Abstract
Objectives
Despite extensive research on procalcitonin (PCT)-guided therapy in lower respiratory tract infections, the association between PCT and bacterial pneumonia remains unclear.
Methods
We evaluated retrospectively the performance of PCT in patients presenting with lower respiratory tract infection symptoms and grouped by seven diagnoses. All patients had microbial testing, chest imaging, and CBC counts within 1 day of PCT testing.
Results
Median PCT level in patients diagnosed with bacterial pneumonia was significantly higher than in patients diagnosed with other sources of infections or those not diagnosed with infections. Median PCT levels were not different among patients grouped by type or quantity of pathogen detected. They were significantly higher in patients with higher pathogenicity scores for isolated bacteria, those with abnormal WBC count, and those with chest imaging consistent with bacterial pneumonia. A diagnostic workup that included imaging, WBC count, and Gram stain had an area under the receiver operating characteristic curve of 0.748, and the addition of PCT increased it to 0.778.
Conclusions
PCT was higher in patients diagnosed with bacterial pneumonia. Less clear is its diagnostic ability to detect bacterial pneumonia over and above imaging and laboratory data routinely available to clinicians.
Keywords: Procalcitonin, Bacteria, Viruses, Pneumonia, Diagnosis, Imaging
Key Points.
Procalcitonin levels correlate with an abnormal workup for and a diagnosis of bacterial pneumonia but do not appear to add much discriminatory information for the detection of this disease.
Evidence-based protocols are needed for procalcitonin test implementation in clinical laboratories.
Antibiotics are commonly prescribed for lower respiratory tract infections (LRTIs).1 However, this practice is inappropriate or unnecessary in a significant number of the cases2-4 and partly occurs due to the difficulty in distinguishing bacterial from nonbacterial origins of infection. This has spurred the exploration of serum biomarkers that can discriminate with better accuracy bacterial from viral LRTIs, guide antibiotic stewardship, or both. Procalcitonin (PCT), the 116–amino acid precursor to calcitonin, is considered such a candidate for adults with suspected LRTI and critically ill patients with any type of infection.5-9
PCT is synthesized in the thyroid neuroendocrine cells under normal conditions10 and is strongly induced in almost all tissues by bacterial11,12 but not viral infections.13 A recent systemic review9 found that PCT-guided algorithms were associated with reduced antibiotic use in the absence of significant adverse events in acute respiratory tract infections. While this has also been confirmed under “real-life” conditions,14 PCT-guided algorithms have not been extensively tested outside Europe. In fact, a recent clinical trial conducted in the United States failed to demonstrate reductions in antibiotic use in patients with suspected LRTIs with PCT-guided management.15 On the other hand, the US Food and Drug Administration has recently cleared the expanded use of PCT for the guidance of antibiotic use in acute respiratory illnesses and sepsis.16 At the center of this issue lies the lack of definitive data for the diagnostic ability of PCT to distinguish bacterial from other sources of pneumonia with accuracy. Therefore, we set out to evaluate plasma PCT values in LRTIs and compare their performance with available clinical data in a retrospective review of an inpatient population at Yale New Haven Hospital.
Materials and Methods
This study was approved by the Institutional Review Board of the Human Investigation Committee of the Yale University Human Research Protection Program.
Test Methods
PCT (Vidas Brahms PCT; bioMérieux) testing was performed on plasma. Lower respiratory tract cultures (CXRES) were performed on sputa, endotracheal aspirates (TAs), or bronchial specimens according to standard procedures. All specimens received a Gram stain. Cultures were not performed on sputa or TAs with more than 25 epithelial cells per low-power field. All cultures were assessed for the presence of Staphylococcus aureus, Pseudomonas aeruginosa, and β-hemolytic streptococci in any amount. Other organisms that were predominating were also clinically reported. Samples were plated by conventional four-quadrant streaking. The highest quadrant demonstrating growth of a particular organism was recorded. The highest quadrant demonstrating growth of respiratory microbiota was also recorded. This was used to create the following pathogenicity score:
0 = pathogen less than 1+ with 1+ or more normal microbiota
1 = pathogen equal to normal microbiota
2 = pathogen more than normal microbiota or Streptococcus pneumoniae or Legionella urine antigen positive
While the pathogenicity score is not a published validated score, we believe it reasonably captures how a clinical microbiologist or clinician would interpret the culture results. Respiratory direct fluorescent antigen (DFA) testing (Diagnostic Hybrids; Quidel) was performed as described on nasopharyngeal (NP) swabs17 with all samples tested for influenza A, influenza B, respiratory syncytial virus (RSV), adenovirus, and parainfluenza viruses (PIVs) 1 to 3. Respiratory virus polymerase chain reaction (PCR) testing was performed on NP swabs and bronchial specimens using a panel of laboratory-developed PCR reactions for influenza A, influenza B, RSV, adenovirus, human metapneumovirus, PIVs 1 to 3, and rhinovirus. A detailed breakdown of the types of microorganisms and viruses detected is available in Supplementary Figure 1 (all supplemental materials can be found at American Journal of Clinical Pathology online).
