SUMMARY
Wnt3a-coated beads can induce asymmetric divisions of mouse embryonic stem cells (mESCs), resulting in one self-renewed mESC and one differentiating epiblast stem cell. This provides an opportunity for studying histone inheritance pattern at a single-cell resolution in cell culture. Here, we report that mESCs with Wnt3a-bead induction display non-overlapping preexisting (old) versus newly synthesized (new) histone H3 pattern, but mESCs without Wnt3a-bead have largely overlapping patterns. Furthermore, H4K20me2/3, an old histone-enriched modification, displays a higher instance of asymmetric distribution on chromatin fibers from Wnt3a-induced mESCs than those from non-induced mESCs. These locally distinct distributions between old and new histones have both cellular specificity in Wnt3a-induced mESCs and molecular specificity for histones H3 and H4. Given that post-translational modifications at H3 and H4 carry the major histone modifications, our findings provide a mammalian cell culture system to study histone inheritance for maintaining stem cell fate and for resetting it during differentiation.
Keywords: Histone, asymmetric cell division, mouse embryonic stem cells, Wnt3a beads
INTRODUCTION
Many types of stem cells undergo asymmetric cell division (ACD) to give rise to two daughter cells with distinct cell fates (Clevers, 2005; Kahney et al., 2017; Knoblich, 2010; Morrison and Kimble, 2006; Venkei and Yamashita, 2018). In this process, it is crucial that the self-renewing daughter cell maintains a stemness fate and the differentiating daughter cell is reset for differentiation. As these two cells derive from one mitosis, they carry identical DNA sequences. Therefore, a long-standing question has been whether these two cells could inherit different sequence-independent epigenetic information that contribute to their distinct cell fates.
DNA methylation and histone modifications are the best characterized epigenetic regulations regarding their molecular mechanisms and biological functions (Francis et al., 2009; Minter et al., 2002). Post-translational histone modifications are known for regulating differential gene expression (Allis and Jenuwein, 2016). Most canonical histones such as H3, H4, H2A, H2B, and H1 linker histone incorporate into the chromatin during DNA replication. Among them, H3 and H4 contain the majority of known modifications (Allis and Jenuwein, 2016; Kouzarides, 2007; Young et al., 2010).
During asymmetric divisions of Drosophila male germline stem cells (GSCs), it has been shown that the preexisting (old) H3 is selectively segregated to the self-renewed GSC, whereas newly synthesized (new) H3 is enriched in the differentiating daughter cell (Tran et al., 2013; Tran et al., 2012). This asymmetric pattern is distinct from the symmetrical H3 inheritance in symmetrically dividing progenitor germ cells. Moreover, asymmetric H3 inheritance is important for establishing different cell fates during ACD, as mis-regulation of this process leads to both early germline tumor and GSC loss phenotypes (Xie et al., 2015; Xie et al., 2017). Additionally, H4 has been shown to have a similar asymmetric inheritance pattern to H3, whereas H2A and H2B are inherited symmetrically during ACD of GSCs (Wooten et al., 2019). However, it remains unclear whether the asymmetric H3 and H4 inheritance mode is restricted to organisms with minimal DNA methylation like Drosophila (Lyko et al., 2000; Zhang et al., 2015) or if it is applicable to mammalian cells. In this study, we investigate different histone distribution patterns in mouse embryonic stem cell (mESC) system.
The mESCs are derived from 3.5 days post coitum embryos (Lo et al., 2012), whose “naïve” ground state is maintained by complete medium (CM) + LIF/2i (Del Olmo et al., 2016; Ying et al., 2008). It has been shown that Wnt3a-coated beads can orient the division plane of mESCs to achieve ACDs: the Wnt3a-proximal cell is retained as mESC while the distal cell adopts the mouse epiblast stem cell (mEpiSCs) fate (Habib et al., 2013). Here, we use this system to study histone distribution patterns in mESCs.
RESULTS
Establish a mammalian cell culture system to study histone inheritance at single-cell resolution
We established a system to study histone inheritance by building stable histone-dendra2 mESC lines. We tagged canonical histones H3, H4, H2A, and H2B as well as H3.3 histone variant coding sequences with the dendra2 sequence at their corresponding 3’-end, which encodes the respective Histone-Dendra2 fusion proteins (Figure 1A). Dendra2 is a photoconvertible protein that can irreversibly switch from green to red fluorescence upon exposure to 405nm light. Therefore, old histones were labeled in red and new histones in green (Chudakov et al., 2007; Gurskaya et al., 2006; Ranjan et al., 2019). To study histone inheritance in mESCs at a single-cell resolution, we used Wnt3a-immobilized beads to induce ACD (Habib et al., 2013).
Figure 1: Local non-overlapping old versus new H3 and H4 in Wnt3a-induced mESCs.
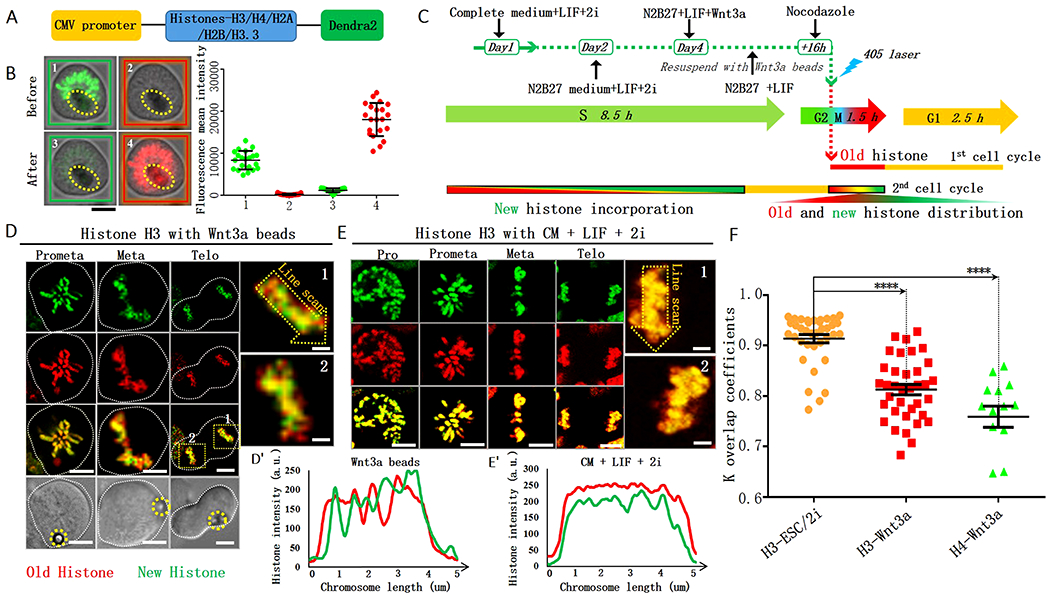
(A) Histone-Dendra2 constructs (STAR Methods). (B) Photoconvert H3-Dendra2-expressing mESCs from green to red fluorescence upon exposure to 405nm light (STAR Methods). (C) Regime for Wnt3a induction and tracing old (red) versus new (green) histones after photoconversion. (D) Non-overlapping subdomains of old versus new H3 during the 2nd mitosis after photoconversion in Wnt3a-induced mESCs. Bottom: DIC (differential interference contrast) images, yellow dotted circles label Wnt3a beads. (E) Overlapping old versus new H3 during the 2nd mitosis after photoconversion in mESCs cultured in CM+LIF+2i. Line plots: non-overlapping H3 pattern in Wnt3a-induced mESCs (D’), overlapping H3 patterns in CM+LIF+2i medium without Wnt3a beads (E’). Deconvolution for (D, E) used Huygens software. (F) Quantification of old versus new H3 and H4 in mitotic mESCs under different conditions. The coefficient k for old versus new H3 in mESCs cultured in CM+LIF+2i: 0.91 ± 0.01 (H3-ESC/2i, N=37), in Wnt3a-induced mESCs: 0.81 ± 0.01 (H3-Wnt3a, N= 36), for old versus new H4 in Wnt3a-induced mESCs: 0.75 ± 0.02 (H4-Wnt3a, N= 14). Average ± SEM, P< 10−4, Student t test. Scale bars in (B, D, E): 5μm, insets for (D, E): 1μm.
First, we confirmed the cellular characteristics of the E14TG2a mESC line used in this study. The dome-shaped colonies with high levels of alkaline phosphatase (AP) expression (validation Figure-A in Mendeley data) showed that the capacity of self-renewal is maintained when cultured in CM+LIF/2i. Consistent with previous findings (Jiang and Berger, 2017), mESCs can be efficiently differentiated into mEpiSCs in N2B27 medium with basic Fibroblast Growth Factor (bFGF) and Activin A for five days with mEpiSC-characteristic flattened morphology, loose compactions, and low AP activity (validation Figure-B in Mendeley data). A flow cytometry analysis of cell cycles showed that the cell doubling time was ~12 hours for mESCs and ~13 hours for mEpiSCs (validation Figure-C in Mendeley data), as reported previously (Fluckiger et al., 2006; Piunti et al., 2014).
