Abstract
BACKGROUND AND PURPOSE:
VBM is increasingly used in the study of neurodegeneration, and recently there has been interest in its potential as a biomarker. However, although it is largely “automated,” VBM is rarely implemented consistently across studies, and changing user-specified options can alter the results in a way similar to the very biologic differences under investigation.
MATERIALS AND METHODS:
This work uses data from patients with HD to demonstrate the effects of several user-specified VBM parameters and analyses: type and level of statistical correction, modulation, smoothing kernel size, adjustment for brain size, subgroup analysis, and software version.
RESULTS:
The results demonstrate that changing these options can alter results in a way similar to the biologic differences under investigation.
CONCLUSIONS:
If VBM is to be useful clinically or considered for use as a biomarker, there is a need for greater recognition of these issues and more uniformity in its application for the method to be both reproducible and valid.
VBM1 involves voxel-wise statistical analysis of structural MR images and is commonly used to infer regions in which brain volume differs between groups or regions in which brain volume is associated with another variable. VBM is increasingly used in the study of neurodegeneration and is a complementary approach to region-of-interest methods because it is automated in many parts and can be applied across the whole brain and thus does not require a priori hypotheses about particular regions of interest. Although VBM has mainly been used to understand structural differences and behavioral correlates, there is increasing interest in the potential use of VBM as a biomarker, both diagnostic2 and also in clinical trials of potentially disease-modifying therapies.3,4 However, although automated in many parts, VBM is rarely implemented consistently across studies, and changing user-specified options can alter the results in a way similar to the biologic differences under investigation.
This article aims to illustrate the above problem by using data from patients with HD, a neurodegenerative disease which has been investigated using VBM. We put this into context with a brief review of the literature on HD, highlighting the wide range of different processing options used in published VBM studies to date. We reference the use of VBM in other areas, including Alzheimer disease, and suggest some changes that could be implemented if this technique is to be considered a useful tool in the context of clinical trials.
The aims of the work were the following: 1) to illustrate that all users need to be aware of these caveats when interpreting results and 2) to show that a more uniform approach to VBM is vital if it is to be considered a robust and valid clinical tool and eventually meet criteria for a biomarker.
Materials and Methods
Subjects
Subjects were recruited from the HD clinics at the National Hospital for Neurology and Neurosurgery, London, and at Addenbrooke's Hospital, Cambridge, UK. All had a CAG repeat length of >39 in the HD gene. Subjects were classified as “early HD” (stages 1 and 2)5 or gene carriers without motor signs (ie, “premanifest”). HD gene carriers with UHDRS diagnostic confidence scores <4 were defined as premanifest subjects (n = 21); those with diagnostic confidence scores of 4 were defined as manifest HD (n = 40).6 Neurologically healthy controls were also recruited (n = 20). These were spouses of patients or subjects from affected families who were known not to carry the HD gene. Subjects gave written informed consent, and the study had local research ethics committee and hospital trust approval. As part of a longitudinal study, all subjects underwent annual assessments including MR imaging and clinical and cognitive evaluations. Baseline MR images were used to determine the impact of VBM parameters on results; details of other findings from the study can be found elsewhere.7,8 Demographic details are shown in Table 1.
Table 1:
Demographic dataa
| Control | Premanifest | Early HD | |
|---|---|---|---|
| (n = 20) | (n = 21) | (n = 40) | |
| Gender (M:F) | 7:13 | 10:11 | 20:20 |
| Age (yr) | 44.9 (10.5) | 37.2 (7.9) | 48.5 (9.6) |
| CAG repeat length | NA | 42.2 (1.8), range, 40–45 | 43.7 (2.4), range, 40–50 |
| Predicted years to onsetb | NA | 18.2 (7.1), range, 9–35 | NA |
| Disease duration (yr since onset) | NA | NA | 4.1 (2.6) |
| UHDRS motorc | 1.1 (0.9) | 3.6 (4.0) | 28.9 (12.6) |
| UHDRS independenced | 100 (0) | 100 (0) | 90.4 (9.6) |
| UHDRS TFCe | 13 (0) | 13 (0) | 10.9 (1.8) |
Data are mean (SD) with the exception of gender and handedness.
Onset was defined as a 60% chance of showing motor signs (a greater chance of showing signs than not, as described in Feigin et al30) and was predicted using the equation of Langbehn et al.31
UHDRS motor is out of 124; higher score indicates more severely impaired.
Independence is a percentage; higher score indicates better function.
TFC is out of 13; higher score indicates better function.
Image Acquisition
Subjects underwent T1-weighted volumetric imaging on a 1.5T Signa scanner (GE Healthcare, Milwaukee, Wisconsin) by using an inversion-recovery prepared Fourier acquired steady-state spoiled gradient-recalled acquisition sequence with a 24 × 18 cm FOV and a 256 × 256 matrix providing 124 contiguous 1.5-mm-thick coronal sections (in-plane voxel dimensions: 0.9375 × 0.9375 mm; acquisition parameters: TR = 13 ms; TE = 5.2 ms; flip angle = 13°; TI = 650 ms; receiver bandwidth = 16 kHz, NEX = 1).
