Abstract
In recent months, a novel virus named Coronavirus has emerged to become a pandemic. The virus is spreading not only humans, but it is also affecting animals. First ever case of Coronavirus was registered in city of Wuhan, Hubei province of China on 31st of December in 2019. Coronavirus infected patients display very similar symptoms like pneumonia, and it attacks the respiratory organs of the body, causing difficulty in breathing. The disease is diagnosed using a Real-Time Reverse Transcriptase Polymerase Chain reaction (RT-PCR) kit and requires time in the laboratory to confirm the presence of the virus. Due to insufficient availability of the kits, the suspected patients cannot be treated in time, which in turn increases the chance of spreading the disease. To overcome this solution, radiologists observed the changes appearing in the radiological images such as X-ray and CT scans. Using deep learning algorithms, the suspected patients’ X-ray or Computed Tomography (CT) scan can differentiate between the healthy person and the patient affected by Coronavirus. In this paper, popular deep learning architectures are used to develop a Coronavirus diagnostic systems. The architectures used in this paper are VGG16, DenseNet121, Xception, NASNet, and EfficientNet. Multiclass classification is performed in this paper. The classes considered are COVID-19 positive patients, normal patients, and other class. In other class, chest X-ray images of pneumonia, influenza, and other illnesses related to the chest region are included. The accuracies obtained for VGG16, DenseNet121, Xception, NASNet, and EfficientNet are 79.01%, 89.96%, 88.03%, 85.03% and 93.48% respectively. The need for deep learning with radiologic images is necessary for this critical condition as this will provide a second opinion to the radiologists fast and accurately. These deep learning Coronavirus detection systems can also be useful in the regions where expert physicians and well-equipped clinics are not easily accessible.
Keywords: COVID-19, Deep learning, Coronavirus, Pandemic
1. Introduction
Coronavirus Disease 2019 (COVID-19) is one of the deadliest viruses found in the world, which has high death rate and spread rate. This has caused a pandemic in the world. Severe Acute Respiratory Syndrome CoronaVirus 2 (SARS-CoV-2) is the name coined by the International Committee on Taxonomy of Viruses (ICTV). The virus was first discovered in Wuhan, Hubei province of China reporting causes similar to pneumonia. The first case was registered on December 31, 2019. By January 3, 2020, the number of patients with similar symptoms was 44, which was reported to the World Health Organization by the national authorities of China. From the reported 44 cases, there were 11 patients with severe illness and 33 patients in stable condition (Wu et al., 2020). The virus spread to almost of China within a span of 30 days. The spread of the virus affects both animals and humans. The number of cases registered in the United States of America was seven on January 20, 2020. The count of the cases quickly escalated to 300,000 by April 5, 2020 (Holshue et al., 2020). The virus spread to many countries including some of the biggest nations such as the USA, Japan, Germany, etc via transportation.
A standard and most common kit is used for the diagnosis of SARS-CoV-2 virus named as RT-PCR (Lee et al., 2020). But, due to the fast spread of this virus, there are very few kits for diagnosing the virus accurately for the suspected patients. This has posed to be one of the greatest threats in avoiding the infection of this virus (Zheng et al., 2020). There seem to be no findings of the virus in the first two days when observed on a lung CT scan. The early symptoms of SARS-CoV-2 are hard to identify as the symptoms of the disease are visible after ten days (Pan et al., 2020). In this case, the carrier of the virus may have contacted other people and infected most of them. To avoid this, there has to be a solution where the suspected patients get faster confirmation of the presence of the disease. This can be provided by imaging modalities such as Chest radiographs (X-rays) or Computed Tomography (CT) scan images. The radiologists can give an opinion by analyzing and applying some image analysis methods to diagnose SARS-CoV-2 (Kanne et al., 2020). The radiologists state that X-ray and CT images contain vital information related to COVID-19 cases. Therefore, combining radiologic images with Artificial Intelligence (AI) such as Deep learning methods, an accurate and faster diagnosis, can be performed to determine COVID-19 (Lee et al., 2020).
The most common symptoms found are usually cough, fever, breathlessness, fatigue, malaise, sore throat, etc. The elderly age group and people with respiratory issues are more prone to getting infected. It starts with pneumonia and Acute Respiratory Distress Syndrome (ARDS), leading to dysfunctioning of multiple organs. Some of the laboratory findings include the normal or low count of white cells, along with an increased amount of C-reactive protein (CRP). The initial step for preventing the spread of disease is to keep the suspected patients in-home quarantine. People with a higher rate of infection are kept under treatment at hospitals with strict infection control measures.
In this paper, five popular deep learning architectures are used to develop the COVID-19 diagnostic system. Various sources are screened for collecting X-ray images. The raw input images usually consist of unnecessary text information, poor quality of X-ray images, and different image dimensions. Before providing these images as input to the classifier, the images need to be preprocessed. Multiple challenges occurred in this step, such as text in the chest X-rays, dimension mismatch, data imbalance, etc. Multiple techniques handle all these issues. After preprocessing of the input images, these preprocessed images of three classes are provided as input to five deep learning architectures, namely, VGG16, DenseNet121, Xception, NASNet, and EfficientNet. The detailed description of the study is discussed in the below sections.
The contributions of this work are stated below:
-
•
A deep learning COVID-19 diagnostic system is developed using state-of-the-art (SoTA) deep learning architectures.
-
•
A detailed analysis of different methods used for detecting COVID-19 is presented in this paper.
-
•
Chest X-rays from different sources are collected to build a robust classification model.
-
•
The developed diagnostic model provided efficient results for larger variety of input images.
-
•
The results produced in this work are validated by an expert radiologist.
The rest of the paper is organized as follows: The recent research performed in COVID-19 identification is illustrated in the literature review section. The limitations and method details are also discussed in this section. Materials and Methods section gives a detailed description of the datasets used, preprocessing of X-ray images, and classification models used in this paper. A detailed analysis and performance comparison of the SoTA models are presented in the Results and Discussion section. Limitations and Future scope are also stated. Finally, a brief conclusion of this work is given in the Conclusion section.
