Abstract
Motivation
The identification of enhancer–promoter interactions (EPIs), especially condition-specific ones, is important for the study of gene transcriptional regulation. Existing experimental approaches for EPI identification are still expensive, and available computational methods either do not consider or have low performance in predicting condition-specific EPIs.
Results
We developed a novel computational method called EPIP to reliably predict EPIs, especially condition-specific ones. EPIP is capable of predicting interactions in samples with limited data as well as in samples with abundant data. Tested on more than eight cell lines, EPIP reliably identifies EPIs, with an average area under the receiver operating characteristic curve of 0.95 and an average area under the precision–recall curve of 0.73. Tested on condition-specific EPIPs, EPIP correctly identified 99.26% of them. Compared with two recently developed methods, EPIP outperforms them with a better accuracy.
Availability and implementation
The EPIP tool is freely available at http://www.cs.ucf.edu/˜xiaoman/EPIP/.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Enhancers play important regulatory roles. They control the expression patterns of their target genes by directly interacting with the promoter regions of those genes (Mora et al., 2016). Even though enhancers may be tens of kilobases (kb) away from their target genes, they get in direct contact with the promoters of their target genes via chromatin looping (Cai et al., 2010; Dekker et al., 2002; De Laat and Duboule, 2013; Zheng et al., 2015). Because of the long range of possible distances [1 kb to several megabases (Mb)] between enhancers and their targeted promoters, it is challenging to predict enhancer–promoter interactions (EPIs) (De Laat and Duboule, 2013). To date, the majority of EPIs under specific experimental conditions have not been discovered yet (Corradin et al., 2014).
Experimental approaches for identifying EPIs are mainly based on chromosome conformation capture (3C) and its variants such as chromatin interaction analysis with paired-end tag sequencing (ChIA-PET) and high throughput genome-wide 3C (Hi-C) (Dekker et al., 2002; Fullwood et al., 2009; Rao et al., 2014). These experimental techniques determine the relative frequency of direct physical contacts between genomic regions and have successfully identified EPIs and other long-range interactions (He et al., 2014). However, the ChIA-PET method still has a low signal-to-noise ratio and most available Hi-C data have a low resolution (Fullwood et al., 2009; Rao et al., 2014). In addition, since certain EPIs are condition-specific, experimental EPI data in one sample cannot always be directly applied to infer EPIs in other samples. Here, a ‘sample’ refers to a cell type, a cell line or a tissue sample under a specific experimental condition. An EPI is called condition-specific, if the interaction only occurs in a specific sample.
As most experimental procedures are expensive, computational methods have been indispensable alternatives for identifying EPIs. These methods employ available genomic and/or epigenomic data to predict EPIs in an inexpensive way. Early methods considered the closest promoter as the only target of an enhancer. However, a study demonstrated that only 40% of enhancers regulate their nearest promoters and one enhancer may regulate multiple genes (Andersson et al., 2014). Later, several computational approaches were developed based on the correlation of epigenomic signals in enhancers and those in promoters (Andersson et al., 2014; Corradin et al., 2014; Ernst et al., 2011; Thurman et al., 2012). One challenge of using these methods is to find a proper threshold of correlations to reduce false EPI predictions (Roy et al., 2015; Whalen et al., 2016). Recently, supervised learning-based methods have been developed, such as IM-PET (He et al., 2014), PETModule (Zhao et al., 2016), Ripple (Roy et al., 2015) and TargetFinder (Whalen et al., 2016). These methods commonly use genomic and epigenomic data such as those from DNase I hypersensitive sites sequencing (DNase-seq) and histone modification-based chromatin immunoprecipitation followed by massive parallel sequencing (ChIP-seq) to extract features for EPI predictions. IM-PET, Ripple and PETModule utilize random forests as their classifier, while TargetFinder is based on boosted trees. These methods either do not consider or have low performance on condition-specific EPI predictions (Roy et al., 2015).
Here, we proposed a computational method for predicting condition-specific EPIs called EPIP. EPIP stands for ‘Enhancer–Promoter Interaction Prediction’. It is a supervised learning-based approach that utilizes functional genomic and epigenomic data to build a robust model to predict shared and condition-specific EPIs. EPIP can work with missing data, different types of datasets and even a dataset with a partial list of features. Tested on experimental data from more than eight samples, EPIP reliably predicted condition-specific EPIs and shared EPIs in different samples with the average area under the receiver operating characteristic curve (AUROC) about 0.95, and the average area under the precision–recall curve (AUPR) about 0.73. In addition, we compared EPIP with two state-of-the-art computational methods for predicting EPIs and showed that EPIP outperformed both.
2 Materials and methods
2.1 Enhancers and promoters
We obtained all 32 693 enhancers annotated by FANTOM from http://slidebase.binf.ku.dk/human_enhancers/results (Andersson et al., 2014). We chose this set of enhancers because this was arguably the largest set of enhancers that were defined with the same criteria and supported by experiments. We next overlapped the FANTOM enhancers with the computationally predicted ChromHMM enhancers (Ernst et al., 2011) for samples that had the ChromHMM data available (GM12878, HeLa, HMEC, HUVEC, IMR90 and NHEK). These ChromHMM enhancers were defined with 15 hidden states (https://genome.ucsc.edu/cgi-bin/hgTrackUi? g=wgEncodeBroadHmm&db=hg19). We considered both strong and weak enhancers (states 4–7) as valid ChromHMM enhancers (Ernst et al., 2011). The FANTOM enhancers overlapping with at least one ChromHMM enhancer in a sample were considered as the enhancers for that sample in the following analyses. Since KBM7 does not have any annotated ChromHMM enhancer, all FANTOM enhancers were used for KBM7. An enhancer was considered ‘active’ in a sample if it overlapped with the H3K27ac ChIP-seq peaks in this sample. The H3K27ac peaks were downloaded from ENCODE (Dunham et al., 2012). With no H3K27ac data available for KBM7, all obtained enhancers were considered as ‘active’ enhancers. In this way, we obtained 7023–32 693 enhancers and 4888–32 693 active enhancers in a sample (Supplementary Table S1).