VBM Analysis
In general, images were normalized and segmented by using standard procedures from SPM5 software and DARTEL (Wellcome Department of Imaging Neuroscience, London, United Kingdom).9 Unless otherwise stated, GM segments were modulated and smoothed at 4-mm FWHM before analysis. At each stage, all segmentations were inspected visually. The main comparison presented in this work is that of controls versus early HD, so most SPMs show regions in which the early HD group has reduced GM volume relative to controls. It is also useful to consider the reverse contrast (where the HD group has increased GM relative to controls) because unpredicted findings in this direction might be an indication of poor registration. Unless otherwise stated, all comparisons controlled for differences in age and head size by including these as covariates. Detailed methods can be found in the supplementary on-line data.
We recognize that VBM can be implemented through other software packages. We have chosen to use SPM5 and DARTEL because they are the latest versions of a commonly used package, but the issues demonstrated here will apply regardless of software type or version. This work should not be interpreted as advocating the use of a particular software package or version.
Results
Varying the Type and Level of Statistical Correction
One of the benefits of VBM is the fact that it examines the whole brain in an unbiased way, but in doing so, many thousands of statistical tests are performed at once. At a standard α level of 0.05, approximately 5000 voxels in an image of 100 000 voxels would be expected to be false-positives. This is often addressed by controlling the FWE rate (ie, controlling the probability of there being at least 1 false-positive voxel in the entire SPM), though this can lack power and hence omit many true-positives10; some authors opt instead to show uncorrected data. This section investigates how variation in the level and type of correction can impact the resulting SPM.
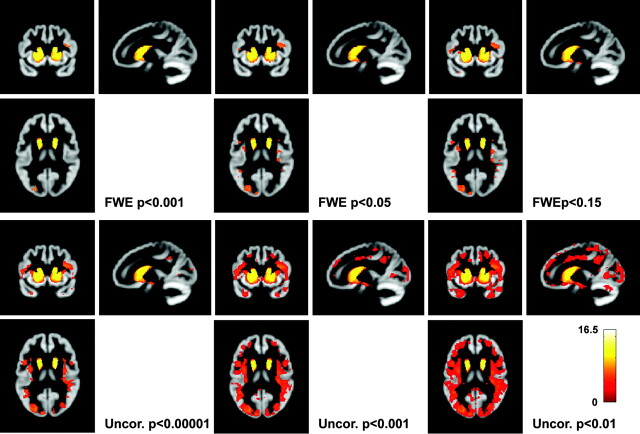
Figure 1 shows regions in which HD subjects have GM loss relative to controls, by using 3 different levels of FWE correction and 3 different levels of voxel-wise correction. At very strict levels, the evidence appears to show atrophy confined to the striatum. At an “exploratory” uncorrected level, most of the GM appears to be involved. Even though the underlying contrast is the same, varying the type and level of correction in this way could mimic the effect of increasing disease stage or the passage of time.
Fig 1.
Effect of type and level of statistical correction. All SPMs show the same contrast: regions in which the early HD group has reduced GM volume relative to controls (this is true throughout the article unless otherwise stated). SPMs are smoothed at 4-mm FWHM. The 3 SPMs in the top panel show various levels of FWE correction, and the 3 SPMs below show various levels of uncorrected SPMs. The color bar shows the t value and is applicable to all figures in this article.
Using Modulated or Unmodulated Data
In the earlier formulations of VBM, normalization aimed to correct for global differences in head position and structure (eg, to align the left superior temporal gyri on all subjects) but not for local differences due to atrophy.1 However in practice, it is likely that normalization results in some atrophy being lost. To correct for this, a modulation step that multiplies the voxel intensity by the Jacobian determinant from the normalization process was introduced.11 The Jacobian determinant is an index of how much a voxel was stretched or contracted during normalization, so modulation, therefore, makes intensity a more accurate representation of volume. With modulated data, one is testing for “regional differences in the absolute amount (volume) of gray matter…,”11 whereas with unmodulated data one is looking at “differences in concentration of gray matter (per unit volume in native space),”1,11 though this is not to be confused with, for example, the histologic attenuation of neurons. More flexible registration methods such as DARTEL intend to recover finer scale differences (eg, due to atrophy), with a greater proportion of the useful information being transferred to the Jacobian, making modulation of greater importance. In the literature, modulation is not always used, but results are often interpreted similarly regardless of whether this step is included. This section investigated how inclusion of the modulation step might affect results.
Figure 2 shows regions of “atrophy” in subjects with HD compared with controls by using both modulated and unmodulated data. With unmodulated data, there is little evidence of putaminal involvement, though both the caudate and insula are shown to be reduced in early HD relative to controls. Using modulated data damage to the insula appears less widespread, while there is much more evidence of caudate and putamen atrophy. The t values are generally higher, indicating that including the information in the Jacobian improves discrimination between the groups.
Fig 2.
