Abstract
Plants in the family Lemnaceae are aquatic monocots and the smallest, simplest, and fastest growing angiosperms. Their small size, the smallest family member is 0.5 mm and the largest is 2.0 cm, as well as their diverse morphologies make these plants ideal for laboratory studies. Their rapid growth rate is partially due to the family’s neotenous lifestyle, where instead of maturing and producing flowers, the plants remain in a juvenile state and continuously bud asexually. Maturation and flowering in the wild are rare in most family members. To promote further research on these unique plants, we have optimized laboratory flowering protocols for 3 of the 5 genera: Spirodela; Lemna; and Wolffia in the Lemnaceae. Duckweeds were widely used in the past for research on flowering, hormone and amino acid biosynthesis, the photosynthetic apparatus, and phytoremediation due to their aqueous lifestyle and ease of aseptic culture. There is a recent renaissance in interest in growing these plants as non-lignified biomass sources for fuel production, and as a resource-efficient complete protein source. The genome sequences of several Lemnaceae family members have become available, providing a foundation for genetic improvement of these plants as crops. The protocols for maximizing flowering described herein are based on screens testing daylength, a variety of media, supplementation with salicylic acid or ethylenediamine-N,N′-bis(2-hydroxyphenylacetic acid) (EDDHA), as well as various culture vessels for effects on flowering of verified Lemnaceae strains available from the Rutgers Duckweed Stock Cooperative.
Keywords: duckweed, Spirodela polyrhiza, Lemna gibba, Lemna minor, Wolffia microscopica, flowering protocols
1. Introduction
The five genera and 36 species comprising the Lemnaceae family, commonly known as duckweeds, are the smallest, fastest growing, most morphologically reduced, and widely distributed family of angiosperms [1,2,3]. These aquatic monocots are found floating and rapidly clonally dividing on still, nutrient rich, waters worldwide. Duckweeds are rare among plants for being a complete protein source, with an amino acid content similar to eggs [4,5]. Wild or greenhouse grown Wolffia species are an especially resource-efficient food source. Protein concentrates from Lemna species are being explored as a scalable and economical way to meet the rapidly increasing demand for plant-based proteins [6,7]. Members of the Lemnaceae are promising industrial crops that can grow at rates of 13–38 dry tons/hectare per year, compared to 6–9 and 2.5–3.5 dry tons/hectare per year for maize and soybean, respectively, reported by the FAO [8,9]. Duckweeds can grow on non-arable land in agricultural or industrial wastewater, recapturing nutrients instead of needing energy intensive fertilizers, without pesticides, while cleaning the water and producing a biomass that can be used as animal feed or biofuel [6,7,8]. Research into this family’s unique biology will result in improving the ways we use these distinctive plants to sustainably provide clean water, food, and energy in the future.
The Lemnaceae plant body generally consists of a small (0.5 mm–2 cm), flat leaf-like structure called a frond. Plants in the Spirodela, Landoltia, and Lemna genera have rhizoid structures, while the smaller, simpler, and more recently evolved Wolffiella and Wolffia genera lack these structures [1]. Wolffia microscopica, one of the smallest flowering plants, has a rootlike projection described in detail in Sree et al., 2015 [10], the function of which remains unknown. Many Lemnaceae species can overwinter in temperate climates by asexually budding a starch rich modified frond known as a turion to create an asexual organ of perenniation. The turion sinks to the bottom of the pond, then rises as a frond in the spring [1,3,11].
Due to their small size, rapid asexual and clonal growth, ease of aseptic cultivation, and simple morphology, the Lemnaceae were, for many years in the past, used as model systems for studying a wide array of biological and biochemical processes such as flowering [12,13,14], hormone and amino acid biosynthesis [15,16,17], the role of the D1 protein and regulation of expression of chlorophyll binding protein genes in photosynthesis [18,19], and many other aspects of plant biology [1,20,21].
In addition, their aquatic lifestyle makes duckweeds particularly useful for phytotoxicity testing, and standardized experimental protocols using duckweeds were developed for testing water quality [22,23,24]. To facilitate rapid and accurate measurement of duckweed growth in toxicity tests, there has been a recent shift from frond counts and fresh or dry weight measurements to image analysis software, which can measure total area, plant health, and possibly flowering. The company Lemnatec sells commercial imaging chambers, with software packages, while researchers have applied the Aphelion software package [25], the NI vision Assistant software [26] in data analysis pipelines, or the free software Image J, with manual assistance in selecting green surface area (P. Fourounjian, unpublished) to take non-destructive measurements of biomass.
Hundreds of diverse duckweed strains from around the world were collected by Elias Landolt [27], and many of these are now curated and stored at the Rutgers Duckweed Stock Cooperative, the RDSC (http://www.ruduckweed.org/ (accessed on 26 January 2021)) Additional collections are found worldwide (listed on the RDSC website), making the morphologically diverse germplasm easily available to researchers. The Landolt strains have retained the original 4 digit identifiers, while newer strains have been assigned a 3 digit code, for example, DWC130. While distinguishing the 36 species based on morphology is challenging, they can be easily identified by genotyping or barcoding at precise intergenic spacers [28].
Renewed interests in the commercialization of members of the Lemnaceae have spurred the publication of draft genomes of: S. polyrhiza strains 7498 [29] and 9509 [30]; L. minor strains 5500 [31] and 8627; L. gibba G3 strain 7741; and W. australiana strain 8730 (https://www.lemna.org/ (accessed on 26 January 2021)). The Landoltia punctata transcriptome was recently described, so 4 of the 5 genera are now under genomic scrutiny [32,33]. A transcriptomic study of turion formation in S. polyrhiza [11]; two maps of miRNAs in S. polyrhiza with cleaved targets, [30,34]; and Spirodela proteome analysis [35] together with several other genomic mapping advances [36,37,38], have resulted in S. polyrhiza strains 7498 and 9509 having the best characterized genomes in the family. The discovery that Spirodela has the least RNA directed DNA methylation (RdDM) of any plant studied to date, yet surprisingly few transposons [29,30,39] aptly illustrates the importance of developing resources with which to explore a wide diversity of plants.
There are 19 published protocols for stable transformation and transient gene expression in Lemnaceae, allowing for genome manipulation in 8 species, in all genera except Wolffiella [40]. In addition, the metagenome, or microbiome of these plants can be readily studied and manipulated. Plant growth promoting bacteria found in natural Lemnaceae microbiomes are being investigated as a means to enhance growth and ability to purify wastewater. As wild duckweeds can be surface sterilized, grown aseptically, and cultures then re-inoculated with a chosen microbe or intentionally engineered microbial communities, the duckweeds are excellent models to study interactions between plant and bacteria, or fungi, and even algae [41,42].
Members of the Lemnaceae grow in a juvenile and asexual manner indefinitely, and then produce flowers in a matter of days if exposed to the correct stimuli. This allows precise study of individual stimuli or genetic pathways which initiate floral development, rather than the earlier or later flowering phenotypes observed in Arabidopsis and other commonly studied annual plants, where regulation of flowering is complicated by multiple active pathways [43,44]. The Lemnaceae were used to search for the mythical “florigen”, first hypothesized in 1937 [45]. Landolt and Kandeler’s multivolume monograph, The Family of Lemnaceae—a monographic study Volumes 1 and 2 [1,3] and a later review by Pieterse [46] describe 112 studies to investigate factors affecting floral regulation using duckweed. Day-length was paramount in every study, while salicylic acid (SA), a plant hormone now understood as important for abiotic stress response and pathogen resistance, was the most studied chemical inducer. Chelating agents such as ethylenediamine-N,N′-bis(2-hydroxyphenylacetic acid) (EDDHA), were also frequent players in those studies.
While many studies of the duckweeds describe ways to induce flowering in the laboratory, there are only two reports of seed production in the laboratory. This is important as the production of seed underlies establishment of breeding protocols to improve agricultural traits. In the 1950s, Maheshwari described the anatomy of W. microscopica and L. paucicostata (now L. aequinoctialis) seed collected from the wild, showing the presence of a cellular endosperm [47]. A study of IAA accumulation in a tissue culture induced large mutant line of L. gibba G3 [16] described that gentle shaking of flowering plants growing on liquid “E” medium resulted in production of seed, which fell to the bottom of the culture flask and germinated when placed in fresh media. Recently, Fu et al., 2017 [48] were the first to produce hybrid duckweed seed by manually crossing under a dissecting microscope, two strains of L. gibba G3 flowering on modified Hoagland’s (MH) medium containing 20 μM SA. In that study, only one of the strains used in the cross, 7741, was able to produce viable pollen. Strain 7741 was manually crossed to a male sterile strain to produce the first hybrid strain created in a laboratory.
The long history of research using duckweeds and the rapidly increasing omics resources available make these plants attractive for use in exploring basic biological questions related to the neotenous, aqueous lifestyle, while simultaneously accelerating the domestication of the fastest growing plants into a sustainable crop species. Yet, one could argue that no duckweed species would ever be a “model organism” unless there are robust flowering protocols and the ability to produce seed. We describe here protocols for reliably obtaining flowering cultures of three genera, that will enable development and floral regulation studies, and facilitate development of protocols for production of improved varieties, whether by classical breeding or gene editing. Such protocols are fundamental to performing the mutant screens, genetic experiments and genome wide association studies that have facilitated research in other model plants, and the commercial breeding of other crops.
2. Results
2.1. Spirodela Polyrhiza
Initial screening experiments involved three media, various SA or EDDHA concentrations and combinations, and several light regimes. Guided by previous work on other Spirodela strains [49,50], we initially screened 3 media: E; Hoagland’s (Hg); and Schenk Hildebrandt (SH) (Table S1) with and without SA and EDDHA over 28 days in 16 h light:8 h dark long days (LD), or continuous light (CL) for induction of flowering (Table S2). Flowering was very sparse or completely absent for both genomically characterized strains 7498 and 9509 on any medium in LD. Neither strain flowered on E (pH 4.6), or E at pH 5.8, although a senescent culture of 9509 flowered on E supplemented with 25 μM EDDHA. In these screens, the highest flowering rate, 4%, was seen with strain 7498 on SH and Hg, supplemented with 1.5 or 2.0 μM SA under CL. By day 14, strain 9509 was senescing on SH, so screening was continued with Hg only, and all later experiments were performed using Hg. A second screen was performed to test whether using different culture vessels affected flowering. Strain 7498 exhibited higher flowering rates than 9509 in any culture vessel. Cultures in Petri dishes and larger flasks marginally outperformed those in small flasks. In this set of screening conditions, the optimal SA concentrations were 1.0 μM and 1.5 μM (Table S2).
