Abstract
The long noncoding RNA WEE2 antisense RNA 1 (WEE2-AS1) plays an oncogenic role in hepatocellular carcinoma and triple negative breast cancer progression. In this study, we investigated the expression and roles of WEE2-AS1 in glioblastoma (GBM). Furthermore, the molecular mechanisms behind the oncogenic actions of WEE2-AS1 in GBM cells were explored in detail. WEE2-AS1 expression was detected using quantitative real-time polymerase chain reaction. The roles of WEE2-AS1 in GBM cells were evaluated by the cell counting kit-8 assay, flow cytometric analysis, Transwell cell migration and invasion assays, and tumor xenograft experiments. WEE2-AS1 expression was evidently enhanced in GBM tissues and cell lines compared with their normal counterparts. An increased level of WEE2-AS1 was correlated with the average tumor diameter, Karnofsky Performance Scale score, and shorter overall survival among GBM patients. Functionally, depleted WEE2-AS1 attenuated GBM cell proliferation, migration, and invasion in vitro, promoted cell apoptosis, and impaired tumor growth in vivo. Mechanistically, WEE2-AS1 functioned as a molecular sponge for microRNA-520f-3p (miR-520f-3p) and consequently increased specificity protein 1 (SP1) expression in GBM cells. A series of recovery experiments revealed that the inhibition of miR-520f-3p and upregulation of SP1 could partially abrogate the influences of WEE2-AS1 downregulation on GBM cells. In conclusion, WEE2-AS1 can adsorb miR-520f-3p to increase endogenous SP1 expression, thereby facilitating the malignancy of GBM. Therefore, targeting the WEE2-AS1–miR-520f-3p–SP1 pathway might be a promising therapy for the management of GBM in the future.
Key words: Glioblastoma (GBM), Competing endogenous RNAs (ceRNAs), Long noncoding RNAs (lncRNAs), MicroRNAs (miRNAs)
INTRODUCTION
Glioma is one of the most common types of primary malignant brain tumors, accounting for approximately 46% of intracranial tumors1. According to the grading system proposed by the World Health Organization (WHO)2, all glioma cases could be classified into four categories: low-grade astrocytomas (grades I–II), anaplastic astrocytomas (grade III), and glioblastomas (GBMs; WHO grade IV). In the past decade, there has been tremendous progress in the development of comprehensive therapies for GBM patients, including surgery, immunotherapy, stereotactic radiotherapy, and chemotherapy3,4. However, the clinical outcomes remain unsatisfactory, with a median survival period of 9–19 months5, and the poor prognosis is primarily caused by the highly aggressive and invasive nature of GBM6. The activation of oncogenes, inactivation of tumor suppressors, and chromosomal abnormalities play key roles in GBM pathogenesis; however, the detailed molecular events are not fully understood7. Hence, further characterization of the mechanisms underlying GBM formation and progression is imperative for developing promising techniques for anticancer therapy.
MicroRNAs (miRNAs) are an enormous group of endogenous and noncoding short RNA molecules with 18–23 nucleotides8. miRNAs function as guide molecules in gene expression that can directly bind to the 3′-untranslated regions (3′-UTRs) of target genes via base pairing, resulting in translational inhibition and/or mRNA degradation9. As endogenous regulators of gene expression, miRNAs directly or indirectly affect almost all physiological and pathological processes, including tumorigenesis and tumor development10,11. Increasing studies have validated the aberrant expression of miRNAs in GBM, which contributes to the occurrence and development of GBM12–14.
Long noncoding RNAs (lncRNAs) refer to a category of transcripts containing more than 200 nucleotides15. They lack protein-coding ability but are involved in numerous cellular processes by controlling gene expression at different levels, including epigenetics, transcription, or posttranscription16. Accumulating studies have demonstrated that changes in lncRNA expression and their dysregulation are closely related to GBM oncogenesis and progression17,18. lncRNAs exert cancer-inhibiting or cancer-promoting activities, and they promote the aggressiveness of GBM by regulating various malignant characteristics19,20. lncRNAs possess miRNA response elements (MREs) and serve as natural miRNA sponges to release target mRNAs from miRNAs21. Hence, lncRNA/miRNA/mRNA regulatory pathways may provide effective targets for diagnosis, prevention, and therapy in GBM.
Recent studies showed that the lncRNA WEE2 antisense RNA 1 (WEE2-AS1) executes an oncogenic role in hepatocellular carcinoma22 and triple negative breast cancer23. However, the expression profile and detailed functions of WEE2-AS1 in GBM and its underlying mechanisms remain poorly defined. In this study, we measured WEE2-AS1 expression in GBM and determined its clinical relevance. Also, the influences of WEE2-AS1 on the malignant behavior of GBM cells were investigated both in vitro and in vivo. More importantly, the mechanisms underlying the effects of WEE2-AS1 on GBM progression were elucidated in detail.
MATERIAL AND METHODS
Clinical Tissues
The Ethics Committees of Shenzhen Second People’s Hospital (Shenzhen, China) approved this study, and written informed consent was obtained from all participants. A total of 59 GBM tissues were collected from patients at Shenzhen Second People’s Hospital. Nineteen normal brain tissues were obtained from patients who died from traffic accidents and craniocerebral trauma. No patient had received any preoperative therapies prior to enrollment in the current study. All tissues were obtained, quickly frozen in liquid nitrogen, and stored in liquid nitrogen prior to RNA extraction.
Cell Culture
A normal human astrocyte (NHA) cell line was purchased from ScienCell Research Laboratories (Carlsbad, CA, USA) and cultured in astrocyte medium (ScienCell Research Laboratories). GBM cell lines T98 and U138 (American Type Culture Collection; Manassas, VA, USA) were maintained in minimum essential media (Gibco, Thermo Fisher Scientific Inc., Waltham, MA, USA) containing 10% (v/v) fetal bovine serum (FBS; Gibco, Thermo Fisher Scientific Inc.), 100 U/ml penicillin, and 100 μg/ml streptomycin (Gibco, Thermo Fisher Scientific Inc.). Two other GBM cell lines, including U251 and SHG-44, were bought from Shanghai Institutes for Biological Sciences Cell Resource Center (Shanghai, China) and maintained in Dulbecco’s modified Eagle’s medium (Gibco, Thermo Fisher Scientific Inc.) and RPMI-1640 medium (Gibco, Thermo Fisher Scientific Inc.), which were both supplemented with 10% (v/v) FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin (Gibco, Thermo Fisher Scientific Inc.). All cells were grown at 37°C in a humidified incubator supplied with 5% CO2.
Cell Transfection
The small interfering RNA (siRNA) targeting WEE2-AS1 (si-WEE2-AS1) and negative control (NC) siRNA (si-NC) were synthesized by RiboBio Technology (Guangzhou, China). miR-520f-3p mimic, NC mimic, miR-520f-3p inhibitor, and NC inhibitor were obtained from GenePharma Co. Ltd. (Shanghai, China). The specificity protein 1 (SP1) overexpression plasmid was established using pcDNA3.1 (pcDNA3.1-SP1; GenePharma Co. Ltd.), with an empty pcDNA3.1 plasmid as the control. GBM cells in the logarithmic growth phase were harvested, counted, and seeded in six-well plates. After overnight incubation, cell transfection was performed using Lipofectamine® 2000 (Invitrogen, Thermo Fisher Scientific Inc.) according to the manufacturer’s protocol.