Patient Selection
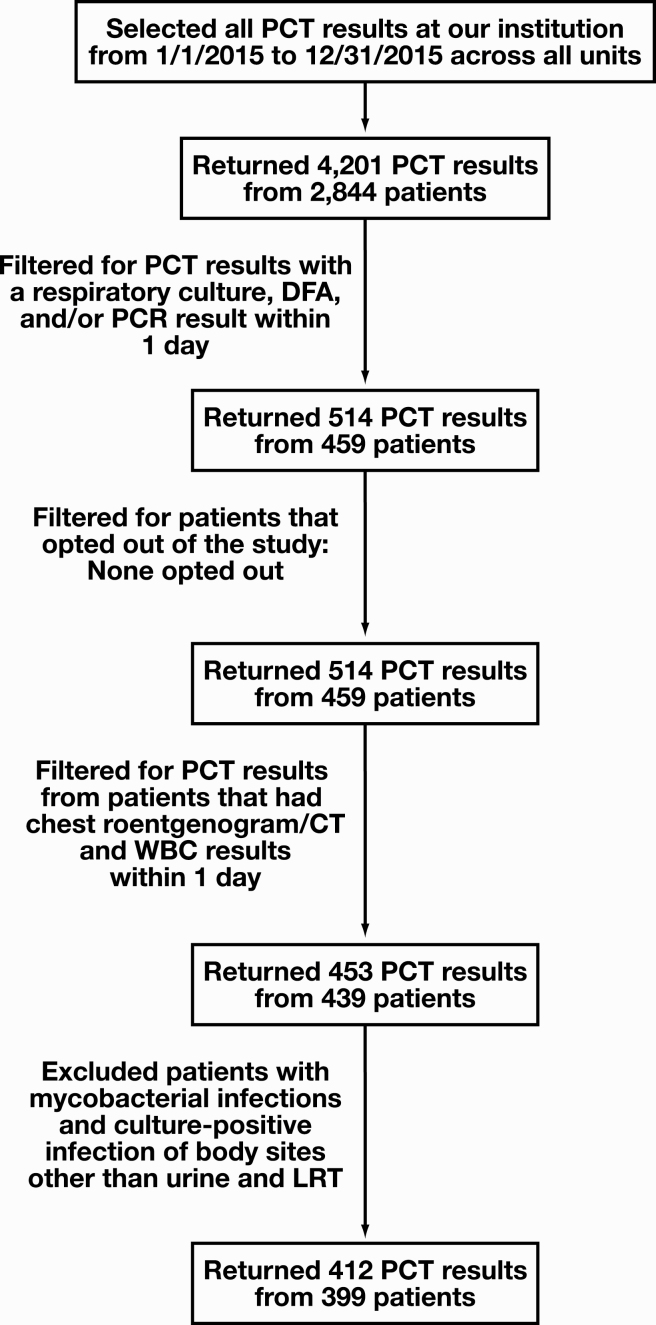
The case-finding approach is described in Figure 1. Laboratory results for PCT, CXRES testing, respiratory virus DFA, and/or respiratory virus PCR testing performed at Yale New Haven Hospital between January 1, 2015, and December 31, 2015, were extracted from the computerized laboratory information system. All appropriate specimen sources were eligible for inclusion. All instances in which CXRES and respiratory virus testing were performed within 1 day of PCT testing were eligible for further analysis. Medical record numbers for these patients and dates of PCT testing were provided to institutional data analysts who (1) verified if patients opted out of medical research, (2) provided results of chest x-ray or thoracic computed tomography studies and WBC counts performed within 1 day of PCT testing, (3) provided 1-month mortality data, and (4) provided admitting and discharge diagnoses. Finally, patients were excluded if they had a positive culture for acid-fast bacilli, a positive blood culture not considered a contaminant, or a positive culture at any site other than the respiratory tract or urine. Pneumococcal and legionella antigens in urine were considered respiratory pathogens and included in the analysis. The discrepancy between total eligible PCT results (412) and total number of patients (399) represents multiple PCT orders for the same patient in 11 patients. These orders were at least 8 days apart, and in every instance, all the inclusion criteria were met.
Figure 1.
Flow diagram of results and patients selected for the study. CT, computed tomography; DFA, direct immunofluorescence assay; LRT, lower respiratory tract; PCT, procalcitonin; PCR, polymerase chain reaction.
Data Categorization and Bacterial Pneumonia Case Definition
Results for PCT below the lower reportable range of 0.05 ng/mL were assigned a value of 0.049 ng/mL to facilitate analysis. This categorization did not affect results, as all analysis was performed using medians.
While no clearly defined criteria have been validated to predict bacterial pneumonia,18 a constellation of clinical signs and symptoms, in combination with radiologic and/or laboratory studies, is generally used to make the diagnosis. To assess how well they correlate with the final diagnoses derived from the electronic health records, we evaluated the following three sets of findings: (1) radiologic study results, (2) peripheral WBC results, and (3) Gram stain results. For the first set, results of imaging studies were reviewed by two individuals and assigned a score of 0 if there was no evidence of pneumonia, 1 if pneumonia could not be ruled out, and 2 if pneumonia was present. Discordant results were blindly adjudicated by a third individual. For the second set, WBC abnormalities were categorized based upon the extracted laboratory information. Patients were assigned a score of 1 if they exhibited leukopenia (<4,000 WBCs/μL) or leukocytosis (≥12,000 WBCs/μL) and a score of 0 if these findings were not present. For the third set, Gram stains were defined as purulent if WBCs were reported at 3+ or 4+. Patients were assigned a score of 1 or 0 based on the presence or absence of purulent Gram stains, respectively.
Final Diagnosis
Admitting and discharge diagnoses for included cases were categorized in the electronic health records as (1) pneumonia (bacterial), (2) respiratory infection (viral), (3) pneumonia (organism unspecified), (4) respiratory infection (other), (5) sepsis, (6) other infection, or (7) no infection. We assigned the final diagnosis by comparing admission diagnoses, discharge diagnoses, date of PCT testing, and duration of admission. For admissions less than 7 days, the discharge diagnoses were used. The discharge diagnoses were also used for admissions of greater than 7 days when the admitting diagnoses were the same. For admissions greater than 7 days with discordant admitting and discharge diagnoses, chart review was performed to ascertain the diagnosis at the time of PCT order. For admissions or visits with multiple diagnoses recorded, the final diagnosis was determined by the rank of the seven categories listed above. Bacterial pneumonia with sepsis was categorized as a final diagnosis of bacterial pneumonia while any other diagnosis with sepsis was categorized as sepsis (Supplementary Table 1). Our reasoning for this categorization was that bacterial pneumonia and sepsis were the only two diagnoses expected a priori to increase PCT levels; therefore, combining both was acceptable, but sepsis needed to be separated from the other five diagnoses.