Although a low red/far-red light ratio accelerates the flowering response in Arabidopsis, [51], screens with cultures growing with supplemental far-red light failed to produce flowers (Table S2). Shifting cultures of strain 7498 from LD or CL to Petri dishes under 12 h light:12 h dark led to 9% of fronds flowering with some dehiscent anthers (Table S2). Replicated testing of strain 7498 with these conditions showed that visible flowering started at day 11, and peaked at day 14 with a maximum rate of 6% independent of whether the inoculant was grown in LD or CL (Figure S1).
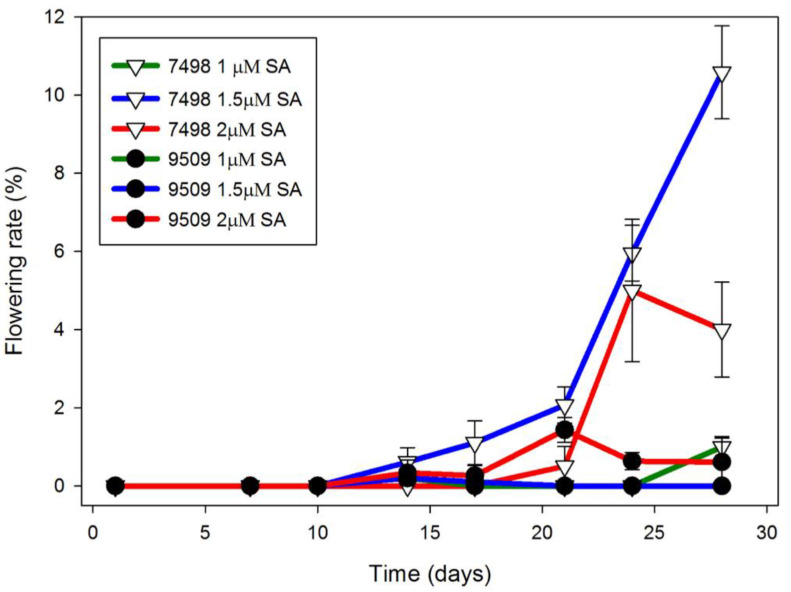
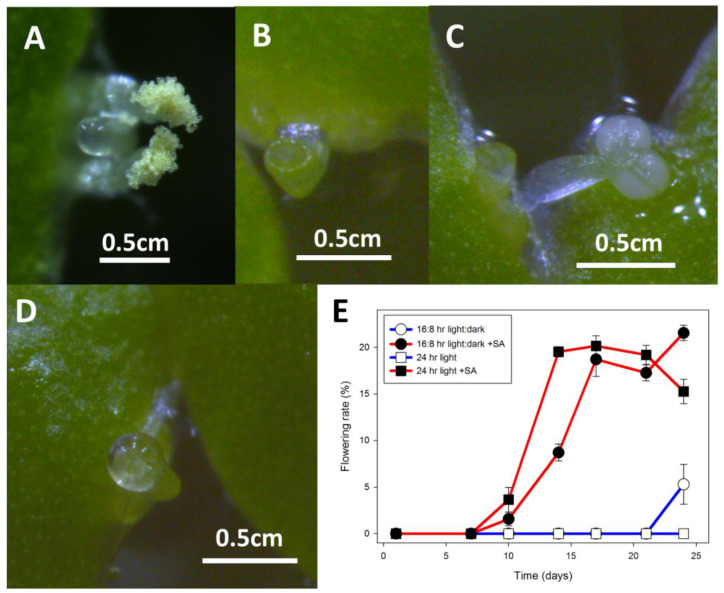
Using 4 week old cultures as a source of inoculant improved flowering rate. In Hg medium, Petri dishes of strain 7498 inoculated with three 3–5 frond colonies from one month old cultures growing in Hg medium, began flowering after 10 days in CL (Figure 1). By day 24, strain 7498 had a significantly higher flowering rate than strain 9509 the 1.5 μM SA condition (p < 0.01). By day 28, when the culture filled the dish, 11 ± 1% of the 7498 fronds growing on media containing 1.5 μM SA were flowering, significantly different from 2.0 μM SA (p < 0.01). Under the same conditions, only 1% of strain 9509 fronds were flowering (Figure 1). Both strains produced flowers with pistils secreting stigmatic fluid as well as flowers with dehiscent anthers, although 7498 did so at a higher rate (Figure 2).
Figure 1.
Flowering rate of S. polyrhiza strain 7498 cultured on Hg was increased to a much greater extent by SA than that of strain 9509. Plants were grown in Petri dishes inoculated with three 3-frond colonies from 4-week-old cultures growing on Hg without SA. Optimal flowering for strain 7498 was observed in the presence of 1.5 μM SA. Data are the average ± SEM (n = 3).
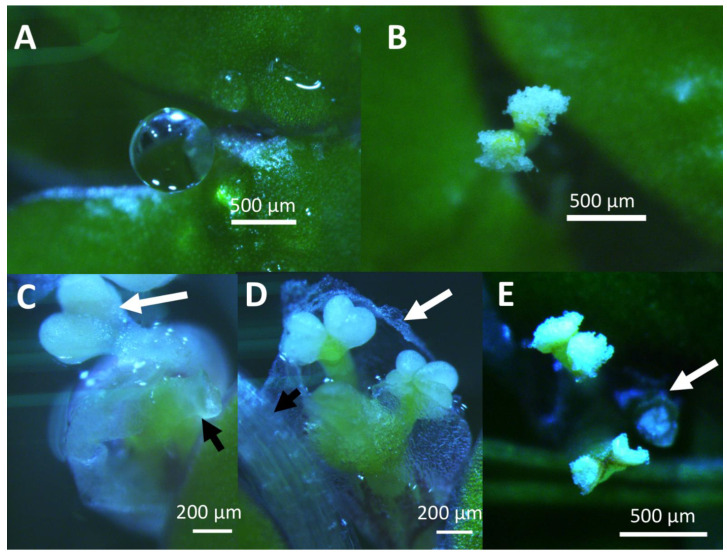
Figure 2.
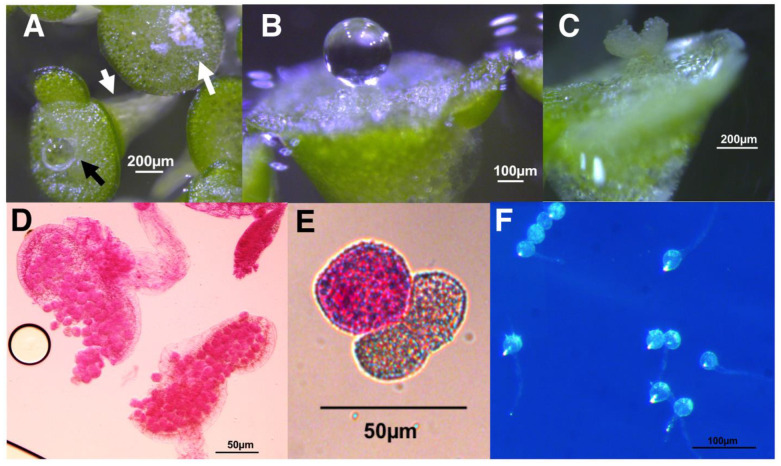
Flowers of S. polyrhiza strains 7498 and 9509. (A) Droplet of stigmatic fluid secreted by an S. polyrhiza 7498 pistil. (B) Dehiscing anthers of strain 7498. (C) Pistil without stigmatic fluid (black arrow) and non-dehiscent anther (white arrow) of S. polyrhiza strain 9509. (D) Two bi-lobed developing anthers and developing pistil of a strain 9509 flower. The transparent membrane known as a spathe (white arrow) has ruptured, but still surrounds the developing flower. The black arrow indicates a stipe, which connects the mother frond to a daughter. (E) Dehiscing anthers of strain 9509 and pistil without stigmatic fluid (white arrow).
Occasionally, abnormal flowers were seen in Petri dish cultures. Unusual kidney shaped fronds of strain 7498 produced flowers that did not appear to emerge from the normal location on the frond (Figure 3A). Strain 9509 occasionally produced flowers with what appeared to be the purple ventral side of the frond overgrowing and covering the anthers (Figure 3B). On this frond, a rhizoid, which would normally grow from the ventral surface of the frond, is growing vertically up out of the medium. Completely detached, aborted developing pistils and anthers enclosed in the spathe were found floating in strain 9509 cultures (Figure 3C).
Figure 3.
Anomalous flowers of S, polyrhiza. (A) An abnormal flower of strain 7498 that appears to have emerged from an abnormal, kidney shaped frond. (B) This inflorescence on strain 9509 appeared to be overgrown by the underside of the frond. The purple pigmentation is similar to that of the ventral surface of this strain. The vertical structure appears rhizoid-like. (C) An aborted inflorescence of strain 9509, consisting of the developing pistil and anthers enveloped in a spathe, found floating in the medium.
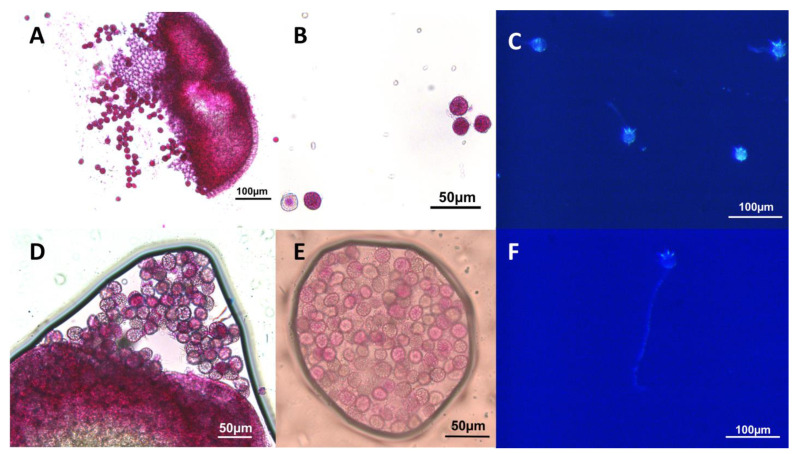
Both strains of Spirodela produced some viable pollen according to the Alexander’s stain [52], that was capable of germinating in vitro (Figure 4). In strain 7498, 69% or 161 of 234 total pollen grains from dehiscing anthers stained as viable, and 48% (47 of 97 total) germinated and produced pollen tubes. Only 26% (207 of 803) of the pollen from strain 9509 dehiscing anthers stained as viable, and only 9.7% (10 of 103) produced pollen tubes.
Figure 4.
S. polyrhiza pollen analysis (A,B) Pollen from dehiscent anthers of strain 7498 stained with modified Alexander’s stain [52] indicating a high percentage of viability. Live pollen is pink or magenta, clear or blue stained pollen is aborted. (C) Pollen tube formation by pollen from dehisced anthers of strain 7498. (D,E) Alexander’s staining showed that anthers of strain 9509 produced viable pollen that was capable of forming pollen tubes (F).