Subcellular Fractionation
The fractionation of nuclear and cytoplasmic RNA was conducted using a PARIS Kit (Invitrogen, Thermo Fisher Scientific Inc.). Cytoplasmic and nuclear RNAs were collected and analyzed by quantitative real-time polymerase chain reaction (qRT-PCR). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6 small nuclear RNA were used as the internal controls for cytoplasmic and nuclear RNA, respectively.
qRT-PCR
After total RNA isolation using TRIzol (Invitrogen, Thermo Fisher Scientific Inc.), the extracted RNA was analyzed using a NanoDrop 2000 spectrophotometer to determine its concentration and purity. To detect miR-520f-3p expression, reverse transcription and quantitative PCR (qPCR) were conducted using an miRcute miRNA First-Strand cDNA Synthesis Kit (Tiangen Biotech, Beijing, China) and miRcute miRNA qPCR Detection Kit SYBR Green (Tiangen Biotech), respectively. U6 small nuclear RNA was used as an endogenous reference to quantify miR-520f-3p expression. For the measurement of WEE2-AS1 and SP1 expression, total RNA was reverse transcribed into complementary deoxyribonucleic acid (cDNA) using a PrimeScript RT reagent kit (Takara Biotechnology Co. Ltd, Dalian, China). The cDNA was then subjected to qPCR using an SYBR Premix Ex Taq kit (Takara Biotechnology Co. Ltd). Expression levels of WEE2-AS1 and SP1 were normalized to the housekeeping gene GAPDH. The 2−ΔΔCt method was used to calculate relative gene expression.
The primers were designed as follows: WEE2-AS1, 5′-ATGGGGCTGTACTGAATACCG-3′ (forward) and 5′-ATACCATGATCACGAAGGTGGTT-3′ (reverse); SP1, 5′-CAGTGAGTCTTCCAAGAATCGC-3′ (forward) and 5′-TTACTTGATACTGAATATTAGGCATCACT-3′ (reverse); GAPDH, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′ (forward) and 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′ (reverse); miR-520f-3p, 5′-TCGGCAGGCCUCUAAAGGGAA-3′ (forward) and 5′-CACTCAACTGGTGTCGTGGA-3′ (reverse); and U6, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ (forward) and 5′-CGCTTCACGAATTTGCGTGTCAT-3′ (reverse).
Cell Counting Kit-8 (CCK-8) Assay
After cultivation for 24 h, transfected cells were harvested, and single-cell suspensions were subsequently prepared. A 100-μl cell suspension containing 3 × 103 cells was inoculated into 96-well plates. Cells were collected 0, 1, 2, and 3 days after seeding. At every time point, 10 μl of CCK-8 solution (Dojindo, Tokyo, Japan) was added to each well, after which cells were incubated at 37°C with 5% CO2 for 2 h. The absorbance was detected at a wavelength of 450 nm, and the growth curves were plotted accordingly.
Flow Cytometric Analysis
Forty-eight hours posttransfection, cells were digested with EDTA-free 0.25% trypsin (Gibco, Thermo Fisher Scientific Inc.) and washed with precooled phosphate-buffered saline. The supernatant was discarded, and the harvested transfected cells were analyzed using the Annexin V-Fluorescein Isothiocyanate (FITC) Apoptosis Detection Kit (BioLegend, San Diego, CA, USA) to evaluate apoptosis. After the cells were resuspended in 100 μl of binding buffer, 5 μl of annexin V-FITC and 5 μl of propidium iodide solution were added to stain the cells. The rate of apoptosis of the cells was detected using a FACScan flow cytometer (BD Biosciences, San Jose, CA, USA).
Transwell Cell Migration and Invasion Assays
To measure cell migration, a single-cell suspension was prepared by mixing 2 × 105 transfected cells with 1 ml FBS-free basal medium. A 200-μl cell suspension was added to the apical chamber of Transwell plates (BD Biosciences), while the basal medium supplemented with 20% FBS was added to the basolateral chambers. The cells were incubated at 37°C with 5% CO2 for 24 h. Before staining with 0.5% crystal violet, the migrated cells were fixed with 95% ethanol at room temperature for 20 min. Cell invasion was examined using the same experimental steps, except that Matrigel (BD Biosciences) diluted in basal medium was added to the apical chamber. The migrated and invaded cells were counted using an inverted microscope (Olympus, Tokyo, Japan).
Tumor Xenograft Experiments
Lentiviral vectors carrying WEE2-AS1 short hairpin RNA (sh-WEE2-AS1) and NC short hairpin RNA (sh-NC) were designed and produced by GenePharma Co. Ltd. U251 cells were infected with the generated lentiviral vectors, after which the cells were treated with puromycin for 2 weeks to obtain stable cell lines expressing sh-WEE2-AS1 or sh-NC.
The animal experiments were approved by the Animal Care and Use Committee of Shenzhen Second People’s Hospital. Four-week-old male BALB/c nude mice were purchased from Beijing HFK Bioscience (Beijing, China) and subcutaneously injected with 2 × 106 U251 cells with WEE2-AS1 stable knockdown. All mice were bred under specific pathogen-free conditions. The width and length of the tumor xenografts were measured using vernier calipers every 4 days. Tumor volumes were calculated using the standard formula: volume = 0.5 × length × width2. At 4 weeks after cell injection, the mice were euthanized, and the tumor xenografts were excised and weighed.
Bioinformatic Analysis
StarBase version 3.0 (http://starbase.sysu.edu.cn/) was used to identify the potential miRNA targets of WEE2-AS1. Three databases were used to search for putative targets of miR-520f-3p: StarBase version 3.0, TargetScan (http://www.targetscan.org/), and miRDB (http://mirdb.org/).
RNA Immunoprecipitation (RIP) Assay
A Magna RIP RNA-binding protein immunoprecipitation kit (Merck Millipore, Darmstadt, Germany) was used to determine the interaction between WEE2-AS1 and miR-520f-3p using RIP assay. GBM cells were harvested via centrifugation and lysed in RIP lysis buffer. The cell extracts were incubated overnight with magnetic beads conjugated with human anti-Argonaute2 (Merck Millipore) or normal immunoglobulin G (IgG) (Merck Millipore) at 4°C. Following the removal of proteins using a proteinase K buffer, qRT-PCR was performed to analyze the enrichment of WEE2-AS1 and miR-520f-3p.
Luciferase Reporter Assay
The WEE2-AS1 wild-type (wt) fragments carrying miR-520f-3p binding sequences and mutant (mut) WEE2-AS1 fragments were cloned into the pmirGLO luciferase reporter plasmid (Promega Corporation, Madison, WI, USA) to generate WEE2-AS1-wt and WEE2-AS1-mut luciferase reporter vectors, respectively. The SP1-wt and SP1-mut reporter plasmids were designed and synthesized via the same experimental procedures. GBM cells were transfected with wt or mut reporter plasmids together with miR-520f-3p mimic or NC mimic using Lipofectamine® 2000. Forty-eight hours later, the collected transfected cells were assayed using a Dual-Luciferase® Reporter Assay System (Promega) for the measurement of luciferase activity.
Western Blotting
To extract total protein, transfected cells were lysed with radioimmunoprecipitation assay (RIPA) solution (Sangon Biotech Co. Ltd., Shanghai, China) supplemented with a protease inhibitor cocktail (Sangon Biotech Co. Ltd.) and phenylmethanesulfonyl fluoride (Sangon Biotech Co. Ltd.). After total protein quantification using a BCA Protein Assay Kit (Sangon Biotech Co. Ltd.), equal amounts of protein were subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) electrophoresis and transferred to polyvinylidene fluoride membranes. The membranes were blocked with 5% skim milk at room temperature for 2 h and subsequently incubated overnight at 4°C with primary antibodies against SP1 (Cat. No. b124804; Abcam, Cambridge, MA, USA) or GAPDH (Cat. No. ab128915; Abcam). The primary antibodies were used with a dilution of 1:1,000. Next, a goat anti-rabbit IgG horseradish peroxidase-conjugated secondary antibody (Cat. No. ab6721; Abcam) was incubated with the membranes at room temperature for 2 h. Finally, the protein signals were visualized using the ECL Prime Western Blotting Detection Reagent (GE Healthcare, Chicago, IL, USA).
