Abstract
Background
This study set out to determine key lncRNAs correlated with sepsis via constructing competing endogenous RNA (ceRNA) network.
Methods
Three septic patients and three healthy controls were recruited to obtain lncRNA profiles in this current study. Combined with the mRNA profiles by RNA‐sequencing, an integrated analysis of mRNA expression profiles downloaded from GEO was performed to obtain the differentially expressed mRNAs (DEmRNAs). Based on differentially expressed lncRNAs (DElncRNAs) and DEmRNAs acquired in this present study and differentially expressed miRNAs (DEmiRNAs) acquired in previous study, a ceRNA network was constructed. Furthermore, LINC00963 was validated in THP‐1 cells by performing loss of function assays.
Results
In this analysis, a total of 290 DEmRNAs and 46 DElncRNAs were detected in sepsis. Parkinson's disease, Oxidative phosphorylation and Cardiac muscle contraction were significantly enriched KEGG pathways in sepsis. XPO1, CUL4A, and NEDD8 were three hub proteins of sepsis‐specific PPI network. A ceRNA network, which contained 16 DElncRNA‐DEmiRNA pairs and 82 DEmiRNA‐DEmRNA pairs, involving 5 lncRNAs, 10 miRNAs, and 60 mRNAs, was obtained. The function experiments indicated that knockdown of LINC00963 could promote cell proliferation, reduce cytokine expression, and suppress inflammasome activation and phagocytosis in LPS‐induced THP‐1 cells.
Conclusion
This study determined key lncRNAs involved in sepsis, which may contribute to the development for novel treatment strategy of sepsis.
Keywords: ceRNA network, GEO, lncRNA, RNA‐sequencing, sepsis

1. INTRODUCTION
The initial definitions, that sepsis is caused by a host's systemic inflammatory response syndrome (SIRS) to infection, was developed in a 1991 consensus conference (Members of the American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference Committee, 1992). In 2016, it is defined as life‐threatening organ dysfunction arising from a dysregulated host response to infection (Singer et al., (2016)). In some cases, many of the components of the innate immune response can lead to cell and tissue damage, and even multiple organ failure, the clinical hallmark of sepsis (Cohen, (2002)). The development of sepsis inflammatory injury is a complex process, in which abnormal structure and expression of many encoding genes or non‐coding genes exist (Sun et al., 2017). Similarly to acute myocardial infarction or stroke, early detection during the first few hours of sepsis improves outcomes (Rhodes et al., 2017). Due to the high mortality of sepsis, many efforts have been made to help understand the dysregulation of the host response in sepsis.
Considerable evidences suggest that long noncoding RNAs (lncRNAs) are RNA species >200 bp in length and frequently polyadenylated (Guttman et al., 2009; Prensner & Chinnaiyan, 2011). Abundant lncRNAs which are associated with various biological functions have been identified with whole genome transcriptomic analyses (Wang, Han, et al., 2014; Wang et al., 2015). LncRNA is closely related to some pathological process, which was reported to exert great influence in the regulation of chondriokinesis, cell‐type determination and tissue homeostasis, pro‐inflammatory cytokines, calcium ion transport, nuclear factor‐κB (NF‐κB) in signal pathway, and other inflammatory lesions (Carpenter et al., 2013; Gupta et al., 2010; Magny et al., 2013; Rapicavoli et al., 2013; Wang, Long, et al., 2014). As per competing endogenous RNA (ceRNA) hypothesis, lncRNAs exert momentous roles in various biological processes by competing miRNAs with mRNAs to attenuate the inhibition ability of miRNAs to mRNAs (Salmena et al., (2011)). Several studies have described the lncRNA profile in sepsis, but the lncRNA landscape in septic patients remains to be explored (Dai et al., 2017; Lin et al., 2015; Mayama et al., 2016).
We previously identified expression profiles of miRNA and mRNA in septic patients by RNA‐sequencing and bioinformatics analysis (Qin et al., 2020). Currently, we aimed to further investigate lncRNAs associated with sepsis by RNA‐sequencing and constructing a ceRNA (lncRNA‐miRNA‐mRNA) network. Combined with the mRNA profiles, we obtained previously, an integrated analysis of mRNA expression profiles downloaded from Gene Expression Omnibus (GEO) database was performed to obtain the differentially expressed mRNAs (DEmRNAs) between septic patients and healthy controls. Based on differentially expressed lncRNAs (DElncRNAs) and DEmRNAs acquired in this present study and differentially expressed miRNAs (DEmiRNAs) acquired in previous study, a ceRNA network was constructed.
2. MATERIALS AND METHODS
2.1. Ethical compliance
This study was approved by the Ethics Committee of The Third Hospital of Hebei Medical University (2018‐021‐1).
2.2. Data sets collection
To acquire mRNA expression profiles of sepsis, data sets in GEO were retrieved. Five data sets, including GSE69528, GSE67652, GSE57065, GSE26378, and GSE26440, were downloaded from GEO (Table 1).
TABLE 1.
List of mRNA study samples from GEO.
| GEO ID | Author | Platform | Samples (N:P) | Country | Year |
|---|---|---|---|---|---|
| GSE69528 | Presnell S | GPL10558Illumina HumanHT−12 V4.0 expression beadchip | 28:73 | USA | 2015 |
| GSE67652 | Reis EM | GPL16699Agilent−039494 SurePrint G3 Human GE v2 8x60 K Microarray 039381 (Feature Number version) | 12:12 | Brazil | 2015 |
| GSE57065 | Cazalis MA | GPL570[HG‐U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array | 25:28 | France | 2014 |
| GSE26378 | Wong HR | GPL570[HG‐U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array | 21:82 | USA | 2011 |
| GSE26440 | Wong HR | GPL570[HG‐U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array | 32:98 | USA | 2011 |
2.3. Patients and samples
Three septic patients and three healthy controls were recruited to obtain lncRNA expression profiles in this current study. Every participant provided written informed consent for use of their samples. A 2.5 ml peripheral whole blood from every individual was collected in PAXgene® RNA blood tubes (PreAnalytiX GmbH) and stored at −80°C prior to processing. RNA isolation was performed with PAXgene blood RNA kit (PreAnalytiX GmbH). Sequencing was performed based on the Illumina Hiseq X‐ten platform (Illumina, Inc.). The GRCh38 was used as the reference genome sequence.
2.4. Identification of DEmRNAs and DElncRNAs
MetaMA was applied to obtain the DEmRNAs based on the mRNA expression profiles downloaded from GEO and mRNAs we identified previously. The threshold for the significance of DEmRNAs was p < .05. As Zhou et al., (2018) described, differentially expressed lncRNAs (DElncRNAs) were acquired with false discovery rate (FDR) <0.01. By using R package “pheatmap,” hierarchical clustering analysis of DElncRNAs and DEmRNAs was performed.