We obtained all annotated transcription start sites (TSSs) from GENCODE V19 (Harrow et al., 2012) and considered the regions between 1 kb upstream and 100 base pairs downstream of the TSSs as ‘promoters’. This resulted in 57 783 promoters. For samples with RNA-Seq data (Dunham et al., 2012) (GM12878, HeLa, HUVEC, IMR90, K562 and NHEK), we defined promoters as ‘active’ if the corresponding genes had at least 0.30 reads per kb of transcript per million mapped reads with the irreproducible discovery rate of 0.1, similarly as previously (Whalen et al., 2016). For samples without RNA-Seq data (HMEC and KBM7), all promoters were considered as active promoters (Supplementary Table S2).
2.2 Training data
To train EPIP, we defined positive and negative enhancer–promoter pairs (EP-pairs) (i.e. interacting and non-interacting EP-pairs) using the normalized Hi-C contact matrices, which were generated with the Knight and Ruiz normalization vectors by Rao et al. (2014). Rao et al. inferred these matrices for the following seven samples: GM12878, HMEC, HUVEC, IMR90, K562, KBM7 and NHEK (GSE63525). They also extracted significant intra-chromosomal chromatin interactions called ‘looplists’ in the above seven samples and the HeLa sample. The number of EP-pairs from the looplists defined at the highest resolution for these samples was too small to train the EPIP model well. We thus defined the positive and negative EP-pairs from their normalized Hi-C contact matrices, as previously (Li et al., 2016; Zhao et al., 2016) (Supplementary Table S3).
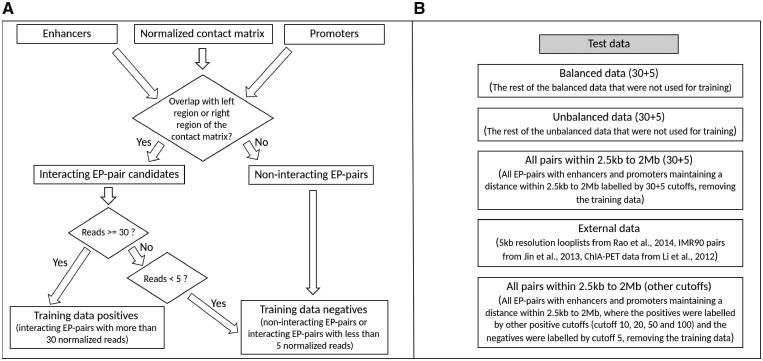
In brief, if an ‘active’ enhancer and an ‘active’ promoter overlapped with a pair of regions that were supported by at least 30 normalized Hi-C reads, we considered this EP-pair as a positive EP-pair. Similarly, an EP-pair was considered as negative if it did not overlap with any pair of regions that were supported by 5 or more normalized Hi-C reads (Fig. 1A). The cutoffs, 30 and 5, were chosen based on our test results with different cutoffs (Supplementary Table S3). In this way, we defined positive and negative EP-pairs for the above seven samples with contract matrices.
Fig. 1.
(A) The flowchart of training data creation. Here, all the read numbers are normalized. An EP-pair with the enhancer overlapping with one of the two interacting regions and the promoter overlapping with the other of the two interacting regions will be considered as an interacting EP-pair candidate. (B) The five test datasets on which we tested EPIP
To train EPIP, we used both balanced and unbalanced models. We randomly chose 30% of positive EP-pairs and the same number of negative EP-pairs in each of the above seven samples. We then combined these positives and negatives from different samples to train a balanced prediction model. We also combined 30% of positive EP-pairs and 10 times randomly chosen negative EP-pairs in each sample to train an unbalanced prediction model. We then combined the two models into the final EPIP model, which predicts an EP-pair as a ‘negative’ pair only when both models predict this pair as a negative pair and predicts an EP-pair as a positive pair otherwise. This strategy was based on the observation that the balanced model had a high sensitivity and the unbalanced model had a high specificity when tested on the training data by cross-validation. For simplicity’s reason, in the remaining of the paper, we called this final EPIP model as ‘EPIP’.
2.3 Testing data
We tested EPIP on a variety of data (Fig. 1B). We tested it on the remaining 70% of positive EP-pairs, together with the same number of randomly selected negative pairs that were not used for training (balanced test data). We also tested it on the remaining 70% of positive EP-pairs together with 10 times randomly selected negative pairs that were not used for training (unbalanced test data). We tested EPIP on all EP-pairs within 2 Mb that were not used for training as well. Moreover, we tested EPIP on the positive EP-pairs defined with normalized Hi-C contact matrices under the cutoffs 10, 20, 30, 50 and 100. Finally, we tested EPIP on EP-pairs collected in other studies (Jin et al., 2013; Li et al., 2012; Rao et al., 2014), which were obtained from the strictly defined interacting regions by the original studies and represented more strictly defined EP-pairs.
2.4 Features of EP-pairs considered
EPIP considers three common features of EP-pairs in every sample. These features are the distance between the enhancer and the promoter in an EP-pair, the conserved synteny score that measures the co-conservation of an EP-pair in five other vertebrate genomes (chicken galGal3, chimpanzee panTro4, frog xenTro3, mouse mm10 and zebrafish zv9) and the correlation of epigenomic signals in the enhancer region and that in the promoter region of an EP-pair across ENCODE Tiers 1 and 2 samples (Zhao et al., 2016). For simplicity’s sake, in the following, these features are called ‘distance’, ‘css’ and ‘corr’, respectively.