Statistical Analysis
All measurement results were presented as the mean ± standard deviation and analyzed with SPSS 21.0 (SPSS, Chicago, IL, USA). The chi-square test was used to test the association between WEE2-AS1 expression and clinicopathological data of patients with GBM. Statistical significance between two groups was determined using Student’s t-test. The differences among multiple groups were assessed using one-way analysis of variance along with Tukey’s post hoc test. Pearson’s correlation coefficient was determined to evaluate the expression correlation between two genes in the 59 GBM tissues. Survival curves were plotted using the Kaplan–Meier method and subsequently analyzed by the log-rank test. A value of p < 0.05 was considered as a statistically significant difference.
RESULTS
WEE2-AS1 Silencing Suppresses Cell Proliferation, Migration, and Invasion and Increases Cell Apoptosis in GBM
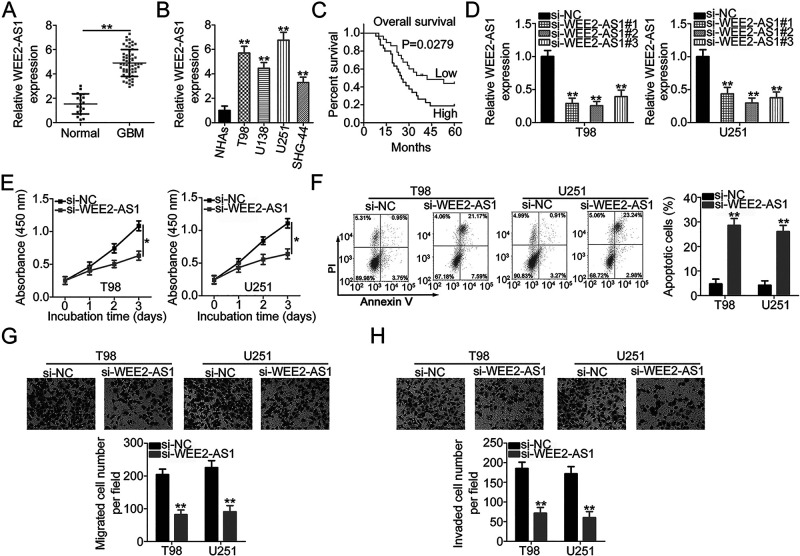
To understand the roles of WEE2-AS1 in GBM, a total of 59 GBM tissues and 19 normal brain tissues were collected and subjected to qRT-PCR for WEE2-AS1 quantification. WEE2-AS1 was upregulated in GBM tissues in comparison with that in normal brain tissues (Fig. 1A). Also, all four GBM cell lines (T98, U138, U251, and SHG-44) showed higher WEE2-AS1 levels compared with that in the NHA cell line, as illustrated by qRT-PCR (Fig. 1B). All GBM patients were arranged by the median value of WEE2-AS1 expression in GBM tissues, and the clinic significance was examined. High WEE2-AS1 expression presented close relationships with the average tumor diameter (p = 0.019) and Karnofsky Performance Scale (KPS) score (p = 0.035) (Table 1). In addition, an increased level of WEE2-AS1 was evidently correlated with shorter overall survival in patients with GBM (p = 0.0279) (Fig. 1C).
Figure 1.
WEE2 antisense RNA 1 (WEE2-AS1) depletion inhibits the malignant characteristics of glioblastoma (GBM) cells. (A) WEE2-AS1 expression in 59 GBM tissues and 19 normal brain tissues was determined using quantitative real-time polymerase chain reaction (qRT-PCR). (B) qRT-PCR analysis was applied to detect WEE2-AS1 expression in four GBM cell lines (T98, U138, U251, and SHG-44) and a normal human astrocyte (NHA) cell line. (C) Kaplan–Meier survival analysis evaluated the correlations between WEE2-AS1 expression and the overall survival of 59 GBM patients (p = 0.0279). (D) The efficiency of si-WEE2-AS1 transfection in T98 and U251 cells was assessed by qRT-PCR. (E) The impact of inhibited proliferation by WEE2-AS1 silencing in T98 and U251 cells relative to that in negative control short hairpin RNA (sh-NC)-transfected cells was examined using the cell counting kit-8 (CCK-8) assay. (F) Flow cytometric analysis was performed to determine the apoptosis of T98 and U251 cells after WEE2-AS1 knockdown. (G, H) Migratory and invasive capacities of WEE2-AS1-deficient T98 and U251 cells were determined using Transwell cell migration and invasion assays. *p < 0.05 and **p < 0.01.
Table 1.
The Correlation Between WEE2-AS1 Expression and Clinicopathological Data in 59 Cases of Glioblastoma (GBM) Tissues
| Clinicopathological Data | WEE2-AS1 | p Value | |
|---|---|---|---|
| High (n = 30) | Low (n = 29) | ||
| Gender | 0.606 | ||
| Male | 17 | 14 | |
| Female | 13 | 15 | |
| Age | 0.795 | ||
| <60 years | 16 | 17 | |
| ≥60 years | 14 | 12 | |
| Average tumor diameter | 0.019 | ||
| <5 cm | 11 | 20 | |
| ≥5 cm | 19 | 9 | |
| KPS score | 0.035 | ||
| ≥60 | 8 | 16 | |
| <60 | 22 | 13 | |
| Extension of resection | 0.796 | ||
| Subtotal | 17 | 15 | |
| Total | 13 | 14 | |
KPS, Karnofsky Performance Scale score. The p value was acquired by chi-square test.
T98 and U251 cell lines exhibited the highest WEE2-AS1 level in the four tested GBM cell lines and were therefore used in the functional experiments. T98 and U251 cells were transfected with three siRNAs targeting WEE2-AS1. si-WEE2-AS1#2 showed the highest efficiency in silencing endogenous WEE2-AS1 expression (Fig. 1D) and was therefore selected for subsequent experiments. The growth curves generated using the results of the CCK-8 assays indicated that loss of WEE2-AS1 substantially decreased the proliferation of T98 and U251 cells (Fig. 1E). Furthermore, the proportion of apoptotic T98 and U251 cells was clearly increased after WEE2-AS1 knockdown (Fig. 1F). Transwell cell migration and invasion assays were employed to determine the impacts of WEE2-AS1 downregulation on the migratory and invasive capacities of T98 and U251 cells. It was obvious that the migration (Fig. 1G) and invasion (Fig. 1H) of WEE2-AS1-deficient T98 and U251 cells were highly impaired compared with that in cells transfected with si-NC. Overall, these data revealed that WEE2-AS1 performed an oncogenic role in GBM cells.