2. Literature review
COVID-19 virus has caused a pandemic in the world, and there is no estimate of its global impact. Due to the unavailability of many testing kits, the number of tests that is being carried out is very less as in comparison to the rate of spread that is caused. In order to overcome this issue, a new system has to be developed to provide an accurate diagnosis of the presence of the virus in the body. This can be done by analyzing the X-ray or CT scan images of the patients with COVID-19. The image analysis, combined with various AI algorithms, can be the best solution for providing a second opinion for radiologists to diagnose the presence of this virus. There are multiple detection systems that have been developed in recent days to diagnose the virus in the human lungs and other organs. The image modalities considered are X-ray and CT scans. Many researchers have made use of deep learning algorithms on the COVID-19 and healthy person data. Table1 provides a brief discussion of popular state-of-the-art COVID-19 diagnostic systems used to identify and classify SARS-CoV-2, non COVID-19 and normal patients. Table content includes the authors’ name, the dataset used by authors, methods used, performance measures, and remarks of the work.
Table 1.
Comparison of various deep learning COVID-19 diagnostic systems.
| Authors | Dataset(s) used | Techniques used | Performance measures | Remarks |
|---|---|---|---|---|
| Apostolopoulos and Mpesiana (2020) | (a) Github repository from Joseph Cohen (b) Radiopaedia, (c) Italian Society of Medical and Interventional Radiology (SIRM), (d) Radiological Society of North America (RSNA), (e) Kermany | Transfer learning using CNNs | Sensitivity = 92.85%, Specificity = 98.75%, Accuracy-2 class = 98.75%, Accuracy-3 class = 93.48% | A multi-class classification using VGGNet with a high performance is achieved. There are few drawbacks of the work. 1. The number of images used for COVID-19 patients is less. 2. Some of the cases considered for pneumonia symptoms are taken from old records. There was no match found among the records and images collected from the COVID-19 patients. |
| Wang and Wong (2020) | (a) COVID-19 Image Data Collection (b) Chest X-ray Dataset Initiative (c) ActualMed COVID-19 Chest X-ray Dataset Initiative (d) RSNA Pneumonia Detection Challenge dataset (e) COVID-19 radiography database | COVID-Net | Accuracy = 93.3% | The architecture proposed uses combinations of 1 × 1 convolution blocks which makes the architecture lighter with fewer number of parameters. Thus, reducing the computational complexity of the network. The model provided better performance in terms of accuracy, but there is still scope in improvising the sensitivity and Positive Predicted Value (PPV) in the model. |
| Narin et al. (2020) | Dr. Joseph Cohen (GitHub repository) | Deep CNN and ResNet50 | Accuracy = 98%, Specificity = 100%, Recall= 96% | In this work, deep architectures such as Deep CNN, Inception, and Inception-ResNet are used. The main drawback in this work is the number of images considered for building and testing the model. Deep learning architectures work well for huge data. In this work, only 50 images of each class have been considered. The scope of different variations of the virus spread or occurrence may not be captured in that limited number of images. |
| Sethy and Behera (2020) | GitHub (Dr. Joseph Cohen) and Kaggle (X-ray images of Pneumonia) | ResNet50 + SVM | Accuracy = 95.38%, FPR = 95.52%, F1-score = 91.41%, Kappa = 90.76% | The model used by the authors is a combination of ResNet and Support Vector Machine (SVM). The obtained accuracy is commendable but the model was built on very few samples. |
| Talaat et al. (2020) | 1. Github repository collected from Joseph Cohen. 2. Collected by a team of researchers from Qatar University in Qatar and the University of Dhaka in Bangladesh along with collaborators from Pakistan and Malaysia medical doctors. | Deep features and fractional-order marine predators algorithm | Accuracy: 98.7; F-score: 99.6 | The method exhibited very promising results using deep features that are extracted from Inception model and the decision is provided by a tree based classifier. However, the drawback of this method is the varying environments used for feature extraction and classification. Also, the method has been tested for very few images. The results may vary in case a larger dataset is fed to the model. |
| Xu et al. (2020) | Private dataset of COVID-19, and Influenza-A pneumonia is collected | ResNet and location based attention | Sensitivity = 98.2%, Specificity = 92.2%, AUC = 0.996 | The performance of the deep learning algorithms proved to provide sufficient results even with few data samples. |
| Oh et al. (2020) | (a) Japanese Society of Radiological Technology (JSRT), (b) Chest posteroanterior (PA) radiographs were collected from 14 institutions including normal and lung nodule cases, (c) SCR database, (d) U.S. National Library of Medicine (USNLM) collected Montgomery Country (MC) dataset | Patch-based CNN | Accuracy = 93.3% | Authors proposed a solution for handling the issue of training deep neural networks on the limited training dataset. Multiple sources are considered for the collection of thoracic lung and chest radiographs. The limitation of this method is the performance of the proposed system in terms of precision, recall, and accuracy. |
| Abbas et al. (2020) | (a) Japanese Society of Radiological Technology (JSRT) Dr. Joseph Cohen Github repository and SARS | Decompose, Transfer, and Compose (DeTraC) | Accuracy = 95.12%, Specificity = 91.87%, Sensitivity = 97.91% | The model provided better performance results. The limited data issue in this work is handled by performing a data augmentation step. But, augmenting the X-ray images may not be a proper solution to handle less data as the location of the presence of the virus spread may never be found correctly. Only frontal images of the chest X-rays are selected and given to the further processing in our work to overcome this problem. |
| Li et al. (2020) | (a) Radiological Society of North America. RSNA pneumonia detection challenge, and (b) GitHub (Dr. Joseph Cohen) and Kaggle (X-ray images of pneumonia) | COVID-MobileXpert | Accuracy = 93.5% | The model takes a noisy X-ray snapshot as an input so that a quick screening can be performed to identify presence of COVID-19. DenseNet-121 architecture is employed to pre-train and fine-tune the network. For on-device COVID-19 screening purposes, lightweight CNNs such as MobileNetv2, SqueezeNet, and ShuffleNetV2 are used. |
| Ahuja et al. (2020) | The database contains 349 CT COVID19 positive CT images from 216 patients and 397 CT images of Non-COVID patients | Data augmentation using stationary wavelets, pre-trained CNN models, abnormality localization | Testing accuracy: 99.4% | In this method, an abnormality localization is implemented along with COVID-19 detection. The results obtained from this method is promising and the use of CT images provided better visibility of images as compared to X-ray images. |
3. Materials and methods
3.1. COVID-19 Dataset
The dataset includes chest X-ray images collected from various private hospitals from Maharashtra and Indore regions from India. The X-ray images are collected from posteroanterior (PA) frontal chest view from the patients. The dataset is divided into three different categories, namely, COVID, normal, and other. The COVID class consists of X-ray images belonging to patients with COVID, normal class consists of X-ray images of healthy patient scans, and other class consists of patients with viral infections or diseases such as effusion, pneumonia, nodule masses, infiltration, hernia etc. The number of X-ray images present in each class is given in Table 2 . The model is trained on 70%, validated on 20% and tested on 10% of the total chest X-ray images respectively.
