Abstract
Diabetes mellitus is the most common chronic disorder and leading cause of renal, neurological, and gastrointestinal manifestations in developed and developing countries. Despite of many drugs and combinational therapies, the complications of diabetes are still listed due to severe consequences of those drugs. In past few years, plant-derived drugs draw special attention due to their higher efficacy and fewer side-effects. Momordica charantia also known as bitter melon is referred as an antidiabetic and hypoglycemic plant in native populations of Asia and East Africa. In current study, an in silico approach was used to evaluate the interactions and binding patterns of plant-derived peptides devised from a hypoglycemic protein adMc1 of M. charantia as potential inhibitor of DPP-IV, SGLT1, and GLUT2 receptor proteins. The study has described a novel approach to investigate hypoglycemic peptides to cure diabetes. A total of eighty tetra-, penta-, and hexapeptides were devised from conserved regions of adMc1 homologs. The molecular docking approach using MOE software was employed to reveal inhibiting potentials of devised peptides against three selected proteins. Out of 30 shortlisted ligands six peptides (i.e. SMCG, DECC, TTIT, RTTI, ARNL and TVEV) accomplished the criteria of being good drug candidates against selected receptor proteins following the drugability assessment test. The overall results are acceptable on the basis of ADMET profiling for being good drug candidates against selected proteins.
1. Introduction
Diabetes mellitus (DM) is a heterogenous group of chronic metabolic disorders associated with irregular glucose homeostasis and results in elevated level of blood glucose and insulin resistance [1]. The rapid increase in diabetes cases is estimated to increase 4.4% in 2030 among all age groups [2]. Currently, there are many synthetic hypoglycemic drugs available in market including sulfonylureas, incretins, α-glucosidase inhibitors, dipeptidyl peptidase-IV (DPP-IV) inhibitors, sodium-dependent glucose transport proteins (SGLTs) inhibitors, and thiazolidinediones but they result in severe consequences. Therefore, owing to severe side-effects of these drugs, there is a need of new class of drugs with much potency and efficacy [3].
The potential target proteins in diabetes include DPP-IV, SGLTs, glucose transporters (GLUT), α-glucosidase inhibitors, and peroxisome proliferator-activated receptors [4]. DPP-IV or adenosine deaminase binding protein is a serine exopeptidase predominantly expressed on surface of cells. DPP-IV selectively involves in N-terminal dipeptide cleavage from a variety of substrates including cytokines, growth factors, and incretin hormones [5]. Since the approval of DPP-IV inhibitors, their importance has been raised clinically to cure DM [6]. DPP-IV inhibitors are a class of oral diabetic drugs commonly used as stable drug candidates which have prospectively been designed using therapeutic agent's strategy focused on deep study on mode of action of incretin peptides [7].
In type 2 diabetes mellitus (T2DM) patients, the insulin resistance leads towards elevated level of glucose and the activation of incretins. Incretins are group of metabolic gut peptides secreted from enteroendocrine cells into the blood after intake of nutrients. Incretins lower the glucose level by stimulating the induction of insulin from islets of Langerhans and also inhibits the glucagon production. Glucagon-like peptide-1 (GLP-1), glucagon-like peptide-2 (GLP-2), and glucose dependent insulinotropic peptide/gastric inhibitory polypeptide (GIP) are two examples of incretin hormones [8].
SGLTs are glucose transporters found in six isoforms (i.e., SGLT1, SGLT2, SGLT3, SGLT4, SGLT5, and SGLT6) and have been found to be scattered across the human body. Among these six isoforms, SGLT1 and SGLT2 are well known and extensively investigated in diabetes. Studies have showed that selective inhibition of SGLT1 can slow postprandial gut uptake of glucose and increase plasma levels of GLP-1 and GIP in healthy volunteers [9]. Glucose transporters are proteins widely distributed in body cells and facilitate in the maintenance of blood glucose level in the human body [2]. Among all the glucose transporters, GLUT2 plays a bidirectional role in specific transportation of glucose in the hepatocytes and absorption and reabsorption of glucose from the enterocytes and the renal tubule particularly [10]. GLUT2 is a sugar carrier that sustains power generation in the cell but can also serve as a receptor for extracellular glucose. GLUT2 is regarded as a competent target in treatment of DM due to its major role in glucose homeostasis.
In developing countries, the plant-based compounds have been used as drugs in 75-80% population to cure various diseases [11–13]. Peptides are generated from specific proteins and then consumed as food ingredients. The bioavailability of these peptides depends on absorption, distribution, metabolism, excretion, and toxicity- (ADMET-) based parameters to reach the target organ [14]. Currently, there are many hypoglycemic drugs in the market but these are correlated with multiple gastrointestinal and renal disorders. In silico targeting of different glucose transporters explores new ways in the management of type 2 diabetes mellitus. In this study, the adMc1 protein of medical plant Momordica charantia was used to prepare ready-to-dock library of 80 ligands. This includes the in silico approach to target DPP-IV, SGLT1, and GLUT2 to gain the structural insight of binding patterns of different ligands with receptor proteins. The main objective of this study was to evaluate the insulin-like activity of Momordica charantia derived peptides using in silico methods and to explore the best inhibitors of DPP-IV, SGLT1, and GLUT2 receptor proteins among prepared ligands.