We next validated all histone-dendra2 mESC lines: (1) All expressed the corresponding Histone-Dendra2 fusion protein (expression Figure-A in Mendeley data). (2) Compared to non-transgenic mESCs, all transgenic lines displayed comparable cell cycle profiles, growth curves, and percentages of S-phase cells (expression Figure B-D in Mendeley data). (3) Active expression of known mESC markers, such as Nanog, Rex1, Oct4, Sox2, and SSEA1, as well as nuclear H3-Dendra2 signals were all readily detectable (expression Figure E in Mendeley data). Together, our results suggest that all transgenic lines express Histone-Dendra2 fusion proteins and have normal cellular properties.
It has been previously shown that mESC can orient its division plane in response to Wnt3a bead (Movie in Mendeley data). The cell proximal to Wnt3a bead has high expression of Wnt signaling pathway components, such as β-catenin and adenomatous polyposis coli (APC), and pluripotency markers. The distal cell downregulates these markers but upregulates mEpiSC marker Claudin6 (Habib et al., 2013). We first investigated the efficiency of Wnt3a beads in inducing ACD of the H3-dendra2 mESCs. In post-mitotic pairs, APC was enriched in 57.80% of the cells proximal to the Wnt3a beads (Figure S1A-B). Conversely, Claudin6 was enriched in 59.35% of the distal cells (Figure S1C-D). Even though the reversed asymmetric and symmetric patterns were detectable, their ratios were lower. In summary, approximately 60% of mESC underwent ACD with Wnt3a beads with the proximal side as the self-renewing mESC and the distal side as the differentiating EpiSC.
Old and new canonical H3 and H4 show non-overlapping patterns in Wnt3a-induced mESCs
Using the photoconvertible feature of Dendra2, abundant green fluorescence (panel 1 in Figure 1B) was seen but red fluorescence was undetectable (panel 2 in Figure 1B) before exposure. After a few short pulses of 405nm light, green fluorescent signals greatly diminished (panel 3 in Figure 1B) while red fluorescent signals significantly increased (panel 4 in Figure 1B). Quantification showed that green fluorescent Histone-Dendra2 decreased more than 90% immediately after exposure, suggesting efficient photoconversion. In subsequent experiments, we only used Histone-Dendra2-expressing cells with >90% photoconversion and analyzed them at a single-cell level.
Next, to visualize the distribution patterns of old versus new histones, we switched mESCs from CM to N2B27 medium with Wnt3a beads. We then arrested mESCs at G2/M using a microtubule depolymerizing drug Nocodazole (NZ). These Wnt3a-induced mESCs were then photoconverted and released from cell cycle arrest by washing away NZ. After the mESCs carrying red fluorescent Histone-Dendra2 underwent the first mitosis and the subsequent G1 and S phases, newly synthesized green fluorescent Histone-Dendra2 proteins were efficiently incorporated into the duplicated genome. We then studied old versus new histone distribution in the subsequent mitosis (2nd mitosis in Figure 1C).
Intriguingly, non-overlapping old versus new H3 can be visualized in mitotic mESCs with Wnt3a beads (Figure 1D-D’; Figure S2A-B). At prometaphase, resolved sister chromatids with both symmetric and asymmetric regions of old versus new H3 could be detected (Figure S2C-D). Using a similar labeling strategy, old and new H4 also displayed non-overlapping patterns in mitotic mESCs with Wnt3a beads (Figure S2E). As a control experiment, we examined old versus new H3 distribution in CM+LIF/2i, where mESCs mainly undergo symmetric divisions to self-renew (Habib et al., 2013; Lecona et al., 2013; Ying et al., 2008), and found that old versus new H3 displayed predominantly overlapping patterns throughout mitosis (Figure 1E-E’).
To quantify the overlapping degree between old and new histones under different conditions, we developed an ‘overlapping coefficient’ k (STAR Methods): If two signals completely overlap, k equals 1; if two signals do not overlap, k equals 0. Using this parameter, the k value for old and new H3 in mESCs cultured in CM+LIF/2i was 0.91. In the presence of Wnt3a beads, the k value reduced to 0.81 for H3 and 0.75 for H4, both significantly lower than that for H3 in mESCs without Wnt3a beads (Figure 1F). Together, these data indicate that the regional non-overlapping patterns between old and new histones are applicable for both H3 and H4. Additionally, this non-overlapping pattern is specific to mESCs with Wnt3a beads, suggesting an association with ACD.
Increased instances of replicative chromatin fibers derived from mESCs with Wnt3a beads display asymmetric distribution of old histone-enriched H4K20me2/3
Established during DNA replication via strand-biased incorporation of old versus new histones (Wooten et al., 2019), asymmetric histone inheritance in asymmetrically dividing Drosophila male GSCs also apply to H3 and H4 (Tran et al., 2012; Wooten et al., 2019). Next, we explored old histone incorporation patterns at the replicative DNA in mESCs with and without Wnt3a beads (flow chart in Mendeley data), using a recently developed SuperResolution of Chromatin Fibers (SRCF) method (Wooten et al., 2020a; Wooten et al., 2019). Here, thymidine analog 5-Ethynyl-2’-deoxyuridine (EdU) was used to mark active DNA replication regions and H4K20me2/3 was used to label old histones (Alabert et al., 2015; Petryk et al., 2018). To obtain strandness information, we immunostained chromatin fibers for lagging strand-enriched Proliferating Cell Nuclear Antigen (PCNA) (Yu et al., 2014).
We then analyzed replicating and/or newly replicated sister chromatid regions on chromatin fibers using two high-spatial resolution microscopy methods, CSU-W1 SoRa (Azuma and Kei, 2015) and Airyscan (Sivaguru et al., 2018). The resolved DAPI-bright and EdU-positive regions into double fiber structures indicate replicating or newly replicated sister chromatids (Figure 2A, 2D). The control fibers from mESCs without Wnt3a beads mainly showed symmetric distribution of H4K20me2/3 between sister chromatids (Figure 2A-B). In contrast, some chromatin fibers from mESCs with Wnt3a beads showed an asymmetric distribution of H4K20me2/3 towards the PCNA-less leading strand (Figure 2D-E). Quantification of these two groups showed that mESCs without Wnt3a beads had a symmetric distribution of H4K20me2/3 (Average ratio= 1.025, Figure 2C, galleries in Mendeley data), but Wnt3a-induced mESCs showed a significantly increased leading-strand bias (Average ratio= 1.657, Figure 2F, galleries in Mendeley data). To further compare, we classified fibers as leading strand enriched, symmetric, and lagging strand enriched [STAR Methods and (Wooten et al., 2019)]: only 10.0% of fibers from mESCs without Wnt3a beads showed leading strand bias, compared to 35.7% of fibers from mESCs with Wnt3a beads, a significant increase (Figure 2G).
Figure 2: Enriched H4K20me2/3 towards the leading strand of replicative chromatin fibers derived from Wnt3a-induced mESCs.
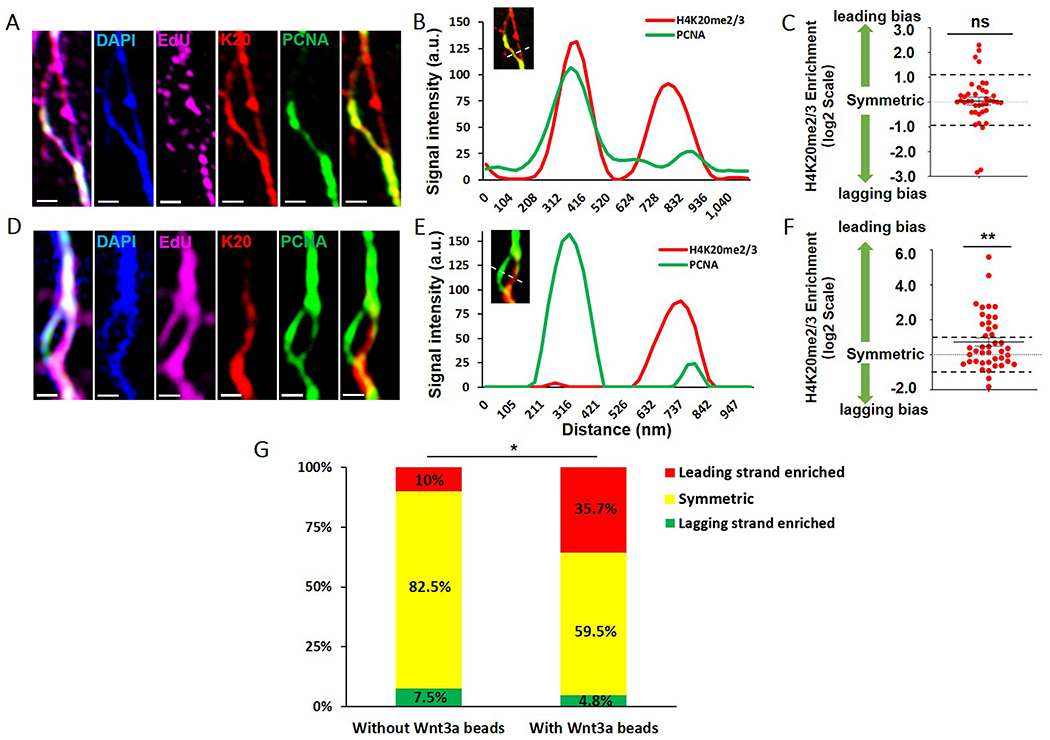
(A) Airyscan images of chromatin fiber from mESCs without Wnt3a beads. (B) Line plot shows comparable H4K20me2/3 between replicative sister chromatids in (A). (C) Quantification of log2 (leading strand H4K20me2/3 / lagging strand H4K20me2/3) using chromatin fibers from mESCs without Wnt3a beads. Log2 ratio= 0.03573 ± 0.1564 (Average ± SEM, N= 40 replicative chromatin fibers): no significant (ns) difference from symmetric distribution (log2 ratio= 0), unpaired t test. (D) Deconvoluted SoRa images of chromatin fiber from Wnt3a-induced mESCs. (A, D): EdU (magenta), H4K20me2/3 (red), DAPI (blue), PCNA (green), scale bars: 500 nm. (E) Line plot shows PCNA-less leading strand-enriched H4K20me2/3 between replicative sister chromatids in (D). (F) Quantification of log2 (leading strand H4K20me2/3 / lagging strand H4K20me2/3) using chromatin fibers from Wnt3a-induced mESCs. Log2 ratio= 0.7282 ± 0.2407 (Average ± SEM, N= 42 replicative chromatin fibers): significantly different from symmetric distribution (log2 ratio= 0). ** P<0.01, unpaired t test. Two dash lines in (C, F) indicate the symmetric range for H4K20me2/3 distribution between sister chromatids (STAR Methods). (G) The percentages of leading strand enriched (log2 ratio >1.025, ratio>2.035), symmetric (−0.953< log2 ratio <1.025, 0.517<ratio<2.035) and lagging strand enriched (log2 ratio <−0. 953, ratio<0.517) using data from (C, F). *P<0.05, Chi-square test.
