Abstract
Liver cancer is a serious disease. It is ranked as the cancer with the second highest number of cancer-related deaths worldwide. Hepatocellular carcinoma (HCC), which arises from transformed hepatocytes, is the major subtype of liver cancer. It accounts for 85% of total liver-cancer cases. An important aspect of HCC that has been actively studied is its metabolism. With the liver as the primary site of numerous metabolic processes in the body, it has been shown that the metabolism of HCC cells is highly dysregulated compared to that of normal hepatocytes. It is therefore crucial to understand the metabolic alterations caused by HCC and the underlying mechanisms for these alterations. This deeper understanding will allow diagnostic and therapeutic advancements in the treatment of HCC. In this review, we will summarize the current literature in HCC metabolic alterations, induced vulnerabilities, and potential therapeutic interventions.
Keywords: hepatocellular carcinoma (HCC), metabolism, metabolic vulnerability, targeted therapy
Introduction
The liver is the site of major metabolic processes in the body, including detoxification of blood, production of bile for the breakdown of fats in the digestive track, the storage of glucose in the form of glycogen, and the synthesis of amino-acid precursors that make up proteins. Hepatocytes, which make up ∼85% of the total mass of the liver [1], are responsible for the majority of these metabolic processes. It is perhaps unsurprising that hepatocellular carcinoma (HCC)—the liver-cancer subtype that originates from the hepatocytes—results in the dysregulation of a large number of metabolic processes to fuel tumorigenesis [2–4]. Liver cancer is the second leading cause of cancer deaths worldwide [5]. HCC accounts for 85% of total liver-cancer cases and risk factors include chronic hepatitis B virus (HBV) and hepatitis C virus (HCV) infection [6], alcohol intake, diabetes, obesity [7, 8], and non-alcoholic fatty liver disease (NAFLD) [9]. While metabolic dysregulation has also been characterized in other primary liver-cancer subtypes such as cholangiocarcinoma and angiosarcoma [10], this review will primarily focus on metabolic alterations and induced vulnerabilities caused by these alterations in HCC.
In HCC, metabolic processes ranging from glucose metabolism and energy generation in the form of adenosine triphosphate (ATP) to amino-acid and fatty-acid metabolism are significantly altered in the disease. These alterations serve to augment the ability of the tumor to thrive, proliferate, and metastasis. However, the increased dependency of the tumor on some of these pathways can lead to metabolic vulnerabilities, which can be exploited by inhibitors of these pathways to therapeutically target HCC. Thus far, a significant number of metabolic pathways have been examined in detail in HCC to better understand their roles in tumorigenesis. However, there remain some processes, such as nucleotide metabolism, for which some preliminary evidence suggests a role in HCC, but detailed tumorigenic mechanisms have yet to be elucidated.
Glycolysis and glycogen metabolism
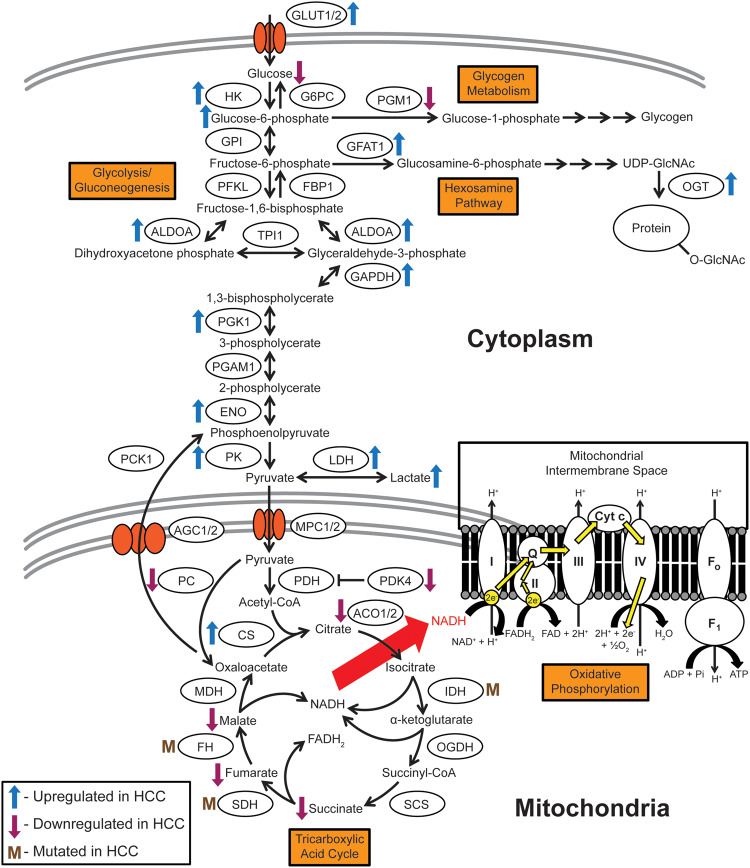
The liver plays a major role in glucose metabolism. It metabolizes glucose through glycolysis for energy but can also store excess glucose in the form of glycogen. The liver releases glucose to the rest of the body through the breakdown of glycogen stores (glycogenolysis) and generation of glucose from other proteins and lipids (gluconeogenesis) [11]. The dysregulation of and dependency on glucose metabolism is one of the earliest metabolic alterations described in cancer. Otto Warburg initially proposed that cancer cells favor glycolysis as a quick source of energy, in the form of ATP, for rapid growth and proliferation over the more energy-efficient oxidative phosphorylation pathway [12]. Glycolysis enzymes are primarily upregulated in HCC, while the glycogen metabolism enzyme phosphoglucomutase 1 (PGM1) is downregulated, suggesting a preference for glucose to undergo glycolysis instead of being stored as glycogen [2–4] (Figure 1).
Figure 1.
Schematic of glycolysis, gluconeogenesis, glycogen metabolism, the hexosamine pathway, oxidative phosphorylation, and the TCA cycle. Glycolysis and its opposing pathway, gluconeogenesis, serve as a central hub for the utilization of glucose in multiple pathways such as glycogen metabolism, the hexosamine pathway, the TCA cycle, and downstream oxidative phosphorylation. Tumors require energy to grow and proliferate, and the glycolysis–TCA cycle–oxidative phosphorylation axis provides significant amounts of cellular energy in the form of ATP. These pathways feature significantly in HCC and their dysregulation provides insight into tumorigenic mechanisms that can be potentially targeted with precision therapeutics. Metabolite nomenclature: ADP, adenosine diphosphate; ATP, adenosine triphosphate; CoA, coenzyme A; FAD, flavin adenine dinucleotide; FADH2, reduced flavin adenine dinucleotide; NAD, nicotinamide adenine dinucleotide. Enzyme nomenclature: I-IV, mitochondrial complexes I-IV; ACO1/2, aconitase 1/2; AGC1/2, aspartate/glutamate carrier 1/2; ALDOA: aldolase A, fructose bisphosphate; CS, citrate synthase; Cyt c, cytochrome c; ENO, enolase; F0, F0 subunit of ATP synthase (mitochondrial complex V); F1, F1 subunit of ATP synthase (mitochondrial complex V); FBP1, fructose bisphosphatase 1; FH, fumarate hydratase; G6PC, glucose-6-phosphatase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GFAT1, glutamine: fructose-6-phosphate transaminase 1; GLUT1/2, glucose transporter type 1/2; GPI, glucose-6-phosphate isomerase; HK, hexokinase; IDH, isocitrate dehydrogenase; LDH, lactate dehydrogenase; MDH, malate dehydrogenase; MPC1/2, mitochondrial pyruvate carrier 1/2; OGDH, oxoglutarate dehydrogenase; OGT, O-linked N-acetylglucosamine (O-GlcNAc) transferase; PC, pyruvate carboxylase; PCK1, phosphoenolpyruvate carboxykinase 1; PDH, pyruvate dehydrogenase; PDK4, pyruvate dehydrogenase kinase 4; PFKL, phosphofructokinase liver type; PGAM1, phosphoglycerate mutase 1; PGM1, phosphoglucomutase 1; PGK1, phosphoglycerate kinase 1; Pi, inorganic phosphate; PK, pyruvate kinase; Q, coenzyme Q; SCS, succinyl coenzyme A synthetase; SDH, succinate dehydrogenase; TPI1, triosephosphate isomerase.
Before glucose can be metabolized, it has to enter cells. Glucose transporters GLUT1 [13, 14] and GLUT2 [15, 16], which are critical for glucose uptake into cells, show increased levels in HCC (Figure 1). This upregulation might be related to oncogene MYC expression, since GLUT1 is transcriptionally upregulated in mouse models where Myc is overexpressed in the liver [17].
Once glucose is transported into the cytosol, hexokinase (HK) enzymes convert it into glucose-6-phosphate (G6P) as the first step in glycolysis (Figure 1). G6P levels are elevated in HCC compared to those in normal liver [18]. This might be due to hexokinase HK2 isoform upregulation in HCC, which contributes to an increased rate of G6P conversion from glucose [19–21]. Silencing HK2 in HCC cells suppresses their cell growth and proliferation in culture and in vivo [21]. The lack of HK2 activity upregulates oxidative phosphorylation, sensitizing HCC cells to the oxidative phosphorylation inhibitor metformin [21]. The synergistic effects of HK2 ablation and metformin in HCC cells suggest that the development of clinical hexokinase inhibitors in combination with oxidative phosphorylation inhibitors could potentially target these metabolic vulnerabilities successfully.
