Abstract
As the global COVID-19 pandemic progresses, it is paramount to gain knowledge on adaptive immunity to SARS-CoV-2 in children to define immune correlates of protection upon immunization or infection. We analyzed anti-SARS-CoV-2 antibodies and their neutralizing activity (PRNT) in 66 COVID-19-infected children at 7 (±2) days after symptom onset. Individuals with specific humoral responses presented faster virus clearance and lower viral load associated with a reduced in vitro infectivity. We demonstrated that the frequencies of SARS-CoV-2-specific CD4+CD40L+ T cells and Spike-specific B cells were associated with the anti-SARS-CoV-2 antibodies and the magnitude of neutralizing activity. The plasma proteome confirmed the association between cellular and humoral SARS-CoV-2 immunity, and PRNT+ patients show higher viral signal transduction molecules (SLAMF1, CD244, CLEC4G). This work sheds lights on cellular and humoral anti-SARS-CoV-2 responses in children, which may drive future vaccination trial endpoints and quarantine measures policies.
Keywords: SARS-CoV-2, COVID-19, pediatric COVID-19, anti-SARS-CoV-2 antibodies, Ab-mediated neutralization activity, antigen-specific CD4 T cells, antigen-specific B cells, proteomic profiling
Graphical abstract
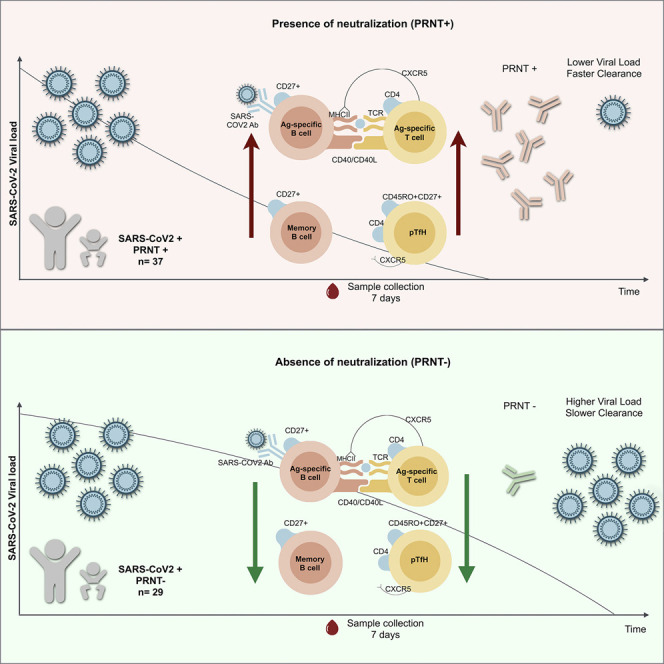
Cotugno et al. show that neutralizing antibodies affect SARS-CoV-2 viral load in infected children. Antigen-specific B and T cells are positively associated with virus neutralization. This information provides a basis for defining SARS-CoV-2 adaptive responses in children.
Introduction
The severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2), first emerged in Wuhan, China in December 2019 (Huang et al., 2020), and spread to Europe and, in particular, the northern regions of Italy in the beginning of February 2020. On March 12, 2020, with >100 countries reporting an increasing number of cases, the World Health Organization (WHO) declared a global pandemic (World Health Organization, 2020). Despite the unprecedented scientific effort, the impact of antigen (Ag)-specific cellular immune responses (Altmann and Boyton, 2020) and neutralizing antibodies (Zhang et al., 2020) is only partially known, particularly in children. Antibody (Ab)-mediated neutralizing activity was shown 21 days after infection onset in the minority (2 of 45) of specific monoclonal Abs isolated from Ag-specific B cells (Seydoux et al., 2020) of an infected adult. In particular, the most effective Ab showed a specific binding for the receptor-binding domain (RBD) and was able to prevent viral entry (Seydoux et al., 2020). The contribution of SARS-CoV-2-specific CD4+ T cells to the production of effective neutralizing Abs has been recently shown in adults (Grifoni et al., 2020; Meckiff et al., 2020; Weiskopf et al., 2020), but no data on its impact on the virus nor its association to Ag-specific B cells have been shown.
In the context of pediatric infection, data on the SARS-CoV-2 specific immunity are still scarce. Few studies have shown that children are able to develop a robust neutralizing Ab (NAb) response after infection (Liu et al., 2020; Yonker et al., 2020; Zhang et al., 2020). However, SARS-CoV-2-specific T and B cell immunity needs to be defined in relation to NAbs and viral control. Overall, children experience a milder clinical course of coronavirus disease 2019 (COVID-19) as compared to adults (Gupta et al., 2020), and it is paramount to define the impact of adaptive SARS-CoV-2-specific immunity on viral spread and clinical course in pediatric cases (Brodin, 2020).
In this work, we aim to define the humoral and cellular responses in SARS-CoV-2-infected children. We investigated the NAb activity in SARS-CoV-2-infected children and its impact on the viral load in nasopharyngeal (NP) swabs. We further explored Ag-specific T and B cells defined as CD4+CD40L+ and SARS-CoV-2 Spike (S1+S2)-positive switched B cells. Finally, we provided a comprehensive proteomic profile focusing on the differences between SARS-CoV-2-infected individuals with differential neutralizing ability.
Results
Clinical and standard laboratory profile of SARS-CoV-2-infected children compared to SARS-CoV-2−
Standard laboratory analysis and demographic characteristics were explored to define differences among SARS-CoV-2-infected children (SARS-CoV-2+, n = 66), and those admitted for the clinical suspicion of SARS-CoV-2 but tested negative by both virological and serological methods (SARS-CoV-2−, n = 11). By study design, we aimed to study humoral and cellular responses in SARS-CoV-2+ children on samples collected 7 days (± 2 days) after symptoms onset (mean overall cohort = 6.63 days). For asymptomatic patients, the symptom onset of the family member or direct contact who tested positive for SARS-CoV-2 was used. In Table 1 , we further stratified the cohort according to the presence of NAbs measured by the plaque reduction neutralization test (PRNT). Overall, 37/66 (56%, PRNT+) developed NAbs, whereas 29/66 (44%, PRNT−) did not. No significant differences in terms of age, gender, and lymphocyte count was found between PRNT+ and PRNT− and SARS-CoV-2−. Higher white blood cell (WBC) count was found in SARS-CoV-2− compared to PRNT+ (p = 0.007) and PRNT− (p = 0.041), as well as a trend for higher hemoglobin (Hb) in PRNT− versus PRNT+ (p = 0.04). This result is in line with the higher C-reactive protein (CRP) levels found in SARS-CoV-2− compared to both PRNT+ and PRNT− (p < 0.0001 and p = 0.002, respectively; Table 1), supporting an alternative infectious/inflammatory condition in SARS-CoV-2−. The development of Abs was not associated with age and did not differ between age groups: infant (<1 year), toddler (1–2 years), preschool (3–5 years), school age (6–12 years), and teenagers (13–17 years) (Table 1). At admission, PRNT+ and PRNT− patients did not differ in regard to disease severity, which can be noted as 0 = asymptomatic, 1 = mild, and 2 = moderate, according to the WHO classification (https://www.who.int/publications/i/item/clinical-management-of-covid-19) (Tables 1 and S1).
Table 1.
Patients' characteristics
| SARS-CoV-2+ (N = 66) |
SARS-CoV-2− (N = 11) | p value | ||
|---|---|---|---|---|
| PRNT+ (N = 37) | PRNT− (N = 29) | |||
| Age, y, mean (SD) | 6.9 (5.4) | 6.6 (5.3) | 6.2 (5.5) | n.s. |
| Male N (%) | 27/37 (65) | 14/29 (48) | 5/11 (45) | n.s. |
| Platelets, mean 103/μL (SD) | 277.2 (90.1), n = 36 | 321.8 (158.6) | 390.9 (201.5) | PRNT+ versus SARS-CoV-2−, p = 0.079 |
| WBC count, mean 103/μL (SD) | 6.4 (2.5) | 6.9 (2.7), n = 26 | 9.4 (3.8) | PRNT+ versus SARS-CoV-2− p = 0.007; PRNT− versus SARS-CoV-2−, p = 0.041 |
| Neutrophils, mean 103/μL (SD) | 2.7 (1.8), n = 36 | 2.6 (1.6) | 4.8 (3.1) | PRNT+ versus SARS-CoV-2−, p = 0.005; PRNT− versus SARS-CoV-2−, p = 0.015 |
| Lymphocytes, mean 103/μL (SD) | 3.4 (2) | 3.8 (2.2) | 3.4 (1.3) | n.s. |
| Hb, mean g/dL (SD) | 12.2 (1.6), n = 36 | 12.4 (1.8) | 12.1 (1.4) | PRNT+ versus PRNT−, p = 0.04 |
| CRP, mean mg/dL (SD) | 1.1 (2.5), n = 31 | 1.9 (4.8), n = 20 | 6.5 (6.8), n = 11 | PRNT+ versus SARS-CoV-2−, p < 0.0001; PRNT− versus SARS-CoV-2−, p = 0.002 |
| Duration of symptoms at analysis,a mean (SD) | 7.1 (3.5), n = 30; min = 2, max = 16 | 5.8 (2.1), n = 19; min = 2, max = 10 | NA | n.s. |
| Symptoms (0/1/2) | (8/24/16) | (7/14/8) | NA | n.s. |
Continuous data were presented as means ± SDs. Non-parametric Mann-Whitney test was used for comparisons. When data were missing, the total N was clearly indicated. Symptomatology: 0 = asymptomatic; 1 = mild; 2 = moderate, according to WHO classification (https://www.who.int/publications/i/item/clinical-management-of-covid-19). CRP, C-reactive protein; Hb, hemoglobin; NA, not applicable; n.s., not significant; SD, standard deviation; WBC, white blood cell.