Statistical Analysis
Data were compiled with Excel (Microsoft). Statistical analysis was performed using GraphPad Prism Version 8 (GraphPad Software). PCT measures were presented on a logarithmic scale. Comparisons of two medians were performed using the Mann-Whitney test. Comparisons of three or more medians were analyzed using a Kruskal-Wallis test followed by a Dunn’s multiple comparisons test. Logistic regression was performed using JMP version 15 (SAS Institute). P < .05 was considered significant.
Results
Demographics of Patients and Sample Collection Sources Included in the Study
The patient demographics, admission units, and sample sources collected in the study are summarized in Table 1. A total of 399 patients were included in the study with a total of 412 eligible instances of PCT testing. There was a similar distribution of males and females in the study (females = 208, males = 191). The distribution according to race was 69.2% white, 18.8% black, and 12.0% other. The mean age was similar for females and males, with an overall mean age of 66.2 years. The youngest patient included in the study was 8 months old and the oldest patient in the study was 101 years. Patients were distributed among the emergency department (26.2%), intensive care unit (32.5%), and inpatient facilities (40.5%). Most respiratory cultures came from sputa (85.7%) followed by endotracheal aspirates (13.8%).
Table 1.
Patient Demographics
| Characteristic | No. (%) | Median Agea (95% CI), y |
|---|---|---|
| Sexb | ||
| Female | 208 (52.1) | 70.0 (65.5-72.1) |
| Male | 191 (47.9) | 66.8 (62.1-69.4) |
| Raceb | ||
| Black | 75 (18.8) | |
| Other | 48 (12.0) | |
| White | 276 (69.2) | |
| Patient locationa | ||
| Emergency department | 108 (26.2) | |
| Intensive care unit | 134 (32.5) | |
| Inpatient | 167 (40.5) | |
| Outpatient | 3 (0.7) | |
| Respiratory culture sourcea | ||
| Bronchial specimen | 2 (0.5) | |
| Tracheal aspirate | 57 (13.8) | |
| Sputum | 353 (85.7) |
CI, confidence interval.
aCalculated from total number of procalcitonin results.
bCalculated from total number of patients.
PCT in Patients With Suspected LRTIs
To investigate PCT elevation in patients with suspected LRTIs, we compared median PCT levels among the final diagnoses of pneumonia (bacterial), respiratory infection (viral), pneumonia (organism unspecified), respiratory infection (other), sepsis, other infection, and no infection in our study population Table 2. Patients who were not diagnosed with any type of infection represented the largest group (26% of total) and had one of the lowest PCT medians at 0.08 ng/mL (95% confidence interval [CI], 0.05-0.19 ng/mL). Patients with viral respiratory infections, other respiratory infections, and other infections also had low PCT medians of 0.10 ng/mL (95% CI, 0.06-0.21 ng/mL), 0.05 ng/mL (95% CI, 0.05-0.15 ng/mL), and 0.09 ng/mL (95% CI, 0.05-0.34 ng/mL), respectively. All of these medians were significantly different from the PCT median for patients with a final diagnosis of bacterial pneumonia. They were also consistent with published cutoffs for PCT-guided antibiotic therapy in which bacterial infection is deemed highly unlikely at PCT levels less than 0.1 ng/mL.19 The median PCT for patients diagnosed with bacterial pneumonia was 0.68 ng/mL (95% CI, 0.29-1.09 ng/mL), which was also consistent with the interpretation of highly likely bacterial infection at levels more than 0.5 ng/mL.19 However, this group included patients who were also septic. The median PCT for patients diagnosed with bacterial pneumonia without sepsis was 0.29 ng/mL (95% CI, 0.10-0.68 ng/mL), which still meets the cutoff of 0.25 ng/mL for likely bacterial infection.19 The median PCT for patients diagnosed with sepsis excluding those with bacterial pneumonia was 0.41 ng/mL (95% CI, 0.23-0.75 ng/mL), and this was not significantly different from the median PCT level for the bacterial pneumonia group.
Table 2.
Procalcitonin Levels Across Final Diagnoses in Study Population
| Diagnosis | PCT Median (95% CI), ng/mL | No. (%) |
|---|---|---|
| Pneumonia (bacterial) | 0.68 (0.29-1.09) | 65 (15.8) |
| Respiratory infection (viral) | 0.10a (0.06-0.21) | 34 (8.3) |
| Pneumonia (NOS) | 0.19a (0.11-0.29) | 68 (16.5) |
| Respiratory infection (other) | 0.05b (0.05-0.15) | 33 (8.0) |
| Sepsis | 0.41 (0.23-0.75) | 81 (19.7) |
| Other infection | 0.09a (0.05-0.34) | 23 (5.6) |
| No infection | 0.08b (0.05-0.19) | 108 (26.2) |
CI, confidence interval; NOS, organism not specified; PCT, procalcitonin.
a P < .01 in a Kruskal-Wallis test vs bacterial pneumonia.
b P < .0001 in a Kruskal-Wallis test vs bacterial pneumonia.