2.5. Functional enrichment of DEmRNAs
With GeneCodis3 (http://genecodis.cnb.csic.es/analysis), Gene Ontology (GO) classification and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of DEmRNAs were conducted. Statistical significance was defined as p < .05.
2.6. Protein‐protein interaction (PPI) network construction
All DEmRNAs were searched with the BioGrid (http://www.uniprot.org/database/DB‐0184), and PPI network was visualized with Cytoscape software (version 3.6.1, http://www.cytoscape.org).
2.7. Construction of the ceRNA (DElncRNA‐DEmiRNA‐DEmRNA) regulatory network
Based on miRTarBase, DEmiRNA‐DEmRNA pairs were acquired. With starBase, DElncRNA‐DEmiRNA interaction pairs were obtained as well. According to ceRNA theory, the ceRNA (DElncRNA‐DEmiRNA‐DEmRNA) regulatory network was constructed with Cytoscape 3.5.1 (http://cytoscape.org/), by combining lncRNA‐miRNA pairs and miRNA‐mRNA pairs.
2.8. Cell culture and transfection
Human monocytes THP‐1 was purchased from National Infrastructure of Cell Line Resource (Beijing, China) and cultured in Roswell Park Memorial Institute (RPMI)‐1640 medium supplemented with 10% fetal bovine serum (FBS) in a 5% CO2 incubator at 37℃. Cells were treated by 10 μg/mL LPS for 24 h to induce sepsis cell model. siRNAs against LINC00963 and negative controls were purchased from GenePharma. Cell transfection was conducted using Lipofectamine 2000 reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's protocol. Culture medium was replaced after transfection 4–6 h. After incubation for 24 h, they were collected for further analyses.
2.9. CCK‐8 assay
The cell proliferation was examined using the CCK‐8 assay kit according to the manufacturer's instructions. THP‐1 cells in the logarithmic growth phase were selected and planted into 96‐well plates at a density of 5 × 10(Cohen, 2002) cells per well with three replicates per condition. At 0, 24, 48, and 72 h, 10 µl CCK‐8 solution was added to each well. After incubation with CCK‐8 solution for 4 h, the absorbance value at 450 nm was measured using a microplate reader (Thermo Fisher Scientific).
2.10. Quantitative reverse transcription PCR assay
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Quantitative reverse transcription PCR (qRT‐PCR) was performed using a SYBR Green QPCR Supermix (Bio‐rad). The primer sequences were as follows: tumor necrosis factor‐α (TNF‐α) F: 5'‐CAGCCTCTTCTCCTTCCTGA‐3', R: 5'‐GGAAGACCCCTCCCAGATAGA‐3'; CCL2 F: 5'‐GCTCATAGCAGCCACCTCATTC‐3', R: 5'‐CCGCCAAATAACCGATGTGATAC‐3'; interleukin (IL)‐1β F: 5'‐GCAACTGTTCCTGAACTCAACT‐3', R: 5'‐ATCTTTTGGGGTCCGTCAACT‐3'; NLRP3 F: 5'‐CTGCAGCGGCCGCGATGGCAAGCACCCGCTGC‐3', R: 5'‐CGTTTGGATCCCTACCAAGAAGGCTCAA‐3'; Caspase1 (CASP1) F: 5'‐ACGCCTTGCCCTCATAAT‐3', R: 5'‐TCTAATACATCTGGGACTTCTT‐3'. GAPDH (F: 5'‐GGAGGGAGATCCCTCCAAAAT‐3', R: 5'‐GGCTGTTGTCATACTTCTCATGG‐3') was used as an internal reference gene. The result of each sample was normalized by GAPDH and data were calculated by 2−ΔΔCt method.
2.11. ELISA
ELISA was performed to detect the concentration of TNF‐α, CCL2, IL‐1β, NLRP3, and CASP1 in the culture medium of THP‐1 cells using ELISA Kit (Abcam) according to the manufacturer's instructions.
2.12. Phagocytosis assay
According to the manufacturer's instructions, phagocytosis assay was performed. THP‐1 cells suspension was prepared by centrifugation after trypsin digestion. Drop 400 μl cell suspension onto a coverslip in a six‐well plate. Then, 3 ml RPMI‐1640 medium supplemented with 2% FBS was added after 4 h, followed by adding a fluorescent microsphere after 8 h and incubated for 24 h. Coverslips were washed with PBS three times, fixed with Carnoy's Fluid for 20 min, washed with PBS three times, and permeabilized with 0.1% Triton X‐100 for 20 min. After washing with PBS, coverslips were treated with 1 mmol/l Dil solution for 20 min. Then, coverslips were mounted with Antifade Mounting Medium and visualized with a Confocal Microscope.
3. RESULTS
3.1. DEmRNAs and DElncRNAs between septic patients and healthy controls
Compared to healthy controls, a total of 290 DEmRNAs (117 upregulated and 173 downregulated DEmRNAs) and 46 DElncRNAs (39 upregulated and 7 downregulated DElncRNAs) were acquired in sepsis. The top 10 up‐ and downregulated DEmRNAs and DElncRNAs were exhibited in Tables 2 and 3, respectively. Hierarchical clustering analysis of DElncRNAs was shown in Figure 1.
TABLE 2.
Top 10 up‐ and downregulated DEmRNAs between patients with sepsis and normal controls.
| ID | Symbol | Combined. ES | p‐value | FDR | Regulation |
|---|---|---|---|---|---|
| 56943 | ENY2 | 1.090987 | <2.22E‐16 | <1.15E‐15 | Up |
| 9961 | MVP | 1.289563 | <2.22E‐16 | <1.15E‐15 | Up |
| 25953 | PNKD | 0.987258 | <2.22E‐16 | <1.15E‐15 | Up |
| 59286 | UBL5 | 1.255282 | <2.22E‐16 | <1.15E‐15 | Up |
| 146712 | B3GNTL1 | 1.171034 | <2.22E‐16 | <1.15E‐15 | Up |
| 1349 | COX7B | 1.001812 | <2.22E‐16 | <1.15E‐15 | Up |
| 10093 | ARPC4 | 1.067445 | <2.22E‐16 | <1.15E‐15 | Up |
| 84316 | NAA38 | 1.022699 | <2.22E‐16 | <1.15E‐15 | Up |
| 6286 | S100P | 1.79302 | <2.22E‐16 | <1.15E‐15 | Up |
| 10170 | DHRS9 | 1.616565 | <2.22E‐16 | <1.15E‐15 | Up |
| 90 | ACVR1 | −1.06486 | <2.22E‐16 | <1.15E‐15 | Down |
| 23301 | EHBP1 | −1.19787 | <2.22E‐16 | <1.15E‐15 | Down |
| 375484 | SIMC1 | −1.14006 | <2.22E‐16 | <1.15E‐15 | Down |
| 116843 | SLC18B1 | −1.08408 | <2.22E‐16 | <1.15E‐15 | Down |
| 116842 | LEAP2 | −0.97889 | <2.22E‐16 | <1.15E‐15 | Down |
| 23421 | ITGB3BP | −1.00718 | <2.22E‐16 | <1.15E‐15 | Down |
| 79830 | ZMYM1 | −1.22112 | <2.22E‐16 | <1.15E‐15 | Down |
| 8881 | CDC16 | −1.30005 | <2.22E‐16 | <1.15E‐15 | Down |
| 55833 | UBAP2 | −1.08297 | <2.22E‐16 | <1.15E‐15 | Down |
| 27099 | SND1‐IT1 | −1.23699 | <2.22E‐16 | <1.15E‐15 | Down |
Abbreviations: DEmRNAs, differentially expressed mRNAs; FDR, false discovery rate.
