Abstract
Background
Two primary histologic subtypes, superficial spreading melanoma (SSM) and nodular melanoma (NM), comprise the majority of all cutaneous melanomas. NM is associated with worse outcomes, which have been attributed to increased thickness at presentation, and it is widely expected that NM and SSM would exhibit similar behavior once metastasized. Herein, we tested the hypothesis that primary histologic subtype is an independent predictor of survival and may impact response to treatment in the metastatic setting.
Methods
We examined the most recent Surveillance, Epidemiology, and End Results (SEER) cohort (n = 118 508) and the New York University (NYU) cohort (n = 1621) with available protocol-driven follow-up. Outcomes specified by primary histology were studied in both the primary and metastatic settings with respect to BRAF-targeted therapy and immunotherapy. We characterized known driver mutations and examined a 140-gene panel in a subset of NM and SSM cases using next-generation sequencing. All statistical tests were two-sided.
Results
NM was an independent risk factor for death in both the SEER (hazard ratio [HR] = 1.55, 95% confidence interval [CI] = 1.41 to 1.70, P < .001) and NYU (HR = 1.47, 95% CI = 1.05, 2.07, P = .03) cohorts, controlling for thickness, ulceration, stage, and other variables. In the metastatic setting, NM remained an independent risk factor for death upon treatment with BRAF-targeted therapy (HR = 3.33, 95% CI = 1.06 to 10.47, P = .04) but showed no statistically significant difference with immune checkpoint inhibition. NM was associated with a higher rate of NRAS mutation (P < .001), and high-throughput sequencing revealed NM-specific genomic alterations in NOTCH4, ANK3, and ZNF560, which were independently validated.
Conclusions
Our data reveal distinct clinical and biological differences between NM and SSM that support revisiting the prognostic and predictive impact of primary histology subtype in the management of cutaneous melanoma.
The two most common histologic subtypes of newly diagnosed melanoma are superficial spreading melanoma (SSM), which comprises approximately 70% of cases, and nodular melanoma (NM), which accounts for 20% (1,2). Histologically, SSM is characterized by a predominantly epidermal component with slow, horizontal growth. In contrast, NM is usually thicker than SSM due to the lack of clinically significant intraepidermal involvement and is characterized by rapid, vertical growth (3). These two subtypes are representative descriptions of melanoma progression, which continues to be perceived as a stepwise process that begins with normal melanocytes at the dermal-epidermal junction acquiring genetic mutations that lead to radial growth phase (RGP) melanoma to vertical growth phase (VGP) melanoma and, eventually, metastasis (4). However, clinical, pathologic, and epidemiologic evidence from our group and others suggest that SSM and NM may progress independently (3,5–12), arguing against the linear progression model.
Unlike other solid malignancies, such as lung or breast carcinomas (13–17), primary histologic subtypes of melanoma are frequently unreported in pathology reports (10,18). Furthermore, histologic subtype has not been included in the American Joint Committee on Cancer (AJCC) staging system (19), as it has been assumed (20) that any increased risk associated with nodular histology is confounded by increases in thickness and ulceration. This assumption is rooted primarily in a multivariable analysis of a small, single-institution cohort from the 1970s (21). More recently, The Cancer Genome Atlas (TCGA) has provided melanoma histologic subtype and mutational data, but only for a minority of cases (31/478) (17). Additionally, most of the available data on the melanoma histologic subtype–specific mutational landscape pertains mainly to a small set of known oncogenes (22–24). Remarkably, there is no published report that we are aware of rigorously examining the role primary histologic subtype has on prognosis or the impact it has on response of metastatic melanoma to currently available targeted and immunotherapies. This may be due to the prevalent belief that melanomas of different histologic subtypes converge in their biologic behavior once they metastasize.
Here, we tested the hypothesis that primary histologic subtype independently predicts recurrence and survival in cutaneous melanoma and may affect the response to systemic therapies in the metastatic setting.
Methods
Patient Cohorts
Surveillance, Epidemiology, and End Results Cohort
Surveillance, Epidemiology, and End Results (SEER) is a US population-based database sponsored by the National Cancer Institute (NCI) that records cancer statistics among specific demographic registries representing 28% of the US population (25). We queried the most recent SEER database for stage I–III melanoma patients diagnosed from 1973 to 2012. SEER*Stat, version 8.2.1 (NCI, Bethesda, MD), was used to identify all SSM and NM cases based on the International Classification of Diseases for Oncology, 3rd edition, codes (M8720–8790). Available patient information includes age, sex, thickness, ulceration, and stage at diagnosis, but not tumor-infiltrating lymphocytes (TILs), mitotic index (MI), or driver mutational status.
NYU Cohort
Melanoma patients presenting to NYU Langone Health for diagnosis and/or treatment of melanoma are enrolled in the NYU Interdisciplinary Melanoma Cooperative Group’s Institutional Review Board–approved clinicopathological database and biorepository, and they are actively followed up on a protocol-driven schedule. This protocol enables the collection of prospective follow-up clinical, pathological, and demographic data. All patients provided written informed consent.
Clinical and Pathologic Data
Among the SEER cohort of NM and SSM, clinicopathologic variables included age, sex, thickness, ulceration, AJCC stage, and survival status. Among the patients enrolled in the NYU cohort, the primary histologic subtype was determined by central review by an attending pathologist (FD) in accordance with well-established diagnostic criteria (26). In brief, as per World Health Organization classification, SSM was defined by a proliferation of atypical melanocytes in which the intraepidermal component appears as irregular nests or single units at and above the dermal-epidermal junction and the invasive dermal component appears as variably sized nests and single cells that fail to mature. NM was defined by a cohesive nodule or small nests of tumor cells in the dermis that have pushing or expansile pattern of growth (vertical growth phase) with no or limited intraepidermal tumor cells confined to three epidermal rete ridges beyond the dermal component. Clinical data included age, sex, thickness, ulceration, mitotic index (MI), AJCC 7th edition staging (7th edition was the most up to date at the time of initial data collection), tumor-infiltrating lymphocytes (graded as previously reported [27]), primary anatomic location, dates of recurrence and follow-up, and status. Long-term follow-up data in the NYU database were used to calculate recurrence-free survival (RFS) and melanoma-specific survival (MSS).