WEE2-AS1 Works by Adsorbing miR-520f-3p in GBM Cells
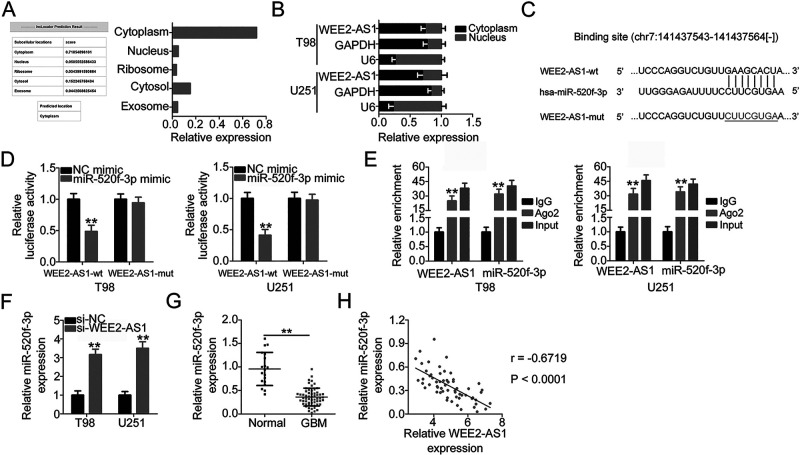
To elucidate the molecular mechanisms underlying the oncogenic role of WEE2-AS1 in GBM, lncLocator (lncRNA subcellular localization predictor) analysis was conducted to identify the subcellular localization of WEE2-AS1. lncLocator predicted that WEE2-AS1 was mostly distributed in the cytoplasm (Fig. 2A), which was further confirmed by subcellular fractionation assay (Fig. 2B). This result suggested that WEE2-AS1 may modulate gene expression at the posttranscriptional level. Recently, several studies revealed that lncRNAs exert biological roles in human cancers via acting as competing endogenous RNAs (ceRNAs) by sponging certain miRNAs and consequently regulating the expression of their target mRNAs24–26. StarBase version 3.0 was utilized to identify the potential miRNAs that can interact with WEE2-AS1. Since miR-520f-3p was complementary to WEE2-AS1 at position chr7:141437479-141437678 (Fig. 2C), it was selected for further confirmation based on its antioncogenic actions in carcinogenesis and cancer progression27–29.
Figure 2.
WEE2-AS1 functions as a molecular sponge for miR-520f-3p in GBM cells. (A) lncLocator was used to predict the subcellular localization of WEE2-AS1. (B) The subcellular distribution of WEE2-AS1 in T98 and U251 cells was tested by the subcellular fractionation assay. (C) The binding site between WEE2-AS1 and miR-520f-3p was predicted by StarBase version 3.0. The mutant sequences are shown. (D) Luciferase activity was detected in T98 and U251 cells that were cotransfected with miR-520f-3p mimic or NC mimic and WEE2-AS1-wild type (wt) or WEE2-AS1-mutant (mut). (E) RNA immunoprecipitation (RIP) assay was carried out in T98 and U251 cells, and the relative RNA level of WEE2-AS1 and miR-520f-3p in the immunoprecipitates was analyzed by qRT-PCR. (F) qRT-PCR was applied to measure miR-520f-3p expression in T98 and U251 cells after si-WEE2-AS1 or si-NC transfection. Transfection of si-WEE2-AS1 increased miR-520f-3p expression in T98 and U251 cells. (G) The miR-520f-3p expression in 59 GBM tissues and 19 normal brain tissues was examined by qRT-PCR. (H) Pearson’s correlation coefficient showed a negative correlation between miR-520f-3p and WEE2-AS1 expression in the 59 GBM tissues (r = −0.6719, p < 0.0001). **p < 0.01.
The luciferase reporter assay was conducted to test the direct binding between WEE2-AS1 and miR-520f-3p in GBM cells. Exogenous miR-520f-3p expression resulted in a significant reduction in luciferase activity of WEE2-AS1-wt, whereas the luciferase activity of WEE2-AS1-mut was unaltered in T98 and U251 cells after miR-520f-3p upregulation (Fig. 2D). In addition, WEE2-AS1 and miR-520f-3p showed enrichment in the complex precipitated by Ago2, which is the critical component of the RNA-induced silencing complex mediated by miRNAs, as demonstrated by the RIP assay (Fig. 2E). Furthermore, the regulatory action of WEE2-AS1 on miR-520f-3p was evaluated in GBM cells after si-WEE2-AS1 or si-NC transfection. Interference of WEE2-AS1 expression caused a notable increase in miR-520f-3p expression in T98 and U251 cells (Fig. 2F). Furthermore, miR-520f-3p expression in GBM and its correlation to WEE2-AS1 expression were determined. The expression of miR-520f-3p was downregulated in GBM tissues compared with that in normal brain tissues (Fig. 2G). More importantly, miR-520f-3p presented an inverse correlation with WEE2-AS1 expression in the 59 GBM tissues (r = −0.6719, p < 0.0001) (Fig. 2H). Taken together, WEE2-AS1 physically interacted with miR-520f-3p and acted as a molecular sponge for miR-520f-3p in GBM cells.
SP1 Is a Direct Target of miR-520f-3p in GBM Cells
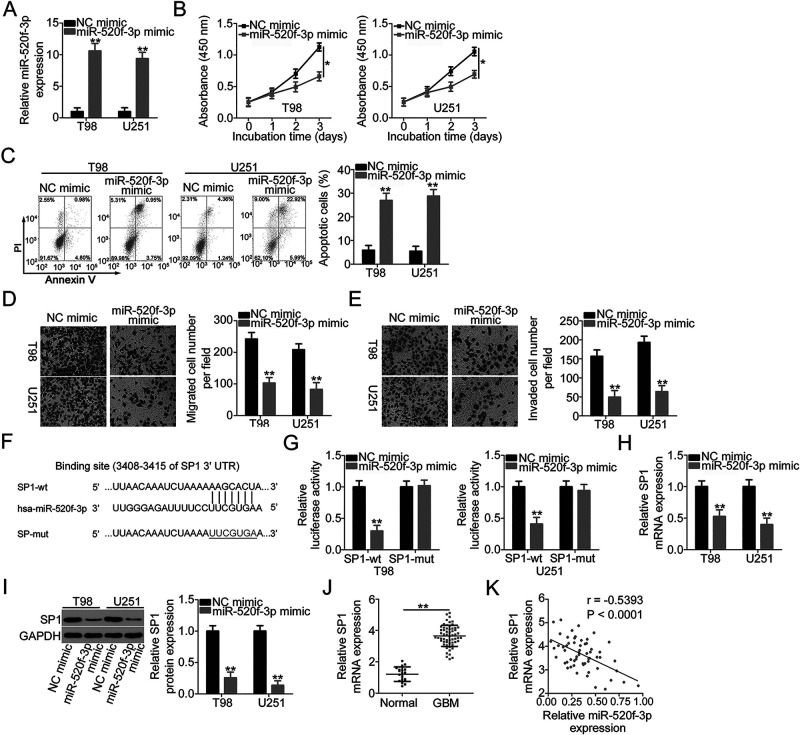
To clarify the functions of miR-520f-3p in GBM cells, miR-520f-3p mimic or NC mimic was transfected into T98 and U251 cells, and functional experiments were performed. First, the efficiency of miR-520f-3p mimic transfection was assessed by qRT-PCR, and the results confirmed the dramatic upregulation of miR-520f-3p expression in miR-520f-3p mimic-transfected T98 and U251 cells (Fig. 3A). Ectopic miR-520f-3p expression distinctly inhibited T98 and U251 cell proliferation (Fig. 3B) and facilitated cell apoptosis (Fig. 3C). Additionally, miR-520f-3p overexpression impaired T98 and U251 cell migration (Fig. 3D) and invasion (Fig. 3E) as it effectively decreased the number of migrated and invaded cells.
Figure 3.
miR-520f-3p exerts tumor-suppressing actions in GBM and directly targets SP1. (A) miR-520f-3p levels in T98 and U251 cells transfected with miR-520f-3p mimic or NC mimic were determined by qRT-PCR. (B, C) Proliferation and apoptosis in T98 and U251 cells after overexpression of miR-520f-3p were analyzed via CCK-8 assay and flow cytometric analysis. (D, E) Transwell cell migration and invasion assays were conducted to detect the migration and invasion of T98 and U251 cells after miR-520f-3p upregulation. (F) The predicted wild-type and mutant miR-520f-3p binding sites in the 3′-untranslated region (3′-UTR) of SP1. (G) The relationship between miR-520f-3p and SP1 was analyzed by detecting the luciferase activity of SP1-wt or SP1-mut after cotransfection with miR-520f-3p mimic or NC mimic. (H, I) qRT-PCR and Western blotting were conducted to measure SP1 mRNA and protein expression in miR-520f-3p-overexpressed T98 and U251 cells. (J) SP1 mRNA expression in 59 GBM tissues and 19 normal brain tissues was analyzed by qRT-PCR. (K) The correlation between SP1 mRNA and miR-520f-3p expression in the 59 GBM tissues was evaluated by Pearson’s correlation coefficient (r = −0.5393, p < 0.0001). *p < 0.05 and **p < 0.01.