Duration of symptoms was calculated as the number of days between symptoms onset and date of analysis; for asymptomatic patients, the symptom onset of the family member or direct contact who tested positive for SARS-CoV-2 was used.
SARS-CoV-2-specific immunoglobulin G (IgG) and Ab neutralizing activity associate with faster virus clearance, lower viral load, and reduced in vitro infectivity
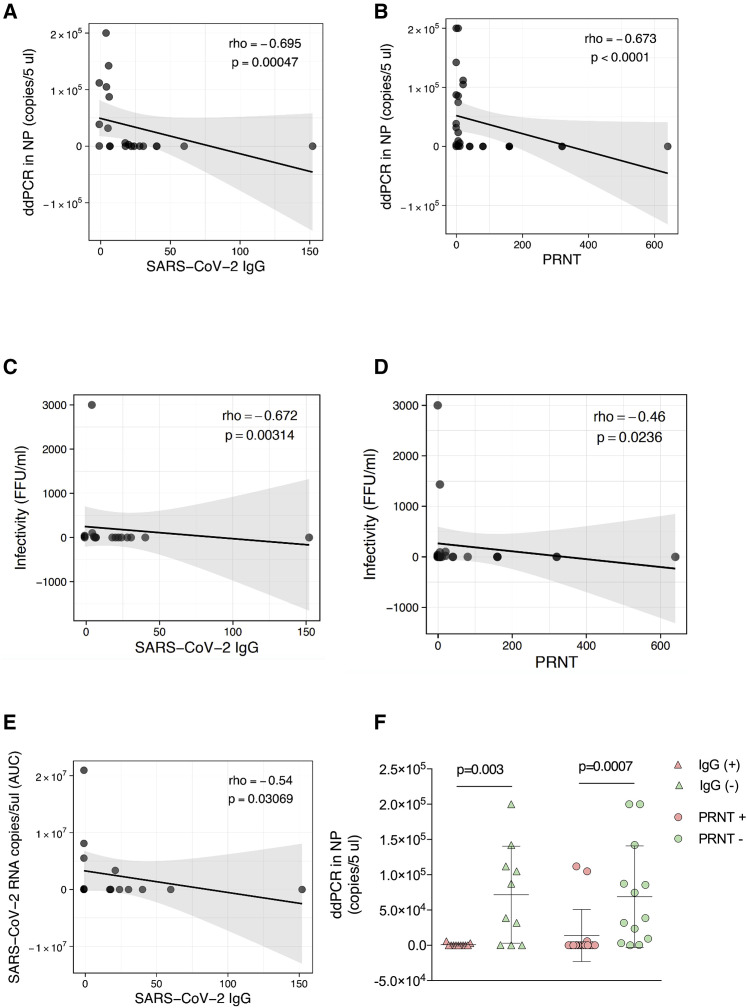
In NP, we evaluated the viral load by droplet digital PCR (ddPCR) and RT-PCR in association with SARS-CoV-2 humoral responses. Our data show an inverse correlation between viral load in NP and both S1/S2 SARS-CoV-2 IgG (p = 0.0005; ρ= −0.69) (Figure 1 A) and PRNT (p < 0.0001; ρ= −0.67) (Figure 1B). In vitro infectivity, measured in foci forming units (FFUs), showed an inverse correlation with both S1/S2 SARS-CoV-2 IgG (p = 0.003; ρ= −0.67) (Figure 1C) and PRNT (p = 0.023; ρ= −0.46) (Figure 1D). Longitudinal analysis confirmed the impact of humoral responses on virus clearance. We calculated the area under the curve (AUC) of the viral load from NP collected every 48 h up to undetectable viral load, and we found an inverse correlation with SARS-CoV-2 Ab (p = 0.031; ρ= −0.54) (Figure 1E). We further compared the levels of SARS-CoV-2 viral load in seropositive and seronegative individuals. Our results show a significantly higher viral load in seronegative patients expressed in terms of both S1/S2 SARS-CoV-2 IgG (p = 0.003) and PRNT (p = 0.0007) (Figure 1F). Overall, these data confirm the role of SARS-CoV-2-specific humoral responses in controlling virus replication, expected infectivity, and virus clearance. As previously reported (GeurtsvanKessel et al., 2020), we found S1/S2 SARS-CoV-2 IgG levels to positively correlate with PRNT titers (p = 0.0002; ρ= 0.65) (Figure S1).
Figure 1.
SARS-CoV-2-specific IgG and Ab neutralization activities were inversely associated with viral load, virus clearance, and infectivity
(A–E) Correlation plots show in (A) association between ddPCR and SARS-CoV-2 IgG titers (n = 21); (B) association between ddPCR and PRNT (n = 29); (C) association between infectivity in vitro (FFU) and SARS-CoV-2 IgG titers (n = 17); (D) association between infectivity in vitro (FFU) and PRNT (n = 24); and (E) association between SARS-CoV-2 IgG and SARS-CoV-2 RNA copies/mL (AUC) (n = 16). Solid lines define linear regression, and grey areas show 95% confidence interval.
Non-parametric Spearman correlation was used in (A)–(E). Non-parametric Mann-Whitney test was used in (F), means with SD are shown in the plot. AUC, area under the curve; ddPCR, digital droplet PCR; FFU, foci forming unit; NP, nasopharyngeal swab. Light purple triangles, SARS-CoV-2 IgG+; light green triangles, SARS-CoV-2 IgG−; light purple solid circle, PRNT+; light green solid circle, PRNT−.
SARS-CoV-2-specific B cells positively associate with SARS-CoV-2-specific IgG and PRNT
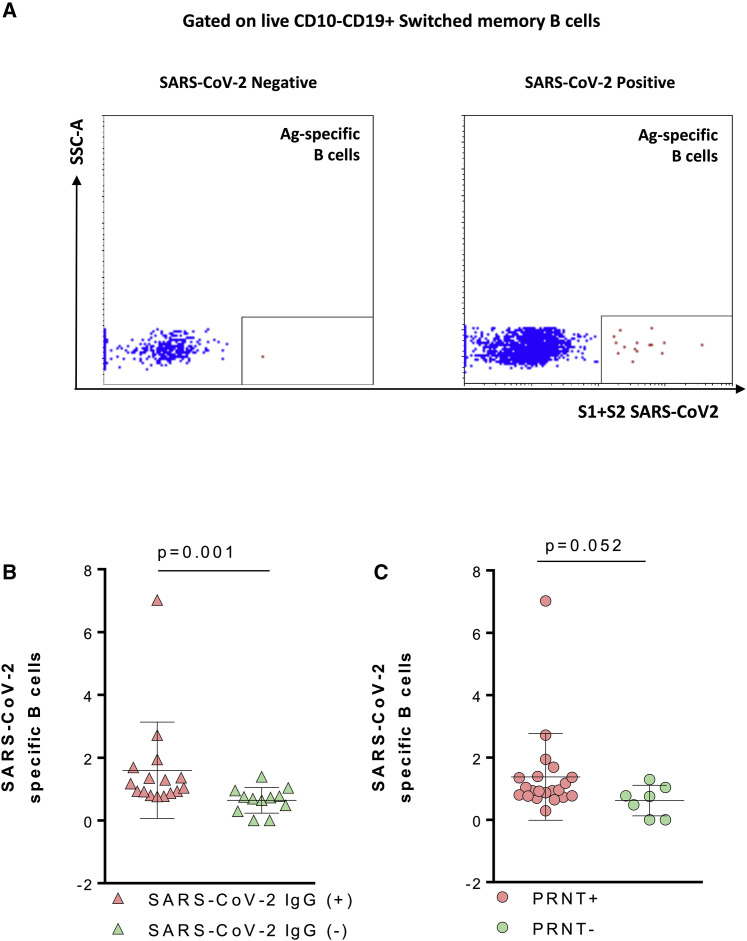
The B cell compartment and the frequency of SARS-CoV-2-specific B cells were analyzed by fluorescence-activated cell sorting (FACS) using an in-house fluorescently labeled probe expressing S1+S2 SARS-CoV-2 proteins (gating strategy in Figures 2 A and S2A). We analyzed the SARS-CoV-2-specific IgD-CD27+ switched B cells (Figure 2A) and found a positive association with both S1/S2 SARS-CoV-2 IgG (p = 0.003; ρ= 0.54) and PRNT (p = 0.008; ρ= 0.49) (not shown). This association was also confirmed when Ag-specific B cells were re-analyzed in SARS-CoV-2+ children stratified by their serological status. A higher frequency of Ag-specific B cells was found in seropositive compared to seronegative SARS-CoV-2 children (p = 0.001) (Figure 2B). We also observed a trend for a higher frequency of Ag-specific B cells in PRNT+ (p = 0.052) (Figure 2C).