TABLE 3.
Top 10 up‐ and downregulated DElncRNAs between patients with sepsis and normal controls.
| ID | Symbol | log2FC | p‐value | FDR | Regulation |
|---|---|---|---|---|---|
| 105372578 | LOC105372578 | 5.08547 | 5.00E‐05 | 0.000929853 | Up |
| LOC105369942 | 4.66767 | .00015 | 0.00235192 | Up | |
| LOC105377068 | 4.12439 | 5.00E‐05 | 0.000929853 | Up | |
| 105375634 | LOC105375634 | 3.75795 | 5.00E‐05 | 0.000929853 | Up |
| 285154 | CYP1B1AS1 | 3.37181 | 5.00E‐05 | 0.000929853 | Up |
| 105376569 | LOC105376569 | 3.02187 | .0007 | 0.00795777 | Up |
| ST3GAL4AS1 | 2.74941 | .00065 | 0.00750607 | Up | |
| 105376332 | LOC105376332 | 2.58516 | .0001 | 0.00168388 | Up |
| 105373262 | LOC105373262 | 2.51971 | 5.00E‐05 | 0.000929853 | Up |
| 101927759 | LOC101927759 | 2.31102 | 5.00E‐05 | 0.000929853 | Up |
| 399715 | LOC399715 | −3.04928 | .0005 | 0.006072 | Down |
| LOC105374771 | −1.82507 | 5.00E‐05 | 0.00093 | Down | |
| 105375328 | LOC105375328 | −1.78289 | .0003 | 0.004062 | Down |
| LOC105376834 | −1.71815 | .0002 | 0.002946 | Down | |
| 105371028 | LOC105371028 | −1.31767 | .0004 | 0.005098 | Down |
| 100506328 | LINC01127 | −1.3044 | .00045 | 0.005597 | Down |
| 101929707 | LOC101929707 | −1.24464 | 5.00E‐05 | 0.00093 | Down |
Abbreviations: DElncRNAs, differentially expressed lncRNAs. FC, fold change. FDR, false discovery rate.
FIGURE 1.
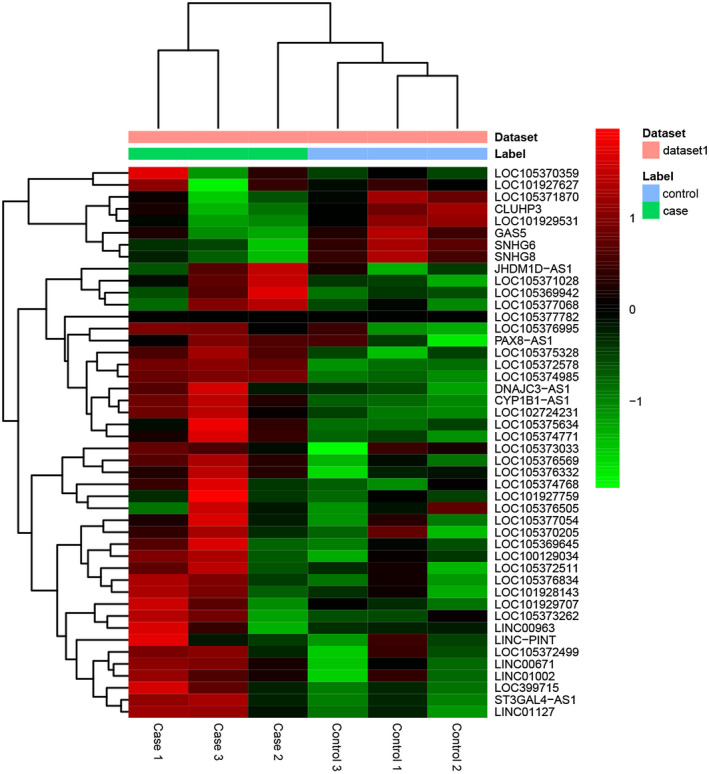
The heatmap of DElncRNAs between sepsis and healthy controls. Row and column represented DElncRNAs and tissue samples, respectively. The color scale represented the expression levels.
3.2. Functional enrichment of DEmRNAs
Respiratory electron transport chain (p = 1.66E‐08), G1/S transition of mitotic cell cycle (p = 1.50E‐06), nucleus (p = 1.26E‐20), protein binding (p = 4.58E‐21), and nucleic acid binding (p = 2.06E‐07) were significantly enriched GO terms in sepsis (Figure 2a‐c). Parkinson's disease (p = 2.63E‐10), Oxidative phosphorylation (p = 2.88E‐10), and Cardiac muscle contraction (p = 5.50E‐09) were significantly enriched KEGG pathways in sepsis (Figure 2d).
FIGURE 2.
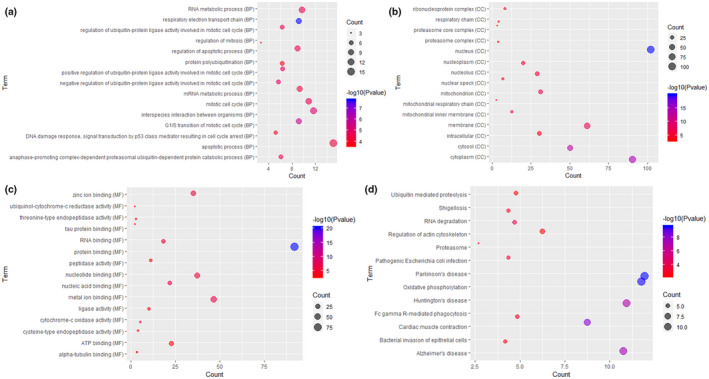
Significantly enriched GO terms and KEGG pathways of DEmRNAs between sepsis and healthy controls. (a) BP, biological process; (b) CC, cellular component; (c) MF, molecular function; (d) KEGG pathways. The x‐axis shows counts of DEmRNAs enriched in GO terms or KEGG pathways and the y‐axis shows GO terms or KEGG pathways. The color scale represented ‐lg p‐value.
