SUMMARY:
Texture analysis describes a variety of image-analysis techniques that quantify the variation in surface intensity or patterns, including some that are imperceptible to the human visual system. Texture analysis may be particularly well-suited for lesion segmentation and characterization and for the longitudinal monitoring of disease or recovery. We begin this review by outlining the general procedure for performing texture analysis, identifying some potential pitfalls and strategies for avoiding them. We then provide an overview of some intriguing neuro-MR imaging applications of texture analysis, particularly in the characterization of brain tumors, prediction of seizures in epilepsy, and a host of applications to MS.
Conventionally, radiologists produce diagnoses on the basis of a combination of their training, experience, and individual judgment. Radiologists perceive and recognize image patterns and associate or infer a diagnosis consistent with those patterns. It follows that there will be an inevitable degree of variability in image interpretation as long as it relies primarily on human visual perception.1,2 Tools for automated pattern recognition and image analysis can provide objective information to support clinical decision-making and may serve to reduce this variability. Texture analysis, for example, describes a wide range of techniques that enable quantification of the gray-level patterns, pixel interrelationships, and the spectral properties of an image. An inherent property of all surfaces, “texture” provides us with a vocabulary to describe the variation in surface intensity or patterns, including some that are imperceptible to the human visual system. Texture analysis was initially developed for the assessment of aerial photographs,3,4 with the first reported applications to medical image interpretation appearing shortly thereafter.5–7 Although initially slow to build clinical interest, there has been a sort of “texture renaissance” during the past decade, coincident with the steep increase in computational and digital storage capability, as well as a growing comfort with (and demand for) automatic or semiautomatic image analysis tools.8
The emerging hypothesis is that by examining the nature of gray-level transitions in medical images, we can extract a subset of textural features, or what Tourassi8 referred to as a “texture signature,” that will best characterize the pathology or disease process of interest. Texture analysis has already demonstrated considerable potential in neuro-MR imaging as an objective strategy for lesion segmentation and characterization, particularly in cases in which the lesions are inseparable on the basis of standard T1, T2, proton-attenuation, or diffusion-weighted imaging, as will be discussed in this review. Additionally, texture may assist in scenarios in which robustness is particularly desirable, such as in monitoring disease progression or in the longitudinal evaluation of emerging therapies. We begin this review by providing a recipe for performing texture analysis, followed by a brief survey of its applications to neuro-MR imaging.
Strategies for Texture Analysis
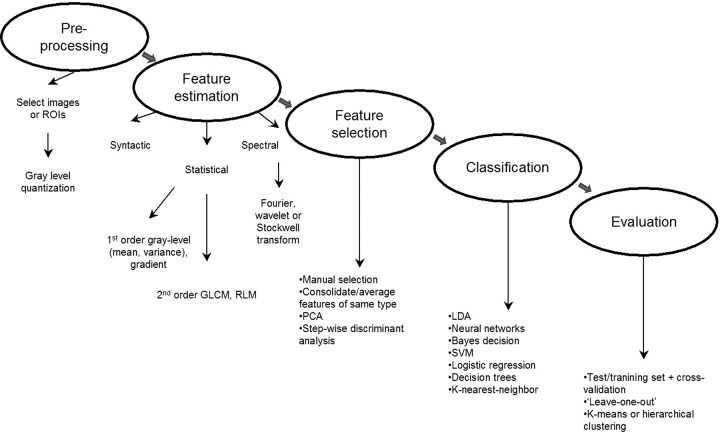
Texture analysis is tremendously versatile and can be applied to virtually any digital image. If the spatial extent of the lesion can be identified by an independent means, then the application of texture analysis can be restricted to a set of predefined regions of interest (eg, lesion versus “normal” or contralateral reference). In the selection of image region of interest or image size, the investigator or analyst will have to balance the need to capture sufficient textural information for classification purposes with the desire to avoid including objects that span multiple tissue categories.9 The flowchart provided in Fig 1 outlines the general strategy for implementing texture analysis, regardless of whether regions of interest or whole images are selected.
Fig 1.
Overview of the steps involved in texture analysis of medical images.
Feature Estimation
Numerous approaches to the quantification and characterization of image texture have been proposed, with most textural features falling under 3 general categories: syntactic, statistical, and spectral. Syntactic texture analysis identifies fundamental or “primitive” elements of the image, which are then linked through syntax.10 Although it appears to show potential for brain surface mapping and volumetry,11 to the best of our knowledge, there have been very few reported applications of syntactic texture analysis to neuro-MR imaging, so we will focus primarily on statistical and spectral approaches. Fortunately, virtually all of the features discussed herein can be evaluated with freely accessible software or programming tools, (eg, the OpenGL tool MaZda; Piotr M. Szczypiński, Institute of Electronics, Technical University of Lodz, Lodz, Poland).
Statistical Features.
Formulas for all of the statistical features discussed herein are provided in the Appendix. Readers will immediately recognize the first-order statistical features (On-line Table 1, Appendix),9 particularly MGL and VGL, because these are routinely quantified directly on the console and are self-explanatory. The remaining 3 first-order features refer to gradient parameters that characterize local gray-level differences.