Figure 2.
SARS-CoV-2-specific B cells are positively associated with anti-SARS-CoV-2 IgG and Ab neutralization activity
(A) Gating strategy for antigen-specific B cells.
(B and C) SARS-CoV-2 Mann-Whitney test (n = 28); means and SD are shown in the plots.
Light purple triangles, SARS-CoV-2 IgG+; light green triangles, SARS-CoV-2 IgG−; light purple solid circle, PRNT+; light green solid circle, PRNT−.
We further explored the standard B cell phenotype and found no differences in terms of total B cells (CD19+CD10− B cells) in PRNT+ compared to PRNT− (Figure S2B). B cell memory compartment analysis, according to the expression of IgD and CD27, showed a higher frequency of switched memory B cells in PRNT+ compared to PRNT− (Figure S2C) despite no differences in age between the groups. According to the expression of CD21 and CD27, a lower frequency of tissue-like memory (TLM) B cells resulted in PRNT+ compared to PRNT− (Figure S2D).
SARS-CoV-2-specific CD4 T cells are positively associated with PRNT
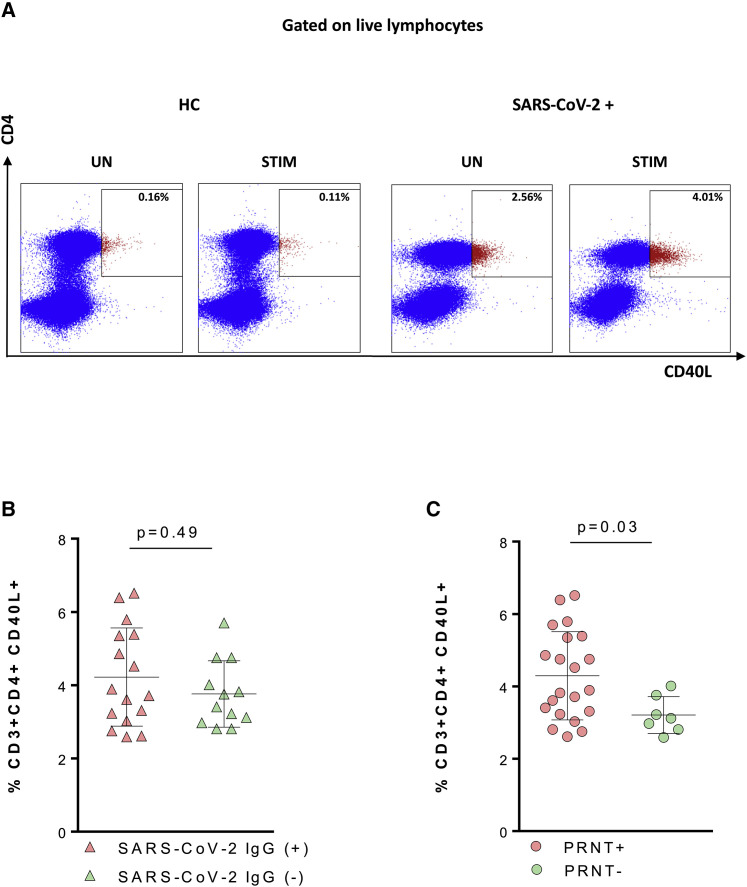
To further characterize the immunological features of patients with differential neutralization activity against SARS-CoV-2, we investigated CD3+CD4+ general phenotype and frequency of SARS-CoV-2-specific CD4 T cells (gating strategy in Figures 3 A and S3A). Ag-specific CD4 T cells were defined through the analysis of CD40L after in vitro stimulation with SARS-CoV-2 Ags using a previously validated methodology (de Armas et al., 2017; Cotugno et al., 2017, 2020a, 2020b; Chattopadhyay et al., 2006). Pairwise correlation analysis showed a positive association between Ag-specific T cells and Ab-mediated neutralization activity (p = 0.039; ρ= 0.39) (not shown). Furthermore, PRNT+ revealed a higher frequency of SARS-CoV-2-specific T cells compared to PRNT− (p = 0.03; Figure 3C). No differences in terms of Ag-specific T cells emerged in patients with differential SARS-CoV-2 IgG serostatus. Overall, these results may suggest that Ag-specific T cells may correlate with NAb classes other than IgG such as IgA in turn having a higher correlation with mucosal and tissue resident antibodies (Hsueh et al., 2004).
Figure 3.
SARS-CoV-2-specific CD4 T cells are positively associated with SARS-CoV-2-specific IgG and Ab-neutralization activity
(A) Gating strategy for antigen-specific T cells (CD40L+). Left-side panels show CD40L+CD4+ T cells for unstimulated (UN) and stimulated (STIM) samples in a healthy control (HC). Right-side panels show the same conditions for a SARS-CoV-2-infected patient.
(B and C) Twenty-eight patients split according to SARS-CoV-2 IgG (B) and PRNT (C) were compared by the Mann-Whitney test; means and SD are shown in the plots.
Light purple triangles, SARS-CoV-2 IgG+; light green triangles, SARS-CoV-2 IgG−; light purple solid circle, PRNT+; light green solid circle, PRNT−.
We also explored the standard T cell phenotype, and we could benefit from a cohort of healthy age- and gender-matched children examined in the pre-COVID-19 era. Whereas no differences emerged in terms of total CD3+CD4+ between PRNT+, PRNT−, and SARS-CoV-2−, both SARS-CoV-2+ groups (PRNT+ and PRNT−) showed a higher CD3+CD4+ frequency compared to healthy control (HC) (p = 0.001 and p = 0.017, respectively) (Figure S3B). Maturational subsets were analyzed according to the surface expression of CD45RO and CD27. PRNT+ expressed a significantly lower frequency of terminally differentiated effector memory (TEMRA) T cells compared to both PRNT− and SARS-CoV-2− patients (Figure S3C). Through the expression of CXCR5 on live CD3+CD4+CD45RO+CD27+ central memory T cells, we investigated peripheral follicular T helper cells (pThf). Our analysis showed a lower frequency of pThf in both PRNT+ and PRNT− patients compared to HC (p = 0.004 and p = 0.004, respectively) (Figure S3D). We also found that PRNT− had lower pTfh compared to SARS-CoV-2− children (p = 0.040) (Figure 3D), suggesting how the frequency of this subset can play a crucial role in the context of SARS-CoV-2 infection.
Proteomics characteristics of SARS-CoV-2 patients with differential neutralizing activity
We explored plasma proteomic correlates of immune responses in patients with SARS-CoV-2 neutralization activity. We could benefit from previous data generated with the Olink platform on plasma samples collected in our cohort at diagnosis and before treatment initiation (Consiglio et al., 2020). We obtained 121 unique plasma proteins using 2 Olink panels associated with inflammation pathways filtering out proteins with >30% measurements below the threshold of detection. Principal-component analysis (PCA) analyzed for proteomics revealed no clear clusters between PRNT+ and PRNT− (data not shown), suggesting a similar pro-inflammatory pattern in both SARS-CoV-2 groups, which were similar overall in terms of symptomatology (see Tables 1 and S1). The full dataset of correlations is available in Figure S4.