To elucidate the mechanisms underlying the activities of miR-520f-3p in GBM cells, bioinformatic analysis was conducted to determine candidates for miR-520f-3p. SP1 (Fig. 3F), a tumor-promoting gene that is reported to be highly expressed in GBM30–34, attracted our attention and was chosen for further study. The luciferase reporter assay was applied to confirm this prediction. The results revealed that the upregulation of miR-520f-3p could reduce the luciferase activity of SP1-wt in T98 and U251 cells, while the suppression of luciferase activity was abrogated when the binding sequences were mutated (Fig. 3G). Furthermore, upregulated miR-520f-3p expression inhibited SP1 mRNA (Fig. 3H) and protein (Fig. 3I) expression in T98 and U251 cells. In addition, SP1 was evidently overexpressed in GBM tissues when compared with that in normal brain tissues (Fig. 3J). In the 59 GBM tissues, the mRNA level of SP1 was negatively associated with miR-520f-3p expression (r = −0.5393, p < 0.0001) (Fig. 3K). All of the above results provided sufficient evidence that SP1 is a direct target of miR-520f-3p in GBM cells.
WEE2-AS1 Positively Regulates SP1 Expression in GBM Cells via Sponging miR-520f-3p
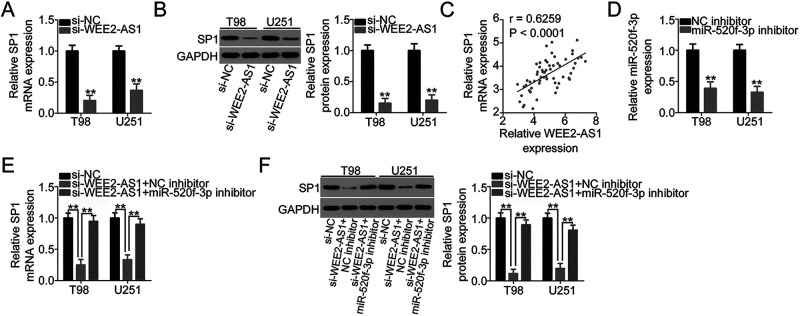
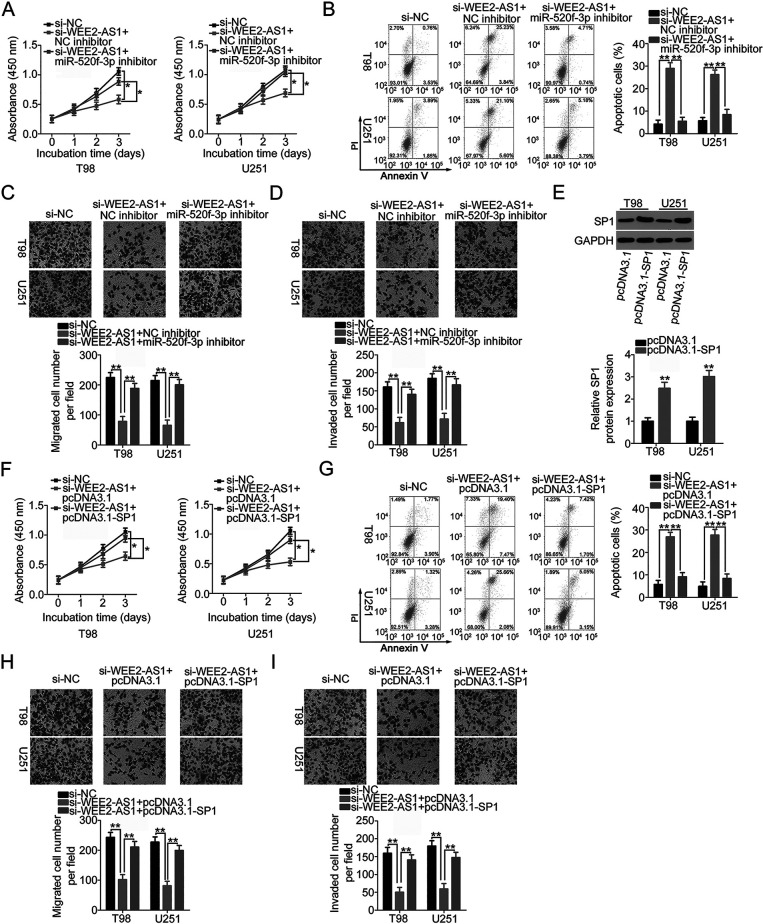
Our study demonstrated that WEE2-AS1 works by adsorbing miR-520f-3p in GBM cells and that SP1 is a direct target of miR-520f-3p. Accordingly, we next examined whether SP1 expression is affected by WEE2-AS1 knockdown in GBM cells via sponging miR-520f-3p. To this end, WEE2-AS1-depleted T98 and U251 cells were subjected to qRT-PCR and Western blotting. A reduction in WEE2-AS1 expression obviously suppressed the amount of SP1 mRNA (Fig. 4A) and protein (Fig. 4B) in T98 and U251 cells. In addition, the expression levels of WEE2-AS1 and SP1 mRNA were positively correlated in the 59 GBM tissues (r = 0.6259, p < 0.0001) (Fig. 4C), as identified by Pearson’s correlation coefficient. Next, si-WEE2-AS1, together with miR-520f-3p inhibitor or NC inhibitor, was introduced into T98 and U251 cells, and the change in SP1 expression was measured. The transfection efficiency of miR-520f-3p inhibitor in T98 and U251 cells was verified using qRT-PCR (Fig. 4D). qRT-PCR and Western blotting revealed that loss of WEE2-AS1 significantly decreased SP1 mRNA (Fig. 4E) and protein (Fig. 4F) levels in T98 and U251 cells, while cotransfection with miR-520f-3p inhibitor nearly reversed this trend. In short, WEE2-AS1 worked as a molecular sponge for miR-520f-3p and consequently increased SP1 expression in GBM cells.
Figure 4.
WEE2-AS1 positively regulates SP1 expression in GBM cells via sponging miR-520f-3p. (A, B) The mRNA and protein levels of SP1 in T98 and U251 cells were measured by qRT-PCR and Western blotting after the transfection of si-WEE2-AS1 or si-NC. (C) Pearson’s correlation coefficient was determined to assess the correlation between WEE2-AS1 and SP1 mRNA expression in the 59 GBM tissues (r = 0.6259, p < 0.0001). (D) qRT-PCR was conducted to detect miR-520f-3p expression in T98 and U251 cells after miR-520f-3p inhibitor or NC inhibitor transfection. (E, F) si-WEE2-AS1 was cotransfected with miR-520f-3p inhibitor or NC inhibitor into T98 and U251 cells. The cotransfected cells were collected and subjected to the measurement of SP1 mRNA and protein expression via qRT-PCR and Western blotting. **p < 0.01.