Using Airyscan on fibers from Wnt3a-induced mESCs, we obtained similar results of asymmetric H4K20me2/3 distribution towards the PCNA-less leading strand (Average ratio= 1.356, Figure S3A-C, galleries in Mendeley data). With the same criteria, 25.7% of fibers showed leading strand bias. Data from both imaging methods suggest ~26%-36% of fibers showed asymmetric H4K20me2/3 at the replicative chromatin fibers derived from Wnt3a-induced mESCs (Figure 2G, Figure S3D), significantly higher than that of the control fibers. Furthermore, it should be considered that less than 60% of mESCs respond to Wnt3a induction for ACD (Figure S1B, S1D). In summary, leading-strand biased old histone distribution was detected at the replicative region using chromatin fibers and this asymmetry increased with Wnt3a beads.
Largely overlapping old and new histone distribution when mESCs are cultured with inactivated Wnt3a beads or in differentiation-promoting EpiSC medium
To further confirm that the non-overlapping old versus new histone distribution patterns are specific to Wnt3a-induced mESCs, we applied two methods to either abolish or compromise Wnt3a activity. First, we disrupted the crucial disulfide bonds of Wnt3a proteins using Dithiothreitol (DTT). When treated with DTT-inactivated Wnt3a beads, only 11.6% of the mESC divisions were asymmetric by APC localization (Figure S4A-B), significantly lower than the results using active Wnt3a beads (57.8%, Figure S1B). Second, mESCs can be induced to differentiate into mEpiSCs by bFGF+Activin A (Martinez and Cavalli, 2006; Pasini et al., 2004) (validation Figure B-C in Mendeley data). We checked the ACD occurrence after culturing mESCs with bFGF+Activin A in EpiSC medium for 24 hours, when mESCs transit from self-renewal to differentiation. We found that asymmetrically dividing mESCs decreased to 20.6% by Claudin 6 localization (Figure S4C-D), and 36.0% by APC localization (Figure S4E-F). In summary, these results demonstrate that inactivating Wnt3a or using the EpiSC medium with bFGF+Activin A can abrogate Wnt3a-induced ACD.
We next investigated how the above circumstances affect non-overlapping histone distribution. Upon inactivating Wnt3a, old and new H3 largely overlapped in mitotic mESCs (Figure 3A). The overlapping coefficient k value 0.90 is significantly higher than that in mESCs with active Wnt3a beads (Figure 3C), but is not significantly different from the k value for mESCs with CM+LIF+2i culturing condition (Figure 3C).
Figure 3: Compromised local non-overlapping old versus new H3 with DTT-inactivated Wnt3a beads or using EpiSC medium.
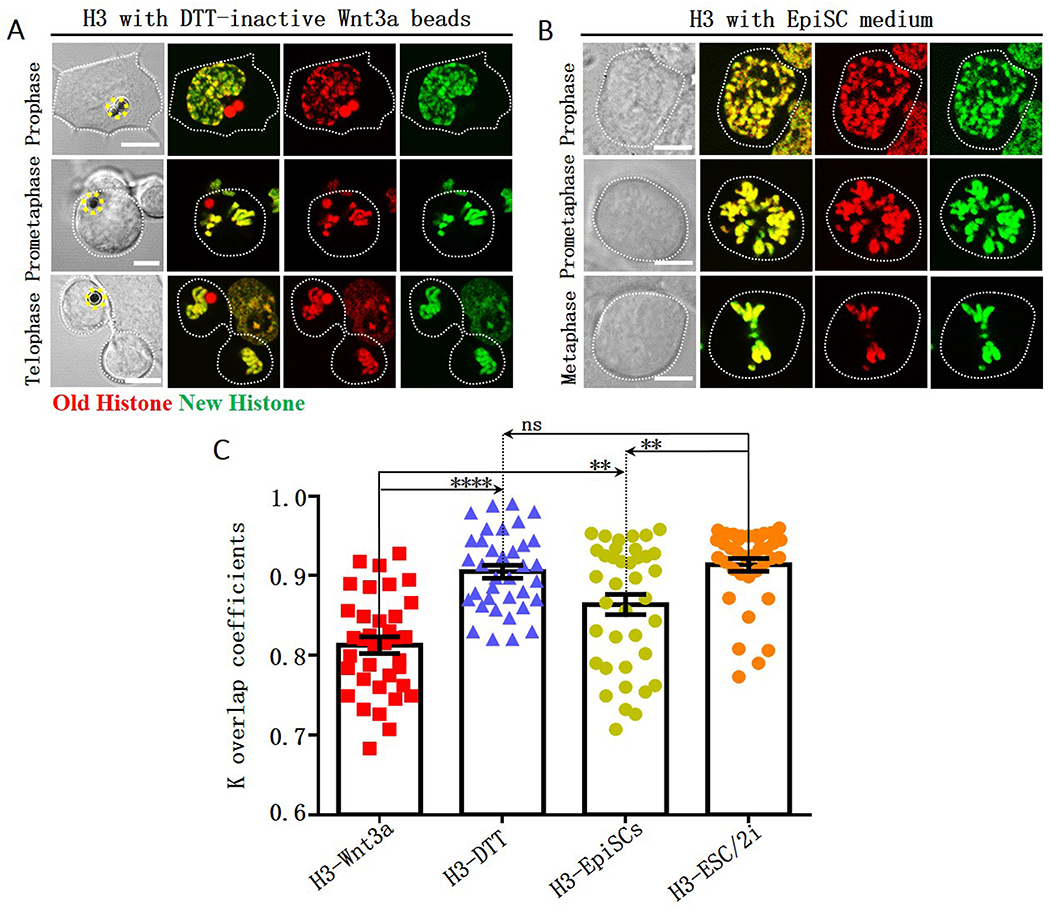
Largely overlapping old (red) versus new (green) H3 during the 2nd mitosis of mESCs after photoconversion (Figure 1C) with DTT-treated Wnt3a beads (A, yellow dotted circles label Wnt3a beads) or cultured with EpiSC medium (B). Scale bars: 5μm. (C) Quantification of old versus new H3 distribution in mitotic mESCs under different conditions. The coefficient k for old versus new H3 in mESCs with DTT-treated Wnt3a beads: 0.90 ± 0.01 (H3-DTT, N= 36), in mESCs cultured with EpiSC medium: 0.86 ± 0.01 (H3-EpiSCs, N= 38). The H3-Wnt3a and H3-ESC/2i data are from Figure 1F for direct comparison. Average ± SEM; **** P< 10−4, ** P< 0.005, ns: no significance, Student t test.
Furthermore, old and new H3 displayed more instances of overlapping in mitotic mESCs using EpiSC medium (Figure 3B). The overlapping coefficient k showed a range of distribution (Average= 0.86, Figure 3C), which is significantly higher than that of mESCs with Wnt3a beads but lower than that of mESCs with CM+LIF+2i (Figure 3C). Notably, more ACDs were detected using EpiSC medium than using DTT-inactivated Wnt3a beads (Figure S4D, S4F versus S4B). Since EpiSC medium promotes differentiation, both asymmetric and symmetric divisions can occur in the first 1-2 cell cycles. Therefore, old versus new H3 could have mixed non-overlapping and overlapping patterns.
Collectively, the local non-overlapping old versus new H3 distribution could be closely associated with the Wnt3a-induced ACD of mESCs. The culture condition that promotes either predominantly symmetric self-renewal in the CM+LIF+2i medium or differentiation in the EpiSC medium greatly attenuates this local non-overlapping pattern.