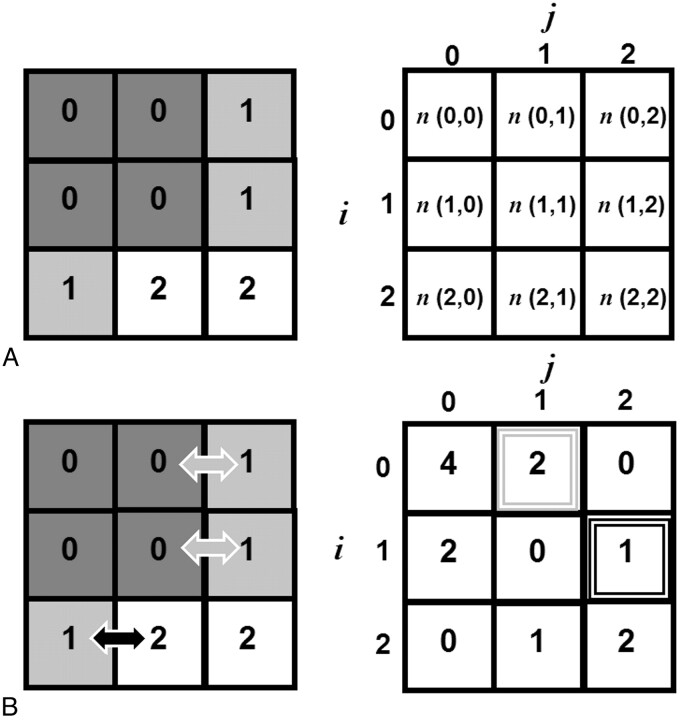
The second-order statistical features are extracted from GLCMs and RLMs, respectively. In brief, a GLCM is constructed by systematically considering the relationship between pixel pairs and tabulating the frequency of various gray-level combinations within an image or within a region of interest (On-line Table 2, Appendix).9 The maximum number of gray levels considered for each image or region of interest is typically scaled down to 32 or 64 levels, rather than using the full dynamic range of, for example, 65,536 gray levels (16 bits/pixel). This quantization step is required to avoid a large number of zero-valued entries in the co-occurrence matrix (ie, sparse matrices).12 Figure 2 provides a schematic guide to the construction of a single GLCM of a hypothetic 3 × 3 pixel image. Given an image f(x,y) with a set of Ng discrete gray levels, the matrix hdθ(i,j) is defined such that entry (i,j) is equal to the number of times that f(x1, y1) = i and f(x2, y2) = j, where (x2, y2) is the sum of (x1, y1) and (dcosθ, dsinθ). A GLCM is computed for each distance, d = 1, …, n pixels in each direction, θ (eg, θ = 0°, 45°, 90°, and 135°). Fourteen textural features can be extracted from each 2D GLCM for each combination of d and θ (On-line Table 2, Appendix).
Fig 2.
A, Construction of a simple GLCM from a stylized 3 × 3 pixel image with 3 gray levels (Ng = 3); distance, d = 1 pixel; and direction, θ = 0° (ie, left or right only). We begin by identifying the reference pixel, with gray-level i, and counting the number of neighboring pixels with gray-level j. B, For example, looking at the second entry in the first row of the GLCM and counting n (0,1) we see 2 pixels with gray level = 0 that have left- or right-handed neighbors with gray level = 1. If we look at the entry for (i,j) =(1,2) we count 1 pixel with gray level = 1, which has a left- or right-handed neighbor with gray level = 2.
In a similar manner, run-length features can be computed to evaluate the coarseness of a texture in a predetermined direction.13 A gray-level run consists of a set of consecutive collinear pixels in a given direction, θ. The RLM is constructed such that Pθ(i,j) is the number of gray-level runs j pixels long for a given gray level, i. The usefulness of run-length features relative to GLCM or even first-order features has been the subject of some debate. Early comparison studies14,15 suggested that run-length features are the least efficient texture features relative to GLCM, gray-level difference features, and features derived from spatial-frequency power spectra. A more recent comparison, however, found that run-length features performed comparably well with those derived from GLCM and were superior to wavelet features for the identification of 16 images from the VisTex texture data base (Massachusetts Institute of Technology, Media Laboratory, Cambridge, Massachusetts).16 Formulae for run-length features are provided in On-line Table 3 of the Appendix.
With the possible exception of features that are extracted from parametric T1 or T2 maps, we would strongly advise that GLCM and RLM features undergo normalization at this stage (eg, dividing each entry in the GLCM or RLM by the total number of matrix entries).17 This step will facilitate interscan comparisons, not only between subjects but also for longitudinal evaluations of the same subjects.
Spectral Features.
Co-occurrence or run-length features may lack the sensitivity to identify larger scale or more coarse changes in spatial frequency.18 Wavelet functions, for example, can be designed to evaluate spatial frequencies at multiple scales and have found a natural application to texture analysis.19 Readers will recognize the close relative of the wavelet tranform, the Fourier transform, which can identify the spatial frequencies present in a signal intensity but cannot delineate temporal changes in frequency content and presumes that all signals reflect a superposition of sinusoids. Some time localization can be imparted to Fourier analysis by means of the windowed or “short-time” method, which allows for the Fourier transform to be performed on sequential portions of the entire signal intensity, each of a set length or “window.”20
The wavelet transform provides even more flexibility by enabling us to trade some degree of spatial-frequency resolution for the ability to localize this frequency content in time. An intuitive way to think of the wavelet transform is to imagine a window of constant area but fluid dimensions, its width (time) narrowing with increasing length (spatial frequency) and vice versa, making it equally suitable for identifying fine texture (short bursts of high spatial frequency) and coarse texture (slow waves of low-frequency content). Unlike the Fourier transform, the wavelet transform does not require sinusoidal basis functions. In fact, “wavelet” refers to the generalized basis function used to compute the transform, which involves scaling and translating a core or “mother” wavelet function and comparing each resultant “daughter” wavelet with the signal intensity and computing a coefficient reflecting the strength of this similarity (wavelet coefficients).21 As such, the wavelet transform is an inherently multiscale analysis method. Due to the computational complexity of the continuous wavelet transform, however, there is a substantial disincentive for implementing this type of analysis in the clinical setting. Others have proposed more parsimonious approaches such as the discrete orthonormal Stockwell transform, which can be computed in a clinically realistic timeframe.22
Feature Selection and Extraction: Avoiding “Fishing Expeditions”
The next priority is to select the subset of features most likely to distinguish 1 tissue class or patient diagnosis from another. The challenge here is that even a modest GLCM approach with 3 d and 4 θ values can produce many more textural features (df) than are suitable for the number of positive cases that will be ultimately subjected to classification. If measures are not taken to reduce the number of features before classification, then the statistical model will better reflect the noise or random error than the underlying data (so-called “overfitting”).23 Fortunately, there are a number of strategies available for dimensionality reduction, beginning with simple consolidation or averaging of a given feature over all directions.12 One can also manually select a subset of features a priori, particularly if previous work has convincingly isolated the features most relevant to the hypothesis being tested. Scatterplots and linear regression can be used to evaluate correlations between pairs of textural features and, therefore, identify potentially redundant features.17 More systematic techniques can be implemented to search for feature subsets, such as those based on the Fisher criterion24 or by means of PCA, which we have previously used to extract mutually orthogonal features from the larger consolidated feature space (or from the entire original feature space).25
Feature Classification and Evaluation
The classification of textural features is analogous to a radiologist's clinical interpretation and involves partitioning the streamlined feature space according to tissue class or diagnostic category. Classification is typically accomplished by using a decision or discriminant function. Provided that the feature space is multivariate normal, supervised classification can be performed by using the Bayesian decision theory or linear discriminant analysis with a distance classifier (often the Mahalanobis distance26). Supervised classification of nonparametric data can be accomplished by using decision trees, k-nearest neighbor,27 support vector machines,28 or neural network29,30 techniques. The accuracy or success of our feature classification strategy is generally evaluated by cross-validation, beginning with dividing the data into training and testing subsets, performing the classification on the training set, and then validating the results of the classification on the testing set. If we then re-partition the original cases and repeat the procedure, we can ultimately use the average classification accuracy as our overall validation metric. We can also construct a logistic regression model by assigning the most discriminating features as predictors and either tissue class or diagnosis as the outcome measure.31 The classification accuracy is then calculated by measuring the area under the ROC curve. We should also note that unsupervised classification techniques including K-means or hierarchical clustering32 are suitable for scenarios in which there is no prior knowledge of how the feature space is organized; therefore, all cases belong to a single testing set.