In addition, depending on the types of data available in a sample, EPIP considers features from 14 additional types of data. These include DNase-seq data, ChIP-seq data for nine types of histone modifications (H3K4me1, H3K4me2, H3K4me3, H3K27ac, H3K27me3, H3K36me3, H3K79me2, H3K9ac and H4K20me1) and four types of chromatin factors (CTCF, POL2, RAD21 and SMC3). These data are shown to provide important indicators for predicting EPIs (Roy et al., 2015). For each of the 14 types of data, EPIP generates two features that correspond to its signals in enhancer regions and its signals in promoter regions (Supplementary Table S4). The value of a feature for a region corresponds to the ‘peak strength’ value of this feature in its signal peak that overlapped with this region. If a region overlaps with multiple peaks of a feature signal, we considered the average signal value in these overlapping peaks as the feature value for this region. For instance, when H3K4me1 ChIP-seq data is available for an enhancer in a sample, the H3K4me1 feature value for this enhancer is the average peak strength of all H3K4me1 ChIP-seq peaks overlapping with this enhancer. The feature signal peaks and their signal strength are downloaded from ENCODE (Dunham et al., 2012). Due to the difference of available types of data in different samples, we could consider 31, 25, 27, 31, 31, 3 and 27 features in GM12878, HMEC, HUVEC, IMR90, K562, KBM7 and NHEK, respectively, including the three common features (Supplementary Table S4).
2.5 Partitioning feature space to handle missing data
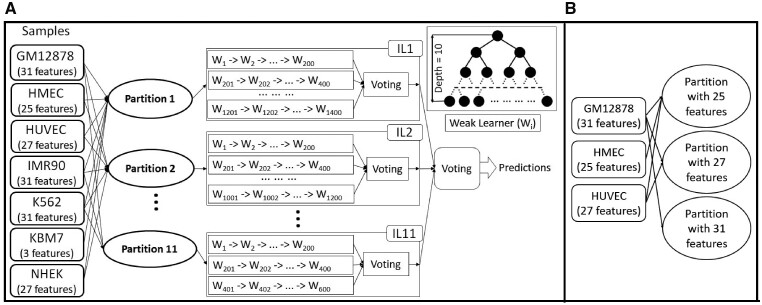
EPIP groups features into 11 partitions or overlapping feature sets (Fig. 2A, Supplementary Table S5). Partitions with overlapping features are used, because in this way, (i) EPIP can be trained and tested on various samples, no matter whether the samples have data for a large or small number of features; (ii) such partitions enable more samples to be used to train each partition and thus likely produce more accurate predictors; and (iii) the trained EPIP model can be used to make predictions in more samples. For instance, samples with a large number of features will benefit from a large number of partitions, while EPIP can still make predictions for samples with a small number of features.
Fig. 2.
(A) The training process of EPIP. There are three types of partitions and in total 11 partitions used. Samples with the features required by a partition are used to train the corresponding IL for this partition. Each IL trains a maximum 200 weak learners (W) for a sample. The weak learners trained from all available samples then vote to make the predictions for the corresponding IL. The prediction of all ILs determines the final prediction with another voting process. (B) An example of the third type of partitions from three samples. The 25 features in HMEC are included in the 27 features in HUVEC, which are included in the 31 features in GM12878
In brief, EPIP considers the three common features: distance, css and corr, as a partition, which shows the static genomic information of EPIs. Moreover, EPIP considers the above three common features together with features from each of the following feature groups as a different partition: H3K4me1; DNase-seq; H3K4me1 and H3K27ac; DNase-seq and H3K27ac; H3K4me1 with H3K27ac and H3K4me3; DNase-seq with H3K27ac and H3K4me3; H3K4me1-3 together with H3K27ac and DNase-seq (Supplementary Table S5). Note that for every histone modification mark, EPIP considers its signal values in enhancers and in promoters. Therefore, the above partitions have two features for every histone modification mark (such as H3K4me1_P and H3K4me1_E for the mark H3K4me1 in promoters and enhancers, respectively). Finally, EPIP considers the largest sets of features that are not considered above and occur in at least one training sample as additional partitions (Fig. 2B).
2.6 An ensemble approach to predict condition-specific EPIs under a variety of conditions
We developed an ensemble method called EPIP to distinguish positive from negative EP-pairs (Fig. 2A). The method uses the training data to train the model iteratively, based on the idea of AdaBoost (Polikar et al., 2000). The details are in the following paragraphs.
With the above partitions of features, EPIP trains an incremental learner (IL) for every partition. An IL consists of a number of weak learners trained on the training data. The weak learners in EPIP are decision tree classifiers. We set a maximum allowed depth of 10 for the decision trees to avoid overfitting, after testing on several depth options. For a given partition and a corresponding sample, EPIP trains 200 weak learners, because 200 is the smallest number that gives EPIP the highest AUROC and AUPR scores (Supplementary Fig. S1). With weak learners trained on data from all samples with the required features by the corresponding partition, the corresponding IL combines predictions from all its weak learners through a majority voting. EPIP then combines the predictions by the trained ILs from all partitions by a majority voting (Fig. 2A).