PCT and Other Workup Suggestive of LRTIs
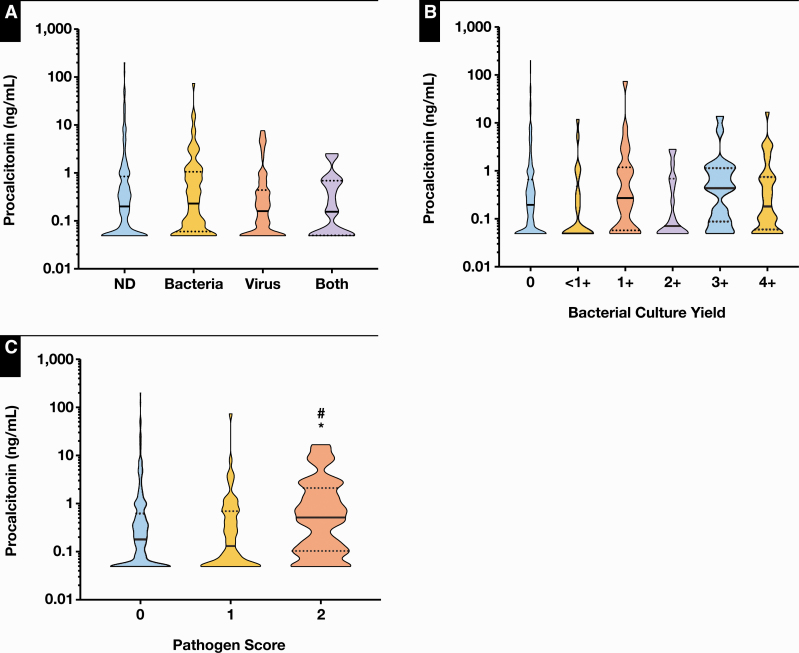
We then determined the association between PCT and other diagnostic testing commonly ordered in the workup of LRTIs. First, we assessed the relationship between PCT levels and respiratory pathogens detected by bacterial culture and/or respiratory virus DFA/PCR. Median PCT results were not significantly different among patients (1) negative for the presence of any pathogens (n = 226), (2) positive for the presence of bacteria only (n = 81), (3) positive for the presence of virus only (n = 87), or (4) positive for the presence of both bacteria and viruses (n = 18; P = .506; Figure 2A). Median PCT results were also not significantly different when stratified according to culture quantification results from 0 (n = 290), less than 1+ (n = 22), 1+ (n = 26), 2+ (n = 19), 3+ (n = 22), to 4+ (n = 33; P = .119; Figure 2B). Interestingly, when patients were stratified by their pathogen score, which was calculated based on the presence or absence of normal microbiota as well as specific bacterial pathogens Figure 2C, median PCT results were significantly higher in the patient group with the highest pathogen score of 2 (n = 40) compared with those with the lower score of 1 (n = 62) or 0 (n = 310; P < .05). The association between PCT levels and organism quantification as well as pathogen score demonstrated a similar trend when we limited the analysis to patients with bacterial isolates only (Supplementary Figure 2). This suggests that PCT levels correlate with recovery of bacteria in culture that may be causative agents of LRTI.
Figure 2.
Association between procalcitonin (PCT) results and respiratory isolates. A, Comparison of median PCT levels in patients in whom no pathogens were detected (0.20 ng/mL; 95% confidence interval [CI], 0.14-0.29 ng/mL), only bacteria were detected (0.23 ng/mL; 95% CI, 0.10-0.44 ng/mL), only viruses were detected (0.16 ng/mL; 95% CI, 0.09-0.25 ng/mL), and both types detected (0.16 ng/mL; 95% CI, 0.05-0.68 ng/mL) revealed no significant differences. B, Comparison of median PCT levels in patients in whom bacterial culture yielded 0 (0.20 ng/mL; 95% CI, 0.14-0.25 ng/mL), less than 1+ (0.05 ng/mL; 95% CI, 0.05-0.37 ng/mL), 1+ (0.27 ng/mL; 95% CI, 0.08-0.97 ng/mL), 2+ (0.07 ng/mL; 95% CI, 0.05-0.68 ng/mL), 3+ (0.43 ng/mL; 95% CI, 0.09-1.09 ng/mL), and 4+ (0.18 ng/mL; 95% CI, 0.10-0.67 ng/mL) organisms revealed no significant differences. C, Comparison of PCT levels stratified according to pathogen score revealed that patients with a pathogen score of 2 (normal microbiota or Streptococcus pneumonia or Legionella urine antigen positive) had median PCT levels (0.51 ng/mL; 95% CI, 0.16-1.09 ng/mL) that were significantly higher than both those with pathogen score of 1 (normal microbiota) (0.13 ng/mL; 95% CI, 0.06-0.31 ng/mL) and 0 (<6 colony-forming units of a particular organism with ≥1 normal microbiota) (0.18 ng/mL; 95% CI, 0.13-0.24 ng/mL). Black bars in the violin plots represent the median. Dotted black lines represent the upper and lower quartiles. *P < .05 compared with group 0. #P < .05 compared with group 1. ND, none detected.