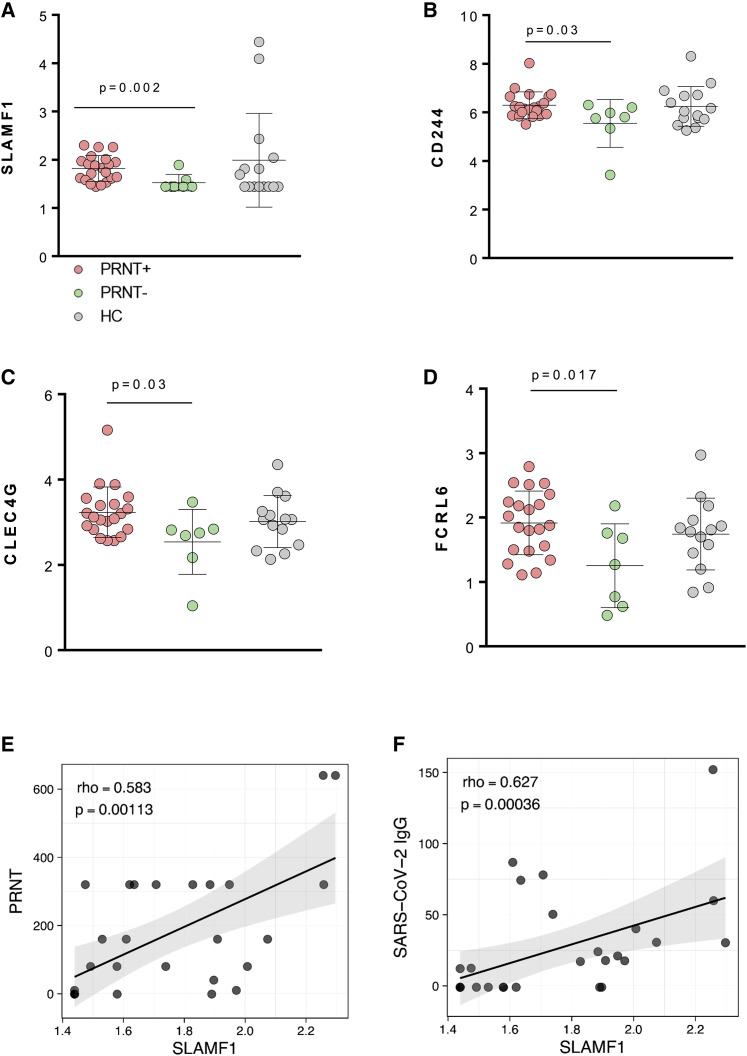
We further assessed whether the presence of neutralizing activity was associated with the differential expression of specific plasma factors. Two different proteins belonging to the signaling lymphocytic activation molecule pathway (SLAMF1) (Figure 4 A) and CD244 (Figure 4B) were significantly higher in PRNT+ compared to PRNT− (p = 0.002 and p = 0.03, respectively). Such molecules were previously shown to be present on activated T and B cells and to be essential receptor and signaling transductors for viral entry (Koethe et al., 2012). Proteomic analysis also showed that PRNT+ had higher levels of two other factors associated with signal transduction and viral entry. First, there is CLEC4G (p = 0.017; Figure 4C), which is a glycan-binding receptor already described as acting as a recognition and uptake molecule for multiple viruses, including SARS coronaviruses (Gramberg et al., 2005). Second, there is fragment crystallizable (Fc) receptor-like 6 (FCRL6) (p = 0.017; Figure 4D), which can act as the major histocompatibility complex II receptor (MHC II) mediating virus entry (Schreeder et al., 2010). Finally, correlation analysis confirmed that SLAMF1 was positively associated with PRNT (p = 0.001; Figure 4E), SARS-CoV-2 IgG (p = 0.0003; Figure 4F), and Ag-specific B cells (p = 0.028) (not shown). These data collectively confirm a close relation between cellular and neutralizing activity in SARS-CoV-2 immunity.
Figure 4.
Proteomic analysis revealing distinct patterns in patients with differential anti-SARS-CoV-2 Ab-neutralization activity
(A–D) Non-parametric Mann-Whitney test was used to compare proteomics analysis of 7 PRNT−, 21 PRNT, and 15 HC; means and SD are shown in the plots.
(E and F) Correlation analysis between PRNT (E) and SARS-CoV-2 IgG versus SLAMF1 (F, n = 28). Non-parametric Spearman correlation test was used in (E) and (F); solid lines define linear regression, and grey area shows 95% confidence interval.
Light purple solid circle, PRNT+; light green solid circle, PRNT−; gray solid circle, HC.
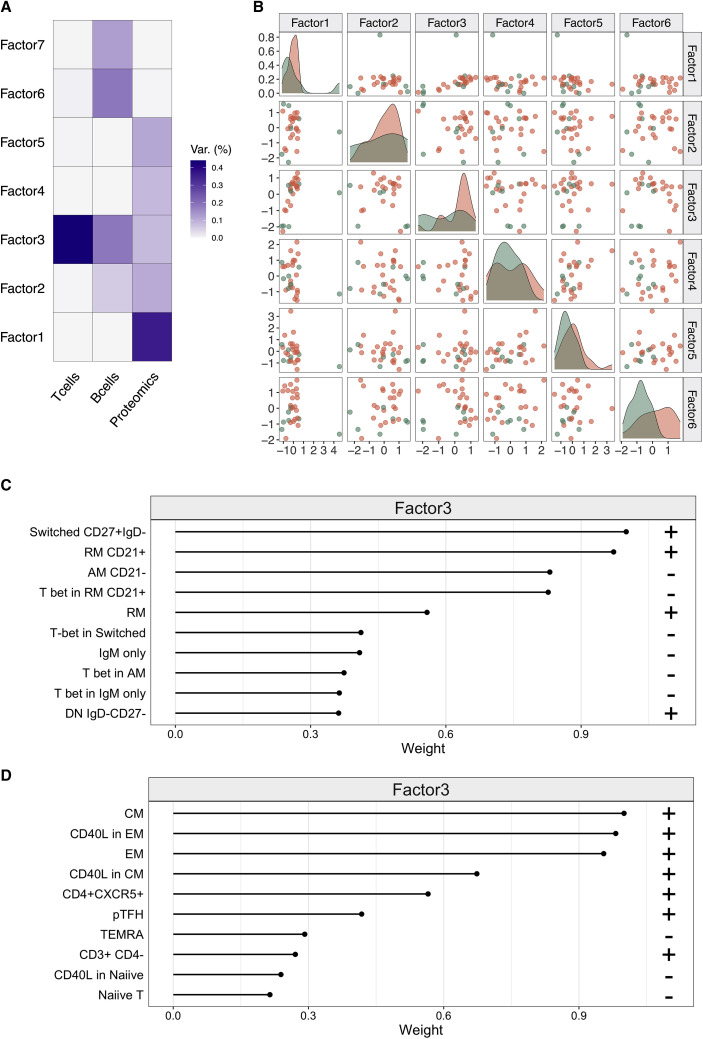
Multiomics factor analysis (MOFA) revealing predictors of anti-SARS-CoV-2 neutralization activity
To provide a comprehensive immunologic description of patients with the differential ability to produce neutralizing antibodies, we applied MOFA (Argelaguet et al., 2019). In this analysis, we used as input phenotypic features describing T and B cells, Ag-specific cell frequencies, and plasma proteins. We focused on the top seven factors that were identified as major contributors toward the neutralization activity (Figure 5 A). Among those, the plasma proteome did not show a relevant contribution (factor 1 in Figures 5A and S5A). Conversely, factors 3 and 6, mainly driven by the frequencies of B and T cell subsets, were able to discriminate between PRNT+ and PRNT− (Figure 5C). This analysis proved that resting memory B cells (IgD−CD27+CD21+) and activated memory B cells (IgD−CD27+CD21−) in factor 3 (Figure 5C), along with activated T-Bet+ B cell subsets (Figure S5B) in factor 6, represented the main loading features for the identification of PRNT+/PRNT− groups. For the T cell subsets in factor 3, the highest weight for group discrimination was mainly provided by the frequency of central memory (CM), effector memory (EM), and CD40L+ memory T cells (Figure 5D). Overall, these data suggest that the development of NAbs is favored by the presence of an intact memory B and T cell compartment.
Figure 5.
Multiomics factor analysis (MOFA) showing clusterization factors of patients with Ab-neutralization activity
(A) Fraction of variance explained by 7 latent factors is shown for 28 patients.
(B) Samples distributed across the top 6 latent factors.
(C and D) Top contributing features and loading direction (+ and −) for factor 3 in B cell (C) and T cell (D) populations.
AM, activated memory; CM, central memory; DN, double negative; EM, effector memory; RM, resting memory; TEMRA, terminally differentiated effector memory; TLM, tissue-like memory.
Discussion
This work provides a virological and immunological characterization of SARS-CoV-2-infected children presenting differential Ab-mediated neutralizing activity. It further shows that children with NAbs present a reduced viral load, faster virus clearance, and lower in vitro infectivity. These data provide information that can drive vaccination endpoints and quarantine measures policies. In particular, we explored SARS-CoV-2-specific humoral and cellular responses in peripheral blood mononuclear cells (PBMCs) and plasma collected ∼1 week after symptoms onset. We correlated such results with a quantitative analysis of SARS-CoV-2 viral load and in vitro infectivity. We stratified the cohort according to humoral responses and found that the SARS-CoV-2 viral load was lower in individuals with both SARS-CoV-2 IgG and PRNT, showing the impact of Ab-mediated responses on the SARS-CoV-2 viral load in NP in children. Confirming previous in vitro findings (Qin et al., 2020; Walls et al., 2020), we showed that children presenting higher anti-Spike protein IgG levels and higher concentrations of NAbs are the ones with lower viral burdens and faster virus clearance. This information further confirms that pediatric patients are able to mount a humoral response relatively early after symptoms onset (Yonker et al., 2020), which affects both viral load and virus clearance. Although there is as yet no consensus that a higher viral load in NP correlates with a higher infectivity of patients, here, we show the impact of humoral immunity on virus replication, in vitro infectivity, and, presumably, virus spread. Hence, SARS-CoV-2-infected children able to mount in vitro neutralization could benefit from less restrictive quarantine measures.
Furthermore, we studied SARS-CoV-2-specific cellular responses that are still unknown in the pediatric setting. Recent works have focused mainly on adult groups and demonstrated a clear association between the presence of SARS-CoV-2 specific CD4+ T cells and the development of effective neutralizing activity (Grifoni et al., 2020; Weiskopf et al., 2020; Altmann and Boyton, 2020; Rydyznski Moderbacher et al., 2020). In the present study, we confirmed a positive association between humoral responses and SARS-CoV-2-specific CD4+ T cells in children. In particular, individuals with elevated levels of SARS-CoV-2-specific CD4+ T cells also showed elevated levels of effective neutralizing activity, associated with reduced viral load. We further revealed that Ag-specific B cells were positively associated with SARS-CoV-2 neutralization activity and IgG titers. These results suggest Ag-specific B cell frequency as a cellular marker for Ab-mediated neutralization.