Genomic Sequencing
Sanger sequencing for known BRAF and NRAS driver mutations was performed on a large set of 808 NM and SSM tumors, as previously described (28,29). To further explore the molecular differences between NM and SSM, we designed a panel of 140 genes of biologic interest (see Supplementary Table 1, available online, for a list of genes), which included known melanoma drivers and exploratory genes. High-throughput targeted hybrid-capture sequencing was performed on a subset cohort of 65 NM and SSM patient-derived tissues from the NYU cohort utilizing the Illumina platform. Single nucleotide variants (SNVs) were identified and filtered against each matched germline, generating tumor-specific lists of SNVs. A threshold of 15% allelic frequency was utilized to identify biologically significant SNVs. Each gene was examined for SNVs within protein-coding regions and deemed either mutant or wild-type for each gene of interest. A subset of 10 genes showing differences between NM and SSM was further validated in an independent NYU cohort (n = 34).
Systemic Treatment Response
We analyzed systemic response to treatment among patients in the NYU cohort who developed metastatic disease during active follow-up in the new era of targeted and immunotherapy. Specifically, survival outcomes were assessed in patients treated with checkpoint blockade immunotherapy (n = 154) and BRAF-targeted therapy (n = 56), categorized by the histologic subtype of the primary tumor. Patients were evaluated for extent and duration of response to systemic therapy. Best responses were defined by the treating medical oncologists (AP, MW) based on interpretation of radiographic, laboratory, and clinical data documented in the medical records. Responses were graded as complete response (CR), partial response (PR), stable disease (SD) or mixed response, and progression of disease.
Statistical Methods
Descriptive statistics were performed on both the NYU and SEER data to characterize the presentation of each histologic subtype, and the chi-square test was used to compare patient cohorts. Kaplan-Meier curves and the log-rank test were utilized to compare survival between histologic subtypes. Multivariable Cox proportional hazard models were used to calculate adjusted hazard ratios (HRs) and 95 % confidence intervals (CIs). The proportional hazard assumption was checked using statistical tests (30) and graphical diagnostics based on the scaled Schoenfeld residuals. The adjusted prognostic factors in the multivariable models included melanoma stage at diagnosis, thickness, ulceration, and MI. All statistical tests were two-sided, and a P value of less than .05 was considered statistically significant.
Results
Impact of Primary Melanoma Histologic Subtype on Survival
SEER cohort data spanned 40 years, from 1973 to 2013. A total of 118 508 patients were identified, with 97 169 SSM and 21 399 NM cases. In the NYU cohort, a total of 1111 SSM and 510 NM patients were accrued from 2002 to 2016 (Table 1). The NYU cohort was characterized by a higher frequency of NM (31.5% in NYU vs 18.0% in SEER), which is expected at a tertiary referral center (12). Despite the differences in the histologic subtype and years of accrual, both SEER and NYU cohorts exhibit similar AJCC stage distributions for both NM and SSM.
Table 1.
Baseline characteristics of nodular melanoma and superficial spreading melanoma patients at the time of presentation and survival status among both SEER and NYU cohorts
| Characteristics | SEER cohort |
NYU cohort |
||||
|---|---|---|---|---|---|---|
| NM No. (%) | SSM No. (%) | P* | NM No. (%) | SSM No. (%) | P* | |
| Distribution | 21 399 (18.0) | 97 169 (82.0) | 510 (31.5) | 1111 (68.5) | ||
| Year of diagnosis | ||||||
| <2000 | 7369 (34.4) | 33 055 (34.0) | .02 | 20 (4.1) | 38 (3.4) | .85 |
| 2001–2010 | 10 247 (47.9) | 47 482 (48.9) | 336 (65.9) | 728 (65.5) | ||
| >2010 | 3783 (17.7) | 16 632 (17.1) | 154 (30.2) | 345 (31.1) | ||
| Age (SD), y | 62.1 (17.8) | 54.6 (16.7) | <.001 | 61.2 (17.2) | 57.3 (16.9) | <.001 |
| Sex | ||||||
| Female | 8333 (38.9) | 45 886 (47.2) | <.001 | 211 (41.4) | 496 (44.6) | .24 |
| Male | 13 066 (61.1) | 51 283 (52.8) | 299 (58.6) | 615 (55.4) | ||
| Thickness, mm | ||||||
| <1.01 | 1539 (15.3) | 36 057 (79.2) | <.001 | 36 (7.1) | 821 (73.9) | <.001 |
| 1.01–2.0 | 2359 (23.4) | 6409 (14.1) | 140 (27.5) | 219 (19.7) | ||
| 2.01–4.0 | 3150 (31.3) | 2241 (4.9) | 181 (35.5) | 61 (5.5) | ||
| >4.0 | 3023 (30.0) | 830 (1.8) | 153 (30.0) | 10 (0.9) | ||
| Thickness, median (IQR), mm | 2.6 (1.4–4.7) | 0.6 (0.4–0.9) | <.001 | 2.7 (1.7–4.5) | 0.6 (0.4–1.1) | <.001 |
| Ulceration | ||||||
| Absent | 5250 (52.4) | 41764 (92.5) | <.001 | 247 (48.7) | 1029 (93.0) | <.001 |
| Present | 4777 (47.6) | 3384 (7.5) | 260 (51.3) | 77 (7.0) | ||
| Mitotic index† | ||||||
| Absent | − | − | − | 40 (8.1) | 592 (53.8) | <.001 |
| Few | − | − | − | 111 (22.4) | 335 (30.5) | |
| Moderate | − | − | − | 130 (26.3) | 108 (9.8) | |
| Many | − | − | − | 214 (43.2) | 65 (5.9) | |
| AJCC stage | ||||||
| I | 2657 (27.5) | 38 947 (88.9) | <.001 | 116 (22.7) | 961 (86.5) | <.001 |
| II | 4730 (49.0) | 2943 (6.7) | 224 (43.9) | 89 (8.0) | ||
| III | 2270 (23.5) | 1916 (4.4) | 170 (33.3) | 61 (5.5) | ||
| Anatomic site | ||||||
| Axial | 6761 (31.8) | 37 955 (39.3) | <.001 | 197 (38.6) | 434 (39.1) | .56 |
| Extremity | 9617 (45.2) | 45 553 (47.1) | 236 (46.3) | 531 (47.8) | ||
| Head and neck | 4905 (23.0) | 13 129 (13.6) | 77 (15.1) | 146 (13.1) | ||
| TIL† | ||||||
| Absent | − | − | − | 110 (33.7) | 302 (37.2) | .047 |
| Nonbrisk | − | − | − | 98 (30.1) | 187 (23.0) | |
| Brisk | − | − | − | 118 (36.2) | 323 (39.8) | |
| Status | ||||||
| Alive | 10 708 (50.0) | 75 973 (78.2) | <.001 | 306 (60.0) | 956 (86.0) | <.001 |
| Died of melanoma | 5524 (25.8) | 5507 (5.7) | 160 (31.4) | 85 (7.7) | ||
| Died of other Cause | 5167 (24.1) | 15 689 (16.1) | 44 (8.6) | 70 (6.3) | ||
| Recurrence† | ||||||
| No | − | − | − | 268 (52.5) | 959 (86.3) | <.001 |
| Yes | − | − | − | 242 (47.5) | 152 (13.7) | |
Continuous variables were tested by two-sample t test, and categorical variables were tested by χ2 test. AJCC = American Joint Committee on Cancer; IQR = interquartile range; NM = nodular melanoma; SEER = Surveillance, Epidemiology, and End Results; SSM = superficial spreading melanoma; TIL = tumor-infiltrating lymphocyte.