Canonical H2A and H2B, as well as the histone variant H3.3, show largely overlapping patterns in mESCs with Wnt3a beads
Previously, it has been shown that the globally asymmetric histone inheritance mode is specific to H3 and H4, as neither H2A, H2B (Wooten et al., 2019), nor H3.3 (Tran et al., 2012) exhibit the same pattern in Drosophila male GSCs. We next examined whether the local non-overlapping distribution patterns of H3 and H4 in Wnt3a-induced mESCs have a similar molecular specificity. We performed parallel tagging and photoconversion experiments to trace old versus new H2A, H2B and H3.3 distribution patterns in Wnt3a-induced mESCs (Figure 1A, 1C): Old versus new H2A (Figure 4A) and old versus new H2B (Figure 4B) displayed largely overlapping patterns in the mitotic mESCs with Wnt3a beads. The coefficient k for old versus new H2A and old versus new H2B were both 0.90, significantly higher than that of H3 (Figure 4D).
Figure 4: Local overlapping old versus new H2A, H2B and H3.3 in Wnt3a-induced mESCs.
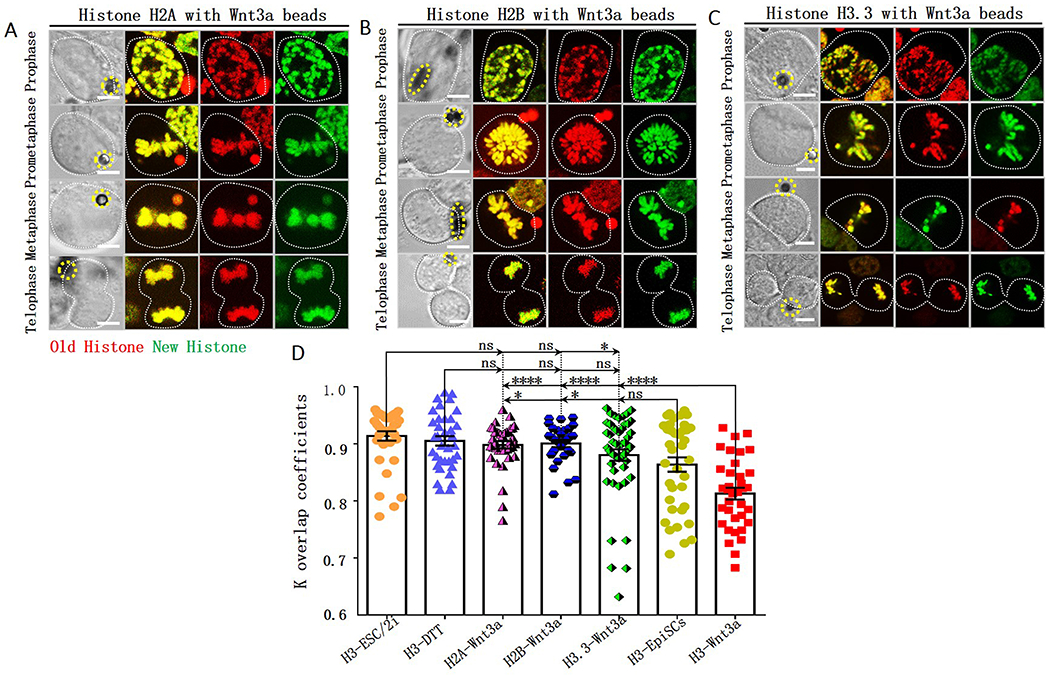
Overlapping old (red) versus new (green) H2A (A), H2B (B) and H3.3 (C) during the 2nd mitosis after photoconversion (Figure 1C) in Wnt3a-induced mESCs, yellow dotted circles label Wnt3a beads. Scale bars: 5μm. (D) Quantification of old versus new histone distribution in mitotic mESCs with Wnt3a beads. The coefficient k for old versus new H2A: 0.90 ± 0.01 (H2A-Wnt3a, N=38), for old versus new H2B: 0.90 ± 0.01 (H2B-Wnt3a, N=29), for old versus new H3.3: 0.88 ± 0.01 (H3.3-Wnt3a, N=52 with two subpopulations: 90.38% mESCs have k = 0.90 ± 0.01 while 9.62% mESCs have k = 0.69 ± 0.02). The H3-Wnt3a and H3-ESC/2i data are from Figure 1F; the H3-DTT and H3-EpiSC data are from Figure 3C for direct comparison. Average ± SEM; **** P< 10−4, * P< 0.05, ns: no significance, Student t test.
Overall, old and new H3.3 showed an overlapping pattern in mitotic mESCs with Wnt3a beads (Average k = 0.88, Figure 4C). However, the overlapping coefficient k showed a bimodal distribution: The majority of cells (90.4%) displayed largely overlapping patterns (k= 0.90) but a small population (9.6%) showed non-overlapping subdomains (k= 0.69, Figure 4D). Given the well-defined roles of H3.3 in regulating transcription (Ahmad and Henikoff, 2002; Tagami et al., 2004), the non-overlapping old versus new H3.3 likely reflects distinct transcriptional activities at different genomic regions. For example, transcriptionally active regions could have a faster turn-over carrying more new H3.3 than silenced regions (schematic model-D in Mendeley data).
DISCUSSION
Here, our study utilizes a cell culture system where Wnt3a beads induce ACD of mESCs (Habib et al., 2013). Combining this system with a photoconvertable Dendra2 to distinguish old versus new histones, we found that the distribution of old versus new H3 and H4 in Wnt3a-induced mESCs have distinct enriched regions during the 2nd mitosis after color ‘switch’ (schematic model-A in Mendeley data). Chromatin fiber assay further suggests that this non-overlapping histone distribution is likely established during DNA replication. In contrast, symmetrically dividing mESCs without or with inactivated Wnt3a beads do not display such a pattern (schematic model-B in Mendeley data). Moreover, this pattern is not for H2A, H2B (schematic model-C in Mendeley data), or the histone variant H3.3 (schematic model-D in Mendeley data). In conclusion, the differential old versus new histone distribution pattern is specific to H3 and H4 in Wnt3a-induced mESCs and can serve as a new system in studying histone inheritance during ACD in cell culture.
Although the cellular specificity to ACD and molecular specificity to H3/H4 are similar between Wnt3a-induced mESCs and Drosophila male GSCs (Tran et al., 2012; Wooten et al., 2019), there are differences between these two systems. The separation observed between old and new H3/H4 is local in mESCs but global in Drosophila male GSCs. When considering that ACD leads to two cells with distinct fates, the degree of differential gene expression between these two daughter cells could be different. For example, in the induced mESC system, the self-renewed ESC and the differentiation-poised EpiSC are regulated by a localized Wnt3a signal. It has been shown that only 852 genes are differentially expressed while 14346 genes share similar expression between mESCs and mEpiSCs (Factor et al., 2014). The ACD that generates two cells with a subset of differentially expressed genes could have an asymmetry at the corresponding genomic loci but a symmetric outcome in the rest of the genome. In Drosophila male germline, transcriptome changes dramatically to prepare for meiosis and terminal differentiation (Chen et al., 2011; Gan et al., 2010; Shi et al., 2020; Terry et al., 2006). It is possible that asymmetric inheritance of old versus new H3/H4 occurs at a substantial number of genes, leading to more globally separable signals. During stem cell ACD, the strength of the signals from the niche may also play an important role. For example, Wnt3a beads serve as a weaker niche signal as compared to the endogenous Drosophila testicular niche with multiple signaling pathways (Losick et al., 2011; Matunis et al., 2012). This is consistent with the observation that ~60% of mESCs undergo ACD with Wnt3a beads [Figure S1, (Habib et al., 2013)], whereas almost all Drosophila male GSCs undertake ACD (Yamashita et al., 2003; Yamashita et al., 2007). The different histone distribution patterns could also be due to the heterogeneity of mESCs (Chambers et al., 2007; Hayashi et al., 2008; Kalmar et al., 2009; Toyooka et al., 2008), which makes them respond to Wnt3a induction differently. Other cell type-specific features such as cell cycle progression may also contribute. For example, we detected old versus new histone signals during the 2nd mitosis after photoconversion. This way, new histones are efficiently incorporated into the duplicated genome during the previous S phase. However, post-replication chromatin maturation during S and G2 phases could also lead to differences in histone distribution pattern in M phase. The cell cycle of Drosophila male GSCs is unique, with elongated G2 phase and almost negligible G1 phase (Cheng et al., 2008; Sheng and Matunis, 2011; Yadlapalli et al., 2011; Yamashita et al., 2003; Yamashita et al., 2007). In contrast, mESCs have a longer and more detectable G1 phase that is critical for maintaining stemness (Koledova et al., 2010).
