Abstract
Tau and amyloid β (Aβ), 2 key pathogenic proteins in Alzheimer’s disease (AD), reportedly spread throughout the brain as the disease progresses. Models of how these pathogenic proteins spread from affected to unaffected areas had been proposed based on the observation that these proteins could transmit to other regions either through neural fibers (transneuronal spread model) or through extracellular space (local spread model). In this study, we modeled the spread of tau and Aβ using a graph theoretical approach based on resting-state functional magnetic resonance imaging. We tested whether these models predict the distribution of tau and Aβ in the brains of AD spectrum patients. To assess the models’ performance, we calculated spatial correlation between the model-predicted map and the actual map from tau and amyloid positron emission tomography. The transneuronal spread model predicted the distribution of tau and Aβ deposition with significantly higher accuracy than the local spread model. Compared with tau, the local spread model also predicted a comparable portion of Aβ deposition. These findings provide evidence of transneuronal spread of AD pathogenic proteins in a large-scale brain network and furthermore suggest different contributions of spread models for tau and Aβ in AD.
Keywords: Alzheimer’s disease, amyloid β, positron emission tomography, resting-state functional magnetic resonance imaging, tau
Introduction
Alzheimer’s disease (AD) is a neurodegenerative disorder characterized by the extracellular deposition of amyloid β (Aβ) and intracellular aggregation of hyperphosphorylated tau (Braak and Braak 1991a, 1991b). Postmortem and in vivo positron emission tomography (PET) imaging studies have demonstrated that during disease progression, tau and Aβ spread throughout the brain with characteristic spreading patterns (Braak and Braak 1991a, 1991b; Cho et al. 2016). Tau aggregates are first found in the transentorhinal cortex of the anteromedial temporal lobe and then in other parts of the temporal lobe, including entorhinal and hippocampal areas. Subsequently, tau spreads to limbic and association areas (Braak and Braak 1991a, 1991b). In contrast, patterns of Aβ plaques follow essentially the opposite direction: Aβ spreads from neocortical areas to the brainstem and cerebellum (Thal et al. 2002).
The mechanism by which these 2 pathogenic proteins spread from affected to unaffected areas has been studied, and several hypothetical models have been proposed. First, the “transneuronal spread model” suggests that pathogenic proteins spread along neuronal connections, exhibiting prion-like behavior (Goedert et al. 2017). This model is based on previous reports showing that regions with high tau and Aβ levels exhibit higher synaptic connections with each other than with regions with low tau and Aβ levels (Brettschneider et al. 2015). At the local circuit level, studies using animal models and cell culture have shown that locally infused or formed Aβ (Nath et al. 2012; Song et al. 2014) and tau (de Calignon et al. 2012; Liu et al. 2012; Wu et al. 2016) are transmitted to other neurons through neural fibers. Alternatively, observations that Aβ (Rajendran et al. 2006) and tau (Simón et al. 2012) are released into the extracellular space either in a free form or within vesicles, and uptaken by nearby neurons or glial cells, have suggested that pathogenic proteins spread to physically adjacent regions. This is termed “local spread model” in this study.
Recent neuroimaging studies have shown that the distribution of brain atrophy, hypometabolism, or Aβ can be predicted by brain connectivity, supporting the transneuronal spread of pathogenic proteins (Seeley et al. 2009; Raj et al. 2012; Zhou et al. 2012; Mutlu et al. 2017). To date, however, there has been no quantitative comparison of these 2 spread models in the brains of AD patients, or evaluation of whether tau and Aβ follow different spread models.
In this study, we modeled the spread of pathogenic proteins using a graph theoretical approach based on resting-state functional magnetic resonance imaging (rs-fMRI) data. We tested whether the models could predict the distribution of tau and Aβ in the brains of AD spectrum patients. We postulated that this could provide insight into the mechanism of spreading of tau and Aβ on a large scale, and thus the progression of the disease.
Material and Methods
Two data sets using different radiotracers for tau and amyloid PET were used in this study.
Data Set 1
Participants
First data were collected from the dementia clinic at Samsung Medical Center (Center 1) and Asan Medical Center (Center 2) in Seoul, Republic of Korea. Subjects aged 60–90 years, who were either cognitively healthy or were diagnosed with mild cognitive impairment (MCI) or AD, were recruited between January 2016 and July 2017. For inclusion as a cognitively healthy elderly subject, subjects required a normal neurological examination, a clinical dementia rating of 0, and a Mini-Mental State Examination (MMSE) score higher than 27. All participants with MCI met Petersen’s criteria (Petersen et al. 1999) and those with AD satisfied the criteria for a clinical diagnosis of AD according to the National Institute of Neurological and Communicative Disorders and Stroke and Alzheimer’s Disease and Related Disorders Association (McKhann et al. 2011). All recruited subjects underwent tau (18F-THK5351) and amyloid (18F-florbetaben) PET; in addition, cognitively healthy elderly subjects underwent rs-fMRI within 60 days of PET imaging. Among the 87 recruited subjects, we included cognitively healthy elderly subjects with negative amyloid pathology (brain amyloid plaque load score <2; Supplementary Table S1) and MCI and AD patients with positive amyloid pathology (brain amyloid plaque load score ≥2) based on amyloid (18F-florbetaben) PET (Seibyl et al. 2016). We excluded subjects with atypical AD or non-amnestic MCI. Furthermore, we inspected all imaging data for apparent artefacts and for motion artefacts, that is, for movements exceeding 2 mm on any axis or 2° on rotation, but no subject had to be excluded. Eventually, 23 cognitively healthy elderly subjects and 35 AD spectrum patients (14 MCI and 21 AD) were available for analysis.
The study protocol was approved by the institutional review boards of both Samsung Medical Center and Asan Medical Center, and informed consent was obtained from each participant according to the guidelines outlined in the Declaration of Helsinki.
MRI Acquisition and Preprocessing
T1 and T2* MR images were acquired using a 3.0-T MR scanner, Philips Intera Achieva (Philips Healthcare, Eindhoven, The Netherlands). T1-weighted anatomical MR images were obtained with repetition time = 9.9 ms, echo time = 4.6 ms, flip angle = 8°, voxel size = 1.0 × 1.0 × 0.5 mm3 for Center 1 and repetition time = 6.8 ms, echo time = 3.1 ms; flip angle = 9°, voxel size = 1.11 × 1.11 × 1.2 mm3 for Center 2. T2*-weighted MR images were obtained using a gradient echo planar imaging pulse sequence (repetition time = 3000 ms, echo time = 35 ms, flip angle = 90°, voxel size = 1.72 × 1.72 × 4 mm3, slice number = 35, frame number = 100 for Center 1 and repetition time = 3000 ms, echo time = 30 ms, flip angle = 90°, voxel size = 3.3 × 3.3 × 3.3 mm3, slice number = 47, frame number = 140 for Center 2).
