NTD mAbs bound efficiently to NTD variants carrying either a 69/70 deletion or the A222V substitutions, indicating the site i-targeting mAbs would not be affected by these common SARS-CoV-2 mutations (Hodcroft et al., 2020) (Figures 4A and 4B). The R246A substitution decreased binding of S2L28, S2M28, and S2X333 (to various extents), in agreement with the extensive interactions formed by the R246 side chain with aromatic and hydrophobic residues found in the HCDR3 of each mAb. This substitution also markedly affected the binding of S2X28 and of the previously described mAb 4A8 (Chi et al., 2020) (Figure 4B). The L18F, D80A, D253G/Y, or S255F variants only abrogated binding of S2L28 to the NTD, which is relevant considering the L18F substitution is one of the most prevalent mutants sequenced to date and is present in both the B.1.351 and P.1 lineages (Tegally et al., 2020) (Figure 4B). Conversely, the Y144 deletion abrogated binding to S2M28, S2X28, S2X333, and 4A8, but not S2L28, explaining the lost ability of these mAbs to neutralize the B.1.1.7 S pseudovirus, which contains this deletion (Collier et al., 2021; Davies et al., 2020) (Figure 4B). The H146Y mutant reduced binding of S2M28, S2X28, and in particular of 4A8 (Figure 4A). Binding of all site i-specific NTD mAbs to the B.1.351 NTD was abrogated, in agreement with a recent study showing that 4A8 did not recognize this NTD variant (Wibmer et al., 2021). However, binding of the non-neutralizing S2L20 NTD-specific mAb was not affected by any of these variants, confirming retention of proper folding of the purified NTD mutants (Figure 4A). Collectively these data show that S2M28, S2X28, S2X333, and 4A8 have a similar reactivity profile to these NTD mutants, whereas S2L28 has a distinct and complementary pattern. These data demonstrate that several currently circulating variants will partially or completely escape neutralization mediated by mAbs targeting the antigenic supersite (site i), including the B.1.1.7, B.1.351, and P.1 lineages.
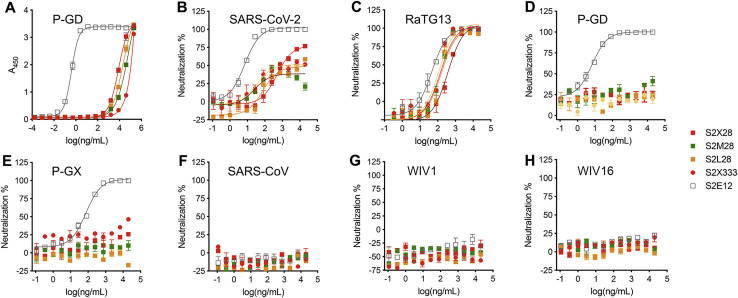
To investigate the cross-reactivity of NTD-specific mAbs with sarbecovirus S glycoproteins, we probed individual mAb binding to ExpiCHO cells expressing full-length S from representative members of the three clades of the sarbecovirus subgenus. All mAbs tested bound to RaTG13 S, which is the most closely related S glycoprotein and NTD to SARS-CoV-2 S (Figure 4C). S2X333 and S2M28 weakly bound to S from the Pangolin Guanxi 2017 (P-GX) isolate, whereas S2L28 and S2M28 weakly bound to S from the Pangolin Guangdong 2019 (P-GD) isolate (Figure 4C). Consistently, weak mAb binding to P-GD S was confirmed by ELISA for S2L28, S2M28, S2X333, and S2M24 (Figure S4 A). We next evaluated the neutralization breadth of S2L28, S2M28, S2X28, and S2X333 against SARS-CoV-2- and SARS-CoV-related sarbecoviruses using a vesicular stomatitis virus (VSV) pseudotyped neutralization assay. All mAbs tested neutralized RaTG13 S pseudotypes with comparable potency to SARS-CoV-2 S pseudotyped virus but none of them neutralized the more distantly related P-GX S, P-GD S, SARS-CoV S, WIV1 S, or WIV16 S pseudotyped viruses (Figures S4B–S4H). These findings are in line with the low conservation of the NTD supersite (site i) in phylogenetically distant viruses and indicate that NTD mAbs could be useful countermeasures against a subset of viruses closely related to SARS-CoV-2 but not against divergent viruses. Both species-specific functions and host immunity may play a role in the antigenic diversification of this region compared to other S domains that are more conserved.
Figure S4.
NTD neutralizing mAbs inhibit S-mediated entry of the closely related RaTG13 but not of more distant sarbecoviruses, related to Figure 4
(A) Binding of NTD- and RBD-specific mAbs to immobilized P-GD S ectodomain trimer analyzed by ELISA.
(B–H) VSV pseudovirus neutralization assays in the presence of varying concentrations of the NTD-specific mAbs S2L28, S2M28, S2X58, S2X333 or the RBD-specific mAb S2E12.
SARS-CoV-2 S neutralization escape mutants reveal unconventional mechanisms of NTD mAb resistance
To investigate potential viral escape from neutralization mediated by the potent neutralizing NTD mAbs, we passaged a replication competent VSV-SARS-CoV-2 chimera in which the native glycoprotein was replaced by the SARS-CoV-2 S Wuhan-1 isolate gene in the presence of S2L28, S2M28, S2X28, and S2X333, as previously described (Case et al., 2020; Liu et al., 2020b). Selective pressure provided by each mAb led to emergence of several escape mutants that clustered within the antigenic supersite i.e., the NTD N terminus, the supersite β-hairpin, and the supersite loop as well as surrounding residues (Figures 5A and 5B). Similar to what was previously described for RBD mAbs (Starr et al., 2020a, 2020b), all viral escape mutants resulted from a single non-synonymous nucleotide mutation.
Figure 5.
Analysis of SARS-CoV-2 S neutralization escape mutants
(A) Escape mutations selected with each mAb (black background white font) and residues that remained unchanged (white background) upon passaging a VSV-SARS-CoV-2 S chimeric virus.
(B) S2L28, S2M28, S2X28, S2X333, 4A8, and S2L20 binding to selected SARS-CoV-2 NTD escape mutants analyzed by ELISA and displayed as a heatmap. The native signal peptide was introduced using the wild-type sequence (SP) or with the S12P mutation (SP+S12P). EC50s were calculated as mean of duplicates from two technical replicates.
(C–F) Deconvoluted mass spectra of purified NTD constructs, including the NTD construct with an optimized signal peptide (C, NTD), the NTD construct with an optimized signal peptide and the C136Y mutation (D, NTD), the NTD construct with the native signal peptide (E, NTD), and the NTD construct with the native signal peptide carrying the S12P mutation (F, NTD). The empirical mass (black) and theoretical mass (red) are shown beside the corresponding peak. Additional 119 Da were observed for the C136Y and SP+S12P NTDs corresponding to cysteinylation of the free cysteine residue in these constructs (as L-cysteine was present in the expression media). The cleaved signal peptide (black text white background) and subsequent N-terminal sequence (white text black background) are also shown based on the mass spectrometry (MS) results; C15 is highlighted in light red. Peptides identified by MS/MS analysis consistent with the mass of N-terminal peptides are shown above the N-terminal sequence (black horizontal lines).
See also Data S1.
The four mAbs selected for many shared escape mutations but with a distinct pattern for S2L28, in agreement with the structural observations indicating distinct residues are important for recognition of this antibody. For instance, S2L28 selected for and was uniquely sensitive to the D80N and D253G mutations, whereas it still bound to the K147T mutation, which abrogated binding by S2M28, S2X28, S2X333, and 4A8. S2X28 selected for Y144C/N substitutions, both S2M28 and S2X28 selected for R246G substitutions, and all four mAbs selected for L18P substitutions (Figure 5A). Mutations including L18F, D80A, deletion of residue Y144, R246I/T/K, and D253G/Y have been detected in SARS-CoV-2 clinical isolates, most of them in the B.1.1.7, B.1.351, or P.1 lineages, demonstrating the ability of this system to recapitulate variants at naturally occurring S residues (Davies et al., 2020; Tegally et al., 2020).
Besides residue substitutions that are expected to impact the epitope/paratope interface directly, we observed escape mutants in the signal peptide region and at cysteine residues 15 and 136. The latter two residues are jointly engaged in a disulfide bond that staples the NTD N terminus against the galectin-like β sandwich (Figures 2G–2I). Alteration of this bond (via C15S or C136Y substitutions) abrogates recognition by S2L28, S2M28, S2X28, S2X333, and 4A8 (Figure 5B), likely via dislodging this key supersite component, explaining the observed phenotype. These data further suggest that this mechanism of escape is likely to be effective with all mAbs targeting the NTD antigenic supersite.
The predicted SARS-CoV-2 S signal peptide cleavage site occurs between residues T13 and Q14. Signal peptide cleavage site prediction using SignalP5.0 (Almagro Armenteros et al., 2019) suggests that the selected S12P escape mutation alters cleavage to occur between residues C15 and V16, thereby eliminating the C15-C136 disulfide bond—similarly to escape mutants at positions C15 and C136. To test the effect of the S12P substitution with purified NTD, we introduced the native signal peptide in the presence or absence of S12P. Similar to direct disruption of the C15-C136 disulfide bond, the S12P signal peptide mutation was found to markedly reduce binding to S2L28, S2M28, S2X28, S2X333, and 4A8 (Figure 5B). Mass spectrometry analysis of the S12P mutant was consistent with signal peptide cleavage occurring immediately after residue C15 (Figures 5C–5F). The S12F mutation observed in clinical isolates, however, did not affect binding of the NTD-specific mAbs tested (Figure 4B). Collectively these findings indicate the involvement of the S signal peptide in immune evasion and warrant closer monitoring of NTD variants.
Mechanism of action of NTD-specific neutralizing mAbs
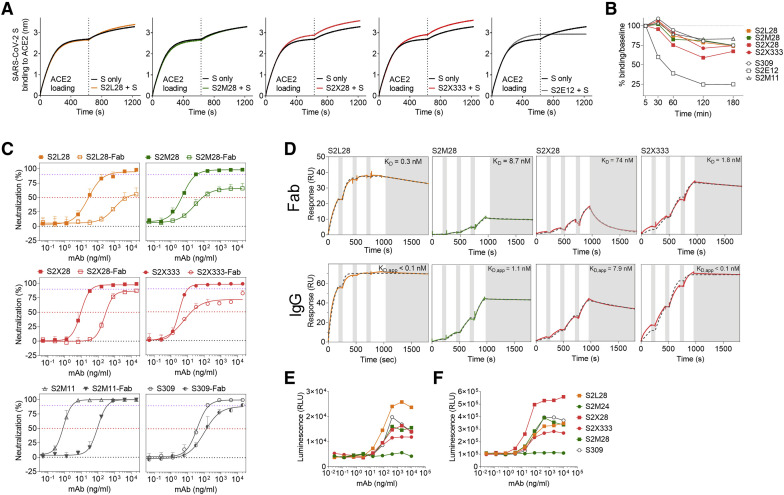
To understand NTD-specific mAb-mediated inhibition of viral entry, we evaluated the ability of these mAbs to block ACE2 binding as this step correlates with neutralization titers in SARS-CoV-2-exposed individuals (Piccoli et al., 2020). None of the site i-targeting mAbs (S2L28, S2M28, S2X28, and S2X333) blocked binding of SARS-CoV-2 S to immobilized human recombinant ACE2 as measured by biolayer interferometry (Figure 6 A), excluding interference with engagement of the main entry receptor as mechanism of action. Moreover, these mAbs did not promote shedding of the S1 subunit from cell-surface-expressed full-length SARS-CoV-2 S (Figure 6B), ruling out the possibility of premature S triggering, as previously shown for a SARS-CoV and several SARS-CoV-2 RBD-specific mAbs (Huo et al., 2020; Piccoli et al., 2020; Walls et al., 2019; Wec et al., 2020; Wrobel et al., 2020a). However, S2L28, S2M28, S2X28, and S2X333 blocked SARS-CoV-2 S-mediated cell-cell fusion, indicating they could prevent interaction with an auxiliary receptor, proteolytic activation, or membrane fusion (Figure S1G).
Figure 6.
Mechanism of action of NTD-specific neutralizing mAbs
(A) Competition of S2L28, S2M28, S2X28, and S2X333 with ACE2 to bind to SARS-CoV-2 S as measured by biolayer interferometry. ACE2 was immobilized at the surface of the biosensors before incubation with the S ectodomain trimer alone or pre-complexed with mAbs. The vertical dashed line indicates the start of the association of free S or S/mAb complexes with solid-phase ACE2. The anti-RBD S2E12 mAb was included as positive control.
(B) mAb-mediated S1 subunit shedding from cell surface expressed SARS-CoV-2 S as determined by flow cytometry. The anti-RBD S2M11 and S2E12 mAbs were included as negative and positive controls, respectively.