2. Materials and Methods
2.1. Retrieval of the 3D Structures of Receptor Proteins
In this study, the 3D structures of three target molecules were employed to reveal hypoglycemic activities of different peptides which were companionable with the properties of the target binding site in the molecular docking study. The three-dimensional structures of human dipeptidyl peptidase-IV (PDB ID: 4A5S) and sodium-dependent glucose cotransporter (PDB ID: 3DH4) were retrieved from the RCSB Protein Data Bank (https://www.rcsb.org/).The protein sequence of GLUT2 was retrieved from the protein database of NCBI (accession number: P11168.1). The 3D structure of GLUT1 (PDB ID: 5EQG) was used as a template to predict the 3D structure of the GLUT2 protein. MODELLER 9.21 was used for homology modeling, and the best model was selected as a receptor protein for further analysis [15, 16].
2.2. Ligand Selection and Receptor Optimization
The protein sequence of the adMc1 protein from Momordica charantia was retrieved from the protein database of NCBI (accession No: CDG50933.1). MEME Suite was used to predict motifs from the selected protein homologs [17, 18]. Tetra-, penta-, and hexapeptides were devised from the predicted motifs, and their 3D structures were obtained using ChemSketch in MOL format [19]. Optimization of receptor was done by removing water molecules, addition of hydrogen atoms, energy minimization, and 3D protonation for the perfect and accurate docking. The minimized structures were then docked against devised peptides.
2.3. Molecular Docking
The prediction of the active site is the most crucial step in in silico drug discovery as it has a direct impact on the reliability of the results. Molecular docking of 80 ligands was performed at the active site of DPP-IV, SGLT1, and GLUT2 receptor proteins separately. Site finder tool of Molecular Operating Environment (MOE) was used to predict active sites in the selected receptor proteins. The parameters (i.e., rescoring 1: London dG, retain: 10, refinement: force field and rescoring 1: London dG, retain: 10) were set to their default to calculate the interactions of ligands with the binding residues of receptor proteins. All devised peptides were docked against the receptor proteins using MOE. Most appropriate interactions and bindings between ligands and receptor proteins were selected on the basis of the best S-score, root mean squared deviation (RMSD), and energy validation rankings.
2.4. Drug Scan and ADMET Profiling
The drug-likeness properties were determined following the Lipinski's rule of five using SwissADME [20]. The Lipinski's rule of five is based on five parameters (i.e., molecular mass: ≤500 Dalton, molar refractive index: 40-130, partition coefficient (log P): ≤5, hydrogen bond donors: <5, and hydrogen bond acceptors: <10). Only the molecules that accomplish all these parameters could be accepted as potential drug candidates. The online bioinformatic server admetSAR was used for ADMET-based chemical screening of selected drug candidates [21].
3. Results
3.1. Devising Tetra-, Penta-, and Hexapeptides from adMc1
The MEME Suite was used to explore five motifs in ten selected homologs of the adMc1 protein. The motifs were used to devise tetra-, penta-, and hexapeptides to be docked as ligands against DPP-IV, SGLT1, and GLUT2.
3.2. Molecular Docking
Molecular docking provides discernment into structural interactions of inhibitors with the receptor proteins. The prepared library of 80 peptides devised from the adMc1 protein of M. charantia was docked against DPP-IV, SGLT1, and GLUT2 receptor proteins using the docking algorithm of MOE software. MOE aligned the suitable confirmations and ranked the ligands based on four factors (i.e., the binding pocket with minimum energy structure, strength of hydrogen bond, root mean squared deviation value, and S-score). The results of computational docking showed the potency of adMc1 peptides as good inhibitors of DPP-IV, SGLT1, and GLUT2. In this study, three different receptor proteins were selected due to their major roles in diabetes and upregulation of glucose homeostasis. Top 10 conformations were selected in each analysis based on binding patterns and energy validations.
3.3. Interaction Analysis
For the receptor protein DPP-IV, top 10 conformations have been selected based on their structural interactions and S-scores. In literature, Glu205, Glu206, Tyr547, and Tyr662 have been reported as major amino acids of catalytic pocket of DPP-IV [22]. In this study, the selected top 10 peptides also showed interactions with these reported amino acids (Table 1). The peptide MRGID showed the best interaction (binding score: -17.8389) with the receptor protein, and Glu206, Tyr547, Arg356, and Pro550 were found to be the leading interactive residues in these interactions. All the remaining ligands also showed strong interactions with Glu205, Glu206, Tyr547, and Tyr662 which are also the active amino acid residues of DPP-IV. These ligands have been revealed as maximum pocket shareholders by interacting with the active amino acids of catalytic triad.