The next significantly altered glycolytic step in HCC is the conversion of phosphoenolpyruvate to pyruvate by the pyruvate kinase (PK) enzyme (Figure 1). The PKLR and PKM genes code for four PK splice isoforms: PKL, PKR, PKM1, and PKM2 [22–24]. PKL is expressed in normal liver [23]. PKM2, however, is upregulated in HCC, while PKM1 and PKL levels remain unchanged, and PKR is undetectable [25]. In mouse models, Myc induction lowers PKL levels [26]. High PKM2 expression correlates with poor prognosis in HCC patients [27, 28]. PKM2 also shows higher enzymatic activity in HCC cells compared to that in adjacent normal tissue [28]. On the contrary, murine PKM2 knockouts promote HCC [29], suggesting a more complicated mechanism for how PKM2 influences HCC tumorigenesis. Myc mouse tumors reflect an increase in PKM1/2 levels [26]. The interplay among PK isoforms in HCC remains unclear and should be further investigated.
In anaerobic respiration, pyruvate is converted into lactate instead of acetyl-coenzyme A (acetyl-CoA) that enters the tricarboxylic acid (TCA) cycle (Figure 1). This conversion is catalysed by lactate dehydrogenase (LDH). High levels of LDH observed in HCC patients simultaneously raises lactate levels [30] and is a risk factor for HCC recurrence [31]. Sorafenib-treated patients with high serum levels of LDH showed decreased progression-free survival [32]. Since the LDH A subunit (LDHA) is upregulated in a range of different cancers and LDHA-targeting therapeutics are available [33], it is important to study this gene’s impact on HCC in greater detail.
A number of factors have been shown to influence glycolysis and gluconeogenesis through the upstream gene regulation of metabolic enzymes. Transmembrane glycoprotein CD147 has been shown to upregulate glycolysis through p53-dependent upregulation of GLUT1 and PFKL, the liver-specific isoform of phosphofructokinase [34]. CD147 also downregulates mitochondrial biogenesis genes such as peroxisome proliferator-activated receptor gamma co-activator 1-alpha (PGC1α) and transcription factor A, mitochondrial, suggesting a reverse effect on mitochondrial energetic processes such as the TCA cycle and oxidative phosphorylation [34]. HCV infection in primary human hepatocytes upregulates glycolysis through the activation of transcription factor hepatocyte nuclear factor 4-alpha (HNF4α), which in turn transcriptionally upregulates glycolytic genes such as PKLR [35]. Interestingly, HCV infection in a HCC cell line has been shown to upregulate gluconeogenesis through the regulation of gluconeogenic transcription factors such as FoxO1 by histone deacetylase 9 (HDAC9) [36]. The upstream regulatory mechanisms of glucose metabolism gene regulation in HCC are not as well characterized and require greater understanding.
In terms of studies on drugging glucose metabolism to treat HCC, there have been some encouraging results. The administration of the diabetic drug metformin, which lowers the amount of sugar produced in the liver and sensitizes muscle cells to insulin, has been shown to decrease HCC risk [37] and is associated with reduced recurrence in increased overall HCC patient survival post hepatic resection [38]. In addition, a novel compound combining metformin and rosiglitazone, the latter a compound that blocks peroxisome proliferator-activated receptors in fat cells to make them more responsive to insulin, has been shown to suppress HCC [39]. With further research efforts, there is potential for the development of drugs targeting glucose metabolism in HCC.
TCA cycle
The TCA cycle utilizes pyruvate from glycolysis to generate reduced nicotinamide adenine dinucleotide (NADH) and reduced flavin adenine dinucleotide (FADH2)—cofactors that channel electrons to oxidative phosphorylation for downstream energy generation (Figure 1). TCA metabolic intermediates such as succinate, fumarate, and malate are reduced in HCC [18].
In addition, TCA enzyme expression levels are also altered in HCC [2–4]. Pyruvate dehydrogenase (PDH) converts pyruvate from glycolysis into acetyl-CoA, which enters the TCA cycle (Figure 1). Downregulation of pyruvate dehydrogenase kinase 4 (PDK4), which inhibits PDH by phosphorylation, is associated with poor prognosis in HCC [40]. Interestingly, the knockout of PDK4 did not affect oxidative phosphorylation and glycolysis, but instead upregulated lipogenesis [40]. Succinate dehydrogenase (SDH), which converts succinate into fumarate, and fumarate hydratase (FH), which converts fumarate into malate (Figure 1), potentially function as tumor suppressors, since they tend to gain loss-of-function mutations [41, 42]. As a result, the build-up of succinate and fumarate stabilizes transcription factor hypoxia-inducible factor 1-alpha (HIF-1α), transcriptionally activating glycolysis and angiogenesis [42]. The SDH B isoform is decreased in HCC and this change is associated with tumorigenic phenotypes [43]. Mutations in isocitrate dehydrogenase (IDH), the enzyme that converts isocitrate into α-ketoglutarate (Figure 1), are rampant in multiple cancer types but are rarely observed (∼2%) in HCC [10]. The upstream regulatory mechanisms that alter TCA cycle enzyme expression and activity in HCC have yet to be thoroughly explored.
Oxidative phosphorylation
Oxidative phosphorylation is the major energy-producing metabolic process in the cell, generating large amounts of ATP from NADH and FADH2 produced in the TCA cycle (Figure 1). Electrons from the oxidation of NADH and FADH2 are transferred through membrane-bound complexes in the mitochondrial inner membrane, with the energy derived utilized to activate proton pumps. These pumps create a potential gradient across the mitochondrial inner membrane by transporting positively charged hydrogen ions from the mitochondrial matrix into the intermembrane space. Oxidative phosphorylation is therefore also known as the electron-transport chain.
Impaired oxidative phosphorylation is associated with increased tumorigenicity in HCC [44, 45]. HCV infection downregulates oxidative phosphorylation-associated protein subunit expression, reminiscent of the Warburg effect [46]. Mitochondrial microRNAs have also been shown to downregulate oxidative phosphorylation gene expression in HCC, thereby promoting glucose metabolism [47]. Rab3A, a Ras-like GTPase important in membrane trafficking, can also dysregulate oxidative phosphorylation in HCC [48]. Rab3A is upregulated at both the transcript and protein levels in HCC tumor tissue [48]. Unlike in other cancers, where it acts as an oncogene, Rab3A inhibits HCC metastasis by enhancing oxidative phosphorylation, resulting in migration and invasion [48]. However, modification of Rab3a with N-acetylglucosamine (O-GlcNAc) attenuates these effects [48]. Since HCC tumors are commonly hyper O-GlcNAcylated, the tumor-suppressive function of Rab3A is likely diminished in HCC [48].
Upstream signaling and transcription factors have also been linked to modulating oxidative phosphorylation in HCC. Cytokine transforming growth factor beta (TGFβ), which induces migration and invasion in HCC, reduces oxidative phosphorylation with no alteration in glycolysis [45]. The stem-cell homeobox transcription factor NANOG is upregulated in alcohol and obesity-HCV-induced mouse models of HCC and in human tumor-initiating cells [49, 50]. NANOG in HCC tumor-initiating cells suppresses oxidative phosphorylation to support self-renewal and drug resistance [50]. Interestingly, as opposed to NANOG, stem-cell transcription factor spalt like transcription factor 4 (SALL4), which is upregulated in a subset of HCC, promotes oxidative phosphorylation through an increase in mitochondrial gene expression [51]. Mitochondrial oxidative phosphorylation inhibitors were shown to be particularly effective in suppressing SALL4-expressing HCC tumorigenesis in culture and in vivo [51]. The effectiveness of these mitochondrial inhibitors and aforementioned metformin, which both modulates glucose metabolism and inhibits oxidative phosphorylation, suggests the therapeutic potential of targeting oxidative phosphorylation in HCC patients.
Hexosamine pathway
The hexosamine biosynthetic pathway (HBP) is required for the synthesis of uridine diphosphate N-acetylglucosamine (UDP-GlcNAc) for post-translational modification of proteins on their serine or threonine residues [52, 53] (Figure 1). This O-GlcNAc modification of proteins, catalysed by the O-GlcNAc transferase (OGT) enzyme, is a response to nutrient sensing and aberrant O-GlcNAcylation has been implicated in a number of diseases such as diabetes and cancer [54]. A high expression of glutamine: fructose-6-phosphate amidotransferase (GFAT1), the rate-limiting enzyme that converts fructose-6-phosphate into glucosamine-6-phosphate (Figure 1), correlates with poor HCC patient prognosis [55]. GFAT1 overexpression in HCC cell lines increases the tumorigenic phenotypes of proliferation and migration in culture [55]. Expression levels of the OGT enzyme are also increased in tumor tissue compared to those in non-cancerous controls [56]. Global O-GlcNAcylation levels are significantly increased in HCC tumors compared to those in healthy liver tissues, as well as in tumor tissues of HCC patients with recurring disease post liver transplantation [57].
In terms of mechanism, both OGT and the O-GlcNAc modification have been shown to influence the functionalities of various oncogenic proteins and processes. OGT upregulation has also been shown to regulate lipid metabolism and activate oncogenic pathways through increased oncogenic protein levels in HCC [58]. Increased O-GlcNAcylation leads to HCC tumorigenesis and metastasis through modification of oncogenic transcription factors such as c-Jun [59, 60]. O-GlcNAcylation of the receptor for activated C kinase 1 (RACK1) stabilizes it so that it can interact with the protein kinase C βII isoform, activating the kinase to phosphorylate eukaryotic translation initiation factor 4E (eIF4E) to translate oncogenes [61]. As mentioned in the oxidative phosphorylation section, tumor suppressor Rab3A is inactivated when modified with O-GlcNAc in HCC [48]. Both RACK1 and Rab3A O-GlcNAcylation promote HCC progression and metastasis [48, 61]. Interestingly, eIF4E itself can be O-GlcNAcylated, protecting it from proteasomal degradation, and subsequently promoting HCC cell-line proliferation and tumor-sphere formation [56]. More detailed studies of the effect of the HBP on the proteome in HCC could yield greater mechanistic and therapeutic insight into the disease.