(C) Neutralization of authentic SARS-CoV-2 (SARS-CoV-2-Nluc) by S2L28, S2M28, S2X28, and S2X333 IgG or Fab. S309 and S2M11 Fab and IgG were also included as controls. Symbols are means ± SD of triplicates. Dotted lines indicate IC50 and IC90 values.
(D) SPR analysis of mAbs binding to the SARS-CoV-2 S ectodomain trimer. Gray dashed line indicates a fit to a 1:1 binding model. The equilibrium dissociation constants (KD) or apparent equilibrium dissociation constants (KD, app) are indicated. White and gray stripes indicate association and dissociation phases, respectively.
(E and F) Activation of FcγRIIa H131 (E) and FcγRIIIa V158 (F) induced by NTD-specific mAbs. The anti-RBD mAb S309 was included as positive control. One independent experiment out of at least two is shown.
See also Figures S1 and S5 and Table S1.
To assess whether steric hindrance plays a role in the neutralization activity of S2L28, S2M28, S2X28, and S2X333, we evaluated the neutralization potency of each mAb, in both Fab and IgG formats, against authentic SARS-CoV-2-Nluc (Figure 6C). NTD-specific Fabs displayed a marked potency reduction, both in terms of IC50 values and maximal neutralization plateau reached (Table S1), as compared to IgGs, possibly due to reduced avidity as observed by surface plasmon resonance (Figure 6D). Since the Fabs could still partially neutralize SARS-CoV-2, at least part of the observed neutralization activity results from direct interaction with their respective epitopes. It is possible that NTD-specific mAb-mediated neutralization further relies on the steric hindrance provided by Fc positioning, similar to what was observed for anti-hemagglutinin influenza A virus neutralizing mAbs (Xiong et al., 2015).
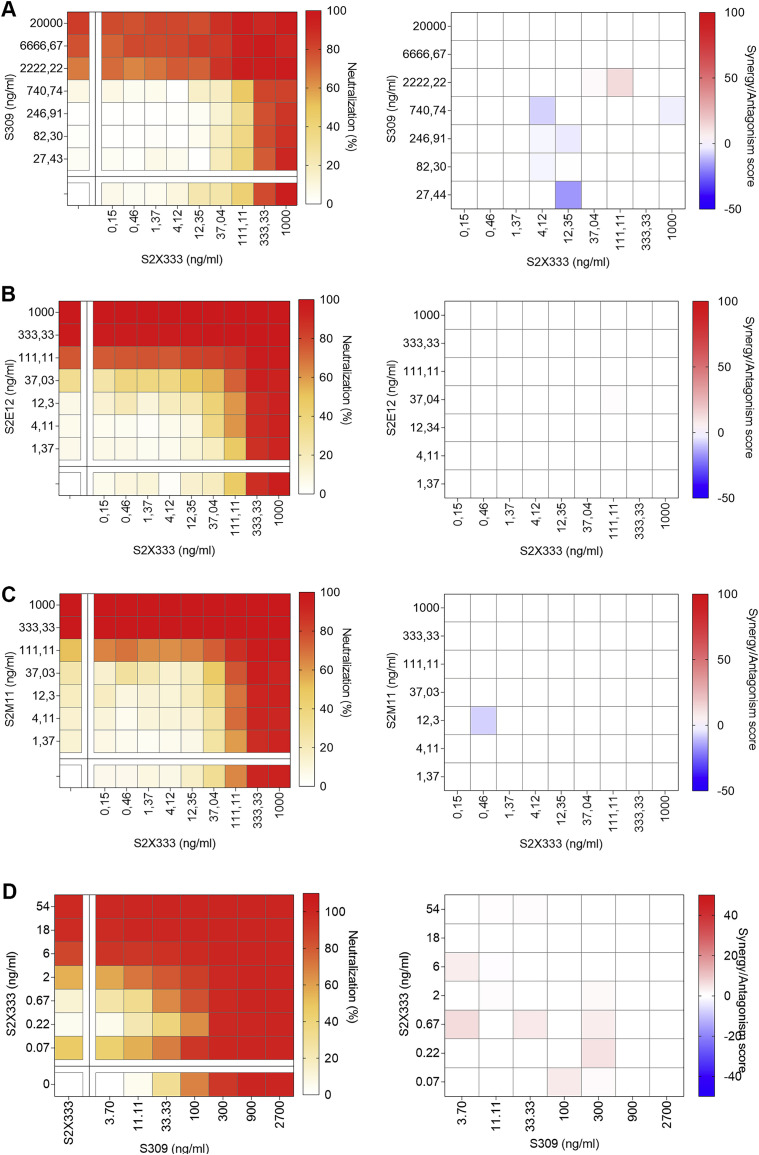
We next examined potential additive, antagonistic, or synergistic effects of NTD- and RBD-targeting mAbs, as mAb synergy was previously described for SARS-CoV and SARS-CoV-2 neutralization (Pinto et al., 2020; ter Meulen et al., 2006). Cocktails of S2X333 with S309, S2E12, or S2M11 additively prevented entry of SARS-CoV-2 S-MLV pseudotyped virus in Vero E6 cells ( Figures S5 A–S5C). This additive effect was also observed between S2X333 and S309 using authentic SARS-CoV-2 at 24 h post-infection in Vero E6 cells (Figure S5D). These results are consistent with RBD- and NTD-targeting mAbs mediating inhibition by distinct mechanisms and demonstrate that they could be used as cocktails for prophylaxis or therapy.
Figure S5.
Neutralizing activity of SARS-CoV-2 NTD- and RBD-targeting mAb cocktails, related to Figures 1 and 6
(A–C) SARS-CoV-2-MLV pseudotypes neutralization (left) and synergy score (right) measured combining S2X333 with the RBD-targeting mAb S309 (A), S2E12 (B), or S2M11 (C).
(D) Neutralization matrix to assess the synergistic activity of S2X333 and S309 mAb cocktails in vitro with authentic SARS-CoV-2-Nluc. Data for authentic SARS-CoV-2-Nluc are from one representative experiment performed in triplicate each.
Since Fc-mediated effector functions contribute to protection by promoting viral clearance and anti-viral immune responses in vivo (Bournazos et al., 2020, 2016; Schäfer et al., 2021; Winkler et al., 2020), we evaluated the ability of site i-targeting mAbs to trigger activation of FcγRIIa and FcγRIIIa as a proxy for Ab-dependent cellular phagocytosis (ADCP) and Ab-dependent cellular cytotoxicity (ADCC), respectively. S2L28, S2M28, S2X28, and S2X333 promoted dose-dependent FcγRIIa- and FcγRIIIa-mediated signaling to levels comparable to those of the highly effective mAb S309 (Pinto et al., 2020) (Figures 6E and 6F). In contrast, the non-neutralizing site vi-targeting S2M24 mAb did not promote FcγR-mediated signaling, possibly due to the different orientation relative to the membrane of the effector cells in comparison to site i-specific mAbs (Figures 6E and 6F). These findings suggest that besides their neutralizing activity, mAbs recognizing site i can exert a protective activity via promoting Fc-mediated effector functions.
NTD neutralizing mAbs protect against SARS-CoV-2 challenge in hamsters
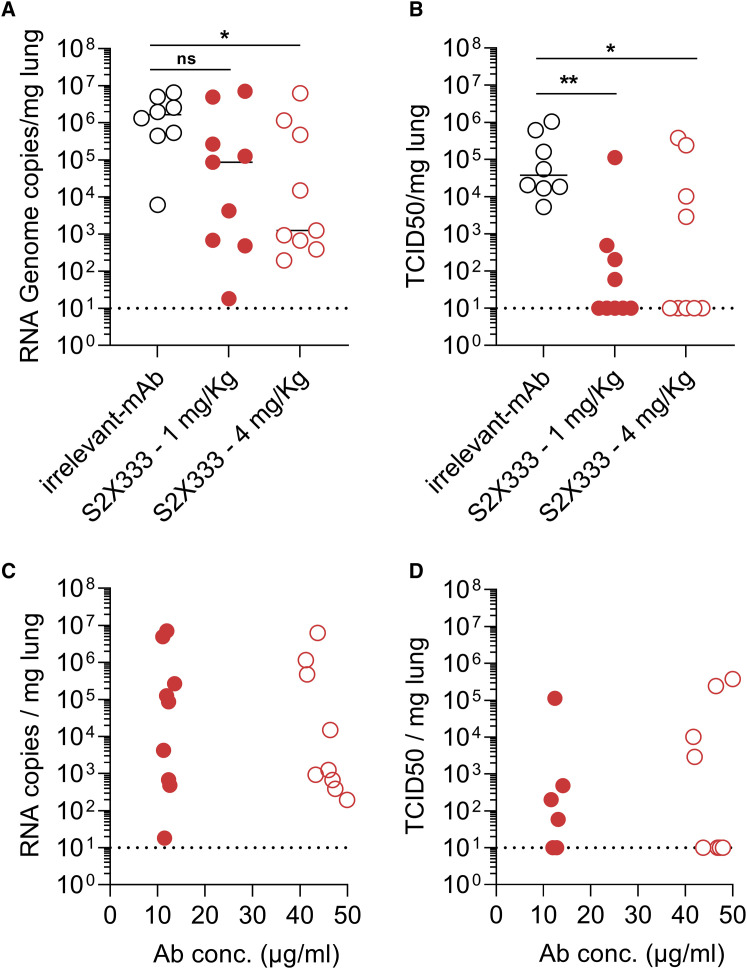
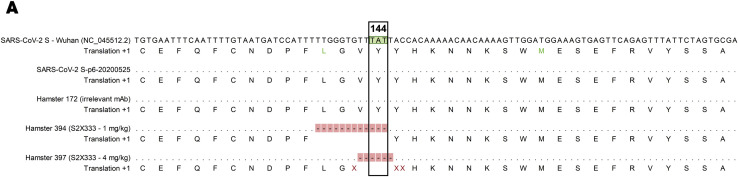
To evaluate the protective efficacy of NTD-directed mAbs against SARS-CoV-2 challenge in vivo, we selected the S2X333 mAb for a prophylactic study in a Syrian hamster model (Boudewijns et al., 2020). The mAb was administered at 4 and 1 mg/kg via intraperitoneal injection 48 h before intranasal SARS-CoV-2 challenge. Four days later, lungs were collected for the quantification of viral RNA and infectious virus titers. Prophylactic administration of S2X333 at a dose of 4 mg/kg decreased the amount of viral RNA detected in the lungs by ∼3 orders of magnitude, compared to hamsters receiving a control mAb (Figure 7 A). Viral replication in the lungs was completely abrogated for most of the animals at both doses tested (Figure 7B). However, although all animals had similar serum mAb concentrations within each group, infectious viral titers were not decreased relative to the control group for 1 and 4 hamsters administered with S2X333 at 1 and 4 mg/kg, respectively (Figures 7C and 7D). Based on the aforementioned variability and mutation tolerance of the SARS-CoV-2 NTD, we hypothesized that S2X333 escape mutants might have been selected in these animals. Sequencing of the viral RNA extracted from the lungs of one outlier from each group administered with S2X333 revealed an S deletion encompassing position Y144 (within site i) in comparison to the input virus or the animal from the control group. The deletion corresponds to 4 and 2 amino acids for the animals treated with 1 (LGVY144) and 4 mg/kg (VY144), respectively (Figure S6 ). These results show that potent NTD mAbs targeting the antigenic supersite (site i) impose a selection pressure driving viral evolution to escape neutralization. Overall, these data show that low doses of anti-NTD mAbs provide remarkable prophylactic activity in vivo, comparable to best-in-class RBD-specific mAbs S2E12 and S2M11 (Tortorici et al., 2020), consistent with their potent in vitro neutralizing activity. We anticipate that the protection efficacy of S2X333 (and related NTD mAbs) would be further enhanced in humans due to an optimal matching with human Fcγ receptors. However, our results also indicate that potent NTD mAbs targeting the antigenic supersite (site i) impose a selection pressure driving viral evolution to escape neutralization.
Figure 7.
S2X333 provides robust in vivo protection against SARS-CoV-2 challenge but selects for escape mutants
Syrian hamsters were injected with the indicated amount of mAb 48 h before intra-nasal challenge with SARS-CoV-2.
(A) Quantification of viral RNA in the lungs 4 days post-infection.
(B) Quantification of replicating virus in lung homogenates harvested 4 days post-infection using a TCID50 assay.
(C and D) Viral RNA loads and replicating virus titers in the lung 4 days post-infection plotted as a function of serum mAb concentrations before infection (day 0). Mann-Whitney test was used. ∗p < 0.05, ∗∗p < 0.01. Data from two independent experiment are presented. (Irrelevant mAb n = 8; S2X333 1 mg/kg n = 9; S2X333 4 mg/kg n = 9).
See also Figure S6.
Figure S6.