During rs-fMRI scanning, subjects were instructed to remain awake with their eyes closed. The following preprocessing steps were performed on the rs-fMRI data, using Statistical Parametric Mapping software 12 (SPM, http://www.fil.ion/uc.ac.uk/spm) and our custom codes, running on MATLAB (MathWorks, Natrick, MA): 1) slice-timing correction, 2) realignment with rigid-body transformation, 3) linear detrending, 4) regressing-out of nuisance variables (12 parameters of rigid body head motion, signals averaged over deep white matter and lateral ventricles, and signals averaged over the whole brain), 5) normalization to a Montreal Neurological Institute (MNI) space (voxel size 2 × 2 × 2 mm3), 6) spatial smoothing with a Gaussian kernel of 6-mm full-width at half-maximum (FWHM), and 7) temporal filtering at 0.01–0.1 Hz. After removing the first 5 volumes for T1-equilibration effects, 95 (285 s) and 135 volumes (405 s) were used for constructing the connectivity map for each center, respectively. Because performing global signal regression during preprocessing is still controversial (Murphy et al. 2009), we analyzed our data both with and without global signal regression (see Supplementary Result).
PET Acquisition and Preprocessing
We used a Discovery STE PET/CT scanner at Center 1 and Discovery 690, 710, and 690 Elite PET/CT scanner at Center 2 (GE Healthcare, Milwaukee, WI, USA). The same imaging and reconstruction protocol was used in both centers. Amyloid PET images were acquired for 20 min, starting at 90 min after the intravenous bolus injection of 300 ± 30 MBq of 18F-florbetaben (Barthel et al. 2011). Tau PET images were acquired for 20 min, starting at 50 min after the intravenous bolus injection of 185 ± 18.5 MBq of 18F-THK5351, which binds to the aggregated tau in paired helical filaments (Harada et al. 2016). Prior to the PET scan, we applied a head holder to minimize head motion and acquired brain CT images for attenuation correction. Using the ordered-subsets expectation maximization algorithm (iteration = 4, subset = 16), 3D PET images were reconstructed with a voxel size of 2.0 × 2.0 × 3.27 mm3. To increase data uniformity between different PET scanners, we applied a 3D Hoffman phantom-based PET harmonization method, as described by Joshi et al. (2009). We calculated the standardized uptake value (SUV) by normalizing the tissue radioactivity concentration by the injected dose (MBq) and body weight (kg). We corrected the partial volume effects (PVE) for SUV images using the geometric transfer matrix approach (Rousset et al. 1998; Gonzalez-Escamilla et al. 2017). In PVE correction, we used brain parcellation based on automated anatomical labeling (AAL), which is typically used in functional neuroimaging research, comprising 116 cortical and subcortical regions (Tzourio-Mazoyer et al. 2002). Subsequently, SUV images were coregistered to T1-weighted anatomical images. After T1 image segmentation, the brain parcellation atlas in MNI space was normalized to T1-coregistered PET image, and region-wise PVE corrected data were obtained. Finally, we calculated the regional SUV ratio (SUVR), using the cerebellar cortex value as a reference. We measured both PVE corrected and uncorrected data and primarily used the former for the analysis; we also confirmed the results using PVE uncorrected data (see Supplementary Results).
For tau PET, we only included cortical regions in the analysis, considering “off-target” binding of tau tracer at subcortical structures (Betthauser et al. 2017). After excluding cerebellar regions, 82 and 90 regions were analyzed for tau and Aβ, respectively (Supplementary Table S2).
Because tau and Aβ are also found in the brains of cognitively healthy elderly subjects, we computed the regional t values of PET between the cognitively healthy subjects and AD spectrum patients, to focus on AD-related tau and Aβ distribution. The t value was calculated from 2-sample t-test after controlling for age, sex, and level of education. Thus, a higher positive t value signified a higher pathogenic protein burden in AD spectrum patients than in cognitively healthy subjects. Negative t values were regarded as noise and were therefore set to zero. As this study aimed to evaluate the distribution (spatial pattern) of pathogenic protein deposition, we rescaled the t value of tau and Aβ PET from 0 to 1 using the min–max normalization before the analysis in order to have a similar scale for tau and Aβ.
Constructing the Connectivity Map
For constructing the connectivity map, we used the rs-fMRI data of cognitively healthy elderly subjects. We used AAL parcellation to define the node. The averaged rs-fMRI time series were extracted for each region after masking it with individual gray matter probability maps higher than 0.5. Pearson’s correlation coefficients between the rs-fMRI time series of each region were calculated to determine functional connectivity. We only included positive functional connections after false discovery rate multiple comparison correction (at a q level of <0.01) in the connectivity map, to exclude spurious connections. Furthermore, we transformed the correlation value with Fisher’s z normalization for normality of the data. A group-averaged functional connectivity map was used in the analysis.
Identification of Pathogenic Protein Epicenters
We assumed that regions with a higher burden of pathogenic proteins, tau or Aβ, would more likely be the sites of initial deposition, acting as epicenters for pathogenic protein spreading. Furthermore, we considered that, if spreading occurred via a diffusion process, molecules would spread from an area of high to an area of low concentration. Thus, we identified regions as epicenters if they contained high levels of tau or Aβ (above a certain threshold of normalized t value), as determined using the PET images. To avoid arbitrariness, we applied a wide range of thresholds, from 0.1 to 0.9 (increasing by steps of 0.1), when determining these epicenters. Therefore, the number of epicenters varied according to the applied thresholds. We assessed whether results were consistent across the thresholds.
Transneuronal Spread Model
We modeled transneuronal spreading of pathogenic proteins as a process of diffusion from the epicenters to other regions, through the brain network. Thus, we hypothesized that the regional levels of pathogenic proteins depended on the functional network distance from the epicenters. For the transneuronal spread model, we used a linear regression model composed of a spread variable and intercept, expressed as follows:
| (1) |
where Pi is the predicted level of pathogenic protein at the ith region, Li,Ej is the shortest weighted path length between the ith region and the jth epicenter (Ej), CEj is the level of pathogenic protein at the jth epicenter, and e is the total number of epicenters. Β and ɑ are the coefficients for spread variable and intercept, respectively. The shortest weighted path length is a value that minimizes the sum of all edges between 2 regions (Rubinov and Sporns 2010) and is expressed as follows:
| (2) |
where f is a function that inverses connection weights, Wuv, and is the shortest weighted path between the ith and jth regions. We set the self-connection to zero. Therefore, the level of pathogenic protein at one epicenter was predicted based on its network distance from other epicenters. We weighted the contribution of each epicenter by its pathogenic protein levels (CEj), considering that an epicenter with higher levels of pathogenic protein would be a site of earlier deposition and have a higher contribution to the spread of the protein. Therefore, according to the model, the regional level of pathogenic protein was predicted by the normalized sum of pathogenic protein, afferent from all epicenters.
Local Spread Model
We developed a “local spread model,” under the hypothesis that the regional levels of pathogenic proteins depend on the physical distances from epicenters. The local spread model uses the Euclidean distances between regions, instead of network distances and is expressed as follows:
| (3) |
where Di,Ej is the Euclidean distance between the ith region and thejth epicenter. We used the coordinates of the center of mass given for each brain region to calculate the Euclidean distances, which is expressed as follows:
| (4) |
where (i1, i2, i3) and (j1, j2, j3) are the coordinates for the center of mass of the ith region and jth region.
Graph theoretical measurements of the shortest path length and Euclidean distance were calculated using the Brain Connectivity Toolbox (http://www.brain-connectivity-toolbox.net). The results were visualized using the BrainNet viewer (http://www.nitrc.org.projects.bnv).