EPIP trains weak learners iteratively. Given a partition and a sample with the required features by this partition, the first weak learner is trained with equal weight to all data points (i.e. all EP-pairs) in the training data in this sample. The second weak learner is then trained with the same input data points but with increased weights to the misclassified data points and decreased weight to the correctly classified data points by the first weak learner. The modified weights to the data points are calculated according to the AdaBoost algorithm, based on the prediction errors and the average weights of the misclassified data points by the previous weak learners (Freund and Schapire, 1997). Similarly, the third weak learner is trained with the same input data points but with increased weights to the misclassified data points and decreased weight to the correctly classified data points by the first two weak learners. This process is repeated again and again until the 200th weak learner is trained.
With a new sample that has at least a subset of the aforementioned 14 types of data, EPIP is able to learn from this sample without retraining the whole model. EPIP identifies the partitions applicable to the new sample. For these partitions, similarly as above, the corresponding ILs learn the new training sample by an additional set of 200 weak learners iteratively trained with the new training data. EPIP then combines the predictions from the updated ILs and the originally trained ILs by the majority voting.
2.7 Comparison with TargetFinder and Ripple
TargetFinder predicted EPIs in six samples (GM12878, HeLa, HUVEC, IMR90, K562 and NHEK) (Whalen et al., 2016). It provided positive and negative EP-pairs used, together with its prediction by four classifiers (https://github.com/shwhalen/targetfinder). One of the classifiers, gradient boosting (gbm), showed a better precision and recall than the other three classifiers. We thus compared EPIP with TargetFinder by running EPIP and TargetFinder (gbm) on both the TargetFinder data and EPIP data.
Ripple uses a combination of random forests and group LASSO in a multi-task learning framework (Roy et al., 2015). It uses different types of data such as DNase-seq, ChIP-seq and RNA-Seq data to extract features. Ripple is trained on multiple samples and is capable of predicting condition-specific interactions in a new sample. Its training and test data were based on the 5C (GSE39510) and Hi-C (GSE63525) datasets. We compared EPIP with Ripple by running them on EPIP data and the above TargetFinder data in three shared samples (GM12878, HeLa and K562). We did not compare them on the Ripple data, as (i) the Ripple data is balanced, which does not represent the reality well, where we often have much more negatives than positive EP-pairs; (ii) the resolution of the Ripple data is low, where a promoter within 2.5 kb of a pair of interacting regions may be considered as the targets of one of the regions; and (iii) the data barely overlap with any FANTOM enhancer.
We used the 10-fold cross-validation method to train and test EPIP, TargetFinder and Ripple, similarly as that in the TargetFinder study and in the Ripple study. We used the generate_training.py in TargetFinder to generate TargetFinder features for EP-pairs. Then we used the procedure mentioned in its readme file to apply the 10-fold cross-validation on the training data using GradientBoostingClassifier (GBM). In terms of Ripple, we used the genFeatures tool in Ripple to generate features for EP-pairs. Then we used its runAllfeatures_crosscellline.m Matlab code to apply 10-fold cross-validation on the training data.
3 Results
3.1 EPIP reliably predicts EPIs
We tested EPIP on the balanced test data, unbalanced test data, all EP-pairs within 2.5 kb to 2 Mb, EP-pairs defined with different normalized Hi-C contact number cutoffs and EP-pairs from other studies. EPIP reliably predicted untrained EPIs in all datasets, with a high AUROC, AUPR and/or F1 score (Table 1 and Supplementary Tables S6–S8). The AUROC, AUPR and the F1 score were calculated using the scikit-learn libraries (Pedregosa et al., 2011).
Table 1.
The performance of EPIP on all pairs within 2.5 kb and 2 Mb, balanced and unbalanced test data
| Cell line | AUROC | AUPR | F1 | Precision | Sensitivity/recall |
|---|---|---|---|---|---|
| GM12878 | 0.9007 (0.994, 0.9941) | 0.8502 (0.9939, 0.9541) | 0.9337 (0.9942, 0.9862) | 0.8772 (0.9979, 0.979) | 0.9979 (0.9906, 0.9936) |
| HMEC | 0.9872 (0.9962, 0.9981) | 0.9038 (0.9963, 0.9929) | 0.4287 (0.9893, 0.9415) | 0.2733 (0.9873, 0.8955) | 0.9938 (0.9913, 0.9925) |
| HUVEC | 0.9842 (0.9943, 0.9934) | 0.8904 (0.9952, 0.9807) | 0.3081 (0.9844, 0.9369) | 0.1824 (0.987, 0.8978) | 0.9915 (0.9818, 0.9796) |
| IMR90 | 0.9963 (0.9985, 0.9981) | 0.9894 (0.9989, 0.995) | 0.6365 (0.9933, 0.9577) | 0.4672 (0.9921, 0.9247) | 0.9985 (0.9946, 0.9932) |
| K562 | 0.9947 (0.9985, 0.9986) | 0.9874 (0.9988, 0.9939) | 0.5589 (0.9884, 0.9486) | 0.3887 (0.9909, 0.9141) | 0.9938 (0.9859, 0.9859) |
| KBM7 | 0.9856 (0.9856, 0.9861) | 0.9285 (0.9876, 0.9509) | 0.5317 (0.9864, 0.9328) | 0.3629 (0.9876, 0.8905) | 0.994 (0.9852, 0.9793) |
| NHEK | 0.9942 (0.9979, 0.9981) | 0.9394 (0.9982, 0.9939) | 0.4485 (0.9892, 0.9406) | 0.2897 (0.9899, 0.8969) | 0.9926 (0.9885, 0.9887) |
Note: The scores for all pairs within 2.5 kb to 2 Mb are shown as the first value. The scores for balanced and unbalanced data are shown in parentheses.