Table 1.
The interactions of top ten devised peptides with DPP-IV as the receptor protein.
| Sr. no. | Peptide | S-score | Interacting residues |
|---|---|---|---|
| 1 | MRGID | -17.8389 | Glu206, Tyr547, Arg356, Pro550 |
| 2 | TTVEV | -15.8132 | Glu206, Tyr662, Ser209, Arg669 |
| 3 | LRQQSR | -15.7212 | Glu206, Tyr547, Arg356, Ser209, Arg358 |
| 4 | TVEV | -15.0490 | Glu205, Tyr662, Asn710, His126 |
| 5 | FDECC | -14.6527 | Glu206, Tyr547 |
| 6 | ECCRE | -14.3605 | Tyr547, Tyr662, Tyr666, Pro550, Arg358, Ser209 |
| 7 | MRGIEN | -14.3088 | Glu206, Tyr662 |
| 8 | RCRQ | -13.5417 | Tyr547, Val207 |
| 9 | TTIT | -13.5247 | Glu205, Ser209 |
| 10 | EECR | -13.3807 | Tyr662, Ser209 |
The protein sodium-dependent glucose cotransporter 1 (SGLT1) is a major contributor in the reabsorption of glucose in proximal tubules of nephrons. The binding pocket of the SGLT1 receptor protein contains Ser372 and Ser368 as main interacting amino acids. Out of 80 prepared ligands in this study, the top 10 ligands with best S-scores and interactions with these active amino acids were selected (Table 2). The peptides EEQRQA, FDEC, and ITTVE with docking scores of -21.0223, -21.0722, and -20.0948 showed strong interactions with Ser372 and Ser368.
Table 2.
The interactions of top ten devised peptides with SGLT1 as receptor protein.
| Sr. no. | Peptide | S-score | Interacting residues |
|---|---|---|---|
| 1 | EEQRQA | -21.0223 | Ser372, Ser368 |
| 2 | FDEC | -21.0722 | Ser372, Ser368, Ser365, Ser364, Tyr138, Tyr263, Asp189, Ala63, Gln268 |
| 3 | ITTVE | -20.0948 | Ser368, Ser365, Ser364, Asn267, Asp189 |
| 4 | AREEQR | -19.3288 | Ser368, Ser365, Tyr138, Ala361, Ala63, Asp189 |
| 5 | YAYRTT | -18.9325 | Ser368, Tyr263, Asn142, Asn267 |
| 6 | EEQR | -18.5788 | Ser372, Ser368, Ser364, Tyr138, Tyr176 |
| 7 | SMCG | -17.4970 | Ser372, Tyr138, Tyr269, Gln428 |
| 8 | DECC | -17.3894 | Ser368, Ser365, Asn267 |
| 9 | ERCR | -17.1217 | Ser368, Asn142, Thr375 |
| 10 | TTIT | -16.9639 | Ser372, Ser368, Tyr138, Asn142 |
Glucose transporters play major role in renal and intestinal glucose absorption and reabsorption. In current study, the Lys288 showed strong interactions with the GLUT2 protein and revealed as a major interacting amino acid. Among the selected ligands, the peptide REEQR with S-score of -17.4161 exhibited potential of being a good drug candidate with the best binding affinity with Lys288, Glu286, Ser284, Arg124, and Ser112. The remaining ligands could also be foremost inhibitors of GLUT2 with the best binding affinity and S-scores and could be potential drug candidates (Table 3).
Table 3.
The interactions of top ten devised peptides with GLUT2 as the receptor protein.
| Sr. no. | Peptide | S-score | Interacting residues |
|---|---|---|---|
| 1 | REEQR | -17.4161 | Lys288, Glu286, Ser284, Arg124, Ser112 |
| 2 | YAYRTT | -17.1186 | Lys288, Glu282, Arg124 |
| 3 | ITTVEV | -16.9540 | Lys288, Glu286, Ala283, Ser169 |
| 4 | LEEIA | -15.8995 | Lys288, Ser169 |
| 5 | YLRQ | -14.8063 | Lys288 |
| 6 | RCRQ | -13.5942 | Lys288 |
| 7 | RTTI | -13.5003 | Lys288, Phe113, Val289 |
| 8 | ARNL | -12.9365 | Lys288 |
| 9 | FDEC | -12.8992 | Lys288 |
| 10 | TITTVE | -15.6597 | Ser169, Ser112 |
3.4. Drugability and ADMET Profiling
The Lipinski's rule of five evaluates the drug-like properties of proposed drug candidates based on five parameters. The rule illustrates the drugability and behavior of selected drug candidate as it distinguishes the drug-like and nondrug-like nature of biologically active ligands. Only the molecules that accomplish all these parameters could be accepted as potential drug candidates. In this study, total 30 ligands have been shortlisted on the bases of their best interactions with active amino acids, S-scores, and energy validations. Out of 30 selected ligands used in this study, three tetrapeptides (SMCG, DECC, and TTIT) were found to be effective inhibitors of SGLT1, two tetrapeptides (RTTI and ARNL) were found effective inhibitors of GLUT2, and one tetrapeptide (TVEV) was found to be an efficient inhibitor of DPPI-IV as all these peptides only violated one parameter of Lipinski's rule of five (Table 4). The structures of these six peptides are given in Figure 1. On the basis of Lipinski's rule, these peptides are expected for having reasonable oral bioavailability. The interactions and binding patterns of best peptides each against SGLT1 (i.e., TTIT), GLUT2 (i.e., RTTI), and DPP-IV (i.e., TVEV) have been shown in Figure 2.