We then moved on to explore whether neutralization activity was associated with particular B and T cell phenotype characteristics that could favor humoral responses in SARS-CoV-2-infected children. Our data showed that neutralization activity was associated with lower levels of exhausted B and T cell phenotypes. TLM B cell frequencies, previously shown to be higher in chronic infections (Meffre et al., 2016; Moir et al., 2008), were lower in PRNT+ patients. Accordingly, memory B cells were positively associated with both SARS-CoV-2 IgG and PRNT titers. Therefore, these B cell phenotype characteristics can be useful to estimate the ability of the patients to produce anti SARS-CoV-2 humoral responses. Of note, these results are in line with a recent report in adults showing that the majority (>90%) of Ag-specific cells able to produce effective NAbs present a memory phenotype (Seydoux et al., 2020). This observation may further support the use of convalescent plasma containing high doses of NAbs (Tonn et al., 2020), as in previous attempts to support faster virus clearance in critical cases of COVID-19 (ClinicalTrials.gov: NCT04391101 and NCT04374370) (Li et al., 2020; Valk et al., 2020).
The characterization of children with differential anti-SARS-CoV-2 neutralizing activity was enriched by the analysis of plasma proteomic profiling. We found that few proteins involved in viral signal transduction were differentially expressed in PRNT+ and PRNT−. In particular, SLAMF1 and CD244 were significantly higher in PRNT+ patients as compared to PRNT− patients. The reduction of these molecules in plasma may reflect an intracellular consumption in a phase of higher viral load, such as the one found in PRNT−. These data support the idea that an anti-SARS-CoV-2 immune response is being triggered in PRNT−. These molecules are crucial in activated lymphocytes to transduce the signal of an MHC-processed antigen, such as measles virus (Lin and Richardson, 2016). In line with this, Walls et al. (2020) showed the homologous protein domains of measles virus and coronaviruses, including SARS-CoV-2. It was recently argued that adaptive protection may be provided by the measles, mumps, and rubella vaccine and bacille Calmette-Guerin (BCG) (Mantovani and Netea, 2020; Sidiq et al., 2020). Furthermore, CLEC4G, already described as acting as a recognition and uptake molecule for multiple viruses, including Ebola, West Nile, and SARS coronaviruses (Gramberg et al., 2005), was found to be higher in PRNT+ compared to PRNT−. Overall, these data confirm the close relation between cellular and humoral responses to control SARS-CoV-2 burden in children.
From a clinical perspective, our cohort was characterized by scarce symptoms variability, with the majority of children presenting with mild symptoms, as also observed by others (Brodin, 2020). However, in accordance with other findings (Weisberg et al., 2021), we found no association between disease severity and anti-SARS-CoV-2 Ab production. It is important to note that the asymptomatic patients (n = 14) included in our cohort and the 11 multi-inflammatory syndrome associated with COVID-19 (MIS-C) patients, previously published (Consiglio et al., 2020), showed no major differences in terms of SARS-CoV-2 serology and neutralization activity compared to mild COVID-19 cases (Consiglio et al., 2020). These data support the hypothesis that inflammation dynamics rather than adaptive immunity orchestrates the clinical course of the disease and specifically the development of MIS-C. In line with this, plasma inflammation markers were not able to discriminate between PRNT+ and PRTN− patients, supporting the hypothesis that the ability to produce neutralizing antibodies does not directly associate with grade of inflammation, but rather with the status of the immune system.
This work provides a description of SARS-CoV-2-specific humoral and cellular immunity in children and its relation to virus load, virus clearance, and infectivity. Both Ag-specific T and B cells positively correlate with anti-SARS-CoV-2 antibody titers and neutralization activity, suggesting cell correlates of SARS-CoV-2 immunity in this population. Public health restrictive measures should take into account such results to reconsider the timing of confinement and NP retesting in pediatric patients presenting strong humoral responses. Additional confirmatory studies involving adult cohorts as well as larger cohorts of children with COVID-19 and with a higher symptom variability are needed.
Limitations of study
This study presents some limitations: (1) the small sample sizes within each comparison; (2) the fact that these patients are from one region reduces the ability to translate the impact of this information on arising viral strains; (3) the lack of adjustment for multiple comparisons, as this was a descriptive study of a unique cohort; and (4) the inability to accurately estimate timing of infection in asymptomatic patients.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD154-PE (CD40L, clone TRP1) | BD | Cat#555700; RRID: AB_396050 |
| CD3PE-CF594 (clone UCHT1) | BD | Cat#562280; RRID: AB_11153674 |
| CD4 BV510 (SK3) | BD | Cat#562970; RRID: AB_2744424 |
| CD27 V450 (M-T271) | BD | Cat#561408; RRID: AB_10682577 |
| CD45RO PE-Cy5 (UCHL1) | Biolegend | Cat#304208; RRID: AB_314424 |
| CD185 BV605 (CXCR5, RF8B2) | BD | Cat#740379; RRID: AB_2740110 |
| CD197 BV786 (CCR7, 3D12) | Biolegend | Cat#563710; RRID: NA |
| IL-21 APC (3A3-N2) | Biolegend | Cat#513008; RRID: AB_11150407 |
| IL-2 PE-Cy7 (MQ1-17H12) | Biolegend | Cat#500326; RRID: NA |
| IFNγ FITC | BD | Cat#552887; RRID: NA |
| TNFα AF700 (Mab11) | Biolegend | Cat#502928; RRID: NA |
| CD10 Pe-Cy7(HI10a) | BD | Cat#312214; RRID: AB_400216 |
| CD19 APC-R700(SJ25C1) | BD | Cat#659121; RRID: AB_2870470 |
| CD21 APC (B-Ly4) | BD | Cat#559867; RRID: AB_2085309 |
| CD27 FITC(M-T271) | BD | Cat#555440; RRID: AB_395833 |
| IgD BV421(IA6-2) | BD | Cat#565940; RRID: AB_2739404 |
| IgM PE-CF594(G20-127) | BD | Cat#562539; RRID: AB_2737641 |
| IgG BV605(G18-145) | BD | Cat#563246; RRID: AB_2738092 |
| Double Strand RNA (dsRNA J2) | Scicons | J2 produced in mouse |
| Peroxidase-labeled goat anti-mouse | Thermofisher | Cat#G-21040 |
| True Blue (KPL) | SeraCare | Cat#5510-0052 |
| Biological samples | ||
| Peripheral venous EDTA blood | OPBG | n.a. |
| Swab-preserving media | OPBG | n.a. |
| Chemicals, peptides, and recombinant proteins | ||
| BD FACS Permeabilizing Solution 2 | BD | Cat# 347692 |
| BD Golgi Stop | BD Biosciences | Cat#554724 |
| Anti-CD28 (1 μg/ml) | BD | Cat#555725 |
| Bovine Albumin Fraction V (7.5% solution) | Thermofisher | Cat#15260037 |
| Dimethyl sulfoxide | Sigma-Aldrich | Cat#D8418 |
| Dulbecco’s Phosphate buffered saline | EuroClone | Cat#ECB4004L |
| Fetal Bovine Serum | GIBCO | Cat#10270-106 |
| Ficoll-Paque PLUS | GE Healthcare | Cat#17-1440-03 |
| PepTivator SARS-CoV-2 Prot_S | Miltenyi Biotec | Cat#130-126-700 |
| S1+S1 SARS-CoV-2 Prot S | Sino Biological | Cat# 40589-V08B1 |
| RPMI 1640 | EuroClone | Cat#ECM9106L |
| Dulbecco’s modified Eagle’s medium | GIBCO | Cat#11995065 |
| Triton X-100 | Sigma | Cat# T8787 |
| Penicillin-Streptomycin | Thermofisher | Cat# 15140122 |
| Paraformaldehyde (PFA) 4% solution | Sigma | Cat# 1004968350 |
| Carbo xymethyl cellulose salt | Sigma | Cat# C5013-500G |
| MEM | Thermofisher | Cat# 11095080 |
| Tween-80 | Sigma | Cat#P1754 |
| Critical commercial assays | ||
| Live/dead Fixable Near-IR Dead cell Stain kit | Invitrogen | Cat#L34976 |
| Lightning-Link® R-PE Conjugation Kit | (Innova Biosciences) | Cat#703-0030 |
| Deposited data | ||
| Full datasets | Mendeley | http://doi.org/10.17632/stxwzbyfsc.1 |
| Experimental models: cell lines | ||
| Vero E6 cells | ATCC | CRL 1586 |
| Oligonucleotides | ||
| Primers for SARS-CoV-2 E gene: forward ACAGGTACGTTAATAGTTAATAGCGT | Corman et al., 2020 | N/A |
| Primers for SARS-CoV-2 E gene: reverse ATATTGCAGCAGTACGCACACA | Corman et al., 2020 | N/A |
| Probe: FAM-ACACTAGCCATCCTTACTGCGCTTCG-BBQ | Corman et al., 2020 | N/A |
| Housekeeping GAPDH | PE Applied Biosystems | Cat#402869 |
| GAPDH Kit (PE Applied Biosystems) | N/A | N/A |
| Software and algorithms | ||
| Kaluza software | Beckman Coulter | https://www.beckman.com/flow-cytometry/software/kaluza?country=US |
| R software (v 3.6.2) | R Foundation | https://www.r-project.org/ |
| QuantaSoftTM Analysis Software 1.7.4.0917 | Bio-Rad | Cat#1864011 |
| R scripts to reproduce the analyses | GitHub | https://github.com/GRP92/PRNT_manuscript |
| Other | ||
| Droplet Generation Oil for Probes | Bio-Rad | Cat#1863005 |
| One-Step RT-ddPCR Adv Kit | Bio-Rad | Cat#1864022 |
| QX200TM Droplet Generator | Bio-Rad | Cat#1864002 |
| 96 well plate | Bio-Rad | Cat#HSS9641 |
| Eppendorf Mastercycler | Eppendorf | Cat#6313000018 |
| QX200TM Droplet Reader | Bio-Rad | Cat#1864003 |
| DG8 Cartridges | Bio-Rad | Cat#1864008 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Paolo Palma (paolo.palma@opbg.net).