Data for the SEER cohort were not available.
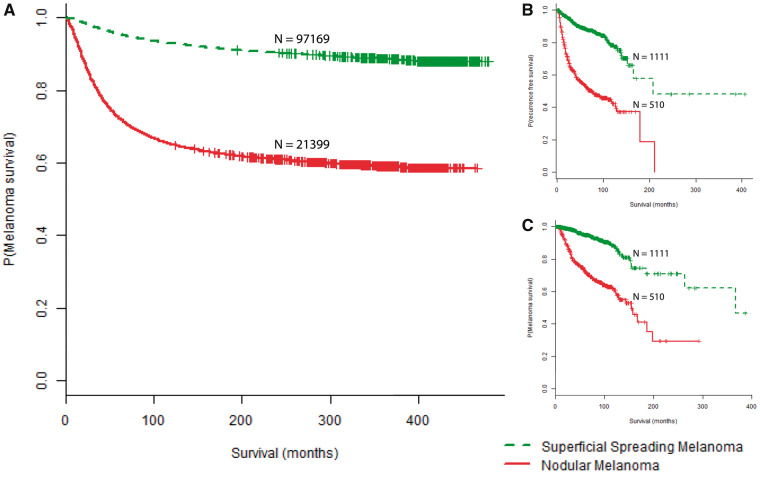
Follow-up time for NM was grossly comparable between SEER and NYU, with median follow-up durations of 68 and 73 months, respectively. We observed a longer median follow-up time for SSM in SEER (89 months) relative to SSM follow-up at NYU (69 months). In the SEER cohort, NM was a statistically significant risk factor for death from melanoma, with a hazard ratio (HR) of 6.01 (95% CI = 5.79 to 6.24, P < .001) (Table 2, Figure 1A). In the multivariable analysis, NM continued to be a statistically significant risk factor for death from melanoma, with a hazard ratio of 1.55 (95% CI = 1.41 to 1.70, P < .001) among the SEER cohort, while adjusting for thickness, ulceration, and AJCC stage at diagnosis (Table 2). The shorter survival of NM was subsequently recapitulated in the NYU cohort, with NM being statistically significantly associated with shorter RFS (HR = 4.63, 95% CI = 3.78 to 5.68, P < .001) (Table 2, Figure 1B) and MSS (HR = 4.77, 95% CI = 3.66 to 6.22, P < .001) (Table 2, Figure 1C). After controlling for thickness, ulceration, MI, and AJCC stage at diagnosis in the multivariable analysis, NM remained an independent risk factor for recurrence (HR = 1.45, 95% CI = 1.11 to 1.89, P = .01) and death from melanoma (HR = 1.47, 95% CI = 1.05 to 2.07, P = .03) (Table 2).
Table 2.
Univariate and multivariable Cox proportional hazard models for SEER (melanoma-specific survival) and NYU (recurrence-free survival and melanoma-specific survival)
| Variable | SEER cohort |
NYU cohort |
||||
|---|---|---|---|---|---|---|
| Melanoma-specific survival |
Melanoma-specific survival |
Recurrence-free survival |
||||
| HR (95% CI) | P* | HR (95% CI) | P* | HR (95% CI) | P* | |
| Univariate Cox proportional hazards model | ||||||
| NM vs SSM | 6.01 (5.79 to 6.24) | <.001 | 4.77 (3.66 to 6.22) | <.001 | 4.63 (3.78 to 5.68) | <.001 |
| Multivariable Cox proportional hazard model | ||||||
| NM vs SSM | 1.55 (1.41 to 1.70) | <.001 | 1.47 (1.05 to 2.07) | .03 | 1.45 (1.11 to 1.89) | .01 |
| Age at diagnosis† | 1.25 (1.22 to 1.28) | <.001 | 1.09 (1.00 to 1.18) | .04 | 1.08 (1.02 to 1.15) | .01 |
| Male | 1.37 (1.26 to 1.48) | <.001 | 1.30 (0.98 to 1.72) | .07 | 1.27 (1.02 to 1.57) | .03 |
| Year of diagnosis | 0.88 (0.81 to 0.95) | .00 | 0.89 (0.77 to 1.04) | .14 | 0.73 (0.66 to 0.81) | <.001 |
| Thickness | 1.16 (1.14 to 1.18) | <.001 | 1.04 (1.01 to 1.06) | .01 | 1.03 (1.00 to 1.05) | .02 |
| Ulceration | 1.60 (1.46 to 1.74) | <.001 | 1.92 (1.39 to 2.66) | <.001 | 1.60 (1.24 to 2.07) | <.001 |
| Mitotic index | − | − | 1.54 (1.01 to 2.33) | .04 | 1.92 (1.39 to 2.65) | <.001 |
| Stage II | 3.01 (2.65 to 3.42) | <.001 | 1.53 (0.97 to 2.42) | .07 | 1.83 (1.31 to 2.56) | <.001 |
| Stage III | 8.56 (7.56 to 9.69) | <.001 | 4.76 (3.10 to 7.32) | <.001 | 4.36 (3.14 to 6.05) | <.001 |
All statistical tests are two-sided. CI = confidence interval; HR = hazard ratio; NM = nodular melanoma; SEER = Surveillance, Epidemiology, and End Results; SSM = superficial spreading melanoma.