Table 2.
Number of images used to train, validate, and test the models.
| Dataset | Normal | COVID | Others |
|---|---|---|---|
| Total | 6000 | 5634 | 5000 |
| Training | 4200 | 3944 | 3500 |
| Validation | 1200 | 1127 | 1000 |
| Testing | 600 | 563 | 500 |
3.2. Preprocessing
Preprocessing the input images is one of the important prerequisites in developing a better performing detection system. The raw input images consisted of unnecessary text information such as the name of the person, age, name of the hospital where the scan was taken, etc. This information may cause a problem in the training process. To avoid this, the images are cropped by using a Masked R-CNN (He et al., 2017) technique. Mask Region-based CNN is the next version of Faster R-CNN model. The main functionality of this model is to predict a mask for each detected region or object in an image. Mask R-CNN performs both object detection and localization in an image. The difference between detection and localization is rather subtle meaning in localization is to predict the object in an image as well as its boundaries. Localization aims to locate the most visible object in an image, whereas the detection of all the objects and their corresponding boundaries are identified. The dataset included different image formats such as “.jpg”, “.jpeg” and “.png”. Therefore, a model was developed to detect X-ray images using You Only Look Once (YOLO) model, and the annotations of the image are also provided. The images considered for the dataset are only of frontal view. COVID-19 X-rays are included in the dataset only after getting it cross-checked by an expert radiologist to avoid false scans to be included in the training set. The images are stored in RGB format to get maximum information as possible. A three-class classification is performed in this study.
3.3. Classification
The classification is performed using five different deep learning architectures, namely, VGG16, DenseNet121, Xception, NASNet and EfficientNet. These models are trained using transfer learning. Every model was trained for 100 epochs. The models are pre-trained using ImageNet weights, and these are also used for fine-tuning the model. The detailed explanation of the models is mentioned in the following sections.
3.3.1. VGG16
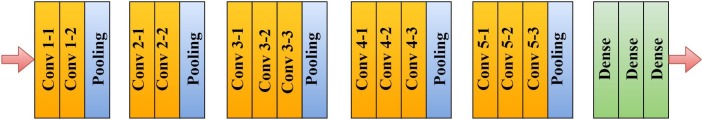
VGG16 is a CNN model that is developed by Visual Geometry Group (VGG) at Oxford university (Simonyan and Zisserman, 2014). The network is a successor of AlexNet, which was developed in 2012. VGG16 consists of 11 layers, including eight convolution blocks, three fully connected layers, five max-pooling layers and, one softmax layer. The architecture was developed for the ImageNet challenge. The convolution blocks’ width is set to a small number (i.e., starting from 64 in the initial layer). The width parameter is increased by two until it reaches 512 after each max-pooling operation is carried out. The model architecture is shown in Fig. 1 . The details of the architecture are as follows. The input image size fed to the VGG16 is of 224 × 224. The kernel size was set to 3 × 3 with a stride of 1. By performing spatial padding, the spatial resolution of the image was preserved. The pool size of the max-pooling operation was set to 2 × 2 with a stride of 2. In the fully connected layers, the first two layers‘ size is 4096 and the last fully connected layer of 1000. The last layer was set to 1000 because of 1000 classes in the ImageNet classification. In this work, the last dense layer is set to 3, because the number of classes used in this work are 3. Finally, the last layer was a softmax function. VGG16 network is made available for public use so that similar tasks can be performed using this model. The model can also be used for Transfer learning as the pre-trained weights are available in some of the frameworks such as Keras, so these can be used to develop own models by performing slight modifications accordingly.
Fig. 1.
Architecture of VGG16 model.
3.3.2. DenseNet121
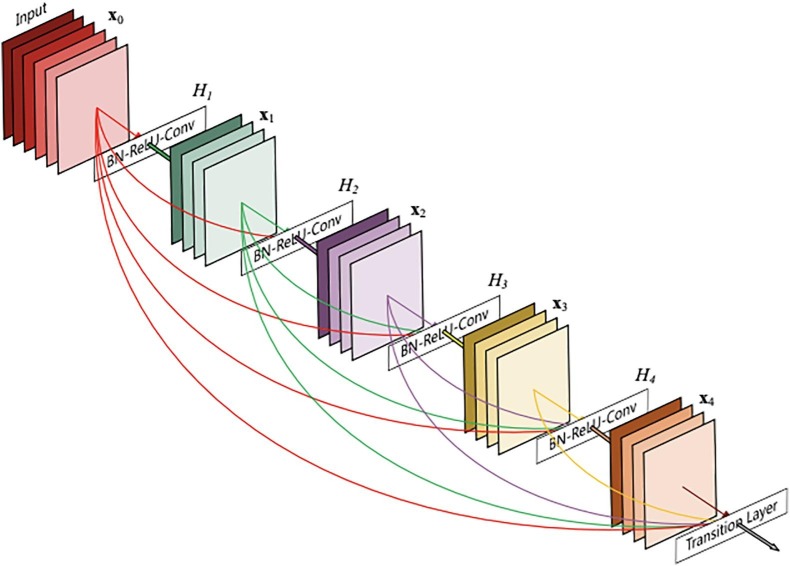
Densely connected Convolutional Networks are called as DenseNets (Huang et al., 2016). This is another way of increasing the depth of the deep convolutional networks without having issues such as exploding gradients and vanishing gradients. These issues are solved by connecting every layer directly with each other, which allows the passing of maximum information and gradient flow. The main key here is to explore the feature reuse instead of drawing representational power from extensive deep or wide CNN architectures. As compared to traditional CNN. DenseNets require less number or an equivalent number of parameters because, in DenseNets, the feature maps are not learned redundantly. There are some versions of ResNets also, which have contributed barely, and those layers can be dropped. DenseNet layers add only a small set of new feature-maps, and layers are narrow, i.e., the number of filters is very few. As there is a mentioned flow of information and gradients in the deep neural networks, the issue arises in training the input. DenseNets can solve this issue by providing direct access to the gradients from the original input and the loss function. The network structure of the DenseNet is progressively hierarchical as the feature maps from the (i-1)th layer is the input of the ith layer. The DenseNet can be stated as a generalizable network as the input of the ith layer can be of (i-1)th, (i-2)th) or even (i-n)th layer (where n must be less than the total number of layers). The diagrammatic representation of a DenseNet architecture can be illustrated in Fig. 2 (Huang et al., 2016).