Table 4.
Pharmacokinetic parameters important for bioavailability of compounds drug-likeness properties of selected peptides.
| Peptides | Target | Molecular properties† | ||||||
|---|---|---|---|---|---|---|---|---|
| MW | HBD | HBA | Nrotb | Log P | A | Violations | ||
| SMCG | SGLT1 | 396.48 | 6 | 7 | 15 | -1.86 | 94.77 | 1 |
| DECC | 468.50 | 7 | 10 | 17 | -2.39 | 107.10 | 1 | |
| TTIT | 434.48 | 8 | 9 | 15 | -2.06 | 105.61 | 1 | |
| RTTI | GLUT2 | 489.57 | 10 | 9 | 19 | -2.17 | 123.16 | 1 |
| ARNL | 472.54 | 9 | 8 | 19 | -2.38 | 118.94 | 1 | |
| TVEV | DPP-IV | 446.50 | 7 | 9 | 16 | -0.93 | 109.86 | 1 |
| FDEC | 512.53 | 7 | 10 | 18 | -1.40 | 123.66 | 2 | |
†Molecular properties were calculated using SwissADME an online tool. MW: molecular weight; HBD: number of hydrogen bond donors; HBA: number of hydrogen bond acceptors; nrotb: number of rotatable bonds; log P: the logarithm of octanol/water partition coefficient; A: molar refractivity.
Figure 1.
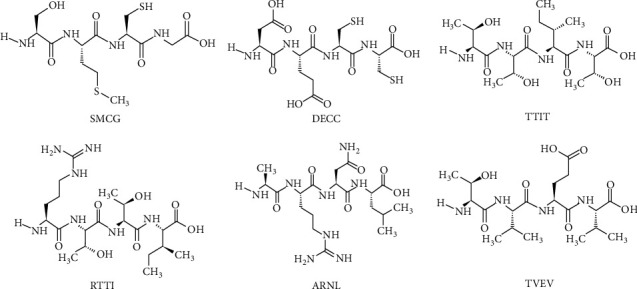
Structures of best selected peptides.
Figure 2.
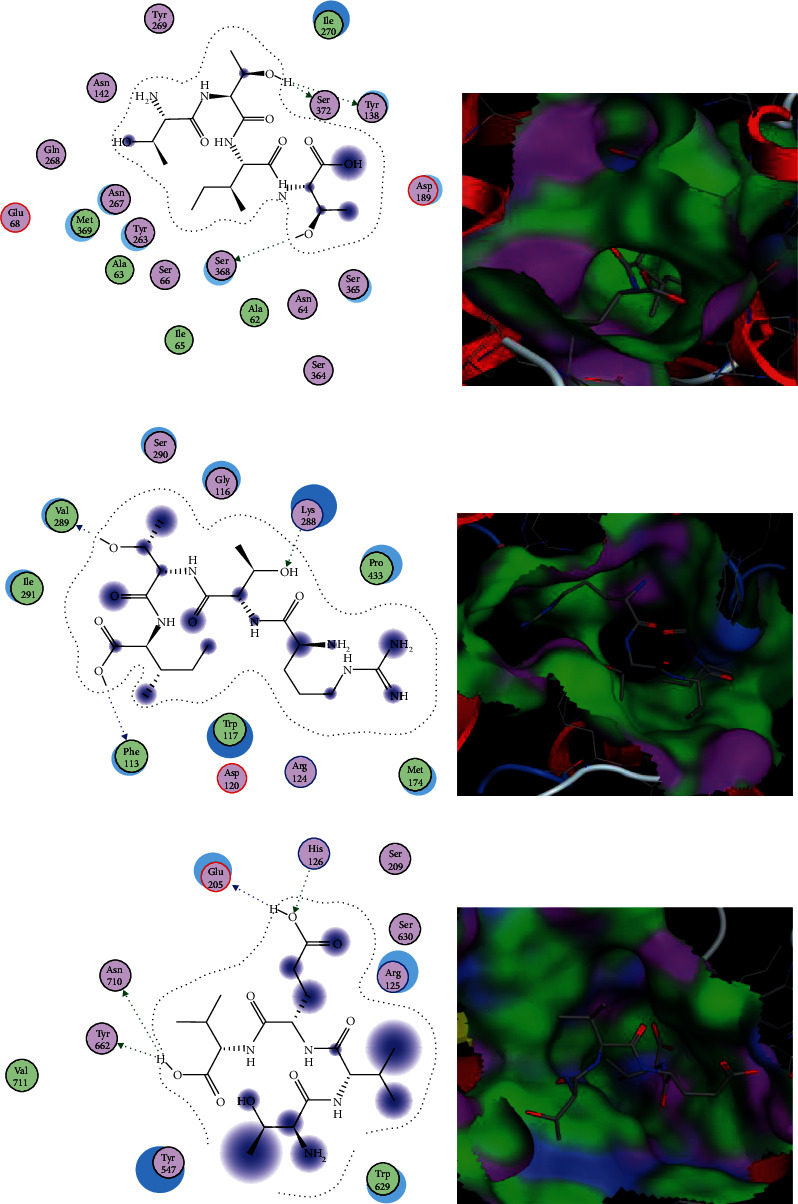
Interactions (on left) and binding patterns (on right) of best selected peptides. (a, b) Interactions and binding patterns of TTIT with SGLT1. (c, d) Interactions and binding pattern of RTTI with GLUT2. (e, f) Interaction and binding pattern of TVEV with DPP-IV, respectively.