Pentose-phosphate pathway
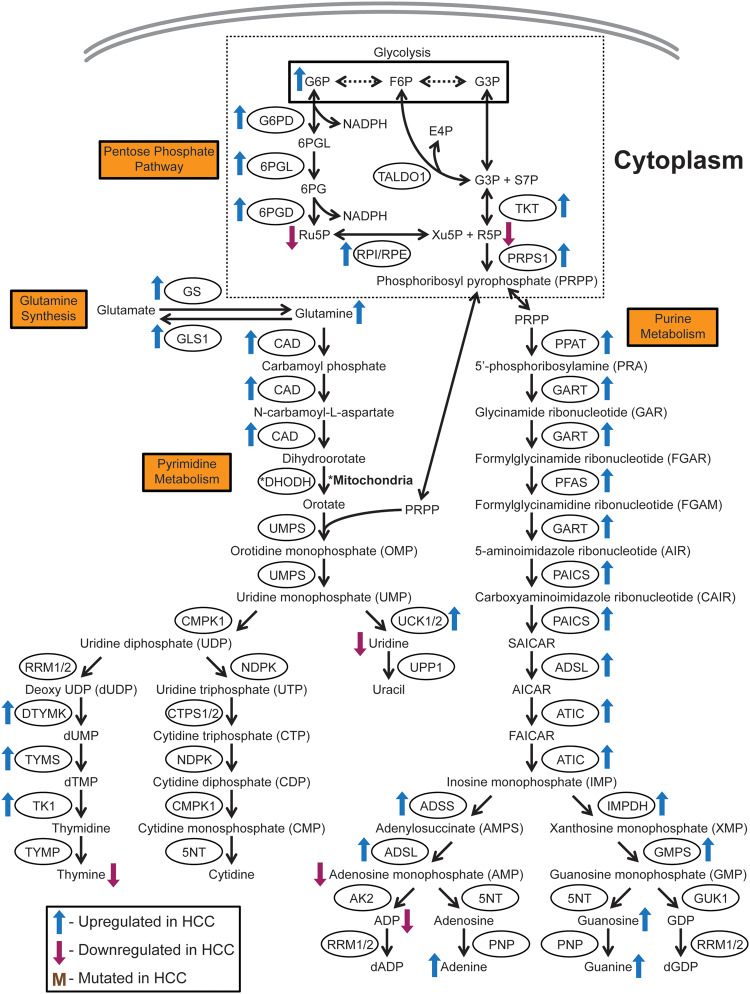
The pentose-phosphate pathway (PPP) generates the ribose-5-phosphate backbone required for the synthesis of nucleotides, the precursors of DNA and RNA (Figure 2). The PPP also generates the cofactor-reduced nicotinamide adenine dinucleotide phosphate (NADPH) for lipid biosynthesis, maintaining glutathione and thioredoxin in reduced states, and for its antioxidant properties [62] (Figure 2). Nearly all PPP enzymes are transcriptionally upregulated [2–4], while PPP metabolites ribulose-5-phosphate (Ru5P) and ribose-5-phosphate (R5P) are downregulated [10, 18] in HCC. This suggests that the PPP is highly activated in HCC as metabolites are rapidly used up by the larger number of enzyme active sites available, possibly to feed precursor molecule phosphoribosyl pyrophosphate (PRPP) into nucleotide biosynthesis (Figure 2).
Figure 2.
Schematic of glutamine synthesis, nucleotide biosynthesis, and the pentose-phosphate pathway. Nucleotide metabolism and its supporting processes such as the pentose-phosphate pathways and glutamine synthesis are significantly altered in HCC. Nucleotides are important precursors for DNA and RNA, as well as the energy currency ATP, which are needed for cell growth and proliferation. The exact mechanisms by which these pathways promote HCC tumorigenesis remain unclear however. Metabolite nomenclature: 6PG, 6-phosphogluconate; 6PGL, 6-phosphogluconolactone; ADP, adenosine diphosphate; AICAR, 5-aminoimidazole-4-carboxamide ribonucleotide; dADP, deoxyadenosine diphosphate; dGDP, deoxyguanosine diphosphate; dTMP, deoxythymidine monophosphate; dUMP, deoxyuridine monophosphate; E4P, erythrose-4-phosphate; F6P, fructose-6-phosphate; FAICAR, 5-formamidoimidazole-4-carboxamide ribonucleotide; G3P, glyceraldehyde-3-phosphate; G6P, glucose-6-phosphate; GDP, guanosine diphosphate; NADPH, reduced nicotinamide adenine dinucleotide phosphate; R5P, ribose-5-phosphate; Ru5P, ribulose-5-phospahte; S7P, sedoheptulose-7-phosphate; SAICAR, succinyl 5-aminoimidazole-4-carboxamide ribonucleotide; Xu5P, xylulose-5-phospahte. Enzyme nomenclature: 5NT, 5’-nucleotidase; 6PGD, 6-phosphogluconate dehydrogenase; 6PGL, 6-phosphogluconolactonase; ADSL, adenylosuccinate lyase; ADSS, adenylosuccinate synthetase; AK2, adenylate kinase 2; ATIC, 5-aminoimidazole-4-carboxamiade ribonucleotide formyltransferase/IMP cyclohydrolase; CAD, carbamoyl phosphate synthetase 2 aspartate transcarbamylase, and dihydroorotase; CMPK1, cytidine/uridine monophosphate kinase 1; CTPS1/2, cytidine triphosphate synthase 1/2; DHODH, dihydroorotate dehydrogenase; DTYML, deoxythymidylate kinase; G6PD, glucose-6-phosphate dehydrogenase; GART, glycinamide ribonucleotide synthetase, aminoimidazole ribonucleotide synthetase, and glycinamide ribonucleotide transformylase; GLS1, glutaminase 1; GMPS, guanine monophosphate synthase; GS, glutamine synthetase; GUK1, guanylate kinase 1; IMPDH, inosine monophosphate dehydrogenase; NDPK, nucleoside diphosphate kinase; PAICS, phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazole succinocarboxamide synthase; PFAS, phosphoribosylformylglycinamide synthase; PNP, purine nucleoside phosphorylase; PPAT, phosphoribosyl pyrophosphate amidotransferase; PRPS1, phosphoribosyl pyrophosphate synthetase 1; RPE, ribulose-5-phosphate-3-epimerase; RPI, ribose-5-phosphate isomerase; RRM1/2, ribonucleoside diphosphate reductase 1/2; TALDO1, transaldolase 1; TK1, thymidine kinase 1; TKT, transketolase; TYMP, thymidine phosphorylase; TYMS, thymidylate synthase; UCK1/2, uridine-cytidine kinase 1/2; UMPS, uridine monophosphate synthetase; UPP1, uridine phosphorylase 1.
The conversion of G6P to 6-phosphogluconolactone (6PGL), catalysed by glucose-6-phosphate dehydrogenase (G6PD), is the first rate-limiting step of the PPP (Figure 2). Increased G6PD expression is associated with migration and invasion [63, 64], chemoresistance [65], metastasis, higher tumor grade, and decreased survival [66] in HCC. O-GlcNAcylation of G6PD induces tumorigenic activity, suggesting a synergistic interaction between the HBP and PPP in HCC [67]. HBV protein X (HBx) stimulates G6PD expression through oxidative stress transcription factor nuclear factor erythroid 2-related factor 2 (Nrf2) activation, implying a role for the PPP in HBV-induced HCC [66, 68].
Interestingly, Nrf2 overexpression itself has been shown to drive HCC [69]. Nrf2 also activates PPP enzyme transketolase (TKT), which converts glyceraldehyde-3-phosphate (G3P) and sedoheptulose-7-phosphate (S7P) into xylulose-5-phosphate (Xu5P) and R5P, in non-oxidative PPP to promote HCC by increasing antioxidants and purine biosynthesis, protecting cancer cells from reactive oxygen species toxicity [70]. In HCV-positive HCC, cell death and survival protein p62 phosphorylation prevents adaptor protein Kelch-like ECH-associated protein 1 (KEAP1) from recruiting the E3 ubiquitin ligase complex to ubiquitinate Nrf2, allowing active Nrf2 to activate the PPP [71]. A chemical inhibitor of KEAP1 that blocks its interaction with phosphorylated p62 facilitates Nrf2 degradation to suppress tumor-cell proliferation and increase sorafenib sensitivity in HCC cell lines [71]. The mechanisms by which the PPP promotes HCC have not been fully characterized and the fact that the PPP is crucial for NADPH generation and nucleotide biosynthesis suggests critical effects on downstream processes in tumorigenesis that should be examined in more detail.
Nucleotide metabolism
Nucleotides form the building blocks of DNA and RNA molecules, as well as being used as the major energy currency in the cell in terms of ATP. Nucleotide biosynthesis requires R5P generated by the PPP and amino acids (Figure 2). Nucleotide metabolism genes involved in both purine and pyrimidine metabolism are transcriptionally upregulated in HCC [2–4, 72]. The levels of a number of nucleotides and nitrogenous bases are also altered [10, 18]. As mentioned previously, the PPP is also upregulated to supply metabolite PRPP that makes up the backbone of nucleotides. This suggests a greater demand for nucleotides by HCC, although the reasons for this are not well understood.
The carbamoyl phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase (CAD) gene, which codes for a multi-domain trifunctional enzyme that catalyses the first three steps of pyrimidine biosynthesis, is upregulated in HCC [72, 73]. The upregulation of a number of rate-limiting enzymes in pyrimidine biosynthesis such as deoxythymidylate kinase (DTYMK), thymidylate synthase (TYMS), and thymidine kinase 1 (TK1) in HCC is associated with cancer stemness and poor prognosis [3, 4, 74]. These three enzymes form a continuous cascade of reactions, in which DTYMK converts deoxyuridine diphosphate (dUDP) into deoxyuridine monophosphate (dUMP), TYMS converts dUMP into deoxythymidine monophosphate (dTMP), and TK1 converts dTMP into thymidine (Figure 2). Nearly all purine biosynthesis enzymes are upregulated in HCC [2–4]. Limited mechanistic insight is currently available on how nucleotide biosynthesis contributes to HCC. However, inhibiting nucleotide biosynthesis has shown therapeutic potential in acute myeloid leukemia and melanoma, suggesting that it is prudent to study this process in greater detail in HCC [75–77].