Reported hazard ratios calculated per 10 years of age.
Figure 1.
Survival curves stratified by primary melanoma histologic subtype. A) Kaplan-Meier survival curves for melanoma-specific survival (P < .001) in the Surveillance, Epidemiology, and End Results cohort. B) Recurrence-free survival (P < .001) in the NYU cohort. C) Melanoma-specific survival (P < .001) in the NYU cohort. All P values were calculated using a two-sided log-rank test.
Though the SEER cohort lacks information on MI and TIL, these data were available for the NYU cohort. Among the subset of tumors evaluated for TIL (n = 1138), brisk TIL was more common among SSM than NM (P < .05) (Table 1). NM primaries also exhibited higher MI (P < .001) (Table 1).
Differences in the Somatic Mutational Patterns of Nodular and Superficial Spreading Melanomas
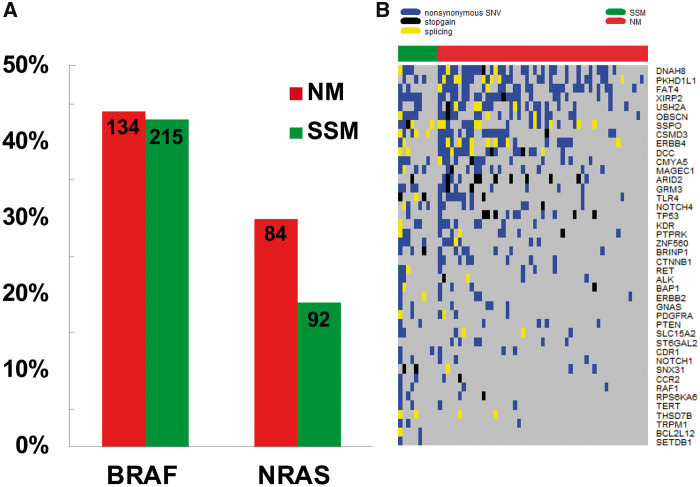
In a large subset of cases with available tissues, NM and SSM cases were analyzed for common driver mutation status, including 808 for BRAF (303 NM, 505 SSM) and 763 for NRAS (276 NM, 487 SSM). NM was found to have a statistically significantly higher frequency of NRAS mutation vs SSM (30% vs 19%, P < .001), whereas no difference was observed with respect to the presence of BRAF mutation among NM and SSM tumors (44% vs 43%) (Figure 2A). High-throughput targeted sequencing was performed on an initial cohort of 65 cases (54 NM and 11 SSM). Two NM samples and one SSM sample did not pass quality control and were subsequently excluded. Within the 140-gene panel, NM exhibited a lower prevalence of mutated genes relative to SSM (P < .001). We subsequently characterized the somatic alterations among NM and SSM (Figure 2B), focusing on SNVs with an expected impact on protein coding. Eight genes were found to be statistically significantly undermutated in NM relative to SSM: NOTCH4, RPS6KA6, BCL2L12, ERBB3, TERT, SNX31, SSPO, and ZNF560 (all P < .05) (Supplementary Table 2, available online, for a list of genes). In a second cohort of NM and SSM patient-derived samples (n = 34), we further explored the protein-coding regions of 10 of the most differentially mutated genes between NM and SSM. We identified three NM-specific SNVs present in both the first and second cohort. These SNVs include nonsynonymous SNVs in ANK3 (exon16:c.G1774A:p.D592N) and NOTCH4 and a synonymous SNV in ZNF560 (exon10:c.G1116A:p.G372G). Interestingly, the nonsynonymous SNV in NOTCH4 in exon 21 (c.G3587A:p.G1196E) has been previously identified in both renal cell carcinoma and melanoma (previously no subtype specified).
Figure 2.
Genomic sequencing of nodular and superficial spreading melanoma. A) Prevalence of known BRAF and NRAS melanoma driver mutations among nodular melanoma and superficial spreading melanoma (P < .001 from two-sided χ2 test for NRAS). B) Distribution of nonsynonymous, nonsense, and splice-site single nucleotide variants among the most commonly mutated genes identified in nodular melanoma and superficial spreading melanoma. NM = nodular melanoma; SNV = single nucleotide variant; SSM = superficial spreading melanoma.
