Abstract
Background
N6‐methyladenosine (m6A) modification is one of the critical gene regulatory mechanisms implicated in cancer biology. However, the roles of m6A regulators in ovarian cancer are still poorly understood.
Methods
We integrated multiple databases including Gene Expression Omnibus (GEO), ROC Plotter, Kaplan‐Meier Plotter, and Tumor Immune Estimation Resource (TIMER) to explore clinicopathological significance of m6A regulators in ovarian cancer.
Results
We showed that alterations in the expression of m6A regulators were related to the malignancy and poor prognosis of ovarian cancer. We found decreased YTHDC1 and increased RBM15 expressions were associated with ovarian cancer cell metastases and HNRNPC was a predictor of paclitaxel resistance. Moreover, dysregulated m6A regulators were enriched in the activation of cancer‐related pathways. Our results further demonstrated that the level of immune cell infiltration and the expression of various immune gene markers were closely associated with the expressions of specific m6A regulators (RBM15B, ZC3H13, YTHDF1, and IGF2BP1).
Conclusions
Our study establishes a new prognostic profile of ovarian cancer patients based on m6A regulators, and highlights the potential roles of m6A regulators in ovarian cancer development.
Keywords: immune infiltration, m6A regulators, ovarian cancer, survival
m6A regulators play important roles in ovarian cancer cell biology.

1. INTRODUCTION
Ovarian cancer is the first leading cause for death of gynecological cancers worldwide, with an estimated 295,000 new cases and 185,000 deaths in 2018 (Bray et al., 2018). Although the therapy modalities have been greatly improved, more than 70% of patients with advanced stages still have tumor recurrence, and the 5‐year overall survival rate of ovarian cancer patients is still very low (Lheureux et al., 2019). Tumor‐related immune modulation plays an important role in ovarian cancer. Tumor‐infiltrating lymphocytes (TILs), including CD8+ T cells, macrophages, neutrophils, and dendritic cells affect the prognosis and efficacy of immunochemotherapy (Santoiemma & Powell, 2015). Therefore, it is an urgent need to find new biomarkers and immune‐related targets for the prognosis and treatment of ovarian cancer.
N6‐methyladenosine (m6A) is the most common post‐transcriptional modification in mRNA. It affects RNA metabolism, such as alternative splicing, translation, and degradation (Roundtree et al., 2017). The modification of m6A is catalyzed by different types of regulators, including m6A methyltransferases (METTL3/14, RBM15/15B, VIRMA, WTAP, and ZC3H13, termed as ‘writers’), demethylases (FTO and ALKBH5, termed as ‘erasers’), and RNA binding proteins (HNRNPA2B1, HNRNPC, IGF2BP1/2/3, YTHDC1/2, YTHDF1/2/3, and RBMX, termed as ‘readers’) (Meyer & Jaffrey, 2017; Zaccara et al., 2019). The dynamic modification in m6A mediated by these regulators not only plays important roles in the development of oocytes and cerebellum but also plays essential roles in regulating cell proliferation and migration, leading to the malignant progression of various cancers and treatment resistance (Chen et al., 2019; Lan et al., 2019). The latest studies also revealed the connection between m6A regulators and tumor immune‐cell infiltration (Han et al., 2019; Li et al., 2020; Wang et al., 2019, 2020; Winkler et al., 2019). For example, inhibition of METTL3/14 promoted IFN‐γ‐STAT1‐IRF1 signaling and enhanced response to anti‐PD‐1 treatment in colorectal cancer (Wang et al., 2020). ALKBH5 inhibitor could heighten the efficacy of cancer immunotherapy (Li et al., 2020). In recent years, comprehensive analysis of the clinical relevance and molecular characteristics of m6A regulators across several cancer types has been reported (Chai et al., 2019; Kwok et al., 2017; Li et al., 2019; Su et al., 2019; Zhou et al., 2019). However, their roles in ovarian tumorigenesis remain unclear.
Here, we systematically assessed the expression pattern, clinicopathological, and prognostic relevance of m6A regulators through extensive bioinformatics analyses. We revealed the predictive value and clinical significance of m6A regulators in ovarian cancer. Importantly, our results also indicated that the level of immune cell infiltration and the expression of various immune gene markers were closely related to the expression of specific m6A regulators.
2. MATERIALS AND METHODS
2.1. Data acquisition
The TCGA‐OV dataset used in our study were downloaded from The Cancer Genome Atlas (TCGA) data portal (https://cancergenome.nih.gov/). Genetic data were obtained from cBioPortal (https://www.cbioportal.org/) (Cerami et al., 2012; Gao et al., 2013). Nine sets of microarrays (GSE14407, GSE12470, GSE69428, GSE84829, GSE28979, GSE9891, GSE73168, GSE30587, and GSE51373) were extracted from the Gene Expression Omnibus (GEO) datasets (http://www.ncbi.nlm.nih.gov/geo/).
2.2. Selection of RNA m6A methylation regulators
We collated a list of 20 m6A regulators from recently published literature, including 11 readers, 7 writers, and 2 erasers (Yang et al., 2018). We extracted the available mRNA expression data in GEO datasets of these genes and the clinicopathological information of the samples for subsequent bioinformatics analysis.
2.3. Bioinformatic analysis of expression profiles
Genetic status data available at TCGA database were assessed using the cBioPortal to investigate the genomic profiling of m6A regulators in ovarian cancer. The GEO datasets were used to evaluate the expression alterations of m6A regulators in normal and tumor tissues. GSE14407 evaluated the differential gene expression between 12 laser capture microdissected serous ovarian cancers and 12 ovarian surface epithelial cells. GSE12470 evaluated the differential gene expression between 43 serous ovarian cancer and 10 normal peritoneum samples. GSE69428 compared gene expression profiles of high‐grade serous ovarian cancer (HGSOC) and paired normal oviduct samples from 10 independent patients. GSE84829 assessed gene expression patterns in 3 ascitic fluid‐isolated mesothelial cell samples obtained from stage III/IV ovarian serous carcinoma patients and 3 control peritoneal mesothelial cell samples isolated from omentum obtained from non‐oncologic patients. GSE28979 assessed gene expression patterns in 3 normal mouse fallopian tube oviduct and 3 early tumors from fallopian tubes of Dicer/PTEN knockout mice. GSE9891 analyzed the correlation between pathological grades/stages and expression level of m6A regulators in 285 ovarian cancer samples. GSE73168 evaluated the differential gene expression between 12 HGSOC primary tumor cells and 12 HGSOC ascites tumor cells. GSE30587 assessed gene expression patterns in 9 matched pairs of primary ovarian tumors and metastases from the omentum. GSE51373 evaluated the differential gene expression between 12 chemotherapy‐resistant and 16 chemotherapy‐sensitive HGSOC samples.
2.4. Receiver operating characteristics (ROC) Plotter
The ROC Plotter online platform (http://www.rocplot.org/) was used to identify specific m6A regulators which predicts benefit from chemotherapy (Fekete & Győrffy, 2019). The platform integrates multiple gene expression datasets at transcriptome level and contains 2369 ovarian cancer patients with treatment and response data.
2.5. Kaplan‐Meier Plotter analysis
Kaplan‐Meier plotter database (http://kmplot.com/analysis/) was used to investigate the prognostic value of m6A regulators in patients with ovarian cancer (Nagy et al., 2018). The hazard ratio (HR) with 95% confidence intervals (CI) and log‐rank p‐value were estimated.