Fig. 2.
Diagrammatic representation of DenseNet architecture.
One thing to be taken care of for the concatenation of the features maps is that the size of the feature maps must be consistent, which means that the convolutional layer’s output must be of the same size as the input. The working of densely concatenated convolutions can be represented in the following Eq. 1.
| (1) |
where represents the layer, represents the output of the layer. In the above equation, each layer’s input is fed to output of all frontal layers. To achieve a normalize the input of the layer, a Batch normalization (Ioffe and Szegedy, 2015) step in included in the network, which reduces the absolute difference between data and takes relative difference into consideration.
3.3.3. Xception
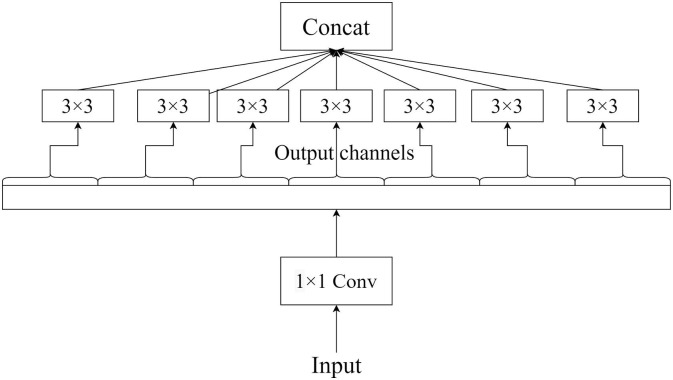
The Xception network is a successor of the Inception network. The name Xception stands for “eXtreme Inception”. The Xception network consists of depth-wise separable convolution layers instead of conventional convolution layers (Chollet, 2017). The schematic representation of a block in Xception is demonstrated in Fig. 3 . Xception involves mapping of spatial correlations and cross-channel correlations which can be entirely decoupled in CNN feature maps. Xception performed than the underlying Inception architecture. The Xception model consists of 36 convolution layers which can be divided into 14 different modules. Each layer has a linear residual connection around them by removing the initial and final layer. In short, Xception is the stacking of depth-wise separable convolution layers in a linear manner consisting of residual connections. To capture the cross-channel correlation in an input image, the input data is mapped to spatial correlations for each output channel separately. After performing this operation, a depth-wise 1 × 1 convolution operation is carried out. The correlations can be looked at as a 2D + 1D mapping instead of a 3D mapping. In Xception, 2D space correlations are performed initially and then followed by 1D space correlation.
Fig. 3.
Diagrammatic representation of a module in Xception.
3.3.4. NASNet
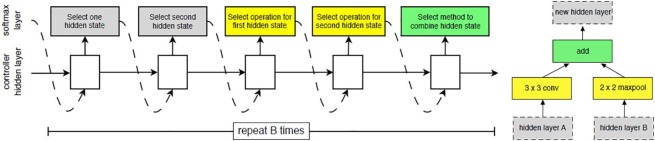
Neural Architectural Search (NAS) Network was developed by the Google ML group. The network architecture is based on reinforcement learning. In this network, the efficiency of the child block is reviewed by the parent block, and based on these changes, the network adjustment is made. The components of the network include a Controller Recurrent Neural Network (CRNN) and a CNN block. The schematic illustration of the NASNet architecture blocks is shown in Fig. 4 (Zoph et al., 2018). To achieve the best performance from the network, various modifications were done to the architecture such as weights, layers, regularization methods, optimizer functions, etc. To choose the best candidates, reinforced evolutionary algorithms are implemented. The different versions of NASNet, such as A, B, and C algorithms, are used to select the best cells using the reinforcement learning method (Zoph et al., 2018). The worst performing cells are eliminated using tournament selection algorithms.
Fig. 4.
Diagrammatic representation of a NASNet architecture.
The performance of the cell structure is optimized by optimizing the child fitness function and by performing reinforcement mutations. The smallest unit in the architecture is known as a block and the combination of these blocks is known as a cell. The network’s search space is done by factorizing it into cells and then dividing the cells into blocks. The number of cells and blocks is decided by the type of dataset are is not fixed. The operations that are performed in a block are convolutions, pooling, mapping, etc. In this work, NASNet-A is chosen for identifying COVID-19 patients and normal patients because of the transferable learning technique adopted in this network. This helps to provide better decisions with minimum network architecture as the network structure is optimized according to the dataset.
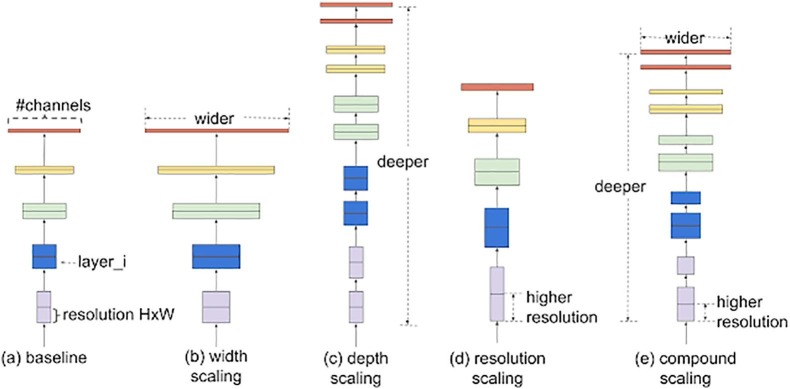
3.3.5. EfficientNet
One of the critical issues in using CNNs is the scaling of the model. We are aware of the fact that the depth of the model increases the performance of the system. But, to decide the depth of the model is a difficult task as a manual hit and trial method must be done to choose a better performing model. Therefore, to resolve this issue, ‘EfficientNets’ was introduced by Google research group (Tan and Le, 2019). MBConv is the primary building block of the EfficientNet models. To this block, a squeeze-and-excitation optimization block is added. The working of the MBConv block is similar to that of MobileNet v2 inverted residual blocks. A shortcut connection in between the start and end of a convolution block is formed. The output feature maps’ channels are reduced by performing 3 × 3 Depth-wise convolutions and point-wise convolutions. The narrow layers are connected using shortcut connections, whereas the wider layers are kept in between the skip connections. The size of the model as well as all number of the operations in the structure are reduced in this architecture. The illustration of the EfficientNet architecture can be demonstrated in Fig. 5 (Tan and Le, 2019). In this work, to classify COVID-19 patients and healthy patients, EfficientNet-B7 is used.
