Abstract
Summary
Here we introduce Lipid Mini-On, an open-source tool that performs lipid enrichment analyses and visualizations of lipidomics data. Lipid Mini-On uses a text-mining process to bin individual lipid names into multiple lipid ontology groups based on the classification (e.g. LipidMaps) and other characteristics, such as chain length. Lipid Mini-On provides users with the capability to conduct enrichment analysis of the lipid ontology terms using a Shiny app with options of five statistical approaches. Lipid classes can be added to customize the user’s database and remain updated as new lipid classes are discovered. Visualization of results is available for all classification options (e.g. lipid subclass and individual fatty acid chains). Results are also visualized through an editable network of relationships between the individual lipids and their associated lipid ontology terms. The utility of the tool is demonstrated using biological (e.g. human lung endothelial cells) and environmental (e.g. peat soil) samples.
Availability and implementation
Rodin (R package: https://github.com/PNNL-Comp-Mass-Spec/Rodin), Lipid Mini-On Shiny app (https://github.com/PNNL-Comp-Mass-Spec/LipidMiniOn) and Lipid Mini-On online tool (https://omicstools.pnnl.gov/shiny/lipid-mini-on/).
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The examination of lipidomics data for pattern recognition and biological interpretation remains highly manual. In addition, researchers with limited background in lipidomics (e.g. classification and structure) often have difficulty in knowing where to begin examining the data. Ontology enrichments are a powerful way to identify shared characteristics of biomolecules. Numerous tools are available to assess the enrichment on sets of proteins and genes (Huang da et al., 2009; Yu et al., 2015) however, such tools for lipids remain sparse. Here we introduce an R-based, open-source, lipid enrichment tool suite, Lipid Mining and Ontology (Lipid Mini-On) that is database independent. Lipid Mini-On uses a text-mining process partitioning individual lipid species into multiple ontology groups based on LipidMaps classification (Sud et al., 2007) and other molecular characteristics (e.g. chain length and number of double bonds). As such, it can perform enrichment analysis on any lipid based on the LipidMaps annotation scheme [e.g. PC(16: 0/18: 1)]. This approach enables the enrichment of classes of lipids currently absent from databases or those yet to be discovered.
2 Implementation
Lipid Mini-On was developed using R 3.4.0. (Team, 2013). It is composed of an R package, Rodin, and of a Shiny-based graphical user interface (Chang et al., 2018) that does not require knowledge of statistical programing. The parsing function of Rodin [i.e. ‘lipid.miner()’] uses regular expressions to extract details from the lipid names into three R objects named ‘intact’, ‘chains’ and ‘allchains’ (Fig. 1A). The current Lipid Mini-On database contains 6 lipid categories, 41 main classes and 107 subclasses (Supplementary Table S1) and can easily be extended to include additional groupings.
Fig. 1.
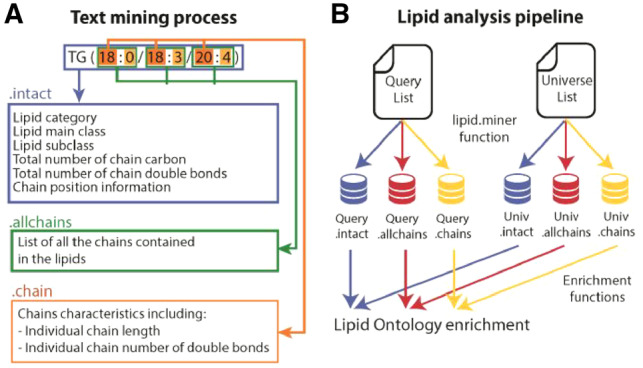
Lipid Mini-On text-mining pipeline. (A) Text-mining process parses lipid names into three distinct objects entitled .intact [e.g. TG(18:0/18:3/20:4)], .allchains (e.g. 56:7) and .chains (e.g. 18:0). (B) Based on the classification in (A) LO terms are mined from the query and universe files and compared for LO enrichment
Lipid Mini-On (i) creates lipid ontology (LO) terms associated with lists of lipids; (ii) compares lipid lists (e.g. query versus universe) using statistical methods for over-representation of LOs (Fig. 1B); (iii) generates a graphical representation of results and (iv) creates an editable network of relationships between individual lipids and their associated LOs. Five types of statistical approaches to identify the enriched LO terms in a ‘Query’ list compared to a ‘Universe’ list are available (Supplementary Material).