2.6. TIMER database analysis
The TIMER online tool (https://cistrome.shinyapps.io/timer/) is a comprehensive resource for systematic analysis of immune infiltrates and contains 10,897 samples across 32 cancer types from TCGA (Li et al., 2016; Li, Fan, et al., 2017). It was used to analyze the correlation of m6A regulators with the abundance of immune cell infiltrates, including B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells. Additionally, correlations between the expression of m6A regulators and various immune gene markers were explored via correlation modules. The gene expression level was displayed with log2 RSEM.
2.7. Gene set enrichment analysis
The biological functions potentially regulated by m6A regulators in ovarian cancer were evaluated by GSEA v3.0 software (Mootha et al., 2003; Subramanian et al., 2005). Hallmark gene sets and KEGG gene sets deposited in the GSEA Molecular Signatures Database v7.0 (MSigDB) were used.
2.8. Statistical analysis
One‐way ANOVA was used to compare the expression level of normal and tumor samples in GEO dataset. Student paired t test was used to compare the expression level in ovarian cancer for grade and stage. Chi‐square tests were used to compare the distribution of grade and stage between high‐ and low‐expression level groups. The expression of m6A regulators and therapy response were compared using ROC and Mann–Whitney tests. Survival rates were assessed using Kaplan–Meier curves and the log‐rank test. The correlation of m6A regulators with immune infiltration level and various immune gene markers was determined by Spearman's correlation. The data were analyzed using GraphPad Prism version 6.01 (GraphPad Software, Inc.) and presented as mean ± SD. p‐values <0.05 were considered statistically significant.
3. RESULTS
3.1. Expression profiles and clinical relevance of m6A regulators in ovarian cancer
In light of the crucial biological functions of m6A regulators in tumorigenesis, we systematically explored the genetic status and expression profile of each individual m6A regulator in ovarian cancer. We selected 20 well‐characterized m6A regulatory genes for analysis in current study, including 11 readers, 7 writers, and 2 erasers (Figure 1a). The genetic alteration of m6A regulators was first determined in the ovarian cancer patient cohort from TCGA database using cBioPortal. We found that IGF2BP2, YTHDF1, and VIRMA showed higher percentage of amplification, whereas the other m6A regulators had the lower frequency of overall mutation, ranging from 0.9 to 8.0% (Figure 1b‐e). In contrary to the relatively rare genetic mutations, more than half of m6A regulators showed significant alterations in mRNA expression level between normal and cancer samples.
FIGURE 1.
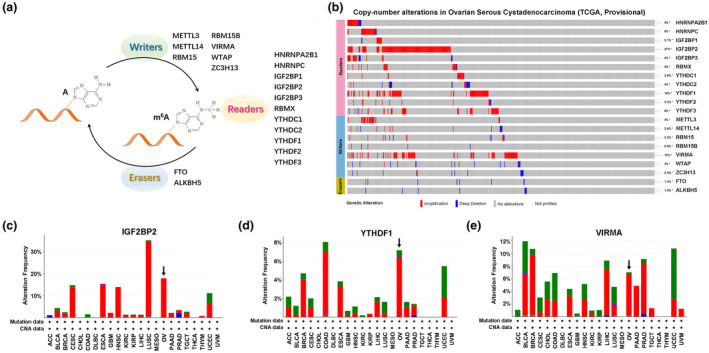
Genetic profiles of m6A regulators in ovarian cancer. (a) Diagram of m6A regulators analyzed in current study. (b) Genetic alterations of m6A regulators in ovarian cancer available at TCGA database by using cBioPortal (http://cbioportal.org). (c–e) Genetic alterations of IGF2BP2 (c), YTHDF1 (d), and VIRMA (e) across 23 cancer types. ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangio carcinoma; COAD, colon adenocarcinoma; DLBC, diffuse large B‐cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PRAD, prostate adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UVM, uveal melanoma
Two GEO databases demonstrated that 7 readers (HNRNPC, IGF2BP1, IGF2BP2, IGF2BP3, RBMX, YTHDC2, and YTHDF2) and 3 writers (METTL3, RBM15, and RBM15B) were more highly expressed in ovarian cancer than in ovarian surface epithelium or normal peritoneum tissues (GEO14407 and GEO12470, Figure 2a,b). Besides that, HNRNPA2B1, YTHDC1, METTL14, WTAP, and ZC3H13 were downregulated in cancer tissues, whereas VIRMA had opposite alterations between these two databases (Figure 2a,b). Given the theory implicating the distal oviduct as a common source for epithelial ovarian cancer, we analyzed the GSE69428 data and showed that the expression of IGF2BP2, IGF2BP3, RBMX, YTDHF1, and RBM15 was also higher in ovarian cancer than in normal oviduct (Figure 2c). Moreover, IGF2BP1, IGF2BP2, and ALKBH5 were upregulated in ascitic fluid isolated mesothelial cells than in normal peritoneal mesothelial cell (GSE84829, Figure 2d). In addition, tumors from fallopian tubes of Dicer/PTEN knockout mice revealed alterations in expression of 7 readers (HNRNPA2B1, HNRNPC, IGF2BP3, RBMX, YTHDC2, YTHDF2, and YTHDF3), 2 writers (RBM15 and WTAP) and 2 erasers (FTO and ALKBH5) in comparison with normal mouse fallopian tube oviduct (GSE28979, Figure 2e).
FIGURE 2.