Statistical Analysis
We calculated the spatial correlation between the model-predicted and actual measured map (from tau and Aβ PET) to evaluate the spatial similarity between the 2 maps and, thus, assess the prediction performance of the model. In order to infer the statistical significance of the model, we performed a permutation test. Under the assumption that the accuracy of the model prediction depended on the particular organization of the brain, we randomly scrambled the network, as well as a Euclidian distance map (10 000 permutations). From these permutations, R values between the predicted map, based on the random map, and the actual measured map were calculated and their null distribution was evaluated. We set R values higher than the highest 5% of the 10 000 permutations as significant (permutation P value < 0.001).
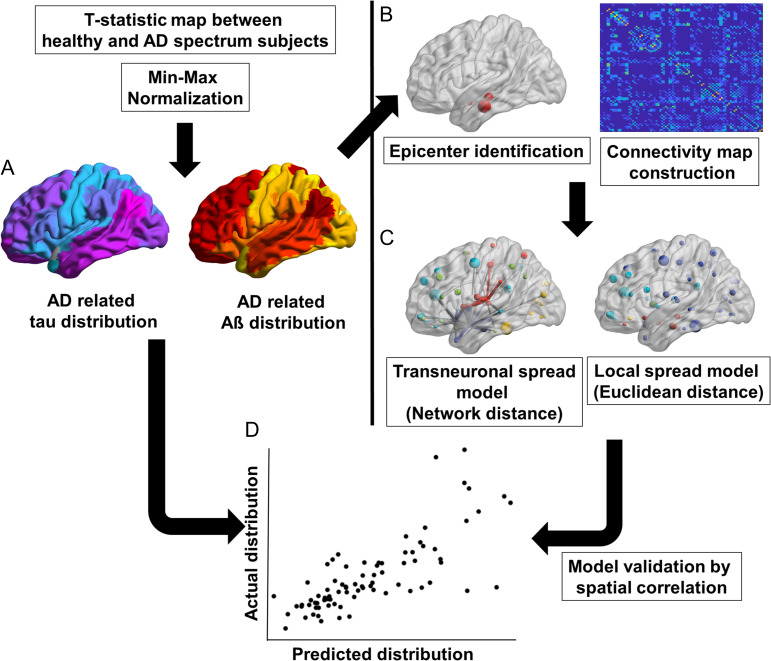
To compare the performance between the 2 models (transneuronal spread model and local spread model), we calculated Fisher’s R to z transformation, a tool that measures whether 2 given Pearson’s correlation coefficients (R value) are statistically significantly different. We used 2-tailed tests, with statistical significance set at P < 0.05. Lastly, to evaluate the robustness of our results to intersubject variability, we performed a bootstrap analysis. The analysis was repeated 1000 times using random bootstrap samples of cognitively healthy subjects and patients with AD spectrum disorders. The 95% confidence interval of the results were calculated. For all statistical analyzes, MATLAB was used. The overall study design is illustrated in Figure 1.
Figure 1.
Schematic diagram of the analysis steps. Steps included: (A) calculation of the t-statistical map for tau and Aβ between healthy subjects and AD spectrum patients, (B) identification of tau and Aβ epicenters from PET images and brain network construction from rs-fMRI data, (C) construction of the predicted maps using the transneuronal spread model and local spread model, and (D) model validation by calculating the spatial correlation between the predicted and actually measured map. Aβ, amyloid β; AD, Alzheimer’s disease; rs-fMRI, resting-state functional magnetic resonance imaging; PET, positron emission tomography.
Replication in AD Neuroimaging Initiative Cohort (Data Set 2)
Since we aimed to evaluate the model that predicts the distribution of tau and Aβ in patients with AD spectrum disorders, we tested whether our findings could be replicated with data collected using other tau and Aβ PET radiotracers. For this purpose, we used tau (18F-AV1451) and amyloid (18F-florbetapir) PET data extracted from AD Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu), as a replication cohort. Since different PET tracers were used for generating these data, our results would be more generalizable if we could replicated them in this cohort. We applied the same inclusion and exclusion criteria as for data set 1 and eventually analyzed 49 cognitively healthy subjects and 45 AD spectrum patients (Supplementary Table S3). Detailed description of participants and PET data is provided in Supplementary Methods.
Analysis Using a Different Brain Parcellation Scheme
In order to demonstrate that the performance of the 2 models was invariant to the brain parcellation scheme used, we performed an additional analysis using different brain parcellation. We created non-overlapping spherical masks of 3-mm radius, according to the coordinates described by Power et al. (2011) and performed the same analysis (see Supplementary Methods).
Results
Participants
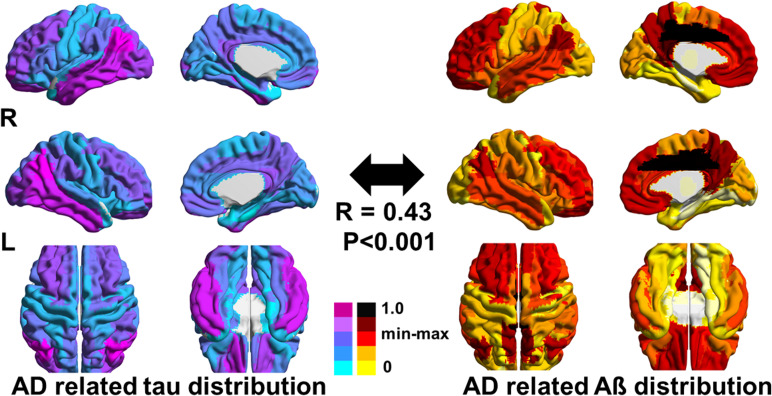
Table 1 shows the demographics of the study participants. As expected, AD spectrum patients performed worse on the MMSE than did cognitively healthy elderly subjects. The t-statistic maps of PET data showed a spatially distinct distribution of tau and Aβ. Tau was mainly deposited in the inferior temporal and parietal areas, while Aβ was deposited mainly in the medial frontal and parietal areas (Fig. 2).
Table 1.
Study population demographics
| Demographics | Healthy (N = 23) | AD spectrum (N = 35) | P value |
|---|---|---|---|
| Age, mean (SD), years | 69.73 (5.72) | 71.74 (5.48) | 0.18 |
| Onset age, mean (SD), years | 67.06 (5.86) | ||
| Female, N (%) | 18 (78) | 24 (68) | 0.41 |
| Education, mean (SD), years | 11.56 (4.37) | 10.85 (4.43) | 0.55 |
| K-MMSE, median (IQR), score | 29 (28–30) | 23 (20–26.5) | <0.001 |
| CDR 0/0.5/1, N | 23/0/0 | 35/0/0 | |
| Presence of ε4 allele of APOE, N (%)a | 5 (21) | 9 (25) | 0.73 |
| Hypertension, N (%) | 9 (39) | 11 (31) | 0.54 |
| Diabetes, N (%) | 1 (4) | 5 (14) | 0.22 |
| Family history of dementia, N (%) | 4 (17) | 4 (11) | 0.51 |
| Center 1/Center 2, N | 10/13 | 13/22 | 0.62 |
P values and statistics were calculated using Student t-test, Mann–Whitney test, or chi-squared test (2-tailed). aSubject has one or more ε4 alleles.