Materials availability
No new reagents were developed during the study. Patient samples can be made available within the limits of the approved Ethical permits. Such requests should be directed to the lead contact author.
Data and code availability
The published article includes all dataset generated or analyzed during this study.
Scripts to reproduce the analyses presented in each figure of the paper are available at https://github.com/GRP92/PRNT_manuscript. Full datasets are available at https://data.mendeley.com/datasets/stxwzbyfsc/1.
Experimental model and subject details
Study participants
Sixty six SARS-CoV-2 infected children (SARS-CoV-2 +) (see Table 1) and 11 SARS-CoV-2 negative controls (SARS-CoV-2 neg) were enrolled in the CACTUS (Immunological studies in Children AffeCTed by COVID and acUte reSpiratory diseases) study from March to April 2020 at Bambino Gesù Children’s Hospital in Rome (see Table S2). Local ethical committee approved the study and written informed consent was obtained from all participants or legal guardians. Age, gender, clinical and routine laboratory characteristics of the cohort are described in Table 1. SARS-CoV-2 + were patients tested positive for SARS-CoV-2 real-time reverse transcriptase–polymerase chain reaction (RT-PCR) tests (GeneXpert, Cepheid, Sunnyvale, CA; 250 copies/mL sensitivity, 100% specificity) in nasopharingeal swab (NP). SARS-CoV-2 + were stratified according to presence (PRNT+) or absence (PRNT-) of neutralizing antibodies. Controls enrolled in the studies (SARS-CoV-2 neg) were children admitted to our hospital with suspected SARS-CoV-2 infection, due to fever or respiratory symptoms, but tested negative for 2 consecutive nasopharyngeal swabs (performed 24 hours apart) and for SARS-CoV-2 S1/S2 IgG test. Longitudinal blood samples were collected at admission, after 48 hours and approximately 7 days after admission. Viral load in NP was performed in SARS-CoV-2 + every 48 hours up to undetectable viral load. Patients with clinical records of primary or acquired immune compromising conditions, recent or current administration of immune suppressive therapies, or other diseases affecting the immune system were excluded from the study. Duration of symptoms was calculated as the number of days between symptoms onset and date of analysis; for asymptomatic patients, the symptom onset of the family member or direct contact who resulted positive for SARS-CoV-2 was used. A control group of age, gender and ethnicity individuals (N = 12) was included to analyze the T cell phenotype. A previously described control group was used for proteomic analysis (Consiglio et al., 2020).
Method details
Sample collection and storage
Venous blood was collected in EDTA tubes and processed within 2 hours. Plasma was isolated from blood and stored at −80°C. Peripheral blood mononuclear cells (PBMCs) were isolated from blood of all patients with Ficoll density gradient and cryopreserved in FBS 10% DMSO until analysis, in liquid nitrogen. Nasopharyngeal swab preserving media was stored at −80°C until use.
Detection and quantification of SARS-CoV-2 RNA in nasopharyngeal swab (NP)
SARS-CoV-2 viral load was evaluated using both RT-PCR and ddPCR. For the RT-PCR, the qualitative SARS-CoV-2 real-time reverse transcriptase–polymerase chain reaction (RT-PCR) tests was used according to manufacturer instruction (Seegene’s Allplex 2019-nCoV Assay, Republic of Korea). The assay is designed to allow for detection of both E and RdRp genes, which showed comparable specificity for SARS-CoV-2 (Corman et al., 2020). The system is designed to provide an auto-interpretation of the data alongside a Ct value resulting from the amplification of each gene.
An in-house multiplex quantitative assay based on One-Step RT- digital droplet (dd) PCR was used to quantitate SARS-CoV-2 viral load. Total RNA was isolated from NP preserving media and eluted in a final volume of 100 μl. The reaction mixture consisted of 5 ul of supermix (Bio-Rad, CA, USA), 2 μL of reverse transcriptase; 2 μL of DTT final concentration 300mM; forward and reverse primers of SARS-CoV-2 E gene to a final concentration of 400 nM each and probe to a final concentration of 200 nM (Corman et al., 2020) and 5 μL of nucleic acids eluted from nasapharyngeal swab samples into a final volume of 20 μl. Housekeeping GAPDH, employed to control the quality of extracted RNA, was amplified under the same conditions using the GAPDH Kit (PE Applied Biosystems); 5 μL of nucleic acids eluted from nasapharyngeal swab samples into a final volume of 20 μl. Each well of the prepared mix was loaded into the 8-channel cartridge to generate the droplets in the oil suspension according to manufacturer (Bio-Rad). Reverse transcription and PCR cycles were run as follow: reverse transcription at 42-50°C for 60 min; enzyme activation at 95°C for 10 min; denaturation of cDNA at 95°C for 30sec and annealing/extension at 60°C for 1 min; these last two passages were repeated for 40 cycles followed by enzyme deactivation at 98°C for 10 min. The droplets were then read by the QX200™ Droplet Reader (Bio-Rad) and the results were analyzed with the QuantaSoft™ Analysis Software 1.7.4.0917 (Bio-Rad). Wells with less than 10000 droplets were discarded from the analysis. Each sample was run at least in duplicate. The final results were expressed as SARS-CoV-2 copies/5 μl. We compared the results obtained by both ddPCR and RT-PCR and we found a significant inverse linear correlation between the Ct values of RT-PCR and the viral copy number provided by the ddPCR (r = −0.906, p < 0.0001). The derived formula y = −3.40 x+ 36.88 was employed to convert the Ct values in viral copies for samples tested only by RT-PCR.
AUC
The SARS-CoV-2 viral load measurements for the gene E in NP were used to calculate the AUC as follow. The viral load was calculated at multiple time points to calculate the area under the curve for each patient. The curves were drawn plotting on the y axis the viral copies and on the x axis the number of days between the date of swab collection and the date of onset of symptoms for symptomatic patients and the hospital admission date for non-symptomatic ones. The average value was considered for the association with PRNT and SARS-CoV-2 IgG.
Virus titration by focus forming assay (FFA)
Vero E6 cells (ATCC® CRL 1586) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, GIBCO) supplemented with 10% FBS, penicillin (100 U/ml) and streptomycin (100U/ml), at 37°C in a humidified CO2 incubator. To titrate virus infectivity, nasopharyngeal swab preserving media was filtered through a 0.22 μm filter and serially diluted in DMEM supplemented with 2% FBS, penicillin (100 U/ml) and streptomycin (100U/ml). Dilutions were incubated on confluent monolayers of Vero E6 cells, in 96-well plates, for 1 hour. After infection, the inoculum was removed and an overlay of MEM, 2% FBS, penicillin (100 U/ml) and streptomycin (100U/ml) and 0.8% carboxymethyl cellulose was added. After 27 hours, the overlay medium was removed and cells were fixed in PBS 4% PFA, for 30 minutes at 4°C. Upon removal, cells were permeabilized with a 0.5% Triton X-100 solution for 10 minutes. Immunostaining was performed by incubation of the J2 anti-dsRNA monoclonal antibody (1:10,000; Scicons) for 1 hour, followed by incubation with peroxidase-labeled goat anti-mouse antibodies (1:1000; DAKO) for 1 hour and a 7 min incubation with the True Blue (KPL) peroxidase substrate. Immuno-reagents were prepared in a solution of 1% BSA and 0.05% Tween-80 in PBS. After each antibody incubation, cells were washed 4 times with a 0.05% Tween-80 PBS solution. Focus forming units per ml (FFU/ml) were counted after acquisition of pictures at a high resolution of 4800 × 9400dpi, on a flatbed scanner.