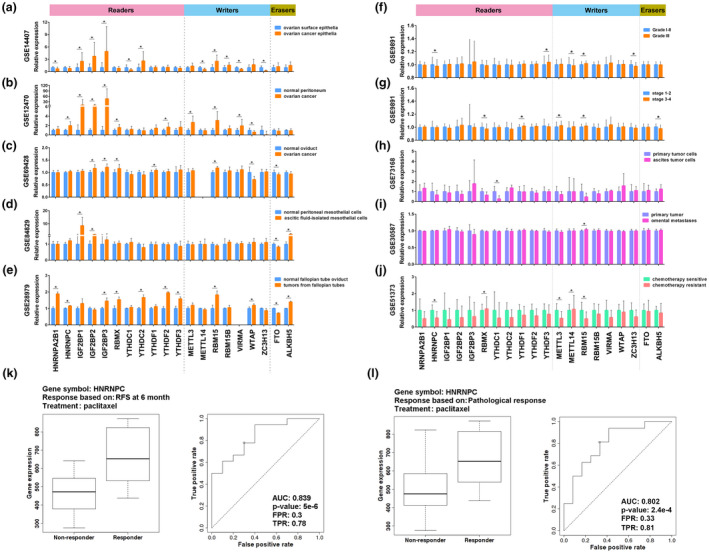
Expression profiles and clinical relevance of m6A regulators in ovarian cancer. (a–j) Analysis of differential gene expression of m6A regulators between ovarian cancer and ovarian surface epithelial cells in GSE14407 (a), between ovarian cancer and normal peritoneum samples in GSE12470 (b), between HGSOC and paired normal oviduct samples in GSE69428 (c), between ascitic fluid‐isolated mesothelial cells and normal peritoneal mesothelial cells in GSE84829 (d), between early tumors from fallopian tubes of Dicer/PTEN knockout mice and normal mouse fallopian tube oviduct in GSE28979 (e), between different grades in GSE9891 (f), between different stages in GSE9891 (g), between primary ovarian cancer cells and ascites tumor cells in GSE73168 (h), between primary ovarian tumors and metastases from the omentum in GSE30587 (i), and between chemotherapy‐resistant and ‐sensitive samples in GSE51373 (j). (k and l) ROC curves and box‐plots of HNRNPC validated for paclitaxel resistance based on RFS at 6 months (k) and pathological response (l). Error bar ± SD, *p < 0.05
To determine the clinical relevance of m6A regulators in ovarian cancer, we analyzed the relationship between expression alteration of m6A regulators and ovarian cancer clinicopathological features. As the pathological grade increased, the expression of YTHDF3 enhanced, while HNRNPC and ZC3H13 decreased (GSE9891, Figure 2f; see also File S1). The significant correlation between pathological stages and expression levels of YTHDF1 and RBM15 was confirmed in GSE9891 data (Figure 2g; see also Table S1). We also noticed that decreased expression of YTHDC1 and increased expression of RBM15 were correlated with the status of ovarian cancer cell metastasis (GSE73168 and GSE30587, Figure 2h,i). Moreover, as the combination of platin plus paclitaxel is the standard first‐line chemotherapy for patients with ovarian cancer, we analyzed the relationship between expression pattern of m6A regulators and chemotherapy sensitivity. We found that HNRNPC, METTL3, and RBM15 were downregulated in chemotherapy‐resistant group, while RMBX and METTL14 were increased (Figure 2j). Importantly, the ROC curve showed that increased expression of HNRNPC could perfectly predict response to paclitaxel for ovarian cancer patients based on relapse‐free survival (RFS) at 6 months (AUC =0.839, p = 5.0e‐6, Figure 2k and Table 1), RFS at 12 months (AUC =0.802, p = 2.4e‐4, see also Table S2) and pathological response (AUC =0.803, p = 2.4e‐4, Figure 2l), while decreased expression of YTHDC1 could predict response to paclitaxel on RFS at 6 months (AUC =0.707, p = 1.5e‐3, Table 1). These data indicated that m6A regulators may play critical roles in ovarian tumorigenesis and function as a predictor of metastasis and chemoresponsiveness.
Table 1.
Predictive value of m6A regulators in response to chemotherapy in ovarian cancer based on relapse‐free survival at 6 months
| Paclitaxel | Platin | |||||
|---|---|---|---|---|---|---|
| AUC |
ROC p‐value |
Mann–Whitney test p‐value | AUC |
ROC p‐value |
Mann–Whitney test p‐value | |
| HNRNPA2B1 | 0.556 | 0.31 | 0.65 | 0.535 | 0.17 | 0.32 |
| HNRNPC | 0.839 | 5.0e‐6 | 0.0037 | 0.502 | 0.48 | 0.95 |
| IGF2BP1 | 0.619 | 0.15 | 0.31 | 0.539 | 0.13 | 0.27 |
| IGF2BP2 | 0.578 | 0.13 | 0.25 | 0.508 | 0.39 | 0.79 |
| IGF2BP3 | 0.565 | 0.19 | 0.34 | 0.546 | 0.062 | 0.11 |
| RBMX | 0.61 | 0.052 | 0.11 | 0.548 | 4.2e‐02 | 0.094 |
| YTHDC1 | 0.707 | 1.5e‐03 | 0.0024 | 0.636 | 5.8e‐06 | 1.6e‐06 |
| YTHDC2 | 0.634 | 3.0e‐02 | 0.049 | 0.625 | 4.5e‐06 | 1.1e‐05 |
| YTHDF1 | 0.571 | 0.16 | 0.3 | 0.599 | 2.5e‐04 | 4.8e‐04 |
| YTHDF2 | 0.566 | 0.15 | 0.33 | 0.657 | 3.6e‐09 | 3.2e‐08 |
| YTHDF3 | 0.66 | 1.3e‐02 | 0.019 | 0.579 | 3.9e‐03 | 0.0057 |
| METTL3 | 0.648 | 2.1e‐02 | 0.03 | 0.669 | 3.1e‐08 | 1.4e‐06 |
| METTL14 | 0.544 | 0.36 | 0.72 | 0.502 | 0.47 | 0.94 |
| RBM15 | 0.517 | 0.41 | 0.8 | 0.575 | 3.6e‐03 | 0.0085 |
| RBM15B | 0.663 | 1.5e‐03 | 0.017 | 0.577 | 2e‐03 | 0.0071 |
| VIRMA | 0.522 | 0.43 | 0.87 | 0.527 | 0.24 | 0.45 |
| WTAP | 0.543 | 0.27 | 0.52 | 0.569 | 5.4e‐03 | 0.015 |
| ZC3H13 | 0.511 | 0.43 | 0.87 | 0.545 | 4.9e‐02 | 0.11 |
| FTO | 0.606 | 3.8e‐02 | 0.12 | 0.563 | 1.2e‐02 | 0.027 |
| ALKBH5 | 0.636 | 0.12 | 0.24 | 0.502 | 0.48 | 0.97 |
3.2. Prognostic value of m6A regulators in ovarian cancer
Next, we sought to evaluate the predictive value of m6A regulators for prognosis in ovarian cancer. Kaplan‐Meier log‐rank analysis revealed that high expressions of YTHDF1 (p = 0.0024, HR = 1.23, 95% CI = 1.08–1.41), YTHDF2 (p = 0.0006, HR = 1.26, 95% CI = 1.10–1.43), WTAP (p = 1.5e‐6, HR = 1.39, 95% CI = 1.21–1.59), FTO (p = 0.001, HR = 1.26, 95% CI = 1.10–1.44), and ALKBH5 (p = 0.0003, HR = 1.48, 95% CI = 1.20–1.83) were significantly correlated with poor overall survival (OS) (Figure 3a). Besides these genes, high expressions of HNRNPA2B1 (p = 0.0018, HR = 1.36, 95% CI = 1.12–1.65), IGF2BP1 (p = 6.9e‐6, HR = 1.53, 95% CI = 1.27–1.85), YTHDC1 (p = 0.0001, HR = 1.28, 95% CI = 1.13–1.46), YTHDF3 (p = 0.001, HR = 1.24, 95% CI = 1.09–1.41), METTL3 (p = 1.4e‐5, HR = 1.32, 95% CI = 1.16–1.5), RBM15B (p = 0.008, HR = 1.31, 95% CI = 1.07–1.6), and VIRMA (p = 0.0015, HR = 1.37, 95% CI = 1.13–1.66) were correlated with worse progression‐free survival (PFS) (Figure 3b). We also evaluated the prognostic value for each m6A regulator in ovarian cancer patients with different clinicopathologic features. Among these m6A regulators, YTHDC1, YTHDF3, WTAP, FTO, and ALKBH5 were risk prognostic factors with HR >1 for both OS and PFS in patients with TP53 mutation (Figure 3c,d). HNRNPC, YTHDF1, YTHDF2, YTHDF3, and WTAP were correlated with poor OS and PFS with the status of CA125 level below lower quartile (Figure 3e,f). In addition, several risk factors of m6A regulators for OS and PFS in ovarian cancer patients with different grade, stage, and chemotherapy were assessed (Figure 3g). For instance, compared with YTHDF2low group, YTHDF2high group had shorter OS and PFS in patients treated with platin or Taxol, whereas no differences were found in patients with low pathological stage. Besides, high expression of ALKBH5 was a risk factor for OS and PFS in patients with high pathological grade and stage (Figure 3g). These results highlighted potential roles of m6A regulators as prognostic markers in ovarian cancer patients.