On average, strain 7498 formed 518 ± 98 turions per flask over a 2 month culture period, and an average of 309 ± 66 turions were found per flask of strain 9509 over the same time period. After 2 months, no fruit or seed were found at the bottom of flasks or plates of either strain. Gentle rotary shaking of both strains, and manually self-pollinating 6 fluid secreting pistils of 7498 with dehiscent anthers failed to result in seed formation.
2.2. Wolffia Microscopica
A screen in which plants were cultured on Hg, with daylength, chelating agent EDDHA, and sucrose addition varied, clearly showed that LD were better for flowering than CL, provided that sucrose and EDDHA were present (Table S3). EDDHA enhanced frond multiplication when sucrose is present, although plants were capable of multiplying without either sucrose or EDDHA. However, cultures without sucrose and EDDHA senesced earlier than those with sucrose but without EDDHA. Overall, the screen indicated that in LD, Hg supplemented with sucrose and either 25 or 75 μM of EDDHA resulted in the highest flowering rates (Table S3).
Further screens with E and Hg media in flasks and Petri dishes exhibited very high variability, but overall greater flowering was seen with E, and on this medium no influence of EDDHA on flowering rate was observed (Table S4). This screen indicated that when growing on E, W. microscopica was greatly affected by the type of culture vessel.
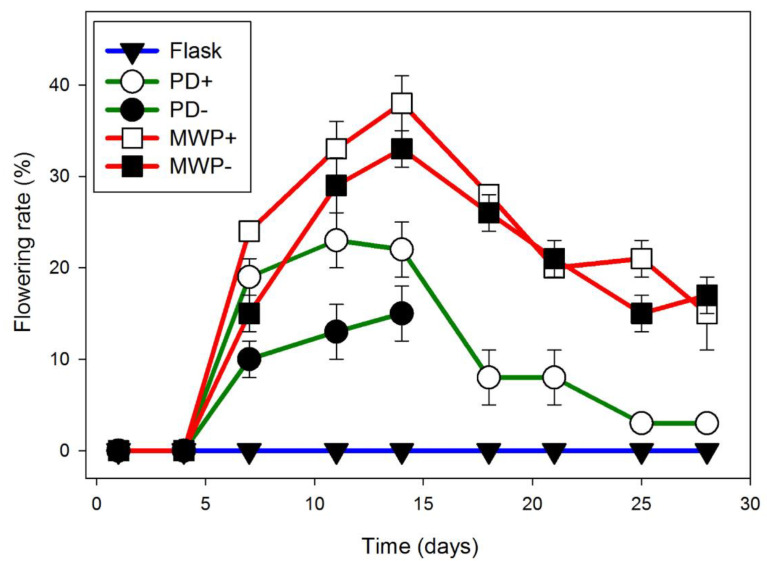
No flowering was observed if Wolffia was grown in cotton stoppered flasks containing 100 mL of E, whereas if the plants were grown in 10 mL of E in a 6-well plate, 38 ± 2% of the fronds were flowering over the same time period (Figure 5). After day 7, flowering was significantly greater in parafilm sealed 6-well plates (red) than in parafilm sealed Petri dishes (green) (p < 0.05) and no significant differences were found between 6-well plates sealed or not sealed with parafilm (Figure 5). Under these conditions, the abundant pistils were easily observed due to a drop of stigmatic fluid, which was almost the size of the frond, and dehiscent anthers were also abundant (Figure 6). Pollen from these anthers stained as 56% (94 of 169) viable, and 28% of the pollen grains formed germination tubes (65 of 235) (Figure 6). Despite plentiful flowering, none of the cultures, either gently shaken or stationary, were observed to produce seed.
Figure 5.
W. microscopica flowering rate is affected by culture vessel. Flowering rates of W. microscopica growing on E media in cotton stoppered flasks (blue), Petri dishes (PD) (green), or multi-well plates (MWP) (red). Petri dishes or 6-well plates were sealed with parafilm (open symbols) or left unsealed (closed symbols). Maximal flowering was observed at day 15 after inoculation in sealed 6-well plates. Data are the average ± SEM (n = 3).
Figure 6.
W. microscopica 2005 flowering and pollen analysis (A) Aerial view of mature W. microscopica 2005 fronds showing the pseudoroot (white arrowhead) extending from the ventral surface of a frond that is asexually reproducing a daughter frond at the same time it has produced a pistil. The pistil emerged from a furrow on the dorsal surface and has secreted stigmatic fluid (black arrow). A dehiscing anther has emerged from the furrow of a different frond (white arrow). (B) Side view of mature pistil with secreted fluid. (C) Side view of a dehiscing bilobed anther. Morphology is as described in Sree et al. [10]. Ruptured anthers releasing pollen (D) and pollen grains (E) stained with modified Alexander’s stain indicating that W. microscopica 2005 is producing viable pollen in culture. (F) 28% of W. microscopica 2005 pollen produced pollen tubes.
2.3. Lemna
No flowering was observed in our initial experiments with DWC130, in contrast to what was reported for this strain in Slovin and Cohen 1988 [16]. As of 2017, there were 5 strains of Lemna gibba G3 listed in the RDSC inventory: DWC114 (7741), collected in Sicily, which is the pollen producing strain described by Fu et al. [48], DWC130 and DWC131, labeled as a self-fertile parental line and a larger auxin mutant jsR1 regenerated from tissue culture [16], DWC132, which was contributed by Biolex Corporation, and DWC128 from the Waksman Collection, which was not included in this study. Contrary to expectations, DWC114 was, in fact, the largest of the 4 strains we grew. DNA barcoding using chloroplast atpH-F primers [28] unequivocally showed that strain DWC114 is L. gibba G3, and revealed that the other 3 strains are, in fact, L. minor (Table 1). The DWC114 barcode sequence is an exact match to both L. gibba 5504 and L. gibba 7741 described by Fu et al. [48]. DWC132 was further barcoded using matK primers, which clarified its identity as L. minor. Barcoding revealed a high degree of similarity among the three L. minor strains, DWC130, 131, and 132, with all closely matching L. minor strain 7210. The auxin mutant jsR1 appears to have been lost in one of the collection’s multiple transfers, while DWC114 appears to be the L. gibba G3 originally described in Hillman 1961 [53]. Based on the results, our colleagues at the RDSC updated the database as part of their curation of the global collection.
Table 1.
DNA Barcoding results showed that only DWC114 is L. gibba and that DWC130, DWC131, and DWC132 are L. minor. DNA barcoding results are given as the strain identification (genus and species, with ID in parentheses) and accession numbers for the first and second matches from BLAST [51] searches with the chloroplast atpH-atpF intergenic region from each strain.
| Strain | 1st Match | Accession | 2nd Match | Accession |
|---|---|---|---|---|
| DWC114 | Lemna gibba (5504) | KX212889.1 | Lemna gibba (7741) | KX212887.1 |
| DWC130 | Lemna minor (7210) | KX212888.1 | Lemna minor | DQ400350.1 |
| DWC131 | Lemna minor (7210) | KX212888.1 | Lemna minor | DQ400350.1 |
| DWC132 | Lemna japonica (0216) | KJ921747.1 | Lemna minor (7210) | KX212888.1 |
As preliminary screening for flowering rate, all 4 strains were grown in flasks of E with 0–100 μM SA in CL, and also in E with either 20 μM SA or 20 μM SA + 75 μM EDDHA in either LD or CL (Figure S2). Overall, this screen showed that DWC114 had the highest flowering rate of the 4 Lemna, that 24 h of light promoted flowering, that the optimum SA concentration for flowering was 20 μM, and that EDDHA had no obvious additive effect on flowering.
Under these conditions, L. gibba DWC114 produced perfect flowers, with a translucent pistil often topped with a drop of stigmatic fluid and two dehiscent anthers (Figure 7A). L. minor DWC130 (Figure 7B) produced small pistils observable at 16× magnification with no stigmatic fluid, but no anthers. L. minor strains DWC131 (Figure 7C) and DWC132 (Figure 7D) produced large pistils and large drops of stigmatic fluid, with two white but non-dehiscent anthers which were prone to detaching and sinking to the bottom of the flask.
Figure 7.
Flowering of L, gibba G3 and L. minor strains growing on E medium supplemented with 20 μM SA. (A) L. gibba DWC114 flower with the pistil having secreted a drop of clear stigmatic fluid and pollen on both dehiscent anthers. (B) L. minor DWC130 pistil with no stigmatic fluid (C) L. minor DWC131 pistil and mature non-dehiscent anther. (D) L. minor DWC132 pistil with a drop of stigmatic fluid. (E) Flowering of L. gibba G3 DWC114 on E supplemented with 20 μM SA is independent of LD or CL lighting conditions. Data are the average ± SEM (n = 3).
To determine if flowering of L. gibba G3, DWC114, was affected by growth in constant light, plants were cultured, in triplicate, in cotton stoppered flasks containing E supplemented with no SA or 20 μM SA, in LD or CL (Figure 7E). The results show that the presence of SA was required to produce flowering fronds until very late in the culture cycle. In the presence of SA, flowering under CL was significantly greater than in LD at day 14 (p < 0.01) but the appearance of flowers was essentially equal thereafter under either light condition until the end of the culture cycle.
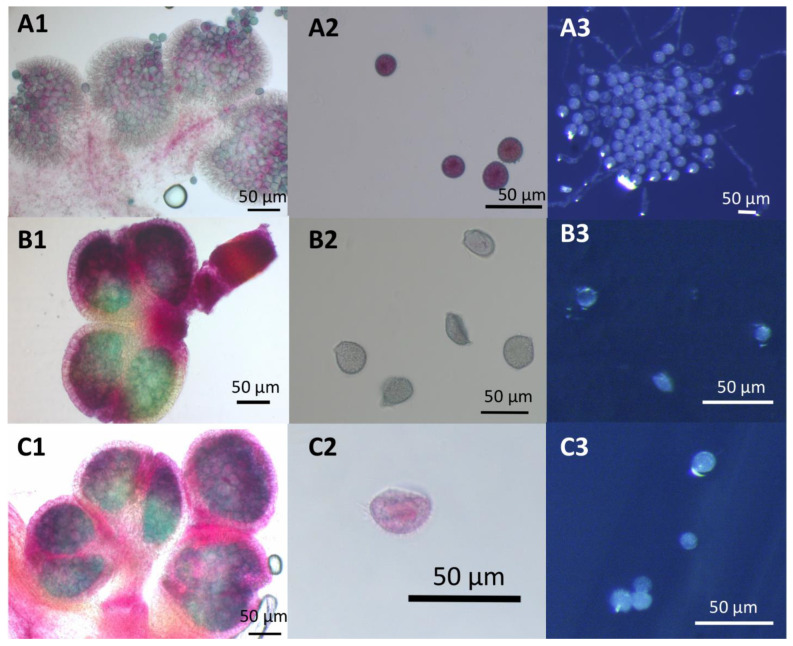
The production of viable pollen by DWC114, DWC131, and DWC132 was assessed with modified Alexander’s stain [52] (Figure 8A,B) and by pollen germination assays (Figure 8C). Pollen grains from dehisced anthers of L. gibba G3 DWC114 were largely viable as assessed by staining (83% viability) (Figure 8(A1,A2)), with 43% capable of forming a pollen tube (59 of 131) (Figure 8(A3)). The non-dehiscent anthers of L. minor strains DWC131 and DWC132 contained mostly non-viable pollen, with 1% (3 of 227), and 2% (11 of 454), respectively, of pollen grains staining as viable (Figure 8(B1,B2,C1,C2)). None of the pollen from these anthers produced germination tubes (Figure 8(B3,C3)).