3.3. PPI network construction
The sepsis‐specific PPI network included 114 nodes and 131 edges. XPO1 (degree = 26), CUL4A (degree = 10), and NEDD8 (degree = 9) were three hub proteins of sepsis‐specific PPI network (Figure 3).
FIGURE 3.
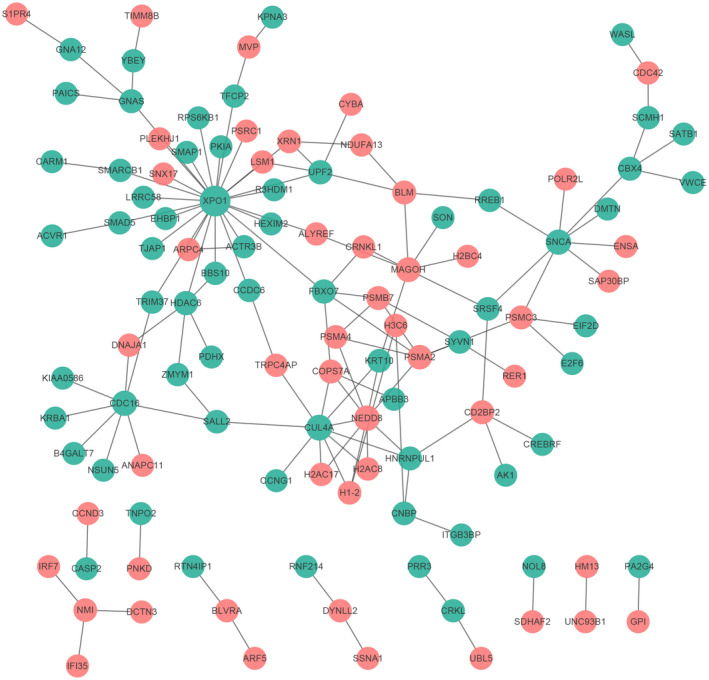
Sepsis‐specific PPI network. The red and blue ellipses represented proteins encoded by up‐ and downregulated DEmRNAs between sepsis and healthy controls.
3.4. CeRNA regulatory network
By combining DElncRNA‐DEmiRNA pairs and DEmiRNA‐DEmRNA pairs as mentioned above, the ceRNA network, which contained 16 DElncRNA‐DEmiRNA pairs and 82 DEmiRNA‐DEmRNA pairs, involving 5 lncRNAs, 10 miRNAs, and 60 mRNAs, was obtained (Figure 4).
FIGURE 4.
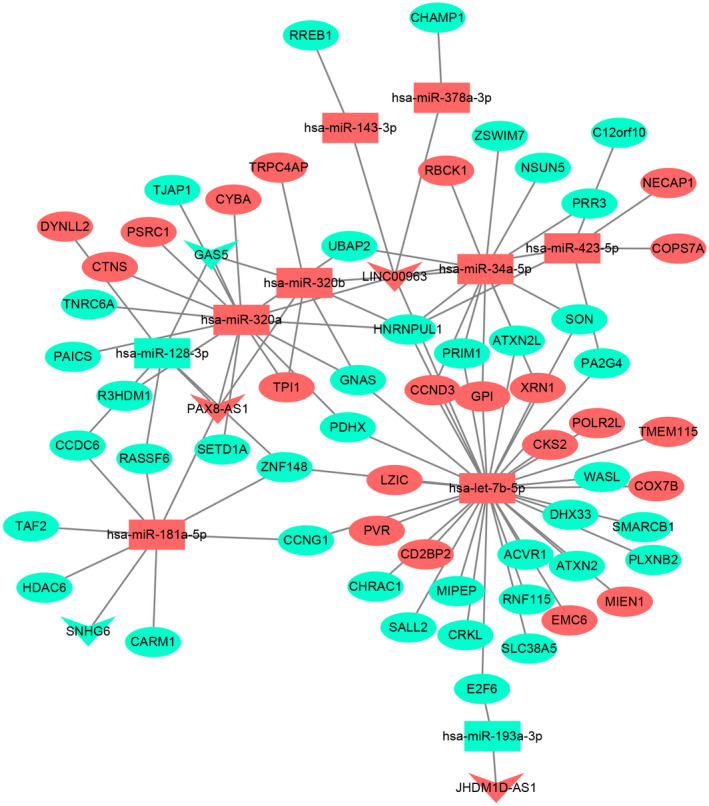
CeRNA (DElncRNA‐DEmiRNA‐DEmRNA) regulatory network. The inverted triangles, rectangle, and elliptical nodes indicate DElncRNAs, DEmiRNAs, and DEmRNAs, respectively. Red and green color represent upregulation and downregulation, respectively.
3.5. Knockdown of LINC00963 promotes cell proliferation in THP‐1 cells
To validate the analysis results, we chose an lncRNA termed LINC00963 for further investigation. In this study, LINC00963 was detected to be upregulated in sepsis. Then, LINC00963 was suppressed by siRNA in THP‐1 cells. The effect of a reduced LINC00963 level on cell viability was determined by using CCK8 assay. THP‐1 cells that were transfected with siRNA targeting LINC00963 showed significant increase in cell viability compared to the negative controls (Figure 5). This result demonstrated that knockdown of LINC00963 may promote cell proliferation in THP‐1 cells.
FIGURE 5.

CCK8 assay showed that LINC00963 knockdown by siRNA promoted the proliferation of THP‐1 cells.
3.6. Knockdown of LINC00963 reduces cytokine expression and suppresses inflammasome activation and phagocytosis in LPS‐induced THP‐1 cells
Compared to the control group, the mRNA levels of cytokine TNF, CCL2, and IL1β were significantly upregulated in LPS group (Figure 6a‐c). Knockdown of LINC00963 had obvious influences on these cytokines, significantly suppressing their mRNA levels compared to the LPS group (Figure 6a‐c). Then, the concentration of cytokine TNF, CCL2, and IL1β were detected by ELISA as well. The results showed the markedly increase of these cytokines in the LPS group compared to the control group, as well as the significantly suppressed levels by knockdown of LINC00963 (Figure 7a‐c). Thus, knockdown of LINC00963 could influence the concentration of TNF, CCL2, and IL1β, which may indicate that knockdown of LINC00963 inhibited the expression of TNF, CCL2, and IL1β in LPS‐induced THP‐1 cells. Subsequently, the degree of inflammasome activation, as indicated by the levels of NLRP3 and CASP1, was assessed. Both qRT‐PCR and ELISA showed these two factors were significantly upregulated in the LPS group and significantly suppressed by knockdown of LINC00963, which suggested that knockdown of LINC00963 may suppress inflammasome activation in LPS‐induced THP‐1 cells (Figures 6d‐e and 7d‐e). Finally, the phagocytosis activity of THP‐1 cells was examined. As showed in Figure 8, the numbers of cells ingesting fluorescent microsphere was increased in LPS group, and decreased by knockdown of LINC00963. Hence, we speculated knockdown of LINC00963 could inhibit the phagocytosis activity of monocytes in the cell model of sepsis.