Fig. 5.
Diagrammatic representation of an EfficientNet architecture.
4. Results and discussion
An alternative method of prevention of the SARS-Cov-2 virus spread is essential in the pandemic situation like this. The models used in this paper can be useful in providing a second opinion to the radiologists to differentiate between the radiologic images obtained from the suspected patients. This can lessen the time of the testing of the patient using RT-PCR kits and getting laboratory results in a short duration of time.
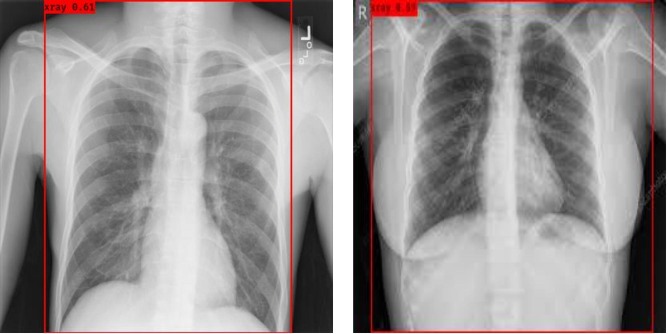
As mentioned earlier in the preprocessing section, only X-ray images are detected from the raw input images excluding the text and other unnecessary details on the images. This operation is performed using YOLO. The illustration of this preprocessing is illustrated in Fig. 6 .
Fig. 6.
Illustration of X-ray images region selection from raw input images.
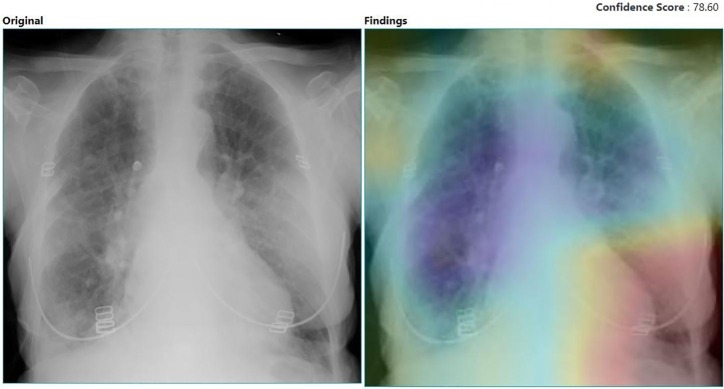
The models were run on NVIDIA Tesla K80 machine. All the models were run for 100 epochs. The parameters used for training models are presented in Table 3 . The accuracy achieved in training and testing of the learning process is stated in the table. A probability score of an X-ray image is given indicating the class which the X-ray image belongs to. In Fig. 7 , the heatmap of an X-ray image and its original image extracted from Efficient model is provided. The heatmap is extracted using GRAD-CAM method. With respect to the activation map in the model, the gradient of the most dominant logit is computed for a heatmap. A channel-wise pooling of these gradients is performed, and the activation channels are weighted with their corresponding gradients. This results in a collection of weighted activation channels. When these channels are inspected, it can be decided that which of the channels play a significant role in the decision of the class. The probability values obtained from the heatmap are: it belongs to COVID class with a probability of 0.0%, normal class with a probablity of 3.11% and to others class with a probability of 100.0%.
Table 3.
Parameters used for training the models.
| Performance measures | VGG16 | DenseNet121 | Xception | NASNet | EfficientNet |
|---|---|---|---|---|---|
| Batch size | 32 | 32 | 32 | 32 | 32 |
| Image dimension | 512 × 512 | 512 × 512 | 512 × 512 | 512 × 512 | 512 × 512 |
| Optimizer | Adam | Stochastic Gradient Descent | Adam | Adam | Adam |
| Learning rate | 1e−4 | 1e−4 | 1e−4 | 1e−4 | 1e−3 |
| Decay rate | 1e−5 | 1e−5 | 1e−5 | 1e−5 | 1e−4 |
| Activation function | Softmax | Softmax | Softmax | Softmax | Softmax |
| Loss function | Weighted binary cross entropy | Weighted binary cross entropy | Weighted binary cross entropy | Categorical cross entropy | Categorical cross entropy |
| Training accuracy | 80.32% | 92.98% | 90.84% | 89.99% | 97.17% |
| Testing accuracy | 79.01% | 89.96% | 88.03% | 85.03% | 93.48% |
Fig. 7.
Heatmap extracted from EfficientNet model.