Figure 8.
Pollen viability and germination assays for Lemna DWC114, DWC131, and DWC132 (A) L. gibba G3 DWC114 anthers and pollen (B) L. minor DWC131 anthers and pollen (C) L. minor DWC132 anthers and pollen (1 and 2) Anthers and pollen were stained with modified Alexander’s stain. (3) Pollen tube formation after 1 h of germination on E media.
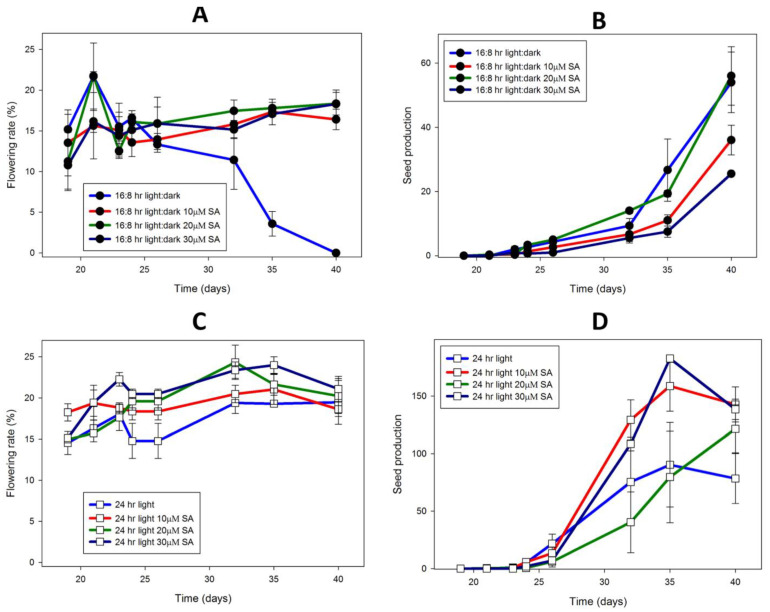
The effect of SA on seed production by L. gibba G3, DWC114 (Figure 9) was determined under two light regimes and at four concentrations of SA (10–30 μM SA) over a 40 day culture period. Although similar rates of flowering were observed in LD if SA was present, after 25 days in culture flowering percent dropped to zero by 40 days if SA was not present (Figure 9A). In contrast, the number of seed produced remained low until 25 day of culture, and increased thereafter until 40 days, independent of whether SA was present or not (Figure 9B). In LD, plants growing on E with 30 μM SA produced the fewest seed, an average of 25 ± 0.3 seed per flask. Under CL conditions, flowering remained high throughout the 40 day culture period with small increases attributable to the presence of SA (Figure 9C). In comparison to cultures growing in LD, seed production by plants in CL was substantially higher, with cultures containing 30 μM SA producing almost 150 seed per flask by day 35 (Figure 9D).
Figure 9.
L. gibba G3 DWC114 flowering is not affected by daylength but requires SA, whereas seed production is greatly increased under constant light, and is dependent on SA concentration. Flowering rate is described as the percent of flowering fronds per 100 fronds. Seed production was measured as the number of seed per flask. Flowering (A) and seed production (B) In LD with various concentrations of SA. Flowering (C) and seed production (D) under CL with various concentrations of SA. Data are the average ± SEM (n = 3).
Additional media supplements, such as yeast extract and bactopeptone (Eye) with 75 μM EDDHA, were tested for their reported ability to enhance seed production [16]. Addition of 20 μM SA slightly improved flowering (Figure S3A), however, seed production was decreased substantially with SA in the enriched medium (Figure S3B). In comparison, plants growing on Eye + 75 μM EDDHA produced fewer seed than plants growing on E supplemented with 30 μM SA.
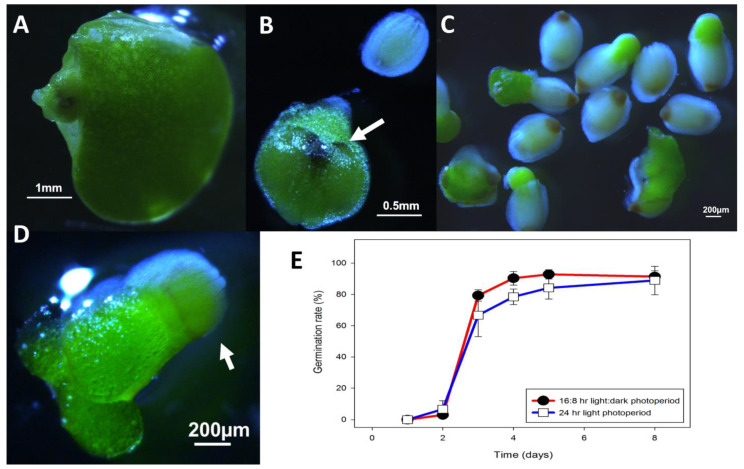
Developing seed of L. gibba G3 DWC114 were visible within the frond due to the highly pigmented operculum at one end (Figure 10A). Occasionally, developing seed within the ovary (Figure 10B) were seen detached from the mother frond, although most seed became apparent when they became free of the ovary and dropped to the bottom of the flask (Figure 10B). Seeds could then be harvested using a Pasteur pipette and stored dry, or in the cold in diluted E. Seeds germinated if transferred to fresh E (Figure 10C), achieving a germination rate by day 3 of 65% or higher, independent of LD or CL light conditions (Figure 10).
Figure 10.
L. gibba G3 DWC114 seed development and germination (A) Developing seed growing in the meristematic pouch, (B) Detached fruit with 2 developing seed. Note the two hyperpigmented spots, which are the opercula (white arrow). The operculum is visible on the free seed, which shows the typical ribbing on the seed coat. (C) Germinating seed showing emergence of the new green frond together with ungerminated seed at the bottom of the culture vessel. (D) A seedling floating on the surface of the medium still attached to the seed coat (white arrow), and already undergoing asexual budding. (E) Germination percent of seed over eight days in E in multi-well plates in LD or CL lighting conditions. Data are the average ± SEM (n = 3).
3. Discussion
The majority of the vast body of flowering research using members of the Lemnaceae was conducted before the sequencing of the human and Arabidopsis genomes [54,55,56] and before the first miRNA was discovered in Caenorhabditis [57] or miRNAs were found in Arabidopsis [58]. Today’s technology, which now allows for determination of the genetic mechanisms involved in flowering in these neotenic aquatic monocots, is still dependent on being able to have a convenient and reliable means of inducing flowering in the laboratory. While previous publications on flowering almost always focused on a single genus of the Lemnaceae, we developed protocols for obtaining flowering dependably in 3 different genera, and applied lessons between them, as a means of enabling the further study of the genetic mechanisms regulating floral induction and progressing toward breeding protocols in this plant family.
The first step in developing reliable flowering protocols was the standardization of the number and the birth order of the inoculating fronds. In Lemna, first daughter fronds are more robust that those produced subsequently [59,60,61]. Birth order also influences production of turions by S. polyrhiza [62]. We consistently used 4-frond colonies of Lemna and 3–5 frond colonies of Spirodela as inoculant to help standardize growth and flowering rate. In this way, a colony would have a grandmother frond, with an attached mother frond, and a developing daughter frond, which would be the first daughter. Depending on environmental conditions, a first daughter frond of Lemna gibba is capable of producing 12–14 daughters of its own before senescing, although the production of a flower significantly reduces the number of possible daughter fronds [63].
It is, therefore, worth considering that while the number of daughter fronds produced by a particular meristem before it can produce a flower is likely to be genetically determined, flowering may also be influenced epigenetically through the frond’s mother. A first daughter will already have its first daughter developing in the meristematic pouch, with at least one generation of internally developing fronds in L. minor and S. polyrhiza [64], and W. microscopica containing up to three unseen internal generations of daughter fronds nested within each other [10]. Evolutionarily conserved epigenomic stress responses such as DNA methylation and histone modification that influence flowering time in rice, sugar beets, Arabidopsis, and other plants [65,66], are likely to be active through these internal generations and perhaps future generations, and may help explain the long term effects of previous culture conditions on flowering rates. Comparing the results in early Wolffia screens (Table S4) to experiments where inoculants came from fully acclimated cultures (Figure 5) underscores the importance of acclimating cultures for 4 weeks to fully eliminate variation before running experiments, as suggested in Duckweed Forum issue 8 [67].
3.1. Spirodela Polyrhiza
We focused on flowering of S. polyrhiza strains 7498 and 9509 because of the number of recent genomic and transcriptomic studies on them [11,29,30,34,35,37,38,68]. An abundance of molecular evidence illustrates the neotenous nature of Spirodela that must be overcome to induce flowering. Genome sequencing revealed a strong enrichment of genes for floral repressors, and a reduction in the number of members of the floral promoter gene families in both Spirodela strains compared to Arabidopsis [29,30]. About 9 members of the juvenile marker miR156 gene family were found, compared to 5 members of the adult marker miR172 gene family in the 7498 and 9509 genomes [29,30,68]. Sequencing of small RNAs in strain 7498 growing in 8 different stress conditions or hormonal applications, found that the ratio of miR156 to miR172 ranged from 71–408 [34]. These two highly conserved microRNAs regulate the abundance of the key floral promoters which integrate flowering responses to day length, stress, and accumulated sugar states [43,69,70,71].