For further validations of drug-like behavior of selected ligands, all six ligands were subjected to admetSAR server for assessment of five parameters of ADMET profiling (absorption, distribution, metabolism, excretion, and toxicity). From the results, it has been revealed that all the selected peptides are non-AMES toxic and noncarcinogens (Table 5). The overall results of ADMET drug scanning of these lead ligands were tolerable, and these peptides could be accepted as efficient drug candidates against selected receptor proteins.
Table 5.
ADMET profiling of best selected peptides.
| SMCG | DECC | TTIT | RTTI | ARNL | TVEV | |
|---|---|---|---|---|---|---|
| Absorption | ||||||
| Blood-brain barrier | BBB+ | BBB+ | BBB+ | BBB+ | BBB+ | BBB+ |
| Human intestinal absorption | HIA- | HIA- | HIA- | HIA- | HIA+ | HIA- |
| Caco-2 permeability | Caco-2- | Caco-2- | Caco-2- | Caco-2- | Caco-2- | Caco-2- |
| P-Glycoprotein substrate | Substrate | Nonsubstrate | Nonsubstrate | Nonsubstrate | Substrate | Nonsubstrate |
| P-Glycoprotein inhibitor | Noninhibitor | Noninhibitor | Noninhibitor | Noninhibitor | Noninhibitor | Noninhibitor |
| Renal organic Cation transporter | Noninhibitor | Noninhibitor | Noninhibitor | Noninhibitor | Noninhibitor | Noninhibitor |
| Metabolism | ||||||
| CYP3A4 substrate | Substrate | Nonsubstrate | Nonsubstrate | Nonsubstrate | Substrate | Nonsubstrate |
| CYP2C9 substrate | Nonsubstrate | Nonsubstrate | Nonsubstrate | Nonsubstrate | Substrate | Nonsubstrate |
| CYP2D6 substrate | Nonsubstrate | Nonsubstrate | Nonsubstrate | Nonsubstrate | Nonsubstrate | Nonsubstrate |
| CYP3A4 inhibition | Noninhibitor | Noninhibitor | Noninhibitor | Noninhibitor | Noninhibitor | Noninhibitor |
| CYP2C9 inhibition | Noninhibitor | Noninhibitor | Noninhibitor | Noninhibitor | Noninhibitor | Noninhibitor |
| CYP2C19 inhibition | Noninhibitor | Noninhibitor | Noninhibitor | Noninhibitor | Noninhibitor | Noninhibitor |
| CYP2D6 inhibition | Noninhibitor | Noninhibitor | Noninhibitor | Noninhibitor | Noninhibitor | Noninhibitor |
| CYP1A2 inhibition | Noninhibitor | Noninhibitor | Noninhibitor | Noninhibitor | Noninhibitor | Noninhibitor |
| Toxicity | ||||||
| AMES toxicity | Non-AMES toxic | Non-AMES toxic | Non-AMES toxic | Non-AMES toxic | Non-AMES toxic | Non-AMES toxic |
| Carcinogens | Noncarcinogens | Noncarcinogens | Noncarcinogens | Noncarcinogens | Noncarcinogens | Noncarcinogens |
4. Discussion
Diabetes mellitus is a heterogeneous group of diseases characterized by irregular glucose homeostasis and chronic hyperglycemia due to flaw in insulin secretion and activity [23]. Type 1 diabetes mellitus (insulin dependent) and type 2 diabetes mellitus (noninsulin dependent) are referred as two peculiar forms of diabetes. Type 1 or insulin dependent DM is characterized by total destruction of pancreatic β-cells and shutting down the glucose metabolism. The type 2 or noninsulin-dependent diabetes is reported as the most common form due to abnormal insulin secretion and insulin resistance. In type 2 DM, the continuous exposure of body to glucose causes the severity of the disease leading to neurological, gastrointestinal, renal, and cardiovascular syndromes [24]. A vast variety of natural compounds have been reported with medicinal perspectives for the treatment of various diseases [25]. In this study, tetra-, penta-, and hexapeptides (80 in total) were devised from the adMc1 protein of Momordica charantia, and their interactions with different receptor proteins that play role in glucose homeostasis were checked through the molecular docking approach. Molecular docking is an elaborative method to evaluate the best possibilities of ligand interactions against different targets for in silico drug designing [26].