We studied the performance of EPIP on the balanced test data, the unbalanced test data and all EP-pairs within 2.5 kb to 2 Mb (Section 2). No EP-pair in these test data was used for training. With five sets of randomly chosen training data and the corresponding test data, on average, EPIP had an AUROC of 0.96, 0.96 and 0.95; an AUPR of 0.96, 0.92 and 0.73 and an F1 score of 0.99, 0.95 and 0.51 for the balanced, unbalanced and all EP-pairs within 2.5 kb to 2 Mb test data, respectively (Table 1 and Supplementary Table S6). Note that the F1 score on the third test data was not bad, given the fact that the number of negatives was around 13 times the number of positives here. In this test dataset, the recall in all samples was higher than 0.92, although KBM7 had no epigenomic data. The average precision was 0.34 in these samples, with the largest precision in GM12878, where the Hi-C sequencing depth was the highest. The much higher precision and F1 score in GM12878 suggest that the estimated precision and F1 scores may be underestimated in other samples, as the lower sequencing depth in other samples that may prevent from labeling many true positive pairs as positives while these positives were indeed predicted as positives by EPIP. We also tested EPIP on condition-specific EP-pairs within 2.5 kb to 2 Mb. EPIP predicted 12 455 (99.26%) of the 12 548 condition-specific EP-pairs in the seven samples that were not used for training.
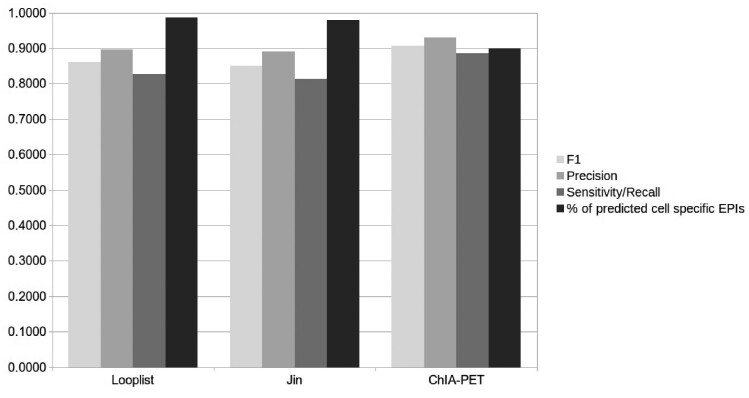
Since the above test datasets were based on the cutoffs 30 and 5, which were not rigorously determined, we tested EPIP on more strictly defined EP-pairs. We tested EPIP on EP-pairs defined with the looplists at 5 kb resolution Hi-C data by Rao et al. (2014), the Hi-C data for IMR90 published by Jin et al. (2013) and the ChIA-PET data for K562 and MCF7 (Li et al., 2012). The EP-pairs were similarly obtained by overlapping ‘active’ enhancers and ‘active’ promoters with the strictly defined interacting regions in these studies. On average, EPIP had a precision of 0.90, 0.89 and 0.93, respectively; a recall of 0.83, 0.81 and 0.89, respectively; and an F1 score of 0.86, 0.85 and 0.91, respectively (Fig. 3 and Supplementary Table S7). Note that although EPIP was not trained on MCF7, EPIP correctly predicted 89.70% of EPIs in MCF7.
Fig. 3.
The overall performance of EPIP on external datasets
We also tested EPIP on all EP-pairs within 2.5 kb to 2 Mb with positives defined by different cutoffs (Supplementary Table S8). We found that when the cutoff was increasing, overall, the AUROC was increasing while the AUPR and the F1 scores were decreasing. This was due to the fact that the number of negatives was the same for different cutoffs, while the number of positive EP-pairs was decreased with the larger cutoffs. Under all cutoffs, the recall (sensitivity) was larger than 0.92 and was increasing with the increment of the cutoff, suggesting that the trained EPIP model was robust and reliable to predict true positive EP-pairs. This was because although EPIP was trained with data under the cutoff 30, it predicted the vast majority of ‘positive’ EP-pairs under different cutoffs and became more accurate when tested on more ‘strictly defined’ positive EP-pairs. The average specificity was 0.80 and unchanged under different cutoffs, since the negative EP-pairs were the same. The average precision was decreasing from 0.76 at the cutoff 10 to 0.09 at the cutoff 100. The dramatically decreasing precision suggested that the larger cutoffs such as 50 and 100 were likely too stringent and the cutoff 30 may be more proper to define positives, given the fact that EPIP had good precision and recall on more strictly defined EP-pairs from the above three previous studies (Fig. 3).
In summary, EPIP reliably predicted EPIs especially condition-specific ones. Its high precision, recall and F1 scores were demonstrated with the published datasets from previous studies. The more realistic measurement of its AUROC and AUPR may come from the test data with all EP-pairs defined with the cutoffs 30 and 5, supported by our analyses with the published datasets and different cutoffs. In this case, EPIP on average had an AUROC of 0.95 and an AUPR of 0.73.
3.2 EPIP reliably predicts condition-specific EPIs in new samples
We studied the performance of EPIP on predicting EP-pairs especially condition-specific ones in new samples. We trained one EPIP model on EP-pairs in every group of six samples and then tested the model on the remaining sample, for each of the seven samples with a normalized Hi-C contact matrix. Following the same way we obtained the EPIP model, we defined the training positive and negative EP-pairs with the cutoffs 30 and 5, and obtained the EPIP model with the combination of the balanced and unbalanced models trained on the corresponding six samples.
Trained similarly as above on positive pairs and two sets of negative pairs from six samples, on average, EPIP had an AUROC of 0.96, an AUPR of 0.89, on the seventh sample, when tested on all EP-pairs within 2.5 kb to 2 Mb based on the cutoffs 30 and 5 (Table 2 and Supplementary Table S8). In terms of condition-specific EP-pairs, which only occurred in the seventh sample, on average, EPIP predicted 5498 (97.66%) of 5630 condition-specific EP-pairs in seven samples except for GM12878. EPIP predicted only 31.77% of condition-specific EP-pairs in GM12878 (Table 2).