Despite these differences, the non-overlapping old versus new H3/H4 in Wnt3a-induced mESCs suggest that differential inheritance of H3 and H4 could serve as a more general mechanism during ACD. Because H4 is incorporated as a tetramer with H3, the similar asymmetric inheritance patterns of old H3 and H4 suggest that the old (H3-H4)2 tetramers did not split and were inherited as a whole unit, consistent with previous reports (Xu et al., 2010). Given the more stable feature of the (H3-H4)2 tetramers in interphase, their non-overlapping patterns on mitotic chromosomes suggest that it is established prior to M phase and could be initiated during S phase (schematic model-E in Mendeley data). Conversely, H2A and H2B are incorporated as dimers (Schuettengruber et al., 2017) and exhibit a more dynamic behavior during cell cycle (Kimura, 2005). In Drosophila male GSC, we proposed a two-step model that the (H3-H4)2 asymmetry is established during DNA replication (Step 1) and segregated during mitosis (Step 2) (Tran et al., 2013; Wooten et al., 2020b; Xie et al., 2017). Furthermore, the stem cell-specific “mitotic drive” regulating Step 2 can be disrupted by the microtubule depolymerizing drug NZ (Ranjan et al., 2019). We used NZ to synchronize mESCs, a necessary step to use photoconversion to distinguish old versus new histones in the context of cell cycle. Thus, the results from this study reflect distinct histone incorporation during replication and/or maturation during S/G2 phase, but not necessarily their segregation pattern during M phase.
Several improvements to the mESC system will help better understand the relationship between non-overlapping old versus new H3/H4 and distinct cell fates derived from ACD of mESCs. (1) More reagents are needed to address the histone inheritance mode at a single-cell resolution. For example, using the localization of APC or Claudin6 to distinguish asymmetric versus symmetric division modes for individual Wnt3a-induced mESCs turned to be technically challenging, as both proteins showed diffused patterns during mitosis especially at prophase and prometaphase. (2) We found increased instances of asymmetric H4K20me2/3 at replication regions in chromatin fibers derived from Wnt3a-induced mESCs (Figure 2, S3). However, because of the induction efficiency of Wnt3a beads, those chromatin fibers are derived from a mixed asymmetrically and symmetrically dividing cell population rather than pure asymmetrically dividing cells. Therefore, new methods are needed to isolate the asymmetric population. (3) To pinpoint the genomic region displaying old versus new H3/H4 asymmetry, more technologies are needed to study gene-specific histone inheritance pattern.
In conclusion, our findings that mESCs with Wnt3a beads exhibit more instances of local non-overlapping old versus new H3/H4 expand our understanding of how stem cells distinguish histone information, thus providing a cell culture system to investigate the molecular properties of pluripotent and lineage committed cells.
STAR ★ METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests of reagents should be directed to and will be fulfilled by the Lead Contact, Xin Chen (xchen32@jhu.edu). Materials Transfer Agreement (MTAs) required.
Materials Availability
Histone-Dendra2 Plasmids
The Dendra2-H3.3-N-14 vector was purchased from Addgene (Plasmid #57725). We amplified the full-length coding sequence of H3, H4, H2A and H2B from genomic DNA of mESCs and then subcloned them into Dendra2-H3.3-N-14 vector to replace H3.3 sequence. Related to Figure 1A. All Histone-Dendea2 plasmids generated in this study have been deposited to Addgene.
Data and Code Availability
The published article includes all datasets and supplemental tables and figures generated or analyzed in this study. The quantification code used for k overlap coefficient measurement and additional supplemental data (including figures, raw image galleries, quantification tables and movie) were all deposited on the Mendeley website:
https://data.mendeley.com/datasets/pc8z47jzm5/draft?a=7e80bf22-2723-423b-8bea-445e629c5ec8
EXPERIMENTAL MODELS AND SUBJECT DETAILS
mESC Cell Line
E14TG2a embryonic stem cells (provided by Dr. Yixian Zheng, Carnegie Institute, Baltimore, USA) were cultured at 37 °C and 5% CO2 on gelatin-coated petri dishes in Dulbecco’s modified Eagle’s solution (Cat# 10829018, Thermo Fisher) supplemented with 15% fetal bovine serum (Cat# 10082-147, Gibco), 50 uM β-mercaptoethanol (Cat# 21985023, Thermo Fisher), 1 mM Sodium Pyruvate (Cat# 11360070, Thermo Fisher), 1× MEM non-essential amino acids (Cat# 11140050, Thermo Fisher), 100 U/mL penicillin, 100 μg/mL streptomycin (Cat#15140122, Thermo Fisher), 1000 U/mL recombinant mouse LIF (Cat# ESG1107, Millipore), 1 μM PD032501 (Cat# Axon 1408, Axon Medchem), and 3 μM CHIR99021 (Cat# Axon 1386, Axon Medchem).
Histone-Dendra2 transgenic mESC lines
The transfection of E14TG2a mESCs was performed using Lipofectamine 2000 (Cat# 12566014, Invitrogen) according to the manufacturer’s instructions. Incubate cells with each Histone-Dendra2 plasmid for 1-3 days at 37°C. Then purify the transfected cells with manual selection or flow cytometry using the green fluorescence of Dendra2. The final stable mESC lines each carries one of the Histone-Dendra2 transgenes, respectively.
METHOD DETAILS
Preparation of the Wnt3a coated beads
The mouse Wnt3a protein was produced in Drosophila S2 cells and purified by Blue Sepharose affinity and gel filtration chromatography. To immobilize the Wnt3a protein onto M-270 Dynabeads® (Cat# 14305D, Invitrogen), we first use carbodiimide and N-hydroxyl succinimide (50mg/ml in 25mM cold MES buffer pH 5) to activate the carboxylic acid groups and then incubate the activated beads and purified Wnt3a protein at room temperature for one hour. Finally store the Wnt3a coated beads in PBS/ 1% BSA buffer at 4°C.
Asymmetric cell division induction of mESCs with Wnt3a beads
First culture the Histone-Dendra2 and WT mESCs in complete medium with LIF and 2i for 1 day; then change it with N2B27 mediumconsisting of Neurobasal medium (Cat# 21103049, Thermo Fisher), DMEM/F12 (Cat# 11320033, Thermo Fisher), N2-Supplement (Cat# 17502048, Thermo Fisher)), B27-Supplement (Cat# 17504044, Thermo Fisher), 7.5% BSA (Cat# 15260037, Thermo Fisher), GlutaMAX (Cat# 35050061, Thermo Fisher), 100 U/mL penicillin, 100 μg/mL streptomycin (Cat# 15140122, Thermo Fisher) with LIF and 2i for 1-2 days (Ying et al., 2008), until seeing the middle sized colonies; then transfer it to the N2B27 medium with LIF and soluble Wnt3a protein for 1 more day; finally use N2B27/LIF medium to resuspend the single cells and Wnt3a beads into Fluorodish (Cat# FD3510-100, WPI) with an appropriate ratio to induce the asymmetric cell division (Habib et al., 2013). Related to Figure 1C.
Photoconversion with Dendra2 fusion protein
For the photoconversion with Dendra2 fusion protein, the 20× objective of LSM 780 microscope was selected; Then draw the region of interest (ROI) using the tools in ‘Regions’, those mitotic green cells were selected; the 405 nm laser at 6%–10% power for 30s-pulse and 100-200 iterations were used to photoswitch Dendra2 protein from green fluorescence to red fluorescence. Related to Figure 1B.
Immunofluorescence and Alkaline Phosphatase (ALP) Detection of mESCs
Immunofluorescence staining of mESCs was performed using standard procedures (Hime et al., 1996). Primary antibodies were rabbit anti-APC (1:150, Santa Cruz sc-7930), goat anti-Claudin 6 (1:150, Santa Cruz sc-17669), rabbit anti-Sox2 (1:200, Active Motif 39823), rat anti-Nanog (1:200, Active Motif 61627), rabbit anti-Rex1 (1:200, Thermo Fisher# PA5-27567), rabbit anti-Oct4 (1:200, Abcam# ab181557), mouse anti-SSEA-1 (1:50, Thermo Fisher# MC-480). Secondary antibodies were the Alexa Fluor-conjugated series (1:600, Molecular Probes). ESCs and EpiSCs were also stained with an Alkaline Phosphatase Detection Kit (SCR004, Millipore) following the manufacturer’s instructions. Culture the cells for 5 days prior to analyzing ALP activity. Then fix the cells with 4% paraformaldehyde (PFA) in PBS for 1-2 mins. Aspirate the fixative and rinse with 1× Rinse buffer for twice. Prepare the Alkaline Phosphatase staining solution by mixing Fast Red Violet (FRV) with Napthol AS-BI phosphate solution and water in a 2:1:1 ratio. Incubate the cells in dark at room temperature for 30 mins. Aspirate the staining solution and rinse with 1× Rinse buffer for twice. Cover the cells with PBS and then image the colonies with ALP activity. Related to validation Figure A-B and expression Figure E in Mendeley data.