Most strains of S. polyrhiza were classified as day neutral plants [1,72]. We found flowering in 12 h days, (Figure S1), although growth in CL produced the highest flowering rates (Figure 1). If inoculant was taken from 4-week-old cultures growing in CL in flasks containing 100 mL of Hg media, and used to inoculate Petri dishes containing 50 mL of Hg + 1.5 μM SA, we obtained optimal flowering of 11% for 7498 and 1% for 9509 (Figure 1). While both 7498 and 9509 produced pistils with stigmatic fluid and pollen capable of producing germination tubes (Figure 2 and Figure 4), gentle shaking, which was found to promote seed production in L. gibba G3 [16] failed to produce seed in S. polyrhiza. Our attempts to manually self-pollinate 7498 by touching dehiscent anthers to pistils, as done with L. gibba by Fu et al. [48] were also unsuccessful.
Diverse types of stress can influence flowering in plants through SA signaling [73,74]. In Spirodela, a more common stress response than flowering and seed formation may be dormancy through the production of the starch-rich turions. In our optimal conditions, a 4 weeks old culture could have up to 39 mature and developing flowers, while at the same time having an average of 240 turions for strain 7498 and 134 turions for strain 9509. After two months, up to 500 turions could be found per flask of strain 7498, suggesting that genetic or environmental manipulation of turion production could dramatically influence the production of flowers, and vice versa.
3.2. Wolffia Microscopica
W. microscopica flowers abundantly in the wild. These diminutive plants were used for many early studies on induction of flowering [75,76], and the morphology and anatomy of seed obtained from the wild have been studied [47]. In addition to being a promising crop plant which provides a complete protein [4], Wolffia is a prime candidate for molecular studies into how a tiny monocot manages to maintain both a vegetative and sexual reproductive cycle. The consistent laboratory protocol for maximum flowering induction we describe supports efforts to maximize research on, and use of, these plants.
Our initial screen validated the reported abundant flowering of these plants, although there was a high degree of variability of replicates within a trial, and variability between replicated experiments (Figure S4). As with Spirodela and Lemna, variability in flowering was decreased when inoculants came from acclimated cultures (Figure 5). Culture inoculant size and age, EDDHA, and hormones like SA, are known to play roles in regulation of flowering of the Lemnaceae. We found that under our conditions, EDDHA had no effect on flowering rate (Table S4), unlike what was reported earlier for a different strain of W. microscopica [77], or S. polyrhiza SP20 [78] and L. gibba G3 [79]. In addition to its chelating effects, EDDHA breaks down in a matter of weeks to form SA and SA-like molecules in sunlight [80], yet this transition from EDDHA to SA may not have occurred in our experiments where EDDHA was added to the medium after autoclaving.
The differences we observed in flowering when plants were grown under the same conditions but in different culture vessels (Table S4) suggests that a physical factor, such as rate of evaporation or accumulation of volatiles, surface to volume ratio, or even quorum sensing influences flowering in Wolffia and perhaps other duckweed genera. To test whether evaporation, or exchange of gases such as ethylene may be responsible, we tested flowering rate in Petri dishes and 6-well plates with and without parafilm. We found no significant difference due to parafilm, suggesting that surface to volume ratio or crowding may play a large role in flowering (Figure 5), although the mechanism for increased flower induction remains ambiguous, it may be SA independent.
3.3. Lemna
While gene editing and transformation present avenues for production of new duckweed varieties for basic and applied research, L. gibba G3 (strain DWC114) is currently the ideal strain for attempts to breed new varieties through sexual reproduction. This strain flowers readily, produces abundant dehiscent anthers, and self-pollinates to readily produce seed [16,48], (Figure 7 and Figure 10). Two different protocols to produce flowers and seed in L. gibba G3 have been described [16,48]. In the first, L. gibba G3 was grown on liquid E media in flasks in CL to produce flowers. Gentle shaking on a rotary shaker for 1 h twice a day was required to produce seed [16]. In the second protocol, the plants are grown in Petri dishes containing semi-solidified Modified Hoagland’s media supplemented with 20 μM SA in LD [48]. Flowers were either self or cross-pollinated manually under a stereoscope to produce seed [48]. It is possible that flowering under CL is influenced by a decrease in Circadian Clock Associated 1, which oscillates every 24 h under LD or SD, but is transcriptionally silenced when L. gibba G3 is grown under CL for 48 h or more [81]. Our best seed production protocol incorporated aspects of both previous protocols to both strongly induce flowering, and easily pollinate flowers with a gentle stir twice per week.
The need for supplementary SA to induce flowering if plants are grown in LD as found by Fu et al. [48] as compared to CL without SA [16] may also be explained if CL is acting as an abiotic stressor and thus increases endogenous levels of SA. Many plants respond to abiotic and biotic stress by the production of SA [73]. Intriguingly, L. gibba grown under CL exhibited increased auxin turnover [82]. Additionally, tight control of auxin levels is mediated through conjugation by GH3 acyl acid amido synthases, one of which, atGH3.5, has been shown to have a role in both auxin and SA homeostasis [83]. SA interacts with the flowering developmental pathway through Flowering Locus D. Flowering Locus D demethylates the histones associated with Flowering Locus C and thus inhibits transcription of floral promoters [84,85,86,87,88].
Fortunately, for a mechanistic understanding of floral regulation, we are no longer reliant on comparisons to other plant species, but can now analyze the transcriptome of flowering L. gibba G3 recently published by Fu et al., 2020 [89]. This study revealed differential expression of 1501 genes throughout the flowering process, and highlighted the role of photoperiod and SA. In LD L. gibba had very low levels of transcripts of two genes, LgCO and LgGI, which in Arabidopsis are responsible for integrating circadian rhythms. Fu et al. [89] found a lack of endogenous SA production, suggesting that flowering is being activated through exogenous SA from the medium. Surprisingly, this transcriptome suggested a flowering pathway distinctly different from other plant models, with a lack of expression of the floral regulators LgCO, LgGI, LgSOC1, and LgFD, and a co-expression of FT with LgTEM1 and LgSVP, which act as floral inhibitors in Arabidopsis [89]. Clearly there is a need for flowering transcriptome data from Spirodela, Wolffia, and the improperly developing Lemna minor strains, with which to build a larger picture of floral regulation strategies across this family.
Under optimal conditions for floral induction, L gibba DWC114 produced pistils with stigmatic fluid and both dehiscent and apparently non-dehiscent anthers, L. minor DWC130 produced pistils but no stigmatic fluid, L. minor DWC131 and L. minor DWC132 pistils produced stigmatic fluid but anthers remained non-dehiscent (Figure 8). Under our conditions, none of the L. minor strains produce seed, whereas L. gibba readily does. Over 70 attempts to manually fertilize L. minor DWC131 and DWC132 with L. gibba pollen were unsuccessful, indicating that the two species may be too diverged to form an interspecific hybrid (data not shown). However, the marked differences in floral development and fertility of these genetically related strains of L. minor and L. gibba provides an opportunity to identify critical genes in floral development.
L. gibba seed can be left in a Petri dish to dry overnight, wrapped in parafilm, and stored dry at 4 °C for long-term storage. Seed can also be stored in diluted E medium, although light must be blocked to prevent germination. Of 234 seed stored in diluted E medium for 15 months, 36% germinated when placed in fresh E medium in the light. L. gibba seeds collected in the wild germinated at 70% efficiency after being stored in water for 2 years at room temperature, but germination dropped to 1% after 3 years of wet storage [90]. Only 5.6% of seed germinated after dry storage [90]. To eliminate possible fungal or bacterial contamination, seeds were surface sterilized with 5% bleach for 3 min or 10 min, then washed 3 times with sterile water, or more conveniently, rinsed with 70% ethanol and air dried. Ninety seven percent of one month old seed treated with any of the three methods germinated with no contamination. Single seed germinated within 4 days in as little as 5 mL of E medium in a 6-well plate, making it easy to rapidly obtain progeny from a cross.
Similar to many plant species that rely on protogyny (the sequential development of the pistil followed by the anther) to promote cross-fertilization while ensuring fruit production [91], like most family members the three genera of Lemnaceae we studied are protogynous [3,92,93,94]. Due to the clonal nature of Lemnaceae populations, protogyny would be substantially less effective at promoting cross-pollination and reducing deleterious in breeding than other self-incompatibility (SI) strategies [3]. Mechanisms involving the determinants at the S locus are diverse, and have evolved at least 35 times in the angiosperms, yet very little is known about them in monocots [95], and certain monocot orchids appear to have a novel SI mechanism [96]. Observations of flowering and seed production in the wild led to the suggestion that 22 species of duckweed are likely self-compatible [3]. Historical reports stated that W. welwitschii was able to self-pollinate [97], and that two L. minor populations were self-incompatible but capable of cross fertilization [98]. This work is summarized in Landolt and Kandeler (1987) [1]. Although Maheshwari (1956) [47] reported seed production by W. microscopica in the wild, it is not clear whether the wild populations were clonal or heterogeneous. We were unable to obtain seed from our W. microscopica and S. polyrhiza cultures even though pollen was capable of forming germination tubes, suggesting self-incompatibility in these species. It is therefore likely that SI is variable across the family, and co-cultivation of genotypes within a species may be required to develop breeding protocols. It will be interesting to investigate the relationship of SI mechanisms to genetic diversity in native habitats and the ability of a species to produce seed or develop turions. Both of these organs are capable of becoming dormant and surviving desiccation, and thus their formation might represent alternative stress-response strategies.
4. Summary
The Lemnaceae can be seen as new model plants ideally suited to probing important questions in plant biology, and for developing a new crop that can be a part of sustainably providing humanity with clean water, food, and fuel. To make future floral and breeding experiments easier we developed culturing protocols for obtaining flowering plants in the laboratory.
In CL or LD, L. gibba G3 flowers and sets seed in E medium supplemented with 20 μM SA. Occasional movement of the culture increases seed production.
W. microscopica flowers conveniently under LD in E medium in 6-well plates.
S. polyrhiza flowers in Petri dishes with 50 mL of Hg under CL when 1.5 μM SA is added.
Neither Wolffia or Spirodela produced seeds under these conditions, suggesting that self-incompatibility may be at play, and requiring further testing. With these culture conditions and modern molecular tools, it is now possible to determine the genetic mechanisms leading to flowering in W. microscopica, and determine if flowering in S. polyrhiza can reveal new insights in transposon mobility and DNA methylation. The Lemnaceae family is both exciting in its own right and an excellent basal monocot model with which to study the genetic mechanisms of floral regulation, or the mechanisms behind self-incompatibility in clonal populations.
5. Materials and Methods
5.1. Plant Material and Culture Media
All strains of S. polyrhiza, L. gibba, L. minor and W. microscopica described are available from the RDSC (http://www.ruduckweed.org/ accessed on 26 January 2021). W. microscopica 2005 was kindly provided by Professor K.J. Appenroth, Friedrich Schiller University, Jena, Germany. DWC114, DWC130, DWC131, and DWC132 were barcoded at the chloroplast ATPase subunit I gene atpF-atpH intergenic spacer, according to the PCR protocol described in [28]. DWC132 was also barcoded using matK primers [28].