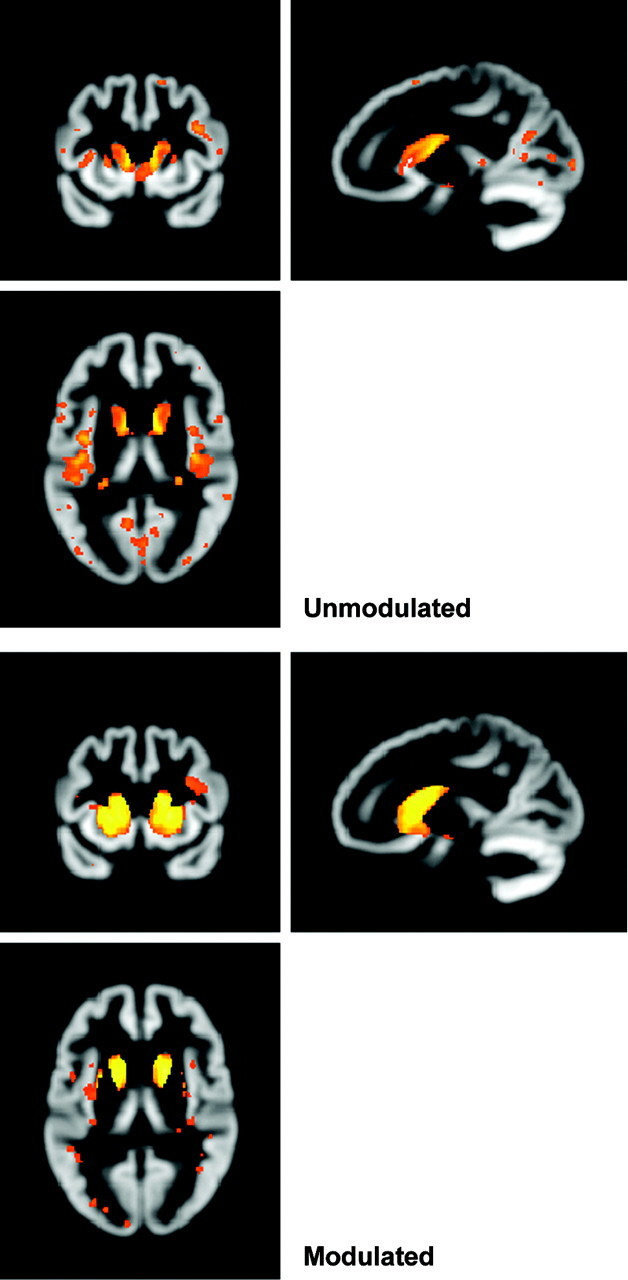
Effect of using modulated or unmodulated data. Both SPMs show the same contrast of early HD versus controls, corrected at FWE P < .05, smoothed at 4-mm FWHM.
Changing the Size of the Smoothing Kernel
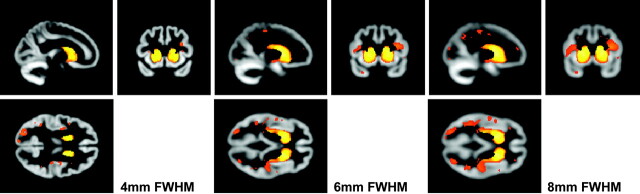
A final preprocessing option is the smoothing kernel. Data are convolved with a 3D Gaussian kernel so that voxel intensities become a weighted average of the surrounding voxels; the size of this kernel is user-defined. Smoothing is required to render the data more normally distributed and to correct for some error in the registration process.1 A range of smoothing kernel sizes has been used in the literature, and this section compares the effect of 3 different smoothing kernels on a single dataset.
Regions in which HD has significantly reduced GM relative to controls are shown in Fig 3 for 3 different smoothing kernels (4-, 6-, and 8-mm). As the kernel size increases, so does the extent of the findings, with, for example, the insula and posterior cortical regions becoming increasingly involved. Elsewhere in the work presented here, a kernel of 4 mm was chosen because the increased accuracy of the DARTEL registration algorithm means that smaller kernels should be sufficient to correct for misalignment.
Fig 3.
Effect of smoothing kernel size. All SPMs show early HD versus controls, corrected at FWE P < .05. The SPMs are smoothed at 4-, 6-, and 8-mm FWHM.
Adjusting for Brain Volume
Apart from the effects of pathology, total brain volume in healthy subjects is known to vary with both head size12 and sex,13 and it has been shown that adjusting whole-brain volume for TIV eliminates differences due to sex.14 It is common for volumetric studies to include an adjustment for some index of head size to ensure that these differences are not influencing findings.15,16
However, few VBM studies of neurodegeneration include an index or measure of TIV as a covariate, though many adjust for total GM volume. In healthy subjects, total GM volume is likely to correlate with TIV, though it will decrease with age.17 If one adjusts for age, covarying for total GM volume approximates an adjustment for TIV and allows investigation of differences in GM volume that are not caused by differences in overall head size. However in subjects with a neurodegenerative disease, total GM volume will almost certainly decrease with the duration or severity of the disease; hence, adjusting for it is likely to mask some disease-related effects (Fig 4). At an extreme level, if degeneration proceeded uniformly throughout the brain, then a comparison between healthy controls and patients that was adjusted for total GM volume would find no evidence of group differences.
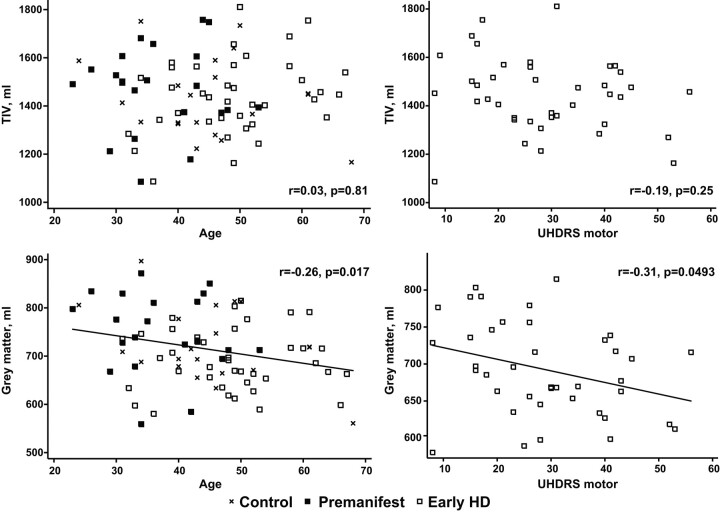
Fig 4.