Sequence alignment of SARS-CoV-2 S vRNA from input virus, control group, and selected outlier animals administered with S2X333, related to Figure 7
Nucleotide alignment of a region of interest reveals a deletion encompassing amino acid at position 144 only in S2X333-treated animals. SARS-CoV-2 S Wuhan = reference sequence (NCBI Reference Sequence NC_045512.2); SARS-CoV-2 S -p6-20200525 = virus stock; hamster 172 = animal from the control group; hamster 394 = outlier (TCID50) from the group administered with S2X333 at 1 mg/kg; hamster 397 = outlier (TCID50) from the group administered with S2X333 at 4 mg/kg. Alignment was performed using CLC Main Workbench 21 (QIAGEN)
Discussion
The data herein demonstrate that neutralizing NTD-targeting mAbs represent a key aspect of immunity to SARS-CoV-2 and account for 5%–20% of SARS-CoV-2 S-specific mAbs cloned from memory B cells isolated from the PBMCs of three COVID-19 individuals. Analysis of a large panel of neutralizing and non-neutralizing mAbs led us to define an antigenic map of the heavily glycosylated SARS-CoV-2 NTD, in which six antigenic sites (i–vi) were identified. We found that all the neutralizing mAbs from the three donors investigated, along with the mAbs 4A8 (Chi et al., 2020), FC05 (Zhang et al., 2020), and CM25 (Voss et al., 2020), target the same antigenic supersite (site i) using various germline V genes, thereby providing examples of convergent solutions to NTD-targeted mAb neutralization. We show that a highly potent NTD mAb provides prophylactic protection against SARS-CoV-2 challenge of Syrian hamsters showcasing that this class of mAbs can be a critical barrier to infection and are promising clinical candidates against SARS-CoV-2. Likewise, previous work on Middle East respiratory syndrome coronavirus (MERS-CoV) established that the NTD is the target of several potent and protective neutralizing mAbs and an attractive target for vaccine design (Chen et al., 2017; Jiaming et al., 2017; Wang et al., 2018, 2019; Zhou et al., 2019).
Our cryo-EM structures of SARS-CoV-2 S bound to the ultrapotent RBD-specific mAb S2M11 and to S2X333, S2M28, or S2L28, together with evidence of additive neutralizing effect of RBD- and NTD-targeted mAbs, provide proof of concept for implementing countermeasures using mAb cocktails targeting both the NTD and the RBD and potentially also for vaccine design. As several examples of single amino acid mutations reducing or completely abrogating neutralization by immune sera have been reported (Li et al., 2020; Liu et al., 2020b; Weisblum et al., 2020), we propose that combinations of mAbs targeting distinct domains might reduce the likelihood of emergence of escape mutants, as previously demonstrated for MERS-CoV (Wang et al., 2018).
Here, we show that site i-targeting NTD neutralizing mAbs efficiently activate FcγRIIa and FcγRIIIa in vitro. Fc-mediated effector functions can be affected by the epitope specificity of the mAbs (Piccoli et al., 2020), highlighting the importance of the orientation of the S-bound Fc fragments for efficient FcγR cross-linking and engagement. The site vi-targeting NTD mAb S2M24 did not activate either FcγRIIa or FcγRIIIa, presumably due to the different angle of approach. The contribution of Fc-mediated effector functions could further enhance the prophylactic activity of potent NTD-specific mAbs against SARS-CoV-2 in humans as demonstrated for two RBD-specific SARS-CoV-2 neutralizing mAbs (Schäfer et al., 2021; Winkler et al., 2020).
The emergence and fixation of SARS-CoV-2 D614G yielded a virus mutant with enhanced ACE2 binding, transmissibility, and replication but without marked changes of antigenicity (Hou et al., 2020; Korber et al., 2020; Plante et al., 2020; Yurkovetskiy et al., 2020). In contrast, we observed that several variants detected in circulating SARS-CoV-2 clinical isolates reduced or abrogated recognition by some NTD-specific neutralizing mAbs. For instance, the L18F substitution, which has a prevalence of 10% among sequenced SARS-CoV-2 genomes and is present in the B.1.351 and P.1 lineages initially identified in South Africa and Brazil (Tegally et al., 2020), would escape S2L28-mediated neutralization. The deletion of residue Y144 found in 13% of circulating SARS-CoV-2 clinical isolates, including the B.1.1.7 lineage originally detected in the UK before being identified in numerous other countries (Davies et al., 2020), would circumvent neutralization by S2M28, S2X28, S2X333, and 4A8 (Collier et al., 2021). Using selection of viral escape mutants and structural analysis, we identified additional key residues for binding of neutralizing mAbs, such as S12 (indirectly) and R246, for which natural variants are reported and are thus expected to participate in neutralization escape. The finding that multiple circulating SARS-CoV-2 variants map to the NTD, including several of them in the antigenic supersite (site i), suggests that the NTD is under selective pressure from the host humoral immune response. This is further supported by the identification of NTD deletions within the antigenic supersite (site i) in immunocompromised hosts with prolonged infections (Avanzato et al., 2020; Choi et al., 2020; McCarthy et al., 2021), the in vitro selection of SARS-CoV-2 S escape variants with NTD mutations that decrease the neutralization potency of COVID-19 convalescent patient sera (Andreano et al., 2020; Weisblum et al., 2020), and the escape mutants reported here. Collectively, these data indicate that the mutations and deletions accumulated in the NTDs of the B.1.1.7, B.1.351, and P.1 lineages are a direct result of mAb-mediated selective pressure in infected hosts. We propose that mAb cocktails comprising S2L28 plus S2M28, S2X28, or S2X333 could provide broad protection against some SARS-CoV-2 variants as these mAbs are susceptible to distinct sets of SARS-CoV-2 S mutations, as previously proposed for RBD-specific mAbs (Greaney et al., 2020).
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial Strains | ||
| E. coli DH10B Competent Cells | ThermoFisher Scientific | Cat# EC0113 |
| Deposited Data | ||
| S + S2M11 + S2X333 Cryo-EM Map | https://www.ebi.ac.uk/pdbe/emdb/ | EMD-23579 |
| S + S2M11 + S2X333 Cryo-EM Model | https://www.rcsb.org | PDB 7LXY |
| S + S2M11 + S2X333 Focused Cryo-EM Map | https://www.ebi.ac.uk/pdbe/emdb/ | EMD-23577 |
| S + S2M11 + S2X333 Focused Cryo-EM Model | https://www.rcsb.org | PDB 7LXW |
| S + S2M11 + S2M28 Cryo-EM Map | https://www.ebi.ac.uk/pdbe/emdb/ | EMD-23582 |
| S + S2M11 + S2M28 Cryo-EM Model | https://www.rcsb.org | PDB 7LY2 |
| S + S2M11 + S2M28 Focused Cryo-EM Map | https://www.ebi.ac.uk/pdbe/emdb/ | EMD-23581 |
| S + S2M11 + S2M28 Focused Cryo-EM Model | https://www.rcsb.org | PDB 7LY0 |
| S + S2M11 + S2L28 Cryo-EM Map | https://www.ebi.ac.uk/pdbe/emdb/ | EMD-23580 |
| S + S2M11 + S2L28 Cry-oEM Model | https://www.rcsb.org | PDB 7LXZ |
| S + S2M11 + S2L28 Focused Cryo-EM Map | https://www.ebi.ac.uk/pdbe/emdb/ | EMD-23578 |
| S + S2M11 + S2L28 Focused Cryo-EM Model | https://www.rcsb.org | PDB 7LXX |
| 2P-DS-S + S2X28 Cryo-EM Map | https://www.ebi.ac.uk/pdbe/emdb/ | EMD-23584 |
| S + S2M11 + S2L20 Cryo-EM Map | https://www.ebi.ac.uk/pdbe/emdb/ | EMD-23586 |
| S + S2M11 + S2X316 Cryo-EM Map | https://www.ebi.ac.uk/pdbe/emdb/ | EMD-23585 |
| S + S2M11 + S2M24 Cryo-EM Map | https://www.ebi.ac.uk/pdbe/emdb/ | EMD-23583 |
| S NTD + S2M28 Crystal Structure | https://www.rcsb.org | PDB 7LY3 |
| Experimental Models: Cell Lines | ||
| FreeStyle™ 293-F Cells | ThermoFisher Scientific | Cat# R79007 |
| Expi293F™ Cells | ThermoFisher Scientific | Cat# A14527 |
| HEK293T | ATCC | Cat# CRL-11268 |
| Vero-E6 | ATCC | CRL-1586 |
| Expi-CHO cells | ThermoFisher Scientific | Cat# A29127 |
| Expi-CHO cells stably expressing SARS-CoV-2 | This study | N/A |
| Jurkat cells stably expressing FcγRIIIa receptor (V158 variant) and NFAT-driven luciferase gene (effector cells) | Promega | Cat# G7018 |
| Jurkat cells stably expressing FcγRIIa receptor (H131 variant) and NFAT-driven luciferase gene | Promega | Cat# G9995 |
| Oligonucleotides | ||
| NTD_fwd: TATAATGGTACCGCCACCATGGGCAT | Integrated DNA Technologies, Inc. | N/A |
| NTD_rev: TATAATAAGCTTTCAGTGGTGGTGGTGGTGGTGAT GATGCGCGGTAAAGCTCTTCGCTGTACACTT |
Integrated DNA Technologies, Inc. | N/A |
| N149Q_fwd: AAAAGTCCTGGATGGAGTCTGAGT | Integrated DNA Technologies, Inc. | N/A |
| N149Q_rev: GGTTCTTGTGATAGTACACGCCC | Integrated DNA Technologies, Inc. | N/A |
| D253G_fwd: GCTCCTCTAGCGGATGGAC | Integrated DNA Technologies, Inc. | N/A |
| D253G_rev: CGCCTGGTGTCAGGTAGG | Integrated DNA Technologies, Inc. | N/A |
| D253Y_fwd: ACTCCTCTAGCGGATGGAC | Integrated DNA Technologies, Inc. | N/A |
| D253Y_rev: AGCCTGGTGTCAGGTAGG | Integrated DNA Technologies, Inc. | N/A |
| T19A_fwd: ACAAGGACCCAGCTGCC | Integrated DNA Technologies, Inc. | N/A |
| T19A_rev: GGCCAGGTTCACGCACTG | Integrated DNA Technologies, Inc. | N/A |
| R246A_fwd: CATCCTACCTGACACCAGGC | Integrated DNA Technologies, Inc. | N/A |
| R246A_rev: CGTGCAGGGCCAGCAG | Integrated DNA Technologies, Inc. | N/A |
| L18F_fwd: CTTTACCACAAGGACCCAGC | Integrated DNA Technologies, Inc. | N/A |
| L18F_rev: TTCACGCACTGTGTGCC | Integrated DNA Technologies, Inc. | N/A |
| H146Y_fwd: ACAAGAACAATAAGTCCTGGATGGAGTC | Integrated DNA Technologies, Inc. | N/A |
| H146Y_rev: AATAGTACACGCCCAGGAATGGAT | Integrated DNA Technologies, Inc. | N/A |
| A222V_fwd: TCCTGGAGCCACTGGTG | Integrated DNA Technologies, Inc. | N/A |
| A222V_rev: CGGAGAATCCCTGTGGCAG | Integrated DNA Technologies, Inc. | N/A |
| SP_rev: AGGACGAGGAAGACGAACATGGTGGCGGTA CCTCCG |
Integrated DNA Technologies, Inc. | N/A |
| SP_fwd: GCTCCCTCTGGTTTCTTCCCAGTGCGTGAAC CTGACCA |
Integrated DNA Technologies, Inc. | N/A |
| SP_S12P_fwd: GCTCCCTCTGGTTCCATCCCAGTGCGT GAACCTGACCA |
Integrated DNA Technologies, Inc. | N/A |
| Y144del_fwd: CACAAGAACAATAAGTCCTGGATGGAGT | Integrated DNA Technologies, Inc. | N/A |
| Y144del_rev: GTACACGCCCAGGAATGGATC | Integrated DNA Technologies, Inc. | N/A |
| S254F_fwd: CTCTAGCGGATGGACCGCA | Integrated DNA Technologies, Inc. | N/A |