The media and culture vessels used for this study are described in Table 2 and Table 3. The chemical compositions of each of the media tested are provided in Supplementary Table S1, while the instructions for making stock solutions for, and preparing, E medium are in Document S1. All media were supplemented with 29 mM sucrose unless specified otherwise with a minus (−) sign after the medium abbreviation. Additions of yeast extract and bactopeptone to E (Eye media) were made before autoclaving. SA was prepared as a 100 mM stock solution in ethanol. EDDHA was prepared as a 100 mM stock solution in water, and filter sterilized. SA and EDDHA were added to cooled media at the concentrations indicated. Flasks were fitted with cotton stoppers and loose aluminum foil covers. Petri dishes and 6-well plates were wrapped with strips of parafilm to maintain sterility and reduce evaporation.
Table 2.
Media.
| Media | Abbreviation | Reference | pH | Supplier |
|---|---|---|---|---|
| E a | E | [60,99] | 4.6 | Chemicals from Sigma-Aldrich, St. Louis, MO, USA |
| Hoagland’s a | Hg | [100] | 5.8 | Cassion Labs, Smithfield, UT, USA |
| Shenk Hildebrandt a | SH | [101] | 5.8 | Sigma-Aldrich |
a The chemical composition of each of these media along with the other common medium, Steinberg [102], can be found in Table S1. All media were adjusted for pH with KOH, supplemented with 29 mM sucrose, and autoclaved. All flasks had cotton stoppers with loose aluminum foil covers unless stated otherwise.
Table 3.
Containers.
| Container | Container Size | Volume of Media | Supplier |
|---|---|---|---|
| Flask | 250 mL | 100 mL | VWR, Bridgeport, PA, USA |
| Small flask | 125 mL | 50 mL | VWR |
| Petri dish | 100 × 15 mm | 50 mL | Kord-Valmark, Bridgeport PA, USA |
| Multi-well Plate | 6 well | 10 mL | Corning, NY, USA |
All plants were grown at 24 °C, under General Electric Daylight 6500 K fluorescent bulbs or an LED panel. Plants in 16:8 h light:dark photoperiod (LD) were under a PPFD (Photosynthetic Photon Flux Density, μmol photons (400–700 nm)/m2/s) of 71–141, and a red (655–665 nm):far-red (725–735 nm) ratio of (2.54), while plants in continuous light (CL) or 12 h light:12 h dark days received a lower PPFD of 21–56, and a red:far-red ratio of (2.36). Supplemental far-red light experiments used custom red and blue LED panels providing PPFD of 362–516 and a red:far-red ratio of 1.69–1.84.
New cultures of W. microscopica were inoculated with a single 1.5 × 1.5 cm mesh loopful of floating fronds from a 3–5 week old mature culture growing in a 250 mL Erlenmeyer flask containing 100 mL E medium. Spirodela experiments were started with three 3–5 frond colonies from a 1, 2, or 4 week old culture grown in a flask of Hg medium. For L. gibba and L. minor, all experiments were started with 3 four-frond colonies from a 7 or 14 day old population growing in E medium in flasks.
5.2. Flowering, Seed Production and Seed Storage
Flowers were counted if they were developing or mature. An observable pistil, anthers, or a perfect flower (pistil with 2 anthers) were counted as one flower. Flowering rate is expressed as the percent of developing and mature flowers per 100 fronds. The number of fronds scored was either every frond in the culture vessel, or ≥100 fronds from any one culture. Flower or seed production data are presented as the average ± SEM (standard error of the mean, (n ≥ 3). Statistically significant difference between variables were calculated by Student’s t-test. Seed production is expressed as the total number per culture vessel. Seeds were stored dry, or in a flask of depleted media diluted 1:2 with sterile water at 4 °C. Unless otherwise specified, flowers were counted on days 7, 10, 14, 17, 21, 24, 28 after subculture.
5.3. Pollen Viability and Fertility
Pollen from all strains that produced anthers was tested for viability with a modified Alexander’s stain [52] and for the ability to form pollen tubes by gently spreading anthers on E solidified with agar. For staining, pollen from 5–10 anthers was stained, and at least 169 grains per anther were scored as viable or dead. Pollen tube formation was observed at 20× using a Leica DM550B microscope 1 h later. Fertility is described as the percent of pollen grains with germination tubes. At least 97 grains from 4–7 anthers were observed.
Acknowledgments
P.F. and J.S. dedicate this manuscript to Joachim Messing, who conceived and guided the study “to investigate the neotenous nature of the Spirodela genome”, yet passed away before its completion. P.F. thanks K. Friedman for helping conduct some of the experiments and J. Cohen for assistance with plotting data. P.F. thanks S. Xu, S. Sree, and K. Appenroth for their insightful discussions during the ICDRA 2019 conference.
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/22/5/2733/s1, Figure S1: Flowering rates of Spirodela polyrhiza in LD and CL. Figure S2: Screen for the effects of 0–100 µM SA and 75 µM EDDHA in E on flowering rate of L. gibba G3 and 2 strains of L. minor in LD or CL. Figure S3: Flowering rate and seed production in strain DWC114. Table S1: Media compositions. Table S2: Summary of Spirodela screens and experiments. Table S3: Screening for optimal flowering of W. microscopica 2005. Table S4: Summary of W. microscopica flowering rates in different containers and media, with and without EDDHA. Document S1: E medium protocol.
Author Contributions
Conceived, guided, and supervised the research, J.M.; designed and conducted experiments, P.F.; contributed expertise to the project, J.S.; wrote and edited the manuscript, P.F. and J.S. All authors have read and agreed to the published version of the manuscript.
Funding
Was provided by The Selman Waksman Chair in Molecular Genetics to P.F., and by USDA CRIS 8042-21220-254-00D.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data presented in article and Supplementals.
Conflicts of Interest
The authors report no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Landolt E., Kandeler R. Biosystematic Investigations in the Family of Duckweeds (Lemnaceae) ETH Zurich; Zurich, Switzerland: 1987. The Family of Lemnaceae—Monographic Study Volume 2. [DOI] [Google Scholar]
- 2.Ziegler P., Adelmann K., Zimmer S., Schmidt C., Appenroth K.J. Relative in vitro growth rates of duckweeds (Lemnaceae)—The most rapidly growing higher plants. Plant Biol. 2015;17:33–41. doi: 10.1111/plb.12184. [DOI] [PubMed] [Google Scholar]
- 3.Landolt E. Biosystematic Investigations in the Family of Duckweeds (Lemnaceae) ETH Zurich; Zurich, Switzerland: 1986. The Family of Lemnaceae—Monographic Study Volume 1. [DOI] [Google Scholar]
- 4.Appenroth K.-J., Sree K.S., Bog M., Ecker J., Seeliger C., Böhm V., Lorkowski S., Sommer K., Vetter W., Tolzin-Banasch K., et al. Nutritional Value of the Duckweed Species of the Genus Wolffia (Lemnaceae) as Human Food. Front. Chem. 2018;6:483. doi: 10.3389/fchem.2018.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Appenroth K.-J., Sree K.S., Böhm V., Hammann S., Vetter W., Leiterer M., Jahreis G. Nutritional value of duckweeds (Lemnaceae) as human food. Food Chem. 2017;217:266–273. doi: 10.1016/j.foodchem.2016.08.116. [DOI] [PubMed] [Google Scholar]
- 6.Cao H.X., Fourounjian P., Wang W. Handbook of Environmental Materials Management. Springer International Publishing; Cham, Switzerland: 2018. The Importance and Potential of Duckweeds as a Model and Crop Plant for Biomass-Based Applications and Beyond; pp. 1–16. [Google Scholar]
- 7.Fourounjian P., Fakhoorian T., Cao X.H. Compendium of Plant Genomes. Springer International Publishing; Berlin/Heidelberg, Germany: 2020. Importance of Duckweeds in Basic Research and Their Industrial Applications; pp. 1–17. [Google Scholar]
- 8.Skillicorn P., Spira W., Journey W. Duckweed Aquaculture: A New Aquatic Farming System for Developing Countries. World Bank; Washington, DC, USA: 1993. [Google Scholar]
- 9.Food and Agriculture Organization . FAOP Statistics. Food and Agriculture Organization; Quebec City, QC, Canada: 2021. [Google Scholar]
- 10.Sree K.S., Maheshwari S.C., Bóka K., Khurana J.P., Keresztes Á., Appenroth K.-J. The duckweed Wolffia microscopica: A unique aquatic monocot. Flora Morphol. Distrib. Funct. Ecol. Plants. 2015;210:31–39. doi: 10.1016/j.flora.2014.10.006. [DOI] [Google Scholar]
- 11.Wang W., Wu Y., Messing J. RNA-Seq transcriptome analysis of Spirodela dormancy without reproduction. BMC Genom. 2014;15:60. doi: 10.1186/1471-2164-15-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka O., Cleland C.F. Influence of Ammonium on the Ability of Salicylic Acid to Induce Flowering in the Short-Day Plant Lemna paucicostata 6746. Plant Cell Physiol. 1981;22:597–602. doi: 10.1093/oxfordjournals.pcp.a076202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Micheli P.A. Nova Plantarum Genera Iuxta Tournefortii Methodum Disposita Quibus Plantae Recensentur, Scilicet Fere Nondum Observatae, Reliquae Suis Sedibus, Florentiae. Typis Bernardi Paperini; Florentiae, Italy: 1729. [Google Scholar]
- 14.Hicks L. Flower Production in the Lemnaceae. Ohio State University; Columbus, OH, USA: 1932. [Google Scholar]