The Potential Clinical Role for Texture Analysis
Textural features may complement the macrotexture information already used by radiologists, such as the organization of lesions within normal brain parenchyma. In the following section, we will attempt to illustrate how texture analysis has been applied to the diagnosis and characterization of brain tumors, epilepsy, and MS.
Brain Tumors
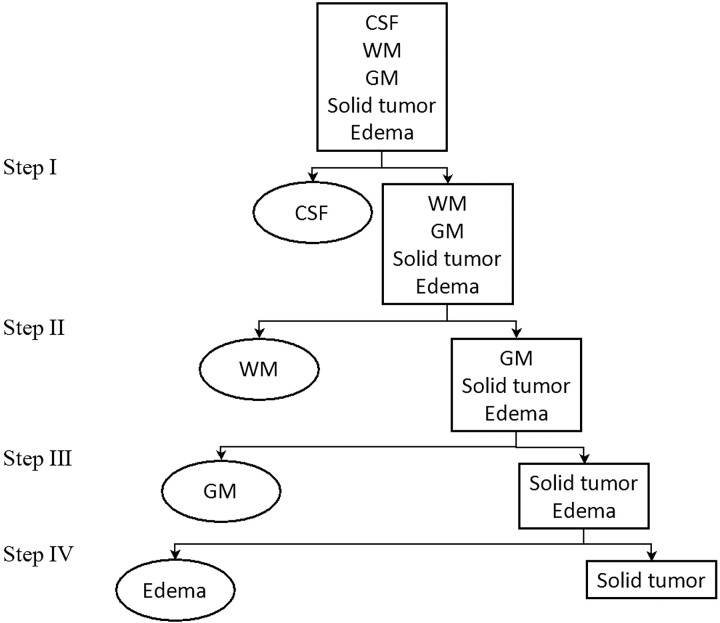
One of the earliest neuro-MR imaging applications of statistical texture analysis was the characterization of brain tumors.17,33 This remains a particularly important problem because there can be substantial intersection between the T1 and T2 of benign and malignant brain tumors,34–36 which complicates lesion characterization with conventional MR imaging. Lerski et al17 were the first to report an MR imaging texture analysis procedure for identifying tumor constituents in their 1993 feasibility study of 12 patients with intracranial tumors. The authors combined first-order histogram, gradient, and second-order GLCM features (f1, f2, f3, and f9) as extracted from T1- and T2-weighted spin-echo MR images into a 4- layer hierarchical decision tree with stepwise discriminant analysis applied at each level to identify the features most capable of discriminating between and among tissue and tumor constituents (ie, CSF, WM, GM, solid tumor, and edema). The results of the 4-layer classification tree are depicted in Fig 3.
Fig 3.
Schematic illustration of the 4-layer hierarchical decision tree used by Lerski et al,17 in which first- and second-order statistical features extracted from T1- and T2-weighted spin-echo MR images are used to delineate CSF, WM, GM, solid tumor, and edema. Stepwise discriminant analysis is applied at each of the 4 levels: At step I, the analysis reveals that the MGL of T2-weighted images is best for discriminating CSF and the rest of the regions. At step II, the MGL of T1-weighted and the f9 of T2-weighted images are best for discriminating WM and the rest of the non-CSF regions; at step III, the MGL of both T2- and proton-attenuation-weighted images and the f9 of T1-weighted images are best for discriminating GM and the combined edema and tumor regions. At step IV, the MGR of T2-weighted images and the f3 of T1-weighted images and the run-length distance of proton-attenuation images are best for performing the final decision, that which discriminates between edema and tumor regions.
Mahmoud-Ghoneim et al37 were able to further stratify tumor constituents, identifying a separate texture class corresponding to peritumoral WM. The authors discovered that the specificity of GLCM features for the identification of tumor constituents could be improved by extending the evaluation of in-plane 2D interpixel relationships to include through-plane 3D relationships (ie, in a multiple-section volume). Compared with their 2D equivalents, the 6 GLCM features extracted from 3D noncontrast T1-weighted gradient-echo images were superior for discriminating between necrosis and solid tumor (sensitivity and specificity were each 100% for 3D versus 75% and 60% for 2D) and between solid tumor and edema (sensitivity and specificity were each 82% for 3D versus 60% and 55% for 2D).