FIGURE 3.

Prognostic value of m6A regulators in ovarian cancer. (a and b) The distribution of hazard ratios of OS and PFS across m6A regulators in ovarian cancer patients from Kaplan–Meier plotter database (http://kmplot.com/analysis/). (c and d) The distribution of hazard ratios of OS and PFS in patients with TP53 mutation. (e and f) The distribution of hazard ratios of OS and PFS in patients with the status of CA125 level below lower quartile. (g) The distribution of hazard ratios of OS and PFS in patients with different grades, stages, and chemotherapy
3.3. Oncogenic pathways regulated by m6A regulators in ovarian cancer
To better understand the functions of m6A regulators in ovarian cancer, we first analyzed the correlation among these regulators. As shown in Figure 4a, the expressions of m6A regulators were not only correlated with several regulators in the same functional type but also among different types. For example, the expression of HNRNPA2B1 was positively correlated with the expressions of IGF2BP3, YTHDC1, YTHDF2, RBM15, and ZC3H13 in ovarian cancer. Similarly, the expression of HNRNPC was positively correlated with RBMX and METTL3, and negatively correlated with YTHDC2 and ZC3H13.
FIGURE 4.
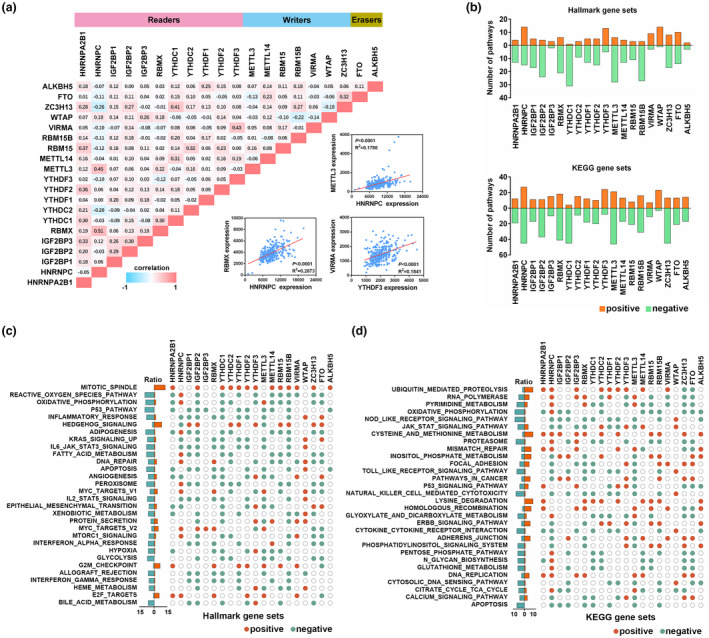
Oncogenic pathways regulated by m6A regulators in ovarian cancer. (a) Correlation among the expression of m6A regulators. The scatter plot shows the correlation between METTL3 and HNRNPC, RBMX and HNRNPC, and VIRMA and YTHDF3. (b) The number of Hallmark gene sets (upper panel) and KEGG gene sets (lower panel) is correlated with individual m6A regulators. (c) The correlation between m6A regulators and Hallmark gene sets. (d) The correlation between m6A regulators and KEGG gene sets
Then, we investigated the oncogenic pathways potentially regulated by m6A regulators in ovarian cancer. GSEA analysis demonstrated that the expressions of METTL3, YTHDC1, RBM15B, HNRNPC, IGF2BP2, RBMX, and ZC3H13 were correlated with a higher number of multiple Hallmark pathways in ovarian cancer (Figure 4b). Upregulated expressions of m6A regulators were enriched in the activation of several cancer‐related pathways, such as mitotic spindle, Hedgehog signaling, MYC targets, G2M checkpoint, and E2F target, whereas reactive oxygen species pathway, oxidative phosphorylation, p53 pathway, inflammatory response, adipogenesis, IL6/JAK/STAT3 signaling, fatty acid metabolism, apoptosis, and peroxisome were negatively correlated with the expression of m6A regulators (Figure 4c). We also performed KEGG pathway enrichment to recognize biological processes regulated by m6A regulators. Similarly, the expressions of HNRNPC, METTL3, RBMX, ZC3H13, YTHDC1, IGF2BP2, and RBM15B were correlated with a higher number of KEGG pathways (Figure 4b). Our results also indicated that upregulated m6A regulators were positively enriched in ubiquitin‐mediated proteolysis, cysteine and methionine metabolism, lysine degradation, and homologous recombination, whereas oxidative phosphorylation, NOD‐like receptor signaling pathway, proteasome, natural killer cell‐mediated cytotoxicity, pyrimidine metabolism, and Toll‐like receptor signaling pathway were negatively correlated with the expression of m6A regulators (Figure 4d).
3.4. Correlation between immune cell infiltration and the expression of m6A regulators in ovarian cancer
Infiltration of lymphocytes is an independent predictor of ovarian cancer patient survival and chemoresistance. Hence, we explored whether the expression of m6A regulators was correlated with immune infiltration levels in ovarian cancer. The results showed that several m6A regulators, including 4 readers (IGF2BP1, IGF2BP2, YTHDF1, and YTHDC2), 3 writers (ZC3H13, RBM15B, and WTAP), and 1 eraser (ALKBH5) had significant correlations with immune cell infiltration levels (Figure 5a). In particular, IGF2BP1 expression level had significant negative correlation with infiltrating of B cells (Cor = −0.167, p = 2.4e‐4), CD8+ T cells (Cor = −0.192, p = 2.3e‐5), neutrophils (Cor = −0.220, p = 1.2e‐6), and dendritic cells (Cor = −0.232, p = 2.7e‐7) (Figure 5b). RBM15B expression level had significantly negative correlation with infiltrating of CD8+ T cells (Cor = −0.156, p = 6.2e‐4), macrophages (Cor = −0.280, p = 4.2e‐10), neutrophils (Cor = −0.220, p = 1.2e‐6), and dendritic cells (Cor = −0.180, p = 7.5e‐5) (Figure 5c). Similarly, the expression of ZC3H13 was significantly negatively correlated with infiltrating level of CD8+ T cells (Cor = −0.184, p = 4.9e‐5), CD4+ T cells (Cor = −0.121, p = 8.2e‐3), macrophages (Cor = −0.199, p = 1.1e‐5), neutrophils (Cor = −0.322, p = 4.5e‐13), and dendritic cells (Cor = −0.265, p = 3.6e‐9) (Figure 5d). The expression of YTHDF1 was significantly negatively correlated with infiltrating level of CD8+ T cells (Cor = −0.263, p = 5.2e‐9), neutrophils (Cor = −0.182, p = 6.0e‐5), and dendritic cells (Cor = −0.198, p = 1.2e‐5) (Figure 5e). Moreover, similar correlations were also observed across different types of cancers (see Figure S1). These findings propose that the expressions of specific m6A regulators may be correlated with immune cell infiltration in ovarian cancer.