- 15.Baldi B.G., Maher B.R., Slovin J.P., Cohen J.D. Stable Isotope Labeling, in vivo, of d-and l-Tryptophan Pools in Lemna gibba and the Low Incorporation of Label into Indole-3-Acetic Acid. Plant Physiol. 1991;95:1203–1208. doi: 10.1104/pp.95.4.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slovin J.P., Cohen J.D. Levels of Indole-3-Acetic Acid in Lemna gibba G-3 and in a Large Lemna Mutant Regenerated from Tissue Culture. Plant Physiol. 1988;86:522–526. doi: 10.1104/pp.86.2.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giovanelli J., Veluthambi K., Thompson G.A., Mudd S.H., Datko A.H., Jameel S., Reddy V.M., Rhodes W.G., McFadden B.A. Threonine Synthase of Lemna paucicostata Hegelm. 6746. Plant Physiol. 1984;76:285–292. doi: 10.1104/pp.76.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mattoo A.K., Hoffman-Falk H., Marder J.B., Edelman M. Regulation of protein metabolism: Coupling of photosynthetic electron transport to in vivo degradation of the rapidly metabolized 32-kilodalton protein of the chloroplast membranes. Proc. Natl. Acad. Sci. USA. 1984;81:1380–1384. doi: 10.1073/pnas.81.5.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slovin J.P., Tobin E.M. Synthesis and turnover of the light-harvesting chlorophylla/b-protein in Lemna gibba grown with intermittent red light: Possible translational control. Planta. 1982;154:465–472. doi: 10.1007/BF01267815. [DOI] [PubMed] [Google Scholar]
- 20.Zhao H., Appenroth K., Landesman L., Salmeán A.A., Lam E. Duckweed rising at Chengdu: Summary of the 1st International Conference on Duckweed Application and Research. Plant Mol. Biol. 2012;78:627–632. doi: 10.1007/s11103-012-9889-y. [DOI] [PubMed] [Google Scholar]
- 21.Hillman W.S. The Lemnaceae, or duckweeds. Bot. Rev. 1961;27:221–287. doi: 10.1007/BF02860083. [DOI] [Google Scholar]
- 22.Oláh V., Hepp A., Vaca N.Y.G., Tamás M., Mészáros I. Retrospective analyses of archive phytotoxicity test data can help in assessing internal dynamics and stability of growth in laboratory duckweed cultures. Aquat. Toxicol. 2018;201:40–46. doi: 10.1016/j.aquatox.2018.05.022. [DOI] [PubMed] [Google Scholar]
- 23.OECD . OECD Guidelines for the Testing of Chemicals, Revised Proposal for a New Guideline 221, Lemna sp. Growth Inhibition Test. OECD; Paris, France: 2006. [Google Scholar]
- 24.ISO Water quality determination of the toxic effect of water constituents and waste water on duckweed (Lemna minor)—Duckweed growth inhibition test. ISO. 2005;3:23. [Google Scholar]
- 25.Mazur R., Szoszkiewicz K., Lewicki P., Bedla D. The use of computer image analysis in a Lemna minor L. bioassay. Hydrobiologia. 2018;812:193–201. doi: 10.1007/s10750-016-2972-7. [DOI] [Google Scholar]
- 26.Haffner O., Kučera E., Drahoš P., Cigánek J., Kozáková A., Urminská B. Lemna minor Bioassay Evaluation Using Computer Image Analysis. Water. 2020;12:2207. doi: 10.3390/w12082207. [DOI] [Google Scholar]
- 27.Sree K.S., Appenroth K.-J. Compendium of Plant Genomes. Springer International Publishing; Berlin/Heidelberg, Germany: 2020. Worldwide Genetic Resources of Duckweed: Stock Collections; pp. 39–46. [Google Scholar]
- 28.Wang W., Wu Y., Yan Y., Ermakova M., Kerstetter R., Messing J. DNA barcoding of the Lemnaceae, a family of aquatic monocots. BMC Plant Biol. 2010;10:205. doi: 10.1186/1471-2229-10-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang W., Haberer G., Gundlach H., Gläßer C., Nussbaumer T., Luo M., Lomsadze A., Borodovsky M., Kerstetter R., Shanklin J.D., et al. The Spirodela polyrhiza genome reveals insights into its neotenous reduction fast growth and aquatic lifestyle. Nat. Commun. 2014;5:3311. doi: 10.1038/ncomms4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michael T.P., Bryant D., Gutierrez R., Borisjuk N., Chu P., Zhang H., Xia J., Zhou J., Peng H., El Baidouri M., et al. Comprehensive definition of genome features in Spirodela polyrhiza by high-depth physical mapping and short-read DNA sequencing strategies. Plant J. 2017;89:617–635. doi: 10.1111/tpj.13400. [DOI] [PubMed] [Google Scholar]
- 31.Van Hoeck A., Horemans N., Monsieurs P., Cao H.X., Vandenhove H., Blust R. The first draft genome of the aquatic model plant Lemna minor opens the route for future stress physiology research and biotechnological applications. Biotechnol. Biofuels. 2015;8:1–13. doi: 10.1186/s13068-015-0381-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W., Messing J. Status of duckweed genomics and transcriptomics. Plant Biol. 2014;17:10–15. doi: 10.1111/plb.12201. [DOI] [PubMed] [Google Scholar]
- 33.Fang Y., Du A., Tan L., He K., Jin Y., Ding Y., Guo L., Zhao H. The Transcriptome in Landoltia punctata. In: Cao X.H., Fourounjian P., Wang W., editors. The Duckweed Genomes. Springer International Publishing; Berlin/Heidelberg, Germany: 2020. pp. 125–131. [Google Scholar]
- 34.Fourounjian P., Tang J., Tanyolac B., Feng Y., Gelfand B., Kakrana A., Tu M., Wakim C., Meyers B.C., Ma J., et al. Post-transcriptional adaptation of the aquatic plant Spirodela polyrhiza under stress and hormonal stimuli. Plant J. 2019;98 doi: 10.1111/tpj.14294. [DOI] [PubMed] [Google Scholar]
- 35.Harkess A., McLoughlin F., Bilkey N., Elliott K., Emenecker R., Mattoon E., Miller K., Czymmek K., Vierstra R., Meyers B.C., et al. A new Spirodela polyrhiza genome and proteome reveal a conserved chromosomal structure with high abundances of proteins favoring energy production. bioRxiv. 2020 doi: 10.1101/2020.01.23.909457. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y., Wang X., Yang R., Niu L., Wang W. Comparison of protein extraction methods for 2DE-based proteomic analysis of duckweed Spirodela polyrhiza, a small aquatic model plant. Aquat. Bot. 2020;163:103216. doi: 10.1016/j.aquabot.2020.103216. [DOI] [Google Scholar]
- 37.Hoang P.N., Michael T.P., Gilbert S., Chu P., Motley S.T., Appenroth K.J., Schubert I., Lam E. Generating a high-confidence reference genome map of the Greater Duckweed by integration of cytogenomic, optical mapping, and Oxford Nanopore technologies. Plant J. 2018;96:670–684. doi: 10.1111/tpj.14049. [DOI] [PubMed] [Google Scholar]
- 38.An D., Zhou Y., Li C., Xiao Q., Wang T., Zhang Y., Wu Y., Li Y., Chao D.-Y., Messing J., et al. Plant evolution and environmental adaptation unveiled by long-read whole-genome sequencing of Spirodela. Proc. Natl. Acad. Sci. USA. 2019;116:18893–18899. doi: 10.1073/pnas.1910401116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fourounjian P. Repetitive Sequences: Impacts and Uses in the Spirodela Genome. In: Cao X.H., Fourounjian P., Wang W., editors. The Duckweed Genomes. Springer International Publishing; Berlin/Heidelberg, Germany: 2020. pp. 87–90. [Google Scholar]
- 40.Yang J., Hu S., Li G., Khan S., Kumar S., Yao L., Duan P., Hou H. Transformation Development in Duckweeds. In: Cao X.H., Fourounjian P., Wang W., editors. The Duckweed Genomes. Springer; Berlin/Heidelberg, Germany: 2020. pp. 143–155. [Google Scholar]
- 41.Ishizawa H., Kuroda M., Morikawa M., Ike M. Evaluation of environmental bacterial communities as a factor affecting the growth of duckweed Lemna minor. Biotechnol. Biofuels. 2017;10:62. doi: 10.1186/s13068-017-0746-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishizawa H., Kuroda M., Inoue D., Morikawa M., Ike M. Community dynamics of duckweed-associated bacteria upon inoculation of plant growth-promoting bacteria. FEMS Microbiol. Ecol. 2020;96:101. doi: 10.1093/femsec/fiaa101. [DOI] [PubMed] [Google Scholar]
- 43.Spanudakis E., Jackson S. The role of microRNAs in the control of flowering time. J. Exp. Bot. 2014;65:365–380. doi: 10.1093/jxb/ert453. [DOI] [PubMed] [Google Scholar]
- 44.Van Dijk A.D.J., Molenaar J. Floral pathway integrator gene expression mediates gradual transmission of environmental and endogenous cues to flowering time. PeerJ. 2017;2017:1–18. doi: 10.7717/peerj.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chaïlakhyan M.K. Hormonal Theory of Plant Development. Akademii Nauk SSSR (Academy of Sciences of the USSR); Moscow, Russia: 1937. [Google Scholar]
- 46.Pieterse A.H. Is flowering in Lemnaceae stress-induced? A review. Aquat. Bot. 2013;104:1–4. doi: 10.1016/j.aquabot.2012.08.002. [DOI] [Google Scholar]
- 47.Maheshwari S.C. Endosperm and Seed of Wolffia. Nat. Cell Biol. 1956;178:925–926. doi: 10.1038/178925b0. [DOI] [Google Scholar]
- 48.Fu L., Huang M., Han B., Sun X., Sree K.S., Appenroth K.-J., Zhang J. Flower induction, microscope-aided cross-pollination, and seed production in the duckweed Lemna gibba with discovery of a male-sterile clone. Sci. Rep. 2017;7:3047. doi: 10.1038/s41598-017-03240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khurana J.P., Maheshwari S.C. Some effects of salicylic acid on growth and flowering in Spirodela polyrrhiza SP20. Plant Cell Physiol. 1980;21:923–927. doi: 10.1093/oxfordjournals.pcp.a076066. [DOI] [Google Scholar]
- 50.Lacor M.A.M. Flowering of Spirodela Polyrhiza (L.) Schleiden. Acta Bot. Neerl. 1968;17:357–359. doi: 10.1111/j.1438-8677.1968.tb00139.x. [DOI] [Google Scholar]
- 51.Halliday K.J., Koornneef M., Whitelam G.C. Phytochrome B and at Least One Other Phytochrome Mediate the Accelerated Flowering Response of Arabidopsis thaliana L. to Low Red/Far-Red Ratio. Plant Physiol. 1994;104:1311–1315. doi: 10.1104/pp.104.4.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peterson R., Slovin J.P., Chen C. A simplified method for differential staining of aborted and non-aborted pollen grains. Int. J. Plant Biol. 2010;1:13. doi: 10.4081/pb.2010.e13. [DOI] [Google Scholar]