| S254F_rev: AAGTCGCCTGGTGTCAGGTAG | Integrated DNA Technologies, Inc. | N/A |
| K147T_fwd: CGAACAATAAGTCCTGGATGGAGTCT | Integrated DNA Technologies, Inc. | N/A |
| K147T_rev: TGTGATAGTACACGCCCAGGAAT | Integrated DNA Technologies, Inc. | N/A |
| C136Y_fwd: ATAATGATCCATTCCTGGGCGTGTA | Integrated DNA Technologies, Inc. | N/A |
| C136Y_rev: AAAACTGGAACTCGCACACCTTG | Integrated DNA Technologies, Inc. | N/A |
| L19P_fwd: GGGTTCACGCACTGTG | Integrated DNA Technologies, Inc. | N/A |
| L19P_rev: GACCACAAGGACCCAGCT | Integrated DNA Technologies, Inc. | N/A |
| C15S_fwd: CTCTGTGTGCCGGTCTCTG | Integrated DNA Technologies, Inc. | N/A |
| C15S_rev: CGTGAACCTGACCACAAGGAC | Integrated DNA Technologies, Inc. | N/A |
| G142D_fwd: CCAGGAATGGATCATTACAAAACTGGAAC | Integrated DNA Technologies, Inc. | N/A |
| G142D_rev: ACGTGTACTATCACAAGAACAATAAGTCCTGG | Integrated DNA Technologies, Inc. | N/A |
| S255F_fwd: AAGGAGTCGCCTGGTGTCA | Integrated DNA Technologies, Inc. | N/A |
| S255F_rev: TAGCGGATGGACCGCAG | Integrated DNA Technologies, Inc. | N/A |
| S12F_fwd: TTCCCAGTGCGTGAACCTGA | Integrated DNA Technologies, Inc. | N/A |
| S12F_rev: AAAACCAGAGGGAGCAGGACG | Integrated DNA Technologies, Inc. | N/A |
| D80N_fwd: CAATCCCGTGCTGCCTTTTAAC | Integrated DNA Technologies, Inc. | N/A |
| D80N_rev: TTGAACCGCTTTGTGCCATTG | Integrated DNA Technologies, Inc. | N/A |
| D80A_fwd: CCAATCCCGTGCTGCCTTTTAAC | Integrated DNA Technologies, Inc. | N/A |
| D80A_rev: CGAACCGCTTTGTGCCATTG | Integrated DNA Technologies, Inc. | N/A |
| 69/70del_2fwd: TCCGGCACCAATGGCACA | Integrated DNA Technologies, Inc. | N/A |
| 69/70del_2rev: GATGGCGTGGAACCAGGTCAC | Integrated DNA Technologies, Inc. | N/A |
| Spike-cDNA-R: CAAGAATCTCAAGTGTCTGTGG | Microsynth AG | N/A |
| Spike-SEQ1-F: CACTAGTCTCTAGTCAGTGTG | Microsynth AG | N/A |
| Spike-SEQ1-R: GACTCAGTAAGAACACCTGTGC | Microsynth AG | N/A |
| Spike-SEQ2-F: AGTGCGTGATCTCCCTCAGG | Microsynth AG | N/A |
| Spike-SEQ3-F: AGTCAGACAAATCGCTCCAGG | Microsynth AG | N/A |
| Spike-SEQ-NTD-R: AGCACCAGCTGTCCAACCTG | Microsynth AG | N/A |
| Spike-SEQ2-NTD-R: CATAAGAAAAGGCTGAGAGA | Microsynth AG | N/A |
| Recombinant DNA | ||
| pCMV::SARS-CoV-2_S_ecto_hexapro | Ref: PMID: 32577660 | N/A |
| pCMV::SARS-CoV-2_S_ecto_2P_DS | Ref PMID: 32753755 | N/A |
| pCMV::SARS-CoV-2_S_NTD | This study | N/A |
| pCMV::P-GD_S_ecto | This study | N/A |
| pCMV::SARS-CoV-2_S_ecto_avi | PMID: 32972994 | N/A |
| pcDNA3.1::SARS-CoV-2-S-D19 | (Ou et al., 2020) | N/A |
| phCMV1::SARS-CoV-2 | PMID: 32972994 | N/A |
| phCMV1::RatG13_S | This study | N/A |
| phCMV1::P-GX_S | This study | N/A |
| phCMV1::P-GD_S | This study | N/A |
| phCMV1::ZC45_S | This study | N/A |
| phCMV1::ZXC21_S | This study | N/A |
| phCMV1::YN2013_S | This study | N/A |
| phCMV1::SARS-CoV_S | This study? | N/A |
| phCMV1::RmYN02_S | This study | N/A |
| phCMV1::Bt-kY72_S | This study | N/A |
| phCMV1::BM48-31_S | This study | N/A |
| pSARS-CoV-2-Nluc | (Xie et al., 2020) | N/A |
| Biological Samples | ||
| PE-Cy7 anti-CD19 antibody | BD Bioscience | Cat# 341113 |
| Anti-PE MicroBeads | Miltenyi Biotec | Cat# 130-048-801 |
| LS separation cloumns | Miltenyi Biotec | Cat# 130-042-401 |
| PE anti-human IgM | Lucerna-Chem AG | Cat# 314508 |
| PE anti-human IgD | BD Biosciences | Cat# 555779 |
| PE anti-human IgA | Southern Biotech | Cat# 2050-09 |
| PE anti-human CD14 | BD Biosciences | Cat# 562691 |
| PEI MAX | Polysciences | Cat# POL24765-1 |
| 4-Nitrophenyl phosphate disodium salt hexahydrate (pNPP) | Sigma-Aldrich | Cat# N2765-100TAB |
| Tween 20 | Sigma-Aldrich | Cat# 93773 |
| Bovine Serum Albumine (BSA) | Sigma-Aldrich | Cat# 3059 |
| Blocker Casein | ThermoFisher Scientific | Cat# 37528 |
| EZ-Link™ NHS-PEG solid phase biotinylation kit | ThermoFisher Scientific | Cat# 21450 |
| Zeba ™ Spin Desalting Columns | ThermoFisher Scientific | Cat# 89892 |
| TPCK treated Trypsin | Worthington Biochem | Cat# LS003750 |
| Alexa Fluor 647-AffiniPure F(ab’)2 Fragment Goat Anti-Human IgG, Fcg Fragment Specific |
Jackson ImmunoResearch | Cat# 109-606-098 |
| Goat Anti-Human IgG-AP | Southern Biotech | Cat# 2040-04 |
| Streptavidin, Alexa Fluor 647 conjugate | Life technologies | Cat# S21374 |
| Streptavidin, alkaline-phospatase conjugate | Jackson ImmunoResearch | Cat# 016-050-084 |
| Bio-Glo™ Luciferase Assay System | Promega | Cat# G7940 |
| ExpiFectamine™ CHO Transfection Kit | Life technologies | Cat# A29130 |
| ADCC Reporter Bioassays, F Variant | Promega | Cat# G9798 |
| ADCC Reporter Bioassays, V Variant | Promega | Cat# G7018 |
| FcgammaRIIa-H ADCP Reporter Bioassay | Promega | Cat# G9995 |
| 10% Igepal CA-630 | Sigma-Aldrich | Cat# I8896 |
| 25 mM dNTPs | Cytiva | Cat# 28406560 |
| RNase OUT | Life Technologies | Cat# 10777-019 |
| SupeScript III RT | Life Technologies | Cat# 18080-044 |
| DNase/RNase free water | Life Technologies | Cat# 10977-035 |
| 10 mM dNTPs mix | Cytiva | Cat# 28406564 |
| Q5 high fidelity DNA polymerase | Bioconcept | Cat# M0493S |
| AgeI-HF | New England Biolabs | Cat# R3552 |
| SalI-HF | New England Biolabs | Cat# R3138 |
| XhoI | New England Biolabs | Cat# R0146 |
| BsiWI | New England Biolabs | Cat# R3553 |
| NotI | New England Biolabs | Cat# R0189 |
| KpnI | New England Biolabs | Cat# R0142 |
| Cutsmart | New England Biolabs | Cat# B7204S |
| T4 DNA Ligase | New England Biolabs | Cat# M0202 |
| T4 Ligase buffer | New England Biolabs | Cat# B0202S |
| RNeasy Mini Kit | QIAGEN | Cat# 74104 |
| OneStep RT-PCR Kit | QIAGEN | Cat# 210210 |
| iTaq Universal One-Step RT-qPCR Kits | Biorad | Cat# 1725150 |
| NucleoSpin Plasmid, Mini kit for plasmid DNA | Macherey-Nagel | Cat# 740588.50 |
| GFX PCR DNA and Gel Band Purification Kit | Cytiva | Cat# 28903470 |
| Software and Algorithms | ||
| cryoSPARC v3.0.1 | (Punjani et al., 2017) | https://cryosparc.com |
| Relion v3.0 | (Zivanov et al., 2018) | https://www3.mrc-lmb.cam.ac.uk/relion |
| Coot | (Casañal et al., 2019) | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Phenix-Refine | (Adams et al., 2010) | https://www.phenix-online.org/download/ |
| Phenix-Phaser | (McCoy et al., 2007) | https://www.phenix-online.org/download/ |
| XDS | (Kabsch, 2010) | http://xds.mpimf-heidelberg.mpg.de |
| Prism 8 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Biacore Evaluation software | Cytiva | http://www.cytivalifesciences.com/en/us/shop/protein-analysis/spr-label-free-analysis/software/biacore-insight-evaluation-software-p-23528 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David Veesler (dveesler@uw.edu).
Materials availability
Materials generated in this study will be made available on request and may require a material transfer agreement.
Data and code availability
The cryo-EM maps, X-ray diffraction data and atomic models have been deposited at the Electron Microscopy Data Bank and the PDB with accession codes listed in the Key Resources Tables.
Experimental model and subject details
Cell lines
Cell lines used in this study were obtained from ATCC (HEK293T and Vero-E6) or ThermoFisher Scientific (Expi CHO cells, FreeStyle 293-F cells and Expi293F cells).
Sample donors
Samples from three SARS-CoV-2 recovered individuals, designates as donors L (male, 34 years-old), M (female, 50 years-old), and X (male, 52 years-old), were obtained under study protocols approved by the local Institutional Review Boards (Canton Ticino Ethics Committee, Switzerland, the Ethical committee of Luigi Sacco Hospital, Milan, Italy). All donors provided written informed consent for the use of blood and blood components (such as PBMCs, sera or plasma).
Samples were collected 14 and 52 days after symptoms onset for donor L and M, respectively. Blood drawn from donor X was obtained at day 36, 48, 75 and 125 after symptoms onset.
Method details
Cloning and mutant generation
SARS-CoV-2 NTD was sub-cloned with E. coli DH10B Competent Cells into pCMV using primers NTD_fwd and NTD_rev. The resulting construct was mutated by PCR mutagenesis to generate C15S, 69/70del, D80A/N, G142D, N149Q, D253G/Y, T19A, R246A, L18F/P, H146Y, A222V, Y144del, S254F, S255F, K147T, C136Y, and the NTD construct with native signal peptide with and without S12P/F, using the eponymously named primers (Key Resources Table). The genes encoding for the Sarbecovirus S proteins tested were cloned in the phCMV1 or pcDNA.3 vectors, and the gene for the C-terminally his-tagged ectodomain of P-GD S was cloned into pCMV (Key Resources Table). Plasmid sequences were verified by Genewiz sequencing facilities (Brooks Life Sciences).
Recombinant ectodomains production
All SARS-CoV-2 S spike ectodomains were produced in 500 mL cultures of FreeStyle 293-F cells (ThermoFisher Scientific) grown in suspension using FreeStyle 293 expression medium (ThermoFisher Scientific) at 37°C in a humidified 8% CO2 incubator rotating at 130 rpm Cells grown to a density of 2.5 million cells per mL were transfected using PEI (9 μg/mL) and pCMV::SARS-CoV-2_S_ecto_hexapro, pCMV::SARS-CoV-2_S_ecto_2P_DS, pCMV::P-GD_S_ecto, pCMV::SARS-CoV-2_S_ecto_avi, pCMV::SARS-CoV-2_S_D614G_ecto_avi and cultivated for 4 days. The supernatant was harvested and cells were resuspended for another three days, yielding two harvests. S ectodomains were purified from clarified supernatants using a Cobalt affinity column (Cytiva, HiTrap TALON crude), washing with 20 column volumes of 20 mM Tris-HCl pH 8.0 and 150 mM NaCl and eluted with a gradient of 600 mM imidazole. The same protocol was followed for P-GD spike ectodomain purification, except that 25 mM sodium phosphate pH 7 and 300 mM sodium chloride were used instead of 20 mM Tris-HCl pH 8.0 and 150 mM NaCl. At this stage, SARS-CoV-2 S with the avi tag (from pCMV::SARS-CoV-2_S_ecto_avi) was biotinylated (BirA biotin-protein ligase standard reaction kit, Avidity) and further purified by size exclusion chromatography (Superose6, GE Healthcare). All purified proteins were then concentrated using a 100 kDa centrifugal filter (Amicon Ultra 0.5 mL centrifugal filters, MilliporeSigma), residual imidazole was washed away by consecutive dilutions in the centrifugal filter unit with 20 mM Tris-HCl pH 8.0 and 150 mM NaCl, and finally concentrated to 5 mg/mL and flash frozen.
