Abstract
The natural history of tuberculosis (TB) is characterized by a large inter-individual outcome variability after exposure to Mycobacterium tuberculosis. Specifically, some highly exposed individuals remain resistant to M. tuberculosis infection, as inferred by tuberculin skin test (TST) or interferon-gamma release assays (IGRAs). We performed a genome-wide association study of resistance to M. tuberculosis infection in an endemic region of Southern Vietnam. We enrolled household contacts (HHC) of pulmonary TB cases and compared subjects who were negative for both TST and IGRA (n = 185) with infected individuals (n = 353) who were either positive for both TST and IGRA or had a diagnosis of TB. We found a genome-wide significant locus on chromosome 10q26.2 with a cluster of variants associated with strong protection against M. tuberculosis infection (OR = 0.42, 95%CI 0.35–0.49, P = 3.71×10−8, for the genotyped variant rs17155120). The locus was replicated in a French multi-ethnic HHC cohort and a familial admixed cohort from a hyper-endemic area of South Africa, with an overall OR for rs17155120 estimated at 0.50 (95%CI 0.45–0.55, P = 1.26×10−9). The variants are located in intronic regions and upstream of C10orf90, a tumor suppressor gene which encodes an ubiquitin ligase activating the transcription factor p53. In silico analysis showed that the protective alleles were associated with a decreased expression in monocytes of the nearby gene ADAM12 which could lead to an enhanced response of Th17 lymphocytes. Our results reveal a novel locus controlling resistance to M. tuberculosis infection across different populations.
Author summary
There is strong epidemiological evidence that a proportion of highly exposed individuals remain resistant to M. tuberculosis infection, as shown by a negative result for Tuberculin Skin Test (TST) or IFN-γ Release Assays (IGRAs). We performed a genome-wide association study between resistant and infected individuals, which were carefully selected employing a household contact design to maximize exposure by infectious index patients. We employed stringently defined concordant results for both TST and IGRA assays to avoid misclassifications. We discovered a locus at 10q26.2 associated with resistance to M. tuberculosis infection in a Vietnamese discovery cohort. This locus could be replicated in two independent cohorts from different epidemiological settings and of diverse ancestries enrolled in France and South Africa.
Introduction
Tuberculosis (TB) remains a major public health threat worldwide [1]. An estimated 10 million people developed TB disease in 2018, of whom 1.45 million died. The causative agent of TB is Mycobacterium tuberculosis which is transmitted by aerosol from contagious TB patients. However, not all persons encountering infectious aerosols will become infected with M. tuberculosis, defining the first line of human resistance against TB [2,3]. Infection is inferred from the presence of anti-mycobacterial immunoreactivity, as shown by a positive result in tuberculin skin test (TST) and/or interferon-gamma (IFN-γ) release assay (IGRA). TST is done in vivo and consists of an intradermal injection of purified protein derivative (PPD) that provokes a delayed hypersensitivity reaction at the site of injection. IGRAs are performed ex vivo and measure the secretion of IFN-γ by leukocytes in response to M. tuberculosis-specific antigens. Both tests have their own limitations and results are not fully concordant [4–6]. Individuals who score positive by TST and/or IGRA are considered to suffer from asymptomatic latent TB infection (LTBI). Conversely, persons who score negative despite documented exposure to M. tuberculosis are considered resistant to infection. Based on TST and/or IGRA results, the intensity of exposure or the duration of follow-up, from 7% to 25% of subjects display the M. tuberculosis infection resistance phenotype [2,7–9].
The large inter-individual variability in exposure outcomes supports a major role for human genetic factors [10]. Various genome-wide approaches have confirmed this hypothesis by either considering TST and IGRA results as quantitative traits or relying on TST reactivity (positive/negative) as a surrogate marker for infection[11–14]. Regarding this latter phenotype, persistent TST negativity was linked to loci on 2q21-2q24, further fine-mapped to ZEB2, and 5p13-5q22 in an Ugandan population [14,15]. A major locus, named TST1 on chromosome 11p14, employing stringently defined TST negativity (0 mm vs. > 0 mm) as phenotype, was identified in a linkage analysis conducted in South Africa [11]. TST1 was later replicated in a household contact (HHC) study of French families [16]. A genome-wide association study (GWAS) among highly M. tuberculosis exposed HIV-seropositive individuals from East Africa, identified a locus in the 5q31.1 region near IL9 associated with negative TST [17].
Here, we performed a GWAS of resistance to M. tuberculosis infection using a robust phenotype based on both TST and IGRA information. In addition, we used a HHC study design guaranteeing shared environmental effects and high intensity of exposure to M. tuberculosis. We found a locus on chromosome 10q26.2 associated with resistance to M. tuberculosis infection in an East Asian population from Southern Vietnam. Importantly, this locus was replicated in two other cohorts from France and South Africa, representing different ancestries and epidemiological settings.
Results
Genome-wide association study of resistance to M. tuberculosis infection in Vietnam
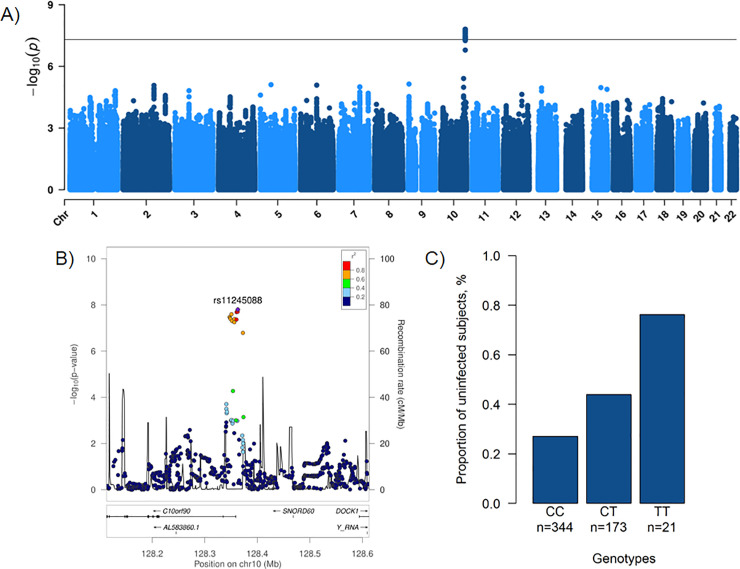
First, we conducted a GWAS in the Vietnamese sample using 185 uninfected and 353 infected subjects, consisting of 201 individuals with positive TST/QuantiFERON-TB Gold In-Tube test (QFT-GIT) results and 152 pulmonary TB (PTB) patients. A total of 5,591,951 high quality variants were tested with a genomic inflation factor (λ) at 0.997, suggesting that effects from the familial study design were well controlled (S1 Fig). The corresponding Manhattan plot is shown in Fig 1A. We observed a genome-wide significant association on chromosome 10q26.2, corresponding to a cluster of 12 variants and 6 additional variants in high linkage disequilibrium (LD) with P < 5 × 10−7 in the intronic regions and upstream of C10orf90 (or FATS, HGNC: 26563) (Fig 1B). The top-associated variant was the imputed rs11245088 (odds ratio (OR) = 0.42, 95% confidence interval (CI) 0.39–0.45, P = 1.58 × 10−8) while the top-associated genotyped variant was rs17155120 (P = 3.71 × 10−8) (Table 1). Each copy of the minor allele T of rs17155120 conferred protection against M. tuberculosis infection with an OR of being infected for CT vs. CC or TT vs. CT at 0.42 (95%CI 0.35–0.49) (Fig 1C). The intensity cluster plot for rs17155120 showed that the genotype calling was of high quality and separated clearly into 3 genotype groups (S2 Fig). Since all 18 variants in the locus were in high LD (S3 Fig), the imputed variants were likely to have a high imputation quality as suggested by their info score (S1 Table).