AD, Alzheimer’s disease; IQR, interquartile range; CDR, clinical dementia rating scale; K-MMSE, Korean version of mini-mental state examination; SD, standard deviation.
Figure 2.
t-Statistical maps of tau and Aβ calculated from PET data. R values and statistics were calculated using Pearson’s correlation test. t Values were rescaled using min–max normalization. Aβ, amyloid β; AD, Alzheimer’s disease; PET, positron emission tomography.
Prediction of Tau Distribution
In the permutation test, the highest 5% cutoff was found at an R value of 0.2 for both the network and Euclidean distance random maps (Supplementary Fig. S1).
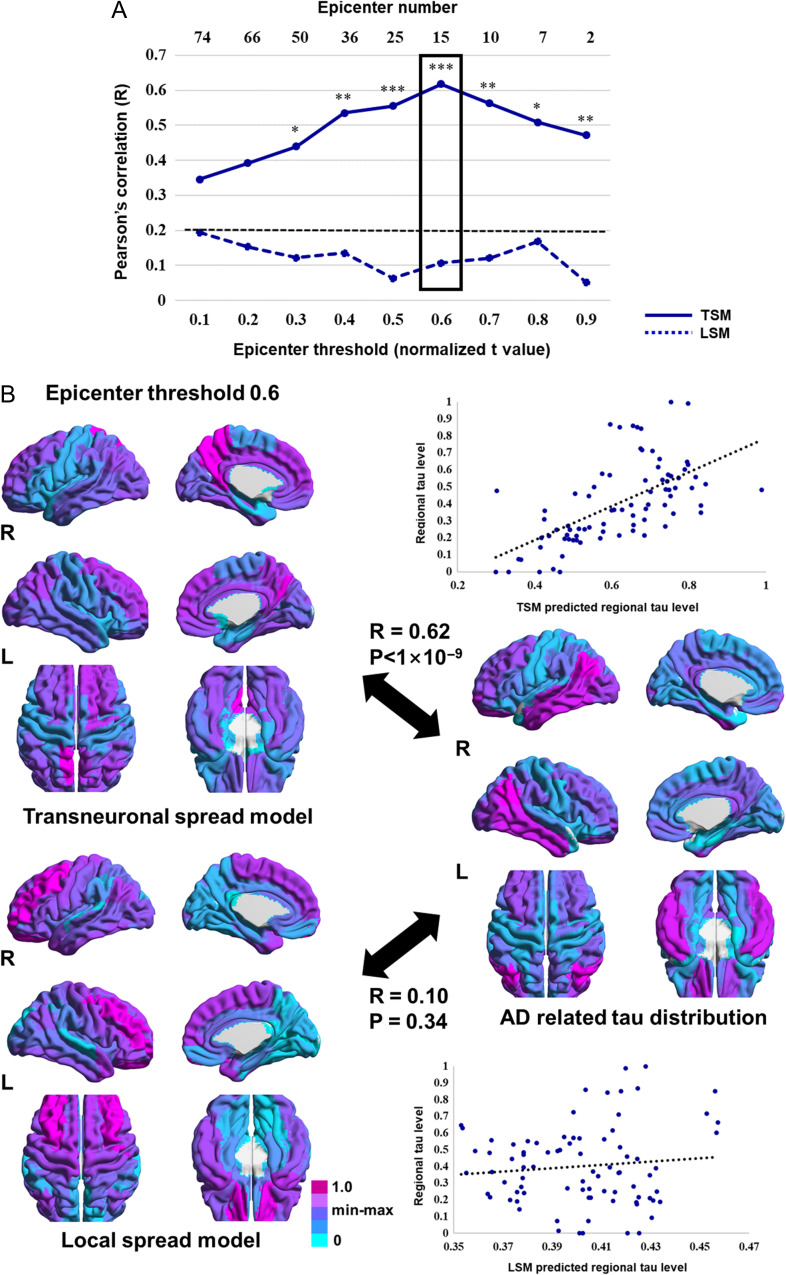
The spatial correlation between the predicted tau map, based on the transneuronal spread model, and the actual measured map was consistently higher than the cutoff value across the epicenter thresholds. Fisher’s R to z transformation showed that correlations obtained from the transneuronal spread model were significantly higher than those obtained from the local spread model (Fig. 3A).
Figure 3.
Comparison between the predicted and actually measured tau map. (A) Spatial correlation between the model-predicted and actually measured tau map across the different epicenter thresholds. Normalized t values were used in epicenter thresholds. The dotted line (black) indicates the highest 5% of R values obtained from the permutation test. The highest correlation was found for the transneuronal spread model at a threshold of 0.6 (box). (B) The predicted tau maps at an epicenter threshold of 0.6 and the actually measured tau map are shown by scatter plots; each dot represents a brain region. The dotted line represents the best-fitted line. R values and statistics were calculated using Pearson’s correlation. Significant differences between 2 R values were calculated using Fisher’s R to z transformation. *P < 0.05; **P < 0.01; ***P < 0.001 (2-tailed). AD, Alzheimer’s disease; LSM, local spread model; TSM, transneuronal spread model.
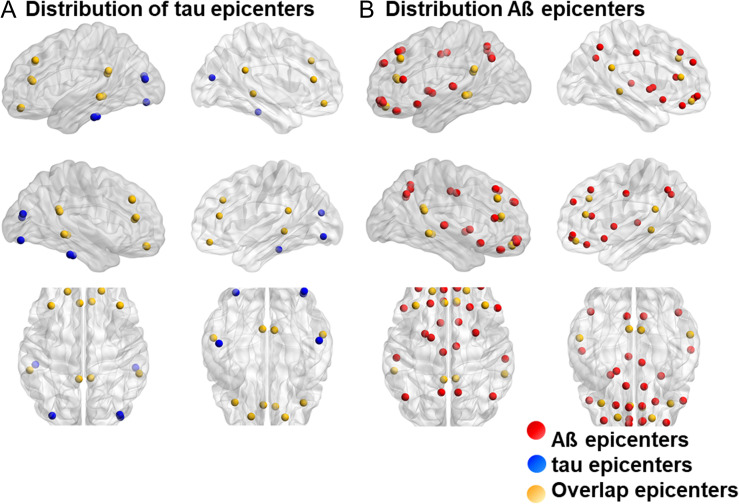
While the performance of each model depended on the number of epicenters, the highest correlation was found for the transneuronal spread model at an epicenter threshold of 0.6. At this threshold, 15 regions, including the bilateral angular, posterior cingulate, middle frontal, superior orbitofrontal, middle occipital, inferior and middle temporal, and right inferior occipital cortices, were identified as epicenters, that is, best-fit epicenters for the model (Fig. 4). Functional distances among these epicenters and other brain regions explained 38% of the tau distribution, while Euclidean distances poorly explained the distribution (Fig. 3B). Statistics and estimated parameters of the models are included in Table 2.
Figure 4.
Map of Aß and tau epicenters (best-fit epicenters of the transneuronal spread model). (A) Tau epicenters at a threshold of 0.6 (blue dots), (B) Aß epicenters at a threshold of 0.5 (red dots), and epicenters overlapping in both tau and Aß (yellow dots) are depicted in the map. Aβ, amyloid β.
Table 2.