The objectivity of texture analysis depends on the assumption that images are acquired, processed, and analyzed under identical conditions. Herlidou-Même et al35 acquired standard T1- and T2-weighted images from 10 healthy volunteers and 63 patients with confirmed intracranial tumors and data acquired by using 3 different scanners to test the robustness of the technique. The authors were able to use the same statistical textural features to segment the tumors, irrespective of the acquisition parameters, scanner, reconstruction, or processing used, and reported highly reproducible results in a head-to-head comparison of second-order features computed from T1- and T2-weighted images of gliomas acquired with 3 different MR imaging scanners.
With respect to multiscale or spectral features, Brown et al38 have suggested that the Stockwell transform is sensitive enough to distinguish among tumor genotypes, such as in the identification of oligodendrogliomas with a genetic signature associated with good outcomes. The spectral analysis of T2-weighted MR images with the Stockwell transform was more accurate than visual assessment, with a sensitivity and specificity of 93% and 96%, respectively, for delineating these particular tumor genotypes (compared with 70% and 63% for visual assessment). The authors cautioned that these results, while impressive, were obtained in a retrospective analysis and the technique will require further validation before clinical implementation. In a similar manner, Stockwell transform analyses of T2-weighted images have shown potential for identifying the patients with high-grade gliomas (glioblastoma multiforme) most amenable to therapy.39 Specifically, Drabycz et al39 compared visual assessment with the analysis of spatial frequency with respect to their capacity to identify the methylation status of a gene promoter associated with good chemotherapeutic outcomes. After examination of the Stockwell spectra for T2, FLAIR, and postcontrast T1 images, the authors found that the spectral features extracted from T2-weighted images were more specific markers of methylation status than visual identification of ring-enhancement (65% versus 39%) but at a loss of sensitivity (79% versus 93% for spectral and visual textural features, respectively).
Epilepsy
Apart from excluding tumors or other lesions that could precipitate seizures, the objectives in epilepsy imaging include the identification of cortical dysplasia and, particularly in the case of patients with temporal lobe epilepsy, detecting evidence of hippocampal sclerosis. Both of these objectives demand high-resolution anatomic images (usually 3D T1-weighted gradient-echo) that are capable of good GM delineation.40 For example, high-resolution cortical images are needed to investigate focal disruption of the cortical lamination, increased cortical thickness,41 among other abnormalities that can pose a serious risk for seizures.42 Bernasconi et al43 developed a strategy for identifying FCD with a combination of morphologic and texture analysis of 3D T1-weighted MR images (16 FCD and 20 healthy controls). The authors produced 3 types of feature maps: 1) a GM thickening map, 2) a gradient map to identify the GM/WM interface, and 3) a relative signal-intensity map. Ratio maps, reflecting information from all 3 maps were then constructed by using the following formula: [(GM Thickness × Relative Signal Intensity)/Gradients]. ROC analysis revealed that the ratio maps were superior to visual assessment, with 87.5% sensitivity and 95% specificity for identifying FCD. For comparison, the conventional visual assessment of MR images achieved 50% sensitivity and 100% specificity. This work was later extended by introducing 2 additional tissue classes: the GM/WM transition class and the GM/CSF class.44
In addition to the morphometric and first-order features used in Bernasconi's work, Antel et al44 also calculated 3D GLCM features. The authors acquired T1-weighted MR images from 18 patients with FCD and 14 healthy controls. Voxels were initially classified as “lesion” by a Bayesian classifier that was based on morphometric and first-order features. Lesion voxels were then subjected to a GLCM-trained classifier constructed by using the Fisher discriminant ratio. The 2 classifiers ultimately selected 3 features (f1, f2, and f11) from among the 9 features evaluated. Although both visual analysis and Antel's twin classifier had perfect specificity, the latter also had a sensitivity of 83% for the identification of FCD lesions compared with 61% when performed by using the standard visual assessment of MR images.
As indicated earlier, the other objective of the MR imaging protocol for epilepsy is to obtain high-resolution images of the hippocampi and the temporal lobes.40 Hippocampal sclerosis is a common finding in temporal lobe epilepsy.45 Identification of hippocampal abnormalities is of substantial interest, however, particularly with respect to surgical planning. Yu et al45 evaluated >200 texture features extracted from proton-attenuation and T2-weighted images of the hippocampi of 23 patients with unilateral temporal lobe epilepsy and in 9 healthy controls. The correct identification of hippocampal sclerosis with proton-attenuation MR images was achieved by using f9 and f3, whereas a total of 9 T2-weighted MR image features were able to correctly classify the hippocampi. In a subsequent study, Sankar et al46 discovered decreased gradient and f9 features in patients with epilepsy, connoting a blurring of the temporopolar WM (65% sensitivity and 100% specificity for hippocampal atrophy versus 17% and 69% with visual assessment).
Using histologic evidence of hippocampal sclerosis as the ground truth, Bonilha et al47 evaluated several statistical textural features extracted from T1-weighted MR images acquired from 19 patients with mesial temporal lobe epilepsy. Multivariate analysis of variance isolated 8 potentially discriminating features that were subsequently submitted to post hoc analysis. Almost all 8 were capable of discriminating between sclerotic and healthy hippocampi. Furthermore, the authors reported no difference between ipsilateral and contralateral hemispheres in the patients with epilepsy. Although we would not recommend discarding features prematurely or arbitrarily, it is clear that the authors could have benefited from further data-reduction techniques (and they acknowledge this limitation). In this context, we would recommend the approach reported by Freeborough and Fox48 in their MR imaging texture study of patients with Alzheimer disease. Their initial set of features was also quite large (n = 260), so the authors first reduced the dimensionality of their feature space by averaging features for all 4 directions. Then, their consolidated features were subjected to stepwise discriminant analysis by using the forward selection method based on the Mahalanobis distance between controls and cases in the training set and iterated until none of remaining features significantly increased the decision distance.