All SARS-CoV-2 S NTD domain constructs (residues 14-307) with a C-terminal 8XHis-tag were produced in 100 mL culture of Expi293F Cells (ThermoFisher Scientific) grown in suspension using Expi293 Expression Medium (ThermoFisher Scientific) at 37°C in a humidified 8% CO2 incubator rotating at 130 rpm) (Walls et al., 2020b). Cells grown to a density of 3 million cells per mL were transfected using pCMV::SARS-CoV-2_S_NTD derivative mutants with the ExpiFectamine 293 Transfection Kit (ThermoFisher Scientific) with and cultivated for five days at which point the supernatant was harvested. His-tagged NTD domain constructs were purified from clarified supernatants using 2 mL of cobalt resin (Takara Bio TALON), washing with 50 column volumes of 20 mM HEPES-HCl pH 8.0 and 150 mM NaCl and eluted with 600 mM imidazole. Purified protein was concentrated using a 30 kDa centrifugal filter (Amicon Ultra 0.5 mL centrifugal filters, MilliporeSigma), the imidazole was washed away by consecutive dilutions in the centrifugal filter unit with 20 mM HEPES-HCl pH 8.0 and 150 mM NaCl, and finally concentrated to 20 mg/mL and flash frozen. For crystallization, the purified NTD was not frozen but was further purified by size exclusion chromatography (Superdex Increase 75 10/300 G, GE Healthcare), concentrated using a new 30 kDa centrifugal filter, and used immediately.
Intact mass spectrometry analysis of purified NTD constructs
The purpose of intact MS was to verify the n-terminal sequence on four constructs. N-linked glycans were removed by PNGase F after overnight non-denaturing reaction at room temperature. 4 μg of deglycosylated protein was used for each injection on the LC-MS system to acquire intact MS signal after separation of protease and protein by LC (Agilent PLRP-S reversed phase column). Thermo MS (Q Exactive Plus Orbitrap) was used to acquire intact protein mass under denaturing condition. BioPharma Finder 3.2 software was used to deconvolute the raw m/z data to protein average mass.
Non-reducing Peptide Mapping mass spectrometry analysis of purified NTD constructs
The purpose of peptide mapping was to identify the post modification on unpaired cysteines. Tryptic digestion was used without adding reducing reagent. 50 μg of deglycosyated protein was denatured (6 M guanidine hydrochloride), alkylated (Iodoacetamide), and buffer exchanged (Zeba spin desalting column) before trypsin digestion. 10 μg of digested peptide was analyzed on the LC-MS system (Agilent AdvanceBio peptide mapping column and Thermo Q Exactive Plus Orbitrap MS) to acquire both MS1 and MS2 data under HCD fragmentation. Peptide mapping data was analyzed on Biopharma Finder 3.2 by searching the possible modifications such as cysteinylation and carbamidomethylation on the cysteines.
Isolation of peripheral blood mononuclear cells (PBMCs), plasma and sera
PBMCs were isolated from blood draw performed using tubes pre-filled with heparin, followed by Ficoll density gradient centrifugation. PBMCs were either used freshly along SARS-CoV2 Spike protein specific memory B cells sorting or stored in liquid nitrogen for later use. Sera were obtained from blood collected using tubes containing clot activator, followed by centrifugation and stored at −80°C.
B cell isolation and recombinant mAb production
Starting from freshly isolated PBMCs or upon cells thawing, B cells were enriched by staining with CD19 PE-Cy7 and incubation with anti-PE beads, followed by positive selection using LS columns. Enriched B cells were stained with anti-IgM, anti-IgD, anti-CD14 and anti-IgA, all PE labeled, and prefusion SARS-CoV-2 S with a biotinylated avi tag conjugated to Streptavidin Alexa Fluor 647 (Life Technologies). SARSCoV-2 S-specific IgG+ memory B cells were sorted by flow cytometry via gating for PE negative and Alexa Fluor 647 positive cells. Cells were cultured for the screening of positive supernatants. Antibody VH and VL sequences were obtained by RT-PCR and mAbs were expressed as recombinant human Fab fragment or as IgG1 (G1m3 allotype) carrying the half-life extending M428L/N434S (LS) mutation in the Fc region. ExpiCHO cells were transiently transfected with heavy and light chain expression vectors as previously described (Pinto et al., 2020).
Affinity purification was performed on ÄKTA Xpress FPLC (Cytiva) operated by UNICORN software version 5.11 (Build 407) using HiTrap Protein A columns (Cytiva) for full length human and hamster mAbs and CaptureSelect CH1-XL MiniChrom columns (ThermoFisher Scientific) for Fab fragments, using PBS as mobile phase. Buffer exchange to the appropriate formulation buffer was performed with a HiTrap Fast desalting column (Cytiva). The final products were sterilized by filtration through 0.22 μm filters and stored at 4°C.
Enzyme-linked immunosorbent assay (ELISA)
To determine specificity of recombinantly produced mAbs, 96 half area well-plates (Corning) were coated over-night at 4°C with of SARS-CoV-2 S, NTD or RBD proteins prepared at 1 μg/mL, 2 μg/mL and 5 μg/mL in PBS pH 7.2, respectively. Plates were then blocked with PBS 1% BSA (Sigma) and subsequently incubated with mAbs serial dilutions for 1 h at room temperature. After 2 washing steps with PBS 0.05% Tween 20 (PBS-T) (Sigma-Aldrich) goat anti-human IgG secondary antibody (Southern Biotech) was added and incubated for 1 h at room temperature. Plates were then washed again with PBS-T and 4-NitroPhenyl phosphate (pNPP, Sigma-Aldrich) substrate added. After 30 min incubation, absorbance at 405 nm was measured by a plate reader (Biotek) and data plotted using Prism GraphPad.
For all other applications reported, the following ELISA procedure was followed: 30 μl of ectodomains (stabilized prefusion trimer) of S or NTD from SARS-CoV-2 were coated on 384 well ELISA plates at 1 ng/μl for 16 h at 4°C. Plates were washed with a 405 TS Microplate Washer (BioTek Instruments) then blocked with 80 μl SuperBlock (PBS) Blocking Buffer (Thermo Scientific) for 1 h at 37°C. Plates were then washed and 30 μl antibodies were added to the plates at concentrations between 0.001 and 100,000 ng/mL and incubated for 1 h at 37°C. Plates were washed and then incubated with 30 μl of 1/5000 diluted goat anti-human Fc IgG-HRP (Invitrogen). Plates were washed and then 30 μl Substrate TMB microwell peroxidase (Seracare) was added for 4 min at room temperature. The colorimetric reaction was stopped by addition of 30 μl of 1 N HCl. A450 was read on a Varioskan Lux plate reader (Thermo Scientific).
MLV-based pseudotyped virus production and neutralization
To generate SARS-CoV-2 S murine leukemia virus pseudotyped virus, HEK293T cells were seeded in 10-cm dishes in DMEM supplemented with 10% FBS. The next day cells were transfected with a SARS-CoV-2 S glycoprotein-encoding plasmid harboring the D19 C-terminal truncation (Ou et al., 2020), an MLV Gag-Pol packaging construct and the reporter vector pTG-Luc, using the X-tremeGENE HP DNA transfection reagent (Roche) according to the manufacturer’s instructions. Cells were then incubated at 37°C with 5% CO2 for 72 h. Supernatant was harvested and cleared from cellular debris by centrifugation at 400 x g, and stored at −80°C.
For neutralization assays, Vero E6 cells were seeded into white 96-well plates (PerkinElmer) at 20,000 cells/well and cultured overnight at 37°C with 5% CO2 in 100 μl DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. The next day, MLV-SARS-CoV-2 pseudovirus was activated with 10 μg/mL TPCK treated-Trypsin (Worthington Biochem) for 1 h at 37°C. Next, recombinant antibodies at various concentrations were incubated with activated pseudovirus for 1 h at 37°C. The Vero E6 cells were then washed with DMEM, and 50 μL of pseudovirus/mAbs mixes were added and incubated for 2 h at 37°C with 5% CO2. After incubation, 50 μl of DMEM containing 20% FBS and 2% penicillin/streptomycin was added and the cells were incubated 48 h at 37°C with 5% CO2. Following these 48 h of infection, culture medium was removed from the cells and 50 μl/well of Bio-Glo (Promega) diluted 1:2 with PBS with Ca2+Mg2+ (Thermo Fisher) was added to the cells and incubated in the dark for 15 min before reading on a Synergy H1 Hybrid Multi-Mode plate reader (Biotek). Measurements were done in duplicate, RLU values were converted to percentage of neutralization and plotted with a nonlinear regression curve fit in Graph Prism.
VSV-based pseudotype virus production and neutralization assay
SARS-CoV-2 S (YP 009724390.1), RaTG13 S (QHR63300.2), Pangolin-Guangdong S(QLR06867.1), Pangolin-Guanxi S (EPI ISL 410539), SARS-CoV S (YP 009825051.1), WIV1 S (AGZ48831.1) and WIV16 S (ALK02457.1) pseudotyped VSV viruses were prepared using 293T cells seeded in 10-cm dishes. Briefly, cells in DMEM supplemented with 10% FBS, 1% PenStrep were transfected with the plasmid encoding for the corresponding S glycoprotein using lipofectamine 2000 (Life Technologies) following manufacturer’s indications. One day post-transfection, cells were infected with VSV(G∗ΔG-luciferase) and after 2 h, infected cells were washed four times with DMEM before adding medium supplemented with anti-VSV-G antibody (I1- mouse hybridoma supernatant diluted 1 to 50, from CRL- 2700, ATCC). Particles were harvested 18 h post-inoculation, clarified from cellular debris by centrifugation at 2,000 x g for 5 min and concentrated 10 times using a 30 kDa cut off membrane and used for neutralization experiments, aliquoted and frozen at −80°C until use in neutralization experiments.
For neutralization, stable 293T cells expressing ACE2 (Crawford et al., 2020) in DMEM supplemented with 10% FBS, 1% PenStrep were seeded at 40,000 cells/well into clear bottom white walled 96-well plates and cultured overnight at 37°C. Twelve-point 3-fold serial dilutions of the corresponding mAb were prepared in DMEM and pseudotyped VSV viruses were added 1:1 to each mAb dilution in the presence of anti-VSV-G antibody from I1- mouse hybridoma supernatant diluted 50 times. After 45 min incubation at 37 °C, 40 μl of the mixture was added to the cells and 2 h post-infection, 40 μL DMEM was added to the cells. After 17-20 h 50 μL/well of One-Glo-EX substrate (Promega) was added to the cells and incubated in the dark for 5-10 min prior reading on a Varioskan LUX plate reader (ThermoFisher). Measurements were done in duplicate with two independent productions of pseudotyped viruses and RLU values were converted to percentage of neutralization and plotted with a nonlinear regression curve fit in Graph Prism.
Neutralization of authentic SARS-CoV-2-Nluc virus
Neutralization of authentic SARS-CoV-2 by entry-inhibition assay Neutralization was determined using SARS-CoV-2-Nluc, an infectious clone of SARSCoV-2 (based on strain 2019-nCoV/USA_WA1/2020) which encodes nanoluciferase in place of the viral ORF7 and demonstrated comparable growth kinetics to wildtype virus (Xie et al., 2020). Vero E6 cells were seeded into black-walled, clear-bottom 96-well plates at 2 × 104 cells/well and cultured overnight at 37°C. The next day, 9-point 4-fold serial dilutions of mAbs were prepared in infection media (DMEM + 10% FBS). SARS-CoV-2-Nluc was diluted in infection media at a final MOI of 0.1 or 0.01 PFU/cell, added to the mAb dilutions and incubated for 30 min at 37°C. Media was removed from the Vero E6 cells, mAb-virus complexes were added and incubated at 37°C for 6 or 24 h. Media was removed from the cells, Nano-Glo luciferase substrate (Promega) was added according to the manufacturer’s recommendations, incubated for 10 min at room temperature and the luciferase signal was quantified on a VICTOR Nivo plate reader (Perkin Elmer).
Binding and affinity determination by Biolayer Interferometry (BLI)
BLI measurements were performed using an Octet Red96 (ForteBio). All reagents were prepared in kinetics buffer (PBS plus 0.01% BSA) at the indicated concentrations.
BLI was used to assess antibody binding affinity to SARS-CoV-2 NTD. IgG antibodies were prepared at 2.7 μg/mL and captured on pre-hydrated Protein A biosensors (Sartorius) for 1 min. The biosensors with immobilized antibodies were moved into kinetics buffer with SARS-CoV-2 NTD (concentrations tested: 333.3, 166.6, 83.3, 41.7, 20.8, 10.4, 5.2 nM) for 5 min (i.e., association). The dissociation of the SARS-CoV-2 NTD was then recorded for 9 min in wells containing kinetics buffer. Affinity constants were calculated using a global fit model and results were plotted using GraphPad Prism.
BLI was also used to assess antibody competition studies to define the NTD antigenic map. Biotinylated SARS-CoV-2 S protein was prepared at 10 μg/mL in kinetics buffer and loaded on pre-hydrated High Precision Streptavidin SAX Biosensors (Sartorius) for 3 min. NTD mAbs at 20 μg/mL in kinetics buffer were then sequentially added to observe binding competition and signal recorded for 5 min (or 7 min). Given the low responses detected for binding of S2L11 and S2X176 to the S ectodomain trimer, mapping of these mAbs was performed by capturing the purified NTD to BLI biosensors. NTD was prepared at 8 μg/mL and assay performed using the same kinetics conditions described above.