Linear regression analysis of tau and Aß distribution for the transneuronal spread model and local spread model
| Tau distribution | ||||
|---|---|---|---|---|
| At threshold 0.6 | Coefficient | SE | t value | P value |
| Transneuronal spread model | ||||
| Intercept (ɑ) | −0.60 | 0.14 | −4.17 | <0.001 |
| Spread variable (ß) | 5.44 | 0.77 | 7.03 | <0.001 |
| R2/R | 0.38/0.62 | |||
| P value | <0.001 | |||
| Local spread model | ||||
| Intercept (ɑ) | 0.59 | 0.19 | 2.97 | 0.004 |
| Spread variable (ß) | 16.926 | 17.62 | 0.96 | 0.33 |
| R2/R | 0.01/0.10 | |||
| P value | 0.34 | |||
| Aß distribution | ||||
|---|---|---|---|---|
| At threshold 0.5 | Coefficient | SE | t value | P value |
| Transneuronal spread model | ||||
| Intercept (ɑ) | −0.21 | 0.09 | −2.18 | 0.03 |
| Spread variable (ß) | 3.64 | 0.54 | 6.71 | <0.001 |
| R2/R | 0.34/0.58 | |||
| P value | <0.001 | |||
| Local spread model | ||||
| Intercept (ɑ) | 0.12 | 0.11 | 1.13 | 0.25 |
| Spread variable (ß) | 29.01 | 9.82 | 2.95 | 0.004 |
| R2/R | 0.09/0.30 | |||
| P value | <0.004 | |||
Coefficients and statistical values were calculated using linear regression analysis.
Aß, amyloid ß; SE, standard error.
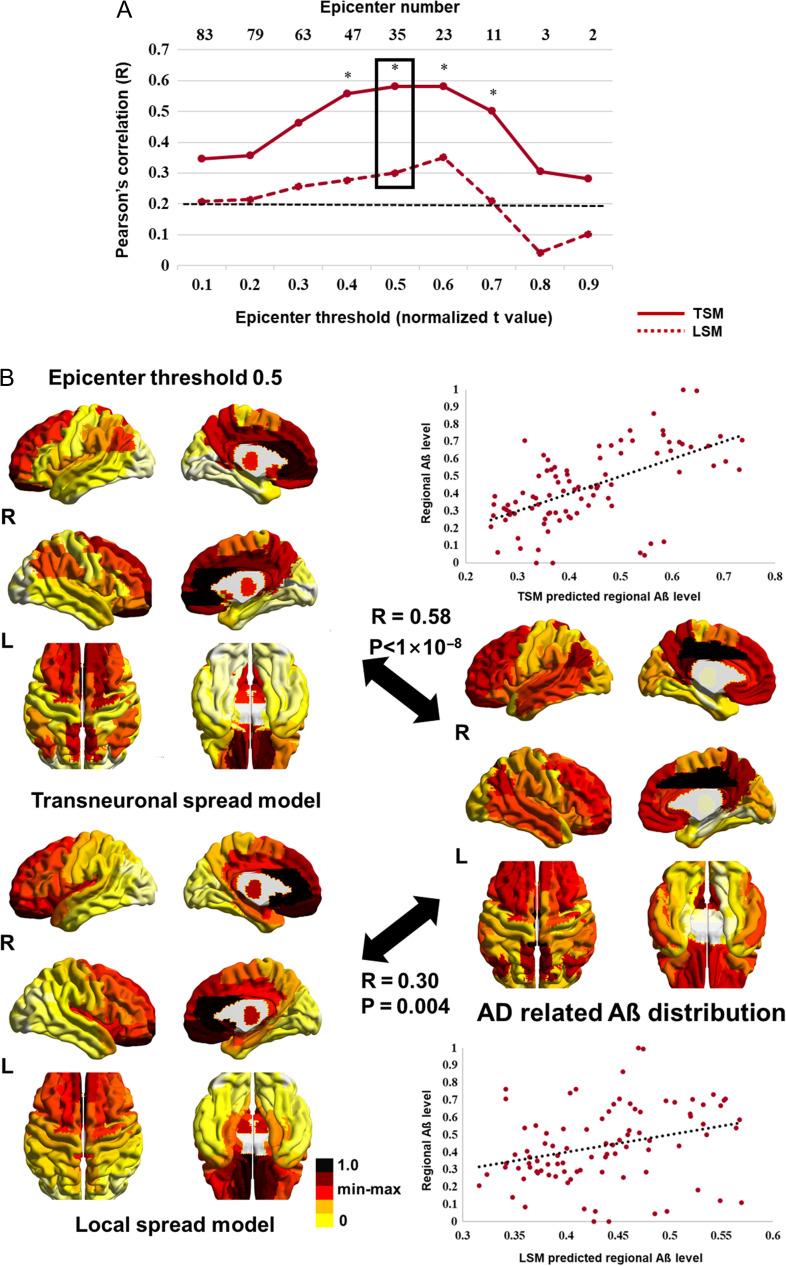
Prediction of Aβ Distribution
The correlation between the predicted Aβ map, based on both models, and the actual measured map was consistently high across different epicenter thresholds. The correlations obtained from the transneuronal spread model were significantly higher than those obtained from the local spread model at thresholds ranging from 0.4 to 0.7 (Fig. 5A). The highest correlation was found for the transneuronal spread model at an epicenter threshold of 0.5. At this threshold, 35 regions were identified as epicenters, mostly located in the frontoparietal area (Fig. 4). With these epicenters, the transneuronal spread model explained 34% of the Aβ distribution, while the local spread model explained 9% of the distribution (Fig. 5B). Statistics and estimated parameters of the models are included in Table 2 (for estimated parameters using different epicenter thresholds, see Supplementary Table S4).
Figure 5.
Comparison between the predicted and the actually measured Aβ map. (A) Spatial correlation between the model-predicted and the actually measured Aβ map across the different epicenter thresholds. Normalized t values were used in epicenter thresholds. The dotted line (black) indicates the highest 5% of R values obtained from the permutation test. The highest correlation was found for the transneuronal spread model at a threshold of 0.5 (box). (B) The predicted Aβ maps at an epicenter threshold of 0.5 and the actually measured Aβ map are shown in scatter plots, in which, each dot represents a brain region. The dotted line represents the best-fitted line. R values and statistics were calculated using Pearson’s correlation test. Significant differences between 2 R values were calculated using Fisher’s R to z transformation. *P < 0.05 (2-tailed). Aβ, amyloid β; AD, Alzheimer’s disease; LSM, local spread model; TSM, transneuronal spread model.