Impact of Primary Histologic Subtype on Outcomes in Metastatic Melanoma
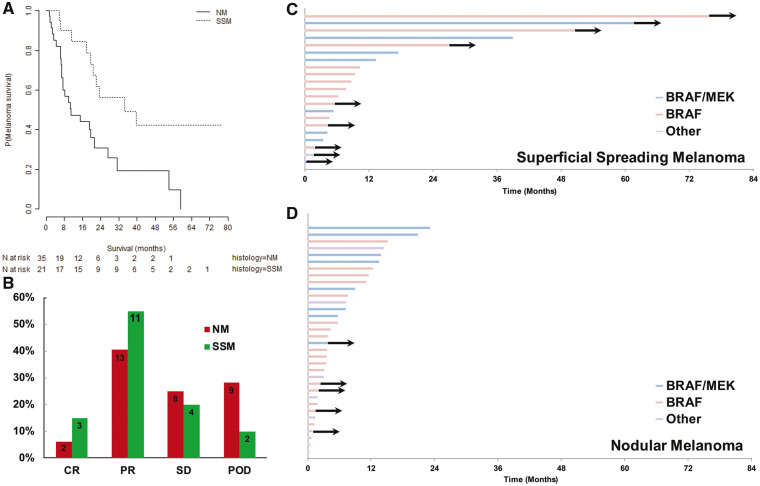
To investigate the impact of primary histologic subtype on clinical outcomes in the metastatic setting, survival analyses were performed on subsets of patients who developed metastatic disease. Of note, no difference in serum lactate dehydrogenase concentration was detected between metastatic NM and SSM. As the NYU cohort accrued between 2002 and 2016, treatment strategies were markedly heterogeneous. We therefore focused on the more recent era of melanoma treatment with checkpoint inhibitor immunotherapy (105 NM, 49 SSM) and BRAF-directed targeted therapy (35 NM, 21 SSM). Among the targeted therapy cohort with evaluable response data, NM exhibited a shorter median survival relative to SSM (10.9 months, 95% CI = 7.3 to 31.2 months, vs 34.4 months, 95% CI = 20.6 months to NA, P = .004) (Figure 3A) and a lower response rate relative to SSM (CR = 6.3%, 95% CI = −2.1% to 14.6%, vs. 15.0%, 95% CI = −0.6% to 30.6%, PR = 40.6%, 95% CI = 23.6% to 57.6%, vs. 55.0%, 95% CI = 33.2% to 76.8%, P = 0.02) Figure 3B). In the Cox proportional hazard models, NM was a statistically significant predictor of worse MSS (HR = 2.89, 95% CI = 1.36 to 6.14, P = .006) (Table 3). After controlling for other prognostic variables like thickness and stage, NM remained an independent risk factor for death from melanoma (HR = 3.33, 95% CI = 1.06 to 10.47, P = .04) (Table 3).
Figure 3.
Clinical outcomes of BRAF-targeted therapy in metastatic melanoma stratified by histologic subtype. A) Kaplan-Meier survival curves for metastatic nodular melanoma and metastatic superficial spreading melanoma from the initiation of BRAF-targeted therapy (log-rank P = .003). B) Distribution of responses to BRAF-targeted therapy between nodular melanoma (NM) and superficial spreading melanoma (SSM; P = .02). C, D) Swimmer plots demonstrating the duration of treatment among SSM and NM (P = .01), with arrows indicating continuation of therapy. All P values were calculated using a two-sided log-rank test. CR = complete response; NM = nodular melanoma; POD = progression of disease; PR = partial response; SD = stable disease; SSM = superficial spreading melanoma.
Table 3.
Univariate and multivariable Cox proportional hazard models for the NYU BRAF-targeted therapy cohort in metastatic melanoma
| Variable | Melanoma-specific survival |
|
|---|---|---|
| HR (95% CI) | P* | |
| Univariate Cox proportional hazard model | ||
| NM vs SSM | 2.89 (1.36 to 6.14) | .006 |
| Multivariable Cox proportional hazard model | ||
| NM vs SSM | 3.33 (1.06 to 10.47) | .04 |
| Age at primary diagnosis† | 1.20 (0.95 to 1.53) | .13 |
| Sex | 1.29 (0.52 to 3.18) | .58 |
| Primary thickness | 1.04 (0.95 to 1.14) | .37 |
| Primary ulceration | 0.84 (0.30 to 2.36) | .74 |
| Primary mitotic index | 7.42 (0.92 to 59.51) | .06 |
| Primary Dx stage II | 0.76 (0.14 to 4.22) | .76 |
| Primary Dx stage III | 1.41 (0.40 to 4.93) | .59 |
All statistical tests are two-sided. CI = confidence interval; HR = hazard ratio; NM = nodular melanoma; SSM = superficial spreading melanoma.
Reported HRs calculated per 10 years of age.
The majority of NM and SSM patients received BRAF inhibitor monotherapy (54% vs 57%, respectively). The more favorable outcomes among SSM patients treated with targeted therapy are demonstrated by longer treatment duration among SSM relative to NM patients (Figure 3, C and D). NM patients remained on treatment for a median of only 7.17 months (95% CI = 3.8 to 13.6 months) before progression, whereas SSM patients continued on treatment for a median of 10.33 months (95% CI = 7.67 months to NA, P = .01). Notably, all five patients remaining on treatment after two years are among the SSM cohort, and the majority of these five were treated with BRAF-inhibitor monotherapy (Figure 3C).
Among the immunotherapy cohort of 154 patients (anti-CTLA-4 n = 114; anti-PD-1 n = 29; and combination therapy n = 11), no difference was noted in MSS from the time of treatment initiation (median = 16.7 months, 95% CI = 12.7 to 37.3 months, for NM vs 22.3 months, 95% CI = 14.1 months to NA, for SSM, respectively, P = .56). A majority of these patients were treated with ipilimumab monotherapy (76, 72%, NM; 38, 78%, SSM) in place of anti-PD-1, a trend that was exaggerated in the SSM cohort, in which only seven (14%) patients received anti-PD-1, compared with 22 (21%) patients in the NM cohort.
Discussion
To the best of our knowledge, our analysis of both the population-based SEER cohort, with more than 100 000 patients, and our institution registry, comprised of more than 1600 patients with prospective protocol-driven follow-up data, constitutes the first report demonstrating histologic subtype to be an independent predictor of survival in melanoma. In addition, these data represent one of the broadest attempts to characterize the histologic subtype–specific somatic mutational landscape of melanoma and reveal divergent clinical behavior between NM and SSM in the metastatic setting with respect to BRAF-targeted therapy.
Although recent studies describe the rising incidence of lentigo maligna (melanoma in situ on sun-exposed skin) or lentigo maligna melanoma in specific population subsets (31), overall, NM and SSM remain the two most common histologic subtypes among invasive tumors (32–35). Within cases of invasive melanoma, the poor prognosis of NM relative to SSM has been very well established among many cohorts, ranging from small institutional studies (36,37) to large regional and national data sets (38–41). However, controversy remains as to what extent this effect is driven by NM’s known associations with other histopathologic parameters, especially thickness (20,42,43). In an earlier study, data from our group (12) demonstrated that primary SSM tumors have become thinner over time between the 1970s and 2000s, without evidence of stage migration in the presentation of NM, arguing against the canonical linear progression model. Furthermore, although a marker to distinguish NM from the vertical growth phase of SSM has been elusive, emerging molecular studies show that the two subtypes harbor distinct molecular signatures, promising the development of biomarkers not only for histologic classification but also for prognostic stratification (44). In contrast to the generally held assumption that NM exhibits shorter recurrence-free and disease-specific survival merely because of its thicker primary tumors, our present data demonstrate that the risk of NM is, at least in part, mediated by inherently more aggressive biological processes. In fact, the increased hazard ratio associated with nodular histology is substantial, with a 54% increased risk of death in SEER and a 43% increased risk among the NYU cohort. An increased hazard ratio of this magnitude is comparable with known prognostic variables such as thickness, ulceration, and MI.