- 53.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 54.Kaul S., Koo H.L., Jenkins J., Rizzo M., Rooney T., Tallon L.J., Feldblyum T., Nierman W., Benito M.I., Lin X., et al. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 55.Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 56.Consortium I.H.G.S. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 57.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 58.Reinhart B.J., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barks P.M., Laird R.A. Senescence in duckweed: Age-related declines in survival, reproduction and offspring quality. Funct. Ecol. 2015;29:540–548. doi: 10.1111/1365-2435.12359. [DOI] [Google Scholar]
- 60.Barks P.M., Laird R.A. A multigenerational effect of parental age on offspring size but not fitness in common duckweed (Lemna minor) J. Evol. Biol. 2016;29:748–756. doi: 10.1111/jeb.12823. [DOI] [PubMed] [Google Scholar]
- 61.Mejbel H.S., Simons A.M. Aberrant clones: Birth order generates life history diversity in Greater Duckweed, Spirodela polyrhiza. Ecol. Evol. 2018;8:2021–2031. doi: 10.1002/ece3.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cleland C.F., Briggs W.R. Flowering responses of the long-day plant Lemna gibba G3. Plant Physiol. 1967;42:1553–1561. doi: 10.1104/pp.42.11.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lemon G.D., Posluszny U. Comparative Shoot Development and Evolution in the Lemnaceae. Int. J. Plant Sci. 2000;161:733–748. doi: 10.1086/314298. [DOI] [Google Scholar]
- 64.Basu S. Rice Research for Quality Improvement: Genomics and Genetic Engineering. Springer International Publishing; Berlin/Heidelberg, Germany: 2020. An Insight into the Factors Regulating Flowering in Rice: From Genetics to Epigenetics; pp. 233–247. [Google Scholar]
- 65.Yaish M.W., Colasanti J., Rothstein S.J. The role of epigenetic processes in controlling flowering time in plants exposed to stress. J. Exp. Bot. 2011;62:3727–3735. doi: 10.1093/jxb/err177. [DOI] [PubMed] [Google Scholar]
- 66.Hébrard C., Peterson D.G., Willems G., Delaunay A., Jesson B., Lefèbvre M., Barnes S., Maury S. Epigenomics and bolting tolerance in sugar beet genotypes. J. Exp. Bot. 2016;67:207–225. doi: 10.1093/jxb/erv449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Appenroth K.-J. Useful Methods 1. Determination of Growth Rates in Duckweed. Int. Conf. Duckweed Res. Appl. 2015;3:34–80. [Google Scholar]
- 68.Fourounjian P. Small RNAs in Duckweeds. In: Cao X.H., Fourounjian P., Wang W., editors. The Duckweed Genomes. Springer; Berlin/Heidelberg, Germany: 2020. pp. 157–164. [Google Scholar]
- 69.Srikanth A., Schmid M. Regulation of flowering time: All roads lead to Rome. Cell. Mol. Life Sci. 2011;68:2013–2037. doi: 10.1007/s00018-011-0673-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu G., Park M.Y., Conway S.R., Wang J.W., Weigel D., Poethig R.S. The sequential actions of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138:750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aukerman M.J., Sakai H. Regulation of Flowering Time and Floral Organ Identity by a MicroRNA and Its APETALA2-Like Target Genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krajnčič B., Devidé Z. Flower development in Spirodela polyrrhiza (Lemnaceae) Plant Syst. Evol. 1979;132:305–312. doi: 10.1007/BF00982392. [DOI] [Google Scholar]
- 73.Takeno K. Stress-induced flowering: The third category of flowering response. J. Exp. Bot. 2016;67:4925–4934. doi: 10.1093/jxb/erw272. [DOI] [PubMed] [Google Scholar]
- 74.Cho L.-H., Yoon J., An G. The control of flowering time by environmental factors. Plant J. 2016;90:708–719. doi: 10.1111/tpj.13461. [DOI] [PubMed] [Google Scholar]
- 75.Hegelmaier F. The Lemnaceen: A Monographic Study. Utersuchung; Liepzig, Germany: 1868. [Google Scholar]
- 76.Khurana J., Maheshwari S. Floral Induction in Wolffia microscopica by Salicylic Acid and Related Compounds under Non-inductive Long Days. Plant Cell Physiol. 1983;24:907–912. doi: 10.1093/oxfordjournals.pcp.a076594. [DOI] [Google Scholar]
- 77.Maheshwari P.S. Induction of flowering in Wolffia microscopica by iron salt of ethylenediamine-di-o-hydroxyphenylacetic acid (Fe-EDDHA) Z. Pflanzenphysiol. 1966;55:89–95. [Google Scholar]
- 78.Khurana J., Maheshwari S. A Comparison of the Effects of Chelates, Salicylic Acid and Benzoic Acid on Growth and Flowering of Spirodela polyrrhiza. Plant Cell Physiol. 1986;27:919–924. doi: 10.1093/oxfordjournals.pcp.a077179. [DOI] [Google Scholar]
- 79.Pieterse A.H., Müller L.J. Induction of flowering in Lemna gibba G3 under short-day conditions. Plant Cell Physiol. 1977;18:45–53. doi: 10.1093/oxfordjournals.pcp.a075427. [DOI] [Google Scholar]
- 80.Hernández-Apaolaza L., Lucena J.J. Influence of irradiation time and solution concentration on the photochemical degradation of EDDHA/Fe3+: Effect of its photodecomposition products on soybean growth. J. Sci. Food Agric. 2011;91:2024–2030. doi: 10.1002/jsfa.4414. [DOI] [PubMed] [Google Scholar]
- 81.Miwa K., Serikawa M., Suzuki S., Kondo T., Oyama T. Conserved Expression Profiles of Circadian Clock-related Genes in Two Lemna Species Showing Long-day and Short-day Photoperiodic Flowering Responses. Plant Cell Physiol. 2006;47:601–612. doi: 10.1093/pcp/pcj027. [DOI] [PubMed] [Google Scholar]
- 82.Tam Y.Y., Slovin J.P., Cohen J.D. Continuous light alters indole-3-acetic acid metabolism in Lemna gibba. Phytochemistry. 1998;49:17–21. doi: 10.1016/S0031-9422(97)00853-4. [DOI] [Google Scholar]
- 83.Westfall C.S., Sherp A.M., Zubieta C., Alvarez S., Schraft E., Marcellin R., Ramirez L., Jez J.M. Arabidopsis thaliana GH3.5 acyl acid amido synthetase mediates metabolic crosstalk in auxin and salicylic acid homeostasis. Proc. Natl. Acad. Sci. USA. 2016;113:13917–13922. doi: 10.1073/pnas.1612635113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.He Y., Michaels S.D., Amasino R.M. Regulation of Flowering Time by Histone Acetylation in Arabidopsis. Science. 2003;302:1751–1754. doi: 10.1126/science.1091109. [DOI] [PubMed] [Google Scholar]
- 85.Liu F., Quesada V., Crevillén P., Bäurle I., Swiezewski S., Dean C. The Arabidopsis RNA-Binding Protein FCA requires a Lysine-specific Demethylase 1 Homolog to Downregulate FLC. Mol. Cell. 2007;28:398–407. doi: 10.1016/j.molcel.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 86.He Y., Amasino R.M. Role of chromatin modification in flowering-time control. Trends Plant Sci. 2005;10:30–35. doi: 10.1016/j.tplants.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 87.Singh V., Roy S., Giri M.K., Chaturvedi R., Chowdhury Z., Shah J., Nandi A.K. Arabidopsis thaliana FLOWERING LOCUS D Is Required for Systemic Acquired Resistance. Mol. Plant Microbe Interact. 2013;26:1079–1088. doi: 10.1094/MPMI-04-13-0096-R. [DOI] [PubMed] [Google Scholar]
- 88.Singh V., Roy S., Singh D., Nandi A.K. Arabidopsis Flowering Locus D influences systemic-acquired-resistance-induced expression and histone modifications of WRKY genes. J. Biosci. 2014;39:119–126. doi: 10.1007/s12038-013-9407-7. [DOI] [PubMed] [Google Scholar]
- 89.Fu L., Tan D., Sun X., Ding Z., Zhang J. Transcriptional analysis reveals potential genes and regulatory networks involved in salicylic acid-induced flowering in duckweed (Lemna gibba) Plant Physiol. Biochem. 2020;155:512–522. doi: 10.1016/j.plaphy.2020.08.001. [DOI] [PubMed] [Google Scholar]
- 90.Rejmánková E. Germination of seeds of Lemna gibba. Folia Geobot. Phytotaxon. 1976;11:261–267. doi: 10.1007/BF02909473. [DOI] [Google Scholar]
- 91.Bertin R.I. Incidence of Monoecy and Dichogamy in Relation to Self-Fertilization in Angiosperms. Am. J. Bot. 1993;80:557–560. doi: 10.1002/j.1537-2197.1993.tb13840.x. [DOI] [PubMed] [Google Scholar]
- 92.Ruiz L.A. La floración de Lemna gibba L. y Lemna parodiana Giard. (Lemnaceae) en Mendoza. Rev. Fac. Cienc. Agrar. 1951;3:1. [Google Scholar]
- 93.Maheshwari S.C. The embryology of Wolffia. Phytomorphology. 1954;4:355–365. [Google Scholar]
- 94.Brooks J.S. The Cytology and Morphology of the Lemnaceae. Cornell University; Ithaca, NY, USA: 1940. [Google Scholar]
- 95.Fujii S., Kubo K.-I., Takayama S. Non-self- and self-recognition models in plant self-incompatibility. Nat. Plants. 2016;2:16130. doi: 10.1038/nplants.2016.130. [DOI] [PubMed] [Google Scholar]
- 96.Niu S.-C., Huang J., Zhang Y.-Q., Li P.-X., Zhang G.-Q., Xu Q., Chen L.-J., Wang J.-Y., Luo Y.-B., Liu Z.-J. Lack of S-RNase-Based Gametophytic Self-Incompatibility in Orchids Suggests That This System Evolved after the Monocot-Eudicot Split. Front. Plant Sci. 2017;8:8. doi: 10.3389/fpls.2017.01106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Witte K.E. Zur Kultur von Wolffiella welwitschii Monod. Aqua Planta. 1985;10:7. [Google Scholar]
- 98.Caldwell O.W. On the Life-History of Lemna minor. Int. J. Plant Sci. 1899;27:37–66. doi: 10.1086/327786. [DOI] [Google Scholar]
- 99.Hillman W.S. Experimental control of flowering in Lemna III. A relationship between medium composition and the opposite photoperiodic responses of L. perpusilla 6746 and L. gibba G3. Am. J. Bot. 1961;48:413–419. doi: 10.1002/j.1537-2197.1961.tb11659.x. [DOI] [Google Scholar]
- 100.Hoagland D.R., Arnon D.I. The water-culture method for growing plants without soil. Circ. Calif. Agric. Exp. Stn. 1938;347:32. [Google Scholar]
- 101.Shenk R.U., Hildebrandt A.C. Production of Protoplasts from Plant Cells in Liquid Culture Using Purified Commercial Cellulases 1. Crop. Sci. 1969;9:629–631. doi: 10.2135/cropsci1969.0011183X000900050036x. [DOI] [Google Scholar]
- 102.Steinberg R.A. Mineral Requirements of Lemna minor. Plant Physiol. 1946;21:42–48. doi: 10.1104/pp.21.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data presented in article and Supplementals.