FIGURE 6.
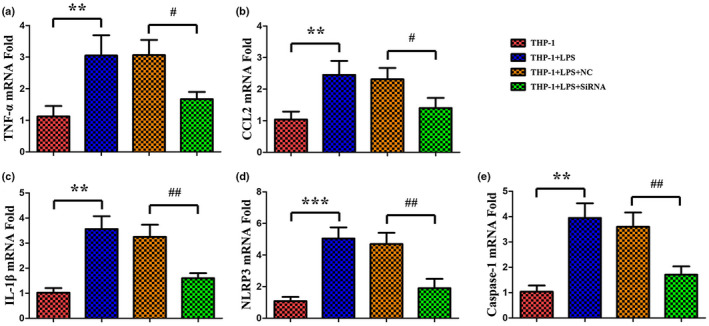
QRT‐PCR analysis of TNF, CCL2, IL1β, NLRP3, and CASP1 in LPS‐induced THP‐1 cells.
FIGURE 7.
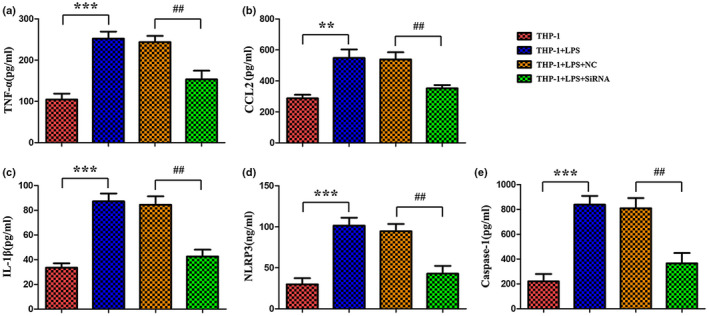
ELISA assay of TNF, CCL2, IL1β, NLRP3, and CASP1 in LPS‐induced THP‐1 cells.
FIGURE 8.
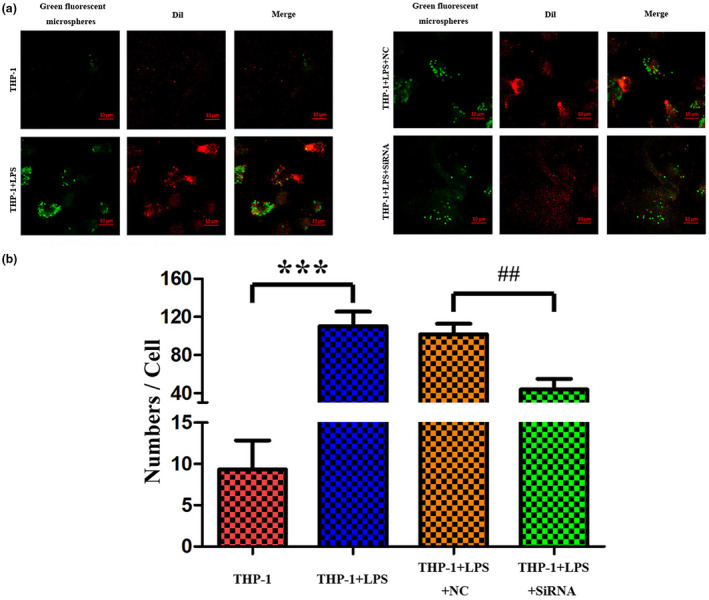
The phagocytosis activity of monocytes in LPS‐induced THP‐1 cells is inhibited by knockdown of LINC00963.
4. DISCUSSION
Sepsis, a catastrophically amplified and dysregulated induction of pro‐inflammatory cytokines often eventuating in multiorgan failure and death, is a major public health problem and a significant burden on the global healthcare system (Cohen, 2002; Edbrooke et al., 1999). Hence, it is of great importance to investigate key lncRNAs associated with the pathological mechanism of sepsis.
UBAP2 (HGNC: 14185) encodes the protein ubiquitin‐associated protein 2, a protein containing a UBA (ubiquitin‐associated) domain characteristic of proteins that function in the ubiquitination pathway (Latonen et al., 2016). Abnormal UBAP2 was detected in different cancers, such as hepatocellular carcinoma (HCC) and prostate cancer (PC; Bai et al., 2016; Latonen et al., 2016). Substantial evidences suggest that the ubiquitin‐proteasome pathway (UPP) may play roles in regulating intermediary metabolism and cell function in catabolic disease states, including sepsis (Patel et al., 2007). It is reported that UPP is a major protein degradation pathway that is modulated during sepsis (Jamart et al., 2014). However, to our best knowledge, there is no research linked UBAP2 with sepsis. In current study, UBAP2 was significantly downregulated in septic patients. Hence, we speculated that UBAP2 may involve in the process of sepsis.
Activin A receptor type 1 (ACVR1, HGNC: 171), also named as ALK2, is a bone morphogenetic protein (BMP) type I receptor and an important member of the BMP signaling pathway (Wang et al., 2018). Fibrodysplasia ossificans progressiva (FOP) is the most known ACVR1‐related disorder (Rafati et al., 2016). In addition, it has been reported that ACVR1 not only functions in BMP signaling pathway, but also Wnt signaling pathway and plays critical roles in many diseases including breast cancer (Wang et al., 2018). In this study, ACVR1 was the most significantly downregulated DEmRNA and it may play key roles in sepsis.
Cytochrome c oxidase (COX) consists of three mitochondrial encoded proteins and 10 nuclear‐encoded proteins, the latter of which including cytochrome c oxidase subunit 7B (COX7B, HGNC: 2291), to form the mature holocomplex in humans (Fontanesi et al., 2006). Over‐expressed COX7B was detected in multiple diseases, such as, nasopharyngeal carcinoma, ovarian cancer, and Alzheimer's disease (L'Esperance et al., 2006; Xiong et al., 2011; Zhang et al., 2015). As we all know, oxidative phosphorylation (OXPHOS) is an important biological process in mitochondria, and closely correlated to multiple neurodegenerative diseases, such as Parkinson's disease, Alzheimer's disease, and Huntington's disease (Singh et al., 2019). Recently, mitochondrial dysfunction is increasingly recognized as an accomplice in most of the common human diseases, not only including cancer, neurodegeneration, but also diabetes, and sepsis (Lee & Huttemann, 2014). Substantial evidences suggest that the mitochondrial OXPHOS system is a primary site of action during acute inflammation and inflammatory responses are key for the outcome of sepsis (Lee & Huttemann, 2014). Our study also indicated that COX7B is significantly upregulated and significantly enriched in Parkinson's disease, oxidative phosphorylation, Alzheimer's disease, and Huntington's disease. Hence, we speculated that COX7B might involve in sepsis through OXPHOS.