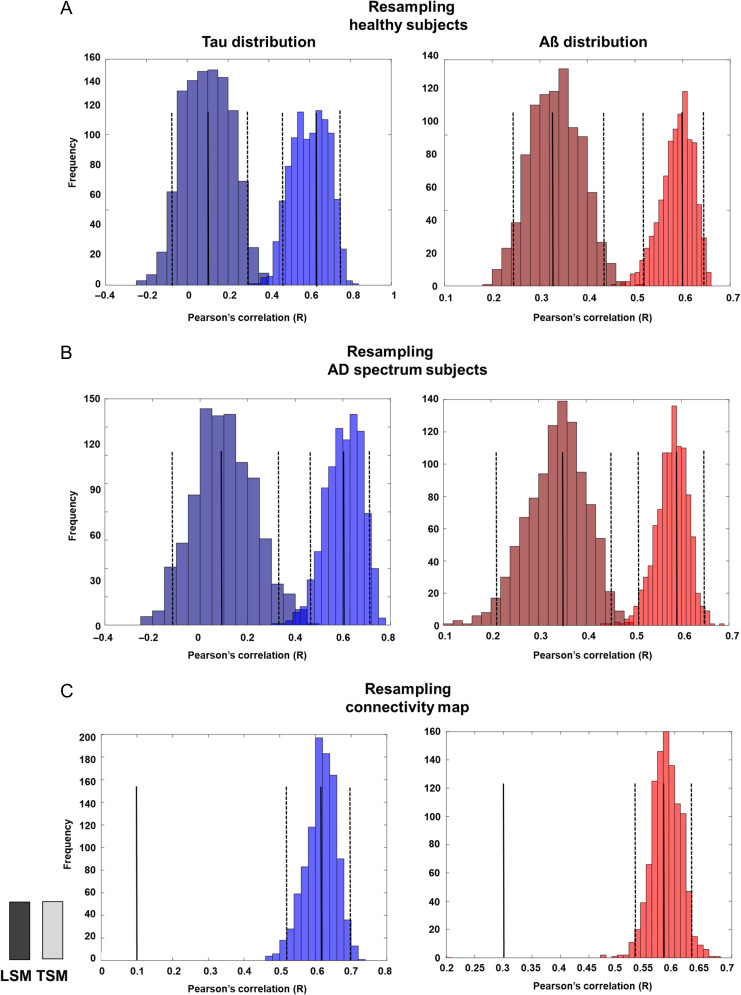
Evaluating Robustness Using Bootstrap Analysis
First, 1000 bootstrap samples, with replacement of 23 healthy subjects’ PET images (tau and amyloid PET), were evaluated. Using the bootstrap samples, 1000 R values were calculated from the 2 models (transneuronal spread model and local spread model) at epicenter threshold of 0.6 and 0.5 for tau and Aβ, respectively. The histogram of R values demonstrated that R values differed significantly between the 2 models (Fig. 6A).
Figure 6.
Bootstrap histogram of R values. We used 1000 bootstrap samples to calculate R values from the 2 models (transneuronal spread model and local spread model) at an epicenter threshold of 0.6 and 0.5 for tau and Aβ, respectively. (A) Distribution of R values from bootstrap samples, after replacing the PET images (tau and amyloid PET) of 23 healthy subjects. (B) Distribution of R values from bootstrap samples, after replacing PET images of 35 AD spectrum patients. (C) Distribution of R values from bootstrap samples, after replacing rs-fMRI data (connectivity map) of 23 healthy subjects. The solid line represents the median, and the dotted line represents the 95% confidence interval for each histogram. Aβ, amyloid β; AD, Alzheimer’s disease; LSM, local spread model; PET, positron emission tomography; rs-fMRI, resting-state functional magnetic resonance imaging; TSM, transneuronal spread model.
We then again evaluated bootstrap samples with replacement of 35 AD spectrum patients’ PET images. The bootstrap results showed that R values differed significantly between the 2 models (Fig. 6B).
Lastly, bootstrap samples, with replacement of 23 healthy subjects’ rs-fMRI data (connectivity map), were evaluated. This demonstrated that R values calculated from the transneuronal spread model were significantly higher than those calculated from the local spread model for tau and Aβ (Fig. 6C).
Replication in the ADNI Cohort (Data Set 2)
When different data were used in the analysis, we obtained results very similar to our primary results (Supplementary Figs S2 and S3). The transneuronal spread model predicted the distribution of tau and Aβ deposition with significantly higher accuracy than the local spread model, but the local spread model also predicted a comparable portion of the Aβ deposition.
Analysis Using Different Brain Parcellation Scheme
While the use of the different brain parcellation scheme gave results that were similar to our primary results, the overall performance of the local spread model improved across the epicenter thresholds for both tau and Aβ (Supplementary Fig. S4).
Discussion
In this study, we modeled the spreading of pathogenic proteins using a graph theoretical approach and validated 2 currently considered models, using imaging data of AD spectrum patients. Overall, our results revealed that the transneuronal spread model outperformed the local spread model in predicting the distribution of the pathogenic proteins, tau and Aβ.
Previous studies have demonstrated the transneuronal spreading of tau and Aβ in AD, both in vitro and in vivo (Liu et al. 2012; Nath et al. 2012; Song et al. 2014; Wu et al. 2016). The results of the current study indicate that the transneuronal spread model is also valid in large-scale brain networks. Similarly, previous studies have shown that brain atrophy follows a network pattern, which might reflect the transneuronal spreading of pathogenic proteins (Raj et al. 2012; Zhou et al. 2012; Mutlu et al. 2017). In the current study, we provide direct evidence for pathogenic protein spreading, using tau and amyloid PET imaging. Furthermore, a previous study by Mutlu et al. (2017) showed that the distribution of Aβ can be predicted based on the functional connectivity of brain areas with the epicenter of Aβ deposition. However, while the study of Mutlu et al. used one seed region as an epicenter, we allowed for a larger number of epicenters, which may contribute to the spreading of Aβ.
The best performance was found for the transneuronal spread model, with 15 and 35 epicenters for tau and Aβ, respectively. Recently, study using longitudinal PET data demonstrated “out-degree hubs” for tau and Aβ propagation. Interestingly, epicenters in our study overlap with the hubs, suggesting that epicenters are source of pathogenic protein propagation (Sepulcre et al. 2018). While epicenter distribution differed between tau and Aβ, epicenters overlapped in areas of the angular, posterior cingulate, middle temporal, and middle and superior orbitofrontal cortices (Fig. 4). These regions form part of the default mode network, which is metabolically active and highly connected with the rest of the brain. Considering that such network hubs are more vulnerable to high basal metabolism and oxidative stress (Melov et al. 2007; Vlassenko et al. 2010), they could be sites of tau and Aβ deposition from which the protein spreading is initiated.
Interestingly, while the transneuronal spread model outperformed the local spread model in predicting the distribution of pathogenic proteins, the local spread model also predicted a comparable portion of Aβ deposition. Considering that tau forms neurofibrillary tangles intracellularly, while Aβ forms amyloid plaques extracellularly (Braak and Braak 1991a, 1991b), it can be speculated that, compared with tau, a larger proportion of Aβ could diffuse through the extracellular space, in addition to spreading transneuronally. Thus, our findings might reflect the different contribution of spreading model for tau and Aβ in AD.
In this study, we used a t-statistical map of PET data between cognitively healthy subjects and AD spectrum patients, to focus on AD-related tau and Aβ distribution, as well as their spreading mechanism. The tau t-statistical map showed high levels of tau in the angular gyrus, inferior temporal, and posterior cingulate cortices but relatively low levels in the medial temporal cortex. This result is consistent with the results of previous studies showing that aggregated tau in the medial temporal lobe (area of Braak stages I and II) is prevalent in cognitively healthy elderly subjects, whereas isocortical accumulation of tau is associated with AD progression (Markesbery 1997; Braak et al. 2011; Schöll et al. 2016). This condition, termed primary age-related tauopathy (Crary et al. 2014; Jellinger et al. 2015), has been repeatedly demonstrated in both neuropathological (Davis et al. 1999; Bennett et al. 2006; Braak et al. 2011) and PET studies (Schöll et al. 2016; Sone et al. 2017).