FIGURE 5.
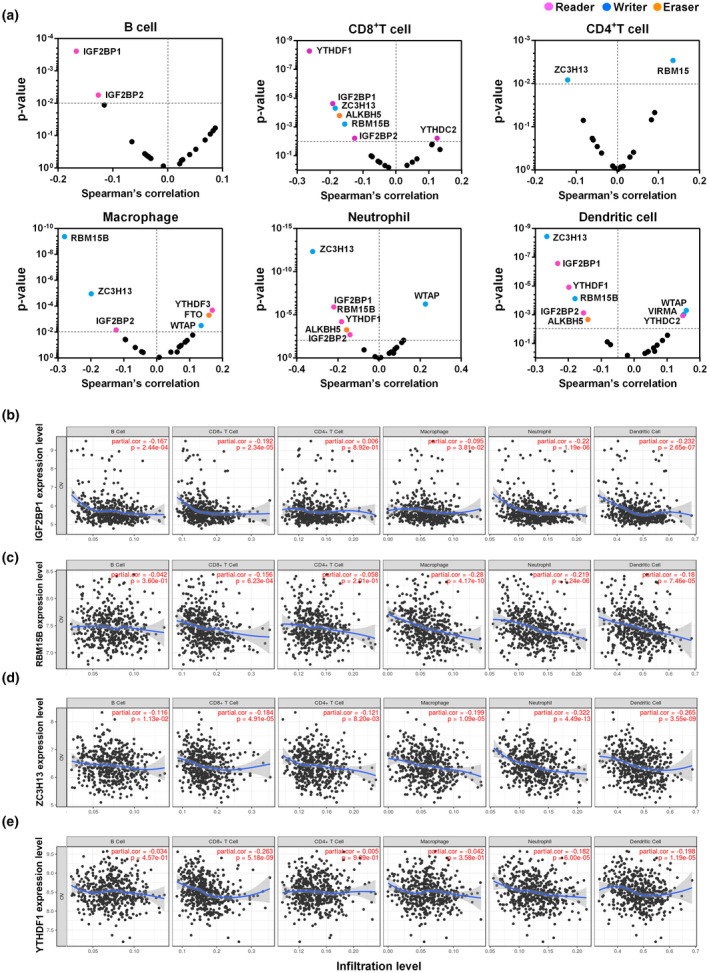
Correlation between immune cell infiltration and the expression of m6A regulators in ovarian cancer. (a) Correlation between the expression of m6A regulators and infiltrating levels of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells in ovarian cancer. (b–e) Correlation between the expression of specific m6A regulators (IGF2BP1, RBM15B, ZC3H13, and YTHDF1) and infiltrating levels of B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells in ovarian cancer
We further investigated the relationship between these immune infiltration‐related m6A regulators and immune marker genes of diverse immune cells, including B cells, T cells (general), CD8+ T cells, Th1 cells, Th2 cells, Treg cells, tumor‐associated macrophages (TAM), M1 and M2 macrophages, neutrophils, natural killer (NK) cells, and dendritic cells in ovarian cancer. As shown in Table 2, the expression of RBM15B was significantly correlated with most immune marker genes of different T cells and various immune cells. ZC3H13 expression had correlation with immune marker genes of CD8+ T cells, TAM, M2 macrophages, and dendritic cells. The expression of YTHDF1 was correlated with immune marker genes of T cells (general), CD8+ T cells, and dendritic cells, whereas IGF2BP1 expression had correlation only with dendritic cells. Our results also showed significant correlations between two immune infiltration‐related m6A regulators (RBM15B and ZC3H13) and marker genes of T‐cell exhaustion, including PDCD1 (PD‐1), CD274 (PD‐L1), CTLA4, LAG3, and GZMB (Table 2). Moreover, we found that the expression of these immune infiltration‐related m6A regulators was also significantly correlated with several interleukins (IL1B, IL7, IL15, and IL18), CC and CXC chemokines (CCL2, CCL5, CXCL10, CXCL11, and CXCL17), and human leukocyte antigens (HLA‐A, HLA‐B, HLA‐C, HLA‐E, and HLA‐F) (Figure 6; see also Table S3). Therefore, these data confirmed the findings that the expressions of specific m6A regulators were associated with tumor immune cell infiltration.
Table 2.
Correlation analysis between m6A regulators and related gene markers of immune cells
| Immune cells | Gene markers | ZC3H13 | RBM15B | IGF2BP1 | YTHDF1 | ||||
|---|---|---|---|---|---|---|---|---|---|
| Cor | p | Cor | p | Cor | p | Cor | p | ||
| B cell | CD19 | −0.02 | 0.73 | −0.02 | 0.69 | 0.19 | ** | 0.10 | 0.08 |
| CD79A | −0.03 | 0.56 | −0.13 | 0.02 | 0.02 | 0.67 | 0.03 | 0.63 | |
| T cell (general) | CD2 | −0.16 | * | −0.31 | *** | −0.14 | 0.14 | −0.20 | ** |
| CD3D | −0.18 | * | −0.31 | *** | −0.13 | 0.02 | −0.22 | ** | |
| CD3E | −0.12 | 0.03 | −0.29 | *** | −0.14 | 0.01 | −0.21 | ** | |
| CD8+ T cell | CD8A | −0.16 | * | −0.32 | *** | −0.10 | 0.08 | −0.15 | * |
| CD8B | −0.16 | * | −0.27 | *** | 0.05 | 0.37 | −0.03 | 0.64 | |
| Th1 | IFN‐γ | −0.15 | 0.01 | −0.30 | *** | −0.09 | 0.13 | −0.15 | * |
| TBX21 | −0.11 | 0.04 | −0.24 | *** | −0.12 | 0.03 | −0.19 | * | |
| TNF‐α | −0.06 | 0.28 | −0.14 | 0.02 | 0.02 | 0.77 | −0.16 | 0.04 | |
| Th2 | GATA3 | 0.07 | 0.22 | −0.13 | 0.03 | 0.03 | 0.65 | −0.09 | 0.12 |
| STAT6 | 0.05 | 0.38 | 0.00 | 0.96 | −0.15 | 0.01 | −0.14 | 0.01 | |
| IL13 | 0.02 | 0.70 | −0.05 | 0.41 | −0.05 | 0.37 | −0.12 | 0.04 | |
| Treg | CCR8 | −0.09 | 0.13 | −0.14 | 0.02 | 0.01 | 0.83 | −0.11 | 0.06 |
| FOXP3 | −0.09 | 0.13 | −0.21 | ** | 0.00 | 0.96 | −0.06 | 0.28 | |
| STAT5B | 0.34 | *** | 0.20 | ** | 0.08 | 0.16 | 0.14 | 0.15 | |
| TAM | CD68 | −0.25 | *** | −0.32 | *** | −0.09 | 0.14 | −0.15 | * |
| CCL2 | −0.21 | ** | −0.25 | *** | −0.15 | 0.01 | −0.23 | *** | |
| IL10 | −0.15 | 0.01 | −0.24 | *** | 0.00 | 0.91 | −0.10 | 0.08 | |
| M1 macrophage | NOS2 | −0.02 | 0.75 | 0.00 | 0.99 | 0.19 | * | 0.10 | 0.10 |
| PTGS2 | 0.04 | 0.52 | −0.02 | 0.72 | 0.03 | 0.63 | −0.09 | 0.13 | |
| IRF5 | −0.15 | 0.01 | −0.22 | ** | −0.03 | 0.66 | −0.06 | 0.28 | |
| M2 macrophage | CD163 | −0.12 | 0.04 | −0.23 | *** | −0.02 | 0.79 | −0.08 | 0.16 |
| MS4A4A | −0.21 | ** | −0.33 | *** | −0.07 | 0.26 | −0.15 | 0.01 | |
| VSIG4 | −0.25 | *** | −0.31 | *** | −0.06 | 0.28 | −0.09 | 0.12 | |
| Neutrophils | CCR7 | −0.07 | 0.24 | −0.19 | ** | −0.05 | 0.36 | −0.20 | ** |
| CEACAM8 | 0.27 | *** | 0.00 | 0.89 | 0.07 | 0.23 | 0.02 | 0.76 | |
| ITGAM | −0.11 | 0.05 | −0.20 | ** | −0.07 | 0.23 | −0.14 | 0.13 | |
| NK cell | KIR2DL1 | 0.03 | 0.64 | −0.10 | 0.08 | −0.01 | 0.91 | −0.09 | 0.13 |
| KIR2DL3 | −0.06 | 0.28 | −0.18 | * | −0.21 | ** | −0.13 | 0.03 | |
| KIR3DL1 | 0.00 | 0.97 | −0.15 | * | −0.12 | 0.04 | −0.07 | 0.20 | |
| KIR3DL2 | −0.02 | 0.74 | −0.15 | 0.01 | −0.10 | 0.10 | −0.13 | 0.02 | |