SP1 Upregulation or miR-520f-3p Inhibition Weakens the Impacts of WEE2-AS1 Depletion on the Malignant Characteristics of GBM Cells
To determine whether WEE2-AS1 regulates the oncogenicity of GBM by targeting the miR-520f-3p/SP1 axis, rescue experiments were performed in T98 and U251 cells after cotransfection with si-WEE2-AS1 and miR-520f-3p inhibitor or NC inhibitor. The results of the CCK-8 assay and flow cytometric analysis suggested that depleted WEE2-AS1 expression significantly impaired T98 and U251 cell proliferation and promoted cell apoptosis. Significantly, inhibition of miR-520f-3p abrogated the antiproliferation (Fig. 5A) and proapoptosis (Fig. 5B) influences of WEE2-AS1 knockdown in T98 and U251 cells. In addition, the results from Transwell cell migration and invasion assays revealed that downregulation of WEE2-AS1 restricted the migration (Fig. 5C) and invasion (Fig. 5D) of T98 and U251 cells, while cotransfection with miR-520f-3p inhibitor almost restored the migration and invasion abilities of T98 and U251 cells that were hindered by WEE2-AS1 knockdown.
Figure 5.
The rescue effects of the miR-520f-3p/SP1 axis on WEE2-AS1 deficiency-mediated inhibition of GBM progression. (A, B) si-WEE2-AS1, in combination with miR-520f-3p inhibitor or NC inhibitor, was cotransfected into T98 and U251 cells. Cellular proliferation and apoptosis were examined via the CCK-8 assay and flow cytometric analysis. (C, D) The migratory and invasive capacities of T98 and U251 cells treated as described above were assessed using Transwell cell migration and invasion assays. (E) The protein levels of SP1 in T98 and U251 cells were detected by Western blotting after the transfection of pcDNA3.1-SP1 or pcDNA3.1. (F–I) T98 and U251 cells transfected with si-WEE2-AS1, together with pcDNA3.1-SP1 or pcDNA3.1, were subjected to the CCK-8 assay, flow cytometric analysis, and Transwell cell migration and invasion assays for the assessment of cell proliferation, apoptosis, migration, and invasion, respectively. *p < 0.05 and **p < 0.01.
Furthermore, the SP1 overexpression plasmid pcDNA3.1-SP1 or empty pcDNA3.1 plasmid, in parallel with si-WEE2-AS1, was transfected into T98 and U251 cells. Western blotting verified that transfection with pcDNA3.1-SP1 efficiency increased SP1 protein expression in T98 and U251 cells (Fig. 5E). The inhibited proliferation (Fig. 5F) and increased apoptosis (Fig. 5G) in T98 and U251cells caused by WEE2-AS1 silencing were abrogated by the reintroduction of SP1. Furthermore, the suppression of migratory (Fig. 5H) and invasive (Fig. 5I) abilities triggered by WEE2-AS1 interference was abated by SP1 upregulation in T98 and U251 cells. These results collectively suggested that WEE2-AS1 performed its cancer-promoting actions in GBM progression via modulation of the miR-520f-3p/SP1 axis.
Downregulation of WEE2-AS1 Impairs Tumor Growth of GBM Cells In Vivo
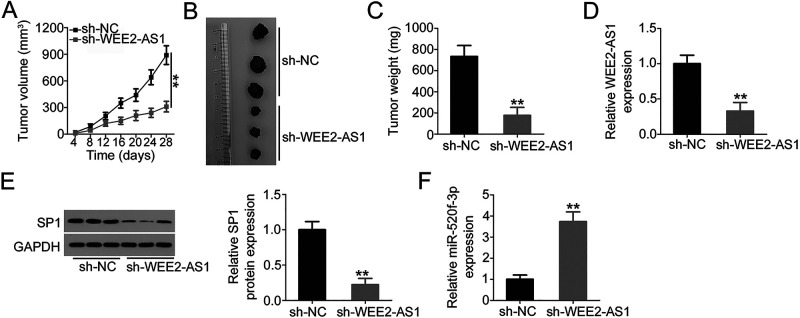
Lastly, tumor xenograft experiments were conducted to examine the impact of WEE2-AS1 depletion on GBM tumor growth in vivo. U251 cells infected with a WEE2-AS1-depleted lentiviral vector or control lentiviral vector were subcutaneously inoculated into nude mice to generate tumor xenografts. The volumes of tumor xenografts in the sh-WEE2-AS1 group were smaller than those in the sh-NC group (Fig. 6A and B). Also, the weights of tumor xenografts of the nude mice injected with sh-WEE2-AS1-transfected U251 cells were decreased in comparison with those of the nude mice injected with sh-NC-transfected cells (Fig. 6C). Four weeks postinjection, tumor xenografts were excised, and the expression levels of WEE2-AS1, miR-520f-3p, and SP1 were determined. The tumor xenografts derived from sh-WEE2-AS1-transfected U251 cells showed lower WEE2-AS1 (Fig. 6D) and SP1 (Fig. 6E) protein expression as well as higher miR-520f-3p expression (Fig. 6F) compared with those derived from sh-NC-transfected U251 cells. In summary, knockdown of WEE2-AS1 inhibited GBM tumor growth in vivo via regulating the miR-520f-3p/SP1 axis.
Figure 6.
Knockdown of WEE2-AS1 impairs GBM tumor growth in vivo. (A) Tumor growth curve after knockdown of WEE2-AS1. (B) Representative photographs of tumor xenografts collected from sh-WEE2-AS1 and sh-NC groups. (C) Weights of tumor xenografts were measured after tumor removal. (D) WEE2-AS1 expression in resected tumor xenografts was measured by qRT-PCR. (E) The expression of SP1 protein in tumor xenografts was detected by Western blot analysis. (F) Total RNA was extracted from resected tumor xenografts and used in qRT-PCR for the determination of miR-520f-3p expression. **p < 0.01.
DISCUSSION
Dysregulated lncRNA expression has been observed in many human cancer types, including GBM35–37. Numerous lncRNAs closely related to GBM pathogenesis have been identified, leading to the development of promising molecular-targeted therapy techniques38–40. Accordingly, targeting oncogenic lncRNAs or restoring antioncogenic lncRNAs is possible and will likely be an effective method to treat GBM. Although the number of identified lncRNAs continuously increases41, their detailed roles are largely unknown and require further investigation. In this study, we first investigated the expression and roles of WEE2-AS1 in GBM. Additionally, the molecular mechanisms underlying the oncogenic actions of WEE2-AS1 in GBM cells were explored in detail.
WEE2-AS1 is upregulated in hepatocellular carcinoma22 and triple negative breast cancer23. A high WEE2-AS1 expression is significantly associated with hepatitis B infection, major vascular invasion, and tumor differentiation in hepatocellular carcinoma22. Hepatocellular carcinoma patients with high WEE2-AS1 expression have shorter overall survival in contrast to those patients with a low WEE2-AS1 level22. Functionally, WEE2-AS1 exerts pro-oncogenic activities in hepatocellular carcinoma22 and triple negative breast cancer23 and is implicated in the control of multiple malignant behaviors. However, to the best of our knowledge, whether WEE2-AS1 is differentially expressed in GBM and contributes to GBM progression has not yet been reported. Here, we found that WEE2-AS1 was upregulated in GBM tissues and cell lines relative to their normal counterparts. High WEE2-AS1 expression was obviously correlated with average tumor diameter, KPS score, and shorter overall survival among patients with GBM. Loss of WEE2-AS1 expression in GBM cells suppressed cell proliferation, promoted cell apoptosis, and hindered migration and invasion in vitro. Additionally, interference of WEE2-AS1 expression inhibited GBM tumor growth in vivo.
Recently, considerable research has demonstrated that lncRNAs, acting as ceRNAs or molecular sponges for miRNAs, formed a huge and complex regulatory network42–44. lncRNAs are capable of competitively binding to MREs and consequently weakening the regulatory effects of miRNAs on their target mRNAs45. Extensive investigation over the past few decades has demonstrated the important functions of lncRNA–miRNA–mRNA pathways in many biological and pathological behaviors46 and suggested that disruption of ceRNA networks may cause several diseases, including cancers47,48.