Table 2.
The performance of cell-specific EPIP models on all pairs within 2.5 kb to 2 Mb
| Cell line | AUROC | AUPR | F1 | Precision | Sensitivity/recall | Number of cell-specific EPIs | % of predicted cell-specific EPIs |
|---|---|---|---|---|---|---|---|
| GM12878 | 0.9873 | 0.9835 | 0.9385 | 0.9946 | 0.8885 | 58765 | 0.9975 |
| HMEC | 0.9951 | 0.9905 | 0.7664 | 0.6256 | 0.9889 | 42 | 0.8333 |
| HUVEC | 0.9920 | 0.9816 | 0.6955 | 0.5387 | 0.9810 | 73 | 0.8767 |
| IMR90 | 0.9979 | 0.9960 | 0.9286 | 0.8714 | 0.9938 | 2781 | 0.9960 |
| K562 | 0.9955 | 0.9921 | 0.9093 | 0.8421 | 0.9881 | 1178 | 0.9677 |
| KBM7 | 0.9892 | 0.9826 | 0.8431 | 0.7377 | 0.9836 | 2004 | 0.9880 |
| NHEK | 0.9942 | 0.9791 | 0.7675 | 0.6275 | 0.9879 | 102 | 0.8235 |
Note: The number and percentage of cell-specific EP-pairs predicted are also shown in the last two columns.
We hypothesized that the low performance in GM12878 was likely due to the fact that the Hi-C sequencing depth was much higher in GM12878 than that in other samples. In other words, the quality of the EP-pairs in other samples was different from that in GM12878. To test this hypothesis, we applied the same EPIP model trained on other six samples based on the cutoffs 30 and 5 to test condition-specific EP-pairs defined with the cutoff 100 in GM12878. We found that EPIP correctly predicted 2396 (78.69%) of 3045 condition-specific EP-pairs in GM12878 (Supplementary Table S9). Therefore, EPIP indeed can reliably predict condition-specific EP-pairs in new samples, with an accuracy of 91.00% (7894 out of 8675 condition-specific EPIs) in all seven samples.
3.3 EPIP performs better than the state-of-the-art methods TargetFinder and Ripple
We compared EPIP with two recently published methods, TargetFinder and Ripple. We compared them on the TargetFinder data and the EPIP all EP-pair test data within 2 kb to 2 Mb. EPIP showed better performance than TargetFinder and Ripple (Table 3).
Table 3.
The performance comparison between EPIP with TargetFinder and Ripple for the two types of data considered
| # Pos | # Neg | F1 | Precision | Sensitivity/recall | ||
|---|---|---|---|---|---|---|
| TargetFinder data | EPIP versus TargetFinder | 1394 | 197 | 0.9018 (0.6433) | 0.9182 (0.997) | 0.8859 (0.4749) |
| EPIP versus Ripple | 67 | 18 | 0.8824 (0.275) | 0.8696 (0.8462) | 0.8955 (0.1642) | |
| EPIP data | EPIP versus TargetFinder | 912 | 197 | 0.9431 (0.2507) | 0.8924 (0.985) | 1 (0.1436) |
| EPIP versus Ripple | 3230 | 79867 | 0.917 (0.2271) | 0.8474 (0.7238) | 0.9991 (0.1347) |
Note: F1, precision and sensitivity/recall columns show the EPIP score first, and TargetFinder or Ripple score in parentheses.
First, we compared EPIP with TargetFinder and Ripple on the data from TargetFinder (Table 3 and Supplementary Table S10). For the six samples in TargetFinder data (GM12878, HeLa, HUVEC, IMR90, K562 and NHEK), on average, EPIP had an AUROC, AUPR, F1, precision, recall and specificity of 0.95, 0.84, 0.64, 0.98, 0.48 and 1.00, respectively. TargetFinder had an AUROC, AUPR, F1, precision, recall and specificity of 0.92, 0.59, 0.50, 0.72, 0.39 and 0.99, respectively (Table 3). Ripple had an AUROC, AUPR, F1, precision, recall and specificity of 0.75, 0.19, 0.02, 0.75, 0.01 and 1.00, respectively (Table 3). The poor performance of Ripple may be due to the fact that Ripple could not deal with unbalanced data well, although the real data are always unbalanced in practice.
Next, we compared EPIP with TargetFinder and Ripple on all EP-pairs test data within 2.5 kb to 2 Mb (Table 3 and Supplementary Table S11). For the five samples shared with the TargetFinder study (GM12878, HUVEC, IMR90, K562, NHEK), on average, EPIP had an AUROC, AUPR, F1, precision, recall and specificity of 1.00, 0.98, 0.99, 0.99, 1.00 and 1.00, respectively. Based on the predictions from its best model, GBM, on average, TargetFinder had an AUROC, AUPR, F1, precision, recall and specificity of 0.96, 0.87, 0.86, 0.94, 0.79 and 0.98, respectively. To find out how Ripple performed on the same dataset, we considered the 23 808 positive and 52 313 negative EP-pairs in two common samples (GM12878 and K562) with the promoters and enhancers used in Ripple predictions. Ripple showed an AUROC, AUPR, F1, precision, recall and specificity of 0.66, 0.39, 0.36, 0.61, 0.25 and 0.93, respectively, while EPIP showed a much better AUROC, AUPR, F1, precision, recall and specificity of 1.00, 1.00, 1.00, 0.99, 1.00 and 1.00, respectively, on the same dataset.