| Dendritic cell | HLA‐DPB1 | −0.31 | *** | −0.26 | *** | −0.30 | *** | −0.26 | *** |
| HLA‐DQB1 | −0.175 | * | −0.21 | ** | −0.23 | *** | −0.19 | ** | |
| HLA‐DRA | −0.36 | *** | −0.32 | *** | −0.31 | *** | −0.27 | *** | |
| ITGAX | −0.08 | 0.16 | −0.22 | ** | −0.11 | 0.05 | −0.18 | * | |
| T‐cell exhaustion | PDCD1 | −0.16 | * | −0.28 | *** | −0.04 | 0.53 | −0.12 | 0.04 |
| CD274 | −0.17 | * | −0.25 | *** | −0.14 | 0.01 | −0.21 | ** | |
| CTLA4 | −0.22 | ** | −0.29 | *** | −0.07 | 0.20 | −0.10 | 0.08 | |
| LAG3 | −0.22 | ** | −0.32 | *** | −0.06 | 0.28 | −0.12 | 0.04 | |
| GZMB | −0.20 | ** | −0.33 | *** | −0.14 | 0.01 | −0.19 | * | |
Abbreviation:TAM, tumor‐associated macrophage; Th, T helper cells; Treg, regulatory T cells; NK, natural killer; Cor, R value of Spearman’s correlation
*(p < 0.01).
**(p < 0.001).
***(p < 0.0001).
FIGURE 6.
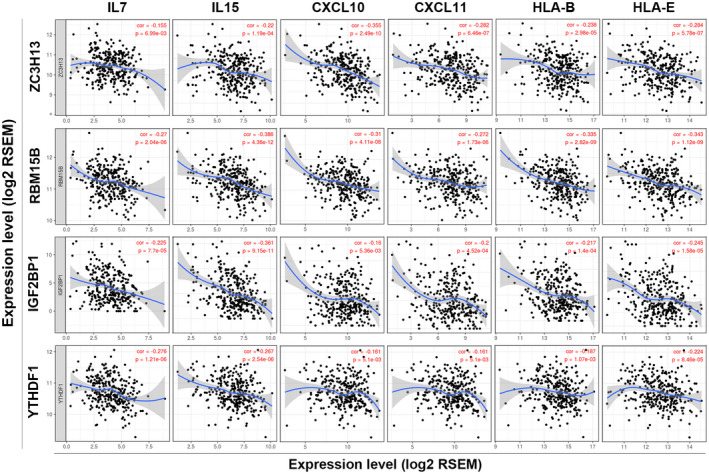
Correlation between the expression of specific m6A regulators (IGF2BP1, RBM15B, ZC3H13, and YTHDF1) and immune‐related factors (IL7, IL15, CXCL10, CXCL11, HLA‐B, and HLA‐E)
4. DISCUSSION
Herein, we demonstrated that changes in m6A regulator expression were associated with malignancy and prognosis of ovarian cancer. Increased expression of YTHDF3, WTAP, FTO, and ALKBH5 was associated with shorter OS and PFS regardless of the status of TP53 mutation. We found that a decrease in YTHDC1 and an increase in RBM15 expressions were correlated with ovarian cancer cell metastases. We also suggested that HNRNPC was a predictor of paclitaxel resistance. In addition, GSEA analysis showed that the mechanism of m6A regulators regulating ovarian cancer was related to a variety of tumor‐related pathways. Importantly, our data showed that immune cell infiltration levels and various immune gene markers were closely associated with the expression of m6A regulators, suggesting that RBM15B, ZC3H13, YTHDF1, and IGF2BP1 might play the role of immune infiltration‐related m6A regulators in ovarian cancer. Thus, our current study provided insights into the value of m6A regulators in the determination of prognosis and understanding of their potential roles in ovarian cancer immunology.
RNA m6A methylation is a widespread modification that regulates selective control of gene expression (Dominissini et al., 2012; Li, Tong, et al., 2017; Yue et al., 2015). Research on the roles of m6A readers, writers, and erasers have improved our understanding of physiological and pathological significance of RNA methylation (Meyer & Jaffrey, 2017). Accumulating evidences suggest that these m6A methylation regulators function as oncogenes or tumor‐suppressor genes and are involved in the proliferation, differentiation, invasion, and metastasis of cancer cells (Lan et al., 2019). Recent years, the clinical relevance and molecular characteristics of m6A regulators in different cancer types have been reported (Chai et al., 2019; Kwok et al., 2017; Li et al., 2019; Su et al., 2019; Zhou et al., 2019). Although some studies have demonstrated that the m6A writer METTL3, reader IGF2BP1, and eraser ALKBH5 are involved in the development of ovarian cancer (Hua et al., 2018; Müller et al., 2019; Zhu et al., 2019), little is known about the role of other m6A regulators in ovarian cancer.
Here, we comprehensively evaluated the expression alterations of 20 m6A regulators in different databases. Compared with normal ovarian surface epithelium, peritoneum, and oviduct tissues, increased or decreased expression of several specific m6A regulators were found in ovarian cancer tissues and ascitic fluid‐isolated cells. The relationship between the expression of m6A regulators and clinicopathological characteristics, such as grading, staging, metastasis,, and chemotherapy response was also confirmed in our study. Moreover, HNRNPC, a member of ubiquitously expressed heterogeneous nuclear ribonucleoproteins (hnRNPs) family, which influences pre‐mRNA processing and mRNA transport and metabolism (Fischl et al., 2019), was downregulated in chemotherapy‐resistant group and upregulated in paclitaxel response group. The ROC/AUC score was also high, indicating its predictive value of paclitaxel response in ovarian cancer. Our study also revealed that m6A regulators might be correlated with several tumor‐related signaling pathways and biological processes in ovarian cancer, including Hedgehog signaling, p53 pathway, Myc‐dependent pathway, reactive oxygen species pathway, IL6/JAK/STAT3 signaling, apoptosis, mitotic spindle, proteolysis, amino acid metabolism, homologous recombination, etc.