Fig 1. Genome-wide association study of resistance to M. tuberculosis infection in Vietnam.
A) Manhattan plot showing results from a genome-wide association study between 185 uninfected subjects (negative for both tuberculin skin test and QuantiFERON-TB Gold In-Tube test) and 353 infected subjects (201 infected individuals positive for both tests and 152 patients with a history of pulmonary tuberculosis) for 5,591,951 variants (minor allele frequency > 5% and info > 0.8) with an unadjusted additive genetic model. The -log10(P value) for each variant (y-axis) is presented according to its chromosomal position (x-axis, build hg19). The dashed line indicates the genome-wide significant threshold at P = 5 × 10−8. B) Locus zoom plot showing association for the 10q26.2 locus, in a 500 kb window surrounding the top imputed variant rs11245088 (purple diamond). Colors represent pairwise linkage disequilibrium (r2) with rs11245088 as calculated for the Vietnamese Kinh population of 1000 Genomes phase 3. C) Proportion of Vietnamese individuals resistant to M. tuberculosis infection by genotype for the variant rs17155120. Each bar represents the proportion of uninfected subjects among CC individuals (n = 93/344), CT individuals (n = 76/173) and TT individuals (n = 16/21) for the variant rs17155120 in Vietnam.
Table 1. Association between an additive genetic effect of variants on chromosome region 10q26.2 and resistance to M. tuberculosis infection in Vietnam, France and South Africa.
| Variant | C10orf90 | EA | Vietnam | France | South Africa | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| EAF | OR (95%CI) | P value | EAF | OR (95%CI) | P value | EAF | OR (95%CI) | P value | |||
| rs11245088 | upstream | C | 0.25 | 0.42 (0.39–0.45) | 1.58×10−8 | - | - | - | 0.44 | 0.99 (0.91–1.07) | 4.75×10−1 |
| rs72163291 | intron | ins | 0.24 | 0.42 (0.35–0.49) | 1.94×10−8 | 0.37 | 0.88 (0.78–0.98) | 3.54×10−1 | - | - | - |
| rs17155143 | upstream | A | 0.20 | 0.39 (0.31–0.47) | 1.98×10−8 | 0.18 | 0.63 (0.50–0.77) | 1.29×10−1 | 0.15 | 0.60 (0.48–0.72) | 1.72×10−2 |
| rs7909756 | upstream | G | 0.24 | 0.42 (0.35–0.49) | 2.03×10−8 | 0.37 | 0.83 (0.73–0.93) | 2.87×10−1 | 0.36 | 0.90 (0.81–0.99) | 2.77×10−1 |
| rs28703703 | intron | G | 0.20 | 0.41 (0.34–0.49) | 2.52×10−8 | 0.20 | 0.59 (0.48–0.69) | 4.95×10−2 | 0.17 | 0.66 (0.55–0.77) | 2.95×10−2 |
| rs56106518 | intron | C | 0.23 | 0.41 (0.33–0.48) | 3.07×10−8 | 0.30 | 0.40 (0.30–0.51) | 2.98×10−3 | - | - | - |
| rs75482972 | intron | A | 0.20 | 0.42 (0.34–0.49) | 3.35×10−8 | 0.20 | 0.58 (0.47–0.68) | 4.38×10−2 | 0.17 | 0.68 (0.57–0.78) | 3.48×10−2 |
| rs17155120 | intron | T | 0.20 | 0.42 (0.35–0.49) | 3.71×10−8 | 0.18 | 0.48 (0.36–0.59) | 1.51×10−2 | 0.16 | 0.62 (0.52–0.73) | 1.74×10−2 |
| rs73370887 | intron | A | 0.20 | 0.40 (0.33–0.48) | 4.05×10−8 | 0.31 | 0.78 (0.71–0.85) | 1.43×10−1 | 0.28 | 0.75 (0.67–0.83) | 3.69×10−2 |
| rs79608098 | intron | T | 0.20 | 0.42 (0.35–0.49) | 4.06×10−8 | 0.20 | 0.58 (0.48–0.69) | 4.50×10−2 | 0.17 | 0.66 (0.55–0.76) | 2.71×10−2 |
| rs61750007 | upstream | C | 0.24 | 0.43 (0.36–0.50) | 4.30×10−8 | 0.24 | 0.53 (0.42–0.64) | 3.49×10−2 | 0.20 | 0.73 (0.62–0.84) | 7.62×10−2 |
| rs77513326 | intron | A | 0.20 | 0.42 (0.34–0.49) | 4.93×10−8 | 0.17 | 0.47 (0.36–0.59) | 1.61×10−2 | 0.16 | 0.63 (0.52–0.75) | 2.35×10−2 |
| rs79918233 | intron | A | 0.20 | 0.41 (0.33–0.49) | 5.61×10−8 | 0.17 | 0.51 (0.39–0.63) | 3.00×10−2 | 0.16 | 0.63 (0.52–0.74) | 1.95×10−2 |
| rs147584264 | upstream | C | 0.19 | 0.41 (0.33–0.49) | 1.21×10−7 | 0.22 | 0.77 (0.62–0.93) | 3.00×10−1 | - | - | - |
| rs191820708 | upstream | A | 0.19 | 0.41 (0.33–0.49) | 1.31×10−7 | 0.19 | 0.63 (0.50–0.77) | 1.40×10−1 | - | - | - |
| rs201178890 | upstream | T | 0.19 | 0.41 (0.33–0.49) | 1.36×10−7 | 0.22 | 0.79 (0.62–0.97) | 3.36×10−1 | - | - | - |
| rs202189321 | upstream | T | 0.20 | 0.41 (0.33–0.49) | 1.37×10−7 | 0.18 | 0.65 (0.52–0.78) | 1.44×10−1 | - | - | - |
| rs118037357 | upstream | A | 0.19 | 0.41 (0.33–0.49) | 1.62×10−7 | 0.15 | 0.47 (0.33–0.60) | 2.83×10−2 | 0.14 | 0.55 (0.43–0.67) | 7.38×10−3 |
CI, confidence intervals; EA, effect allele; EAF, effect allele frequency; OR, odds ratio; ins, insertion
We also performed a GWAS between the 185 uninfected and the 201 infected subjects, excluding the 152 PTB patients (S4 Fig). Despite a smaller sample size, all the 18 variants of the locus were still associated with protection against infection with P < 5.0 × 10−6, with similar ORs (for rs17155120, OR = 0.40, 95%CI 0.32–0.49, P = 2.55 × 10−7) (S2 Table). Similar findings were also observed when considering only the 152 PTB patients as infection reference, with an OR for rs17155120 estimated at 0.50 (95%CI 0.41–0.59, P = 2.10 × 10−4). These results indicate that PTB patients are an appropriate infection reference group in this analysis.