Western Blotting
The cell samples were lysed in RIPA Lysis and Extraction buffer (Cat# 89901; Thermo Fisher) with Protease Inhibitor Cocktail for general use (Cat# P1005; Beyotime) and Phosphatase inhibitor cocktail A (Cat# P1081; Beyotime). The total protein concentration in histone-dendra2 transfected and WT mESCs samples was normalized using the Pierce BCA Protein Assay Kit (Cat# 23225; Thermo Fisher). Equal amounts of protein were resolved by 10% SDS-polyacrylamide gel electrophoresis and transferred to Immun-Blot PVDF Membrane (Cat# 1620177, Bio-Rad). After blocking in 3% bovine serum albumin in TBS with 0.05% Tween-20 (TBST), the membranes were incubated with Rabbit polyclonal anti-Histone H3 (1:4000, Cat# ab1791, Abcam), Rabbit polyclonal anti-Histone H4 (1:4000, Cat# ab10158, Abcam), Rabbit monoclonal anti-Histone H2A (1:2000, Cat# ab18975, Abcam), Mouse monoclonal anti-Histone H2B (1:2000, Cat# ab52484, Abcam), Rabbit monoclonal anti-Histone H3.3 (1:4000, Cat# ab176840, Abcam), Rabbit monoclonal anti-GAPDH antibody (1:2000, Cat# ab181602; Abcam). The secondary antibodies are Goat anti-Rabbit IgG (H+L) HRP (1:3000, Cat# LK2001L, Sungene) and Goat anti-Mouse IgG (H+L) HRP (1:3000, Cat# LK2003L, Sungene). The blots were visualized with Immobilon™ Western chemiluminescence kit (Cat# P90720, Millipore). The images were acquired on the Image Quant LAS 4000 Gel Imager (iBright CL1000, Thermo Fisher). Related to expression Figure-A in Mendeley data.
FACS analysis of cell cycle with propidium iodide staining
Harvest the cells in the appropriate manner and wash in PBS and fix them in cold 70% ethanol for 30 mins at 4°C. Then wash the cells in PBS for twice. Spin at 1500 rpm in the centrifuge (5810R, Eppendorf) and discard the supernatant. Add 2 μL-20 mg/mL stock of RNase (100×) to 50 uL PBS and incubate 5 mins at room temperature. Finally add 50 μL propidium iodide-PI (1mg/mL stock solution) to 450 uL PBS and measure the cell cycle of mESCs carrying different histone-dendra2 transgenes with BD FACS Verse machine. The exported data was analyzed with FlowJo software. Related to expression Figure-B in Mendeley data.
Chromatin fiber preparation with EdU incorporation and immunostaining
Asymmetrically division of H3-dendra2 mESCs was first induced with Wnt3a beads. Then incubate those cells with 10μM EdU analog for 30 mins in a humidified incubator with 37°C, 5% CO2. After the EdU incorporation, mESCs were washed for twice with PBS and then trypsinized to single cell suspensions. Resuspend those cells in 1 mL of 0.8% sodium citrate, vortex and stay still for 15 mins at room temperature. Adjust the cell density to 2×105 cells per tube, then centrifuge (5810R, Eppendorf) 100 uL of cells onto precleaned Superfrost Plus microslides (48311-703, VWR) at 1200 rpm for 4 mins using Single Cytofunnel™ (Cat# 5991040, Shandon). After spinning, place the slides into the lysis buffer (500 mM NaCl, 100mM Tris, 0.5 M Urea and 1% Triton X-100) and stay still for 20 mins at room temperature. Slightly and slowly remove the slides from lysis buffer for 1-2 mins, and dry the bottom of slide on a Kimwipe. Next, fix the slides with 2% PFA for 15 mins at room temperature. Wash the slides with PBS + 0.2% Triton X-100 for 2×5 mins, and then block the slides with blocking buffer (PBS + 0.2% Triton X-100 + 10% Normal donkey serum) for 30 mins at room temperature. Perform H4K20me2/3 (1:800, Cat# ab92552, Abcam) and PCNA (1:800, Cat# ab78517, Abcam) antibody incubations and DAPI staining in a humid chamber overnight at 4°C, with a piece of Parafilm over the slides. Slides were then washed twice with 50 mL PBS and incubated with secondary antibodies for 2 hours at room temperature. EdU detection was performed for 30 mins at room temperature according to Click-iT Plus EdU Cell Proliferation Kit for Imaging, Alexa FluorTM 647 dye (Cat# C10640, Invitrogen). Finally, the slides were washed twice with 50 mL PBS and mounted with Vectashield mounting medium (Cat# H1200, Vector).
Image Acquisition and Airyscan processing
All confocal images of cell samples were processed with the Zeiss LSM 700, Zeiss LSM 780 Multiphoton confocal microscope with 40× or 63× oil immersion objectives and processed using Image J Fiji software. Conventional confocal imaging was performed to show the old vs new histone distribution patterns in Figures 1B, 1D, 1E, 3A, 3B, 4A, 4B, 4C, S1A, S1C, S1E; the mESCs markers for stemness identity in expression Figure E in Mendeley data ; APC and Claudin 6 labelled asymmetric cell division in Figures S2A, S2C, S4A, S4C, S4E.
For the chromatin fibers, Zeiss LSM 880 with Airyscan and GaAsp detectors was used for both conventional confocal and Airyscan superresolution mode with a 63×, Plan Apochromat (1.4 NA) oil objective. Airyscan confocal imaging was performed to resolve the H4K20me2/3 and PCNA distribution along the replicating chromatin fibers in Figures 2A and S3A.
Super-Resolution SoRa Imaging
Super-resolution images were acquired using CSU-W1 SoRa. Chromatin fibers with EdU labeling, H4K20me2/3 and PCNA distribution were imaged with a 100×, 1.4 NA oil objective on a Nikon Spinning Disk confocal microscope. Related to Figure 2D. Deconvolutions of images were then carried out using the Huygens Professional Deconvolution software, which achieved improved calculated/theoretical PSFs that is based on values that are calculated from the metadata of the acquired image.
QUANTIFICATION AND STATISTICAL ANALYSIS
Measurement of Colocalization of old vs new histone in mESCs
The k overlap coefficient is defined using the following calculation:
For each focal plane of the segmented cell, the intensity value of individual voxel for one channel is normalized by percentage of intensity of that channel.
(e.g.)
The k-overlap coefficient for each focal plane, is calculated as the equations described above on normalized intensity values of voxels.
The overall k coefficient of one cell is a weighted average of k-overlap coefficient for each focal plane, based on the green (first) channel.
(e.g.)
Unpaired Student’s t-test was applied to calculate the P-values for the k overlap coefficients of histone distribution patterns. Related to Figures 1F, 3C, 4D.
Measurement of H4K20me2/3 enrichments on PCNA (-) and PCNA (+) Chromatids in DNA Replication regions
The quantification of fluorescent protein level along the chromatin fibers was performed by Fiji (Image J) software. The signal intensity of target protein was measured by subtracting the signal values of the background from the raw signal intensity in the same region size. All quantification of fluorescence signal was done using this method. First select the chromatin fibers with ~200nm diameter, then use EdU to show the DNA replication regions. Then use PCNA (a lagging strand-enriched protein) as a proxy for lagging strands. The classification of asymmetric PCNA distribution used the log2 ratio of criteria (chromatin strand-1/ chromatin strand-2) >0.5 and <-0.5. Then selected the corresponding H4K20me2/3 intensity for the strand enrichment measurement.
To define the leading and lagging enrichment of H4K20me2/3, the criteria are defined with the dataset generated from chromatin fibers without Wnt3a induction. The ranges are set as Average ± 1 × Standard derivation (SD): fibers with the log2 (PCNA(-) strand/ PCNA(+) strand ratio)>1.025 were classified as leading strand-enriched; fibers with the log2 (PCNA(-) strand/ PCNA(+) strand ratio) -0.953~1.025 were classified as symmetric; fibers with the log2 (PCNA(-) strand/ PCNA(+) strand ratio) <−0.953 were classified as lagging strand-enriched. Related to Figures 2C, 2F, 2G, S3C and S3D.