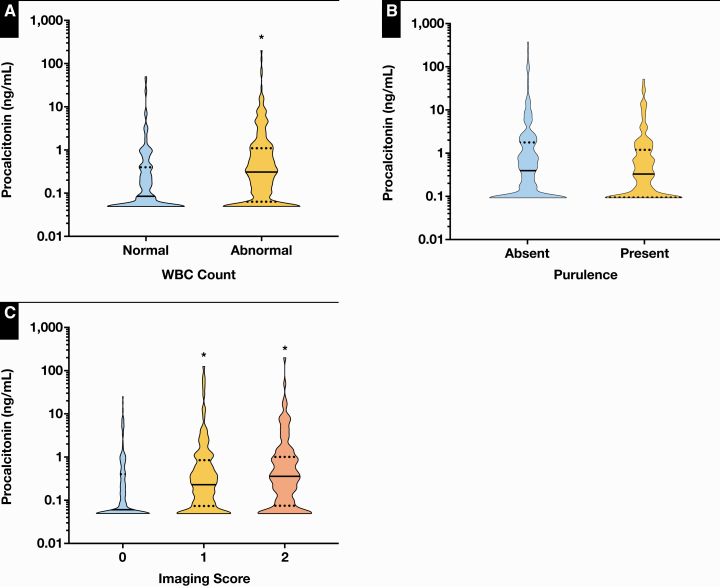
We next assessed the relationship between PCT levels and the results of WBC count, Gram stains, and chest imaging. Leukopenia or leukocytosis was associated with a significantly higher median PCT level (n = 229) compared with WBC counts within the reference interval (n = 183; P < .0001; Figure 3A), but there were no significant differences between purulent (n = 198) vs nonpurulent Gram stains (n = 214; P = .698; Figure 3B). Imaging results that could not rule out (n = 137) and those that were consistent with bacterial pneumonia (n = 125) were significantly associated with higher median PCT levels compared with imaging results that showed no evidence for bacterial pneumonia (n = 150; P < .0001, Figure 3C).
Figure 3.
Association between procalcitonin (PCT) results and other diagnostic workup for lower respiratory tract infections (LRTIs). A, Median PCT levels were significantly higher in patients with abnormal (0.31 ng/mL; 95% confidence interval [CI], 0.23-0.40 ng/mL) vs those with normal WBC count (0.08 ng/mL; 95% CI, 0.07-0.14 ng/mL). B, Median PCT levels were not significantly different in patients with purulent (0.18 ng/mL; 95% CI, 0.12-0.25 ng/mL) vs those with nonpurulent Gram stains (0.21 ng/mL; 95% CI, 0.13-0.31 ng/mL). C, Median PCT levels were significantly higher in patients in whom bacterial pneumonia could not be ruled out (0.25 ng/mL; 95% CI, 0.16-0.35 ng/mL) or was confirmed (0.33 ng/mL; 95% CI, 0.23-0.48 ng/mL) compared with those with no evidence for bacterial pneumonia (0.06 ng/mL; 95% CI, 0.05-0.10 ng/mL) by chest imaging (0, no evidence of pneumonia; 1, cannot rule out bacterial pneumonia; 2, consistent with bacterial pneumonia). Black bars in the violin plots represent the median. Dotted black lines represent the upper and lower quartiles. *P < .0001 compared with group 0.
PCT as a Predictor of Bacterial Pneumonia
PCT correlates with both a final diagnosis of and other workup for bacterial pneumonia. However, this does not necessarily mean that PCT adds valuable diagnostic information over and above what is already available to clinicians. We therefore used logistic regression to predict a final diagnosis of bacterial pneumonia based on abnormal/normal results from imaging studies, WBC count, Gram stain, and PCT analysis. Our binary outcome included bacterial pneumonia (with and without sepsis) vs respiratory infection (viral), pneumonia (organism unspecified), respiratory infection (other), other infection, and no infection combined. We excluded sepsis as a final diagnosis group from this comparison, as it is clearly associated with an elevated median PCT. Importantly, current guidelines do not recommend the use of PCT to initiate antibiotic treatment in patients with sepsis.8 Imaging studies, WBC count, and Gram stain results were categorized as described in the Materials and Methods section, and a PCT of 0.25 ng/mL was used as the diagnostic cutoff for likely vs unlikely bacterial pneumonia.5
Logistic regression revealed a significant association between a final diagnosis of bacterial pneumonia and each of the following variables in a descending order of significance: evidence of bacterial pneumonia by imaging, PCT more than 0.25 ng/mL, and purulent Gram stain Table 3. While the PCT association was significant, the magnitude of the effect was small, as revealed by the receiver operating characteristic curves (Table 3). The area under the curve (AUC) for a diagnostic workup that only included imaging, WBCs, and Gram stain was 0.748, and the addition of PCT increased it to 0.778. In a diagnostic workup that included PCT, WBCs, and Gram stain but excluded imaging, the AUC was 0.705. On the other hand, the AUC for imaging alone was 0.721, suggesting that not much additional diagnostic discrimination is offered by PCT or Gram stain. In Supplementary Table 2, we provide additional diagnostic workup models for comparison.
Table 3.
Association Between Diagnostic Workup and a Diagnosis of Bacterial Pneumonia
| Workup (PCT + Imaging + WBC + GS) | L-R χ2 | Prob > χ2a | ROC AUC |
|---|---|---|---|
| PCT >0.25 ng/mL | 11.222 | <0.001 | 0.778 |
| Bacterial pneumonia by imaging | 24.792 | <0.0001 | |
| Abnormal WBC count | 0.026 | 0.872 | |
| Purulent GS | 4.761 | 0.029 | |
| Workup (imaging + WBC + GS) | |||
| Bacterial pneumonia by imaging | 34.259 | <0.0001 | 0.748 |
| Abnormal WBC count | 0.857 | 0.355 | |
| Purulent GS | 4.006 | 0.045 | |
| Workup (PCT + WBC + GS) | |||
| Bacterial pneumonia by imaging | 20.689 | <0.0001 | 0.705 |
| Abnormal WBC count | 0.580 | 0.446 | |
| Purulent GS | 3.934 | 0.047 | |
| Workup (imaging only) | |||
| Imaging | 18.590 | <0.0001 | 0.721 |
AUC, area under the curve; GS, Gram stain; L-R, likelihood ratio; PCT, procalcitonin; ROC, receiver operating characteristic.
aProbability of a greater χ2 value by chance alone.