The balancing of glucose homeostasis, maintenance of hyperglycemic state to normal level, and shutting of the enzymes that cause irregular glucose homeostasis are the possible ways to control diabetes. Over the past few years, many conventional drug therapies have been in practice but the outcomes are undesirable due to poor efficacy and different side-effects [27]. Thus, scientists are moving towards the use of natural bioactive compounds as antidiabetic agents. The extensive literature survey reveals a large set of targets such as DPP-IV, sodium glucose transporters (SGLTs), α-glucosidase, protein tyrosine phosphatase 1B (PTP1B), glucose transporters (GLUT), and G protein-coupled receptors [28–30].
In this study, DPP-IV, SGLT1, and GLUT2 have been listed as target receptor proteins due to their major roles in glucose homeostasis. DPP-IV is a serine exopeptidase that is associated with incretin deficiency in type 2 DM. Current combinational drug therapies include oral hypoglycemic agents including metformin, komboglyze, janumet, and juvisync but these drugs result in unsustainable outcomes. Therefore, the need of new drugs with more efficacies and less side-effects is urgent in order to treat DM [27]. Citrus flavonoids were taken as ligands against different target proteins (e.g., glycogen synthase kinases 3β, DPP-IV, and glucokinase) [29]. Shen and Lu [29] have reported that the binding pocket of DPP-IV contains Glu205, Glu206, Tyr662, and Ser209 as active amino acids. In this study, Glu205, Ser209, and Tyr662 were also found as active amino acids as they exhibited strong interactions with DPPI-IV with energy validations.
Sodium glucose transporter-1 (SGLT1) is a glucose transporter expressed at the striated border of intestine. In diabetic patients, the overwhelming glucose concentration might be controlled by SGLT1 inhibition [31]. The 3D structure of human SGLT1 was not available; therefore, the 3D structure of SGLT2 from Vibrio parahaemolyticus was taken as a template in this study for the homology modeling of human SGLT1. Both isoforms of sodium glucose transporters share about 60% sequence identity and are highly expressed in S1 segment of border brush membrane [31]. In previous studies, Ser372, Ser50, Asn51, Gly53, Ser54, Gly55, His56, Gln271, Ser368, Ser369, Gln433, and Ser368 of SGLTs have been reported as polar active amino acids [32]. In current study, Ser372 and Ser368 were also found among the interacting residues of SGLT1 that showed strong interactions with the selected ligands.
On the bases of drug-likeness, only 6 peptides have been shortlisted (i.e., SMCG, DECC, TTIT, RTTI, ARNL, and TVEV) followed by Lipinski's rule of five. The remaining 24 ligands violated two or more parameters of Lipinski rule of five. The further assessment of these ligands included ADMET drug scanning to prognosticate the probability of success and failure of potential drug candidates. All the selected ligands passed the criteria of drugability. The efforts and purpose of this study were to target the proteins that play roles in glucose homeostasis to overcome the diabetes mellitus. The mutational changes in certain receptor proteins which are involved in the metabolism and regulation of glucose lead towards severe consequences including the onset of prediabetes and diabetes mellitus. Diabetes mellitus is a group of chronic metabolic diseases that leads towards the onset of many other leading disorders. In the market, many hypoglycemic agents have been in practice to cure diabetes and hyperglycemia but the unsustainable results of these agents lead towards their failure. So, the current need of time is to explore the insulin-like peptides from natural sources with maximum potential and less side-effects.
5. Conclusion
The current study offers conversational and comprehensive computational study towards the discovery of insulin-like hit ligands, screened from ready-to-dock library of peptides as ligand molecules against the DPP-IV, SGLT1, and GLUT2 receptor proteins. In this study, 80 highly conserved tetra-, penta-, and hexapeptides as leading ligands were devised from the adMc1 protein of Momordica charantia, and the in silico approach was applied to check their molecular dynamic validations and ultimately, to explore their antidiabetic potentials. The peptides SMCG, DECC, TTIT, RTTI, ARNL, and TVEV provided valuable results in drug assessment and ADMET drug scanning. Among the 30 ligands selected on the basis of molecular docking studies, these six tetrapeptides passed the drug assessment tests with great ability to be recognized as drug candidates with potential of inhibiting the DPP-IV, SGLT1, and GLUT2 receptor proteins.