MS
MS is a chronic inflammatory disease of the central nervous system that results in demyelination, destruction of oligodendrocytes, and, eventually, long-term functional impairment. MS is a dynamic process with approximately 30%–40% of RRMS deteriorating to SPMS—a phase marked by a continuous clinical worsening during a minimum of 6 months.49 While the inflammation and resultant focal blood-brain barrier disruption associated with RRMS is readily identified by using gadolinium-enhanced T1-weighted MR imaging, the value of these conventional MR imaging markers in the monitoring of SPMS is far less obvious.50,51
Texture analysis has been proposed as an alternative strategy for identifying active MS lesions and monitoring disease progression. For example, Yu et al52 discovered that texture analysis of standard T2-weighted MR images could discriminate between active and nonactive lesions in a study of 8 patients with RRMS (4 with active lesions), suggesting that this technique could be used to minimize or perhaps even obviate gadolinium-based contrast. Specifically, the authors evaluated 42 first- and second-order statistical textural features and performed LDA to classify lesions into active and nonactive groups. Applications of the run-length method have been very limited compared with other methods, yet Yu et al found that RLM features actually outperformed GLCM features in the identification of active MS lesions, with RLM features distinguishing active from inactive lesions with 88% sensitivity and 96% specificity. Conversely, none of the GLCM features provided any discrimination between lesion subtypes.
While acute gadolinium enhancement is currently used as a marker of active inflammation or lesion development, chronic MS lesions are conventionally identified and counted by using T2-weighted imaging.53 However, T2 hyperintensity is not particularly specific for MS and appears to be unsuitable for identifying incremental changes in NAWM with time.54 Zhang et al23 investigated >200 texture features extracted from the T2-weighted MR images of 16 patients with RRMS. The authors selected features on the basis of the greatest difference between tissue classes (ie, MS lesions versus normal WM, MS lesions versus NAWM, and normal WM versus NAWM). The 9 features showing the largest differences and those that were common to all 3 tissue class pairs were subjected to PCA followed by classification by using ANN. Although the combined set of features outperformed the authors' GLCM-only classifier for distinguishing MS lesions from normal WM (100% versus 92%), it was relatively unsuccessful at discriminating normal white matter from NAWM (58% versus 67%).
More recent MR imaging studies by using magnetization transfer imaging and, specifically, the MTR can quantify demyelination severity, because MTR is reduced in WM lesions55 with milder decreases also observed in NAWM and GM.56,57 Tozer et al58 estimated GLCM textural features extracted from magnetization transfer MR images acquired from 23 healthy controls, as well as from 32 patients with RRMS, 3 patients with SPMS, and 38 patients with CIS, and investigated the relationship between textural features and the Expanded Disability Status Scale. While the authors found no differences between the features extracted from controls and patients with CIS, several features differed between patients with MS and the 2 other groups, especially in GM (but also in WM). Disability scores also correlated significantly, if modestly, with GLCM textural features extracted from GM regions. Texture feature abnormalities in MS suggested there might be tissue damage beyond classic WM lesions and that these features show potential for quantifying the severity of demyelination.
Whereas GLCM and other statistical features appear to be better suited for identifying active lesions, recent work suggests that spectral texture features may offer greater sensitivity for monitoring disease progression and evaluating treatment response. Zhang et al54 recently measured the spatial-frequency content of T2-weighted MR images acquired from 20 patients with RRMS who participated in a multicenter clinical trial of oral glatiramer acetate. Participants in this study were scanned every 2 months for 14 months. Low spatial-frequency content, as assessed by a modified Stockwell transform, increased in active lesions, followed by a recovery to normal. This low or “coarse” spatial-frequency content appeared to increase in chronic lesions, coinciding with (but not necessarily reflecting) the fact that inflammation and demyelination continue to be observed in chronic MS lesions.59 The total low spatial-frequency content appears capable of predicting lesion development in disease-prone (normal-appearing) WM tissue.
New Applications of Texture Analysis and Future Directions
AIS
At present, thrombolysis with rtPA is the only treatment available for AIS, yet its use is limited to patients presenting within 4.5 hours of onset, due to the risk of HT.60 Techniques are clearly needed for the early prediction of hemorrhagic complications. Given the findings reported for MS studies and assuming that the blood-brain barrier disruption before HT is similar in degree to that occurring in acute MS,61 we hypothesized that there would be differences in the complexity and homogeneity of HT-prone stroke infarcts. To this end, we evaluated first-order (MGL, VGL) and 4 2D-GLCM texture features (f1, f2, f3, and f9) as extracted from postcontrast T1-weighted spin-echo images acquired from 34 patients with AIS.25 Contrast and correlation (f2 and f3) were the only 2 features capable of predicting HT and were much more sensitive predictors than conventional visual assessment of postcontrast T1-weighted images. Surprisingly, the addition of visual enhancement to either f2 or f3 did not significantly improve accuracy.
4D Texture Analysis
As previously discussed in the context of brain tumor imaging, the specificity of statistical texture analysis can be improved by extending the evaluation of in-plane pixel interrelationships to include through-plane relationships.37 Woods et al62 recently combined 3D GLCM analysis with DCE imaging of breast lesions. By examining the time-evolution of contrast enhancement, in which time constitutes the fourth dimension, the authors reported impressive differentiation of benign and malignant tumors, which may prove instructive in future neuro-MR imaging applications. Like texture analysis, DCE-MR imaging has shown considerable promise in the characterization of blood-brain barrier integrity in brain tumors,63 MS,64,65 and AIS.66,67 Unlike conventional DCE, however, the approach of Woods et al62 requires no pharmacokinetic modeling. Rather, textural features are computed at each movement of the analysis window through the 4D dataset—that is, first from the left to the right of the patient, second from inferior to superior, third from posterior to anterior and finally, the window is moved from pregadolinium injection to the final acquisition in the DCE dataset.
Advanced Pattern-Recognition Techniques
In most of the studies reviewed, the feature classification step was accomplished by some flavor of LDA. While LDA is a reasonable strategy for addressing this issue, further sophistications in feature classification have emerged from the fields of artificial intelligence and machine learning. In the original 4D texture analysis study, Woods et al62 developed an ANN classifier and trained it to classify the DCE data into benign and malignant tissue classes. Apart from 4D texture, others have developed ANN23,29 as well as probabilistic neural network classifiers68 to isolate the most discriminating textural features. Although neural network approaches initially require a great deal of data for training and validation, once this stage has been completed satisfactorily, these procedures could be implemented in a clinically reasonable timeframe. Others have very recently applied support vector machines to discriminate between and among intracranial tumors, delineating metastases from gliomas and high-grade from low-grade gliomas with high diagnostic accuracy,28,69 but a more detailed discussion of this approach is beyond the scope of this review.