PCT and All-Cause Mortality
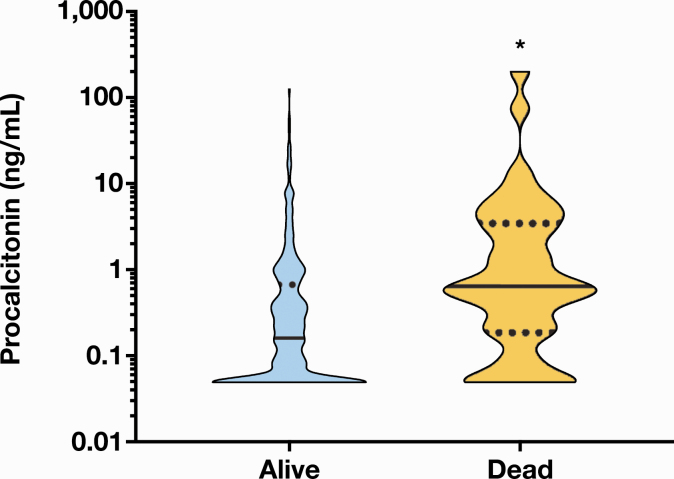
Finally, we looked at PCT levels and all-cause mortality within 1 month of test order. Of the 399 patients in the final analysis, there were no data on whether 30 of the patients were alive or dead. None of the 11 patients with multiple admissions were dead, and their PCT results for each separate admission were included in the median calculation. Of the 369 patients in the analysis, 40 had died within 1 month and had median PCT levels of 0.64 ng/mL (95% CI, 0.40-1.51 ng/mL), which were significantly higher than median PCT levels at 0.16 ng/mL (95% CI, 0.11-0.22 ng/mL) in patients who were still alive (n = 342; P < .0001; Figure 4).
Figure 4.
Association between procalcitonin (PCT) results and all-cause mortality. Median PCT levels were significantly higher in patients who had died within 1 month of test order (0.64 ng/mL; 95% confidence interval [CI], 0.40-1.51 ng/mL) vs patients who were still alive (0.16 ng/mL; 95% CI, 0.11-0.22 ng/mL). Black bars in the violin plots represent the median. Dotted black lines represent the upper and lower quartiles. *P < .0001.
Discussion
We performed a retrospective analysis of patients presenting with suspected LRTIs at our institution between January 1, 2015, and December 31, 2015, who had both lower respiratory tract cultures and respiratory virus DFA/PCR testing within 1 day of initial procalcitonin testing. All of these patients had chest imaging and WBC counts performed within 1 day of procalcitonin testing as well. We chose the 1-day limit to approximate as best as possible the initial diagnostic workup for a suspected bacterial pneumonia. Our main finding is that despite significant elevations in median PCT levels in patients with a final diagnosis of bacterial pneumonia as well as those with elevated WBCs or abnormal imaging consistent with bacterial pneumonia, PCT did not add much discriminatory information to the laboratory and imaging data routinely available to clinicians.
PCT cutoffs for guiding antibiotic therapy in the setting of LRTIs have generally been adapted from early prospective studies.5 PCT levels in different LRTI etiologies have been less studied and could shed light on the discriminatory ability of PCT for bacterial pneumonia. We found no significant differences in median PCT levels in patients with final diagnoses that did not include bacterial pneumonia or sepsis. In patients with no infections or nonbacterial respiratory infections, these medians and the upper limits of the 95% CIs were less than 0.25 ng/mL, which is the generally accepted cutoff for unlikely bacterial infections. These medians were significantly different from those seen with bacterial pneumonia and sepsis. Both the PCT median and the lower limit of the 95% CI were more than 0.25 ng/mL in the bacterial pneumonia group; however, this group included patients who were also diagnosed with sepsis. In the bacterial pneumonia–only group, the median PCT level dropped to 0.29 ng/mL, and the lower limit of the 95% CI was 0.1 ng/mL, indicating that patients with bacterial pneumonia may have normal levels of PCT.
Other groups have assessed the association between median PCT levels and detected pathogens in patients suspected of having LRTIs.20 They found that PCT was significantly elevated with bacterial pathogens compared with viral pathogens. In our data set, there were no significant differences in median PCT levels across all comparison groups, including no pathogens, bacterial pathogens, viral pathogens, and codetected bacterial and viral pathogens. In fact, the median PCT levels were remarkably similar at around 0.20 ng/mL. Comparisons based on pathogen semiquantification by culture also did not reveal any significant differences among median PCT levels that did not even display an increasing trend correlating with higher quantification. Furthermore, median PCT levels were essentially unchanged between patients with purulent and nonpurulent Gram stains. Only a relatively subjective classification of isolated bacterial organisms based on their potential pathogenicity and relative abundance yielded a significant elevation of median PCT levels with higher pathogenicity scores. The presence of a normal microbiome in the respiratory tract makes interpretation of respiratory cultures difficult. Final clinical interpretation relies upon consideration of patient factors as well as the relative abundance of potential pathogens recovered compared with the underlying microbiota. While some organisms are considered true pathogens and always treated if detected, most causative agents of bacterial pneumonia require further clinical interpretation. Nonetheless, we reason that our classification of pathogenicity approximates a clinical interpretation, and the higher median PCT levels with higher pathogenicity scores are in line with the observation that median PCT levels are significantly elevated in patients with final diagnoses of bacterial pneumonia.