In contrast to non–small cell lung cancer and renal cell carcinoma, TCGA data reporting for melanoma generally do not include melanoma primary subtype (17). Because histologic subtype is seldom specified in dermatopathology reports, the TCGA melanoma cohort carries subtype designation for only five SSM and 22 NM specimens. Even fewer specimens are provided with both histological subtype and mutational data. All other specimens in the TCGA database are annotated as melanoma, not otherwise specified. We began characterizing the status of known melanoma driver mutations among a large subgroup of the NYU cohort, comprised of more than 700 NM and SSM patient-derived samples. Although several studies (7,45–47) have reported on the slightly increased rate of BRAF mutations among SSM relative to NM patients, this finding has not been consistently reproducible (48,49). The incidence of BRAF-mutant melanoma observed in our study was approximately 40%–45%, regardless of histologic subtype, and is consistent with the incidence reported in the literature. With regards to the NRAS mutation, we show an increased incidence in NM. We further examined the somatic mutational landscape of NM and SSM and demonstrated in a large panel of cancer-related genes that 1) an overall lower rate of somatic gene mutations was generally seen in NM patients and 2) several key genes were observed to be less frequently mutated in NM relative to SSM. Prior to the present study, such histologic subtype–specific alterations were not widely characterized due to a lack of available histologic subtype data among institutional data sets and the TCGA.
We further explored and validated subtype-specific somatic alterations in a second cohort of NM and SSM samples. The identification of a recurrent SNV in NOTCH4 exon 21 in both the first and second NM cohorts is of interest given its previous identification in renal cell carcinoma and the TCGA melanoma cohort (50). NOTCH4 is a transmembrane notch signaling protein that has been associated with the epithelial-mesenchymal transition of melanoma in vitro (51). Although the NOTCH4 mutation identified among NM patients was previously identified in TCGA, histologic subtype data were unavailable. This mutation is predicted to be pathogenic by Functional Analysis through Hidden Markov Models (FATHMM). Recurrent identification of this SNV in TCGA and, again, in two distinct NYU cohorts of NM patients suggests that it may be biologically significant in the development and progression of a subset of NM and that it warrants further investigation. ANK3 is a large structural protein that has been found to be downregulated in cancer stem cells (52), and ZNF560 is a Zinc finger protein that has also been associated with induced cancer stem cells (53). The aforementioned mutations in ANK3 and ZNF560 are novel mutations, as they have not been reported in other data sets. The targeted panel used in this study, though limited in scope, captures important biologic information regarding the histologic subtype–specific mutational landscape. Additional studies are warranted on whether this will translate to a similar difference in the overall mutational burden.
Based on the evidence of different mutational patterns among NM and SSM, we hypothesized that NM and SSM may respond differently to systemic therapy in the metastatic setting. Among BRAF-mutant patients, we found that metastatic NM exhibited worse outcomes in the context of BRAF-directed targeted therapy, even after controlling for other known risk factors. Because the subgroup of targeted therapy patients is small, continued study of this phenomenon is needed to independently validate and fully characterize the response to BRAF-targeted therapy across histologic subtypes. The lack of primary histologic subtype information on all reported clinical trials of BRAF-targeted therapy prohibits external validation of our findings and supports the inclusion of this information prospectively.
The response rates and survival in the context of checkpoint inhibitor immunotherapy do not statistically significantly differ between NM and SSM patients. However, the majority of patients in our cohort were treated with anti-CTLA-4 monotherapy, known to have only a 20% response rate. It is possible that subtype-specific differences in long-term outcomes might exist in the context of more efficacious anti-PD1 or combination immunotherapy. Of note, the data to support a relationship between mutational burden and response to checkpoint inhibitors are much clearer with respect to anti-PD-1 (54) than anti-CTLA-4 (55).
Our study is not without limitations. First, the reliance on retrospective analyses to examine differences in survival introduces heterogeneity that is perhaps not readily apparent and unaccounted for in these multivariable models. Although we have made every effort to control for confounding variables, these observations all require prospective validation, a limitation that is especially relevant to the survival analyses on the BRAF-targeted therapy cohort because of the relatively small sample size. Second, although the genomic analyses point to interesting mutational differences between these two histologic subtypes, we have not performed functional assays to examine the impact of these somatic variants in vitro, and they do not necessarily constitute clinically significant driver mutations. Finally, although the primary histologic subtype was determined in our study by central review by a single attending dermatopathologist, inter-reader variability may prove a considerable challenge in expanding the applicability of these findings into clinical practice.
Ultimately, the prognostic value of primary histologic subtype must be characterized at the molecular level, particularly with respect to somatic alterations and gene expression. Given the identification of nodular histology as an independent predictor of shorter recurrence-free and overall survival, histologic characterization clearly yields valuable information in the clinical decision-making process without the need for additional serologic or tissue analysis at the time of initial diagnosis. Based on the finding that NM is associated with shorter survival in the national SEER cohort, we subsequently went on to validate this finding in our prospective institutional tumor registry, which has enabled important genomic and treatment-related observations. Given our finding that primary histology might be associated with survival and response to BRAF-targeted therapy in the metastatic setting, more studies are needed to better characterize this phenomenon. Routine incorporation of histologic subtype into prospective clinical trial reporting in melanoma would better ensure balanced patient characteristics across study arms and aid in understanding the impact of primary histologic subtype on response to systemic agents in the clinical trial setting.
Funding
Funding support for the study was provided by the NYU Cancer Center and National Institutes of Health/National Cancer Institute Cancer Center Support Grant P30CA016087, Goldberg Charitable Trust, Wings for Things Foundation, and Clayman Family Foundation to I. Osman.