BLI was also used to assess mAb-mediated inhibition of SARS-CoV-2 S binding to human recombinant ACE2. Before the assay SARS-CoV2 S ectodomain trimer (5 μg/mL) was incubated with tested mAbs (30 μg/mL) or no mAb for 30 min at 37°C. Biotinylated recombinant human ACE2 protein (2 μg/mL) was immobilized on High Precision Streptavidin SAX Biosensors (Sartorius). Next, an association step with S/mAb complexes was performed for 10 min. Results were plotted using GraphPad Prism.
S2X28 blockade-of-binding to NTD
S2X28 mAb was biotinylated using EZ-Link NHS-PEG solid phase biotinylation kit (ThermoFisher Scientific) and binding to NTD tested to set optimal concentration to be used in the assay after sample desalting using Zeba Spin Desalting Columns (ThermoFisher Scietific). Half area 96 well-plates were coated over-night at 4°C with SARS-CoV-2 NTD diluted at 2 μg/mL in PBS. After a blocking step with Blocker Casein (ThermoFisher Scientific), serial plasma dilutions in Blocker Casein were incubated 1 h at room temperature. Biotinylated S2X28 was added at a concentration achieving 70% of maximal binding and the mixture was incubated for 45 min at room temperature. Alkaline-phosphatase conjugated streptavidin (Jackson ImmunoResearch) was diluted at 0.5 μg/mL in Blocker Casein and added on plates previously washed 4 times with PBS 0.05%Tween 20. After 30 min incubation, plates were washed and 4-NitroPhenyl phosphate substrate incubated for 45 min at room temperature. Absorbance at 405 nm was measured and percentage of inhibition was calculated as follows: (1-(OD sample-OD neg ctr)/ (OD pos ctr-OD neg ctr)) x100.
Affinity determination by Surface Plasmon Resonance (SPR)
SPR binding measurements were performed using a Biacore T200 instrument where purified avi-tagged SARS-CoV-2 S D614G ectodomain trimer was captured using anti-AviTag pAb covalently immobilized on a CM5 sensor chip. The running buffer was Cytiva HBS-EP+ pH 7.4; measurements were performed at 25þC. Affinity/avidity determinations were run as single-cycle kinetics, with a 3-fold dilution series of mAb starting from 300 nM, and each concentration injected for 180 s. Double reference-subtracted data were fit to a 1:1 binding model using Biacore Evaluation software. Fit results for IgG yielded apparent equilibrium dissociation constants due to avidity. For dissociation rates that were too slow to fit, equilibrium dissociation constants are reported as an upper limit.
Transient Expression of Sarbecovirus S protein in ExpiCHO-S Cells
Immediately before transfection, ExpiCHO-S cells were seeded at 6 × 106 cells cells/mL in a volume of 5 mL in a 50 mL bioreactor. Spike coding plasmids were diluted in cold OptiPRO SFM, mixed with ExpiFectamine CHO Reagent (Life Technologies) and added to the cells. Transfected cells were then incubated at 37°C with 8% CO2 with an orbital shaking speed of 120 RPM (orbital diameter of 25 mm) for 42 h
Binding to cell surface expressed Sarbecovirus S proteins by Flow Cytometry
Transiently transfected ExpiCHO cells were harvested and washed two times in wash buffer (PBS 1% BSA, 2 mM EDTA). Cells were counted, distributed into round bottom 96-well plates (Corning) and incubated with the NTD antibodies at the final concentration of 5 μg/mL. Alexa Fluor647-labeled Goat Anti-Human IgG secondary Ab (Jackson Immunoresearch) was prepared at 1.5 μg/mL added onto cells after two washing steps. Cells were then washed twice and resuspended in wash buffer for data acquisition at ZE5 cytometer (Biorad).
Fusion inhibition assay
Vero E6 cells were seeded in 96 well plates at 15,000 cells per well in 70 μL DMEM with high glucose and 2.4% FBS (Hyclone). After 16 h at 37°C with 8% CO2, the cells were transfected with SARS-CoV-2-S-D19_pcDNA3.1 as follows: for 10 wells, 0.57 μg plasmid SARS-CoV-2- S-D19_pcDNA3.1 were mixed with 1.68 μl X-tremeGENE HP in 30 μl OPTIMEM. After 15 min incubation, the mixture was diluted 1:10 in DMEM medium and 30 μL was added per well. A 4-fold serial dilution mAbs was prepared and added to the cells, with a starting concentration of 20 μg/mL. The following day, 30 μL 5X concentrated DRAQ5 in DMEM was added per well and incubated for 2 h at 37°C. Nine images of each well were acquired with a Cytation 5 equipment for analysis.
Measurement of Fc-effector functions
mAb-dependent activation of human FcγRs was performed with a bioluminescent reporter assay. ExpiCHO cells stably expressing full-length wild-type SARS-CoV-2 S (target cells) were incubated with different amounts of mAbs. After a 15-min incubation, Jurkat cells stably expressing FcγRIIIa receptor (V158 variant) or FcγRIIa receptor (H131 variant) and NFAT-driven luciferase gene (effector cells) were added at an effector to target ratio of 6:1 for FcγRIIIa and 5:1 for FcγRIIa. Signaling was quantified by the luciferase signal produced as a result of NFAT pathway activation. Luminescence was measured after 20 h of incubation at 37þC with 5% CO2 with a luminometer using the Bio-Glo-TM Luciferase Assay Reagent according to the manufacturer’s instructions (Promega).
Cell-surface mAb-mediated S1 shedding
CHO cells stably expressing wild-type SARS-CoV-2 S were resuspended in wash buffer (PBS 1% BSA, 2 mM EDTA) and treated with 10 μg/mL TPCK-trypsin (Worthington Biochem) for 30 min at 37°C. Cells were then washed and distributed into round bottom 96-well plates (90,000 cells/well). MAbs were added to cells at 15 μg/mL final concentration for 180 min at 37°C. Cells were collected at different time points (5, 30, 60, 120 and 180), washed with wash buffer at 4°C, and incubated with 1.5 mg/mL secondary goat anti-human IgG, Fc fragment specific (Jackson ImmunoResearch) on ice for 20 min. Cells were washed and resuspended in wash buffer and analyzed with ZE5 FACS (Bio-rad).
Selection of SARS-CoV-2 monoclonal antibody escape mutants (MARMS)
VSV-SARS-CoV-2 chimera was used to select for SARS-CoV-2 S monoclonal antibody resistant mutants (MARMS) as previously described (Case et al., 2020; Liu et al., 2020b). Briefly, MARMS were recovered by plaque isolation on Vero cells with the indicated mAb in the overlay. The concentration of mAb in the overlay was determined by neutralization assays at a multiplicity of infection (MOI) of 100. Escape clones were plaque-purified on Vero cells in the presence of mAb, and plaques in agarose plugs were amplified on MA104 cells with the mAb present in the medium. Viral stocks were amplified on MA104 cells at an MOI of 0.01 in Medium 199 containing 2% FBS and 20 mM HEPES pH 7.7 (Millipore Sigma) at 34°C. Viral supernatants were harvested upon extensive cytopathic effect and clarified of cell debris by centrifugation at 1,000 x g for 5 min. Aliquots were maintained at −80°C. Viral RNA was extracted from VSV-SARS-CoV-2 mutant viruses using RNeasy Mini kit (QIAGEN), and S was amplified using OneStep RT-PCR Kit (QIAGEN). The mutations were identified by Sanger sequencing (GENEWIZ). Their resistance was verified by subsequent virus infection in the presence or absence of antibody. Briefly, Vero cells were seeded into 12 well plates overnight. The virus was serially diluted using DMEM and cells were infected at 37°C for 1 h. Cells were cultured with an agarose overlay in the presence or absence of mAb at 34°C for 2 days. Plates were scanned on a biomolecular imager and expression of eGFP is show at 48 h post-infection.
Cryo-EM sample preparation and data collection
2.5 μL of 12 mg/mL S2M11 Fab, 2.5 μL of 12 mg/mL NTD Fab, and 6 μL of 5 mg/mL SARS-CoV-2 S (specifically the hexapro construct (Hsieh et al., 2020)) were incubated together for 30 min at 37°C. Alternatively, 3 μL of 3 mg/mL S2X28 Fab, and 3 μL of 1 mg/mL SARS-CoV-2 2P DS S (McCallum et al., 2020) were incubated together for 30 min at 37°C. Unbound Fab was then washed away with three consecutive dilutions in 400 μL of 20 mM Tris-HCl pH 8.0 and 150 mM NaCl over a 100 kDa centrifugal filter (Amicon Ultra 0.5 mL centrifugal filters, MilliporeSigma). The complex was concentrated to 1.2 mg/mL and 3 μL was applied onto a freshly glow discharged 2.0/2.0 UltraFoil grid (200 mesh), plunge frozen using a vitrobot MarkIV (ThermoFisher Scientific) using a blot force of −1 and 6.5 s blot time at 100% humidity and 23°C.
Data were acquired using the Leginon software (Suloway et al., 2005) to control a FEI Titan Krios or Glacios transmission electron microscope equipped with a Gatan K2 Summit direct detectors and operated at 300 kV with a Gatan Quantum GIF energy filter or at 200 kV, respectively. For both microscopes, the dose rate was adjusted to 8 counts/pixel/s, and each movie was acquired in 50 frames of 200 ms. For the Krios, ∼3000 micrographs were acquired for each session with a super-resolution pixel size of 0.525 Å with a defocus range between −0.8 and −2.0 μm. For the Glacios, ∼100 micrographs with a pixel size of 1.16 Å were collected in a single session with a defocus range between −0.8 and −2.0 μm.
Cryo-EM data processing
Movie frame alignment, estimation of the microscope contrast-transfer function parameters, particle picking and extraction (with a box size of 400 pixels2) were carried out using Warp (Tegunov and Cramer, 2019). At this stage, the pixel size was binned to 1.05 Å for data collected from the Krios. Reference-free 2D classification was performed using cryoSPARC (Punjani et al., 2017) to select well-defined particle images. 3D classification with 50 iterations each (angular sampling 7.5þ for 25 iterations and 1.8þ with local search for 25 iterations) were carried out using Relion (Zivanov et al., 2018) without imposing symmetry to separate distinct SARS-CoV-2 S conformations. 3D refinements were carried out using non-uniform refinement along with per-particle defocus refinement in cryoSPARC (Punjani et al., 2020) before particle images were subjected to Bayesian polishing using Relion (Zivanov et al., 2019). To accommodate the resolution of the S/S2M11/S2X333 dataset, at the polishing stage the box size was adjusted to 600 Å while the pixel size was binned to 0.896 Å. Another round of non-uniform refinement in cryoSPARC was performed, followed by global and per-particle defocus refinement and again non-uniform refinement. Reported resolutions are based on the gold-standard Fourier shell correlation (FSC) of 0.143 criterion and Fourier shell correlation curves were corrected for the effects of soft masking by high-resolution noise substitution (Scheres and Chen, 2012).
Cryo-EM model building and analysis
UCSF Chimera (Goddard et al., 2007) and Coot (Casañal et al., 2019) were used to fit atomic models (PDB 7K43 and 6ZGE) into the cryo-EM maps. The model was then refined into the map using Rosetta (DiMaio et al., 2015; Frenz et al., 2019; Wang et al., 2016), Phenix (Liebschner et al., 2019) and ISOLDE (Croll, 2018). Analysis used MolProbity(Chen et al., 2010), EMringer(Barad et al., 2015), and Phenix (Liebschner et al., 2019). Figures were generated using UCSF ChimeraX( Goddard et al., 2018) and UCSF Chimera (Goddard et al., 2007).
S2M28 Fab and NTD co-crystallization and structure determination
Crystals of S2M28 Fab with SARS-CoV-2 NTD were identified by the sitting-drop vapor diffusion method, set-up by hand with MCSG-1 crystallization screen (Anatrace). Optimized crystals were generated with 1 μl of 5.7 mg/mL Fab and 4 mg/mL NTD in 20 mM HEPES-HCl pH 8.0 and 150 mM NaCl plus 1 μl mother liquor solution containing 0.2 M Ammonium Sulfate, 0.1 M Sodium Citrate pH 4.75, 25% (w/v) PEG4000. Crystals were flash cooled in liquid nitrogen using the mother liquor solution supplemented with 17.5% (w/v) xylitol as a cryoprotectant. Diffraction data were collected on synchrotron beamline 5.0.2 at the Advanced Light Source, and processed with the XDS software package (Kabsch, 2010). Initial phases were obtained by molecular replacement using Phenix-Phaser (McCoy et al., 2007), using the S/S2M11/S2M28 cryoEM structure. Several subsequent rounds of model building and refinement were performed using Coot (Casañal et al., 2019) and Phenix-Refine (Adams et al., 2010).