Replication of variants associated with resistance to M. tuberculosis infection in France and South Africa
We tested the effects of the variants of the 10q26.2 locus in a French multi-ethnic HHC cohort (30 uninfected vs. 157 infected subjects) and an admixed familial sample from South Africa (118 uninfected vs. 136 infected subjects). In the French cohort, 17 variants of the cluster could be genotyped or imputed (including 11 genome-wide significant variants), and 3 were replicated at the P < 0.025 level with effect sizes in the same direction as in the Vietnamese cohort. The most significant associated variant was rs56106518 (OR = 0.40, 95%CI 0.30–0.51, P = 2.98 × 10−3) (Table 1). In the South African population, 12 variants of the locus were genotyped or successfully imputed (including 10 genome-wide significant variants) and 3 of them showed evidence for replication. The most significant associated variant was rs118037357 (OR = 0.55, 95%CI 0.43–0.67, P = 7.38 × 10−3) (Table 1). The top genotyped variant rs17155120 was also replicated in the two cohorts (PFrance = 1.51 × 10−2 and PSouthAfrica = 1.74 × 10−2) (Table 1 and Fig 2A).
Fig 2. Genome-wide association study of resistance to M. tuberculosis infection in 3 cohorts from Vietnam, France and South Africa.
A) Forest plot of the association between an additive genetic effect of rs17155120 on chromosome region 10q26.2 and resistance to M. tuberculosis infection. Odds ratios and 95% confidence intervals derived from a linear mixed model, P values, sample sizes and frequency of the effect allele (EAF) are reported by individual cohort and for the random effects meta-analysis. B) Manhattan plot showing results from a genome-wide association study between 333 uninfected subjects and 616 infected subjects for 3,967,482 variants (minor allele frequency > 5% and info > 0.8) with an unadjusted additive genetic model. The -log10(P value) for each variant (y-axis) is presented according to its chromosomal position (x-axis, build hg19). The dashed line indicates the genome-wide significant threshold at P = 5 × 10−8.
Interestingly, the variants across the two replication cohorts were in high LD with each other presenting similar LD patterns as in Vietnam (S5 Fig). The frequencies of the rs17155120 T allele for all 3 cohorts were also similar, ranging from 0.16 to 0.20. The rs17155120 T allele frequency of 0.20 in the Vietnamese cohort was consistent with the Kinh allele frequency of 0.18 in 1000 Genomes (1000G) phase 3 (S6 Fig [18], and S3 Table). The rs17155120 T allele frequency of 0.18 in the multi-ethnic French cohort was also close to the global frequency of 0.16 in all 1000G populations. However, the frequency of 0.16 for rs17155120 T allele in the Souh African cohort was higher than in any 1000G African population that ranged from 0.05 to 0.11. Discrepancy of allele frequencies between Souh African subjects and African populations of 1000G was confirmed by different patterns of LD in a 30 kb region around rs17155120 (S7 Fig), which could be explained by the specific ethnic origin of the Souh African subjects.
Trans-ethnic meta-analysis
Next, we performed a meta-analysis of resistance to M. tuberculosis infection using the GWAS data of the 3 cohorts. The combined analysis was carried out in 333 uninfected and 616 infected subjects. The only genome-wide significant result was observed with the variants of the 10q26.2 region (Figs 2B and S8). The most significant signal was observed at the genotyped variant rs17155120 (OR = 0.50, 95%CI = 0.45–0.55, P = 1.26 × 10−9) and no heterogeneity was observed across the 3 studies (Phet = 0.31) (Fig 2A and S4 Table).
Functional annotation
The variants of the 10q26.2 region map to the intronic regions and upstream of the tumor suppressor gene C10orf90 (or FATS, for Fragile-site Associated Tumor Suppressor, HGNC: 26563) (Fig 3). According to ENCODE, two associated intronic variants, rs28703703 and rs77513326, are located in a regulatory genomic region characterized by H3K4me1 and H3K27ac histone marks and an active enhancer signature in lymphocytes T helper 17 (Th17). The variant rs77513326 also overlaps ATAC peaks in Th17 cells, memory T cells, natural killer cells and CD8+ T cells. We further explored the variants in various expression quantitative trait loci (eQTLs) databases of relevant tissues for the phenotype and found an association between them and expression of the nearby gene ADAM12. In particular, rs28703703 and the genotyped variant rs17155120 displayed decreasing expression of ADAM12 with each minor allele (having a protective effect against M. tuberculosis infection) in monocytes (P = 2.10 × 10−3 and P = 4.70 × 10−3 respectively) (from https://immunpop.com/kim/eQTL [19]). No association was observed in other immune cell types.
Fig 3. Genomic annotation of the locus on chromosome region 10q26.2.
The upper panel is adapted from Integrative Genomics Viewer (http://igv.org/app/) and the lower panel is adapted from UCSC Genome Browser (http://genome.ucsc.edu/). The 3 vertical grey lines represent the associated variants rs1715120, rs28703703 and rs77513326 (from left to right) that overlap regulatory regions. From top to bottom: H3K4me1 and H3K27ac histone marks from ENCODE, active enhancer in Th17 cells (chromHMM annotation from ROADMAP), chromatin accessibility as represented by ATAC peaks in Th17 cells, memory T cells, NK cells and CD8+ T cells [20].
Discussion
In this study, we explored the genetic determinants of natural resistance to M. tuberculosis infection after intense exposure. There are no direct tests for established infection because TST and IGRAs measure an immune response that does not allow to distinguish between past or present infection with M. tuberculosis bacilli. Nevertheless, our results show a strong genetic effect on resistance to M. tuberculosis infection irrespective of infection being persistent or temporary. TST and IGRA tests have well-known limits [4]. To minimize misclassification of uninfected and infected subjects in a cross-sectional setting, we therefore relied on negative results in both tests and defined stringent cut-offs. As exposure to M. tuberculosis is difficult to quantitate, yet a critical feature of TB studies [21], we focused on individuals at high risk of infection. We recruited household contacts recently exposed for extended periods to a contagious PTB index, some of whom remained infection-free. We discovered a genome-wide significant association between a cluster of variants on chromosome 10q26.2 and resistance to M. tuberculosis infection in well-characterized Vietnamese subjects. Strikingly, this locus could be replicated in two independent cohorts with different epidemiological settings from France and South Africa, resulting in association of the C/T variant rs17155120 across the 3 populations at an estimated combined odds ratio of 0.50 (95%CI 0.45–0.55) for becoming infected for TC vs. CC or TT vs. TC individuals.
The cluster of associated variants overlaps intronic and 5’ regions of C10orf90 (or FATS, HGNC: 26563), a tumor suppressor gene. This gene lies within a common-fragile site, which is an evolutionarily conserved region among mammals and susceptible to DNA damage [22]. The protein encoded by C10orf90 has been shown to promote p53 activation in response to DNA damage through an E3 ubiquitin ligase activity [23,24]. Several E3 ubiquitin ligases have already been shown to participate in the defense against M. tuberculosis, in particular through autophagy[25–27]. The p53 transcription factor is not only a master regulator of autophagy but also activates apoptosis, another key process for host infected cells to limit the spread of pathogens as M. tuberculosis [28,29]. Recent studies demonstrated that p53-induced apoptosis plays a critical role in the inhibition of mycobacteria survival and the macrophage resistance, possibly mediated by IL-17 [30,31]. However, possible role of C10orf90 during M. tuberculosis infection or any related immune process remains unknown.