Previous studies linked LINC00963 (HGNC: 48716) with prostate cancer, hepatocellular carcinoma, and melanoma (Jiao et al., 2018; Wang, Han, et al., 2014; Wu et al., 2018). Chen et al., (2018) reported the effects of LINC00963 on renal interstitial fibrosis and oxidative stress of rats with chronic renal failure (CRF) through the Foxo signaling pathway, which is often complicated by sepsis. To validate our analyses results, the function of LINC00963 was investigated with a series of experiments in THP‐1 cells. CCK8 assay demonstrated that knockdown of LINC00963 promotes cell proliferation in THP‐1 cells. Both qRT‐PCR and ELISA showed knockdown of LINC00963 inhibits cytokine expression and suppresses inflammasome activation in LPS‐induced THP‐1 cells. In addition, phagocytosis assay indicated that knockdown of LINC00963 inhibits the phagocytosis activity of monocytes in the cell model of sepsis. In our analysis, LINC00963 acts as a ceRNA of UBAP2 via sponging hsa‐miR‐34a‐5p, and a ceRNA of ACVR1 and COX7B via sponging hsa‐let‐7b‐5p in ceRNA network, which may suggest that LINC00963 might involve in sepsis by ceRNA mechanism.
Acetylation of histone is an important epigenetic mechanism, regulated by two families of enzymes: histone deacetylases (HDACs) and histone acetyltransferases (HATs; Hawtree et al., 2013). Dysregulated HDAC activity is believed to be associated with the pathogenesis of inflammatory and autoimmune diseases (Hawtree et al., 2013). According to structure and function homology, 18 mammalian HDACs are classified as four different classes, including class I, II, III, and IV. In addition, class II HDACs are further subdivided into class IIa (HDAC4, 5, 7, and 9) and class IIb (HDAC6 and 10; Haberland et al., 2009). As far as we know, HDAC6 (HGNC: 14064) is a unique HDAC and is found primarily in cytoplasm (Valenzuela‐Fernandez et al., 2008). It is reported that HDAC6 promotes inflammation by regulating expression of genes involved in inflammatory responses (Yan et al., 2018). It had been reported that inhibition of HDAC6 resulted in the amelioration of lupus nephritis in lupus‐prone mice (Vieson et al., 2017). In a lethal septic model, Tubastatin A, a selective inhibitor of HDAC6, was demonstrated to significantly improve long‐term survival, which indicated the therapeutic significance of selective inhibition of HDAC6 for sepsis (Li et al., 2015). Recently, Guo et al. demonstrated that HDAC6 promotes sepsis development by impairing PHB1‐mediated mitochondrial respiratory chain function (Guo et al., 2020).
PAX8‐AS1 (HGNC: 49271) is located on chromosome 2q13 in the upstream region of PAX8 (Han et al., 2016). Expression level of PAX8‐AS1 was significantly correlated to thyroid cancer patients overall or recurrence‐free survival time (Lu et al., 2018). Two variants of PAX8‐AS1 were reported to decrease the risk of cervical cancer (Han et al., 2016). SNHG6 (HGNC: 32965), also named as U87HG, which is mapped to chromosome 8q13, has been identified as an oncogene in various cancers, including breast cancer, gastric cancer, and colorectal cancer (Lv et al.,; Xu et al., 2019; Yan et al., 2017). However, to our knowledge, no previous study has linked PAX8‐AS1 and SNHG6 with sepsis. In our study, we found that PAX8‐AS1 and SNHG6 may participate in sepsis by acting as ceRNAs of HDAC6 through sponging hsa‐miR‐181a‐5p in ceRNA network.
In conclusion, a total of 290 DEmRNAs and 46 DElncRNAs in septic patients were obtained. In addition, PAX8‐AS1/SNHG6/hsa‐miR‐181a‐5p/HDAC6, LINC00963/hsa‐miR‐34a‐5p/UBAP2, and LINC00963/hsa‐let‐7b‐5p/ACVR1/COX7B were identified as important interaction pairs which may involve in the process of sepsis. Importantly, a series of function experiments indicated that knockdown of LINC00963 could promote cell proliferation, reduce cytokine expression, and suppress inflammasome activation and phagocytosis in LPS‐induced THP‐1 cells. Hence, we speculated that knockdown of LINC00963 might be effective for the treatment of sepsis. However, there are also a few limitations in the present study. For example, sample size for RNA‐sequencing was small. Only LINC00963 was validated and its underlying molecular mechanism remains unclear. Further researches with large sample size are required to perform to validate these findings.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
AUTHORS’ CONTRIBUTIONS
XG and CZ made substantial contributions to conception and design. YQ and LW performed the experiment. SD and YY collected and analyzed the data. XB interpreted the data. All authors were involved in drafting and revising the manuscript and gave final approval of the manuscript.
ACKNOWLEDGMENTS
None declared.
DATA AVAILABILITY STATEMENT
The data set supporting the conclusions of this article is included within the article.