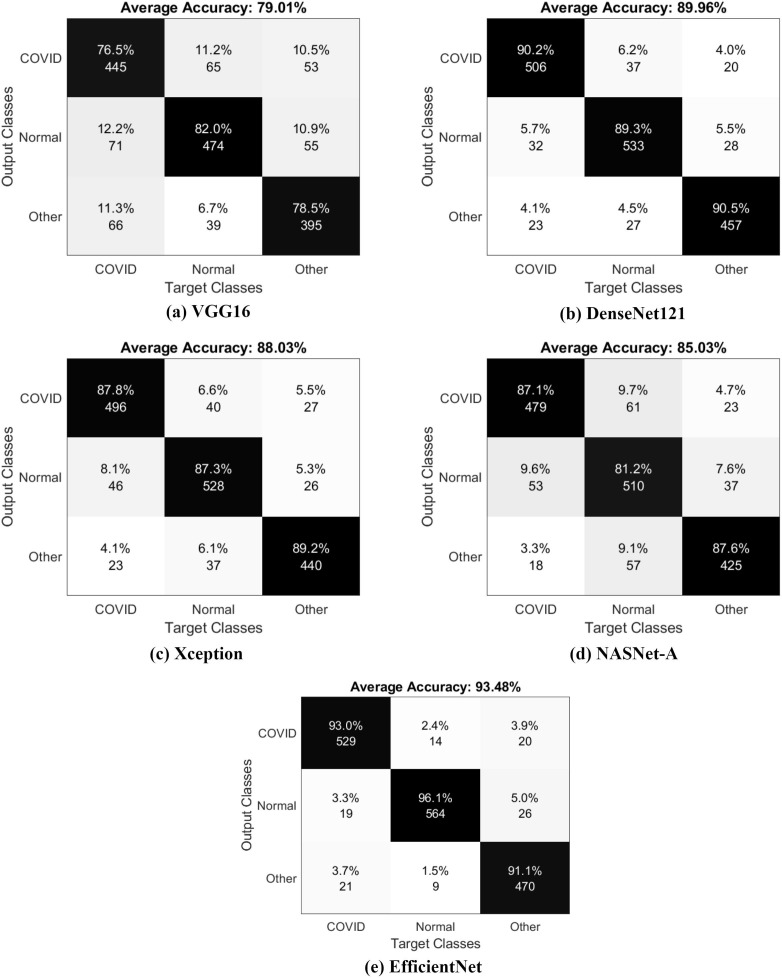
The performance of five architectures used in the study is presented in the confusion matrices illustrated in Fig. 8 . By observing the confusion matrices, the results obtained from testing set are good. Real-time detection of virus’ presence in the human body can be performed using the given models.
Fig. 8.
Confusion matrices obtained for all the models.
Five architectures used in the study are evaluated based on precision, recall and F1 score performance metrics. The class-wise performance of the models is presented in Table 4 . The different classes used in the study are COVID, normal, and others. An observation can be made from results table that the classifiers have performed well in COVID and normal class. The performance has slightly decreased in other class. The reason for this, maybe the combination of multiple illnesses in this class. The number of images present in each illness in “others” class may not be sufficient for the model to learn the images of this class efficiently.
Table 4.
Class-wise Precision, Recall and F1 Score for all the models.
| Performance measures | Class | VGG16 | DenseNet121 | Xception | NASNet | EfficientNet |
|---|---|---|---|---|---|---|
| Precision | COVID | 79% | 90% | 88% | 85% | 93% |
| Precision | Normal | 79% | 90% | 88% | 85% | 94% |
| Precision | Other | 79% | 90% | 88% | 85% | 93% |
| Recall | COVID | 76% | 90% | 88% | 87% | 93% |
| Recall | Normal | 82% | 89% | 87% | 81% | 93% |
| Recall | Other | 79% | 90% | 89% | 88% | 91% |
| F1 Score | COVID | 0.78 | 0.90 | 0.88 | 0.86 | 0.93 |
| F1 Score | Normal | 0.80 | 0.90 | 0.88 | 0.83 | 0.94 |
| F1 Score | Other | 0.79 | 0.90 | 0.89 | 0.86 | 0.93 |
The comparison of various diagnostic systems is provided in Table 5 . The results are compared based on the models developed for virus detection. Many authors have considered multi-class classification. The class considered other than COVID-19 patients and healthy patients are patients with pneumonia.
Table 5.
Comparison of previous works with the proposed work
| Authors | Number of cases | Method/s used | Accuracy |
|---|---|---|---|
| Hemdan et al. (2020) | 25 COVID-19(+) 25 Normal |
COVIDX-Net | 90.0% |
| Zhang et al. (2020) | 100 COVID-19(+) 1431 COVID-19(−) |
18-layer residual CNN | 96% |
| Ozturk et al. (2020) | (a) 125 COVID-19(+) 500 No Findings |
DarkCovidNet | (a) 98.08% |
| (b) 125 COVID-19(+) 500 Pneumonia 500 No Findings |
(b) 87.02% | ||
| Oh et al. (2020) | 8851 Normal 6012 Pneumonia 180 COVID-19(+) |
Patch-based CNN | 88.9% |
| Abbas et al. (2020) | 80 Normal 105 COVID-19 11 SARS |
DeTRAC | 95.12% |
| Proposed method | 795 COVID-19(+) 795 Normal 711 Others |
VGG16, DenseNet121, Xception, NASNet, EfficientNet | 79.01%, 89.96%, 88.03%, 85.03%, 93.48% |
The models considered for classification of COVID-19 and healthy patients mostly include deep learning architectures. Hemdan et al. (2020) have used COVIDX-Net for classifying positive COVID-19 patients and healthy patients. The model has achieved good accuracy, but the limitation of the work is the size of the dataset. The model may or may not work if given more extensive variations in the input images. A residual CNN is used by Zhang et al. (2020) for COVID-19 positive and negative cases. The model performed exceptionally well. Similarly, DarkCovidNet was proposed by Ozturk et al. (2020), who included cases from pneumonia to the dataset. The symptoms of COVID-19 and pneumonia are quite similar. The model achieved good performance for COVID-10 and no findings, but the accuracy of the model decreased when pneumonia cases are added to the dataset.
Oh et al. (2020) proposed a patch-based CNN for COVID-19, pneumonia, and normal patients’ classification. The model performed well for a large number of input images. A Decompose, Transfer, and Compose (DeTRAC) model was used by Abbas et al. (2020) to perform the classification of normal, COVID-19, and SARS patients. The model achieved exceptionally good results for the classification of three classes. Finally, in the proposed method, five SoTA deep learning architectures are used for classification of COVID-19, normal and other cases. The best recognition accuracy is provided by EfficientNet with 93%. The models used in work, outperformed most of the SoTA systems used for COVID-19 classification.
The illustration of some of the misclassified samples are shown in Fig. 9 . Two misclassified samples are presented in the figure. Both the images belong to COVID class but are misclassified to normal class. The reason for the misclassification of the first image is the appearance of breast shadow. The reason for the misclassification of the second image is the over exposure of the X-ray image.
Fig. 9.
Illustration of misclassified X-ray images.