In silico functional annotation of the 10q26.2 associated variants revealed additional regulatory features related to lymphocytes Th17. In particular, the A/G variant rs28703703 was identified as a likely cis-eQTL for ADAM12 in monocytes, with decreased expression of ADAM12 associated with the G allele, protective against M. tuberculosis infection. ADAM12 is located ~280 kb downstream C10orf90, and encodes a matrix metalloprotease linked to a broad range of biological processes [32]. ADAM12 expression correlates with lung inflammation as it is overexpressed in cells issued from asthmatic sputum and in the airway epithelium during allergic inflammatory reaction [33,34]. Previous studies also reported ADAM12 expression in Th17 cells [35,36], and ADAM12 knockdown in human T cells was found to increase Th17 cytokine production (IL-17A, IL-17F, and IL-22) [35]. In South India, TST-negative individuals produced significantly higher levels of Th17 cytokines than TST-positive individuals [37]. Similarly, significant higher levels of Th17 cytokines were observed in persistent negative IGRA individuals as compared with IGRA converters in a recent study conducted in the Gambia [38]. These findings are consistent with our present results showing that the rs28703703 G allele protective against M. tuberculosis infection is associated with lower ADAM12 expression that could lead to higher Th17 cytokine production. Overall, these observations support the view that Th17 cytokines may have a protective role against early stages of M. tuberculosis infection.
The 3 enrolled samples were of modest size. However, the design of the 3 studies, and the definition of a stringent and homogenous phenotype across them enabled us to detect a significant association with large effects. Interestingly, adding the PTB Vietnamese patients, who are by definition infected, increased the power of the analysis. Strikingly, the top associated variants displayed large effects with similar allele frequencies across populations and were in high LD across multiple ancestries. This observation was not expected because of the various epidemiological settings and the genetic diversity of the cohorts that included Kinh Vietnamese, Europeans, North Africans, Sub-Saharan Africans and admixed Western Cape Coloureds. Such populations are usually under-represented in studies of genetic association while they could provide valuable insights to understand complex diseases [39].
In conclusion, we demonstrated that rigorous epidemiological design and phenotype definition with seemingly limited sample sizes can reveal novel genetic factors that offer protection from major pathogens such as M. tuberculosis. We found that C10orf90 and ADAM12 are promising candidate genes involved in the natural resistance to M. tuberculosis. Further investigations are needed to elucidate their role in the process of the initial infection, which could be a major step to provide new opportunities in the fight against TB.
Subjects and methods
Ethics statement
Signed informed consent was obtained from all the participants, and from the parents of enrolled minors. The study was approved by the regulatory authorities in Binh Duong, Vietnam (1366/UBND-VX), the French Consultative Committee for Protecting Persons in Biomedical Research of Henri Mondor Hospital (Créteil, France) and the Stellenbosch University Health Research Ethics Committee (Tygerberg, South Africa).
Study settings and populations
Vietnam
Vietnam is a middle-income country in South-East Asia with a high annual incidence of TB (130/100,000 at the time of the study [40]), high Bacille Calmette-Guérin (BCG) vaccination coverage (with reported rates exceeding 95% [41]) and a low population prevalence of HIV (0.4% in 2015 in the general population and 5% among TB cases [40,42]). From 2010 to 2015, we recruited, in a M. tuberculosis endemic region of Southern Vietnam, index PTB adults with persistent cough before the start of treatment (mean duration of coughing = 3.2 months). PTB diagnosis was assessed by clinical presentation, chest X-ray and/or positive cultures for M. tuberculosis. A total of 1108 HHCs of 466 PTB index cases were invited to participate in the study and underwent both TST and QFT-GIT (S1 Text). HHCs did not undergo any further follow-up.
France
As a first replication cohort, we used household TB contacts studied in Val-de-Marne, a suburban region of Paris, in the context of a general screening procedure. This multi-ethnic cohort has been previously described [12,16]. Val-de-Marne is an area of low TB endemicity, displaying an annual incidence of 22/100,000 at the time of the study [43] and BCG vaccination rates are high [41]. HIV seroprevalence is low in France in the general population (0.3% at the time of the study [44]) and was estimated at 7% in the index cases of the study [45]. For this study, 664 HHCs of 132 PTB index cases were investigated according to national guidelines that required two screening visits. HHCs were individuals sharing residence with an index during the 3 months before diagnosis. Briefly, the first visit (V1) included a physical examination, a chest radiograph, TST and in-house IGRA [12,16] (S1 Text). These investigations, except for IGRA, were repeated 8–12 weeks later (V2) if the contact subject did not meet the criteria for infection at V1.
South Africa
As a second replication cohort, we used a large sample (n = 415) of 153 nuclear families from a suburban area of Cape Town, South Africa, which has been previously described [6,11,12,16]. All individuals belonged to the South African Coloured group, a unique multi-way admixed population [46]. There was no specific requirement for subjects to be HHCs of PTB patients. Indeed, TB is hyper-endemic in this area with an incidence of ~800/100,000 at the time of the study [47] and TB transmission occurs more often outside the household [48]. BCG vaccination at birth is routine in this area [49]. HIV seroprevalence was estimated at 5.2% in the overall population and less than 2% in the pediatric population at the time of the study [11]. In addition, individuals who were known to be HIV positive, pregnant, or using immunomodulatory chemotherapy were excluded at the time of enrollment [11]. TST and in-house IGRA were performed as previously described (S1 Text). Subjects who had clinical TB disease in the two years preceding the study were excluded. Thus, only healthy children and young adults from the area were included and tested for infection.
Genotyping, quality control and imputation
A total of 724 individuals from Vietnam (S9 Fig) and 573 from France were genotyped using the Illumina Infinium OmniExpressExome-8-v1 chip (960,212 single nucleotide polymorphisms, SNPs). For the South African cohort (n = 374), the Illumina HumanOmni2.5–8 BeadChip (~2 million of SNPs) was used. All quality control steps were done in each cohort with PLINK v1.9 [50]. Autosomal SNPs with a minor allele frequency (MAF) > 0.01, a genotype call rate > 0.99 and a Hardy-Weinberg (HWE) equilibrium P > 1.00 × 10−5 were retained. Individuals with a call rate < 95% were excluded (n = 2). Identity-by-descent (IBD) analysis was done to detect duplicated individuals and the members of the pairs with the lower call rate were excluded (n = 1). After the quality control, imputation was performed on 720 individuals and 598,090 variants from Vietnam and 573 individuals and 886,471 variants from France using the Michigan Imputation Server [51] with Eagle2 [52] for the pre-phasing and the 1000G Project as reference panel [53]. For the South African cohort (374 individuals and 1,347,846 variants), the imputation was done on the Sanger Imputation Server with Eagle2 for the pre-phasing and the African Genome Resources as reference panel which includes ~2000 African samples in addition to the individuals from 1000G [54]. Imputed SNPs with an imputation quality info score > 0.8 and MAF > 0.05 were retained for further association analyses (5,591,951 variants in Vietnam, 7,737,070 variants in France and 6,922,541 variants in South Africa).