REFERENCES
- Bai, D. S. , Wu, C. , Yang, L. X. , Zhang, C. , Zhang, P. F. , He, Y. Z. , Cai, J. B. , Song, Z. J. , Dong, Z. R. , Huang, X. Y. , & Ke, A. W. (2016). UBAP2 negatively regulates the invasion of hepatocellular carcinoma cell by ubiquitinating and degradating Annexin A2. Oncotarget, 7(22), 32946–32955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, S. , Atianand, M. , Aiello, D. , Ricci, E. P. , Gandhi, P. , Hall, L. L. , Byron, M. , Monks, B. , Henry‐Bezy, M. , Lawrence, J. B. , & O’Neill, L. A. (2013). A long noncoding RNA induced by TLRs mediates both activation and repression of immune response genes. Science, 341(6147), 789–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W. , Zhang, L. , Zhou, Z.‐Q. , Ren, Y.‐Q. , Sun, L.‐N. , Man, Y.‐L. , Ma, Z.‐W. , & Wang, Z.‐K. (2018). Effects of long non‐coding RNA LINC00963 on renal interstitial fibrosis and oxidative stress of rats with chronic renal failure via the Foxo signaling pathway. Cellular Physiology & Biochemistry, 46(2), 815–828. [DOI] [PubMed] [Google Scholar]
- Cohen, J. (2002). The immunopathogenesis of sepsis. Nature, 420(6917), 885–891. [DOI] [PubMed] [Google Scholar]
- Dai, Y. , Liang, Z. , Li, Y. , Li, C. , & Chen, L. (2017). Circulating Long Noncoding RNAs as Potential Biomarkers of Sepsis: A Preliminary Study. Genetic Testing and Molecular Biomarkers, 21(11), 649–657. [DOI] [PubMed] [Google Scholar]
- Edbrooke, D. L. , Hibbert, C. L. , Kingsley, J. M. , Smith, S. , Bright, N. M. , & Quinn, J. M. (1999). The patient‐related costs of care for sepsis patients in a United Kingdom adult general intensive care unit. Critical Care Medicine, 27(9), 1760–1767. [DOI] [PubMed] [Google Scholar]
- Fontanesi, F. , Soto, I. C. , Horn, D. , & Barrientos, A. (2006). Assembly of mitochondrial cytochrome c‐oxidase, a complicated and highly regulated cellular process. American Journal of Physiology, Cell Physiology, 291(6), C1129–C1147. [DOI] [PubMed] [Google Scholar]
- Guo, S. D. , Yan, S. T. , Li, W. , Zhou, H. , Yang, J. P. , Yao, Y. , Shen, M. J. , Zhang, L. W. , Zhang, H. B. , & Sun, L. C. (2020). HDAC6 promotes sepsis development by impairing PHB1‐mediated mitochondrial respiratory chain function. Aging, 12(6), 5411–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, R. A. , Shah, N. , Wang, K. C. , Kim, J. , Horlings, H. M. , Wong, D. J. , Tsai, M. C. , Hung, T. , Argani, P. , Rinn, J. L. , & Wang, Y. (2010). Long non‐coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature, 464(7291), 1071–1076. 10.1038/nature08975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman, M. , Amit, I. , Garber, M. , French, C. , Lin, M. F. , Feldser, D. , Huarte, M. , Zuk, O. R. , Carey, B. W. , Cassady, J. P. , Cabili, M. N. , Jaenisch, R. , Mikkelsen, T. S. , Jacks, T. , Hacohen, N. , Bernstein, B. E. , Kellis, M. , Regev, A. , Rinn, J. L. , & Lander, E. S. (2009). Chromatin signature reveals over a thousand highly conserved large non‐coding RNAs in mammals. Nature, 458(7235), 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland, M. , Montgomery, R. L. , & Olson, E. N. (2009). The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nature Reviews Genetics, 10(1):32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, J. , Zhou, W. , Jia, M. , Wen, J. , Jiang, J. , Shi, J. , Zhang, K. , Ma, H. , Liu, J. , Ren, J. , & Dai, M. (2016). Expression quantitative trait loci in long non‐coding RNA PAX8‐AS1 are associated with decreased risk of cervical cancer. Molecular Genetics and Genomics, 291(4), 1743–1748. [DOI] [PubMed] [Google Scholar]
- Hawtree, S. , Muthana, M. , & Wilson, A. G. (2013). The role of histone deacetylases in rheumatoid arthritis fibroblast‐like synoviocytes. Biochemical Society Transactions, 41(3), 783–788. [DOI] [PubMed] [Google Scholar]
- Jamart, C. , Gomes, A. V. , Dewey, S. , Deldicque, L. , Raymackers, J. M. , & Francaux, M. (2014). Regulation of ubiquitin‐proteasome and autophagy pathways after acute LPS and epoxomicin administration in mice. BMC Musculoskeletal Disorders, 15, 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, H. , Jiang, S. , Wang, H. , Li, Y. , & Zhang, W. (2018). Upregulation of LINC00963 facilitates melanoma progression through miR‐608/NACC1 pathway and predicts poor prognosis. Biochemical and Biophysical Research Communications, 504(1), 34–39. [DOI] [PubMed] [Google Scholar]
- Latonen, L. , Leinonen, K. A. , Gronlund, T. , Vessella, R. L. , Tammela, T. L. , Saramäki, O. R. , & Visakorpi, T. (2016). Amplification of the 9p13.3 chromosomal region in prostate cancer. Genes, Chromosomes & Cancer, 55(8), 617–625. [DOI] [PubMed] [Google Scholar]
- Lee, I. , & Huttemann, M. (2014). Energy crisis: the role of oxidative phosphorylation in acute inflammation and sepsis. Biochimica Et Biophysica Acta, 1842(9), 1579–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Esperance, S. , Popa, I. , Bachvarova, M. , Plante, M. , Patten, N. , Wu, L. , Têtu, B. , & Bachvarov, D. (2006). Gene expression profiling of paired ovarian tumors obtained prior to and following adjuvant chemotherapy: molecular signatures of chemoresistant tumors. International Journal of Oncology, 29(1), 5–24. [PubMed] [Google Scholar]
- Li, Y. , Zhao, T. , Liu, B. , Halaweish, I. , Mazitschek, R. , Duan, X. , & Alam, H. B. (2015). Inhibition of histone deacetylase 6 improves long‐term survival in a lethal septic model. The Journal of Trauma and Acute Care Surgery, 78(2), 378–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J. , Zhang, X. , Xue, C. , Zhang, H. , Shashaty, M. G. S. , Gosai, S. J. , Meyer, N. , Grazioli, A. , Hinkle, C. , Caughey, J. , Li, W. , Susztak, K. , Gregory, B. D. , Li, M. , & Reilly, M. P. (2015). The long noncoding RNA landscape in hypoxic and inflammatory renal epithelial injury. American Journal of Physiology‐Renal Physiology, 309(11), F901–F913. 10.1152/ajprenal.00290.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, W. , Xu, Y. , Xu, J. , Wang, Z. , & Ye, G. (2018). Identification of differential expressed lncRNAs in human thyroid cancer by a genome‐wide analyses. Cancer Medicine, 7(8), 3935–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv, P. , Qiu, X. , Gu, Y. , Yang, X. , Xu, X. , & Yang, Y. Long non‐coding RNA SNHG6 enhances cell proliferation, migration and invasion by regulating miR‐26a‐5p/MAPK6 in breast cancer. Biomedicine & Pharmacotherapy, 110, 294–301. [DOI] [PubMed] [Google Scholar]
- Magny, E. G. , Pueyo, J. I. , Pearl, F. M. G. , Cespedes, M. A. , Niven, J. E. , Bishop, S. A. , & Couso, J. P. (2013). Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science, 341(6150), 1116–1120. [DOI] [PubMed] [Google Scholar]