Previous studies have shown that alterations in m6A regulators are associated with poor patient outcome (Chai et al., 2019; Kwok et al., 2017; Li et al., 2019; Su et al., 2019; Zhou et al., 2019). In our current study, according to the Kaplan–Meier plotter database, when YTHDF1, YTHDF2, WTAP, FTP, and ALKBH5 were highly expressed in ovarian cancer, they were validated as valuable prognostic risk factors for low OS and PFS with high HR. This observation supports our hypothesis that specific m6A regulators are promising candidate biomarkers for predicting the prognosis of patients with ovarian cancer. Moreover, we also established some m6A regulators for the prognostic value of ovarian cancer with the status of TP53 mutation, CA125 level, different grades/stages, and chemotherapy. Additionally, our analysis showed opposing correlations between members with similar functional directionality and ovarian cancer patient outcomes, indicating the functional diversity of m6A regulators.
Ovarian cancer microenvironment plays a critical role in controlling the cancer cell fate, treatment, and prognosis (Yin et al., 2019). In recent studies, a new concept of immune regulatory function of m6A regulatory factor has been proposed. Han et al. (2019) reported that the loss of the reader YTHDF1 in dendritic cells restricted the expression of lysosomal proteases, promoted cross‐presentation of tumor antigens, improved cross‐priming of CD8+ T cells, and enhanced therapeutic efficacy of PD‐L1 blockade. The writer METTL3 has been revealed to catalyze m6A of membrane co‐stimulatory molecules CD40, CD80, and TLR signaling adaptor TIRAP during dendritic cells maturation, and enhanced their translation for promoting T‐cell activation (Wang et al., 2019). Besides that, METTL3 and YTHDF2 also served as negative regulators of type I interferon response to control the innate immune response (Winkler et al., 2019). Hence, the important aspect of our study is to emphasize the role of m6A regulators in immune cell infiltration and immune escape in ovarian cancer. We demonstrated four immune infiltration‐related m6A regulators in ovarian cancer, including RBM15B, ZC3H13, YTHDF1, and IGF2BP1. Specifically, (1) GSEA analyses revealed that inflammatory response, interferon response, NOD‐like receptor, and Toll‐like receptor pathways were negatively correlated with high expression of these m6A regulators. (2) There was a strong association of the expression level of these m6A regulators with the infiltration level of immune cells (B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells). (3) The expression level of these m6A regulators had a strong correlation with diverse immune marker genes, interleukins, CC and CXC chemokines, and human leukocyte antigens. (4) The increased expression of these m6A regulators correlates with the expression of T‐cell exhaustion markers (PD‐1, CD274, CTLA4, LAG3, and GZMB). Therefore, the cross‐talk between the expressions of m6A regulators and tumor microenvironment might be an important mechanism for the development and progression of ovarian cancer. Nevertheless, more functional and mechanism experiments are needed for further verification.
In summary, our results systematically demonstrated expression alterations and prognostic value of m6A regulators in ovarian cancer. The expressions of several specific m6A regulators were correlated with cancer‐related pathways, tumor metastasis, and chemotherapy resistance. In addition, the expressions of m6A regulators might be involved in the regulation of immune cell infiltration and immune escape. Therefore, our study provides new insights into the role of m6A regulators in ovarian cancer.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
AUTHOR CONTRIBUTIONS
JWZ and QYW contributed to the study conception and design. QYW, QYZ, QXL, and JZ collected and analyzed the data. The first draft of the manuscript was written by QYW, QYZ and JWZ. All the authors read and approved the final manuscript.
Supporting information
Fig S1
Table S1
Table S2
Table S3
ACKNOWLEDGMENT
This study was supported by Grants from the National Natural Science Foundation of China (81802589 and 81502230).
Qingying Wang and Qinyi Zhang contributed equally.
Contributor Information
Qingying Wang, Email: wangqingying@tongji.edu.cn.
Jiawen Zhang, Email: jwzhang929@163.com.
REFERENCES
- Bray, F. , Ferlay, J. , Soerjomataram, I. , Siegel, R. L. , Torre, L. A. , & Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians, 68, 394–424. [DOI] [PubMed] [Google Scholar]
- Cerami, E. , Gao, J. , Dogrusoz, U. , Gross, B. E. , Sumer, S. O. , Aksoy, B. A. , Jacobsen, A. , Byrne, C. J. , Heuer, M. L. , Larsson, E. , Antipin, Y. , Reva, B. , Goldberg, A. P. , Sander, C. , & Schultz, N. et al (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery, 2(5), 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai, R. C. , Wu, F. , Wang, Q. X. , Zhang, S. , Zhang, K.‐N. , Liu, Y.‐Q. , Zhao, Z. , Jiang, T. , Wang, Y.‐Z. , & Kang, C.‐S. (2019). m6A RNA methylation regulators contribute to malignant progression and have clinical prognostic impact in gliomas. Aging (Albany NY), 11, 1204–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. Y. , Zhang, J. , & Zhu, J. S. (2019). The role of m6A RNA methylation in human cancer. Molecular Cancer, 18, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini, D. , Moshitch‐Moshkovitz, S. , Schwartz, S. , Salmon‐Divon, M. , Ungar, L. , Osenberg, S. , Cesarkas, K. , Jacob‐Hirsch, J. , Amariglio, N. , Kupiec, M. , Sorek, R. , & Rechavi, G. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A‐seq. Nature, 485(7397), 201–206. [DOI] [PubMed] [Google Scholar]
- Fekete, J. T. , & Győrffy, B. (2019). ROCplot.org: Validating predictive biomarkers of chemotherapy/hormonal therapy/anti‐HER2 therapy using transcriptomic data of 3,104 breast cancer patients. International Journal of Cancer, 145, 3140–3151. [DOI] [PubMed] [Google Scholar]
- Fischl, H. , Neve, J. , Wang, Z. , Patel, R. , Louey, A. , Tian, B. , & Furger, A. (2019). hnRNPC regulates cancer‐specific alternative cleavage and polyadenylation profiles. Nucleic Acids Research, 47(14), 7580–7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, J. , Aksoy, B. A. , Dogrusoz, U. , Dresdner, G. , Gross, B. , Sumer, S. O. , Sun, Y. , Jacobsen, A. , Sinha, R. , Larsson, E. , Cerami, E. , Sander, C. , & Schultz, N. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science Signaling, 6(269), pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, D. , Liu, J. , Chen, C. , Dong, L. , Liu, Y. I. , Chang, R. , Huang, X. , Liu, Y. , Wang, J. , Dougherty, U. , Bissonnette, M. B. , Shen, B. , Weichselbaum, R. R. , Xu, M. M. , & He, C. (2019). Anti‐tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature, 566, 270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, W. , Zhao, Y. , Jin, X. , Yu, D. , He, J. , Xie, D. , & Duan, P. (2018). METTL3 promotes ovarian carcinoma growth and invasion through the regulation of AXL translation and epithelial to mesenchymal transition. Gynecologic Oncology, 151, 356–365. [DOI] [PubMed] [Google Scholar]