Notes
Affiliations of authors: Department of Medicine (ML, ACP, MAW, JSW, IO), Interdisciplinary Melanoma Cooperative Group (ML, YL, DS, UM, FD, RHK, EH, DP, DH, RS, RB, ACP, MAW, TK, JSW, JZ, IO), The Ronald O. Perelman Department of Dermatology (YL, UM, RHK, DP, ACP, JSW, IO), Department of Population Health (DS, TK, JZ), Department of Pathology (FD, EH, DP, DH), and Department of Surgery (RS, RB), NYU School of Medicine, New York, NY.
The funders did not participate in the study design, data collection, data interpretation, manuscript drafting, manuscript submission, or any other scientific aspect of the study. All contributors to the study are listed as authors. The authors have no conflicts of interest to disclose.
Supplementary Material
References
- 1. Duncan LM. The classification of cutaneous melanoma. Hematology/oncology clinics of North America. 2009;233:501–513, ix. [DOI] [PubMed] [Google Scholar]
- 2. Ossio R, Roldan-Marin R, Martinez-Said H, Adams DJ, Robles-Espinoza CD. Melanoma: A global perspective. Nat Rev Cancer. 2017;177:393–394. [DOI] [PubMed] [Google Scholar]
- 3. Liu W, Dowling JP, Murray WK, et al. Rate of growth in melanomas: Characteristics and associations of rapidly growing melanomas. Arch Dermatol. 2006;14212:1551–1558. [DOI] [PubMed] [Google Scholar]
- 4. Kwong L, Chin L, Wagner SN. Growth factors and oncogenes as targets in melanoma: Lost in translation? Adv Dermatol. 2007;23:99–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Greenwald HS, Friedman EB, Osman I. Superficial spreading and nodular melanoma are distinct biological entities: A challenge to the linear progression model. Melanoma Res. 2012;221:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jaeger J, Koczan D, Thiesen HJ, et al. Gene expression signatures for tumor progression, tumor subtype, and tumor thickness in laser-microdissected melanoma tissues. Clin Cancer Res. 2007;133:806–815. [DOI] [PubMed] [Google Scholar]
- 7. Lee JH, Choi JW, Kim YS. Frequencies of BRAF and NRAS mutations are different in histological types and sites of origin of cutaneous melanoma: A meta-analysis. Br J Dermatol. 2011;1644:776–784. [DOI] [PubMed] [Google Scholar]
- 8. Poliseno L, Haimovic A, Segura MF, et al. Histology-specific microRNA alterations in melanoma. J Invest Dermatol. 2012;1327:1860–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rose AE, Poliseno L, Wang J, et al. Integrative genomics identifies molecular alterations that challenge the linear model of melanoma progression. Cancer Res. 2011;717:2561–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shaikh WR, Dusza SW, Weinstock MA, Oliveria SA, Geller AC, Halpern AC. Melanoma thickness and survival trends in the United States, 1989–2009. J Natl Cancer Inst. 2016;1081:djv294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shen S, Wolfe R, McLean CA, Haskett M, Kelly JW. Characteristics and associations of high-mitotic-rate melanoma. JAMA Dermatol. 2014;15010:1048–1055. [DOI] [PubMed] [Google Scholar]
- 12. Warycha MA, Christos PJ, Mazumdar M, et al. Changes in the presentation of nodular and superficial spreading melanomas over 35 years. Cancer. 2008;11312:3341–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;4897417:519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;5117511:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;5247563:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ciriello G, Gatza ML, Beck AH, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;1632:506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ping Z, Xia Y, Shen T, et al. A microscopic landscape of the invasive breast cancer genome. Sci Rep. 2016;6:27545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell. 2015;1617:1681–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Amin MB, Greene FL, Edge SB, et al. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J Clin. 2017;672:93–99. [DOI] [PubMed] [Google Scholar]
- 20. Balch CM, Buzaid AC, Soong SJ, et al. New TNM melanoma staging system: Linking biology and natural history to clinical outcomes. Semin Surg Oncol. 2003;211:43–52. [DOI] [PubMed] [Google Scholar]
- 21. Balch CM, Murad TM, Soong SJ, Ingalls AL, Halpern NB, Maddox WA. A multifactorial analysis of melanoma: Prognostic histopathological features comparing Clark's and Breslow's staging methods. Ann Surg. 1978;1886:732–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thomas NE, Edmiston SN, Alexander A, et al. Association between NRAS and BRAF mutational status and melanoma-specific survival among patients with higher-risk primary melanoma. JAMA Oncol. 2015;13:359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pracht M, Mogha A, Lespagnol A, et al. Prognostic and predictive values of oncogenic BRAF, NRAS, c-KIT and MITF in cutaneous and mucous melanoma. J Eur Acad Dermatol Venereol. 2015;298:1530–1538. [DOI] [PubMed] [Google Scholar]
- 24. Yaman B, Akalin T, Kandiloglu G. Clinicopathological characteristics and mutation profiling in primary cutaneous melanoma. Am J Dermatopathol. 2015;375:389–397. [DOI] [PubMed] [Google Scholar]
- 25.National Cancer Institute Surveillance Program. List of SEER registries. http://seer.cancer.gov/registries/list.html. Accessed April 1, 2017.