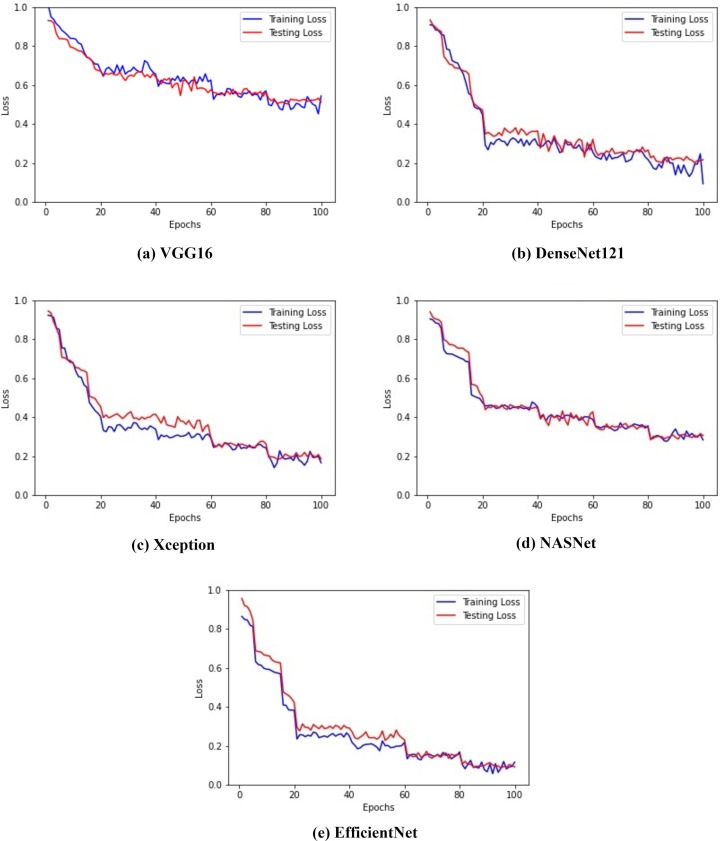
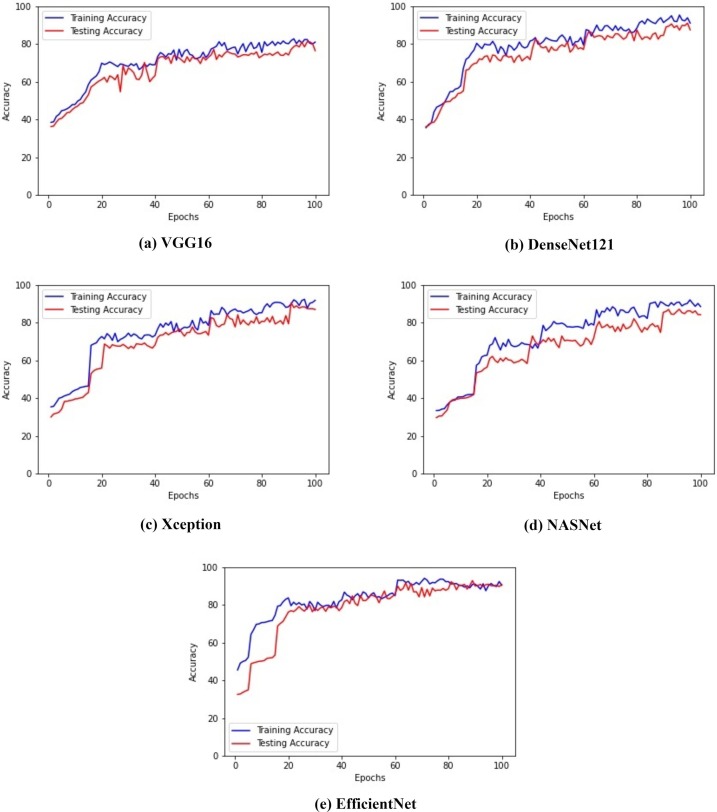
The analysis of the proposed model is presented using epoch versus loss and epoch versus accuracy graphs. The epoch versus loss graph depicts the loss obtained after every epoch. It can be noted that increase in epochs reduces the loss values. In contrast, there is an increase in the model’s accuray as the number of epochs is increased. This signifies that with each epoch, the model is learning the given input very well. Fig. 10 illustrates the epoch versus loss graph obtained for all the five different architectures. The figure shows the depletion of the loss as the epoch is increased. Fig. 11 illustrates the epoch versus accuracy graph for the training and testing accuracy of the five architectures mentioned above.
Fig. 10.
Epochs versus loss graph for all the models.
Fig. 11.
Epochs versus accuracy graph for all the models.
5. Limitations and future scope
In the recent trends, many COVID diagnosis systems have been developed. However, there are still some limitations in the current systems. One of the main obstacles in developing a reliable modes is the availability of chest X-ray dataset for public usage. Since deep learning models are data-driven, there is a need for larger datasets. In order to improve the time complexity of the deep learning models, the model performance can be improvised by using pre-trained weights from the previous SoTA medical chest X-ray diagnostic systems such as CheXpert, CheXnet, CheXNext. Concentrating on the data collection aspect, the data used for training these models are not diverse since medical images for COVID are obtained from the local hospitals. There is a need of huge amount of data for all three classes viz COVID, normal, and others. Due to limited data, to avoid the deep learning models to overfit, the dataset size needs to be increased. Therefore, few augmnetation techniques can be applied on the data. Performing augmentation on medical images is not considered good practice in the field of medical imaging. However, some augmentation techniques such as Affine transformation or noise addition can be used as per requirement. To improve the performance of the COVID diagnosis system, we can also use other SoTA models such as ResNet, MobileNet, Densenet169. In addition, we can use the Classification-Detection pipeline, which has provided good results in various Kaggle competitions. In this work, chest X-ray images are considered. However, CT scans of chest/thoracic regions may be considered to develop the COVID-19 diagnostic system, which can yield better performance.