In vivo mAb testing using a Syrian hamster model
KU LEUVEN R&D has developed and validated a SARS-CoV-2 Syrian Golden hamster infection model (Boudewijns et al., 2020). The SARS-CoV-2 isolate used in this study (BetaCoV/Belgium/GHB-03021/2020-EPI ISL 109 407976|2020-02-03), was recovered from a nasopharyngeal swab taken from a RT-qPCR confirmed asymptomatic patient who returned from Wuhan, China at the beginning of February 2020. A close relatedness with the prototypic Wuhan-Hu-1 2019 SARS-CoV-2 isolate was confirmed by sequencing and phylogenetic analysis. Infectious virus was isolated by serial passaging on Huh7 and Vero E6 cells and passage 6 virus was used for the study described here. The titer of the virus stock was determined by end-point dilution on Vero E6 cells by the Reed and Muench method (Reed and Muench, 1938). This work was conducted in the high-containment A3 and BSL3+ facilities of the KU Leuven Rega Institute (3CAPS) under licenses AMV 30112018 SBB 219 2018 0892 and AMV 23102017 SBB 219 20170589 according to institutional guidelines.
Syrian hamsters (Mesocricetus auratus) were purchased from Janvier Laboratories and were housed per two in ventilated isolator cages (IsoCage N Biocontainment System, Tecniplast) with ad libitum access to food and water and cage enrichment (wood block). Housing conditions and experimental procedures were approved by the ethical committee of animal experimentation of KU Leuven (license P065-2020). 6-10 week-old female hamsters were administered by intraperitoneal injection with S2X333 mAb at 1 mg/kg and 4 mg/kg 48 h before intranasal infection with 1.89 × 106 50% tissue culture infectious dose (TCID50) in 50 μl inoculum. Hamsters were monitored for appearance, behavior and weight. At day 4 post infection hamsters were euthanized by intraperitoneal injection of 500 μL Dolethal (200 mg/mL sodium pentobarbital, Vétoquinol SA). Lungs were collected, homogenized using bead disruption (Precellys) in 350 μL RLT buffer (RNeasy Mini kit, QIAGEN) and centrifuged (10,000 rpm, 5 min, 4°C) to pellet the cell debris. RNA was extracted using a NucleoSpin kit (Macherey-Nagel) according to the manufacturer’s instructions. RT-qPCR was performed on a LightCycler96 platform (Roche) using the iTaq Universal Probes One-Step RTqPCR kit (BioRad) with N2 primers and probes targeting the nucleocapsid (Boudewijns et al., 2020). Standards of SARS-CoV-2 cDNA (IDT) were used to express viral genome copies per mg tissue or per mL serum. To quantify infectious SARS-CoV-2 particles, endpoint titrations were performed on confluent Vero E6 cells in 96-well plates. Viral titers were calculated by the Reed and Muench method (Reed and Muench, 1938) and were expressed as TCID50 per mg tissue.
To detect escape mutants in the in vivo tests, cDNAs were synthesized from 100 ng vRNAs extracted from lung homogenates using Superscript III Reverse Transcriptase (Invitrogen) and Spike-cDNA-R oligonucleotide. A gene-specific fragment of 1664 bp encompassing the NTD region of SARS-CoV-2-S was amplified by PCR using Spike-SEQ1-F and Spike-SEQ1-R primers with Q5® Hot Start High-Fidelity DNA Polymerase (New England Biolabs). Reaction conditions were as follows: 50 μl reaction volume containing 2 μl of cDNA, 1X Q5 Reaction buffer, 200 μM each of four dNTPs (Cytiva Europe GmbH), 0.5 μM of each primer (Microsynth) and 0.02 U/μl of polymerase. PCR conditions: 98°C for 30 s (initial denaturation); 45 cycles of 98°C for 10 s (denaturation), 62°C for 30 s (annealing), 72°C for 60 s (elongation), followed by 1 cycle of 72°C for 2 min (final elongation).PCR products were purified with GFX PCR DNA and Gel Band Purification Kit (Cytiva Europe GmbH), and sequencing was outsourced at Microsynth (Sanger sequencing) with Spike-SEQ2-F, Spike-SEQ3-F, Spike-SEQ-NTD-R, and Spike-SEQ2-NTD-R primers. Sequence analysis was performed with CLC Main Workbench 21 (QIAGEN) using NCBI NC_045512.2 and virus stock SARS-CoV-2 S -p6-20200525 as reference sequences.
Quantification and statistical analysis
Statistical analyses were performed using GraphPad Prism (v9) and Microsoft Excel for Windows 10 (v16.0.13001.20254). Statistical differences were analyzed with Mann-Whitney U-test. Statistical significance was defined as ∗p < 0.05, ∗∗p < 0.01. EC50 and IC50, values were determined by non-linear regression analysis (log(agonist) versus response - Variable slope (four parameters)).
Acknowledgments
We thank Hideki Tani (University of Toyama) for providing the reagents necessary for preparing VSV pseudotyped viruses. The authors would also like to thank Roberto Spreafico for initial SARS-CoV-2 NTD conservation analysis and Marcel Meury for assistance with protein production. This study was supported by the National Institute of General Medical Sciences (R01GM120553 to D.V.), the National Institute of Allergy and Infectious Diseases (DP1AI158186 and HHSN272201700059C to D.V.), a Pew Biomedical Scholars Award (D.V.), Investigators in the Pathogenesis of Infectious Disease Awards from the Burroughs Wellcome Fund (D.V.), Fast Grants (D.V.), the Natural Sciences and Engineering Research Council of Canada (M.M.), the Pasteur Institute (M.A.T.), the University of Washington Arnold and Mabel Beckman cryo-EM center, and BCSB beamline 5.0.2 at the Advanced Light Source, a US DOE Office of Science User Facility under contract no. DE-AC02-05CH11231.
Author contributions
Experiment design, M.M., A.D.M., F.A.L., D.C., M.S.P., and D.V.; donors’ recruitment and sample collection, E.C., F.B., and A.R.; sample processing, A.D.M., D.P., M.B., F.Z., and C.S.F.; protein expression and purification, M.M., S.Z., J.E.B., E.C., and G.S.; isolation and characterization of mAbs, M.M., A.D.M., F.A.L., M.A.T, D.P., A.C.W., M.B., A.C., Z.L., F.Z., S.Z., J.d.I., J.E.B., M.M.-R., J.Z., L.E.R., S.B., B.G., C.S.F., R.A., S.-Y.C.F., P.W.R., L.-M.B., F.B., and E.C.; binding and neutralization assays, M.M. A.D.M., F.A.L., M.A.T, D.P., A.C.W., M.B., A.C., Z.L., F.Z., M.M.-R., and J.Z.; BLI/SPR assays, A.D.M and L.E.R.; cryo-EM data collection, processing, and model building, M.M. and D.V.; bioinformatic analysis of virus variants, J.d.I and A.T.; escape mutants selection and sequencing, Z.L., P.W.R., L.M.B., and S.P.J.W.; hamster model and data analysis, R.A., S.C.F., F.B., J.N., D.C., and M.S.P.; data analysis, M.M., A.D.M, F.A.L., D.C., M.S.P., and D.V; manuscript writing, M.M., A.D.M., A.T., G.S., S.P.J.W., H.W.V., D.C., M.S.P., and D.V.; supervision, F.B., E.C., J.N., A.R., G.S., A.T., S.P.J.W., H.W.V., D.C., M.S.P., and D.V.; funding acquisition, D.V.
Declaration of interests
A.D.M., F.A.L., D.P., M.B., F.Z., J.d.I., M.M.-R., J.Z., L.E.R., S.B., B.G., C.S.F., F.B., E.C., G.S., A.T., H.W.V., D.C., and M.S.P. are employees of Vir Biotechnology Inc. and may hold shares in Vir Biotechnology Inc. D.C. is currently listed as an inventor on multiple patent applications, which disclose the subject matter described in this manuscript. The Neyts laboratories have received sponsored research agreements from Vir Biotechnology Inc. H.W.V. is a founder of PierianDx and Casma Therapeutics. Neither company provided funding for this work or is performing related work. D.V. is a consultant for Vir Biotechnology Inc. The Veesler laboratory has received a sponsored research agreement from Vir Biotechnology Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Published: March 16, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2021.03.028.
Supplemental information
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almagro Armenteros J.J., Tsirigos K.D., Sønderby C.K., Petersen T.N., Winther O., Brunak S., von Heijne G., Nielsen H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019;37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- Andreano E., Piccini G., Licastro D., Casalino L., Johnson N.V., Paciello I., Monego S.D., Pantano E., Manganaro N., Manenti A., et al. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv. 2020 doi: 10.1101/2020.12.28.424451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avanzato V.A., Matson M.J., Seifert S.N., Pryce R., Williamson B.N., Anzick S.L., Barbian K., Judson S.D., Fischer E.R., Martens C., et al. Case Study: Prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised cancer patient. Cell. 2020;183:1901–1912.e9. doi: 10.1016/j.cell.2020.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barad B.A., Echols N., Wang R.Y., Cheng Y., DiMaio F., Adams P.D., Fraser J.S. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods. 2015;12:943–946. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.-M.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. Structural classification of neutralizing antibodies against the SARS-CoV-2 spike receptor-binding domain suggests vaccine and therapeutic strategies. bioRxiv. 2020 doi: 10.1101/2020.08.30.273920. [DOI] [Google Scholar]
- Barnes C.O., West A.P., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell. 2020;182:828–842.e16. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Ajithdoss D., Copin R., Zhou A., Lanza K., Negron N., Ni M., Wei Y., Mohammadi K., Musser B., et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020;370:1110–1115. doi: 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudewijns R., Thibaut H.J., Kaptein S.J.F., Li R., Vergote V., Seldeslachts L., Van Weyenbergh J., De Keyzer C., Bervoets L., Sharma S., et al. STAT2 signaling restricts viral dissemination but drives severe pneumonia in SARS-CoV-2 infected hamsters. Nat. Commun. 2020;11:5838. doi: 10.1038/s41467-020-19684-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S., Wang T.T., Ravetch J.V. The Role and Function of Fcγ Receptors on Myeloid Cells. Microbiol. Spectr. 2016 doi: 10.1128/microbiolspec.MCHD-0045-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S., Corti D., Virgin H.W., Ravetch J.V. Fc-optimized antibodies elicit CD8 immunity to viral respiratory infection. Nature. 2020;588:485–490. doi: 10.1038/s41586-020-2838-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casañal A., Lohkamp B., Emsley P. Current developments in Coot for macromolecular model building of Electron Cryo-microscopy and Crystallographic Data. Protein Sci. 2019;29:1069–1078. doi: 10.1002/pro.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Rothlauf P.W., Chen R.E., Liu Z., Zhao H., Kim A.S., Bloyet L.M., Zeng Q., Tahan S., Droit L., et al. Neutralizing Antibody and Soluble ACE2 Inhibition of a Replication-Competent VSV-SARS-CoV-2 and a Clinical Isolate of SARS-CoV-2. Cell Host Microbe. 2020;28:475–485.e5. doi: 10.1016/j.chom.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Lu S., Jia H., Deng Y., Zhou J., Huang B., Yu Y., Lan J., Wang W., Lou Y., et al. A novel neutralizing monoclonal antibody targeting the N-terminal domain of the MERS-CoV spike protein. Emerg. Microbes Infect. 2017;6:e37. doi: 10.1038/emi.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiodo F., Bruijns S.C.M., Rodriguez E., Li R.J.E., Molinaro A., Silipo A., Di Lorenzo F., Garcia-Rivera D., Valdes-Balbin Y., Verez-Bencomo V., et al. Novel ACE2-Independent Carbohydrate-Binding of SARS-CoV-2 Spike Protein to Host Lectins and Lung Microbiota. bioRxiv. 2020 doi: 10.1101/2020.05.13.092478. [DOI] [Google Scholar]
- Choi B., Choudhary M.C., Regan J., Sparks J.A., Padera R.F., Qiu X., Solomon I.H., Kuo H.H., Boucau J., Bowman K., et al. Persistence and Evolution of SARS-CoV-2 in an Immunocompromised Host. N. Engl. J. Med. 2020;383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., Narayanan A., Majowicz S.A., Kwong E.M., McVicar R.N., et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell. 2020;183:1043–1057.e15. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier D.A., De Marco A., Ferreira I.A.T.M., Meng B., Datir R., Walls A.C., Kemp S S.A., Bassi J., Pinto D., Fregni C.S., et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021 doi: 10.1038/s41586-021-03412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., DiPiazza A.T., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O’Connell S., Bock K.W., Minai M., et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M., et al. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses. 2020;12:513. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll T.I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D Struct. Biol. 2018;74:519–530. doi: 10.1107/S2059798318002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly J.L., Simonetti B., Klein K., Chen K.E., Williamson M.K., Antón-Plágaro C., Shoemark D.K., Simón-Gracia L., Bauer M., Hollandi R., et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J., Pearson C.A.B., Russell T.W., Tully D.C., Abbott S., Gimma A., et al. Estimated transmissibility and severity of novel SARS-CoV-2 Variant of Concern 202012/01 in England. medRxiv. 2020 doi: 10.1101/2020.12.24.20248822. [DOI] [Google Scholar]