Interestingly, when we used smaller brain parcellation, the performance of the local spread model improved for both tau and Aβ. We attributed this finding to the fact that local spreading occurs at a smaller scale in the brain than the transneuronal spreading. Therefore, the local spread model could perform better when using small brain parcellation (Supplementary Fig. S4).
To demonstrate the statistical significance and robustness of our results, we performed a permutation test using random map and bootstrap analysis by resampling from the data. The results obtained from random maps demonstrated that our primary results are less likely to be false positive. Moreover, bootstrap analysis showed that the data used in this study provided self-consistent results, robust to intersubject variability in terms of diagnosis (cognitively healthy and AD spectrum) and image modality (PET and rs-fMRI). Furthermore, we demonstrated that our findings are also valid when using different PET tracers (18F-AV1451 and 18F-florbetapir), as well as brain parcellation schemes.
This study had some limitations. First, while 18F-THK5351 has a high affinity to tau neurofibrillary tangles (Harada et al. 2016), a substantial reduction in its uptake was reported after administration of the monoamine oxidase B (MAO-B) inhibitor, selegiline (Ng et al. 2017). It was found that 18F-THK5351 uptake reflects both tau pathology and increased adjacent MAO activity due to astrogliosis, known as tau-associated neuroinflammation (Harada et al. 2018). In this study, we excluded subcortical structures which express high concentrations of MAO from the analysis, to minimize the effects of off-target binding. In addition, we expected that using t-statistical maps would minimize the confounding effect of the physiological binding of 18F-THK5351 to MAO. Furthermore, we replicated the primary findings in the data using different tau tracer (18F-AV1451), which showed low off-target binding to MAO (Hansen et al. 2018). Nevertheless, our findings should be interpreted with some caution, as they might reflect the tau-associated neuroinflammation. Second, in this cross-sectional study, we could not confirm whether the pathogenic proteins had spread from the affected to the unaffected regions but could only infer their spreading based on their relative level in one region compared with that in other regions (spatial information). Opposite to our results, a recent study by Whittington et al. reported that Aβ accumulates by regional carrying capacities rather than by spreading from a small number of regions (Whittington et al. 2018). Nevertheless, several previous animal and cell studies support our findings, indicating a cell-to-cell transmission of pathogenic proteins (Brettschneider et al. 2015). Therefore, further research, such as a longitudinal study, is needed to solve this issue. Third, as the model was based on the brain network (self-connection was set to zero), local processes that are independent of other brain regions, were not reflected by the model. Lastly, the sample size was too small. Although we have validated our findings in 2 different data sets, they should be interpreted with caution and replicated using a larger data sample. Moreover, a complementary approach to assess connectivity (e.g., structural connectivity), as well as an individual approach, could be tested in future studies.
In conclusion, we compared 2 models to describe the spreading of pathogenic proteins in the brains of AD spectrum patients. Our findings provide evidence for a network-based spreading of both tau and Aβ in large-scale brain networks and further suggest the different contributions of the spread models for tau and Aβ. Lastly, as transneuronal spreading of pathogenic proteins is considered a general process of neurodegenerative disease progression, the present approach can be applied to other neurodegenerative diseases.
Supplementary Material
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report.A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
Funding
Grant (Grant number HI14C2768) from the Korea Health Technology Research and Development Project through the Korea Health Industry Development Institute, funded by the Ministry of Health & Welfare, Republic of Korea.
Notes
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. Conflict of Interest: None declared.
References
- Barthel H, Gertz H-J, Dresel S, Peters O, Bartenstein P, Buerger K, Hiemeyer F, Wittemer-Rump SM, Seibyl J, Reininger J. 2011. Cerebral amyloid-β PET with florbetaben (18 F) in patients with Alzheimer's disease and healthy controls: a multicentre phase 2 diagnostic study. Lancet Neurol. 10:424–435. [DOI] [PubMed] [Google Scholar]
- Bennett D, Schneider J, Arvanitakis Z, Kelly J, Aggarwal N, Shah R, Wilson R. 2006. Neuropathology of older persons without cognitive impairment from two community-based studies. Neurology. 66:1837–1844. [DOI] [PubMed] [Google Scholar]
- Betthauser TJ, Lao PJ, Murali D, Barnhart TE, Furumoto S, Okamura N, Stone CK, Johnson SC, Christian BT. 2017. In vivo comparison of tau radioligands 18F-THK-5351 and 18F-THK-5317. J Nucl Med. 58:996–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Braak E. 1991. a. Demonstration of amyloid deposits and neurofibrillary changes in whole brain sections. Brain Pathol. 1:213–216. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. 1991. b. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82:239–259. [DOI] [PubMed] [Google Scholar]
- Braak H, Thal DR, Ghebremedhin E, Del Tredici K. 2011. Stages of the pathologic process in Alzheimer disease: age categories from 1 to 100 years. J Neuropathol Exp Neurol. 70:960–969. [DOI] [PubMed] [Google Scholar]
- Brettschneider J, Del Tredici K, Lee VM-Y, Trojanowski JQ. 2015. Spreading of pathology in neurodegenerative diseases: a focus on human studies. Nat Rev Neurosci. 16:109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Choi JY, Hwang MS, Kim YJ, Lee HM, Lee HS, Lee JH, Ryu YH, Lee MS, Lyoo CH. 2016. In vivo cortical spreading pattern of tau and amyloid in the Alzheimer disease spectrum. Ann Neurol. 80:247–258. [DOI] [PubMed] [Google Scholar]
- Crary JF, Trojanowski JQ, Schneider JA, Abisambra JF, Abner EL, Alafuzoff I, Arnold SE, Attems J, Beach TG, Bigio EH. 2014. Primary age-related tauopathy (PART): a common pathology associated with human aging. Acta Neuropathol. 128:755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D, Schmitt F, Wekstein D, Markesbery W. 1999. Alzheimer neuropathologic alterations in aged cognitively normal subjects. J Neuropathol Exp Neurol. 58:376–388. [DOI] [PubMed] [Google Scholar]
- de Calignon A, Polydoro M, Suárez-Calvet M, William C, Adamowicz DH, Kopeikina KJ, Pitstick R, Sahara N, Ashe KH, Carlson GA. 2012. Propagation of tau pathology in a model of early Alzheimer’s disease. Neuron. 73:685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M, Masuda-Suzukake M, Falcon B. 2017. Like prions: the propagation of aggregated tau and α-synuclein in neurodegeneration. Brain. 140:266–278. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Escamilla G, Lange C, Teipel S, Buchert R, Grothe MJ, Initiative AsDN . 2017. PETPVE12: an SPM toolbox for partial volume effects correction in brain PET–application to amyloid imaging with AV45-PET. Neuroimage. 147:669–677. [DOI] [PubMed] [Google Scholar]