Graphs demonstrate how TIV and total GM volume vary with age and motor score (an index of HD severity). The top 2 graphs show that the relationship between TIV and both age and motor score is small and not statistically significant. The bottom 2 graphs show that total GM volume decreases with age (r = −0.26, P = .017) and motor score (r = −0.31, P = .0493).
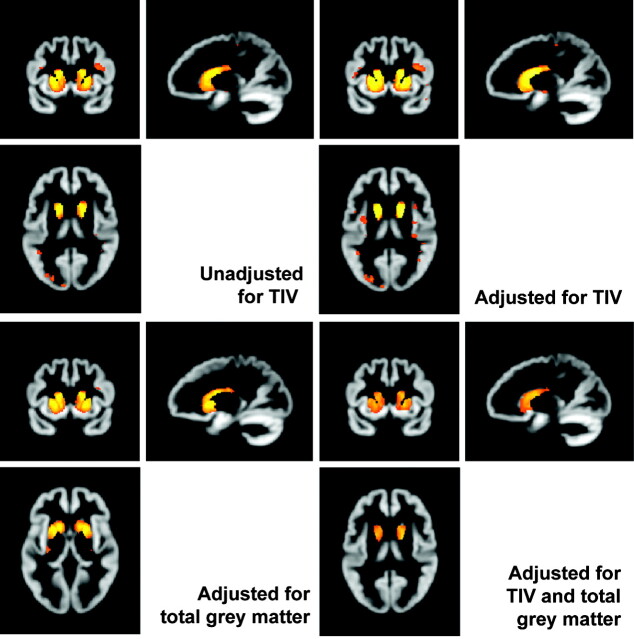
Figure 5 shows the effects of adjusting for TIV and total GM when investigating differences in volume between early HD subjects and controls. In this cohort, there was little effect of adjusting for TIV, though with adjustment, the maximum t value was slightly higher and there was a little more evidence of atrophy in the insula. If one adjusts for GM volume alone, evidence of atrophy outside the striatum almost disappears. When one adjusts for both, there is evidence that striatal atrophy is disproportionately severe (ie, cannot be accounted for by general GM loss or head size).
Fig 5.
Effects of adjusting for TIV with and without including total GM volume. All SPMs show early HD versus controls, corrected at FWE P < .05, smoothed at 4-mm FWHM. The top row shows the effect of including or excluding TIV as a covariate. The bottom row shows the effect of adjusting for total GM volume with and without TIV.
Subgroup Analysis
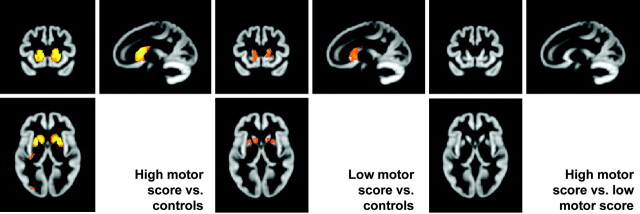
Another common analysis is to use simple regression models to examine the association between a variable of interest and brain volume. While some groups model this as a regression, others chose to compare the outcome of 2 subgroup contrasts (eg, high CAG repeat length versus controls and low CAG repeat length versus controls).18 This section examines a potential pitfall associated with the latter approach by using subgroups of the early HD group (the 12 subjects with the lowest UHDRS motor scores and the 12 subjects with the highest UHDRS motor scores) and a subgroup of 12 controls (Fig 6).
Fig 6.
Subgroup analyses. The left SPM shows regions in which a group of high motor scorers have reduced GM volume relative to matched controls. The center SPM shows regions in which a group of low motor scorers have reduced GM volume relative to controls. The right SPM shows the results when the high and low motor scorer groups are compared directly.
The 2 SPMs showing the contrast of the low motor group and the high motor group with controls show that atrophy in the high motor group is more widespread and perhaps that group differences are larger. However the direct contrast of the low and high motor group shows that there is no evidence that the 2 groups differ from each other (at the same level of statistical correction).
Effect of Software Version and Preprocessing Strategy
Finally, although for consistency all the work in the above sections has been performed by using SPM5 and DARTEL, 2 further points are worth noting. First, these issues will apply to the other software packages available for whole-brain analysis. Second, software package (and version) is a further source of potential variation between studies and, therefore, needs to be taken into account when interpreting and comparing findings. For example, incremental improvements have been made to the SPM software since it was first introduced in the early 1990s. Although a direct comparison of these software versions is beyond the scope of this article, a brief summary of the features relevant to VBM are outlined below. SPM96 had basic 3D spatial normalization by using basis functions and separate tissue segmentation. SPM99 improved the normalization and added MR imaging bias-field correction to the segmentation. This bias-field estimation was enhanced in SPM2, alongside some major changes to the statistical analysis, including restricted maximum likelihood estimation of variance components followed by maximum likelihood (weighted least squares) parameter estimation and the option of controlling the FDR. SPM5 included a unified segmentation approach, which combined the previously separate processes of spatial normalization and tissue classification. In addition, the introduction of DARTEL provided a major advance in the accuracy of spatial alignment of scans. SPM8 (which was released after the completion of our analysis) provides further refinement to the unified segmentation algorithm and a revised FDR procedure.