Amino-acid metabolism
Amino acids are the building blocks of proteins and other important molecules such as heme, nucleotides, and neurotransmitters. A number of genes involved in amino-acid metabolism are significantly altered in HCC [2–4]. The amino acids glutamine and aspartate are upregulated while glycine is downregulated in HCC [10, 18].
Glutamine is a precursor for nucleotide biosynthesis. It is also used to replenish α-ketoglutarate, a key source of glutamate and glutamine for protein synthesis [78]. Lower plasma levels of glutamine have been observed in HCC patients due to increased uptake by tumors [18, 79]. The increased uptake can be explained by the upregulation of glutamine transporter ASCT2 in HCC cells [14]. Glutamine synthetase (GS) is the biosynthetic enzyme that converts glutamate into glutamine (Figure 2). GS is overexpressed and is a diagnostic marker for HCC [80]. HCC tumors in which Wnt signaling transducer β-catenin (CTNNB1) is mutated demonstrate higher GS expression, resulting in higher intracellular glutamine levels that activate the mammalian target of rapamycin complex 1 (mTORC1), a complex that activates protein translation for cell growth and proliferation [81]. This activation induces a vulnerability that can be exploited by mTORC1 inhibitors to target HCC [81]. In addition, a combination of asparaginase and GS inhibitor is able to hinder the growth of CTNNB1-mutated HCC cell-line xenografts [82]. TGFβ expression in HCC increases the levels of glutaminase 1 (GLS1), the enzyme that converts glutamine into glutamate (Figure 2), and upregulates the SLC7A5 glutamine transporter, in addition to its aforementioned effects on oxidative phosphorylation [45]. A possible reason for increased glutamate generation is for the anaplerotic conversion of glutamate to α-ketoglutarate to fuel the TCA cycle [45], although the significance of this is not well understood.
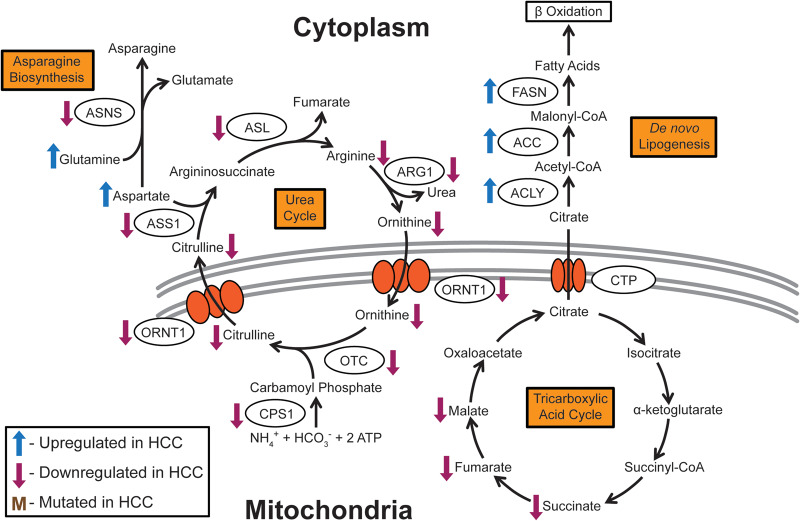
The amino acid asparagine is essential for the formation of glycoproteins, as it serves as a site for sugar-group linkage. Asparagine synthetase (ASNS), which converts aspartate into asparagine (Figure 3), is expressed to a low extent in more malignant HCC [83]. ASNS re-expression can suppress tumorigenic phenotypes [83]. Low ASNS levels result in increased sensitivity to L-asparaginase treatment, which depletes asparagine in the cancer cells [83]. Asparaginase, as well as glutamine biosynthetic enzyme GS inhibitors, has been shown to arrest proliferation and induce apoptosis in HCC [84].
Figure 3.
Schematic of asparagine biosynthesis, de novo lipogenesis, and the urea cycle. The urea cycle and fatty-acid biogenesis pathways, which are linked to important detoxification and lipid-metabolism functions in the liver, are significantly altered in HCC. However, the reasons why alterations in these pathways support tumorigenesis are not well defined. Enzyme nomenclature: ACC, acetyl-CoA carboxylase; ACLY, ATP citrate lyase; ARG1, arginase 1; ASL, argininosuccinate lyase; ASNS, asparagine synthetase; ASS1, argininosuccinate synthase 1; CPS1, carbamoyl phosphate synthetase 1; CTP, citrate transport protein; FASN, fatty-acid synthase; ORNT1, ornithine transporter 1; OTC, ornithine carbamoyltransferase.
Methionine is the amino acid coded for by the initiation codon during translation and is therefore the first amino acid of polypeptides. Methionine metabolism occurs mainly in the liver. It has been shown that the knockout of S-adenosylmethionine synthase isoform type-1 (MAT1A), the enzyme that generates S-adenosylmethionine for the transfer of methyl groups in the cell, in mice causes HCC to develop [85]. The role of a small number of amino acids in HCC has been examined, with the functions of the majority of human amino acids in HCC having yet to be uncovered.
Urea cycle
The liver is the major organ in which the urea cycle takes place. This process enables the body to excrete ammonia, generated from the breakdown of proteins, in the form of urea (Figure 3). Urea-cycle genes are predominantly decreased in HCC [2, 86]. In addition, lower levels of urea-cycle metabolites citrulline, arginine, and ornithine were observed in HCC patients compared to healthy individuals, and a decrease in levels is correlated with a later tumor stage [87]. Interestingly, urea-cycle metabolites are upregulated in the SALL4-expressing subset of HCC, but the reason for this remains unknown [51].
Urea-cycle rate-limiting enzyme carbamoyl phosphate synthetase 1 (CPS1) [73] and argininosuccinate synthase 1 (ASS1) [88] are hypermethylated in HCC, suggesting that they are epigenetically silenced. ASS1 downregulation is associated with cisplatin resistance [88]. CPS1 converts ammonia into carbamoyl phosphate, which enters the urea cycle, while ASS1 converts citrulline into argininosuccinate (Figure 3). Interestingly, arginine auxotrophy has been demonstrated in HCC as a result of ASS1 downregulation. This vulnerability can be exploited with PEGylated arginine deiminase treatment that depletes arginine levels by converting it into citrulline, starving HCC tumors [89, 90]. PEGylated arginase I, a second arginine-depleting recombinant enzyme, is also effective in targeting HCC [91]. However, Phase III clinical trials for arginine deiminase showed that, although it is well tolerated, it did not significantly increase overall survival [92]. More detailed mechanistic studies can shed light on how this downregulation of the urea cycle favors HCC tumorigenesis.
Lipid metabolism
The liver synthesizes, stores, and breaks down lipids. It is therefore unsurprising that lipid metabolism is dysregulated in HCC to provide a steady source of lipids for membrane formation, energy generation, and post-translational modification to support tumorigenesis [93]. In HCC, fatty-acid biosynthesis genes such as fatty-acid synthase (FASN) and ATP citrate lyase (ACLY), cholesterol biosynthesis gene 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), and adipogenesis transcriptional regulators such as sterol regulatory element-binding protein 1 (SREBP1) are upregulated [94]. FASN converts acetyl-CoA and malonyl-CoA into palmitate, ACLY converts citrate into acetyl-CoA (Figure 3), and HMGCR is a rate-limiting enzyme of the mevalonate pathway that converts 3-hydroxy-3-methylglutaryl-CoA into mevalonate for the generation of cholesterol and other isoprenoids. AKT/mTORC growth and proliferation signaling have been demonstrated to be responsible for these gene-expression changes as well as post-transcriptional modifications that increase lipogenesis to promote HCC [94]. Sphingolipids are also upregulated in the serum of HCC patients [95]. Alterations in the carnitine metabolism, which is essential for the transport of fatty acids into the mitochondria for energy generation through β-oxidation, are observed in chronic liver disease and HCC [96]. A high-fat and high-fructose diet in sedentary mice results in HCC [97, 98].
Lipid-metabolism enzymes have been shown to play a role in HCC. The acetyl-CoA carboxylase 1 (ACC1) and acetyl-CoA carboxylase 2 (ACC2) enzymes convert acetyl-CoA into malonyl-CoA, the major substrate for fatty-acid biosynthesis [99] (Figure 3). The knockout of ACC in mice enhances tumorigenesis in a diethylnitrosamine-induced-HCC model [100]. In addition, phospho-mutants of ACC1 and ACC2 result in NAFLD and HCC phenotypes in mice [101, 102]. Phosphorylation of ACC1 and ACC2 by adenosine monophosphate-activated protein kinase (AMPK) inhibits their enzyme activity [101]. FASN is upregulated in chemically and hormonally induced-HCC rat models [103]. However, overexpressing FASN in mouse models is not sufficient to induce tumorigenesis [104]. Mice with a knockout of acyl-CoA oxidase (AOX), which catalyses the first rate-determining step in peroxisomal fatty-acid β-oxidation, develop HCC [105]. This is due to prolonged activation of the transcription factor peroxisome proliferator-activated receptor alpha (PPARα) [105].
Upstream gene-regulatory factors also play a role in altering lipid metabolism in HCC. HBV transgenic mice show altered liver lipid metabolism [106]. Overexpression of HBV protein HBx induces lipid accumulation in cell lines and mouse liver [107]. This is explained by the concomitant increase in the expression levels of SREBP1 and peroxisome proliferator-activated receptor gamma (PPARγ), subsequently upregulating lipogenic and adipogenic enzymes [107]. HCV infection in primary human hepatocytes upregulates ketogenesis, the breakdown of fatty acids and ketogenic amino acids into ketone bodies, by activating transcription factors PPARα and retinoid X receptor (RXR) [35]. Mouse knockouts of Farnesoid X receptor (FXR), a nuclear receptor with essential roles in fatty-acid homeostasis, develop HCC [108]. In human HCC, pro-inflammatory cytokines are upregulated to inhibit transcription factor hepatocyte nuclear factor 1 alpha (HNF1α) activation of FXR, resulting in FXR inactivation reminiscence of the FXR knockout mouse model [109]. Stem-cell transcription factor NANOG activates fatty-acid oxidation in HCC-tumor-initiating cells to support self-renewal and drug resistance [50].