- Hansen AK, Brooks DJ, Borghammer P. 2018. MAO-B inhibitors do not block in vivo flortaucipir ([18 F]-AV-1451) binding. Mol Imaging Biol. 20:356–360. [DOI] [PubMed] [Google Scholar]
- Harada R, Ishiki A, Kai H, Sato N, Furukawa K, Furumoto S, Tago T, Tomita N, Watanuki S, Hiraoka K. 2018. Correlations of 18F-THK5351 PET with postmortem burden of tau and astrogliosis in Alzheimer disease. J Nucl Med. 59:671–674. [DOI] [PubMed] [Google Scholar]
- Harada R, Okamura N, Furumoto S, Furukawa K, Ishiki A, Tomita N, Tago T, Hiraoka K, Watanuki S, Shidahara M. 2016. 18F-THK5351: a novel PET radiotracer for imaging neurofibrillary pathology in Alzheimer disease. J Nucl Med. 57:208–214. [DOI] [PubMed] [Google Scholar]
- Jellinger KA, Alafuzoff I, Attems J, Beach TG, Cairns NJ, Crary JF, Dickson DW, Hof PR, Hyman BT, Jack CR. 2015. PART, a distinct tauopathy, different from classical sporadic Alzheimer disease. Acta Neuropathol. 129:757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A, Koeppe RA, Fessler JA. 2009. Reducing between scanner differences in multi-center PET studies. Neuroimage. 46:154–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Drouet V, Wu JW, Witter MP, Small SA, Clelland C, Duff K. 2012. Trans-synaptic spread of tau pathology in vivo. PLoS One. 7:e31302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markesbery W. 1997. Neuropathological criteria for the diagnosis of Alzheimer’s disease. Neurobiol Aging. 18:S13–S19. [DOI] [PubMed] [Google Scholar]
- McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Kawas CH, Klunk WE, Koroshetz WJ, Manly JJ, Mayeux R. 2011. The diagnosis of dementia due to Alzheimer's disease: Recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 7:263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melov S, Adlard PA, Morten K, Johnson F, Golden TR, Hinerfeld D, Schilling B, Mavros C, Masters CL, Volitakis I. 2007. Mitochondrial oxidative stress causes hyperphosphorylation of tau. PLoS One. 2:e536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Birn RM, Handwerker DA, Jones TB, Bandettini PA. 2009. The impact of global signal regression on resting state correlations: are anti-correlated networks introduced? Neuroimage. 44:893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu J, Landeau B, Gaubert M, de La Sayette V, Desgranges B, Chételat G. 2017. Distinct influence of specific versus global connectivity on the different Alzheimer’s disease biomarkers. Brain. 140:3317–3328. [DOI] [PubMed] [Google Scholar]
- Nath S, Agholme L, Kurudenkandy FR, Granseth B, Marcusson J, Hallbeck M. 2012. Spreading of neurodegenerative pathology via neuron-to-neuron transmission of β-amyloid. J Neurosci. 32:8767–8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KP, Pascoal TA, Mathotaarachchi S, Therriault J, Kang MS, Shin M, Guiot M-C, Guo Q, Harada R, Comley RA. 2017. Monoamine oxidase B inhibitor, selegiline, reduces 18 F-THK5351 uptake in the human brain. Alzheimers Res Ther. 9:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E. 1999. Mild cognitive impairment: clinical characterization and outcome. Arch Neurol. 56:303–308. [DOI] [PubMed] [Google Scholar]
- Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, Vogel AC, Laumann TO, Miezin FM, Schlaggar BL. 2011. Functional network organization of the human brain. Neuron. 72:665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Kuceyeski A, Weiner M. 2012. A network diffusion model of disease progression in dementia. Neuron. 73:1204–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran L, Honsho M, Zahn TR, Keller P, Geiger KD, Verkade P, Simons K. 2006. Alzheimer’s disease β-amyloid peptides are released in association with exosomes. Proc Natl Acad Sci USA. 103:11172–111177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset OG, Ma Y, Evans AC. 1998. Correction for partial volume effects in PET: principle and validation. J Nucl Med. 39:904–911. [PubMed] [Google Scholar]
- Rubinov M, Sporns O. 2010. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 52:1059–1069. [DOI] [PubMed] [Google Scholar]
- Schöll M, Lockhart SN, Schonhaut DR, O’Neil JP, Janabi M, Ossenkoppele R, Baker SL, Vogel JW, Faria J, Schwimmer HD. 2016. PET imaging of tau deposition in the aging human brain. Neuron. 89:971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. 2009. Neurodegenerative diseases target large-scale human brain networks. Neuron. 62:42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibyl J, Catafau AM, Barthel H, Ishii K, Rowe CC, Leverenz JB, Ghetti B, Ironside JW, Takao M, Akatsu H. 2016. Impact of training method on the robustness of the visual assessment of 18F-Florbetaben PET scans: results from a phase-3 study. J Nucl Med. 57:900–906. [DOI] [PubMed] [Google Scholar]
- Sepulcre J, Grothe M, d’Oleire FU, Ortiz-Terán L, Diez I, Yang H, Jacobs H, Hanseeuw B, Li Q, El-Fakhri G. 2018. Neurogenetic contributions to amyloid beta and tau spreading in the human cortex. Nat Med. 10.1038/s41591-018-0206-4/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simón D, García-García E, Gómez-Ramos A, Falcón-Pérez JM, Díaz-Hernández M, Hernández F, Avila J. 2012. Tau overexpression results in its secretion via membrane vesicles. Neurodegener Dis. 10:73–75. [DOI] [PubMed] [Google Scholar]
- Sone D, Imabayashi E, Maikusa N, Okamura N, Furumoto S, Kudo Y, Ogawa M, Takano H, Yokoi Y, Sakata M. 2017. Regional tau deposition and subregion atrophy of medial temporal structures in early Alzheimer’s disease: a combined positron emission tomography/magnetic resonance imaging study. Alzheimers Dement. 9:35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HL, Shim S, Kim DH, Won SH, Joo S, Kim S, Jeon NL, Yoon SY. 2014. β‐Amyloid is transmitted via neuronal connections along axonal membranes. Ann Neurol. 75:88–97. [DOI] [PubMed] [Google Scholar]
- Thal DR, Rüb U, Orantes M, Braak H. 2002. Phases of Aβ-deposition in the human brain and its relevance for the development of AD. Neurology. 58:1791–1800. [DOI] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. 2002. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 15:273–289. [DOI] [PubMed] [Google Scholar]
- Vlassenko AG, Vaishnavi SN, Couture L, Sacco D, Shannon BJ, Mach RH, Morris JC, Raichle ME, Mintun MA. 2010. Spatial correlation between brain aerobic glycolysis and amyloid-β (Aβ) deposition. Proc Natl Acad Sci USA. 107:17763–17767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittington A, Sharp DJ, Gunn RN, Initiative AsDN . 2018. Spatiotemporal distribution of β-amyloid in Alzheimer disease is the result of heterogeneous regional carrying capacities. J Nucl Med. 59:822–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JW, Hussaini SA, Bastille IM, Rodriguez GA, Mrejeru A, Rilett K, Sanders DW, Cook C, Fu H, Boonen RA. 2016. Neuronal activity enhances tau propagation and tau pathology in vivo. Nat Neurosci. 19:1085–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Gennatas ED, Kramer JH, Miller BL, Seeley WW. 2012. Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron. 73:1216–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.