Conclusions
Texture analysis is a potentially valuable and versatile tool in neuro-MR imaging. While we have attempted to emphasize the potential pitfalls, such as statistical overfitting, there are many options for coping with texture datasets. We should also maintain realistic expectations of what is essentially a set of mathematic constructs, and in most applications, the pathophysiologic interpretation of textural features remains an open question. As such, texture analysis will likely play a supportive rather than a comprehensive role in the future of medical image interpretation. In some cases, however, statistical or spectral textural features have outperformed visual assessment in discriminating between or among intracranial tumors, as well as in discerning subtle anatomic changes associated with a high risk of seizures in patients with epilepsy. The robustness of texture analysis makes it particularly attractive for monitoring disease progression or treatment response with time, as demonstrated with MS.
Supplementary Material
Abbreviations
- ABSV
absolute gradient value
- AIS
acute ischemic stroke
- ANN
artificial neural network
- CIS
clinically isolated syndrome
- d
distance
- DCE
dynamic contrast-enhanced
- FCD
focal cortical dysplasia
- FLAIR
fluid-attenuated inversion recovery
- fx
second-order gray-level co-occurrence feature number “x”
- GLCM
gray-level co-occurrence matrix
- GM
gray matter
- HT
hemorrhagic transformation
- LDA
linear discriminant analysis
- MGL
mean gray level
- MGR
mean gradient
- MS
multiple sclerosis
- MTR
magnetization transfer ratio
- NAWM
normal-appearing white matter
- Ng
number of gray levels
- PCA
principal components analysis
- PPMS
primary-progressive MS
- RLM
run-length matrix
- ROC
receiver-operator characteristic
- ROI
region of interest
- RRMS
relapsing-remitting MS
- rtPA
recombinant tissue plasminogen activator
- SPMS
secondary-progressive MS
- SVM
support vector machine
- θ
direction
- VGL
variance of gray levels
- VGR
variance gradient
- WM
white matter
Footnotes
This work was supported by the Canadian Stroke Network, the Canadian Institutes of Health Research, and the Canada Research Chair Program.
Indicates article with supplemental on-line tables.
References
- 1. Anderson RE, Hill RB, Key CR. The sensitivity and specificity of clinical diagnostics during five decades: toward an understanding of necessary fallibility. JAMA 1989;261:1610–17 [PubMed] [Google Scholar]
- 2. Gorovitz S, MacIntyre A. Toward a theory of medical fallibility. In: Engelhardt HT, Jr, Callahan D. eds. The Foundation of Ethics and Its Relationship to Science. Hastings-on-Hudson, New York: The Hastings Center; 1976: 248– 74 [Google Scholar]
- 3. Kaizer H. A quantification of textures on aerial photographs. Technical Note 121, AD 69484. Boston: Boston University Research Laboratory; 1955 [Google Scholar]
- 4. Darling EM, Joseph RD. Pattern recognition from satellite altitudes. IEEE Trans Syst Sci Cybern 1968; 4: 38– 47 [Google Scholar]
- 5. Hall EE, Kruger RP, Dwyer SJ, et al. A survey of preprocessing and feature extraction techniques for radiographic images. IEEE Transactions on Computers 1971;20:1032–44 [Google Scholar]
- 6. Chien YP, Fu KS. Recognition of x-ray picture patterns. IEEE Trans Syst Man Cybern 1974;4:145–56 [Google Scholar]
- 7. Lerski RA, Barnett E, Morley P, et al. Computer analysis of ultrasonic signals in diffuse liver disease. Ultrasound Med Biol 1979;5:341–50 [DOI] [PubMed] [Google Scholar]
- 8. Tourassi GD. Journey toward computer-aided diagnosis: role of image texture analysis. Radiology 1999;213:317–20 [DOI] [PubMed] [Google Scholar]
- 9. Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Tran Syst Man Cybern 1973;3:610–21 [Google Scholar]
- 10. Julesz B, Gilbert EN, Shepp LA. Inability of humans to discriminate between visual textures that agree in second-order statistics: revisited. Perception 1973;2:391–405 [DOI] [PubMed] [Google Scholar]
- 11. Seppä M, Hämäläinen M. Visualizing human brain surface from T1-weighted MR images using texture-mapped triangle meshes. Neuroimage 2005;26:1–12 [DOI] [PubMed] [Google Scholar]
- 12. Mahmoud-Ghoneim D, Alkaabi MK, de Certaines JD, et al. The impact of image dynamic range on texture classification of brain white matter. BMC Med Imaging 2008;8:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Galloway MM. Texture analysis using gray level run lengths. Comp Graph Image Proc 1975;4:172–79 [Google Scholar]
- 14. Weszka JS, Dyer CR, Rosenfeld A. Comparative-study of texture measures for terrain classification. IEEE Trans Syst Man Cybern 1976;6:269–85 [Google Scholar]
- 15. Conners RW, Harlow CA. A theoretical comparison of texture algorithms. IEEE Transactions on Pattern Analysis and Machine Intelligence 1980;2:204–22 [DOI] [PubMed] [Google Scholar]
- 16. Tang X. Texture information in run-length matrices. IEEE Trans Image Proc 1998;7:1602–09 [DOI] [PubMed] [Google Scholar]
- 17. Lerski RA, Straughan K, Schad LR, et al. MR image texture analysis: an approach to tissue characterization. Magn Reson Imaging 1993;11:873–87 [DOI] [PubMed] [Google Scholar]
- 18. Zhu H, Goodyear BG, Lauzon ML, et al. A new local multiscale Fourier analysis for medical imaging. Med Phys 2003;30:1134–41 [DOI] [PubMed] [Google Scholar]