In terms of the role of cholesterol metabolism in HCC, the effect of HMGCR inhibitors, statins, has been examined in HCC. Statin use reduces the risk of HCC in the overall population [110], in chronic HBV-infected patients [111], decreases recurrence in HCC patients post tumor resection [112] and liver transplantation [113], and is associated with reduced mortality when administered post HCC diagnosis [114]. Future studies of the role of lipid metabolism in HCC could therefore yield additional therapeutic insight into the disease.
Metabolic alterations induced by HCC risk factors
Besides understanding the metabolic alternations that occur in HCC, it is also important to consider the metabolic changes, if any, caused by the risk factors for HCC, such as chronic HBV and HCV infections, alcohol intake, diabetes, and NAFLD. This will give insight into whether the metabolic changes that occur during upstream HCC-initiation events are conserved when HCC develops and whether early intervention in suppressing these changes could slow down or prevent the development of HCC.
HBV induces metabolic alterations that are, for the most part, similar to alterations observed in HCC. HBV infection in host cells upregulates hexosamine and membrane phospholipid phosphatidylcholine biosynthesis through upregulating GFAT1 and choline kinase α (CHKA) [115]. Viral replication was shown to be dependent on the presence of these two metabolites, as inhibition of HBP and phosphatidylcholine biosynthesis significantly reduced HBV DNA levels [115]. This is in line with the previously highlighted observation that GFAT1 is upregulated in HCC patients with poor prognosis and that GFAT1 overexpression in cell-line models enhances tumorigenic phenotypes [55]. Similarly to those in HBV-infected cells, phosphatidylcholine levels are significantly upregulated in HCC patients as compared to liver cirrhotic patient controls [116].
HBV also alters lipid metabolism, which is also dysregulated in HCC, as discussed above (Figure 3). The expression of HBV viral proteins in mice alters liver lipid metabolism [106]. HBx overexpression induces lipid accumulation in cell culture and mouse models [107]. SREBP1 and PPARγ, as well as lipogenic and adipogenic enzymes, are upregulated [107]. This increase in lipid-metabolism gene expression is also observed in HCC [94].
Metabolomics performed on serum from HBV patients reveal viral hijacking of the glycerol-phosphate shuttle—the pathway that allows the reducing power of NADH generated by glycolysis to contribute to oxidative phosphorylation [117]. Glycolytic NADH is utilized by cytoplasmic glycerol-3-phosphate dehydrogenase to convert dihydroxyacetone phosphate (DHAP) into glycerol-3-phosphate. Glycerol-3-phosphate is subsequently reconverted into DHAP at the mitochondrial inner membrane via mitochondrial glycerol-3-phosphate dehydrogenase to generate FADH2 from FAD. FADH2 can then be utilized in oxidative phosphorylation (Figure 1). It is currently unknown whether the glycerol-phosphate shuttle is altered in HCC.
The progression of chronic HBV is also linked to an increase in long-chain triglycerides, citrulline, and ornithine from the urea cycle [117]. The increase in triglycerides is analogous to previous studies on the role of increased lipid metabolism in HCC as described. However, the observed increase in urea-cycle intermediates does not correlate with the decrease observed in HCC patients [87] (Figure 3). It remains unclear how the urea cycle factors into HCC pathogenesis and progression.
Similar to those of HBV, metabolic alterations induced by HCV also mirror the alterations observed in HCC. Glycolysis is significantly upregulated in HCC owing to the Warburg effect (Figure 1) and this upregulation is similarly observed in HCV infection in primary human hepatocytes, which upregulates glycolysis through the transcriptional upregulation of glycolytic genes [35]. HCV infection in a HCC cell line also upregulates gluconeogenesis, possibly to fuel energy generation through glycolysis in HCC [36]. HCV infection seems to support the Warburg effect by downregulating oxidative phosphorylation [46, 49] and this could explain the mechanism behind impaired oxidative phosphorylation in HCC [44, 45]. HCV infection in primary human hepatocytes activates lipid-metabolism transcription factor PPARα [35] and this activation is also observed in mouse knockouts of AOX, which develop HCC [105].
The effects of alcohol intake on liver metabolism have not been well characterized, but there is some association with metabolic alterations observed in HCC. CD36 is a gene that facilitates free-fatty-acid uptake and its expression is increased with chronic alcohol consumption [118]. CD36 knockout mice are resistant to liver steatosis when fed alcohol or a high-fat diet [119], implying that CD36 could play a role in lipid metabolism that increases the risk of HCC. Treatment of hepatoma cells with fatty acids and ethanol upregulates SREBP1c and PPARγ, and downregulates SIRT1, leading to impaired fatty-acid oxidation [120]. This is analogous to the upregulation of SREBP1 and PPARγ observed in HCC and HBV infection [94, 107]. It has also been observed that NAD+ levels are reduced with high blood-alcohol levels during binge drinking [121], although the direct consequence of this on HCC induction is unknown.
NAFLD is a HCC risk factor that exhibits lipid-metabolism dysregulation as a key feature since excessive lipid accumulation in the liver, in people with low or no alcohol consumption, is a hallmark of the disease [122, 123]. Fifty-nine percent of NAFLD patients who had biopsies for evaluation demonstrated progression to non-alcoholic steatohepatitis (NASH), owing to the onset of hepatocellular injury and inflammation [124–126]. As previously discussed, a mouse model for NASH fed with a high-fat diet, which leads to the development of steatohepatitis and eventually HCC, showed increased expression of lipid metabolism and insulin-signaling genes in the liver [97]. In HBP, O-GlcNAcylation of Rab3a has been linked to NAFLD-associated HCC since it regulates lipid metabolism [58]. This observation is complemented by the demonstration that Rab3A O-GlcNAcylation promotes HCC progression and metastasis [48].
Other than lipid metabolism, NAFLD also manifests alterations in other metabolic pathways that are reminiscent of HCC metabolism. NAFLD is prevalent among type 2 diabetes patients [127] and is associated with insulin resistance [128, 129]. As discussed, metformin reduces HCC risk and progression so this suggests a link between NAFLD and HCC. Branched-chain amino acids (BCAAs), leucine, isoleucine, and valine are elevated in NAFLD patient blood but the underlying mechanisms remain unknown [130]. Since BCAAs are known to regulate mTOR signaling, one postulation is that the observed elevation of these amino acids alters glucose metabolism [131–133]. While alterations in BCAA metabolism have not yet been reported in HCC, there have been reports of utilizing these amino acids to prevent and treat HCC [134]. Further studies could yield novel insight into how BCAAs play a role in the manifestation and progression of HCC.
NAFLD could also mirror HCC metabolism in terms of mitochondrial metabolism. In NASH patients, mitochondrial abnormalities have been observed [135]. NAFLD patients given 2H and 13C tracers to measure metabolites showed higher rates of lipolysis, gluconeogenesis, anaplerosis, and mitochondrial oxidative metabolism [129]. Alterations in these processes are mostly analogous to what has been observed in HCC. However, there is a need to better understand the underlying mechanisms governing these metabolic changes.
Conclusion
Numerous metabolic processes feature heavily in promoting and supporting HCC tumorigenesis and metastasis. The majority of the major cellular metabolic pathways have been studied and validated to various extents in terms of their roles in HCC. From the current literature, HCC cells predominantly seem to demonstrate the Warburg effect. These cells prefer quick energy generation from glycolysis, through the conversion of glucose to lactate, instead of allowing pyruvate to enter the TCA cycle (Figure 1). As expected, the TCA cycle is downregulated in HCC, promoting the conversion of pyruvate into lactate (Figure 1). Interestingly, the downregulation in the TCA cycle also seems to promote flux of glycolytic metabolites through the PPP to supply precursors for nucleotide metabolism (Figure 2). The observed increase in nucleotide metabolism could provide the DNA and RNA precursors necessary for tumor-cell growth and proliferation, but this hypothesis has yet to be validated. In the case of oxidative phosphorylation, however, there exist genetic subsets of HCC in which this process is either upregulated as an energy source for tumorigenesis or downregulated in favor of glycolysis. The urea cycle, which is the excretion pathway for the byproducts of protein degradation, is downregulated in HCC, but the reasons for this remain unclear (Figure 3). There is also evidence for the upregulation of lipid metabolism (Figure 3) and the biosynthesis of some amino acids to provide energy and precursors that support tumorigenesis. Despite the wealth of information thus far, more has to be done to fully comprehend the range of metabolic alterations responsible for promoting HCC tumorigenesis.
Many of these metabolic processes have been studied in isolation, but it is likely that the cross talk between pathways and synergy among pathways plays an important role in HCC. Examples include the dual roles of TGFβ in reducing oxidative phosphorylation while enhancing glutamine anaplerosis [45], NANOG in repressing oxidative phosphorylation while upregulating fatty-acid oxidation in HCC tumor-initiating cells [50], and SALL4 in upregulating oxidative phosphorylation and urea-cycle intermediates [51]. A more complete understanding of metabolic alterations in HCC will enable a precision-medicine approach, in which patients with HCC metabolic subtypes can be diagnosed and treated with drugs targeting the metabolic vulnerabilities of these subtypes.
The characterization and validation of multiple metabolic vulnerabilities in HCC can also inform the use of combination therapy with metabolic inhibitors, which might elicit better outcomes for patients in the future. There are already a number of small molecules, such as inhibitors of oxidative phosphorylation, GS, and HMG-CoA reductase, available for targeting specific enzymes in the aforementioned metabolic pathways. Based on current and future studies of HCC metabolism, one can envision the future clinical development of combinations of metabolic drugs that can hopefully effectively treat HCC.