Statistics
The statistics analysis in those figures was performed with Prism version 5.0 (GraphPad). Data are presented as average ± SEM and significant difference between two groups were noted by asterisks (* P < 0.05, ** P < 0.01 and **** P < 0.0001). Statistical parameters, including the number of samples (N), type of tests, descriptive statistics and significance are all annotated in the figure legends.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-APC (used at 1: 150) | Santa Cruz | Cat# sc-7930; RRID: AB_2242804 |
| Goat polyclonal anti-Claudin 6 (used at 1:150) | Santa Cruz | Cat# sc-17669; RRID: AB_2083279 |
| Rabbit polyclonal anti-Sox2 (used at 1:200) | Active Motif | Cat# Active Motif 39823; RRID: AB_2793356 |
| Rat monoclonal anti-Nanog (used at 1:200) | Active Motif | Cat# Active Motif 61627; RRID: AB_2793709 |
| Rabbit monoclonal anti-Oct4 (used at 1:200) | Abcam | Cat# ab181557; RRID: AB_2687916 |
| Rabbit polyclonal anti-Rex1 (used at 1:200) | Thermo Fisher | Cat# PA5-27567; RRID: AB_2545043 |
| Mouse monoclonal anti-SSEA-1 (used at 1:50) | Thermo Fisher | Cat# MC-480;RRID: AB_528475 |
| Rabbit monoclonal anti-PCNA (used at 1:800) | Abcam | Cat# ab92552; RRID: AB_10561973 |
| Mouse monoclonal anti-H4K20me2/3 (used at 1:800) | Abcam | Cat# ab78517; RRID: AB_1951279 |
| Rabbit polyclonal anti-Histone H3 (used at 1:4000) | Abcam | Cat# ab1791; RRID: AB_302613 |
| Rabbit polyclonal anti-Histone H4 (used at 1:4000) | Abcam | Cat# ab10158; RRID: AB_296888 |
| Rabbit monoclonal anti-Histone H2A (used at 1:3000) | Abcam | Cat# ab18975; RRID: AB_444713 |
| Mouse monoclonal anti-Histone H2B (used at 1:3000) | Abcam | Cat# ab52484; RRID: AB_1139809 |
| Rabbit monoclonal anti-Histone H3.3 (used at 1:4000) | Abcam | Cat# ab176840; RRID: AB_2715502 |
| Rabbit monoclonal anti-GAPDH (used at 1:3000) | Abcam | Cat# ab181602; RRID: AB_2630358 |
| Goat anti-Rabbit IgG(H+L) HRP | Sungene | Cat# LK2001L |
| Goat anti-Mouse IgG(H+L) HRP | Sungene | Cat# LK2003L |
| Chemicals and Recombinant Proteins | ||
| Dulbecco’s modified Eagle’s solution | Thermo Fisher | Cat# 10829018 |
| Neurobasal™ Medium | Thermo Fisher | Cat# 21103049 |
| DMEM/F-12 | Thermo Fisher | Cat# 11320033 |
| PD032501 | Axon Medchem | Cat# Axon 1408 |
| CHIR99021 | Axon Medchem | Cat# Axon 1386 |
| Activin A, Recombinant Human Protein | Thermo Fisher | Cat# PHC9564 |
| Bfgf, Recombinant Human Protein | Thermo Fisher | Cat# 13256029 |
| Recombinant Mouse Wnt3a protein | R&D Systems | Cat# 1324-WN-002 |
| Fetal Bovine Serum, heat inactivated (FBS) | Gibco | Cat# 10082-147 |
| MEM non-essential amino acids | Thermo Fisher | Cat# 11140050 |
| Sodium Pyruvate | Thermo Fisher | Cat# 11360070 |
| Recombinant mouse LIF | Millipore | Cat# ESG1107 |
| β-mercaptoethanol | Thermo Fisher | Cat# 21985023 |
| GlutaMAX | Thermo Fisher | Cat# 35050061 |
| Trypsin-EDTA (0.25%) | Gibco | Cat# 25200056 |
| 2% Gelatin Solution | Merck | Cat# G1393-20Ml |
| 7.5% BSA | Thermo Fisher | Cat# 15260037 |
| N2-Supplement | Thermo Fisher | Cat# 17502048 |
| B27-Supplement | Thermo Fisher | Cat# 17504044 |
| Fibronectin human plasma | Merck | Cat# F2006-1MG |
| Normal Donkey Serum | Merck | Cat# D9663 |
| Lipofectamine 2000 | Invitrogen | Cat# 12566014 |
| Nocodazole | Sigma | Cat# M1404 |
| Rnase A | TIANGEN | Cat# RT405-02 |
| Dimethyl sulfoxide | Merck | Cat# D2650 |
| Vectashield mounting medium | Vector | Cat# H1200 |
| RIPA Lysis and Extraction buffer | Thermo Fisher | Cat# 89901 |
| Protease Inhibitor Cocktail | Beyotime | Cat# P1005 |
| Phosphatase inhibitor cocktail A | Beyotime | Cat# P1081 |
| Penicillin-Streptomycin | Thermo Fisher | Cat# 15140122 |
| Experimental Models: Cell Lines | ||
| E14TG2a embryonic stem cells | (Ma et al., 2016) | N/A |
| Histone-Dendra2 transgenic mESC lines | This paper | N/A |
| Software and Algorithms | ||
| Matlab | MathWorks | https://www.mathworks.com/products/matlab.html |
| FlowJo | FlowJo LLC | https://www.flowjo.com/ |
| Image J | NIH | https://imagej.nih.gov/ij/ |
| Huygens | Scientific Volume Imaging | The Netherlands, http://svi.nl |
| GraphPad Prism 5.0 | GraphPad | http://www.graphpad.com/scientific-software/prism/ |
| Mendeley data-additional supplemental data including figures, tables and movie | This paper | https://data.mendeley.com/datasets/pc8z47jzm5/draft?a=7e80bf22-2723-423b-8bea-445e629c5ec8 |
| Others | ||
| Dendra2-H3.3-N-14 vector | Addgene | Plasmid# 57725 |
| Dendra2-H3-N-14 vector | This paper | Plasmid# 157814 |
| Dendra2-H4-N-14 vector | This paper | Plasmid# 157815 |
| Dendra2-H2A-N-14 vector | This paper | Plasmid# 157816 |
| Dendra2-H2B-N-14 vector | This paper | Plasmid# 157817 |
| GenElute™ Mammalian Genomic DNA Miniprep Kit | Merck | Cat# G1N10-1KT |
| Alkaline Phosphatase Detection Kit | Millipore | Cat# SCR004 |
| Click-iT Plus EdU Cell Proliferation Kit for Imaging | Invitrogen | Cat# C10640 |
| Immobilon™ Western chemiluminescence kit | Millipore | Cat# P90720 |
| Pierce BCA Protein Assay Kit | Thermo Fisher | Cat# 23225 |
| Immun-Blot PVDF Membrane | Bio-Rad | Cat# 1620177 |
| Fluorodish | WPI, Inc. | Cat# FD3510-100 |
| Superfrost Plus microslides | VWR | Cat# 48311-703 |
| Single Cytofunnel™ | Shandon | Cat# 5991040 |
| Dynabeads™ M-270 Carboxylic Acid | Invitrogen | Cat# 14305D |
ACKNOWLEDGEMENTS
We thank Johns Hopkins Integrated Imaging Center for imaging support, Chengyu Fan and Xiaoming Li for technical assistance and the Molecular Imaging Core Facility at School of Life Science and Technology, ShanghaiTech University for technical support, Qinglan Emily Yu for editorial assistance. Supported by the program of China Scholarships Council (No. 201506230049 to B.M.); Sir Henry Dale Fellowship (102513/Z/13/Z to S.J.H.); Division of Intramural Research, NHLBI/NIH (K.Z.); NIH R21HD084959 and R35GM127075, HHMI Faculty Scholar from Howard Hughes Medical Institute (X.C.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Ahmad K, and Henikoff S (2002). The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell 9, 1191–1200. [DOI] [PubMed] [Google Scholar]
- Alabert C, Barth TK, Reveron-Gomez N, Sidoli S, Schmidt A, Jensen ON, Imhof A, and Groth A (2015). Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev 29, 585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis CD, and Jenuwein T (2016). The molecular hallmarks of epigenetic control. Nat Rev Genet 17, 487–500. [DOI] [PubMed] [Google Scholar]
- Azuma T, and Kei T (2015). Super-resolution spinning-disk confocal microscopy using optical photon reassignment. Opt Express 23, 15003–15011. [DOI] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, and Smith A (2007). Nanog safeguards pluripotency and mediates germline development. Nature 450, 1230–1234. [DOI] [PubMed] [Google Scholar]
- Chen X, Lu C, Prado JR, Eun SH, and Fuller MT (2011). Sequential changes at differentiation gene promoters as they become active in a stem cell lineage. Development 138, 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Turkel N, Hemati N, Fuller MT, Hunt AJ, and Yamashita YM (2008). Centrosome misorientation reduces stem cell division during ageing. Nature 456, 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudakov DM, Lukyanov S, and Lukyanov KA (2007). Tracking intracellular protein movements using photoswitchable fluorescent proteins PS-CFP2 and Dendra2. Nat Protoc 2, 2024–2032. [DOI] [PubMed] [Google Scholar]
- Clevers H (2005). Stem cells, asymmetric division and cancer. Nat Genet 37, 1027–1028. [DOI] [PubMed] [Google Scholar]
- Del Olmo I, Lopez JA, Vazquez J, Raynaud C, Pineiro M, and Jarillo JA (2016). Arabidopsis DNA polymerase recruits components of Polycomb repressor complex to mediate epigenetic gene silencing. Nucleic Acids Res 44, 5597–5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Factor DC, Corradin O, Zentner GE, Saiakhova A, Song L, Chenoweth JG, McKay RD, Crawford GE, Scacheri PC, and Tesar PJ (2014). Epigenomic comparison reveals activation of “seed” enhancers during transition from naive to primed pluripotency. Cell Stem Cell 14, 854–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluckiger AC, Marcy G, Marchand M, Negre D, Cosset FL, Mitalipov S, Wolf D, Savatier P, and Dehay C (2006). Cell cycle features of primate embryonic stem cells. Stem Cells 24, 547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Follmer NE, Simon MD, Aghia G, and Butler JD (2009). Polycomb proteins remain bound to chromatin and DNA during DNA replication in vitro. Cell 137, 110–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Q, Chepelev I, Wei G, Tarayrah L, Cui K, Zhao K, and Chen X (2010). Dynamic regulation of alternative splicing and chromatin structure in Drosophila gonads revealed by RNA-seq. Cell Res 20, 763–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, and Lukyanov KA (2006). Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol 24, 461–465. [DOI] [PubMed] [Google Scholar]
- Habib SJ, Chen BC, Tsai FC, Anastassiadis K, Meyer T, Betzig E, and Nusse R (2013). A localized Wnt signal orients asymmetric stem cell division in vitro. Science 339, 1445–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, de Sousa Lopes SMC, Tang F, Lao K, and Surani MA (2008). Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell 3, 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hime GR, Brill JA, and Fuller MT (1996). Assembly of ring canals in the male germ line from structural components of the contractile ring. J Cell Sci 109 ( Pt 12), 2779–2788. [DOI] [PubMed] [Google Scholar]