- 19. Mallat SG. A theory for multiresolution signal decomposition: the wavelet representation. EEE Transactions on Pattern Analysis and Machine Intelligence 1989;11:674–93 [Google Scholar]
- 20. Gabor D. Theory of communication. Part 3: frequency compression and expansion. J IEEE 1946;93(part III):445–57 [Google Scholar]
- 21. Daubechies I. Ten Lectures on Wavelets. Philadelphia: SIAM; 1992 [Google Scholar]
- 22. Stockwell RG. A basis for efficient representation of the S-transform. Digital Signal Processing 2007; 17: 371– 93 [Google Scholar]
- 23. Zhang J, Tong L, Wang L, et al. Texture analysis of multiple sclerosis: a comparative study. Magn Reson Imaging 2008;26:1160–66 [DOI] [PubMed] [Google Scholar]
- 24. Mayerhoefer ME, Breitenseher MJ, Kramer J, et al. Texture analysis for tissue discrimination on T1-weighted MR images of the knee joint in a multicenter study: transferability of texture features and comparison of feature selection methods and classifiers. J Magn Reson Imaging 2005;22:674–80 [DOI] [PubMed] [Google Scholar]
- 25. Kassner A, Liu F, Thornhill RE, et al. Prediction of hemorrhagic transformation in acute ischemic stroke using texture analysis of postcontrast T1-weighted MR images. J Magn Reson Imaging 2009;30:933–41 [DOI] [PubMed] [Google Scholar]
- 26. Hsu S-Y. The Mahalanobis classifier with the generalized inverse approach for automated analysis of imagery texture data. Computer Graphics and Image Processing 1979;9:117–34 [Google Scholar]
- 27. Cover TM, Hart PE. Nearest neighbor pattern classification. IEEE Transactions on Information Theory 1967;13:21–27 [Google Scholar]
- 28. Georgiadis P, Cavouras D, Kalatzis I, et al. Enhancing the discrimination accuracy between metastases, gliomas and meningiomas on brain MRI by volumetric textural features and ensemble pattern recognition methods. Magn Reson Imaging 2009;27:120–30 [DOI] [PubMed] [Google Scholar]
- 29. Tzacheva AA, Najarian K, Brockway JP. Breast cancer detection in gadolinium-enhanced MR images by static region descriptors and neural networks. J Magn Reson Imaging 2003;17:337–42 [DOI] [PubMed] [Google Scholar]
- 30. Georgiadis P, Cavouras D, Kalatzis I, et al. Improving brain tumor characterization on MRI by probabilistic neural networks and non-linear transformation of textural features. Comput Methods Programs Biomed 2008;89:24–32 [DOI] [PubMed] [Google Scholar]
- 31. Gibbs P, Turnbull LW. Textural analysis of contrast-enhanced MR images of the breast. Magn Reson Med 2003;50:92–98 [DOI] [PubMed] [Google Scholar]
- 32. Jain AK, Duin RPW. Statistical pattern recognition: a review. IEEE Transactions on Pattern Analysis and Machine Intelligence 2000;22:4–37 [Google Scholar]
- 33. Schad LR, Blüml S, Zuna I. MR tissue characterization of intracranial tumors by means of texture analysis. Magn Reson Imaging 1993;11:889–96 [DOI] [PubMed] [Google Scholar]
- 34. Earnest FT, Kelly PJ, Scheithauer BW, et al. Cerebral astrocytomas: histopathologic correlation of MR and CT contrast enhancement with stereotactic biopsy. Radiology 1988;166:823–27 [DOI] [PubMed] [Google Scholar]
- 35. Herlidou-Même S, Constans JM, Carsin B, et al. MRI texture analysis on texture test objects, normal brain and intracranial tumors. Magn Reson Imaging 2003;21:989–93 [DOI] [PubMed] [Google Scholar]
- 36. Provenzale JM, Mukundan S, Barboriak DP. Diffusion-weighted and perfusion MR imaging for brain tumor characterization and assessment of treatment response. Radiology 2006;239:632–49 [DOI] [PubMed] [Google Scholar]
- 37. Mahmoud-Ghoneim D, Toussaint G, Constans JM, et al. Three dimensional texture analysis in MRI: a preliminary evaluation in gliomas. Magn Reson Imaging 2003;21:983–87 [DOI] [PubMed] [Google Scholar]
- 38. Brown R, Zlatescu M, Sijben A, et al. The use of magnetic resonance imaging to noninvasively detect genetic signatures in oligodendroglioma. Clin Cancer Res 2008;14:2357–62 [DOI] [PubMed] [Google Scholar]
- 39. Drabycz S, Roldán G, de Robles P, et al. An analysis of image texture, tumor location, and MGMT promoter methylation in glioblastoma using magnetic resonance imaging. Neuroimage 2009;49:1398–405.Epub 2009 Sep 28 [DOI] [PubMed] [Google Scholar]
- 40. Mikulis DJ, Roberts TP. Neuro MR: protocols. J Magn Reson Imaging 2007;26:838–47 [DOI] [PubMed] [Google Scholar]
- 41. Barkovich AJ, Kuzniecky RI. Neuroimaging of focal malformations of cortical development. J Clin Neurophysiol 1996;13:481–94 [DOI] [PubMed] [Google Scholar]
- 42. Avoli M, Bernasconi A, Mattia D, et al. Epileptiform discharges in the human dysplastic neocortex: in vitro physiology and pharmacology. Ann Neurol 1999;46:816–26 [DOI] [PubMed] [Google Scholar]
- 43. Bernasconi A, Antel SB, Collins DL, et al. Texture analysis and morphological processing of magnetic resonance imaging assist detection of focal cortical dysplasia in extra-temporal partial epilepsy. Ann Neurol 2001;49:770–75 [PubMed] [Google Scholar]
- 44. Antel SB, Collins DL, Bernasconi N, et al. Automated detection of focal cortical dysplasia lesions using computational models of their MRI characteristics and texture analysis. Neuroimage 2003;19:1748–59 [DOI] [PubMed] [Google Scholar]
- 45. Yu O, Mauss Y, Namer IJ, et al. Existence of contralateral abnormalities revealed by texture analysis in unilateral intractable hippocampal epilepsy. Magn Reson Imaging 2001;19:1305–10 [DOI] [PubMed] [Google Scholar]