For each cohort, principal component analysis (PCA) was conducted to evaluate population structure. Genotypes of the individuals from 1000G were used to calculate principal components and data for subjects from the cohorts were projected onto the eigenvectors. The Vietnamese cohort, which was sampled from the Vietnamese Kinh group, was homogenous and clustered with the 1000G East Asian populations (S10 Fig). By contrast, the families in the Val-de-Marne sample showed genetic diversity at the population level with a majority of individuals of European, North African and Sub-Saharan ancestries (S11 Fig). The admixed Souh African subjects, who exhibited genetic diversity at the individual level, were forming a distinct cluster close to the African populations of 1000G (S12 Fig).
Definition of the M. tuberculosis infection phenotype
The definition of the M. tuberculosis infection phenotype relied on both TST and IGRA results. In particular, we used a 5 mm cut-off to determine TST status, based on previous studies in similar settings [7,17] and published guidelines [55]. We explored covariates associated with our infection definition in the entire cohort of enrolled individuals and the subset of those with genotype information were retained for the GWAS (S9 Fig).
For the Vietnamese study, resistance to M. tuberculosis infection was defined by the presence of a negative TST < 5mm and a negative QFT-GIT test result following a protocol suggested by the manufacturer (S1 Text). Infected individuals were defined as subjects presenting both a positive TST ≥ 5mm and a positive QFT-GIT test result. A total of 188 subjects were classified as double negative and 512 as double positive (S13 Fig), among which 185 and 201 subjects were genotyped, respectively. In order to increase the sample size, we also added 152 genotyped PTB patients, consisting of 146 index cases and 6 subjects with a history of PTB, to the infected group (S5 Table). We investigated covariates associated with our infection definition and no significant association was found (S6 Table). Therefore, we conducted an unadjusted genetic association analysis of our binary infection phenotype.
For the French study, which included only HHCs and none of their PTB index cases, contacts could have had one or two screening visits (V1 and V2) with a TST measurement (S14 Fig). A TST was considered negative when the skin induration was (i) < 5 mm at both V1 and V2, (ii) < 5 mm at V1, when only one visit was done. A TST was considered positive when the skin induration was (i) ≥ 5 mm at both V1 and V2, (ii) < 5 mm at V1 and ≥ 10 mm at V2, which reflected true conversions. In-house IGRA was used in this study and provided quantitative levels of IFN-γ production upon early secretory antigenic target 6 (ESAT6) stimulation (S1 Text). A negative IGRA result was defined by a null production of IFN-γ. To determine the optimal positivity cut-off, we built a receiver operating characteristic (ROC) curve with TST status as the observed outcome and the corrected IFN-γ levels (ESAT6 response minus non-stimulated control value) as the predicted outcome among all the contacts enrolled. We selected as positivity threshold the highest sum of sensitivity plus specificity, which was equal to 175 pg/mL (S14 Fig). Then, we defined uninfected subjects as HHCs with a negative TST and a null IFN-γ production (n = 33) and infected subjects as HHCs who presented both a positive TST and a positive IGRA result (IFN-γ production > 175 pg/mL) (n = 147) (S14 Fig). We also looked for covariates associated with our infection definition in this sample of 180 individuals (S7 Table). Age was the only factor significantly associated with our infection definition and was included as covariate in the genetic association analysis that finally included 30 genotyped uninfected and 127 infected subjects (S8 Table).
For the South African study, a 5 mm cut-off was used to identify negative and positive TSTs (S15 Fig). In-house IGRA was also based on the production of IFN-γ upon ESAT6 stimulation (S1 Text). We determined the optimal IGRA positivity cut-off similarly to the French study by building a ROC curve, leading to a threshold of 20.9 pg/mL (S15 Fig). Then, we defined uninfected subjects as HHCs with a negative TST (< 5 mm) and a null IFN-γ production (n = 128), and infected subjects as those with both positive TST and IGRA result (IFN-γ production > 20.9 pg/mL) (n = 152) (S15 Fig). Age was the only factor significantly associated with our infection definition in the whole cohort (S9 Table) and was included as covariate in the genetic association analysis that finally included 118 genotyped uninfected and 136 infected subjects (S10 Table).
Covariates associated with our infection definition were investigated using mixed-effects logistic regression with a random effect per family in each cohort. All the analyses were carried out using R software (version 3.5.2) and related packages “pROC” and “lme4”[56–58].
Genetic association analyses
We conducted genetic association analyses of uninfected vs. infected subjects in the 3 cohorts using a linear mixed-model (LMM) assuming an additive genetic model as implemented in GEMMA v0.98 [59]. To account for the familial relationships, a genetic relatedness matrix (GRM) was used as random effects. In each cohort, the GRM was estimated using centered genotypes after the quality control described above. P values from the likelihood ratio test were reported. For better interpretability, we reported odds ratios (OR) and their 95% confidence intervals (95%CI) after transforming the regression coefficients of the LMM [60]. Manhattan plots of the -log10(P values) and quantile-quantile (QQ) plots were generated using “CMplot” package in R [61]. Regional plots were generated using LocusZoom Standalone v1.4 [62]. Haplotype plots were generated using Haploview [63]. Replication of genome-wide associated variants (P < 5 × 10−8) in the primary cohort from Vietnam was assessed in the two cohorts from France and South Africa. The observed LD in France and South Africa was slightly weaker as compared with Vietnam, and two LD blocks were inferred by Haploview (S5 Fig). We therefore considered variants at a nominal one sided P value < 0.025 and with a consistent direction of the effect size as replicated.
We also conducted a trans-ethnic meta-analysis by using summary statistics (i.e. beta estimates and their standard errors) from the Vietnamese discovery cohort and the two replication datasets. We used the random-effects model of Han and Eskin implemented in METASOFT [64]. This model assumes effect sizes of exactly zero in all the studies (i.e. no heterogeneity) under the null hypothesis of no associations and allows the effect sizes to vary among studies (i.e. heterogeneity) under the alternative hypothesis. The effect size consistency across studies were determined using the Cochran’s Q statistic. Allelic effect estimates were also derived on the log-odds scale.
Functional annotation
We used the UCSC Genome Browser [65] to identify associated variants which may overlap with known regulatory regions: 1) histone marks from the ENCODE project [66], 2) chromatin state annotated by ChromHMM on the basis of ROADMAP [67,68] and 3) chromatin accessibility determined by assay for transposase-accessible chromatin using sequencing (ATAC-seq) from immune cell-types [20]. We also looked at the associated variants in eQTL databases which focused on gene expression in monocytes [19,69], T cells [70], macrophages [71], and various types of immune cells [72].
Supporting information
Immune assays in Vietnam, France and South Africa.
(PDF)
Plot of the expected distribution of association test statistics (x axis) for 5,591,951 variants compared to the observed values (y axis). Red line is the null hypothesis of no association (y = x).
(PDF)
(PDF)
Pairwise LD coefficients r2 are shown in each cell between 18 variants with P < 5.0 × 10–6. Top genotyped variant rs17155120 is second from left.
(PDF)
(PDF)
Pairwise LD coefficients r2 are shown in each cell A) between the 17 variants of the locus in the French cohort, and B) between 12 out of the 18 variants of the locus in the South African cohort. Top genotyped variant rs17155120 is second from left in each figure.