Next, we compared EPIP with TargetFinder and Ripple on condition-specific EPIs in TargetFinder data. EPIP predicted 51.36% of the 8471 condition-specific EP-pairs in the six samples, while TargetFinder predicted 38.85% of the 8471 condition-specific EP-pairs (Supplementary Table S10). Ripple predicted only 0.53% of the 5787 condition-specific EP-pairs in the three samples shared by the Ripple study and the TargetFinder study (GM12878, HeLa and K562), while EPIP predicted 54.42% of the same 5787 EP-pairs (Supplementary Table S10). The accuracy of EPIP on condition-specific EP-pairs here was much lower compared with that on EPIP test data, which may be because the TargetFinder data were not in good quality. For instance, enhancers and promoters used by TargetFinder were from computational predictions (Ernst and Kellis, 2012; Hoffman et al., 2012), which were prone to errors. Moreover, almost 50% of their enhancers and promoters overlap with their promoters and enhancers, respectively. In addition, the negative EP-pairs in TargetFinder data were problematic. TargetFinder labeled an EP-pair ‘negative’, if it did not overlap with Rao et al. looplists of any resolution. Due to the limited sequencing resolution and the limitation of the algorithms to analyze raw Hi-C reads to generate looplists, EP-pairs not identified as looplists are not necessarily negative pairs (Forcato et al., 2017).
Finally, we compared EPIP with TargetFinder and Ripple on condition-specific EPIs on EPIP test data. EPIP predicted 99.99% of the condition-specific EP-pairs in five common samples shared with the TargetFinder study. TargetFinder predicted only 83.91% of these condition-specific EP-pairs (Supplementary Table S11). In the two common samples shared with the Ripple study, EPIP predicted 99.99% of the condition-specific EP-pairs, while Ripple only could predict 27.07% of them (Supplementary Table S11).
4 Discussion
Identifying EPIs is important for the study of gene transcriptional regulation. Although several computational methods are available to predict EPIs, they often cannot predict condition-specific EPIs and their performance is still not satisfactory. We thus developed a computational method, EPIP, to learn the patterns of EPIs and to predict condition-specific EPIs. We demonstrated that, on average, EPIP correctly predicts 99.26% of condition-specific EPIs in different samples. We also showed that EPIP has a much better performance than two state-of-the-art computational methods.
EPIP provides an important framework to integrate useful data for EPI predictions. EPIP has a partitioning method that enables it to use samples with partially available features and samples with abundant types of data. Therefore, it can be trained on various types of samples and thus makes the training model more accurate and more representative. In addition, the learning approach in EPIP provides the opportunity to efficiently train the model when new data become available.
EPIP is trained with different samples. This means that data from different samples are fed to the training model in a specific order. To investigate whether the order of the samples in training has an impact on the performance of EPIP, we considered HUVEC as the testing sample and trained the EPIP model on the remaining six samples in all possible 120 orders. We observed that the order of the samples used in training EPIP does not significantly impact the final performance, as the standard deviation of the AUROC and the F1 score was 0.001 and 0.002, respectively, for all 120 different orders of training in these experiments.
We used FANTOM enhancers to define EP-pairs in this study. The number of FANTOM enhancers is small compared with the known and predicted enhancers in various studies (Ernst and Kellis, 2012). However, FANTOM enhancers arguably represent the largest set of enhancers we have so far that defined with the same criteria and supported by experiments. We further overlapped FANTOM enhancers with ChromHMM enhancers and H3K27ac ChIP-seq peaks to define active enhancers, which is likely to generate more reliable enhancers and more reliable training data. However, the choice of the FANTOM enhancers may have prevented us from testing EPIP more generally, since we only tested EPIP on EP-pairs based on FANTOM enhancers. When there is a larger and more reliable set of experimentally determined enhancers available in the future, it is necessary to test EPIP on the EP-pairs based on the new set of enhancers to make sure that it performs similarly.
We tried to train EPIP on EP-pairs from Rao et al. looplists, which generated suboptimal models due to the much smaller size of training data (Supplementary Tables S12 and S13). Instead, we used the cutoffs 30 and 5 to define positive and negative EP-pairs. We selected this combination of cutoffs based on our testing with different cutoffs and our previous studies (Li et al., 2016; Zhao et al., 2016). Note that these EP-pairs defined by this combination of cutoffs are imperfect. First, available methods to analyze Hi-C contact matrices are still suboptimal (Forcato et al., 2017), which prevents from defining accurate interacting regions. Second, the interacting regions we used are either too strict (such as Rao et al. looplists) or containing false positives and/or negatives (such as those from the cutoff 30), which affects the quality of the obtained EP-pairs. Third, as mentioned above, the FANTOM enhancers only represent a portion of existing enhancers while the ChromHMM enhancers are not so reliable. We chose to use these enhancers together with the H3K27ac peaks to define active enhancers, which may miss true positive EP-pairs. Finally, a fixed cutoff of 30 does not consider the exponential decay of the number of supporting Hi-C reads with the increasing distance between enhancers and promoters, which may miss true positive EP-pairs as well.
Despite these limitations of the enhancers and EP-pairs, we believe that the majority of the positives and negatives in our training and test datasets are true positives and true negatives, respectively. This is because EPIP performed well on more strictly defined EP-pairs based on interacting regions defined by other studies instead of the cutoffs (Supplementary Table S7). Moreover, EPIP always had a high recall/sensitivity to predict the ‘true’ positive EP-pairs when different cutoffs were used to define positive EP-pairs. Finally, EPIP performed well when tested on the remaining 70% of untrained EP-pairs, suggesting that the positive EP-pairs indeed are different from the negatives (Supplementary Table S6).