Rodin incorporates an array of visualization functions utilizing the ggplot2 package (Wickham, 2016). These functions can be grouped in four categories: (i) ‘classification visualization’, (ii) ‘fatty acid characteristics’ that allow the visualization of the chain length and the unsaturation, (iii) ‘specific chains’ that allows the visualization of the chains at the lipid category, main class, and subclass levels. The final (iv) category of visualization functions enables the construction of enrichment networks, allowing the user to visualize the relationships between the LO enriched and their corresponding lipids.
The Shiny application was developed in a tabular fashion. The query and universe list of lipids are uploaded and processed in the ‘Upload’ tab. Once the data have been successfully uploaded, users can perform the ‘Enrichment Analysis’, and then visualize the results in the ‘Visualize’ and ‘Results Network’ tabs (Supplementary Material).
3 Results
To demonstrate the capabilities of Lipid Mini-On, an enrichment analysis was conducted on two contrasting sample types, human lung endothelial cells and peat soil. For the lung, 293 lipids were identified across the four major lung cell types isolated from healthy donors (Supplementary Table S2), of which 74 were significantly modulated (P < 0.05) in endothelial cells (Supplementary Table S3) (Kyle et al., 2018). Using all parameters for the enrichment analysis, 12 LOs were found to be enriched based on a Fisher’s exact test (P < 0.05, Supplementary Table S4). Graphical displays of the results are available in Supplementary Figures S1–S3. Similarly to Gene Ontology, LO terms created by Lipid Mini-On can present a certain level of redundancy. For example, the ceramide [Cer] main class and subclass [Cer(d)] contain the same eight lipids (Supplementary Tables S2–S4). Although both are enriched, the subclass is driving the main class enrichment (Supplementary Fig. S4).
Peat soils were collected from the SPRUCE experiment in Minnesota, USA, Wisconsin (Supplementary Material) where the surface soil were compared to those at 90 cm depth. Of the 162 lipids identified (Kyle et al., 2017), 33 were statically significant at 90 cm depth (Supplementary Tables S5–S6). A total of 18 LOs were significantly modulated (P < 0.05) based on Fisher’s exact tests (Supplementary Table S7). Graphical displays are available in Supplementary Figures S5–S9. Betaine and chloroplast lipids (DGDG and SQDG) were not enriched at depth, as expected. Cer subclasses, coenzyme Q, triacylglycerides with a total number of 47 chain carbons and no double bonds and glycerophospholipids with the chain 19:1 were enriched at depth (Supplementary Table S7). Very long chain fatty acids were also enriched with the network highlighting the direct connection to the sphingolipids (Supplementary Fig. S10).
Lipid Mini-On provides a rapid enrichment analysis using text-mining functions. As biological information can be extracted from the lipid name, this approach provides structural information and directs the users to significant trends. For example, in the lung endothelium, [PI ()] with the chain 20:4 are enriched. PI lipids are a common subclass for obtaining 20:4, as arachidonic acid, for downstream generation of signaling molecules. For the soil, saturated and odd chained lipids are enriched at depth highlighting a more prokaryotic and nutrient deficient system.
Supplementary Material
Acknowledgements
Donor lung tissues were supplied through the United Network for Organ Sharing. We are grateful to the families who have generously given such precious gifts.
Funding
This work was supported by the National Heart Lung Blood Institute [HL122703 to C.A.] of the National Institute of Health. Lipidomics analyses were performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the USA. Department of Energy and located at Pacific Northwest National Laboratory (PNNL) in Richland, WA. PNNL is a multi-program national laboratory operated by Battelle for the DOE under contract DE-AC05-76RLO 1830.
Conflict of Interest: none declared.
References
- Chang W. et al. (2018) Shiny: Web Application Framework for R. R Package Version 1.2.0. https://CRAN.R-project.org/package=shiny.
- Huang da W. et al. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res., 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyle J.E. et al. (2018) Cell type-resolved human lung lipidome reveals cellular cooperation in lung function. Sci. Rep., 8, 13455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyle J.E. et al. (2017) LIQUID: an-open source software for identifying lipids in LC-MS/MS-based lipidomics data. Bioinformatics, 33, 1744–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sud M. et al. (2007) LMSD: lIPID MAPS structure database. Nucleic Acids Res., 35, D527–D532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team R. (2013) R: a Language and Environment for Statistical Computing. https://www.r-project.org/.
- Wickham H. (2016) ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag, New York. [Google Scholar]
- Yu G. et al. (2015) DOSE: an R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics, 31, 608–609. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