As our study shows, improvements in normalization accuracy (and consequently smaller smoothing kernels) and statistical inference can have a noticeable impact on resulting SPMs and, therefore, conclusions about the spatial distribution of atrophy. Software version is 1 source of variation that is beyond the user's control because it is to be expected that users will want to work with the latest versions. However, it does need to be acknowledged if this could (partly) explain differences in findings.
An issue closely intertwined with improvements to the registration and segmentation methods available in different software versions is that of modified pipelines for the combination of these steps. For example, Good et al11 introduced an “optimized” procedure involving generation of “custom templates” and tissue probability maps and normalization of segmentations followed by re-segmentation. The unified segmentation of SPM5 provides a more theoretically grounded version of this iteration, while DARTEL allows registration to the group-wise average space instead of standard or custom templates (though it still typically relies on the initial unified segmentation results). Subject groups that are poorly represented by the individuals used to create the standard tissue probability maps (eg, very young or very old) may not be well segmented by the standard procedure. Wilke et al19 propose a method to statistically generate subject-matched tissue probability maps based on a linear model of the variation of tissues in a separate large cohort of subjects.
Discussion
This study demonstrates that methodologic and biologic differences can appear very similar in VBM analyses, and this finding opens up a risk of misinterpretation of results, as well as making it hard to generalize between studies and, hence, be confident of the robustness of findings.
Very different pictures can be obtained by varying the level and type of correction used. Uncorrected results in which group numbers or effect sizes are small and hence would not survive FWE correction are often published, though this might result in a large number of false-positives. Conversely, stringent control of the FWE rate is likely to lead to under-reporting of true effects. As discussed by Poldrack et al,20 the risk of false-positives in uncorrected data depends on the smoothness, complicating the comparison between different sets of uncorrected results. For this reason, we prefer corrected results with a lower threshold and/or the presentation of unthresholded maps.21 This may help emphasize similarities, rather than differences, between studies.
There were differences between findings with modulated and unmodulated data. These need to be interpreted differently because they are not representing the same phenomena. As more precise registration methods are developed, modulation becomes more important to preserve structural differences. In studies of neurodegeneration, the incorporation of a modulation step is the preferred way of ensuring that intersubject alignment preserves intergroup differences in morphology.22
Ashburner and Friston1 stated, “Whenever possible, the size of the smoothing kernel should be comparable to the size of the expected regional differences between the groups of brains.” New methods of image registration such as DARTEL should have a decreased registration error, and the choice of smaller kernels (eg, 4 or 6 mm) may be sufficient. When studying neurodegeneration, greater smoothing tends to increase sensitivity at the expense of specificity and makes it harder to localize an effect anatomically.23 This again means that inconsistencies between studies in which different kernels have been used might not reflect true differences in the cohorts being studied.
Often the lack of a statistically significant difference between groups in so-called “nuisance covariates” (eg, sex or TIV) is wrongly assumed to imply that these variables are not having a material influence on the results. In this cohort, the groups had, on average, similar head sizes, and including TIV as a covariate did not greatly impact the SPM. However, because GM volume is related to TIV, including TIV as a covariate reduces some of the unexplained variance in the data and, hence, may increase the significance of the contrast of interest. In this cohort, this was reflected in the finding of slightly more atrophy at slightly higher t values when TIV was included as a covariate compared with when it was not.
When total GM volume was also included, it was clear that this had a marked effect on the results. Adjusting for total GM volume allows investigation of the relative loss or preservation of regions, compared with the amount of global loss.11,24,25 This is an interesting question in itself but needs careful interpretation. Some studies seem to equate adjustment for total GM volume with adjustment for head size, but in studies of neurodegeneration in particular, adjusting for the former will get rid of some disease-related effects, whereas adjusting for the latter will not, as the current results demonstrate.
The results also demonstrate that while subgroup comparison can yield interesting SPMs, visual comparison of the 2 resulting statistical maps does not constitute a valid statistical comparison in itself. When one compares each group with a relatively homogeneous control group, the SPMs are not identical, but this is not evidence that the groups differ significantly from each other (see guideline “Report Statistical Tests to Support All Claims” in the recent set of guidelines for reporting functional MR imaging studies20).
Table 2 summarizes some of the processing methods and levels of correction used in a number of previously published VBM studies in HD.