Plaque reduction neutralization test (PRNT)
A high- throughput PRNT method was developed in-house. Briefly, plasma samples were heat- inactivated by incubation at 56°C for 30 minutes and 2-fold dilutions were prepared in Dulbecco modified Eagle medium (DMEM). The dilutions were mixed to a 1:1 ratio with a virus solution containing 20-25 FFUs of SARS-CoV-2 and incubated for 1 hour at 37°C. Fifty microliters of the virus–serum mixtures were added to confluent monolayers of Vero E6 cells, in 96-wells plates and incubated for 1 hour at 37°C, in a 5% CO2 incubator. The inoculum was removed and 100 μL of overlay solution of Minimum essential medium (MEM), 2% fetal bovine serum (FBS), penicillin (100 U/ml), streptomycin (100 U/ml) and 0.8% carboxy methyl cellulose was added to each well. After 26 hours’ incubation, cells were fixed with a 4% paraformaldehyde (PFA) solution. Visualization of plaques was obtained with an immunocytochemical staining method using an anti-dsRNA monoclonal antibody (J2, 1:10,000; Scicons) for 1 hour, followed by 1-hour incubation with peroxidase-labeled goat anti-mouse antibodies (1:1000; DAKO) and a 7-minute incubation with the True BlueTM (KPL) peroxidase substrate. FFUs were counted as described above. The serum neutralization titer was defined as the reciprocal of the highest dilution resulting in a reduction of the control plaque count > 50% (PRNT50). We considered a titer of 1:10 as the seropositive threshold.
Generation of SARS-CoV-2 probe to select B cells antigen-specific cells
The S1+S2 Spike SARS-CoV-2 protein was obtained from MyBiosource and labeled with Lightning-Link® R-PE Conjugation Kit (Innova Biosciences) to obtain a SARS-CoV-2-R-PE.
Ag-specific B cells by flow cytometry
PBMCs underwent surface staining for the B cell phenotype with the following antibodies: CD10 Pe-Cy7(HI10a) from Biolegend, CD19 APC-R700(SJ25C1), CD21 APC(B-Ly4), CD27 FITC(M-T271), IgD BV421(IA6-2), IgM PE-CF594(G20-127), IgG BV605(G18-145) from BD, S1+S2 Sars-CoV-2-R-Phycoerythrin (-R-PE) and Live/Dead (BV510; BD). Following fixing and permeabilization of cells (BD permeabilization solution II 1x), cells were stained with an anti T-bet BV650 (04-46, BD). Cells were acquired using Cytoflex (Beckman Coulter, Brea, CA). Gating strategies are provided in Figure S2. Positive cell gating was set using fluorescence minus one control. Data analyses were performed using Kaluza software (Beckman Coulter).
CD4 Ag-specific T cells by flow cytometry
Thawed PBMC were plated (1.5x106/aliquot/200 ul) in 96 well-plate containing CD154-PE (CD40L, BD PharMingen, Franklin Lakes, NJ, USA), Golgi Stop (SigmaAldrich), anti-CD28 (1 μg/ml) and either 0,4μg/ml PepTivator SARS-CoV-2 Prot_S (Milteny Biotech, Bergisch Gladbach, Germany) or media only. Following 16hrs incubation at 37°C/5% CO2, PBMC were centrifuged and stained with LIVE/DEAD fixable NEAR-IR dead cell stain kit (for 633 or 635 nm excitation, ThermoFisher, Waltham, Massachusetts, US) 1ul per 106cells/ml for 15 minutes at room temperature (RT), protected by light. Antigen specific CD40L+CD4+ T cells were identified according to the gating strategy showed in Figure 3A. Preliminary experiments on unstimulated (UN), SARS-CoV-2 stimulated (STIM), or SEB-stimulated (not shown) healthy controls collected in the pre-COVID-19 era were used to set the gate. Following in vitro stimulation with SARS-CoV-2 antigens, only SARS-CoV-2 + patients showed increase in CD40L+ T cells demonstrating the antigen-specific cellular response (Figure S3A). Surface staining was performed using the following antibodies: CD3 PE-CF594 (UCHT1), CD4 BV510 (SK3), CD27 V450 (M-T271), CD45RO PE-Cy5 (UCHL1), CD185 BV605 (CXCR5, RF8B2) from BD (USA); CD197 BV786 (CCR7, 3D12), from Biolegend (San Diego, CA, USA). Intra-cytokines staining (ICS) was performed following permeabilization using BD Perm sol II 1X (BD) and included IFNγ FITC, from BD; TNFα AF700 (Mab11), IL-2 PE-Cy7 (MQ1-17H12), IL-21 APC (3A3-N2), from Biolegend. Gating strategy for T cell phenotype is provided in Figure S3.
Serum protein profiling
Serum proteins were analyzed using a multiplex technology based upon proximity-extension assays (Lundberg et al., 2011). Briefly, each kit consisted of a microtiter plate for measuring 92 protein biomarkers in all 88 samples and each well contained 96 pairs of DNA-labeled antibody probes. To minimize inter- and intra-run variation, the data were normalized using both an internal control (extension control) and an interplate control, and then transformed using a pre-determined correction factor. The pre-processed data were provided in the arbitrary unit Normalized Protein Expression (NPX) on a log2 scale and where a high NPX represents high protein concentration.
Quantification and statistical analysis
Statistical analyses were performed using GraphPad Prism 8 (GraphPad Software, Inc., San Diego, CA) and R software (version 3.6.2). Statistical significance was set at p < 0.05 and the test were two-tailed. All data were analyzed D’Agostino-Pearson to assess normality. For non-parametric or parametric data, median or mean are presented, respectively and Mann-Whitney non-parametric test or t test were used for the comparisons, as indicated in figure legends. Chi-square test was used for gender analysis. The AUC values were calculated with the MESS package (version 0.5.6) as described above. Spearman’s correlation was used to examine the association between features. In correlation heatmaps only statistically significant correlations were shown. Linear univariate regression analysis were performed with the “lm” function (stats R package version 3.6.2). MOFA was performed with the MOFA2 package (version 1.0) in the R statistical environment (Argelaguet et al., 2019). Graphpad Prism 8 software was used for statistical analysis (of the cells type distribution and serological parameters) for demographic and routine laboratory blood tests.
Acknowledgments
We would like to thank all of the patients and guardians who participated in the study and all of the CACTUS study nurses team of the COVID-19 Center of Bambino Gesù Children’s Hospital. We would like to particularly thank all of the nurses of the Academic Department of Pediatrics who made this study possible. We also thank Jennifer Faudella and Gulia Neccia for their administrative assistance. We thank Francesco Carmona (IOV-IRCCS, Padova, Italy) for help in the preparation of the samples. This work was made possible by support from Ministry of Health (RRC-2020-23669481 to N.C. and P.P.) and Fondazione Cassa di Risparmio di Padova e Rovigo, Progetti di Ricerca Covid-19 (ADR participant). We also thank Dr. Riccardo Truono for providing graphical support.
Author contributions
Conceptualization, N.C., A.R., F.B., and P.P.; data curation, M.R.P., S.Z., G.R.P., P.Z., M.A.D.I., V.S., E.M., D.A., A.C., D.D., and The Cactus Study Team; formal analysis, A.R. and G.R.P.; funding acquisition, P.R., C.G., A.D.R., N.C., and P.P.; project administration, P.R. and P.P.; investigation, N.C., A.R., F.B., M.R.P., S.Z., A.B., M.P., C.C., and G.L.; methodology, F.B., M.R.P., and A.D.R.; resources, P.B., F.B., A.D.R., and P.P.; supervision, P.R., A.D.R., and P.P.; visualization, N.C., A.R., and G.R.P.; writing – original draft, N.C., A.R., and F.B.; writing – review & editing, N.C., A.R., F.B., D.A., P.R., C.G., A.D.R., and P.P.
Declaration of interests
The authors declare no competing interests.
Published: March 16, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.108852.