To investigate whether WEE2-AS1 worked as a miRNA sponge, lncLocator prediction analysis and subcellular fractionation assays were performed to determine the subcellular localization of WEE2-AS1 in GBM cells. WEE2-AS1 was verified to be substantially enriched in the cytoplasm of GBM cells, suggesting that WEE2-AS1 has the potential to directly bind and interact with cytoplasmic miRNAs. Bioinformatic analysis was performed to search for target miRNAs that can interact with WEE2-AS1. miR-520f-3p was predicted to have a high probability of interacting with WEE2-AS1. Using luciferase reporter assay, RIP assay, and qRT-PCR, we found that WEE2-AS1 reduced miR-520f-3p expression in GBM cells through direct binding. In addition, miR-520f-3p expression was low in GBM tissues, which was inversely correlated with WEE2-AS1 expression. In subsequent experiments, our data showed that knockdown of WEE2-AS1 could decrease SP1 expression (a direct target of miR-520f-3p) in GBM cells at both mRNA and protein levels. Furthermore, SP1 was upregulated in GBM tissues and positively correlated with WEE2-AS1 expression. Based on rescue experiments, we confirmed that the regulatory effects of WEE2-AS1 knockdown on SP1 expression in GBM cells were attenuated following miR-520f-3p inhibition. Together, all of these data lead us to the identification of a novel ceRNA regulatory pathway in GBM cells comprising WEE2-AS1, miR-520f-3p, and SP1.
miR-520f-3p is substantially underexpressed in lung cancer27,28 and hepatocellular carcinoma29 and is identified as a tumor-suppressing miRNA. In this study, our data indicated that restoration of miR-520f-3p expression led to a significant decrease in GBM cell proliferation, migration, and invasion and an increase in cell apoptosis. Mechanisms by which miR-520f-3p executed its antioncogenic roles during GBM progression were also investigated. Mechanistic studies confirmed SP1 as a direct downstream target of miR-520f-3p in GBM cells. SP1, located at 12q13.1, is overexpressed in GBM30. Its overexpression is closely associated with WHO grade and poor prognosis in patients with GBM30. Moreover, SP1 is validated as an independent prognostic marker for predicting the survival of patients with GBM30. SP1 plays cancer-promoting roles during GBM oncogenesis and progression and is implicated in the regulation of various malignant processes31–34. Through a series of recovery experiments, our results implied that miR-520f-3p inhibition or SP1 overexpression nearly abrogated the inhibitory impacts of WEE2-AS1 depletion on proliferation, apoptosis, migration, and invasion of GBM cells, suggesting that WEE2-AS1 may perform its actions through regulating the output of the miR-520f-3p/SP1 axis. All of the above findings revealed that WEE2-AS1 can adsorb miR-520f-3p to increase endogenous SP1 expression, thereby facilitating the malignancy of GBM. Therefore, targeting the WEE2-AS1–miR-520f-3p–SP1 pathway might be a promising therapy for the management of GBM in the future.
SP1 sites are numerous in the MGMT promoter and mediate the therapy response in GBM49,50. Accordingly, WEE2-AS1 may be involved in the modulation of temozolomide resistance in GBM. In our study, we did not analyze the effect of WEE2-AS1 on temozolomide resistance in GBM cells, and it was a limitation of our study. We will resolve the limitation in our future experiments.
In summary, WEE2-AS1 knockdown increased miR-520f-3p expression to reduce SP1 expression in GBM cells. This resulted in inhibited cell proliferation, migration, and invasion and increased apoptosis in vitro as well as impaired tumor growth in vivo. Our findings suggest that the WEE2-AS1–miR-520f-3p–SP1 regulatory pathway may provide attractive targets to develop diagnostic and prognostic biomarkers and therapies for treating GBM.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005;109:93–108. [DOI] [PubMed] [Google Scholar]
- 2. Huang BS, Luo QZ, Han Y, Huang D, Tang QP, Wu LX. MiR-223/PAX6 axis regulates glioblastoma stem cell proliferation and the chemo resistance to TMZ via regulating PI3K/Akt pathway. J Cell Biochem. 2017;118:3452–61. [DOI] [PubMed] [Google Scholar]
- 3. Popescu ID, Codrici E, Albulescu L, Mihai S, Enciu AM, Albulescu R, Tanase CP. Potential serum biomarkers for glioblastoma diagnostic assessed by proteomic approaches. Proteome Sci. 2014;12:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–710. [DOI] [PubMed] [Google Scholar]
- 5. Davis ME. Glioblastoma: Overview of disease and treatment. Clin J Oncol Nurs. 2016;20:S2–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rao JS. Molecular mechanisms of glioma invasiveness: The role of proteases. Nat Rev Cancer 2003;3:489–501. [DOI] [PubMed] [Google Scholar]
- 7. Marumoto T, Saya H. Molecular biology of glioma. Adv Exp Med Biol. 2012;746:2–11. [DOI] [PubMed] [Google Scholar]
- 8. Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004;116:281–97. [DOI] [PubMed] [Google Scholar]
- 9. Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Song JL, Nigam P, Tektas SS, Selva E. MicroRNA regulation of Wnt signaling pathways in development and disease. Cell Signal. 2015;27:1380–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nguyen DD, Chang S. Development of novel therapeutic agents by inhibition of oncogenic microRNAs. 2017;19:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jiapaer S, Furuta T, Tanaka S, Kitabayashi T, Nakada M. Potential strategies overcoming the temozolomide resistance for glioblastoma. Neurol Med Chir. (Tokyo) 2018;58:405–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Banelli B, Forlani A, Allemanni G, Morabito A, Pistillo MP, Romani M. MicroRNA in glioblastoma: An overview. Int J Genomics 2017;2017:7639084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Henriksen M, Johnsen KB, Andersen HH, Pilgaard L, Duroux M. MicroRNA expression signatures determine prognosis and survival in glioblastoma multiforme—A systematic overview. Mol Neurobiol. 2014;50:896–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cao J. The functional role of long non-coding RNAs and epigenetics. Biol Proced Online 2014;16:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics 2016;14:42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hu T, Wang F, Han G. LncRNA PSMB8-AS1 acts as ceRNA of miR-22-3p to regulate DDIT4 expression in glioblastoma. Neurosci Lett. 2020;728:134896. [DOI] [PubMed] [Google Scholar]
- 18. Baspinar Y, Elmaci I, Ozpinar A, Altinoz MA. Long non-coding RNA MALAT1 as a key target in pathogenesis of glioblastoma. Janus faces or Achilles’ heal? Gene 2020;739:144518. [DOI] [PubMed] [Google Scholar]
- 19. Han N, Yang L, Zhang X, Zhou Y, Chen R, Yu Y, Dong Z, Zhang M. LncRNA MATN1-AS1 prevents glioblastoma cell from proliferation and invasion via RELA regulation and MAPK signaling pathway. Ann Transl Med. 2019;7:784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hao Y, Li X, Chen H, Huo H, Liu Z, Chai E. Over-expression of long noncoding RNA HOTAIRM1 promotes cell proliferation and invasion in human glioblastoma by up-regulating SP1 via sponging miR-137. Neuroreport 2020;31:109–17. [DOI] [PubMed] [Google Scholar]