Table 2:
Summary of processing methods used by other groupsa
| Study | SPM Version | Normalization | Segmentation | Mod. | Smoothing FWHM (mm) | Correction |
|---|---|---|---|---|---|---|
| Thieben et al32 | 99 | Study-specific GM template, patients and controls | Unspecified | Yes | 10 | SPM uncorrected, p < .005; reported results mostly small-volume-corrected |
| Ho et al33 | 99 | Study-specific GM template, all controls only | Study-specific GM template, all controls only | Yes | 12 | SPM and reported results uncorrected, P < .0001; cluster 10 voxels |
| Kassubek et al18 | 99 | Study-specific template, whole-brain or GM unspecified; subjects unspecified | Unspecified | No | 6 | SPM and reported results FWE P < .001; clusters 54 voxels |
| Kassubek et al34 | 99 | Study-specific template, 50:50 patients:controls; whole-brain or GM unspecified | Study-specific template, 50:50 patients:controls | Yes | 6 | SPM and reported results FWE P < .001 |
| Peinemann et al26 | 99 | Study-specific template, whole-brain or GM only unspecified; subjects unspecified | Unspecified | No | 6 | SPM and reported results FWE P < .05 |
| Douaud et al35 | 2 | Study-specific symmetric GM template, 50:50 patients:controls, from original and mid-plane-reflected images | Study-specific GM template, 50:50 patients:controls, from original and symmetric images | Yes | 8 | SPM and reported results FDR P < .01 |
| Barrios et al27 | Not specified | Standard whole-brain template | Not specified | No | 4 | SPM and reported results uncorrected; P < .01, clusters >10 mm3 |
| Gavazzi et al36 | 2 | Study-specific GM template, subjects unspecified | Study-specific GM template, subjects unspecified | Yes | 10 | SPM and reported results corrected; P < .01 (type unspecified) |
| Jech et al37 | 2 | Study-specific GM template, all patients; no controls in study | Study-specific GM template, all patients; no controls in study | Yes | 10 | SPM uncorrected P < .001; reported results uncorrected in striatum or rolandic area, P < .001, elsewhere, FDR P < .05 |
| Kipps et al38 | 2 | Study-specific template, patients and controls, exact makeup unspecified | Not specified | Yes | 8 | Uncorrected, P < .05 |
| Mühlau et al39 | 2 | Study-specific prior probability maps, subjects and whether used for normalization as well as segmentation unspecified | Yes | 8 | SPM and reported results FWE P <.05, extent P <.05, clusters P < .001 | |
| Mühlau et al25 | 2 | Study-specific prior probability maps, subjects and whether used for normalization as well as segmentation unspecified | Yes | 8 | SPM and reported results FWE P < .05, clusters P < .05 | |
| Ruocco et al40 | 2 | Study-specific GM template, healthy volunteers otherwise unused in study | Study-specific GM template, healthy volunteers otherwise unused in study | Yes | 10 | SPM and reported results FDR P < .05 |
| Wolf et al41 | 2 | Study-specific whole-brain template, 50:50 patients:controls | Study-specific GM templates, 50:50 patients:controls | Yes | 8 | SPM not shown; reported results FWE P < .001 |
| Henley et al7 | 2 | Standard GM template | Standard GM template | Yes | 8 | SPM and reported results small-volume corrected FDR P < .05 |
| Wolf et al42 | 2 | Study-specific whole-brain template, 50:50 patients:controls | Study-specific GM templates, 50:50 patients:controls | Yes | 8 | SPM and reported results FWE P < .001 |
Studies are listed by year and then author.
Within the published VBM studies in HD, there are differences at almost every step. Three of the 17 HD studies that used VBM do not mention modulation18,26,27; hence, the SPMs from these studies may not be showing the same sort of data as the others. The studies cover a wide range of smoothing kernels (from 4- to 12-mm FWHM), which can have a dramatic effect on findings (Fig 3); α levels in these studies range from the conservative 0.001 (controlling the FWE rate) to the more exploratory 0.005 (without correction for multiple comparisons). There is also huge variation between research groups in the covariates they have included in the standard-HD-versus-control comparison: some include age and TIV but some do not. These differences make it hard to interpret the various findings and may mean that results do not generalize to the population as a whole.
Conclusions
The aim of the work presented here was to demonstrate how changes in VBM processing can mimic biologic changes and the potential for misinterpretation that this presents. This can mean that it is hard to generalize findings or to be confident about the robustness of results. This problem is not restricted to VBM or HD, though the methodologic variations in the studies in Table 2 illustrate the difficulties well. In addition, when contradictory results are published, there is a danger that studies are simply repeated; this repetition is a poor use of resources. Image-classification techniques by using VBM-like data have already been used as a diagnostic tool in the early stages of Alzheimer disease2,28 and to measure brain changes in response to antipsychotic treatment in schizophrenia.29 If VBM is to be useful clinically or considered for use as a biomarker, there is a need for more uniformity in its application for the method to be both reproducible and valid.
Supplementary Material
Acknowledgments
We acknowledge Edward Wild, MD, Nicola Hobbs, David MacManus, and Roger Barker, MD, who worked on the London HD study from which these imaging data were obtained. We are also grateful to the patients and controls who took part in that study.
Abbreviations
- CAG
cytosine adenine guanine
- DARTEL
Diffeomorphic Anatomical Registration Through Exponentiated Lie algebra
- EHDN
European Huntington' Disease Network
- FDR
false discovery rate
- FWE
family-wise error
- FWHM
full width at half-maximum
- GM
gray matter
- HD
Huntington disease
- Mod.
modulation
- NA
not applicable
- SPM
statistical parametric mapping/statistical parametric map
- TFC
total functional capacity
- TIV
total intracranial volume
- UHDRS
Unified Huntington Disease Rating Scale
- Uncor.
uncorrected
- VBM
voxel-based morphometry
Appendix
Additional Members of the Euro-HD Imaging Working Group
Stefano Di Donato, Fondazione Istituto Di Ricovero e Cura a Carattere Scientifico, Istituto Neurologico C. Besta, Milan, Italy
Andrea Ginestroni, Radiodiagnostic Section, Department of Clinical Physiopathology, University of Florence, Florence, Italy
Beatriz Gomez-Anson, Clinical Head Neuroradiology and Port d'Informació Científica (Institut de Física d'Altes Energies) Investigator, Hospital Santa Creu i Sant Pau, Autonoma University, Barcelona, Spain
Nicola Z. Hobbs, Dementia Research Centre, University College London Institute of Neurology, London, United Kingdom
Marianne Novak, Wellcome Trust Centre for Neuroimaging, University College London Institute of Neurology, London, United Kingdom
Åsa Petersén, Translational Neuroendocrine Research Unit, Lund University, Lund, Sweden
Carten Saft, Department of Neurology, University of Bochum, St. Josef-Hospital, Bochum, Germany
Edward Wild, Dementia Research Centre, University College London Institute of Neurology, London, United Kingdom
External Member
Hans Johnson, PhD, Department of Psychiatry, The University of Iowa, Iowa City, Iowa
Indicates article with supplemental on-line tables.