(PDF)
The T allele (in blue) is protective against M. tuberculosis infection. Map generated from the Geography of Genetic Variants Browser (http://www.popgen.uchicago.edu/ggv).
(PDF)
Pairwise linkage disequilibrium (r2) in A) Vietnamese, B) French and C) South African study cohorts (left panels) between 60 variants in a 30 kb window around rs17155120 (arrow) as compared to 1000G populations (right panels).
(PDF)
(PDF)
(PDF)
Plot of the first and second principal components of the 720 individuals from the Vietnamese cohort after projection A) on the 1000 Genomes phase 3 population, B) on the East Asian 1000 Genomes Phase 3 populations only.
(PDF)
Plot of the first and second principal components of A) 573 individuals from the French cohort and B) the 157 subjects subsequently analyzed in the GWAS, after projection on the 1000 Genomes phase 3 populations.
(PDF)
Plot of the first and second principal components of 374 individuals from the South African cohort after projection on the 1000 Genomes phase 3 populations.
(PDF)
The dashed line represents a 5 mm cut-off for TST. Uninfected subjects (in yellow) were negative for TST and QuantiFERON-TB Gold In-Tube (QFT-GIT) and infected subjects (in blue) positive for both.
(PDF)
A) Distribution of the tuberculin skin test (TST) induration among 516 HHCs (mean induration when 2 screening visits were done). The dashed line represents a 5 mm cut-off. TST was considered negative when the skin induration was i) < 5 mm at both V1 and V2, ii) < 5 mm at V1, when only one visit was done. TST was considered positive when the skin induration was i) ≥ 5 mm at both V1 and V2, ii) < 5 mm at V1 and ≥ 10 mm at V2. B) Construction of a ROC curve based on the defined TST status to determine the optimal interferon-γ release assay (IGRA) cut-off (175 pg/mL). C) Stacked histogram of the TST distribution stratified by our infection definition. Uninfected subjects (in yellow) presented a negative TST and a null production of IFN-γ. Infected subjects (in blue) presented a positive TST and a positive IGRA (IFN-γ production > 175 pg/mL).
(PDF)
A) Distribution of the tuberculin skin test (TST) induration among 415 participants. The dashed line represents a 5 mm cut-off. B) Construction of a ROC curve based on the TST status to determine the optimal interferon-γ release assay (IGRA) cut-off (20.9 pg/mL). C) Stacked histogram of the TST distribution stratified by our infection definition. Uninfected subjects (in yellow) presented a negative TST and a null production of IFN-γ. Infected subjects (in blue) presented a positive TST and a positive IGRA (IFN-γ production > 20.9 pg/mL).
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We are grateful for the skilled technical assistance of Ho thi Ngoc, Binh Duong Center for Social Diseases Control, and Hanh La, Hospital for Dermato-Venereology, Ho Chi Minh City. We are indebted to the laboratory staff of the Hospital for Dermato-Venereology, Ho Chi Minh City, and the tuberculosis control teams of the Thu dau Mot, Thuận An, Ben Cat, and Di An districts. We sincerely thank all members of the community who participated in this work in South Africa and France and members of the lab of Human Genetics of Infectious Diseases for helpful discussions.
Data Availability
All summary statistics are available in the manuscript or at https://doi.org/10.5281/zenodo.3942125. Because of the IRB restriction on the data from Vietnam, France and South Africa, individual level data are only available in the context of a tuberculosis research project upon request from the VFSA TB infection Access Committee by contacting the committee chair Pr Stanislas Lyonnet (stanislas.lyonnet@inserm.fr), Director of the Imagine Institute in Paris, France. Stanislas Lyonnet was not involved with the research reported in this article.
Funding Statement
This work was supported in part by the French Foundation for Medical Research (FRM) (EQU201903007798; FDM20170637664 to JQ), the Programme Hospitalier de Recherche Clinique (AOR-04-003); the Legs Poix (Chancellerie des Universités de Paris, https://www.sorbonne.fr/en/ to AA), the European Research Council (ERC-2010-AdG-268777 to LA), the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R01AI12434), the French National Research Agency (ANR) under the “Investments for the future” program (grant ANR-10-IAHU-01), the Integrative Biology of Emerging Infectious Diseases Laboratory of Excellence (ANR-10-LABX-62-IBEID), and the TBPATHGEN project (ANR-14-CE14-0007-01 to LA), the SCOR Corporate Foundation for Science (https://www.scor.com/fr/la-fondation-dentreprise-scor-pour-la-science to LA), the Rockefeller University (https://www.rockefeller.edu/), the Institut National de la Santé et de la Recherche Médicale (INSERM, https://www.inserm.fr/), Université de Paris (https://u-paris.fr/en/), the St. Giles Foundation (http://www.stgilesfoundation.org/), the Canadian Institutes of Health Research (CIHR, FDN-143332 to ES) and the Sequella/Aeras Global Tuberculosis Foundation (https://www.aeras.org, to ES). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Global tuberculosis report 2019. World Health Organization; 2019. [cited 25 Oct 2019]. Available: http://www.who.int/tb/publications/global_report/en/
- 2.Verrall AJ, Netea MG, Alisjahbana B, Hill PC, van Crevel R. Early clearance of Mycobacterium tuberculosis: a new frontier in prevention. Immunology. 2014;141: 506–513. 10.1111/imm.12223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simmons JD, Stein CM, Seshadri C, Campo M, Alter G, Fortune S, et al. Immunological mechanisms of human resistance to persistent Mycobacterium tuberculosis infection. Nat Rev Immunol. 2018;18: 575–589. 10.1038/s41577-018-0025-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma SK, Vashishtha R, Chauhan LS, Sreenivas V, Seth D. Comparison of TST and IGRA in Diagnosis of Latent Tuberculosis Infection in a High TB-Burden Setting. PLoS One. 2017;12. 10.1371/journal.pone.0169539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pai M, Menzies D. The new IGRA and the old TST: making good use of disagreement. Am J Respir Crit Care Med. 2007;175: 529–531. 10.1164/rccm.200701-024ED [DOI] [PubMed] [Google Scholar]
- 6.Gallant CJ, Cobat A, Simkin L, Black GF, Stanley K, Hughes J, et al. Tuberculin skin test and in vitro assays provide complementary measures of antimycobacterial immunity in children and adolescents. Chest. 2010;137: 1071–1077. 10.1378/chest.09-1852 [DOI] [PubMed] [Google Scholar]
- 7.Mave V, Chandrasekaran P, Chavan A, Shivakumar SVBY, Danasekaran K, Paradkar M, et al. Infection free “resisters” among household contacts of adult pulmonary tuberculosis. PLoS ONE. 2019;14: e0218034. 10.1371/journal.pone.0218034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verrall AJ, Alisjahbana B, Apriani L, Novianty N, Nurani AC, van Laarhoven A, et al. Early Clearance of Mycobacterium tuberculosis: The INFECT Case Contact Cohort Study in Indonesia. J Infect Dis. 2019. 10.1093/infdis/jiz168 [DOI] [PubMed] [Google Scholar]