Median procalcitonin levels were higher in patients with abnormal WBC counts and chest imaging suggestive of bacterial pneumonia, and the significance was robust. This is not surprising given that PCT is a marker of bacterial infection. Moreover, while bacterial pneumonia is often diagnosed without them, chest radiographic criteria are generally used for defining community-acquired pneumonia.18 Just because PCT levels are elevated in patients with radiographic evidence of or those with a diagnosis of bacterial pneumonia does not necessarily mean that PCT adds valuable diagnostic information over and above what is available to clinicians. In our logistic regression model, we found that purulent abnormal chest imaging, PCT more than 0.25 ng/mL, and purulent Gram stains were significantly associated with distinguishing bacterial pneumonia from diagnoses that included nonbacterial respiratory infections, other infections, or no infections. The rationale for combining all the other diagnoses and excluding sepsis is that these would be conditions that would not in principle require antibiotic therapy. Purulent Gram stains were significantly associated with a diagnosis of bacterial pneumonia, but the P value was more than .01, suggesting a weaker relationship compared with chest imaging, which had a P value less than .0001. PCT more than 0.25 ng/mL was clearly significantly associated with a diagnosis of bacterial pneumonia with a P value less than .001. However, adding PCT to the diagnostic model improved the AUC from 0.748 to 0.778, suggesting that the magnitude of the effect is small. It should be noted that neither model accounts for hidden variables that could correlate with the final diagnosis of bacterial pneumonia, most notably the clinical signs and symptoms. It is also true that no constellation of clinical findings is capable of accurately ruling in bacterial pneumonia.21 Our findings do not suggest that a combination of chest imaging and Gram stains is sufficient for discriminating between bacterial and nonbacterial pneumonia, but PCT adds modest information to what clinicians use in practice for patients with suspected LRTIs. Despite a recent meta-analysis of procalcitonin-guided therapy in patients with acute respiratory infections9 detailing evidence for initiating or withholding antibiotic therapy under certain conditions based on initial PCT levels, others argue against this approach.22,23
Our primary aim did not include PCT as a predictor of mortality in patients who were suspected of having LRTIs; therefore, we did not calculate relative risks for specific PCT cutoffs. However, we found that median PCT levels were significantly higher in patients who had died within 1 month. This is in agreement with previous studies showing that both PCT elevation and nonclearance are associated with an increased risk of all-cause mortality in patients under different clinical settings.24-27
This study has several limitations that constrain the interpretation of the role of PCT testing in patients with suspected LRTIs. First, this is a retrospective analysis of patients who had to meet the combined inclusion criteria of PCT testing, chest imaging, lower respiratory tract cultures, and respiratory virus testing within 1 day of the initial PCT result. This population may not represent the approach to LRTI workup in the community or at other hospitals. Furthermore, it is not possible to separate with certainty the results of this diagnostic workup from the initial clinical judgment that, with the clinical course, likely contributed to the final diagnoses in these patients. Furthermore, these diagnoses were extracted from electronic health records and were not independently adjudicated. It remains possible, however, to draw conclusions regarding the performance and utility of PCT testing in the diagnosis of bacterial pneumonia when the major workup including microbial testing and chest imaging is also performed. While final diagnosis misclassification may underestimate the ability of PCT to predict bacterial pneumonia, our study reflects performance in real life as opposed to ideal situations. Finally, no conclusions can be drawn from our study regarding the utility of PCT testing for the initiation or discontinuation of antibiotic therapy since that data were not obtained and would be difficult to extract in a retrospective analysis without an exhaustive chart review. Furthermore, serial PCT testing was not assessed in this study. Thus, our data cannot exclude that a low or decreasing PCT level may add one more piece of information to clinicians considering discontinuing empiric antibiotics.
In conclusion, our retrospective analysis demonstrates that PCT correlates with other markers of bacterial pneumonia; however, the ability of health care providers to detect this disease using PCT is modest and not greatly improved over and above information provided by chest imaging. Recent studies in the United States have specifically shown the inability of PCT to reliably distinguish between bacterial and viral pneumonia as opposed to its better-documented ability to distinguish between upper respiratory tract infections and LRTIs.20,22 Furthermore, others have shown that PCT-guided management did not result in reduction of antibiotic utilization15 or questioned its utility in settings with improved antibiotic stewardship.28 The lack of definitive cutoffs for the detection of bacterial pneumonia, coupled with the unclear results on antibiotic utilization, has led the American Thoracic Society and Infectious Diseases Society of America to recommend against the use of initial PCT levels in the management of clinically suspected community-acquired pneumonia.18 Our results are in line with a body of evidence in the United States cautioning against the wide implementation of PCT in clinical laboratories in the absence of established evidence-based protocols.29
Supplementary Material
Dr Ayala-Lopez received funding from the National Institutes of Health (NIH-2T32HL007974-16).