- Mayama, T. , Marr, A. K. , & Kino, T. (2016). Differential Expression of Glucocorticoid Receptor Noncoding RNA Repressor Gas5 in Autoimmune and Inflammatory Diseases. Hormone & Metabolic Research., 48(8), 550–557. [DOI] [PubMed] [Google Scholar]
- Members of the American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference Committee . (1992). American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference: definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Critical Care Medicine, 20(6), 864–874. [PubMed] [Google Scholar]
- Patel, M. B. , Earle, S. A. , & Majetschak, M. (2007). Dynamics of tissue ubiquitin pools and ubiquitin‐proteasome pathway component activities during the systemic response to traumatic shock. Physiological Research, 56(5), 547–557. [DOI] [PubMed] [Google Scholar]
- Prensner, J. R. , & Chinnaiyan, A. M. (2011). The emergence of lncRNAs in cancer biology. Cancer Discovery, 1(5), 391–407. 10.1158/2159-8290.CD-11-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, Y. , Guo, X. , Yu, Y. , Dong, S. , Yan, Y. , Bian, X. , & Zhao, C. (2020). Screening key genes and miRNAs in sepsis by RNA‐sequencing. Journal of the Chinese Medical Association, 83(1), 41–47. 10.1097/JCMA.0000000000000209 [DOI] [PubMed] [Google Scholar]
- Rafati, M. , Mohamadhashem, F. , Hoseini, A. , Hoseininasab, F. , & Ghaffari, S. R. (2016). A novel ACVR1 mutation detected by whole exome sequencing in a family with an unusual skeletal dysplasia. European Journal of Medical Genetics, 59(6–7), 330–336. [DOI] [PubMed] [Google Scholar]
- Rapicavoli, N. A. , Qu, K. , Zhang, J. , Mikhail, M. , Laberge, R. M. , & Chang, H. Y. (2013). A mammalian pseudogene lncRNA at the interface of inflammation and anti‐inflammatory therapeutics. Elife, 2(2), e00762. 10.7554/eLife.00762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes, A. , Evans, L. E. , Alhazzani, W. , Levy, M. M. , Antonelli, M. , Ferrer, R. , Kumar, A. , Sevransky, J. E. , Sprung, C. L. , Nunnally, M. E. , & Rochwerg, B. (2017). Surviving SEPSIS CAMPAIGN: International guidelines for management of sepsis and septic shock: 2016. Intensive Care Medicine, 43(3), 304–377. [DOI] [PubMed] [Google Scholar]
- Salmena, L. , Poliseno, L. , Tay, Y. , Kats, L. , & Pandolfi, P. P. (2011). A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell, 146(3), 353–358. 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer, M. , Deutschman, C. S. , Seymour, C. W. , Shankar‐Hari, M. , Annane, D. , Bauer, M. , Bellomo, R. , Bernard, G. R. , Chiche, J. D. , Coopersmith, C. M. , & Hotchkiss, R. S. (2016). The Third international consensus definitions for sepsis and septic shock (Sepsis‐3). JAMA, 315(8), 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, A. , Kukreti, R. , Saso, L. , & Kukreti, S. (2019). Oxidative stress: A key modulator in neurodegenerative diseases. Molecules, 24(8), 1583. 10.3390/molecules24081583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L. , Li, L. , & Yan, J. (2017). Progress in relationship of the long non‐coding RNA and sepsis. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue, 29(2), 181–183. [DOI] [PubMed] [Google Scholar]
- Valenzuela‐Fernandez, A. , Cabrero, J. R. , Serrador, J. M. , & Sanchez‐Madrid, F. (2008). HDAC6: a key regulator of cytoskeleton, cell migration and cell‐cell interactions. Trends in Cell Biology, 18(6), 291–297. [DOI] [PubMed] [Google Scholar]
- Vieson, M. D. , Gojmerac, A. M. , Khan, D. , Dai, R. , van Duzer, J. H. , Mazitschek, R. , Caudell, D. L. , Liao, X. , Luo, X. M. , & Reilly, C. M. (2017). Treatment with a selective histone deacetylase 6 inhibitor decreases lupus nephritis in NZB/W mice. Histology and Histopathology, 32(12), 1317–1332. [DOI] [PubMed] [Google Scholar]
- Wang, K. , Long, B. O. , Zhou, L.‐Y. , Liu, F. , Zhou, Q.‐Y. , Liu, C.‐Y. , Fan, Y.‐Y. , & Li, P.‐F. (2014). CARL lncRNA inhibits anoxia‐induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR‐539‐dependent PHB2 downregulation. Nature Communications, 5(5), 3596. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Han, S. , Jin, G. , Zhou, X. , Li, M. , Ying, X. , Wang, L. E. , Wu, H. , & Zhu, Q. (2014). Linc00963: a novel, long non‐coding RNA involved in the transition of prostate cancer from androgen‐dependence to androgen‐independence. International Journal of Oncology, 44(6), 2041–2049. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Wu, N. , Liu, J. , Wu, Z. , & Dong, D. (2015). FusionCancer: a database of cancer fusion genes derived from RNA‐seq data. Diagnostic Pathology, 10, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Zhang, Z. , & Wang, J. (2018). MicroRNA‐384 inhibits the progression of breast cancer by targeting ACVR1. Oncology Reports., 39(6), 2563–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. H. , Tian, X. Y. , An, Q. M. , Guan, X. Y. , & Hao, C. Y. (2018). LINC00963 promotes hepatocellular carcinoma progression by activating PI3K/AKT pathway. European Review for Medical and Pharmacological Sciences, 22(6), 1645–1652. [DOI] [PubMed] [Google Scholar]
- Xiong, S. , Wang, Q. , Zheng, L. , Gao, F. , & Li, J. (2011). Identification of candidate molecular markers of nasopharyngeal carcinoma by tissue microarray and in situ hybridization. Medical Oncology (Northwood, London, England), 28(Suppl 1), S341–S348. [DOI] [PubMed] [Google Scholar]
- Xu, M. , Chen, X. , Lin, K. , Zeng, K. , Liu, X. , Xu, X. , Pan, B. , Xu, T. , Sun, L. , He, B. , & Pan, Y. (2019). lncRNA SNHG6 regulates EZH2 expression by sponging miR‐26a/b and miR‐214 in colorectal cancer. Journal of Hematology & Oncology. 12(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, B. , Xie, S. , Liu, Y. , Liu, W. , Li, D. , Liu, M. , Luo, H. R. , & Zhou, J. (2018). Histone deacetylase 6 modulates macrophage infiltration during inflammation. Theranostics, 8(11), 2927–2938. 10.7150/thno.25317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, K. , Tian, J. , Shi, W. , Xia, H. , & Zhu, Y. (2017). LncRNA SNHG6 is associated with poor prognosis of gastric cancer and promotes cell proliferation and EMT through epigenetically silencing p27 and sponging miR‐101‐3p. Cellular Physiology and Biochemistry, 42(3), 999–1012. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Guo, X. Q. , Chu, J. F. , Zhang, X. , Yan, Z. R. , & Li, Y. Z. (2015). Potential hippocampal genes and pathways involved in Alzheimer's disease: A bioinformatic analysis. Genetics and Molecular Research., 14(2), 7218–7232. [DOI] [PubMed] [Google Scholar]
- Zhou, Y. , Gu, C. , Li, J. , Zhu, L. , Huang, G. , Dai, J. , & Huang, H. (2018). Aberrantly expressed long noncoding RNAs and genes in Parkinson's disease. Neuropsychiatric Disease and Treatment, 14, 3219–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data set supporting the conclusions of this article is included within the article.