An interesting aspect of HCC metabolism that remains to be fully elucidated is HCC risk-factor-induced metabolic changes. There is evidence that risk factors such as HBV and HCV infections alter the metabolism of liver cells in a similar manner to that observed in HCC. However, there remain unanswered questions on how metabolic changes induced by alcohol intake, diabetes, and NAFLD relate to HCC. Having a complete understanding of how these pre-HCC metabolic changes precondition or encourage cells to progress to HCC might unlock new therapeutic strategies to slow or prevent the progression of these high-risk disease states to HCC. Overall, the study of the HCC metabolism in its entirety is timely and crucial, and will potentially serve as the basis for the development of better HCC therapeutic strategies in the long run.
Funding
This work was supported by the Genome Institute of Singapore Innovation Fellow Award to J.L.T.; the Agency for Science, Technology, and Research A*ccelerate Gap Award [ETPL/18-GAP018-R20H] to J.L.T.; the Singapore Ministry of Health's National Medical Research Council Singapore Translational Research (STaR) Investigator Award to D.G.T.; the Singapore Ministry of Education under its Research Centres of Excellence initiative to D.G.T.; the National Institutes of Health [R35CA197697, P01HL131477] to D.G.T.; and the National Heart, Lung, and Blood Institute at the National Institutes of Health and Xiu Research Fund [P01HL095489] to L.C.
Conflicts of interest
All authors declare no conflict of interest.
Authors’ contribution and Acknowledgments
J.L.T. wrote the manuscript in consultation with D.G.T. and L.C. All authors read and confirmed this paper.
References
- 1. Perkins EJ, Bao W, Guan X et al. Comparison of transcriptional responses in liver tissue and primary hepatocyte cell cultures after exposure to hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine. BMC Bioinformatics 2006;7:S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nwosu ZC, Megger DA, Hammad S et al. Identification of the consistently altered metabolic targets in human hepatocellular carcinoma. Cell Mol Gastroenterol Hepatol 2017;4:303–23.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Uhlen M, Zhang C, Lee S et al. A pathology atlas of the human cancer transcriptome. Science 2017;357:eaan2507. [DOI] [PubMed] [Google Scholar]
- 4. Peng X, Chen Z, Farshidfar F et al. Molecular characterization and clinical relevance of metabolic expression subtypes in human cancers. Cell Rep 2018;23:255–69.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ferlay J, Soerjomataram I, Dikshit R et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359–86. [DOI] [PubMed] [Google Scholar]
- 6. Ng J, Wu J. Hepatitis B- and hepatitis C-related hepatocellular carcinomas in the United States: similarities and differences. Hepat Mon 2012;12:e7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bray F, Ferlay J, Soerjomataram I et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 8. Petrick JL, Thistle JE, Zeleniuch-Jacquotte A et al. Body mass index, diabetes and intrahepatic cholangiocarcinoma risk: the liver cancer pooling project and meta-analysis. Am J Gastroenterol 2018;113:1494–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim GA, Lee HC, Choe J et al. Association between non-alcoholic fatty liver disease and cancer incidence rate. J Hepatol 2017;68:140–6. [DOI] [PubMed] [Google Scholar]
- 10. Satriano L, Lewinska M, Rodrigues PM et al. Metabolic rearrangements in primary liver cancers: cause and consequences. Nat Rev Gastroenterol Hepatol 2019;16:748–66. [DOI] [PubMed] [Google Scholar]
- 11. Petersen MC, Vatner DF, Shulman GI. Regulation of hepatic glucose metabolism in health and disease. Nat Rev Endocrinol 2017;13:572–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Warburg O. On the origin of cancer cells. Science 1956;123:309–14. [DOI] [PubMed] [Google Scholar]
- 13. Amann T, Maegdefrau U, Hartmann A et al. GLUT1 expression is increased in hepatocellular carcinoma and promotes tumorigenesis. Am J Pathol 2009;174:1544–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun HW, Yu XJ, Wu WC et al. GLUT1 and ASCT2 as predictors for prognosis of hepatocellular carcinoma. PloS One 2016;11:e0168907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Paudyal B, Paudyal P, Oriuchi N et al. Clinical implication of glucose transport and metabolism evaluated by 18F-FDG PET in hepatocellular carcinoma. Int J Oncol 2008;33:1047–54. [PubMed] [Google Scholar]
- 16. Kim YH, Jeong DC, Pak K et al. SLC2A2 (GLUT2) as a novel prognostic factor for hepatocellular carcinoma. Oncotarget 2017;8:68381–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Osthus RC, Shim H, Kim S et al. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem 2000;275:21797–800. [DOI] [PubMed] [Google Scholar]
- 18. Huang Q, Tan Y, Yin P et al. Metabolic characterization of hepatocellular carcinoma using nontargeted tissue metabolomics. Cancer Res 2013;73:4992–5002. [DOI] [PubMed] [Google Scholar]
- 19. Pedersen PL, Mathupala S, Rempel A et al. Mitochondrial bound type II hexokinase: a key player in the growth and survival of many cancers and an ideal prospect for therapeutic intervention. Biochim Biophys Acta 2002;1555:14–20. [DOI] [PubMed] [Google Scholar]
- 20. Guzman G, Chennuri R, Chan A et al. Evidence for heightened hexokinase II immunoexpression in hepatocyte dysplasia and hepatocellular carcinoma. Dig Dis Sci 2015;60:420–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. DeWaal D, Nogueira V, Terry AR et al. Hexokinase-2 depletion inhibits glycolysis and induces oxidative phosphorylation in hepatocellular carcinoma and sensitizes to metformin. Nat Commun 2018;9:446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Noguchi T, Inoue H, Tanaka T. The M1- and M2-type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing. J Biol Chem 1986;261:13807–12. [PubMed] [Google Scholar]
- 23. Noguchi T, Yamada K, Inoue H et al. The L- and R-type isozymes of rat pyruvate kinase are produced from a single gene by use of different promoters. J Biol Chem 1987;262:14366–71. [PubMed] [Google Scholar]
- 24. Yamada K, Noguchi T. Nutrient and hormonal regulation of pyruvate kinase gene expression. Biochem J 1999;337:1–11. [PMC free article] [PubMed] [Google Scholar]
- 25. Wong CC, Au SL, Tse AP et al. Switching of pyruvate kinase isoform L to M2 promotes metabolic reprogramming in hepatocarcinogenesis. PloS One 2014;9:e115036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Méndez-Lucas A, Li X, Hu J et al. Glucose catabolism in liver tumors induced by c-MYC can be sustained by various PKM1/PKM2 ratios and pyruvate kinase activities. Cancer Res 2017;77:4355–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen Z, Lu X, Wang Z et al. Co-expression of PKM2 and TRIM35 predicts survival and recurrence in hepatocellular carcinoma. Oncotarget 2015;6:2539–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu Y, Wu H, Mei Y et al. Clinicopathological and prognostic significance of PKM2 protein expression in cirrhotic hepatocellular carcinoma and non-cirrhotic hepatocellular carcinoma. Sci Rep 2017;7:15294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dayton TL, Gocheva V, Miller KM et al. Germline loss of PKM2 promotes metabolic distress and hepatocellular carcinoma. Genes Dev 2016;30:1020–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Budhu A, Roessler S, Zhao X et al. Integrated metabolite and gene expression profiles identify lipid biomarkers associated with progression of hepatocellular carcinoma and patient outcomes. Gastroenterology 2013;144:1066–75.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang ZX, Jiang CP, Cao Y et al. Preoperative serum liver enzyme markers for predicting early recurrence after curative resection of hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int 2015;14:178–85. [DOI] [PubMed] [Google Scholar]
- 32. Faloppi L, Scartozzi M, Bianconi M et al. The role of LDH serum levels in predicting global outcome in HCC patients treated with sorafenib: implications for clinical management. BMC Cancer 2014;14:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Feng Y, Xiong Y, Qiao T et al. Lactate dehydrogenase A: a key player in carcinogenesis and potential target in cancer therapy. Cancer Med 2018;7:6124–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang Q, Li J, Xing J et al. CD147 promotes reprogramming of glucose metabolism and cell proliferation in HCC cells by inhibiting the p53-dependent signaling pathway. J Hepatol 2014;61:859–66. [DOI] [PubMed] [Google Scholar]
- 35. Levy G, Habib N, Guzzardi MA et al. Nuclear receptors control pro-viral and antiviral metabolic responses to hepatitis C virus infection. Nat Chem Biol 2016;12:1037–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen J, Zhang Z, Wang N et al. Role of HDAC9-FoxO1 axis in the transcriptional program associated with hepatic gluconeogenesis . Sci Rep 2017;7:6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen HP, Shieh JJ, Chang CC et al. Metformin decreases hepatocellular carcinoma risk in a dose-dependent manner: population-based and in vitro studies. Gut 2013;62:606–15. [DOI] [PubMed] [Google Scholar]
- 38. Kang WH, Tak E, Hwang S et al. Metformin-associated chemopreventive effects on recurrence after hepatic resection of hepatocellular carcinoma: from in vitro to a clinical study. Anticancer Res 2018;38:2399–407. [DOI] [PubMed] [Google Scholar]
- 39. Liu Y, Hu X, Shan X et al. Rosiglitazone metformin adduct inhibits hepatocellular carcinoma proliferation via activation of AMPK/p21 pathway. Cancer Cell Int 2019;19:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang C, Wang S, Ruan H et al. Downregulation of PDK4 increases lipogenesis and associates with poor prognosis in hepatocellular carcinoma. J Cancer 2019;10:918–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pollard PJ, Brière JJ, Alam NA et al. Accumulation of Krebs cycle intermediates and over-expression of HIF1alpha in tumours which result from germline FH and SDH mutations. Hum Mol Genet 2005;14:2231–9. [DOI] [PubMed] [Google Scholar]