- Kwok, C. T. , Marshall, A. D. , Rasko, J. E. , & Wong, J. J. (2017). Genetic alterations of m6A regulators predict poorer survival in acute myeloid leukemia. Journal of Hematology & Oncology, 10, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, Q. , Liu, P. Y. , Haase, J. , Bell, J. L. , Hüttelmaier, S. , & Liu, T. (2019). The critical role of RNA m6A methylation in cancer. Cancer Research, 79, 1285–1292. [DOI] [PubMed] [Google Scholar]
- Lheureux, S. , Gourley, C. , Vergote, I. , & Oza, A. M. (2019). Epithelial ovarian cancer. The Lancet, 393, 1240–1253. [DOI] [PubMed] [Google Scholar]
- Li, B. , Severson, E. , Pignon, J. C. , Zhao, H. , Li, T. , Novak, J. , Jiang, P. , Shen, H. , Aster, J. C. , Rodig, S. , Signoretti, S. , Liu, J. S. , & Liu, X. S. (2016). Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biology, 17, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. B. , Tong, J. , Zhu, S. , Batista, P. J. , Duffy, E. E. , Zhao, J. , Bailis, W. , Cao, G. , Kroehling, L. , Chen, Y. , Wang, G. , Broughton, J. P. , Chen, Y. G. , Kluger, Y. , Simon, M. D. , Chang, H. Y. , Yin, Z. , & Flavell, R. A. (2017). m6A mRNA methylation controls T cell homeostasis by targeting the IL‐7/STAT5/SOCS pathways. Nature, 548(7667), 338–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, N. , Kang, Y. , Wang, L. , Huff, S. , Tang, R. , Hui, H. , Agrawal, K. , Gonzalez, G. M. , Wang, Y. , Patel, S. P. , & Rana, T. M. (2020). ALKBH5 regulates anti‐PD‐1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proceedings of the National Academy of Sciences of the United States of America, 117, 20159–20170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T. , Fan, J. , Wang, B. , Traugh, N. , Chen, Q. , Liu, J. S. , Li, B. , & Liu, X. S. (2017). TIMER: A web server for comprehensive analysis of tumor‐infiltrating immune cells. Cancer Research, 77, e108–e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Xiao, J. , Bai, J. , Tian, Y. , Qu, Y. , Chen, X. , Wang, Q. , Li, X. , Zhang, Y. , & Xu, J. (2019). Molecular characterization and clinical relevance of m6A regulators across 33 cancer types. Molecular Cancer, 18, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, K. D. , & Jaffrey, S. R. (2017). Rethinking m6A readers, writers, and erasers. Annual Review of Cell and Developmental Biology, 33, 319–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha, V. K. , Lindgren, C. M. , Eriksson, K. F. , Subramanian, A. , Sihag, S. , Lehar, J. , Puigserver, P. , Carlsson, E. , Ridderstråle, M. , Laurila, E. , & Houstis, N. (2003). PGC‐1alpha‐responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genetics, 34(3), 267–273. [DOI] [PubMed] [Google Scholar]
- Müller, S. , Glaß, M. , Singh, A. K. , Haase, J. , Bley, N. , Fuchs, T. , Lederer, M. , Dahl, A. , Huang, H. , Chen, J. , Posern, G. , & Hüttelmaier, S. (2019). IGF2BP1 promotes SRF‐dependent transcription in cancer in a m6A‐ and miRNA‐dependent manner. Nucleic Acids Research, 47, 375–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, Á. , Lánczky, A. , Menyhárt, O. , & Győrffy, B. (2018). Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Scientific Reports, 8, 9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree, I. A. , Evans, M. E. , Pan, T. , & He, C. (2017). Dynamic RNA modifications in gene expression regulation. Cell, 169, 1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoiemma, P. P. , & Powell, D. J. Jr. (2015). Tumor infiltrating lymphocytes in ovarian cancer. Cancer Biology & Therapy, 16, 807–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, Y. , Huang, J. , & Hu, J. (2019). m6A RNA methylation regulators contribute to malignant progression and have clinical prognostic impact in gastric cancer. Frontiers in Oncology, 9, 1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, A. , Tamayo, P. , Mootha, V. K. , Mukherjee, S. , Ebert, B. L. , Gillette, M. A. , Paulovich, A. , Pomeroy, S. L. , Golub, T. R. , Lander, E. S. , & Mesirov, J. P. (2005). Gene set enrichment analysis: A knowledge‐based approach for interpreting genome‐wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America, 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Hu, X. , Huang, M. , Liu, J. , Gu, Y. , Ma, L. , Zhou, Q. , & Cao, X. . (2019). Mettl3‐mediated mRNA m6A methylation promotes dendritic cell activation. Nature Communications, 10, 1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Hui, H. , Agrawal, K. , Kang, Y. , Li, N. , Tang, R. , Yuan, J. , & Rana, T. M. (2020). m6A RNA methyltransferases METTL3/14 regulate immune responses to anti‐PD‐1 therapy. The EMBO Journal, 39, e104514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, R. , Gillis, E. , Lasman, L. , Safra, M. , Geula, S. , Soyris, C. , Nachshon, A. , Tai‐Schmiedel, J. , Friedman, N. , Le‐Trilling, V. T. K. , Trilling, M. , Mandelboim, M. , Hanna, J. H. , Schwartz, S. , & Stern‐Ginossar, N. (2019). m6A modification controls the innate immune response to infection by targeting type I interferons. Nature Immunology, 20, 173–182. [DOI] [PubMed] [Google Scholar]
- Yang, Y. , Hsu, P. J. , Chen, Y. S. , & Yang, Y. G. (2018). Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Research, 28, 616–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, M. , Shen, J. , Yu, S. , Fei, J. , Zhu, X. , Zhao, J. , Zhai, L. , Sadhukhan, A. , & Zhou, J. (2019). Tumor‐associated macrophages (TAMs): A critical activator in ovarian cancer metastasis. OncoTargets and Therapy, 12, 8687–8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, Y. , Liu, J. , & He, C. (2015). RNA N6‐methyladenosine methylation in post‐transcriptional gene expression regulation. Genes & Development, 29, 1343–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccara, S. , Ries, R. J. , & Jaffrey, S. R. (2019). Reading, writing and erasing mRNA methylation. Nature Reviews Molecular Cell Biology, 20, 608–624. [DOI] [PubMed] [Google Scholar]
- Zhou, J. , Wang, J. , Hong, B. , Ma, K. , Xie, H. , Li, L. , Zhang, K. , Zhou, B. , Cai, L. , & Gong, K. (2019). Gene signatures and prognostic values of m6A regulators in clear cell renal cell carcinoma – A retrospective study using TCGA database. Aging (Albany NY), 11, 1633–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, H. , Gan, X. , Jiang, X. , Diao, S. , Wu, H. , & Hu, J. (2019). ALKBH5 inhibited autophagy of epithelial ovarian cancer through miR‐7 and BCL‐2. Journal of Experimental & Clinical Cancer Research, 38, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Table S1
Table S2
Table S3