S.M.D.H., R.I.S., and S.J.T. were funded by CHDI Inc. N.C.F. and G.R.R. were funded by the Medical Research Council. Part of this work was undertaken at University College London Hospital/University College London, which received a proportion of funding from the National Institute for Health Research Biomedical Research Centres funding scheme of the Department of Health. The Dementia Research Centre is an Alzheimer's Research Trust coordinating center.
Paper previously presented in part at: Annual Meeting of the Euro-HD Imaging Working Group, September 5–6, 2008; Lisbon, Portugal.
References
- 1. Ashburner J, Friston KJ. Voxel-based morphometry: the methods. Neuroimage 2000; 11: 805–21 [DOI] [PubMed] [Google Scholar]
- 2. Hirata Y, Matsuda H, Nemoto K, et al. Voxel-based morphometry to discriminate early Alzheimer's disease from controls. Neurosci Lett 2005; 382: 269–74 [DOI] [PubMed] [Google Scholar]
- 3. Landgrebe M, Binder H, Koller M, et al. Design of a placebo-controlled, randomized study of the efficacy of repetitive transcranial magnetic stimulation for the treatment of chronic tinnitus. BMC Psychiatry 2008; 8: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Teipel SJ, Meindl T, Grinberg L, et al. Novel MRI techniques in the assessment of dementia. Eur J Nucl Med Mol Imaging 2008; 3 (suppl 1): S58–S69 [DOI] [PubMed] [Google Scholar]
- 5. Shoulson I, Fahn S. Huntington disease: clinical care and evaluation. Neurology 1979; 29: 1–3 [DOI] [PubMed] [Google Scholar]
- 6. Unified Huntington's Disease Rating Scale: reliability and consistency—Huntington Study Group. Mov Disord 1996; 11: 136–42 [DOI] [PubMed] [Google Scholar]
- 7. Henley SM, Wild EJ, Hobbs NZ, et al. Defective emotion recognition in early HD is neuropsychologically and anatomically generic. Neuropsychologia 2008; 46: 2152–60. Epub 2008 Mar 6 [DOI] [PubMed] [Google Scholar]
- 8. Henley SM, Wild EJ, Hobbs NZ, et al. Relationship between CAG repeat length and brain volume in premanifest and early Huntington's disease. J Neurol 2009; 256: 203–12. Epub 2009 Mar 5 [DOI] [PubMed] [Google Scholar]
- 9. Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage 2007; 38: 95–113. Epub 2007 Jul 18 [DOI] [PubMed] [Google Scholar]
- 10. Genovese CR, Lazar NA, Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage 2002; 15: 870–78 [DOI] [PubMed] [Google Scholar]
- 11. Good CD, Johnsrude IS, Ashburner J, et al. A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage 2001; 14: 21–36 [DOI] [PubMed] [Google Scholar]
- 12. Acer N, Sahin B, Bas O, et al. Comparison of three methods for the estimation of total intracranial volume: stereologic, planimetric, and anthropometric approaches. Ann Plast Surg 2007; 58: 48–53 [DOI] [PubMed] [Google Scholar]
- 13. Good CD, Johnsrude I, Ashburner J, et al. Cerebral asymmetry and the effects of sex and handedness on brain structure: a voxel-based morphometric analysis of 465 normal adult human brains. Neuroimage 2001; 14: 685–700 [DOI] [PubMed] [Google Scholar]
- 14. Whitwell JL, Crum WR, Watt HC, et al. Normalization of cerebral volumes by use of intracranial volume: implications for longitudinal quantitative MR imaging. AJNR Am J Neuroradiol 2001; 22: 1483–89 [PMC free article] [PubMed] [Google Scholar]
- 15. Kassubek J, Bernhard LG, Ecker D, et al. Global cerebral atrophy in early stages of Huntington's disease: quantitative MRI study. Neuroreport 2004; 15: 363–65 [DOI] [PubMed] [Google Scholar]
- 16. Paulsen JS, Magnotta VA, Mikos AE, et al. Brain structure in preclinical Huntington's disease. Biol Psychiatry 2006; 59: 57–63 [DOI] [PubMed] [Google Scholar]
- 17. Scahill RI, Frost C, Jenkins R, et al. A longitudinal study of brain volume changes in normal aging using serial registered magnetic resonance imaging. Arch Neurol 2003; 60: 989–94 [DOI] [PubMed] [Google Scholar]
- 18. Kassubek J, Juengling FD, Kioschies T, et al. Topography of cerebral atrophy in early Huntington's disease: a voxel based morphometric MRI study. J Neurol Neurosurg Psychiatry 2004; 75: 213–20 [PMC free article] [PubMed] [Google Scholar]
- 19. Wilke M, Holland SK, Altaye M, et al. Template-O-Matic: a toolbox for creating customized pediatric templates. Neuroimage 2008; 41: 903–13. Epub 2008 Mar 8 [DOI] [PubMed] [Google Scholar]
- 20. Poldrack RA, Fletcher PC, Henson RN, et al. Guidelines for reporting an fMRI study. Neuroimage 2007; 40: 409–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ridgway GR, Henley SM, Rohrer JD, et al. Ten simple rules for reporting voxel-based morphometry studies. Neuroimage 2008; 40: 1429–35 [DOI] [PubMed] [Google Scholar]