- 9.Stein CM, Zalwango S, Malone LL, Thiel B, Mupere E, Nsereko M, et al. Resistance and Susceptibility to Mycobacterium tuberculosis Infection and Disease in Tuberculosis Households in Kampala, Uganda. Am J Epidemiol. 2018;187: 1477–1489. 10.1093/aje/kwx380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abel L, Fellay J, Haas DW, Schurr E, Srikrishna G, Urbanowski M, et al. Genetics of human susceptibility to active and latent tuberculosis: present knowledge and future perspectives. Lancet Infect Dis. 2018;18: e64–e75. 10.1016/S1473-3099(17)30623-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cobat A, Gallant CJ, Simkin L, Black GF, Stanley K, Hughes J, et al. Two loci control tuberculin skin test reactivity in an area hyperendemic for tuberculosis. J Exp Med. 2009;206: 2583–2591. 10.1084/jem.20090892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jabot-Hanin F, Cobat A, Feinberg J, Grange G, Remus N, Poirier C, et al. Major Loci on Chromosomes 8q and 3q Control Interferon γ Production Triggered by Bacillus Calmette-Guerin and 6-kDa Early Secretory Antigen Target, Respectively, in Various Populations. J Infect Dis. 2016;213: 1173–1179. 10.1093/infdis/jiv757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jabot-Hanin F, Cobat A, Feinberg J, Orlova M, Niay J, Deswarte C, et al. An eQTL variant of ZXDC is associated with IFN-γ production following Mycobacterium tuberculosis antigen-specific stimulation. Sci Rep. 2017;7: 1–8. 10.1038/s41598-016-0028-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stein CM, Zalwango S, Malone LL, Won S, Mayanja-Kizza H, Mugerwa RD, et al. Genome scan of M. tuberculosis infection and disease in Ugandans. PLoS ONE. 2008;3: e4094. 10.1371/journal.pone.0004094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Igo RP, Hall NB, Malone LL, Hall JB, Truitt B, Qiu F, et al. Fine-mapping Analysis of a Chromosome 2 Region Linked to Resistance to Mycobacterium tuberculosis Infection in Uganda Reveals Potential Regulatory Variants. Genes Immun. 2019;20: 473–483. 10.1038/s41435-018-0040-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cobat A, Poirier C, Hoal E, Boland-Auge A, de La Rocque F, Corrard F, et al. Tuberculin skin test negativity is under tight genetic control of chromosomal region 11p14-15 in settings with different tuberculosis endemicities. J Infect Dis. 2015;211: 317–321. 10.1093/infdis/jiu446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sobota RS, Stein CM, Kodaman N, Maro I, Wieland-Alter W, Igo RP, et al. A chromosome 5q31.1 locus associates with tuberculin skin test reactivity in HIV-positive individuals from tuberculosis hyper-endemic regions in east Africa. PLoS Genet. 2017;13: e1006710. 10.1371/journal.pgen.1006710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marcus JH, Novembre J. Visualizing the geography of genetic variants. Bioinformatics. 2017;33: 594–595. 10.1093/bioinformatics/btw643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim-Hellmuth S, Bechheim M, Pütz B, Mohammadi P, Nédélec Y, Giangreco N, et al. Genetic regulatory effects modified by immune activation contribute to autoimmune disease associations. Nat Commun. 2017;8: 1–10. 10.1038/s41467-016-0009-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calderon D, Nguyen MLT, Mezger A, Kathiria A, Müller F, Nguyen V, et al. Landscape of stimulation-responsive chromatin across diverse human immune cells. Nat Genet. 2019;51: 1494–1505. 10.1038/s41588-019-0505-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stein CM, Hall NB, Malone LL, Mupere E. The household contact study design for genetic epidemiological studies of infectious diseases. Front Genet. 2013;4: 61. 10.3389/fgene.2013.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma K, Qiu L, Mrasek K, Zhang J, Liehr T, Quintana LG, et al. Common Fragile Sites: Genomic Hotspots of DNA Damage and Carcinogenesis. Int J Mol Sci. 2012;13: 11974–11999. 10.3390/ijms130911974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan S, Qiu L, Ma K, Zhang X, Zhao Y, Zhang J, et al. FATS is an E2-independent ubiquitin ligase that stabilizes p53 and promotes its activation in response to DNA damage. Oncogene. 2014;33: 5424–5433. 10.1038/onc.2013.494 [DOI] [PubMed] [Google Scholar]
- 24.Song F, Zhang J, Qiu L, Zhao Y, Xing P, Lu J, et al. A functional genetic variant in fragile-site gene FATS modulates the risk of breast cancer in triparous women. BMC Cancer. 2015;15: 559. 10.1186/s12885-015-1570-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, et al. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. 2013;501: 512–516. 10.1038/nature12566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franco LH, Nair VR, Scharn CR, Xavier RJ, Torrealba JR, Shiloh MU, et al. The Ubiquitin Ligase Smurf1 Functions in Selective Autophagy of Mycobacterium tuberculosis and Anti-tuberculous Host Defense. Cell Host Microbe. 2017;21: 59–72. 10.1016/j.chom.2016.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Wu J, Li J, Yang H, Tang T, Liang H, et al. Host-mediated ubiquitination of a mycobacterial protein suppresses immunity. Nature. 2020;577: 682–688. 10.1038/s41586-019-1915-7 [DOI] [PubMed] [Google Scholar]
- 28.Kastenhuber ER, Lowe SW. Putting p53 in Context. Cell. 2017;170: 1062–1078. 10.1016/j.cell.2017.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Behar SM, Martin CJ, Booty MG, Nishimura T, Zhao X, Gan H-X, et al. Apoptosis is an innate defense function of macrophages against Mycobacterium tuberculosis. Mucosal Immunol. 2011;4: 279–287. 10.1038/mi.2011.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim Y-J, Lee J, Choi J-A, Cho S-N, Son S-H, Kwon S-J, et al. M1 macrophage dependent-p53 regulates the intracellular survival of mycobacteria. Apoptosis. 2020;25: 42–55. 10.1007/s10495-019-01578-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cruz A, Ludovico P, Torrado E, Gama JB, Sousa J, Gaifem J, et al. IL-17A Promotes Intracellular Growth of Mycobacterium by Inhibiting Apoptosis of Infected Macrophages. Front Immunol. 2015;6. 10.3389/fimmu.2015.00498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kveiborg M, Albrechtsen R, Couchman JR, Wewer UM. Cellular roles of ADAM12 in health and disease. Int J Biochem Cell Biol. 2008;40: 1685–1702. 10.1016/j.biocel.2008.01.025 [DOI] [PubMed] [Google Scholar]
- 33.Paulissen G, Rocks N, Quesada-Calvo F, Gosset P, Foidart J-M, Noel A, et al. Expression of ADAMs and Their Inhibitors in Sputum from Patients with Asthma. Mol Med. 2006;12: 171–179. 10.2119/2006-00028.Paulissen [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Estrella C, Rocks N, Paulissen G, Quesada-Calvo F, Noël A, Vilain E, et al. Role of A Disintegrin And Metalloprotease-12 in Neutrophil Recruitment Induced by Airway Epithelium. Am J Respir Cell Mol Biol. 2009;41: 449–458. 10.1165/rcmb.2008-0124OC [DOI] [PubMed] [Google Scholar]