6. Conclusion
In this paper, popular and best performing deep learning architectures are used for COVID-19 detection in the suspected patients by analyzing the X-ray images. Radiologic images such as X-ray or CT scans consist of vital information. The models performed efficiently and provided considerable results to many SoTA Coronavirus detection systems. The virus has caused a pandemic and has affected the economy of the entire world. The global impact of the virus is still unknown. The spread rate of the virus is too high, and it can be seen in both humans and animals. The models classify the healthy person, Coronavirus infected person, and to others class which is an illness that is non-COVID19 by analyzing the chest X-ray images. In this work, various SoTA deep learning architectures are used to perform COVID-19 chest X-rays. The highest recognition accuracy is achieved from EfficientNet model, i.e., 93.48%. It is observed that deep learning models provide better and faster results by analysing the image data to identify the presence of COVID in a person. However, the performance of the system can still be improved using various deep learning architectures and also by increasing the size of the dataset. There is an urgent need to diagnose the presence of the virus in the suspected patients as it may prevent it from spreading further. Hence, the proposed system can be used as a tool that can provide a faster and accurate recognition of the virus’s presence.
CRediT authorship contribution statement
Bhawna Nigam: Conceptualization, Methodology, Formal analysis, Investigation, Writing - review & editing, Supervision. Ayan Nigam: Formal analysis, Investigation, Supervision. Rahul Jain: Formal analysis, Investigation, Supervision. Shubham Dodia: Conceptualization, Methodology, Writing - original draft, Writing - review & editing. Nidhi Arora: Methodology. B. Annappa: Formal analysis, Investigation, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Abbas, A., Abdelsamea, M. M. & Gaber, M. M. (2020). Classification of covid-19 in chest x-ray images using detrac deep convolutional neural network. arXiv preprint arXiv:2003.13815. [DOI] [PMC free article] [PubMed]
- Ahuja S., Panigrahi B., Dey N., Rajinikanth V., Gandhi T. Deep transfer learning-based automated detection of covid-19 from lung ct scan slices. Applied Intelligence. 2020:1–15. doi: 10.1007/s10489-020-01826-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolopoulos, I. D. & Mpesiana, T. A. (2020). Covid-19: Automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Physical and Engineering Sciences in Medicine, (p. 1). [DOI] [PMC free article] [PubMed]
- Chollet F. Proceedings of the IEEE conference on computer vision and pattern recognition. 2017. Xception: Deep learning with depthwise separable convolutions; pp. 1251–1258. [Google Scholar]
- He K., Gkioxari G., Dollár P., Girshick R. Proceedings of the IEEE international conference on computer vision. 2017. Mask r-cnn; pp. 2961–2969. [Google Scholar]
- Hemdan, E. E. -D., Shouman, M. A. & Karar, M. E. (2020). Covidx-net: A framework of deep learning classifiers to diagnose covid-19 in X-ray images. arXiv:2003.11055.
- Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K. First case of 2019 novel coronavirus in the united states. New England Journal of Medicine. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G., Sun Y., Liu Z., Sedra D., Weinberger K.Q. European conference on computer vision. Springer; 2016. Deep networks with stochastic depth; pp. 646–661. [Google Scholar]
- Ioffe, S. & Szegedy, C. (2015). Batch normalization: Accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv:1502.03167.
- Kanne J.P., Little B.P., Chung J.H., Elicker B.M., Ketai L.H. Essentials for radiologists on covid-19: An update–radiology scientific expert panel. Radiology. 2020 doi: 10.1148/radiol.2020200527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.Y.P., Ng M.-Y., Khong P.-L. Covid-19 pneumonia: What has ct taught us? The Lancet Infectious Diseases. 2020;20:384–385. doi: 10.1016/S1473-3099(20)30134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Li, C. & Zhu, D. (2020). Covid-mobilexpert: On-device covid-19 screening using snapshots of chest x-ray. ArXiv, abs/2004.03042.
- Narin, A., Kaya, C. & Pamuk, Z. (2020). Automatic detection of coronavirus disease (covid-19) using X-ray images and deep convolutional neural networks. arXiv preprint arXiv:2003.10849. [DOI] [PMC free article] [PubMed]
- Oh Y., Park S., Ye J.C. Deep learning covid-19 features on cxr using limited training data sets. IEEE Transactions on Medical Imaging. 2020:1. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of covid-19 cases using deep neural networks with X-ray images. Computers in Biology and Medicine. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F., Ye T., Sun P., Gui S., Liang B., Li L., Zheng D., Wang J., Hesketh R.L., Yang L., Zheng C. Time course of lung changes on chest ct during recovery from 2019 novel coronavirus (covid-19) pneumonia. Radiology. 2020 doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethy, P. K. & Behera, S. K. (2020). Detection of coronavirus disease (covid-19) based on deep features. Preprints, 2020030300.
- Simonyan, K. & Zisserman, A. (2014). Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556.
- Talaat A., Yousri D., Ewees A., Al-qaness M.A.A., Damasevicius R., Elsayed Abd Elaziz M. Covid-19 image classification using deep features and fractional-order marine predators algorithm. Scientific Reports. 2020;10:15364. doi: 10.1038/s41598-020-71294-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, M. & Le, Q. V. (2019). Efficientnet: Rethinking model scaling for convolutional neural networks. arXiv preprint arXiv:1905.11946.
- Wang, L. & Wong, A. (2020). Covid-net: A tailored deep convolutional neural network design for detection of covid-19 cases from chest radiography images. arXiv preprint arXiv:2003.09871. [DOI] [PMC free article] [PubMed]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., et al. A new coronavirus associated with human respiratory disease in china. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X., Jiang, X., Ma, C., Du, P., Li, X., Lv, S., Yu, L., Chen, Y., Su, J. & Lang, G. (2020). Deep learning system to screen coronavirus disease 2019 pneumonia, arXiv preprint arXiv:2002.09334. [DOI] [PMC free article] [PubMed]
- Zhang, J., Xie, Y., Li, Y., Shen, C. & Xia, Y. (2020). Covid-19 screening on chest X-ray images using deep learning based anomaly detection. arXiv:2003.12338.
- Zheng, C., Deng, X., Fu, Q., Zhou, Q., Feng, J., Ma, H., Liu, W. & Wang, X. (2020). Deep learning-based detection for covid-19 from chest ct using weak label. medRxiv. DOI: 10.1101/2020.03.12.20027185.
- Zoph B., Vasudevan V., Shlens J., Le Q.V. Proceedings of the IEEE conference on computer vision and pattern recognition. 2018. Learning transferable architectures for scalable image recognition; pp. 8697–8710. [Google Scholar]