- Jiang D, and Berger F (2017). DNA replication-coupled histone modification maintains Polycomb gene silencing in plants. Science 357, 1146–1149. [DOI] [PubMed] [Google Scholar]
- Kahney EW, Ranjan R, Gleason RJ, and Chen X (2017). Symmetry from Asymmetry or Asymmetry from Symmetry? Cold Spring Harb Symp Quant Biol 82, 305–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmar T, Lim C, Hayward P, Munoz-Descalzo S, Nichols J, Garcia-Ojalvo J, and Martinez Arias A (2009). Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol 7, e1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H (2005). Histone dynamics in living cells revealed by photobleaching. DNA Repair (Amst) 4, 939–950. [DOI] [PubMed] [Google Scholar]
- Knoblich JA (2010). Asymmetric cell division: recent developments and their implications for tumour biology. Nat Rev Mol Cell Biol 11, 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koledova Z, Kafkova LR, Calabkova L, Krystof V, Dolezel P, and Divoky V (2010). Cdk2 inhibition prolongs G1 phase progression in mouse embryonic stem cells. Stem Cells Dev 19, 181–194. [DOI] [PubMed] [Google Scholar]
- Kouzarides T (2007). Chromatin modifications and their function. Cell 128, 693–705. [DOI] [PubMed] [Google Scholar]
- Lecona E, Rojas LA, Bonasio R, Johnston A, Fernandez-Capetillo O, and Reinberg D (2013). Polycomb protein SCML2 regulates the cell cycle by binding and modulating CDK/CYCLIN/p21 complexes. PLoS Biol 11, e1001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo SM, Follmer NE, Lengsfeld BM, Madamba EV, Seong S, Grau DJ, and Francis NJ (2012). A bridging model for persistence of a polycomb group protein complex through DNA replication in vitro. Mol Cell 46, 784–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick VP, Morris LX, Fox DT, and Spradling A (2011). Drosophila stem cell niches: a decade of discovery suggests a unified view of stem cell regulation. Dev Cell 21, 159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko F, Ramsahoye BH, and Jaenisch R (2000). DNA methylation in Drosophila melanogaster. Nature 408, 538–540. [DOI] [PubMed] [Google Scholar]
- Ma C, Karwacki-Neisius V, Tang H, Li W, Shi Z, Hu H, Xu W, Wang Z, Kong L, Lv R, et al. (2016). Nono, a Bivalent Domain Factor, Regulates Erk Signaling and Mouse Embryonic Stem Cell Pluripotency. Cell Rep 17, 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez AM, and Cavalli G (2006). The role of polycomb group proteins in cell cycle regulation during development. Cell Cycle 5, 1189–1197. [DOI] [PubMed] [Google Scholar]
- Matunis EL, Stine RR, and de Cuevas M (2012). Recent advances in Drosophila male germline stem cell biology. Spermatogenesis 2, 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minter LM, Dickinson ES, Naber SP, and Jerry DJ (2002). Epithelial cell cycling predicts p53 responsiveness to gamma-irradiation during post-natal mammary gland development. Development 129, 2997–3008. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, and Kimble J (2006). Asymmetric and symmetric stem-cell divisions in development and cancer. Nature 441, 1068–1074. [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, and Helin K (2004). Polycomb group proteins in cell cycle progression and cancer. Cell Cycle 3, 396–400. [PubMed] [Google Scholar]
- Petryk N, Dalby M, Wenger A, Stromme CB, Strandsby A, Andersson R, and Groth A (2018). MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science 361, 1389–1392. [DOI] [PubMed] [Google Scholar]
- Piunti A, Rossi A, Cerutti A, Albert M, Jammula S, Scelfo A, Cedrone L, Fragola G, Olsson L, Koseki H, et al. (2014). Polycomb proteins control proliferation and transformation independently of cell cycle checkpoints by regulating DNA replication. Nat Commun 5, 3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan R, Snedeker J, and Chen X (2019). Asymmetric Centromeres Differentially Coordinate with Mitotic Machinery to Ensure Biased Sister Chromatid Segregation in Germline Stem Cells. Cell Stem Cell 25, 666–681.e665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Bourbon HM, Di Croce L, and Cavalli G (2017). Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 171, 34–57. [DOI] [PubMed] [Google Scholar]
- Sheng XR, and Matunis E (2011). Live imaging of the Drosophila spermatogonial stem cell niche reveals novel mechanisms regulating germline stem cell output. Development 138, 3367–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Lim C, Tran V, Cui K, Zhao K, and Chen X (2020). Single-cyst transcriptome analysis of Drosophila male germline stem cell lineage. Development 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaguru M, Urban MA, Fried G, Wesseln CJ, Mander L, and Punyasena SW (2018). Comparative performance of airyscan and structured illumination superresolution microscopy in the study of the surface texture and 3D shape of pollen. Microsc Res Tech 81, 101–114. [DOI] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, and Nakatani Y (2004). Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61. [DOI] [PubMed] [Google Scholar]
- Terry NA, Tulina N, Matunis E, and DiNardo S (2006). Novel regulators revealed by profiling Drosophila testis stem cells within their niche. Dev Biol 294, 246–257. [DOI] [PubMed] [Google Scholar]
- Toyooka Y, Shimosato D, Murakami K, Takahashi K, and Niwa H (2008). Identification and characterization of subpopulations in undifferentiated ES cell culture. Development 135, 909–918. [DOI] [PubMed] [Google Scholar]
- Tran V, Feng L, and Chen X (2013). Asymmetric distribution of histones during Drosophila male germline stem cell asymmetric divisions. Chromosome Res 21, 255–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran V, Lim C, Xie J, and Chen X (2012). Asymmetric division of Drosophila male germline stem cell shows asymmetric histone distribution. Science 338, 679–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkei ZG, and Yamashita YM (2018). Emerging mechanisms of asymmetric stem cell division. J Cell Biol 217, 3785–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooten M, Li Y, Snedeker J, Nizami ZF, Gall JG, and Chen X (2020a). Superresolution imaging of chromatin fibers to visualize epigenetic information on replicative DNA. Nat Protoc 15, 1188–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooten M, Ranjan R, and Chen X (2020b). Asymmetric Histone Inheritance in Asymmetrically Dividing Stem Cells. Trends Genet 36, 30–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooten M, Snedeker J, Nizami ZF, Yang X, Ranjan R, Urban E, Kim JM, Gall J, Xiao J, and Chen X (2019). Asymmetric histone inheritance via strand-specific incorporation and biased replication fork movement. Nat Struct Mol Biol 26, 732–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Wooten M, Tran V, Chen BC, Pozmanter C, Simbolon C, Betzig E, and Chen X (2015). Histone H3 Threonine Phosphorylation Regulates Asymmetric Histone Inheritance in the Drosophila Male Germline. Cell 163, 920–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Wooten M, Tran V, and Chen X (2017). Breaking Symmetry - Asymmetric Histone Inheritance in Stem Cells. Trends Cell Biol 27, 527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Long C, Chen X, Huang C, Chen S, and Zhu B (2010). Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science 328, 94–98. [DOI] [PubMed] [Google Scholar]
- Yadlapalli S, Cheng J, and Yamashita YM (2011). Drosophila male germline stem cells do not asymmetrically segregate chromosome strands. J Cell Sci 124, 933–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita YM, Jones DL, and Fuller MT (2003). Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science 301, 1547–1550. [DOI] [PubMed] [Google Scholar]
- Yamashita YM, Mahowald AP, Perlin JR, and Fuller MT (2007). Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science 315, 518–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, and Smith A (2008). The ground state of embryonic stem cell self-renewal. Nature 453, 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NL, DiMaggio PA, and Garcia BA (2010). The significance, development and progress of high-throughput combinatorial histone code analysis. Cellular and Molecular Life Sciences 67, 3983–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Gan H, Han J, Zhou ZX, Jia S, Chabes A, Farrugia G, Ordog T, and Zhang Z (2014). Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol Cell 56, 551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Huang H, Liu D, Cheng Y, Liu X, Zhang W, Yin R, Zhang D, Zhang P, Liu J, et al. (2015). N6-methyladenine DNA modification in Drosophila. Cell 161, 893–906. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets and supplemental tables and figures generated or analyzed in this study. The quantification code used for k overlap coefficient measurement and additional supplemental data (including figures, raw image galleries, quantification tables and movie) were all deposited on the Mendeley website:
https://data.mendeley.com/datasets/pc8z47jzm5/draft?a=7e80bf22-2723-423b-8bea-445e629c5ec8