- 22. Keller SS, Wilke M, Wieshmann UC, et al. Comparison of standard and optimized voxel-based morphometry for analysis of brain changes associated with temporal lobe epilepsy. Neuroimage 2004; 23: 860–68 [DOI] [PubMed] [Google Scholar]
- 23. Reimold M, Slifstein M, Heinz A, et al. Effect of spatial smoothing on t-maps: arguments for going back from t-maps to masked contrast images. J Cereb Blood Flow Metab 2006; 26: 751–59 [DOI] [PubMed] [Google Scholar]
- 24. Mechelli A, Price CJ, Friston KJ, et al. Voxel-based morphometry of the human brain: methods and applications. Current Medical Imaging Reviews 2005; 1: 105–113 [Google Scholar]
- 25. Mühlau M, Weindl A, Wohlschlager AM, et al. Voxel-based morphometry indicates relative preservation of the limbic prefrontal cortex in early Huntington disease. J Neural Transm 2007; 114: 367–72 [DOI] [PubMed] [Google Scholar]
- 26. Peinemann A, Schuller S, Pohl C, et al. Executive dysfunction in early stages of Huntington's disease is associated with striatal and insular atrophy: a neuropsychological and voxel-based morphometric study. J Neurol Sci 2005; 239: 11–19 [DOI] [PubMed] [Google Scholar]
- 27. Barrios FA, Gonzalez L, Favila R, et al. Olfaction and neurodegeneration in HD. Neuroreport 2007; 18: 73–76 [DOI] [PubMed] [Google Scholar]
- 28. Fan Y, Resnick SM, Wu X, et al. Structural and functional biomarkers of prodromal Alzheimer's disease: a high-dimensional pattern classification study. Neuroimage 2008; 41: 277–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McClure RK, Phillips I, Jazayerli R, et al. Regional change in brain morphometry in schizophrenia associated with antipsychotic treatment. Psychiatry Res 2006; 148: 121–32 [DOI] [PubMed] [Google Scholar]
- 30. Feigin A, Ghilardi MF, Huang C, et al. Preclinical Huntington's disease: compensatory brain responses during learning. Ann Neurol 2006; 59: 53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Langbehn DR, Brinkman RR, Falush D, et al. A new model for prediction of the age of onset and penetrance for Huntington's disease based on CAG length. Clin Genet 2004; 65: 267–77 [DOI] [PubMed] [Google Scholar]
- 32. Thieben MJ, Duggins AJ, Good CD, et al. The distribution of structural neuropathology in pre-clinical Huntington's disease. Brain 2002; 125: 1815–28 [DOI] [PubMed] [Google Scholar]
- 33. Ho AK, Nestor PJ, Williams GB, et al. Pseudo-neglect in Huntington's disease correlates with decreased angular gyrus density. Neuroreport 2004; 15: 1061–64 [DOI] [PubMed] [Google Scholar]
- 34. Kassubek J, Juengling FD, Ecker D, et al. Thalamic atrophy in Huntington's disease co-varies with cognitive performance: a morphometric MRI analysis. Cereb Cortex 2005; 15: 846–53 [DOI] [PubMed] [Google Scholar]
- 35. Douaud G, Gaura V, Ribeiro MJ, et al. Distribution of grey matter atrophy in Huntington's disease patients: a combined ROI-based and voxel-based morphometric study. Neuroimage 2006; 32: 1562–75 [DOI] [PubMed] [Google Scholar]
- 36. Gavazzi C, Nave RD, Petralli R, et al. Combining functional and structural brain magnetic resonance imaging in Huntington disease. J Comput Assist Tomogr 2007; 31: 574–80 [DOI] [PubMed] [Google Scholar]
- 37. Jech R, Klempir J, Vymazal J, et al. Variation of selective gray and white matter atrophy in Huntington's disease. Mov Disord 2007; 22: 1783–89 [DOI] [PubMed] [Google Scholar]
- 38. Kipps CM, Duggins AJ, McCusker EA, et al. Disgust and happiness recognition correlate with anteroventral insula and amygdala volume respectively in preclinical Huntington's disease. J Cogn Neurosci 2007; 19: 1206–17 [DOI] [PubMed] [Google Scholar]
- 39. Mühlau M, Gaser C, Wohlschlager AM, et al. Striatal gray matter loss in Huntington's disease is leftward biased. Mov Disord 2007; 22: 1169–73 [DOI] [PubMed] [Google Scholar]
- 40. Ruocco HH, Bonilha L, Li LM, et al. Longitudinal analysis of regional gray matter loss in Huntington disease: effects of the length of the CAG repeat. J Neurol Neurosurg Psychiatry 2007; 79: 130–35 [DOI] [PubMed] [Google Scholar]
- 41. Wolf RC, Vasic N, Schonfeldt-Lecuona C, et al. Dorsolateral prefrontal cortex dysfunction in presymptomatic Huntington's disease: evidence from event-related fMRI. Brain 2007; 130: 2845–57 [DOI] [PubMed] [Google Scholar]
- 42. Wolf RC, Vasic N, Schonfeldt-Lecuona C, et al. Cortical dysfunction in patients with Huntington's disease during working memory performance. Hum Brain Mapp 2009; 30: 327–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.