Acknowledgments
The authors would like to gratefully acknowledge the Department of Biochemistry, Government College University Faisalabad for providing space and facilities to accomplish this study.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declares that they have no conflicts of interest.
References
- 1.Yasmin A., Bukhari S. A., Zahoor M. K., Mustafa G., Rasul A. Screening of novel phytochemicals as secreted frizzled-related protein 4 inhibitors: an early stage biomarker of type 2 diabetes. Pakistan Journal of Pharmaceutical Sciences. 2020;33(3):1245–1250. doi: 10.36721/PJPS.2020.33.3.SUP.1245-1250.1. [DOI] [Google Scholar]
- 2.Duddela S., Nataraj Sekhar P., Padmavati G., Banerjee A. K., Murty U. Probing the structure of human glucose transporter 2 and analysis of protein ligand interactions. Medicinal Chemistry Research. 2010;19(8, article 9234):836–853. doi: 10.1007/s00044-009-9234-4. [DOI] [Google Scholar]
- 3.Pantaleão S. Q., Maltarollo V. G., Araujo S. C., Gertrudes J. C., Honorio K. M. Molecular docking studies and 2D analyses of DPP-4 inhibitors as candidates in the treatment of diabetes. Molecular BioSystems. 2015;11(11):3188–3193. doi: 10.1039/C5MB00493D. [DOI] [PubMed] [Google Scholar]
- 4.Trang Nguyen N. D., le L. T. Targeted proteins for diabetes drug design. Advances in Natural Sciences: Nanoscience and Nanotechnology. 2012;3(1, article 013001) doi: 10.1088/2043-6262/3/1/013001. [DOI] [Google Scholar]
- 5.Röhrborn D., Wronkowitz N., Eckel J. DPP4 in diabetes. Frontiers in Immunology. 2015;6:p. 386. doi: 10.3389/fimmu.2015.00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamers D., Famulla S., Wronkowitz N., et al. Dipeptidyl peptidase 4 is a novel adipokine potentially linking obesity to the metabolic syndrome. Diabetes. 2011;60(7):1917–1925. doi: 10.2337/db10-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deacon C. F. Peptide degradation and the role of DPP-4 inhibitors in the treatment of type 2 diabetes. Peptides. 2018;100:150–157. doi: 10.1016/j.peptides.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 8.Gupta S., Sen U. More than just an enzyme: Dipeptidyl peptidase-4 (DPP-4) and its association with diabetic kidney remodelling. Pharmacological Research. 2019;147, article 104391 doi: 10.1016/j.phrs.2019.104391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobbins R. L., Greenway F. L., Chen L., et al. Selective sodium-dependent glucose transporter 1 inhibitors block glucose absorption and impair glucose-dependent insulinotropic peptide release. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2015;308(11):G946–G954. doi: 10.1152/ajpgi.00286.2014. [DOI] [PubMed] [Google Scholar]
- 10.Vani M., Ritesh Siddhartha Reddy N., Uma Maheswari Devi P. Advances in Computational and Bio-Engineering: Proceeding of the International Conference on Computational and Bio Engineering, 2019, Volume 1. Vol. 15. Springer; 2020. In silico analysis for detection of glucose transport-2 inhibitors from seagrass; pp. 491–498. (Learning and Analytics in Intelligent Systems). [DOI] [Google Scholar]
- 11.Mustafa G., Arif R., Atta A., Sharif S., Jamil A. Bioactive compounds from medicinal plants and their importance in drug discovery in Pakistan. Matrix Science Pharma. 2017;1(1):17–26. doi: 10.26480/msp.01.2017.17.26. [DOI] [Google Scholar]
- 12.Sharif S., Shahid M., Atta A., Abbas M., Mustafa G. Comparative evaluation of antioxidant and DNA protective potentials of fifteen selected medicinal plants native to Pakistan. Oxidation Communications. 2017;40(2):657–674. [Google Scholar]
- 13.Bukhari S. A., Qasim M., Masoud M., Anwar H., Waqas A., Mustafa G. Evaluation of medicinally important constituents of Cotoneaster afghanicus G. Klotz collected from baluchistan region of Pakistan. Indian Journal of Pharmaceutical Sciences. 2019;81(2):259–265. doi: 10.4172/pharmaceutical-sciences.1000506. [DOI] [Google Scholar]
- 14.Mojica L., Berhow M., Gonzalez de Mejia E. Black bean anthocyanin-rich extracts as food colorants: physicochemical stability and antidiabetes potential. Food Chemistry. 2017;229:628–639. doi: 10.1016/j.foodchem.2017.02.124. [DOI] [PubMed] [Google Scholar]
- 15.Mushtaq A., Ansari T. M., Mustafa G., Shad M. A., Cruz-Reyes J., Jamil A. Isolation and characterization of nprB, a novel protease from Streptomyces thermovulgaris. Pakistan Journal of Pharmaceutical Sciences. 2020;33(5):2361–2369. doi: 10.36721/PJPS.2020.33.5.SUP.2361-2369.1. [DOI] [PubMed] [Google Scholar]