- 42. King A, Selak MA, Gottlieb E. Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene 2006;25:4675–82. [DOI] [PubMed] [Google Scholar]
- 43. Tseng PL, Wu WH, Hu TH et al. Decreased succinate dehydrogenase B in human hepatocellular carcinoma accelerates tumor malignancy by inducing the Warburg effect. Sci Rep 2018;8:3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee YK, Lim JJ, Jeoun UW et al. Lactate-mediated mitoribosomal defects impair mitochondrial oxidative phosphorylation and promote hepatoma cell invasiveness. J Biol Chem 2017;292:20208–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Soukupova J, Malfettone A, Hyroššová P et al. Role of the transforming growth factor-β in regulating hepatocellular carcinoma oxidative metabolism. Sci Rep 2017;7:12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gerresheim GK, Roeb E, Michel AM et al. Hepatitis C Virus downregulates core subunits of oxidative phosphorylation, reminiscent of the Warburg effect in cancer cells. Cells 2019;8:1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhuang X, Chen Y, Wu Z et al. Mitochondrial miR-181a-5p promotes glucose metabolism reprogramming in liver cancer by regulating the electron transport chain. Carcinogenesis 2020;41:972–83. [DOI] [PubMed] [Google Scholar]
- 48. Wu W, Zheng X, Wang J et al. O-GlcNAcylation on Rab3A attenuates its effects on mitochondrial oxidative phosphorylation and metastasis in hepatocellular carcinoma. Cell Death Dis 2018;9:970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen CL, Tsukamoto H, Liu JC et al. Reciprocal regulation by TLR4 and TGF-β in tumor-initiating stem-like cells. J Clin Invest 2013;123:2832–49. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50. Chen CL, Uthaya Kumar DB, Punj V et al. NANOG metabolically reprograms tumor-initiating stem-like cells through tumorigenic changes in oxidative phosphorylation and fatty acid metabolism. Cell Metab 2016;23:206–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tan JL, Li F, Yeo JZ et al. New high-throughput screening identifies compounds that reduce viability specifically in liver cancer cells that express high levels of SALL4 by inhibiting oxidative phosphorylation. Gastroenterology 2019;157:1615–29.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hardivillé S, Hart GW. Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell Metab 2014;20:208–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang X, Qian K. Protein O-GlcNAcylation: emerging mechanisms and functions. Nat Rev Mol Cell Biol 2017;18:452–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ruan HB, Singh JP, Li MD et al. Cracking the O-GlcNAc code in metabolism. Trends Endocrinol Metab 2013;24:301–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li L, Shao M, Peng P et al. High expression of GFAT1 predicts unfavorable prognosis in patients with hepatocellular carcinoma. Oncotarget 2017;8:19205–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cao B, Duan M, Xing Y et al. O-GlcNAc transferase activates stem-like cell potential in hepatocarcinoma through O-GlcNAcylation of eukaryotic initiation factor 4E. J Cell Mol Med 2019;23:2384–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhu Q, Zhou L, Yang Z et al. O-GlcNAcylation plays a role in tumor recurrence of hepatocellular carcinoma following liver transplantation. Med Oncol 2012;29:985–93. [DOI] [PubMed] [Google Scholar]
- 58. Xu W, Zhang X, Wu JL et al. O-GlcNAc transferase promotes fatty liver-associated liver cancer through inducing palmitic acid and activating endoplasmic reticulum stress. J Hepatol 2017;67:310–20. [DOI] [PubMed] [Google Scholar]
- 59. Qiao Y, Zhang X, Zhang Y et al. High glucose stimulates tumorigenesis in hepatocellular carcinoma cells through AGER-dependent O-GlcNAcylation of c-Jun. Diabetes 2016;65:619–32. [DOI] [PubMed] [Google Scholar]
- 60. Lin SH, Liu T, Ming X et al. Regulatory role of hexosamine biosynthetic pathway on hepatic cancer stem cell marker CD133 under low glucose conditions. Sci Rep 2016;6:21184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Duan F, Wu H, Jia D et al. O-GlcNAcylation of RACK1 promotes hepatocellular carcinogenesis. J Hepatol 2018;68:1191–202. [DOI] [PubMed] [Google Scholar]
- 62. Patra KC, Hay N. The pentose phosphate pathway and cancer. Trends Biochem Sci 2014;39:347–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hu H, Ding X, Yang Y et al. Changes in glucose-6-phosphate dehydrogenase expression results in altered behavior of HBV-associated liver cancer cells. Am J Physiol Gastrointest Liver Physiol 2014;307:G611–22. [DOI] [PubMed] [Google Scholar]
- 64. Lu M, Lu L, Dong Q et al. Elevated G6PD expression contributes to migration and invasion of hepatocellular carcinoma cells by inducing epithelial-mesenchymal transition. Acta Biochim Biophys Sin 2018;50:370–80. [DOI] [PubMed] [Google Scholar]
- 65. Yin X, Tang B, Li JH et al. ID1 promotes hepatocellular carcinoma proliferation and confers chemoresistance to oxaliplatin by activating pentose phosphate pathway. J Exp Clin Cancer Res 2017;36:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kowalik MA, Guzzo G, Morandi A et al. Metabolic reprogramming identifies the most aggressive lesions at early phases of hepatic carcinogenesis. Oncotarget 2016;7:32375–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rao X, Duan X, Mao W et al. O-GlcNAcylation of G6PD promotes the pentose phosphate pathway and tumor growth. Nat Commun 2015;6:8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Liu B, Fang M, He Z et al. Hepatitis B virus stimulates G6PD expression through HBx-mediated Nrf2 activation. Cell Death Dis 2015;6:e1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ngo HKC, Kim DH, Cha YN et al. Nrf2 mutagenic activation drives hepatocarcinogenesis. Cancer Res 2017;77:4797–808. [DOI] [PubMed] [Google Scholar]
- 70. Xu IMJ, Lai RKH, Lin SH et al. Transketolase counteracts oxidative stress to drive cancer development. Proc Natl Acad Sci USA 2016;113:E725–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Saito T, Ichimura Y, Taguchi K et al. p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming. Nat Commun 2016;7:12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cancer Genome Atlas Research Network. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 2017;169:1327–41.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Liu H, Dong H, Robertson K et al. DNA methylation suppresses expression of the urea cycle enzyme carbamoyl phosphate synthetase 1 (CPS1) in human hepatocellular carcinoma. Am J Pathol 2011;178:652–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yeh HW, Lee SS, Chang CY et al. Pyrimidine metabolic rate limiting enzymes in poorly-differentiated hepatocellular carcinoma are signature genes of cancer stemness and associated with poor prognosis. Oncotarget 2017;8:77734–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sykes DB, Kfoury YS, Mercier FE et al. Inhibition of dihydroorotate dehydrogenase overcomes differentiation blockade in acute myeloid leukemia. Cell 2016;167:171–86.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tan JL, Fogley RD, Flynn RA et al. Stress from nucleotide depletion activates the transcriptional regulator HEXIM1 to suppress melanoma. Mol Cell 2016;62:34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. White RM, Cech J, Ratanasirintrawoot S et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature 2011;471:518–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wu N, Yang M, Gaur U et al. Alpha-ketoglutarate: physiological functions and applications. Biomol Ther 2016;24:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hirayama C, Suyama K, Horie Y et al. Plasma amino acid patterns in hepatocellular carcinoma. Biochem Med Metab Biol 1987;38:127–33. [DOI] [PubMed] [Google Scholar]
- 80. Di Tommaso L, Franchi G, Park YN et al. Diagnostic value of HSP70, glypican 3, and glutamine synthetase in hepatocellular nodules in cirrhosis. Hepatology 2007;45:725–34. [DOI] [PubMed] [Google Scholar]
- 81. Adebayo Michael AO, Ko S, Tao J et al. Inhibiting glutamine-dependent mTORC1 activation ameliorates liver cancers driven by β-catenin mutations. Cell Metab 2019;29:1135–50.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chiu M, Tardito S, Pillozzi S et al. Glutamine depletion by crisantaspase hinders the growth of human hepatocellular carcinoma xenografts. Br J Cancer 2014;111:1159–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhang B, Dong LW, Tan YX et al. Asparagine synthetase is an independent predictor of surgical survival and a potential therapeutic target in hepatocellular carcinoma. Br J Cancer 2013;109:14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tardito S, Chiu M, Uggeri J et al. L-Asparaginase and inhibitors of glutamine synthetase disclose glutamine addiction of β-catenin-mutated human hepatocellular carcinoma cells. Curr Cancer Drug Targets 2011;11:929–43. [DOI] [PubMed] [Google Scholar]
- 85. Martínez-Chantar ML, Corrales FJ, Martínez-Cruz LA et al. Spontaneous oxidative stress and liver tumors in mice lacking methionine adenosyltransferase 1A. FASEB J 2002;16:1292–4. [DOI] [PubMed] [Google Scholar]
- 86. Chaerkady R, Harsha HC, Nalli A et al. A quantitative proteomic approach for identification of potential biomarkers in hepatocellular carcinoma. J Proteome Res 2008;7:4289–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Chen T, Xie G, Wang X et al. Serum and urine metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Mol Cell Proteomics 2011;10:M110.004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. McAlpine JA, Lu HT, Wu KC et al. Down-regulation of argininosuccinate synthetase is associated with cisplatin resistance in hepatocellular carcinoma cell lines: implications for PEGylated arginine deiminase combination therapy. BMC Cancer 2014;14:621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Feun L, Savaraj N. Pegylated arginine deiminase: a novel anticancer enzyme agent. Expert Opin Investig Drugs 2006;15:815–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ensor CM, Holtsberg FW, Bomalaski JS et al. Pegylated arginine deiminase (ADI-SS PEG20,000 mw) inhibits human melanomas and hepatocellular carcinomas in vitro and in vivo. Cancer Res 2002;62:5443–50. [PubMed] [Google Scholar]