- 26. LeBoit PE. Pathology and Genetics of Skin Tumours. Lyon: IARC; 2006. [Google Scholar]
- 27. Weiss SA, Han SW, Lui K, et al. Immunologic heterogeneity of tumor-infiltrating lymphocyte composition in primary melanoma. Hum Pathol. 2016;57:116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sekine S, Nakanishi Y, Ogawa R, Kouda S, Kanai Y. Esophageal melanomas harbor frequent NRAS mutations unlike melanomas of other mucosal sites. Virchows Arch. 2009;4545:513–517. [DOI] [PubMed] [Google Scholar]
- 29. Farr CJ, Saiki RK, Erlich HA, McCormick F, Marshall CJ. Analysis of RAS gene mutations in acute myeloid leukemia by polymerase chain reaction and oligonucleotide probes. Proc Natl Acad Sci U S A. 1988;855:1629–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fox J, Weisberg S, Fox J. An R Companion to Applied Regression. 2nd ed. Thousand Oaks, CA: SAGE Publications; 2011. [Google Scholar]
- 31. Swetter SM, Boldrick JC, Jung SY, Egbert BM, Harvell JD. Increasing incidence of lentigo maligna melanoma subtypes: Northern California and national trends 1990-2000. J Invest Dermatol. 2005;1254:685–691. [DOI] [PubMed] [Google Scholar]
- 32. Micu E, Juzeniene A, Moan J. Comparison of the time and latitude trends of melanoma incidence in anorectal region and perianal skin with those of cutaneous malignant melanoma in Norway. J Eur Acad Dermatol Venereol. 2011;2512:1444–1449. [DOI] [PubMed] [Google Scholar]
- 33. Baumert J, Plewig G, Volkenandt M, Schmid-Wendtner MH. Factors associated with a high tumour thickness in patients with melanoma. Br J Dermatol. 2007;1565:938–944. [DOI] [PubMed] [Google Scholar]
- 34. Shaikh WR, Xiong M, Weinstock MA. The contribution of nodular subtype to melanoma mortality in the United States, 1978 to 2007. Arch Dermatol. 2012;1481:30–36. [DOI] [PubMed] [Google Scholar]
- 35. Youl PH, Youlden DR, Baade PD. Changes in the site distribution of common melanoma subtypes in Queensland, Australia over time: Implications for public health campaigns. Br J Dermatol. 2013;1681:136–144. [DOI] [PubMed] [Google Scholar]
- 36. Khosrotehrani K, van der Ploeg AP, Siskind V, et al. Nomograms to predict recurrence and survival in stage IIIB and IIIC melanoma after therapeutic lymphadenectomy. Eur J Cancer. 2014;507:1301–1309. [DOI] [PubMed] [Google Scholar]
- 37. van Lanschot CG, Koljenovic S, Grunhagen DJ, Verhoef C, van Akkooi AC. Pigmentation in the sentinel node correlates with increased sentinel node tumor burden in melanoma patients. Melanoma Res. 2014;243:261–266. [DOI] [PubMed] [Google Scholar]
- 38. Mar V, Roberts H, Wolfe R, English DR, Kelly JW. Nodular melanoma: A distinct clinical entity and the largest contributor to melanoma deaths in Victoria, Australia. J Am Acad Dermatol. 2013;684:568–575. [DOI] [PubMed] [Google Scholar]
- 39. Green AC, Baade P, Coory M, Aitken JF, Smithers M. Population-based 20-year survival among people diagnosed with thin melanomas in Queensland, Australia. J Clin Oncol. 2012;3013:1462–1467. [DOI] [PubMed] [Google Scholar]
- 40. Pollack LA, Li J, Berkowitz Z, et al. Melanoma survival in the United States, 1992 to 2005. J Am Acad Dermatol. 2011;65(5 Suppl 1):S78–S86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barnhill RL, Fine JA, Roush GC, Berwick M. Predicting five-year outcome for patients with cutaneous melanoma in a population-based study. Cancer. 1996;783:427–432. [DOI] [PubMed] [Google Scholar]
- 42. Demierre MF, Chung C, Miller DR, Geller AC. Early detection of thick melanomas in the United States: Beware of the nodular subtype. Arch Dermatol. 2005;1416:745–750. [DOI] [PubMed] [Google Scholar]
- 43. Chamberlain AJ, Fritschi L, Giles GG, Dowling JP, Kelly JW. Nodular type and older age as the most significant associations of thick melanoma in Victoria, Australia. Arch Dermatol. 2002;1385:609–614. [DOI] [PubMed] [Google Scholar]
- 44. Faut M, Wevers KP, van Ginkel RJ, et al. Nodular histologic subtype and ulceration are tumor factors associated with high risk of recurrence in sentinel node-negative melanoma patients. Ann Surg Oncol. 2017;241:142–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu W, Kelly JW, Trivett M, et al. Distinct clinical and pathological features are associated with the BRAF(T1799A(V600E)) mutation in primary melanoma. J Invest Dermatol. 2007;1274:900–905. [DOI] [PubMed] [Google Scholar]
- 46. Baiter M, Schuler G, Hartmann A, Schneider-Stock R, Heinzerling L. Pathogenetic implications of BRAF mutation distribution in stage IV melanoma patients. Dermatology. 2015;2312:127–133. [DOI] [PubMed] [Google Scholar]
- 47. Edlundh-Rose E, Egyhazi S, Omholt K, et al. NRAS and BRAF mutations in melanoma tumours in relation to clinical characteristics: A study based on mutation screening by pyrosequencing. Melanoma Res. 2006;166:471–478. [DOI] [PubMed] [Google Scholar]
- 48. Omholt K, Platz A, Kanter L, Ringborg U, Hansson J. NRAS and BRAF mutations arise early during melanoma pathogenesis and are preserved throughout tumor progression. Clin Cancer Res. 2003;917:6483–6488. [PubMed] [Google Scholar]
- 49. Broekaert SM, Roy R, Okamoto I, et al. Genetic and morphologic features for melanoma classification. Pigment Cell Melanoma Res. 2010;236:763–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cosmic. “COSMIC - Catalogue of Somatic Mutations in Cancer.” NRG3 Gene - Somatic Mutations in Cancer, Wellcome Sanger Institute. http://cancer.sanger.ac.uk/cosmic. Accessed July 1, 2017.
- 51. Lin X, Sun B, Zhu D, et al. Notch4+ cancer stem-like cells promote the metastatic and invasive ability of melanoma. Cancer Sci. 2016;1078:1079–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pandit TS, Kennette W, Mackenzie L, et al. Lymphatic metastasis of breast cancer cells is associated with differential gene expression profiles that predict cancer stem cell-like properties and the ability to survive, establish and grow in a foreign environment. Int J Oncol. 2009;352:297–308. [PubMed] [Google Scholar]
- 53. Vencio EF, Nelson AM, Cavanaugh C, et al. Reprogramming of prostate cancer-associated stromal cells to embryonic stem-like. Prostate. 2012;7213:1453–1463. [DOI] [PubMed] [Google Scholar]
- 54. Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;3486230:124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;37123:2189–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.