- 16.Webb B., Sali A. Comparative protein structure modeling using MODELLER. Current protocols in bioinformatics. 2016;54(1) doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bailey T. L., Boden M., Buske F. A., et al. MEME SUITE: tools for motif discovery and searching. Nucleic acids research. 2009;37(Web Server):W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mushtaq A., Mustafa G., Ansari T. M., Shad M. A., Cruz-Reyes J., Jamil A. Antiviral activity of hexapeptides derived from conserved regions of bacterial proteases against HCV NS3 protease. Pakistan Journal of Pharmaceutical Sciences. 2021;34(1):215–223. doi: 10.36721/PJPS.2021.34.1.SUP.217-223.1. [DOI] [PubMed] [Google Scholar]
- 19.Li Z., Wan H., Shi Y., Ouyang P. Personal experience with four kinds of chemical structure drawing software: review on ChemDraw, ChemWindow, ISIS/draw, and ChemSketch. Journal of Chemical Information and Computer Sciences. 2004;44(5):1886–1890. doi: 10.1021/ci049794h. [DOI] [PubMed] [Google Scholar]
- 20.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports. 2017;7(1):1–13. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng F., Li W., Zhou Y., et al. admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. ACS Publications; 2012. [DOI] [PubMed] [Google Scholar]
- 22.Meduru H., Wang Y.-T., Tsai J. J., Chen Y.-C. Finding a potential dipeptidyl peptidase-4 (DPP-4) inhibitor for type-2 diabetes treatment based on molecular docking, pharmacophore generation, and molecular dynamics simulation. International journal of molecular sciences. 2016;17(6):p. 920. doi: 10.3390/ijms17060920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bukhari S. A., Yasmin A., Zahoor M. A., Mustafa G., Sarfraz I., Rasul A. Secreted frizzled-related protein 4 and its implication in obesity and type-2 diabetes. IUBMB Life. 2019;71(11):1701–1710. doi: 10.1002/iub.2123. [DOI] [PubMed] [Google Scholar]
- 24.Kumar M., Verma D. Antidiabetic and antihyperlipidemic effect of Morinda citrofolia and Coccinia indica in alloxan induced diabetic rats. Pharmacology. 2011;2:307–311. [Google Scholar]
- 25.Mustafa G., Ahmed S., Ahmed N., Jamil A. Phytochemical and antibacterial activity of some unexplored medicinal plants of Cholistan desert. Pakistan Journal of Botany. 2016;48(5):2057–2062. [Google Scholar]
- 26.Mustafa G., Majid M., Ghaffar A., Yameen M., Samad H. A., Mahrosh H. S. Screening and molecular docking of selected phytochemicals against NS5B polymerase of hepatitis C virus. Pakistan Journal of Pharmaceutical Sciences. 2020;33(5):2317–2322. doi: 10.36721/PJPS.2020.33.5.SUP.2317-2322.1. [DOI] [PubMed] [Google Scholar]
- 27.Kerru N., Singh-Pillay A., Awolade P., Singh P. Current anti-diabetic agents and their molecular targets: a review. European Journal of Medicinal Chemistry. 2018;152:436–488. doi: 10.1016/j.ejmech.2018.04.061. [DOI] [PubMed] [Google Scholar]
- 28.Balamurugan R., Stalin A., Ignacimuthu S. Molecular docking of γ-sitosterol with some targets related to diabetes. European Journal of Medicinal Chemistry. 2012;47(1):38–43. doi: 10.1016/j.ejmech.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 29.Shen W., Lu Y. H. Molecular docking of citrus flavonoids with some targets related to diabetes. Bangladesh Journal of Pharmacology. 2013;8(2):156–170. doi: 10.3329/bjp.v8i2.14240. [DOI] [Google Scholar]
- 30.Kaushik P., Lal Khokra S., Rana A., Kaushik D. Pharmacophore modeling and molecular docking studies on Pinus roxburghii as a target for diabetes mellitus. Advances in bioinformatics. 2014;2014:8. doi: 10.1155/2014/903246.903246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harada N., Inagaki N. Role of sodium-glucose transporters in glucose uptake of the intestine and kidney. Journal of diabetes investigation. 2012;3(4):352–353. doi: 10.1111/j.2040-1124.2012.00227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng R., Dong L., Wang L., Xu Y., Lu H., Zhang J. Development of sodium glucose co-transporter 2 (SGLT2) inhibitors with novel structure by molecular docking and dynamics simulation. Journal of Molecular Modeling. 2019;25(6, article 4067):1–11. doi: 10.1007/s00894-019-4067-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