Contributor Information
The CACTUS Study Team:
Stefania Bernardi, Lorenza Romani, Paola Pansa, Sara Chiurchiú, Andrea Finocchi, Caterina Cancrini, Laura Lancella, Laura Cursi, Maia De Luca, Renato Cutrera, Libera Sessa, Elena Morrocchi, Chiara Medri, Lorenza Putignani, F.I. Calò Carducci, Patrizia D’Argenio, Marta Ciofi degli Atti, Carmen D’Amore, Livia Piccioni, Martina Di Giuseppe, Alessandro Jenkner, Carmela Giancotta, and Andrzej Krzysztofiak
Supplemental information
References
- Altmann D.M., Boyton R.J. SARS-CoV-2 T cell immunity: specificity, function, durability, and role in protection. Sci. Immunol. 2020;5:eabd6160. doi: 10.1126/sciimmunol.abd6160. [DOI] [PubMed] [Google Scholar]
- Argelaguet R., Clark S.J., Mohammed H., Stapel L.C., Krueger C., Kapourani C.A., Imaz-Rosshandler I., Lohoff T., Xiang Y., Hanna C.W. Multi-omics profiling of mouse gastrulation at single-cell resolution. Nature. 2019;576:487–491. doi: 10.1038/s41586-019-1825-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin P. Why is COVID-19 so mild in children? Acta Paediatr. 2020;109:1082–1083. doi: 10.1111/apa.15271. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay P.K., Yu J., Roederer M. Live-cell assay to detect antigen-specific CD4+ T-cell responses by CD154 expression. Nat. Protoc. 2006;1:1–6. doi: 10.1038/nprot.2006.1. [DOI] [PubMed] [Google Scholar]
- Consiglio C.R., Cotugno N., Sardh F., Pou C., Amodio D., Rodriguez L., Tan Z., Zicari S., Ruggiero A., Pascucci G.R. The Immunology of Multisystem Inflammatory Syndrome in Children With COVID-19. Cell. 2020;183:968–981.e7. doi: 10.1016/j.cell.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotugno N., De Armas L., Pallikkuth S., Rinaldi S., Issac B., Cagigi A., Rossi P., Palma P., Pahwa S. Perturbation of B Cell Gene Expression Persists in HIV-Infected Children Despite Effective Antiretroviral Therapy and Predicts H1N1 Response. Front. Immunol. 2017;8:1083. doi: 10.3389/fimmu.2017.01083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotugno N., Morrocchi E., Rinaldi S., Rocca S., Pepponi I., di Cesare S., Bernardi S., Zangari P., Pallikkuth S., de Armas L., EPIICAL Consortium Early antiretroviral therapy-treated perinatally HIV-infected seronegative children demonstrate distinct long-term persistence of HIV-specific T-cell and B-cell memory. AIDS. 2020;34:669–680. doi: 10.1097/QAD.0000000000002485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotugno N., Zicari S., Morrocchi E., de Armas L.R., Pallikkuth S., Rinaldi S., Ruggiero A., Manno E.C., Zangari P., Chiriaco M. Higher PIK3C2B gene expression of H1N1+ specific B-cells is associated with lower H1N1 immunogenicity after trivalent influenza vaccination in HIV infected children. Clin. Immunol. 2020;215:108440. doi: 10.1016/j.clim.2020.108440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Armas L.R., Cotugno N., Pallikkuth S., Pan L., Rinaldi S., Sanchez M.C., Gonzalez L., Cagigi A., Rossi P., Palma P., Pahwa S. Induction of IL21 in Peripheral T Follicular Helper Cells Is an Indicator of Influenza Vaccine Response in a Previously Vaccinated HIV-Infected Pediatric Cohort. J. Immunol. 2017;198:1995–2005. doi: 10.4049/jimmunol.1601425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GeurtsvanKessel C.H., Okba N.M.A., Igloi Z., Bogers S., Embregts C.W.E., Laksono B.M., Leijten L., Rokx C., Rijnders B., Rahamat-Langendoen J. An evaluation of COVID-19 serological assays informs future diagnostics and exposure assessment. Nat. Commun. 2020;11:3436. doi: 10.1038/s41467-020-17317-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramberg T., Hofmann H., Möller P., Lalor P.F., Marzi A., Geier M., Krumbiegel M., Winkler T., Kirchhoff F., Adams D.H. LSECtin interacts with filovirus glycoproteins and the spike protein of SARS coronavirus. Virology. 2005;340:224–236. doi: 10.1016/j.virol.2005.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans With COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Malhotra N., Gupta N., Agrawal S., Ish P. The curious case of coronavirus disease 2019 (COVID-19) in children. J. Pediatr. 2020;222:258–259. doi: 10.1016/j.jpeds.2020.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh P.R., Huang L.M., Chen P.J., Kao C.L., Yang P.C. Chronological evolution of IgM, IgA, IgG and neutralisation antibodies after infection with SARS-associated coronavirus. Clin. Microbiol. Infect. 2004;10:1062–1066. doi: 10.1111/j.1469-0691.2004.01009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koethe S., Avota E., Schneider-Schaulies S. Measles virus transmission from dendritic cells to T cells: formation of synapse-like interfaces concentrating viral and cellular components. J. Virol. 2012;86:9773–9781. doi: 10.1128/JVI.00458-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zhang Y., Guan Z., Li H., Ye M., Chen X., Shen J., Zhou Y., Shi Z.L., Zhou P., Peng K. SARS-CoV-2 triggers inflammatory responses and cell death through caspase-8 activation. Signal Transduct. Target. Ther. 2020;5:235. doi: 10.1038/s41392-020-00334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L.T., Richardson C.D. The Host Cell Receptors for Measles Virus and Their Interaction with the Viral Hemagglutinin (H) Protein. Viruses. 2016;8:250. doi: 10.3390/v8090250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Cai J., Jia R., Xia S., Wang X., Cao L., Zeng M., Xu J. Dynamic surveillance of SARS-CoV-2 shedding and neutralizing antibody in children with COVID-19. Emerg. Microbes Infect. 2020;9:1254–1258. doi: 10.1080/22221751.2020.1772677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg M., Eriksson A., Tran B., Assarsson E., Fredriksson S. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic. Acids Res. 2011;39:e102. doi: 10.1093/nar/gkr424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A., Netea M.G. Trained Innate Immunity, Epigenetics, and Covid-19. N. Engl. J. Med. 2020;383:1078–1080. doi: 10.1056/NEJMcibr2011679. [DOI] [PubMed] [Google Scholar]
- Meckiff B.J., Ramirez-Suastegui C., Fajardo V., Chee S.J., Kusnadi A., Simon H., Grifoni A., Pelosi E., Weiskopf D., Sette A. Single-Cell Transcriptomic Analysis Of SARS-CoV-2 Reactive CD4 (+) T Cells. bioRxiv. 2020 doi: 10.2139/ssrn.3641939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meffre E., Louie A., Bannock J., Kim L.J., Ho J., Frear C.C., Kardava L., Wang W., Buckner C.M., Wang Y. Maturational characteristics of HIV-specific antibodies in viremic individuals. JCI Insight. 2016;1:e84610. doi: 10.1172/jci.insight.84610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir S., Ho J., Malaspina A., Wang W., DiPoto A.C., O’Shea M.A., Roby G., Kottilil S., Arthos J., Proschan M.A. Evidence for HIV-associated B cell exhaustion in a dysfunctional memory B cell compartment in HIV-infected viremic individuals. J. Exp. Med. 2008;205:1797–1805. doi: 10.1084/jem.20072683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.S. Dysregulation of Immune Response in Patients With Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations With Age and Disease Severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreeder D.M., Cannon J.P., Wu J., Li R., Shakhmatov M.A., Davis R.S. Cutting edge: FcR-like 6 is an MHC class II receptor. J. Immunol. 2010;185:23–27. doi: 10.4049/jimmunol.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux E., Homad L.J., Maccamy A.J., Parks K.R., Hurlburt N.K., Jennewein M.F., Akins N.R., Stuart A.B., Wan Y.H., Feng J. Analysis of a SARS-CoV-2-Infected Individual Reveals Development of Potent Neutralizing Antibodies With Limited Somatic Mutation. Immunity. 2020;53:98–105.e5. doi: 10.1016/j.immuni.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidiq K.R., Sabir D.K., Ali S.M., Kodzius R. Does Early Childhood Vaccination Protect Against COVID-19? Front. Mol. Biosci. 2020;7:120. doi: 10.3389/fmolb.2020.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonn T., Corman V.M., Johnsen M., Richter A., Rodionov R.N., Drosten C., Bornstein S.R. Stability and neutralising capacity of SARS-CoV-2-specific antibodies in convalescent plasma. Lancet Microbe. 2020;1:e63. doi: 10.1016/S2666-5247(20)30037-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valk S.J., Piechotta V., Chai K.L., Doree C., Monsef I., Wood E.M., Lamikanra A., Kimber C., McQuilten Z., So-Osman C. Convalescent plasma or hyperimmune immunoglobulin for people with COVID-19: a rapid review. Cochrane Database Syst. Rev. 2020;5:CD013600. doi: 10.1002/14651858.CD013600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., Mcguire A.T., Veesler D. Structure, Function, and Antigenicity of the Sars-Cov-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg S.P., Connors T.J., Zhu Y., Baldwin M.R., Lin W.H., Wontakal S., Szabo P.A., Wells S.B., Dogra P., Gray J. Distinct antibody responses to SARS-CoV-2 in children and adults across the COVID-19 clinical spectrum. Nat. Immunol. 2021;22:25–31. doi: 10.1038/s41590-020-00826-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., van den Akker J.P.C., Molenkamp R., Koopmans M.P.G., van Gorp E.C.M. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5:eabd2071. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2020. WHO Announces COVID-19 Outbreak a Pandemic.https://www.euro.who.int/en/health-topics/health-emergencies/coronavirus-covid-19/news/news/2020/3/who-announces-covid-19-outbreak-a-pandemic [Google Scholar]
- Yonker L.M., Neilan A.M., Bartsch Y., Patel A.B., Regan J., Arya P., Gootkind E., Park G., Hardcastle M., St John A. Pediatric SARS-CoV-2: Clinical Presentation, Infectivity, and Immune Responses. J. Pediatr. 2020;227:45–52.e5. doi: 10.1016/j.jpeds.2020.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xu J., Jia R., Yi C., Gu W., Liu P., Dong X., Zhou H., Shang B., Cheng S. Protective humoral immunity in SARS-CoV-2 infected pediatric patients. Cell. Mol. Immunol. 2020;17:768–770. doi: 10.1038/s41423-020-0438-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all dataset generated or analyzed during this study.
Scripts to reproduce the analyses presented in each figure of the paper are available at https://github.com/GRP92/PRNT_manuscript. Full datasets are available at https://data.mendeley.com/datasets/stxwzbyfsc/1.