Although this combination of cutoffs 30 and 5 was good, it may be likely that we can do better with sample-specific cutoffs to define positives. We did increase the cutoff to 50, 100 and 150 in GM12878 to define positives. Our rationale was that the sequencing depth in GM12878 was higher, which may result in certain negative pairs with a large number of supporting Hi-C reads. However, it turned out that the trained model based on these EP-pairs (positives defined with the cutoff 100 in GM12878 and 30 in other samples, negatives defined with the cutoff 5 in all samples) did not perform better than the above-trained model based on the cutoffs of 30 and 5 (Supplementary Tables S12 and S13).
We also trained a model with more strictly defined positives (positives and negatives defined with the cutoffs 100 and 5, respectively, in all cell lines). On the three strictly defined test datasets, this model had higher F1 scores than the model based on the combination of 30 and 5, while had worse AUPRC (Supplementary Table S13). The poor AUPRC of this model may explain why it performed much worse than the model trained with the combination of 30 and 5 in cross-validation (Supplementary Table S12). It may also indicate that this set of more strictly defined positives do not represent all positives well.
Although EPIP has better performance compared with the state-of-the-art methods, there is still room for improvement. For instance, the training data used in this study is not perfect. With more accurate and more representative training data in the future, the accuracy of the trained EPIP model should be further improved. Moreover, different experimental methods are available to identify EPIs, such as Hi-C, ChIA-PET and 5C. We used Hi-C for extracting training data. It is worth studying how the performance of EPIP improves if we train EPIP with EPIs from other sources together with Hi-C. In addition, we used chromatin loops extracted by Rao et al. to define significant interactions. Developing a method that is able to extract interactions from raw Hi-C data may help to improve the performance of EPIP. Finally, like all most all existing methods, EPIP considers one EP-pair at a time to predict interacting EP-pairs while considering multiple EP-pairs may help the discovery of true positive EP-pairs, as shown in a previous study (Zhao et al., 2016). In the future, we will work on these directions together with others to further improve the accuracy of the EPI prediction.
Funding
This work has been supported by the National Science Foundation (grants 1356524, 1661414 and 1149955) and the National Institute of Health (grant R15GM123407).
Conflict of Interest: none declared.
Supplementary Material
Contributor Information
Amlan Talukder, Department of Computer Science, University of Central Florida, Orlando, FL, USA.
Samaneh Saadat, Department of Computer Science, University of Central Florida, Orlando, FL, USA.
Xiaoman Li, Burnett School of Biomedical Science, College of Medicine, University of Central Orlando, Orlando, FL, USA.
Haiyan Hu, Department of Computer Science, University of Central Florida, Orlando, FL, USA.
References
- Andersson R. et al. (2014) An atlas of active enhancers across human samples and tissues. Nature, 507, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X. et al. (2010) Systematic identification of conserved motif modules in the human genome. BMC Genomics, 11, 567.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradin O. et al. (2014) Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res., 24, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J. et al. (2002) Capturing chromosome conformation. Science, 295, 1306–1311. [DOI] [PubMed] [Google Scholar]
- De Laat W., Duboule D. (2013) Topology of mammalian developmental enhancers and their regulatory landscapes. Nature, 502, 499–506. [DOI] [PubMed] [Google Scholar]
- Dunham I. et al. (2012) An integrated encyclopaedia of DNA elements in the human genome. Nature, 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J., Kellis M. (2012) ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods, 9, 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J. et al. (2011) Mapping and analysis of chromatin state dynamics in nine human samples. Nature, 473, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forcato M. et al. (2017) Comparison of computational methods for Hi-C data analysis. Nat. Methods, 14, 679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund Y., Schapire R.E. (1997) A decision-theoretic generalization of on-line learning and an application to boosting. J. Comput. Syst. Sci., 55, 119–139. [Google Scholar]
- Fullwood M.J. et al. (2009) An oestrogen-receptor-α-bound human chromatin interactome. Nature, 462, 58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J. et al. (2012) GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res., 22, 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B. et al. (2014) Global view of enhancer-promoter interactome in human cells. Proc. Natl. Acad. Sci. U.S.A., 111, E2191–E2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman M.M. et al. (2012) Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat. Methods, 9, 473–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F. et al. (2013) A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature, 503, 290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G. et al. (2012) Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell, 148, 84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. et al. (2016) Integrative analyses shed new light on human ribosomal protein gene regulation. Sci. Rep., 6, 28619.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora A. et al. (2016) In the loop: promoter-enhancer interactions and bioinformatics. Brief. Bioinform., 17, 980–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F. et al. (2011) Scikit-learn: machine learning in Python. J. Mach. Learn. Res., 12, 2825–2830. [Google Scholar]
- Polikar R. et al. (2000) Acoustics, speech, and signal processing. In Proceedings, 2000 IEEE International Conference on IEEE (ICASSP’00), Vol. 6, pp. 3414–3417. [Google Scholar]
- Rao S.S. et al. (2014) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell, 159, 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S. et al. (2015) A predictive modeling approach for cell line-specific long-range regulatory interactions. Nucl. Acids Res., 43, 8694–8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman R.E. et al. (2012) The accessible chromatin landscape of the human genome. Nature, 489, 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen S. et al. (2016) Enhancer-promoter interactions are encoded by complex genomic signatures on looping chromatin. Nat. Genet., 48, 488–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C. et al. (2016) PETModule: a motif module based approach for enhancer target gene prediction. Sci. Rep., 6, 30043.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y. et al. (2015) Comprehensive discovery of DNA motifs in 349 human cells and tissues reveals new features of motifs. Nucl. Acids Res., 43, 74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.