- 35.Zhou AX, El Hed A, Mercer F, Kozhaya L, Unutmaz D. The Metalloprotease ADAM12 Regulates the Effector Function of Human Th17 Cells. PLoS One. 2013;8. 10.1371/journal.pone.0081146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arlehamn CL, Seumois G, Gerasimova A, Huang C, Fu Z, Yue X, et al. Transcriptional profile of TB antigen-specific T cells reveals novel multifunctional features. J Immunol. 2014;193: 2931–2940. 10.4049/jimmunol.1401151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Babu S, Bhat SQ, Kumar NP, Kumaraswami V, Nutman TB. Regulatory T Cells Modulate Th17 Responses in Patients with Positive Tuberculin Skin Test Results. J Infect Dis. 2010;201: 20–31. 10.1086/648735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coulter F, Parrish A, Manning D, Kampmann B, Mendy J, Garand M, et al. IL-17 Production from T Helper 17, Mucosal-Associated Invariant T, and γδ Cells in Tuberculosis Infection and Disease. Front Immunol. 2017;8. 10.3389/fimmu.2017.01252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gurdasani D, Barroso I, Zeggini E, Sandhu MS. Genomics of disease risk in globally diverse populations. Nat Rev Genet. 2019;20: 520–535. 10.1038/s41576-019-0144-0 [DOI] [PubMed] [Google Scholar]
- 40.Global tuberculosis report 2015. World Health Organization; 2015. Available: http://apps.who.int/iris/handle/10665/191102
- 41.Immunization Country Profile. World Health Organization [cited 1 Oct 2019]. Available: https://apps.who.int/immunization_monitoring/globalsummary/countries?countrycriteria%5Bcountry%5D%5B%5D = VNM&commit = OK
- 42.Prevalence of HIV, total (% of population ages 15–49)—Vietnam | Data. [cited 1 Oct 2019]. Available: https://data.worldbank.org/indicator/SH.DYN.AIDS.ZS?locations = VN
- 43.Antoine D. Les cas de tuberculose maladie déclarés en France en 2006. Bull Epidemiol Hebd. 2008;10–11. [Google Scholar]
- 44.Prevalence of HIV, total (% of population ages 15–49)—France | Data. [cited 20 Nov 2020]. Available: https://data.worldbank.org/indicator/SH.DYN.AIDS.ZS?locations = FR
- 45.Aissa K, Madhi F, Ronsin N, Delarocque F, Lecuyer A, Decludt B, et al. Evaluation of a model for efficient screening of tuberculosis contact subjects. Am J Respir Crit Care Med. 2008;177: 1041–1047. 10.1164/rccm.200711-1756OC [DOI] [PubMed] [Google Scholar]
- 46.de Wit E, Delport W, Rugamika CE, Meintjes A, Möller M, van Helden PD, et al. Genome-wide analysis of the structure of the South African Coloured Population in the Western Cape. Hum Genet. 2010;128: 145–153. 10.1007/s00439-010-0836-1 [DOI] [PubMed] [Google Scholar]
- 47.Naidoo P, Theron G, Rangaka MX, Chihota VN, Vaughan L, Brey ZO, et al. The South African Tuberculosis Care Cascade: Estimated Losses and Methodological Challenges. J Infect Dis. 2017;216: S702–S713. 10.1093/infdis/jix335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mathema B, Andrews JR, Cohen T, Borgdorff MW, Behr M, Glynn JR, et al. Drivers of Tuberculosis Transmission. J Infect Dis. 2017;216: S644–S653. 10.1093/infdis/jix354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hesseling AC, Caldwell J, Cotton MF, Eley BS, Jaspan HB, Jennings K, et al. BCG vaccination in South African HIV-exposed infants–risks and benefits. S Afr Med J. 2009;99: 88–91. [PMC free article] [PubMed] [Google Scholar]
- 50.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet. 2007;81: 559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48: 1284–1287. 10.1038/ng.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loh P-R, Danecek P, Palamara PF, Fuchsberger C, A Reshef Y, K Finucane H, et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet. 2016;48: 1443–1448. 10.1038/ng.3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526: 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48: 1279–1283. 10.1038/ng.3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Standards Diagnostic and Classification of Tuberculosis in Adults and Children. This official statement of the American Thoracic Society and the Centers for Disease Control and Prevention was adopted by the ATS Board of Directors, July 1999. This statement was endorsed by the Council of the Infectious Disease Society of America, September 1999. Am J Respir Crit Care Med. 2000;161: 1376–1395. 10.1164/ajrccm.161.4.16141 [DOI] [PubMed] [Google Scholar]
- 56.R: The R Project for Statistical Computing. [cited 5 Nov 2018]. Available: https://www.r-project.org/
- 57.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12: 77. 10.1186/1471-2105-12-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bates D, Maechler M, Bolker B, Walker S, Christensen RHB, Singmann H, et al. lme4: Linear Mixed-Effects Models using “Eigen” and S4. 2018. Available: https://CRAN.R-project.org/package = lme4 [Google Scholar]
- 59.Zhou X, Stephens M. Genome-wide Efficient Mixed Model Analysis for Association Studies. Nat Genet. 2012;44: 821–824. 10.1038/ng.2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lloyd-Jones LR, Robinson MR, Yang J, Visscher PM. Transformation of Summary Statistics from Linear Mixed Model Association on All-or-None Traits to Odds Ratio. Genetics. 2018;208: 1397–1408. 10.1534/genetics.117.300360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.GitHub—YinLiLin/CMplot: Circular and Rectangular Manhattan Plot. [cited 24 Jun 2020]. Available: https://github.com/YinLiLin/CMplot
- 62.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26: 2336–2337. 10.1093/bioinformatics/btq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21: 263–265. 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 64.Han B, Eskin E. Random-Effects Model Aimed at Discovering Associations in Meta-Analysis of Genome-wide Association Studies. Am J Hum Genet. 2011;88: 586–598. 10.1016/j.ajhg.2011.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haeussler M, Zweig AS, Tyner C, Speir ML, Rosenbloom KR, Raney BJ, et al. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. 2019;47: D853–D858. 10.1093/nar/gky1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ernst J, Kellis M. Chromatin-state discovery and genome annotation with ChromHMM. Nat Protoc. 2017;12: 2478–2492. 10.1038/nprot.2017.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518: 317–330. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nédélec Y, Sanz J, Baharian G, Szpiech ZA, Pacis A, Dumaine A, et al. Genetic Ancestry and Natural Selection Drive Population Differences in Immune Responses to Pathogens. Cell. 2016;167: 657–669.e21. 10.1016/j.cell.2016.09.025 [DOI] [PubMed] [Google Scholar]
- 70.Kasela S, Kisand K, Tserel L, Kaleviste E, Remm A, Fischer K, et al. Pathogenic implications for autoimmune mechanisms derived by comparative eQTL analysis of CD4+ versus CD8+ T cells. PLOS Genetics. 2017;13: e1006643. 10.1371/journal.pgen.1006643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alasoo K, Rodrigues J, Mukhopadhyay S, Knights AJ, Mann AL, Kundu K, et al. Shared genetic effects on chromatin and gene expression indicate a role for enhancer priming in immune response. Nat Genet. 2018;50: 424–431. 10.1038/s41588-018-0046-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schmiedel BJ, Singh D, Madrigal A, Valdovino-Gonzalez AG, White BM, Zapardiel-Gonzalo J, et al. Impact of Genetic Polymorphisms on Human Immune Cell Gene Expression. Cell. 2018;175: 1701–1715.e16. 10.1016/j.cell.2018.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]