- 91. Lam TL, Wong GK, Chong HC et al. Recombinant human arginase inhibits proliferation of human hepatocellular carcinoma by inducing cell cycle arrest. Cancer Lett 2009;277:91–100. [DOI] [PubMed] [Google Scholar]
- 92. Abou-Alfa GK, Qin S, Ryoo BY et al. Phase III randomized study of second line ADI-PEG 20 plus best supportive care versus placebo plus best supportive care in patients with advanced hepatocellular carcinoma. Ann Oncol 2018;29:1402–8. [DOI] [PubMed] [Google Scholar]
- 93. Zaidi N, Lupien L, Kuemmerle NB et al. Lipogenesis and lipolysis: the pathways exploited by the cancer cells to acquire fatty acids. Prog Lipid Res 2013;52:585–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Calvisi DF, Wang C, Ho C et al. Increased lipogenesis, induced by AKT-mTORC1-RPS6 signaling, promotes development of human hepatocellular carcinoma. Gastroenterology 2011;140:1071–83.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Grammatikos G, Schoell N, Ferreirós N et al. Serum sphingolipidomic analyses reveal an upregulation of C16-ceramide and sphingosine-1-phosphate in hepatocellular carcinoma. Oncotarget 2016;7:18095–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zhou L, Wang Q, Yin P et al. Serum metabolomics reveals the deregulation of fatty acids metabolism in hepatocellular carcinoma and chronic liver diseases. Anal Bioanal Chem 2012;403:203–13. [DOI] [PubMed] [Google Scholar]
- 97. Dowman JK, Hopkins LJ, Reynolds GM et al. Development of hepatocellular carcinoma in a murine model of nonalcoholic steatohepatitis induced by use of a high-fat/fructose diet and sedentary lifestyle. Am J Pathol 2014;184:1550–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Hill-Baskin AE, Markiewski MM, Buchner DA et al. Diet-induced hepatocellular carcinoma in genetically predisposed mice. Hum Mol Genet 2009;18:2975–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Wakil SJ, Abu-Elheiga LA. Fatty acid metabolism: target for metabolic syndrome. J Lipid Res 2009;50:S138–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Nelson ME, Lahiri S, Chow JDY et al. Inhibition of hepatic lipogenesis enhances liver tumorigenesis by increasing antioxidant defence and promoting cell survival. Nat Commun 2017;8:14689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Fullerton MD, Galic S, Marcinko K et al. Single phosphorylation sites in Acc1 and Acc2 regulate lipid homeostasis and the insulin–sensitizing effects of metformin. Nat Med 2013;19:1649–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Lally JSV, Ghoshal S, DePeralta DK et al. Inhibition of acetyl-CoA carboxylase by phosphorylation or the inhibitor ND-654 suppresses lipogenesis and hepatocellular carcinoma. Cell Metab 2019;29:174–82.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Evert M, Schneider-Stock R, Dombrowski F. Overexpression of fatty acid synthase in chemically and hormonally induced hepatocarcinogenesis of the rat. Lab Invest 2005;85:99–108. [DOI] [PubMed] [Google Scholar]
- 104. Li L, Pilo GM, Li X et al. Inactivation of fatty acid synthase impairs hepatocarcinogenesis driven by AKT in mice and humans. J Hepatol 2016;64:333–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Fan CY, Pan J, Usuda N et al. Steatohepatitis, spontaneous peroxisome proliferation and liver tumors in mice lacking peroxisomal fatty acyl-CoA oxidase: implications for peroxisome proliferator-activated receptor alpha natural ligand metabolism. J Biol Chem 1998;273:15639–45. [DOI] [PubMed] [Google Scholar]
- 106. Yang F, Yan S, He Y et al. Expression of hepatitis B virus proteins in transgenic mice alters lipid metabolism and induces oxidative stress in the liver. J Hepatol 2008;48:12–9. [DOI] [PubMed] [Google Scholar]
- 107. Kim KH, Shin HJ, Kim K et al. Hepatitis B virus X protein induces hepatic steatosis via transcriptional activation of SREBP1 and PPARgamma. Gastroenterology 2007;132:1955–67. [DOI] [PubMed] [Google Scholar]
- 108. Yang F, Huang X, Yi T et al. Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Res 2007;67:863–7. [DOI] [PubMed] [Google Scholar]
- 109. Liu N, Meng Z, Lou G et al. Hepatocarcinogenesis in FXR-/- mice mimics human HCC progression that operates through HNF1α regulation of FXR expression. Mol Endocrinol 2012;26:775–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Kim G, Jang SY, Nam CM et al. Statin use and the risk of hepatocellular carcinoma in patients at high risk: a nationwide nested case-control study. J Hepatol 2018;68:476–84. [DOI] [PubMed] [Google Scholar]
- 111. Goh MJ, Sinn DH, Kim S et al. Statin use and the risk of hepatocellular carcinoma in patients with chronic hepatitis B. Hepatology 2020;71:2023–32. [DOI] [PubMed] [Google Scholar]
- 112. Kawaguchi Y, Sakamoto Y, Ito D et al. Statin use is associated with a reduced risk of hepatocellular carcinoma recurrence after initial liver resection. Biosci Trends 2017;11:574–80. [DOI] [PubMed] [Google Scholar]
- 113. Cho Y, Kim MS, Nam CM et al. Statin use is associated with decreased hepatocellular carcinoma recurrence in liver transplant patients. Sci Rep 2019;9:1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Thrift AP, Natarajan Y, Liu Y et al. Statin use after diagnosis of hepatocellular carcinoma is associated with decreased mortality. Clin Gastroenterol Hepatol 2019;17:2117–25.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Li H, Zhu W, Zhang L et al. The metabolic responses to hepatitis B virus infection shed new light on pathogenesis and targets for treatment. Sci Rep 2015;5:8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Cotte AK, Cottet V, Aires V et al. Phospholipid profiles and hepatocellular carcinoma risk and prognosis in cirrhotic patients. Oncotarget 2019;10:2161–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Schoeman JC, Hou J, Harms AC et al. Metabolic characterization of the natural progression of chronic hepatitis B. Genome Med 2016;8:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Clugston RD, Jiang H, Lee MX et al. Altered hepatic lipid metabolism in C57BL/6 mice fed alcohol: a targeted lipidomic and gene expression study. J Lipid Res 2011;52:2021–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Clugston RD, Yuen JJ, Hu Y et al. CD36-deficient mice are resistant to alcohol- and high-carbohydrate-induced hepatic steatosis. J Lipid Res 2014;55:239–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Vecchione G, Grasselli E, Compalati AD et al. Ethanol and fatty acids impair lipid homeostasis in an in vitro model of hepatic steatosis. Food Chem Toxicol 2016;90:84–94. [DOI] [PubMed] [Google Scholar]
- 121. French SW. Chronic alcohol binging injures the liver and other organs by reducing NAD+ levels required for sirtuin’s deacetylase activity. Exp Mol Pathol 2016;100:303–6. [DOI] [PubMed] [Google Scholar]
- 122. Chalasani N, Younossi Z, Lavine JE et al. The diagnosis and management of nonalcoholic fatty liver disease: practice guidance from the American Association for the Study of Liver Diseases. Hepatology 2018;67:328–57. [DOI] [PubMed] [Google Scholar]
- 123.European Association for the Study of the Liver (EASL), European Association for the Study of Diabetes (EASD) & European Association for the Study of Obesity (EASO). EASL-EASD-EASO Clinical Practice Guidelines for the management of non-alcoholic fatty liver disease. J Hepatol 2016;64:1388–402. [DOI] [PubMed] [Google Scholar]
- 124. Younossi ZM, Koenig AB, Abdelatif D et al. Global epidemiology of nonalcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016;64:73–84. [DOI] [PubMed] [Google Scholar]
- 125. Bedossa P. Histological assessment of NAFLD. Dig Dis Sci 2016;61:1348–55. [DOI] [PubMed] [Google Scholar]
- 126. Lonardo A, Nascimbeni F, Maurantonio M et al. Nonalcoholic fatty liver disease: evolving paradigms. World J Gastroenterol 2017;23:6571–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Portillo-Sanchez P, Bril F, Maximos M et al. High prevalence of nonalcoholic fatty liver disease in patients with type 2 diabetes mellitus and normal plasma aminotransferase levels. J Clin Endocrinol Metab 2015;100:2231–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Marchesini G, Brizi M, Morselli-Labate AM et al. Association of nonalcoholic fatty liver disease with insulin resistance. Am J Med 1999;107:450–5. [DOI] [PubMed] [Google Scholar]
- 129. Sunny NE, Parks EJ, Browning JD et al. Excessive hepatic mitochondrial TCA cycle and gluconeogenesis in humans with nonalcoholic fatty liver disease. Cell Metab 2011;14:804–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Gaggini M, Carli F, Rosso C et al. Altered amino acid concentrations in NAFLD: impact of obesity and insulin resistance. Hepatology 2018;67:145–58. [DOI] [PubMed] [Google Scholar]
- 131. Chakravarthy MV, Neuschwander-Tetri BA. The metabolic basis of nonalcoholic steatohepatitis. Endocrinol Diabetes Metab 2020:e00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Dodd KM, Tee AR. Leucine and mTORC1: a complex relationship. Am J Physiol Endocrinol Metab 2012;302:E1329–42. [DOI] [PubMed] [Google Scholar]
- 133. Newgard CB, An J, Bain JR et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab 2009;9:311–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Tajiri K, Shimizu Y. Branched-chain amino acids in liver diseases. Transl Gastroenterol Hepatol 2018;3:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Sanyal AJ, Campbell-Sargent C, Mirshahi F et al. Nonalcoholic steatohepatitis: association of insulin resistance and mitochondrial abnormalities. Gastroenterology 2001;120:1183–92. [DOI] [PubMed] [Google Scholar]