- DiMaio F., Song Y., Li X., Brunner M.J., Xu C., Conticello V., Egelman E., Marlovits T., Cheng Y., Baker D. Atomic-accuracy models from 4.5-Å cryo-electron microscopy data with density-guided iterative local refinement. Nat. Methods. 2015;12:361–365. doi: 10.1038/nmeth.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erasmus J.H., Khandhar A.P., O’Connor M.A., Walls A.C., Hemann E.A., Murapa P., Archer J., Leventhal S., Fuller J.T., Lewis T.B., et al. An Alphavirus-derived replicon RNA vaccine induces SARS-CoV-2 neutralizing antibody and T cell responses in mice and nonhuman primates. Sci. Transl. Med. 2020;12:eabc9396. doi: 10.1126/scitranslmed.abc9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenz B., Rämisch S., Borst A.J., Walls A.C., Adolf-Bryfogle J., Schief W.R., Veesler D., DiMaio F. Automatically Fixing Errors in Glycoprotein Structures with Rosetta. Structure. 2019;27:134–139.e3. doi: 10.1016/j.str.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C., Zeng J., Jia N., Stavenhagen K., Matsumoto Y., Zhang H., Li J., Hume A.J., Mühlberger E., van Die I., et al. SARS-CoV-2 Spike Protein Interacts with Multiple Innate Immune Receptors. bioRxiv. 2020 doi: 10.1101/2020.07.29.227462. [DOI] [Google Scholar]
- Goddard T.D., Huang C.C., Ferrin T.E. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., et al. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition. Cell Host Microbe. 2020;29:44–57.e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations to the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human serum antibodies. Cell Host Microbe. 2021;29:463–476.e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.O., Kafai N.M., Dmitriev I.P., Fox J.M., Smith B.K., Harvey I.B., Chen R.E., Winkler E.S., Wessel A.W., Case J.B., et al. A Single-Dose Intranasal ChAd Vaccine Protects Upper and Lower Respiratory Tracts against SARS-CoV-2. Cell. 2020;183:169–184.e13. doi: 10.1016/j.cell.2020.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodcroft E.B., Zuber M., Nadeau S., Crawford K.H.D., Bloom J.D., Veesler D., Vaughan T.G., Comas I., Candelas F.G., Stadler T., et al. Emergence and spread of a SARS-CoV-2 variant through Europe in the summer of 2020. medRxiv. 2020 doi: 10.1101/2020.10.25.20219063. [DOI] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Pöhlmann S. A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell. 2020;78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Mösbauer K., Hofmann-Winkler H., Kaul A., Kleine-Weber H., Krüger N., Gassen N.C., Müller M.A., Drosten C., Pöhlmann S. Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature. 2020;585:588–590. doi: 10.1038/s41586-020-2575-3. [DOI] [PubMed] [Google Scholar]
- Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., 3rd, Leist S.R., Schäfer A., Nakajima N., Takahashi K., et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370:1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.C., Javanmardi K., Le K.C., Wrapp D., Lee A.G., Liu Y., et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo J., Zhao Y., Ren J., Zhou D., Duyvesteyn H.M.E., Ginn H.M., Carrique L., Malinauskas T., Ruza R.R., Shah P.N.M., et al. Neutralization of SARS-CoV-2 by destruction of the prefusion Spike. Cell Host Microbe. 2020;28:445–454.e6. doi: 10.1016/j.chom.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiaming L., Yanfeng Y., Yao D., Yawei H., Linlin B., Baoying H., Jinghua Y., Gao G.F., Chuan Q., Wenjie T. The recombinant N-terminal domain of spike proteins is a potential vaccine against Middle East respiratory syndrome coronavirus (MERS-CoV) infection. Vaccine. 2017;35:10–18. doi: 10.1016/j.vaccine.2016.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B.E., Brown-Augsburger P.L., Corbett K.S., Westendorf K., Davies J., Cujec T.P., Wiethoff C.M., Blackbourne J.L., Heinz B.A., Foster D., et al. LY-CoV555, a rapidly isolated potent neutralizing antibody, provides protection in a non-human primate model of SARS-CoV-2 infection. bioRxiv. 2020 doi: 10.1101/2020.09.30.318972. [DOI] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Yu J., Shan S., Zhou B., Song S., Tang X., Yu J., et al. uman neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech C., Albert G., Cho I., Robertson A., Reed P., Neal S., Plested J.S., Zhu M., Cloney-Clark S., Zhou H., et al. Phase 1-2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. N. Engl. J. Med. 2020;383:2320–2332. doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Sheffield COVID-19 Genomics Group Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell. 2020;182:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D., Afonine P.V., Baker M.L., Bunkóczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J., et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Liu Z., VanBlargan L.A., Rothlauf P.W., Bloyet L.-M., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Ellebedy A.H., et al. Landscape analysis of escape variants identifies SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. bioRxiv. 2020 doi: 10.1101/2020.11.06.372037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., Walls A.C., Bowen J.E., Corti D., Veesler D. Structure-guided covalent stabilization of coronavirus spike glycoprotein trimers in the closed conformation. Nat. Struct. Mol. Biol. 2020;27:942–949. doi: 10.1038/s41594-020-0483-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy K.R., Rennick L.J., Nambulli S., Robinson-McCarthy L.R., Bain W.G., Haidar G., Duprex W.P. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science. 2021;371:1139–1142. doi: 10.1126/science.abf6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado N.B., Zahn R., Wegmann F., Loos C., Chandrashekar A., Yu J., Liu J., Peter L., McMahan K., Tostanoski L.H., et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586:583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Murine Leukemia Virus (MLV)-based Coronavirus Spike-pseudotyped Particle Production and Infection. Biol. Protoc. 2016;6:e2035. doi: 10.21769/BioProtoc.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2020 doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Punjani A., Zhang H., Fleet D.J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods. 2020;17:1214–1221. doi: 10.1038/s41592-020-00990-8. [DOI] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A SIMPLE METHOD OF ESTIMATING FIFTY PER CENT ENDPOINTS12. Am. J. Epidemiol. 1938;27:493–497. [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer A., Muecksch F., Lorenzi J.C.C., Leist S.R., Cipolla M., Bournazos S., Schmidt F., Maison R.M., Gazumyan A., Martinez D.R., et al. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J. Exp. Med. 2021;218:e20201993. doi: 10.1084/jem.20201993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H., Chen S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux E., Homad L.J., MacCamy A.J., Parks K.R., Hurlburt N.K., Jennewein M.F., Akins N.R., Stuart A.B., Wan Y.-H., Feng J., et al. Characterization of neutralizing antibodies from a SARS-CoV-2 infected individual. bioRxiv. 2020 doi: 10.1101/2020.05.12.091298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh W.T., Liu Y., Nakayama E.E., Ono C., Torii S., Nakagami H., Matsuura Y., Shioda T., Arase H. The N-terminal domain of spike glycoprotein mediates SARS-CoV-2 infection by associating with L-SIGN and DC-SIGN. bioRxiv. 2020 doi: 10.1101/2020.11.05.369264. [DOI] [Google Scholar]
- Starr T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., Bloom J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2020;371:850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C., Pulokas J., Fellmann D., Cheng A., Guerra F., Quispe J., Stagg S., Potter C.S., Carragher B. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020 doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- Tegunov D., Cramer P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods. 2019;16:1146–1152. doi: 10.1038/s41592-019-0580-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Meulen J., van den Brink E.N., Poon L.L., Marissen W.E., Leung C.S., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E., et al. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thépaut M., Luczkowiak J., Vivès C., Labiod N., Bally I., Lasala F., Grimoire Y., Fenel D., Sattin S., Thielens N., et al. DC/L-SIGN recognition of spike glycoprotein promotes SARS-CoV-2 trans-infection and can be inhibited by a glycomimetic antagonist. bioRxiv. 2020 doi: 10.1101/2020.08.09.242917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Veesler D. Structural insights into coronavirus entry. Adv. Virus Res. 2019;105:93–116. doi: 10.1016/bs.aivir.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Beltramello M., Lempp F.A., Pinto D., Dang H.V., Rosen L.E., McCallum M., Bowen J., Minola A., Jaconi S., et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss W.N., Hou Y.J., Johnson N.V., Kim J.E., Delidakis G., Horton A.P., Bartzoka F., Paresi C.J., Tanno Y., Abbasi S.A., et al. Prevalent, protective, and convergent IgG recognition of SARS-CoV-2 non-RBD spike epitopes in COVID-19 convalescent plasma. bioRxiv. 2020 doi: 10.1101/2020.12.20.423708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Xiong X., Park Y.J., Tortorici M.A., Snijder J., Quispe J., Cameroni E., Gopal R., Dai M., Lanzavecchia A., et al. Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion. Cell. 2019;176:1026–1039.e15. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Fiala B., Schäfer A., Wrenn S., Pham M.N., Murphy M., Tse L.V., Shehata L., O’Connor M.A., Chen C., et al. Elicitation of Potent Neutralizing Antibody Responses by Designed Protein Nanoparticle Vaccines for SARS-CoV-2. Cell. 2020;183:1367–1382.e17. doi: 10.1016/j.cell.2020.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R.Y., Song Y., Barad B.A., Cheng Y., Fraser J.S., DiMaio F. Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. eLife. 2016;5:e17219. doi: 10.7554/eLife.17219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Shi W., Chappell J.D., Joyce M.G., Zhang Y., Kanekiyo M., Becker M.M., van Doremalen N., Fischer R., Wang N., et al. Importance of Neutralizing Monoclonal Antibodies Targeting Multiple Antigenic Sites on the Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein To Avoid Neutralization Escape. J. Virol. 2018;92:e02002-17. doi: 10.1128/JVI.02002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Rosen O., Wang L., Turner H.L., Stevens L.J., Corbett K.S., Bowman C.A., Pallesen J., Shi W., Zhang Y., et al. Structural Definition of a Neutralization-Sensitive Epitope on the MERS-CoV S1-NTD. Cell Rep. 2019;28:3395–3405.e6. doi: 10.1016/j.celrep.2019.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D., et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C.C., Muecksch F., Rutkowska M., Hoffmann H.-H., Michailidis E., et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife. 2020;9:e61312. doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Lambson B.E., Vermeulen M., van den Berg K., Rossouw T., Boswell M., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. bioRxiv. 2021 doi: 10.1101/2021.01.18.427166. [DOI] [PubMed] [Google Scholar]
- Winkler E.S., Gilchuk P., Yu J., Bailey A.L., Chen R.E., Zost S.J., Jang H., Huang Y., Allen J.D., Case J.B., et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions and monocytes for optimal therapeutic protection. Cell. 2020 doi: 10.1016/j.cell.2021.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel A.G., Benton D.J., Hussain S., Harvey R., Martin S.R., Roustan C., Rosenthal P.B., Skehel J.J., Gamblin S.J. Antibody-mediated disruption of the SARS-CoV-2 spike glycoprotein. Nat. Commun. 2020;11:5337. doi: 10.1038/s41467-020-19146-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel A.G., Benton D.J., Xu P., Roustan C., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat. Struct. Mol. Biol. 2020;27:763–767. doi: 10.1038/s41594-020-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Muruato A.E., Zhang X., Lokugamage K.G., Fontes-Garfias C.R., Zou J., Liu J., Ren P., Balakrishnan M., Cihlar T., et al. A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19. Nat. Commun. 2020;11:5214. doi: 10.1038/s41467-020-19055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X., Corti D., Liu J., Pinna D., Foglierini M., Calder L.J., Martin S.R., Lin Y.P., Walker P.A., Collins P.J., et al. Structures of complexes formed by H5 influenza hemagglutinin with a potent broadly neutralizing human monoclonal antibody. Proc. Natl. Acad. Sci. USA. 2015;112:9430–9435. doi: 10.1073/pnas.1510816112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell. 2020;183:739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Cao L., Gao X.-S., Zheng B.-Y., Deng Y.-Q., Li J.-X., Feng R., Bian Q., Guo X.-L., Wang N., et al. A proof of concept for neutralizing antibody-guided vaccine design against SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.09.23.309294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Chen Y., Zhang S., Niu P., Qin K., Jia W., Huang B., Zhang S., Lan J., Zhang L., et al. Structural definition of a neutralization epitope on the N-terminal domain of MERS-CoV spike glycoprotein. Nat. Commun. 2019;10:3068. doi: 10.1038/s41467-019-10897-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J., Nakane T., Scheres S.H.W. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ. 2019;6:5–17. doi: 10.1107/S205225251801463X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM maps, X-ray diffraction data and atomic models have been deposited at the Electron Microscopy Data Bank and the PDB with accession codes listed in the Key Resources Tables.