References
- 1. Grijalva CG, Nuorti JP, Griffin MR. Antibiotic prescription rates for acute respiratory tract infections in US ambulatory settings. JAMA. 2009;302:758-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fleming-Dutra KE, Hersh AL, Shapiro DJ, et al. Prevalence of inappropriate antibiotic prescriptions among US ambulatory care visits, 2010-2011. JAMA. 2016;315:1864-1873. [DOI] [PubMed] [Google Scholar]
- 3. Kraus EM, Pelzl S, Szecsenyi J, et al. Antibiotic prescribing for acute lower respiratory tract infections (LRTI)—guideline adherence in the German primary care setting: an analysis of routine data. PLoS One. 2017;12:e0174584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Silverman M, Povitz M, Sontrop JM, et al. Antibiotic prescribing for nonbacterial acute upper respiratory infections in elderly persons. Ann Intern Med. 2017;166:765-774. [DOI] [PubMed] [Google Scholar]
- 5. Schuetz P, Christ-Crain M, Thomann R, et al. ; ProHOSP Study Group . Effect of procalcitonin-based guidelines vs standard guidelines on antibiotic use in lower respiratory tract infections: the ProHOSP randomized controlled trial. JAMA. 2009;302:1059-1066. [DOI] [PubMed] [Google Scholar]
- 6. Bouadma L, Luyt CE, Tubach F, et al. ; PRORATA Trial Group . Use of procalcitonin to reduce patients’ exposure to antibiotics in intensive care units (PRORATA trial): a multicentre randomised controlled trial. Lancet. 2010;375:463-474. [DOI] [PubMed] [Google Scholar]
- 7. de Jong E, van Oers JA, Beishuizen A, et al. Efficacy and safety of procalcitonin guidance in reducing the duration of antibiotic treatment in critically ill patients: a randomised, controlled, open-label trial. Lancet Infect Dis. 2016;16:819-827. [DOI] [PubMed] [Google Scholar]
- 8. Rhee C. Using procalcitonin to guide antibiotic therapy. Open Forum Infect Dis. 2017;4:ofw249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schuetz P, Wirz Y, Sager R, et al. Procalcitonin to initiate or discontinue antibiotics in acute respiratory tract infections. Cochrane Database Syst Rev. 2017;10:CD007498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Becker KL, Nylén ES, White JC, et al. Clinical review 167: procalcitonin and the calcitonin gene family of peptides in inflammation, infection, and sepsis: a journey from calcitonin back to its precursors. J Clin Endocrinol Metab. 2004;89:1512-1525. [DOI] [PubMed] [Google Scholar]
- 11. Dandona P, Nix D, Wilson MF, et al. Procalcitonin increase after endotoxin injection in normal subjects. J Clin Endocrinol Metab. 1994;79:1605-1608. [DOI] [PubMed] [Google Scholar]
- 12. Assicot M, Gendrel D, Carsin H, et al. High serum procalcitonin concentrations in patients with sepsis and infection. Lancet. 1993;341:515-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Maruna P, Nedelnikova K, Gurlich R. Physiology and genetics of procalcitonin. Physiol Res. 2000;49(suppl 1):S57-S61. [PubMed] [Google Scholar]
- 14. Albrich WC, Dusemund F, Bucher B, et al. ; ProREAL Study Team . Effectiveness and safety of procalcitonin-guided antibiotic therapy in lower respiratory tract infections in “real life”: an international, multicenter poststudy survey (ProREAL). Arch Intern Med. 2012;172:715-722. [DOI] [PubMed] [Google Scholar]
- 15. Huang DT, Yealy DM, Filbin MR, et al. ; ProACT Investigators . Procalcitonin-guided use of antibiotics for lower respiratory tract infection. N Engl J Med. 2018;379:236-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.US Food and Drug Administration. FDA clears test to help manage antibiotic treatment for lower respiratory tract infections and sepsis. FDA News Release 2017. https://www.fda.gov/news-events/press-announcements/fda-clears-test-help-manage-antibiotic-treatment-lower-respiratory-tract-infections-and-sepsis#:~:text=The%20U.S.%20Food%20and%20Drug,stopped%20in%20patients%20with%20sepsis. Accessed June 18, 2020.
- 17. Landry ML, Ferguson D. Cytospin-enhanced immunofluorescence and impact of sample quality on detection of novel swine origin (H1N1) influenza virus. J Clin Microbiol. 2010;48:957-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Metlay JP, Waterer GW, Long AC, et al. Diagnosis and treatment of adults with community-acquired pneumonia: an official clinical practice guideline of the American Thoracic Society and Infectious Diseases Society of America. Am J Respir Crit Care Med. 2019;200:e45-e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schuetz P, Christ-Crain M, Wolbers M, et al. ; ProHOSP Study Group . Procalcitonin guided antibiotic therapy and hospitalization in patients with lower respiratory tract infections: a prospective, multicenter, randomized controlled trial. BMC Health Serv Res. 2007;7:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Self WH, Balk RA, Grijalva CG, et al. Procalcitonin as a marker of etiology in adults hospitalized with community-acquired pneumonia. Clin Infect Dis. 2017;65:183-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Metlay JP, Kapoor WN, Fine MJ. Does this patient have community-acquired pneumonia? Diagnosing pneumonia by history and physical examination. JAMA. 1997;278:1440-1445. [PubMed] [Google Scholar]
- 22. Kamat IS, Ramachandran V, Eswaran H, et al. Procalcitonin to distinguish viral from bacterial pneumonia: a systematic review and meta-analysis. Clin Infect Dis. 2020;70:538-542. [DOI] [PubMed] [Google Scholar]
- 23. Kamat IS, Ramachandran V, Eswaran H, et al. Low procalcitonin, community acquired pneumonia, and antibiotic therapy. Lancet Infect Dis. 2018;18:496-497. [DOI] [PubMed] [Google Scholar]
- 24. Liu D, Su L, Han G, et al. Prognostic value of procalcitonin in adult patients with sepsis: a systematic review and meta-analysis. PLoS One. 2015;10:e0129450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bloos F, Marshall JC, Dellinger RP, et al. Multinational, observational study of procalcitonin in ICU patients with pneumonia requiring mechanical ventilation: a multicenter observational study. Crit Care. 2011;15:R88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sager R, Wirz Y, Amin D, et al. Are admission procalcitonin levels universal mortality predictors across different medical emergency patient populations? Results from the multi-national, prospective, observational TRIAGE study. Clin Chem Lab Med. 2017;55:1873-1880. [DOI] [PubMed] [Google Scholar]
- 27. Schuetz P, Birkhahn R, Sherwin R, et al. Serial procalcitonin predicts mortality in severe sepsis patients: results from the Multicenter Procalcitonin MOnitoring SEpsis (MOSES) Study. Crit Care Med. 2017;45:781-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hamade B, Huang DT. Procalcitonin: where are we now? Crit Care Clin. 2020;36:23-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. American Society for Clinical Pathology. Don’t perform Procalcitonin testing without an established, evidence-based protocol. 2018. http://www.choosingwisely.org/clinician-lists/ascp-procalcitonin-testing/. Accessed June 18, 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.