- 21. Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19:1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hu Z, Huang P, Yan Y, Zhou Z, Wang J, Wu G. Hepatitis B virus X protein related lncRNA WEE2-AS1 promotes hepatocellular carcinoma proliferation and invasion. Biochem Biophys Res Commun. 2019;508:79–86. [DOI] [PubMed] [Google Scholar]
- 23. Wang R, Huang Z, Qian C, Wang M, Zheng Y, Jiang R, Yu C. LncRNA WEE2-AS1 promotes proliferation and inhibits apoptosis in triple negative breast cancer cells via regulating miR-32-5p/TOB1 axis. Biochem Biophys Res Commun. 2020;526:1005–12. [DOI] [PubMed] [Google Scholar]
- 24. Xue YH, Ge YQ. Construction of lncRNA regulatory networks reveal the key lncRNAs associated with pituitary adenomas progression. Math Biosci Eng. 2020;17:2138–49. [DOI] [PubMed] [Google Scholar]
- 25. Peng S, Yin X, Zhang Y, Mi W, Li T, Yu Y, Jiang J, Liu Q, Fu Y. Competing endogenous RNA network analysis reveals potential long non-coding RNAs as predictive biomarkers of gastric cancer. Oncol Lett. 2020;19:2185–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zheng H, Li BH, Liu C, Jia L, Liu FT. Comprehensive analysis of lncRNA-mediated ceRNA crosstalk and identification of prognostic biomarkers in Wilms’ tumor. Biomed Res Int. 2020;2020:4951692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou Y, Shen S. MiR-520f acts as a biomarker for the diagnosis of lung cancer. Medicine (Baltimore) 2019;98:e16546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cui J, Li W, Liu G, Chen X, Gao X, Lu H, Lin D. A novel circular RNA, hsa_circ_0043278, acts as a potential biomarker and promotes non-small cell lung cancer cell proliferation and migration by regulating miR-520f. Artif Cells Nanomed Biotechnol. 2019;47:810–21. [DOI] [PubMed] [Google Scholar]
- 29. Du X, Fan W, Chen Y. microRNA-520f inhibits hepatocellular carcinoma cell proliferation and invasion by targeting TM4SF1. Gene 2018;657:30–8. [DOI] [PubMed] [Google Scholar]
- 30. Guan H, Cai J, Zhang N, Wu J, Yuan J, Li J, Li M. Sp1 is upregulated in human glioma, promotes MMP-2-mediated cell invasion and predicts poor clinical outcome. Int J Cancer 2012;130:593–601. [DOI] [PubMed] [Google Scholar]
- 31. Li Y, Wu Y, Sun Z, Wang R, Ma D. MicroRNA376a inhibits cell proliferation and invasion in glioblastoma multiforme by directly targeting specificity protein 1. Mol Med Rep. 2018;17:1583–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dong Q, Cai N, Tao T, Zhang R, Yan W, Li R, Zhang J, Luo H, Shi Y, Luan W, Zhang Y, You Y, Wang Y, Liu N. An axis involving SNAI1, microRNA-128 and SP1 modulates glioma progression. PLoS One 2014;9:e98651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Seznec J, Silkenstedt B, Naumann U. Therapeutic effects of the Sp1 inhibitor mithramycin A in glioblastoma. J Neurooncol. 2011;101:365–77. [DOI] [PubMed] [Google Scholar]
- 34. Jiang J, Wei Y, Shen J, Liu D, Chen X, Zhou J, Zong H, Yun X, Kong X, Zhang S, Yang Y, Gu J. Functional interaction of E1AF and Sp1 in glioma invasion. Mol Cell Biol. 2007;27:8770–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang J, Liu X, Yan C, Liu J, Wang S, Hong Y, Gu A, Zhao P. LEF1-AS1, a long-noncoding RNA, promotes malignancy in glioblastoma. Onco Targets Ther. 2017;10:4251–60. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36. Zeng H, Xu N, Liu Y, Liu B, Yang Z, Fu Z, Lian C, Guo H. Genomic profiling of long non-coding RNA and mRNA expression associated with acquired temozolomide resistance in glioblastoma cells. Int J Oncol. 2017;51:445–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li H, Yuan X, Yan D, Li D, Guan F, Dong Y, Wang H, Liu X, Yang B. Long non-coding RNA MALAT1 decreases the sensitivity of resistant glioblastoma cell lines to temozolomide. Cell Physiol Biochem. 2017;42:1192–201. [DOI] [PubMed] [Google Scholar]
- 38. Ma B, Yuan Z, Zhang L, Lv P, Yang T, Gao J, Pan N, Wu Q, Lou J, Han C, Zhang B. Long non-coding RNA AC023115.3 suppresses chemoresistance of glioblastoma by reducing autophagy. Biochim Biophys Acta Mol Cell Res. 2017;1864:1393–404. [DOI] [PubMed] [Google Scholar]
- 39. Zhang L, Wang H, Xu M, Chen F, Li W, Hu H, Yuan Q, Su Y, Liu X, Wuri J, Yan T. Long noncoding RNA HAS2-AS1 promotes tumor progression in glioblastoma via functioning as a competing endogenous RNA. J Cell Biochem. 2020;121:661–71. [DOI] [PubMed] [Google Scholar]
- 40. Argadal OG, Mutlu M, Ak Aksoy S, Kocaeli H, Tunca B, Civan MN, Egeli U, Cecener G, Bekar A, Taskapilioglu MO, Tekin C, Tezcan G, Tolunay S. Long noncoding RNA MALAT1 may be a prognostic biomarker in IDH1/2 wild-type primary glioblastomas. Bosn J Basic Med Sci. 2020;20:63–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Paulmurugan R, Malhotra M, Massoud TF. The protean world of non-coding RNAs in glioblastoma. J Mol Med. (Berl) 2019;97:909–25. [DOI] [PubMed] [Google Scholar]
- 42. Li Y, Liu JJ, Zhou JH, Chen R, Cen CQ. LncRNA HULC induces the progression of osteosarcoma by regulating the miR-372-3p/HMGB1 signalling axis. Mol Med. 2020;26:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang L, Lu R, Wang Y, Wang X, Hao D, Wen X, Li Y, Zeng M, Jiang X. Identification of long noncoding RNA associated ceRNA networks in rosacea. Biomed Res Int. 2020;2020:9705950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu K, Jiang Y, Zhou W, Zhang B, Li Y, Xie F, Zhang J, Wang X, Yan M, Xu Q, Ren Z, Chen W, Cao W. Long Noncoding RNA RC3H2 facilitates cell proliferation and invasion by targeting microRNA-101-3p/EZH2 axis in OSCC. Mol Ther Nucleic Acids 2020;20:97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ye Y, Shen A, Liu A. Long non-coding RNA H19 and cancer: A competing endogenous RNA. Bull Cancer 2019;106:1152–9. [DOI] [PubMed] [Google Scholar]
- 46. Abdollahzadeh R, Daraei A, Mansoori Y, Sepahvand M, Amoli MM, Tavakkoly-Bazzaz J. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: A new look at hallmarks of breast cancer. J Cell Physiol. 2019;234:10080–100. [DOI] [PubMed] [Google Scholar]
- 47. Shuwen H, Qing Z, Yan Z, Xi Y. Competitive endogenous RNA in colorectal cancer: A systematic review. Gene 2018;645:157–62. [DOI] [PubMed] [Google Scholar]
- 48. Renganathan A, Felley-Bosco E. Long noncoding RNAs in cancer and therapeutic potential. Adv Exp Med Biol. 2017;1008:199–222. [DOI] [PubMed] [Google Scholar]
- 49. Chuang JY, Lo WL, Ko CY, Chou SY, Chen RM, Chang KY, Hung JJ, Su WC, Chang WC, Hsu TI. Upregulation of CYP17A1 by Sp1-mediated DNA demethylation confers temozolomide resistance through DHEA-mediated protection in glioma. Oncogenesis 2017;6:e339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yang WB, Chuang JY, Ko CY, Chang WC, Hsu TI. Dehydroepiandrosterone induces temozolomide resistance through modulating phosphorylation and acetylation of Sp1 in glioblastoma. Mol Neurobiol. 2019;56:2301–13. [DOI] [PubMed] [Google Scholar]