- 46. Sankar T, Bernasconi N, Kim H, et al. Temporal lobe epilepsy: differential pattern of damage in temporopolar cortex and white matter. Hum Brain Mapp 2008;29:931–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bonilha L, Kobayashi E, Castellano G, et al. Texture analysis of hippocampal sclerosis. Epilepsia 2003;44:1546–50 [DOI] [PubMed] [Google Scholar]
- 48. Freeborough PA, Fox NC. MR image texture analysis applied to the diagnosis and tracking of Alzheimer's disease. IEEE Trans Med Imaging 1998;17:475–79 [DOI] [PubMed] [Google Scholar]
- 49. Weinshenker BG, Bass B, Rice GPA, et al. The natural history of multiple sclerosis: a geographically based study. I. Clinical course and disability. Brain 1989;112:133–46 [DOI] [PubMed] [Google Scholar]
- 50. Rovaris M, Confavreux C, Furlan R, et al. Secondary progressive multiple sclerosis: current knowledge and future challenges. Lancet Neurol 2006;5:343–54 [DOI] [PubMed] [Google Scholar]
- 51. Wolinsky JS. MRI aspects of secondary progressive multiple sclerosis. Mult Scler 2002;8:85–87 [DOI] [PubMed] [Google Scholar]
- 52. Yu O, Mauss Y, Zollner G, et al. Distinct patterns of active and non-active plaques using texture analysis on brain NMR images in multiple sclerosis patients: preliminary results. Magn Reson Imaging 1999;17:1261–67 [DOI] [PubMed] [Google Scholar]
- 53. Li DK, Paty DW. Magnetic resonance imaging results of the PRISMS trial: a randomized, double-blind, placebo-controlled study of interferon-beta1a in relapsing-remitting multiple sclerosis—Prevention of Relapses and disability by Interferon-beta1a Subcutaneously in Multiple Sclerosis. Ann Neurol 1999;46:197–206 [DOI] [PubMed] [Google Scholar]
- 54. Zhang Y, Zhu H, Mitchell JR, et al. T2 MRI texture analysis is a sensitive measure of tissue injury and recovery resulting from acute inflammatory lesions in multiple sclerosis. Neuroimage 2009;47:107–11 [DOI] [PubMed] [Google Scholar]
- 55. Schmierer K, Scaravilli F, Altmann DR, et al. Magnetization transfer ratio and myelin in postmortem multiple sclerosis brain. Ann Neurol 2004;56:407–15 [DOI] [PubMed] [Google Scholar]
- 56. van Buchem MA, McGowan JC, Kolson DL, et al. Quantitative volumetric magnetization transfer analysis in multiple sclerosis: estimation of macroscopic and microscopic disease burden. Magn Reson Med 1996;36:632–36 [DOI] [PubMed] [Google Scholar]
- 57. Davies GR, Altmann DR, Hadjiprocopis A, et al. Increasing normal-appearing gray and white matter magnetization transfer ratio abnormality in early relapsing-remitting multiple sclerosis. J Neurol 2005;252:1037–44 [DOI] [PubMed] [Google Scholar]
- 58. Tozer DJ, Marongiu G, Swanton JK, et al. Texture analysis of magnetization transfer maps from patients with clinically isolated syndrome and multiple sclerosis. J Magn Reson Imaging 2009;30:506–13 [DOI] [PubMed] [Google Scholar]
- 59. Breij EC, Brink BP, Veerhuis R, et al. Homogeneity of active demyelinating lesions in established multiple sclerosis. Ann Neurol 2008;63:16–25 [DOI] [PubMed] [Google Scholar]
- 60. Hacke W, Kaste M, Bluhmki E, et al. Thrombolysis with alteplase 3 to 4.5 hours after acute ischemic stroke. N Engl J Med 2008;359:1317–29 [DOI] [PubMed] [Google Scholar]
- 61. Waubant E. Biomarkers indicative of blood-brain barrier disruption in multiple sclerosis. Dis Markers 2006;22:235–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Woods BJ, Clymer BD, Kurc T, et al. Malignant-lesion segmentation using 4D co-occurrence texture analysis applied to dynamic contrast-enhanced magnetic resonance breast image data. J Magn Reson Imaging 2007;25:495–501 [DOI] [PubMed] [Google Scholar]
- 63. Roberts H, Roberts T, Brasch R, et al. Quantitative measurement of microvascular permeability in human brain tumors achieved using dynamic contrast-enhanced MR imaging: correlation with histologic grade. AJNR Am J Neuroradiol 2000;21:891–99 [PMC free article] [PubMed] [Google Scholar]
- 64. Tofts P, Kermode A. Measurement of the blood-brain barrier permeability and leakage space using dynamic MR imaging. 1. Fundamental concepts. Magn Reson Med 1991;17:357–67 [DOI] [PubMed] [Google Scholar]
- 65. Larsson H, Stubgaard M, Frederiksen J, et al. Quantitation of blood–brain barrier defect by magnetic resonance imaging and gadolinium-DTPA in patients with multiple sclerosis and brain tumors. Magn Reson Med 1990;16:117–31 [DOI] [PubMed] [Google Scholar]
- 66. Kassner A, Roberts T, Taylor K, et al. Prediction of hemorrhage in acute ischemic stroke using permeability MR imaging. AJNR Am J Neuroradiol 2005;26:2213–17 [PMC free article] [PubMed] [Google Scholar]
- 67. Kassner A, Roberts TPL, Moran B, et al. Recombinant tissue plasminogen activator increases blood-brain barrier disruption in acute ischemic stroke: an MR imaging permeability study. AJNR Am J Neuroradiol 2009;30:1864–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Theocharakis P, Glotsos D, Kalatzis I, et al. Pattern recognition system for the discrimination of multiple sclerosis from cerebral microangiopathy lesions based on texture analysis of magnetic resonance images. Magn Reson Imaging 2009;27:417–22 [DOI] [PubMed] [Google Scholar]
- 69. Zacharaki EI, Wang S, Chawla S, et al. Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn Reson Med 2009;62:1609–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.