Abstract
Recently, it was found that proteomes from poliovirus, measles virus, dengue virus, and severe acute respiratory syndrome-related Coronavirus 2 (SARS-CoV-2) have high molecular mimicry at the heptapeptide level with the human proteome, while heptapeptide commonality is minimal or absent with proteomes from nonhuman primates, that is, gorilla, chimpanzee, and rhesus macaque. To acquire more data on the issue, analyses here have been expanded to Ebola virus, Francisella tularensis , human immunodeficiency virus-1 (HIV-1), Toxoplasma gondii , Variola virus, and Yersinia pestis . Results confirm that heptapeptide overlap is high between pathogens and Homo sapiens , but not between pathogens and primates. Data are discussed in light of the possible genetic bases that differently model primate phenomes, thus possibly underlying the zero/low level of molecular mimicry between infectious agents and primates. Notably, this study might help address preclinical vaccine tests that currently utilize primates as animal models, since autoimmune cross-reactions and the consequent adverse events cannot occur in absentia of shared sequences.
Keywords: infectious agents, peptide sharing, molecular mimicry, cross-reactivity, autoimmunity, nonhuman primates, rhesus macaque, vaccines, preclinical test
Introduction
Beginning in 2000, a high, unexpected level of molecular mimicry between microbial and human proteins has been repeatedly documented; accordingly, the consequent potential cross-reactivity following infections or active immunizations has been highlighted. 1 2 3 4 5 6 7 8 9 10 11 Pathologically, cross-reactions between pathogen and human proteins might lead to thrombocytopenia, altered spermatogenesis, schizophrenia and neuropsychiatric diseases, neurodegeneration, lymphomas, sudden death, microcephaly and Guillain-Barré syndrome, pneumonia, multiple sclerosis, immunodeficiency, developmental disorders, autoinflammatory disease, arthritis, hemochromatosis, myasthenia gravis, and systemic lupus erythematosus. 4 8 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26
As a matter of fact, pathogen-derived immunoreactive epitopes are mostly composed of peptide sequences present in human proteins, 10 18 21 23 26 thus documenting that the immune system does not exert any negative selection of self-reactive lymphocytes. 27 28 Hence, it comes as a logical consequence that peptide sharing between infectious antigens and human proteins can cause cross-reactions in the human host, possibly leading to a multitude of postinfection autoimmune pathologies.
However, as recently underlined, 29 30 in general cross-reactivity and the related potential autoimmune sequelae have not been reported in nonhuman primates following experimental infections or during preclinical trials for vaccine validation. 31 Indeed, preclinical trial reports routinely state that active antipathogen immunization is exempt from adverse events in the animal model par excellence, namely, rhesus macaque ( Macaca mulatta ), which is a nonhuman primate phylogenetically close to humans. 32 33 34 35 36
Therefore, it was hypothesized that if the peptide sharing between pathogens and humans is the primum movens of autoimmune pathologies via cross-reactivity, then different levels of peptide sharing with pathogens should characterize the proteomes of humans and primates. De facto , comparative amino acid (aa) sequence analyses documented that poliovirus, measles virus, dengue virus, and SARS-CoV-2 share peptide sequences almost exclusively with the human and murine proteomes, but not with primate proteomes. 29 30 These data might explain the absence of collateral adverse events in primates during preclinical vaccine tests, since autoimmune cross-reactivity cannot occur in primates in absentia of molecular mimicry.
In this research frame, the present study expands comparative sequence analyses to additional pathogens that, although thoroughly investigated, still remain without safe and efficacious therapies, for example, Yersinia pestis 37 and HIV-1. 38 The results further suggest that primates are animal models unsuitable to show adverse autoimmune pathologic cross-reactions in pre-clinical trials following pathogen administration by infection or active immunization, and indicate that only mice represent animal models suitable to test putative vaccine candidates. The genetic bases that might underlie the low level of heptapeptide sharing between infectious agents and nonhuman primates are discussed.
Materials and Methods
Molecular mimicry analyses were conducted by using heptapeptides as immunobiological units. The analyzed pathogen proteins/proteomes are as follows (with NCBI TaxId in parentheses and further details at http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi ): membrane protein/O-antigen protein from Francisella tularensis , 409 aa (177416); apical membrane antigen 1-like protein from Toxoplasma gondii , 651 aa (432359); surface antigen S from variola virus, 354 aa (587200); virulence-associated V antigen from Yersinia pestis , 326 aa (632); proteome from Ebola virus, 5494 aa (128952); proteome from HIV-1, 3134 aa (11676).
The primary aa sequences of the pathogen proteins/proteomes were dissected into heptapeptides offset by one residue, that is, MIRAYEQ, IRAYEQN, RAYEQNP, and so on. Then, each pathogen heptapeptide was analyzed for occurrences within mammalian reference proteomes, that is, proteomes that have been selected because they cover well-studied model organisms and other organisms of interest for biomedical research and phylogeny ( www.uniprot.org/proteomes ). 39 40 41 Specifically, analyses were conducted on proteomes from the following organisms (with NCBI TaxId in parentheses): Homo sapiens (9606); gorilla, Gorilla gorilla gorilla (9595); chimpanzee, Pan troglodytes (9598); and rhesus macaque, Macaca mulatta (9544). In addition, proteomes from the following mammalian organisms were analyzed as controls: cow, Bos taurus (9913); dog, Canis lupus familiaris (9615); cat, Felis catus (9685); rabbit, Oryctolagus cuniculus (9986); mouse, Mus musculus (10090); rat, Rattus norvegicus (10116); pig, Sus scrofa (9823); and bat, Pteropus alecto (9402).
Heptapeptide matches between pathogen proteins/proteomes and mammalian proteomes were searched using Pir Peptide Match program (research.bioinformatics.udel.edu/peptidematch) 40 and UniProt/Swiss-Prot database that is available at www.uniprot.org 39 and consist of reviewed and annotated protein entries. Protein isoforms were not considered.
Results
We analyzed four protein antigens derived from F. tularensis , T, gondii , variola virus, and Y. pestis , respectively, and two pathogen proteomes, namely, Ebola virus proteome, and HIV-1 proteome for heptapeptide sharing with the mammalian proteomes described under Methods. The heptapeptide sharing is quantitatively reported in Fig. 1 and qualitatively illustrated in Supplementary Tables S1–S6 (online only).
Fig. 1.
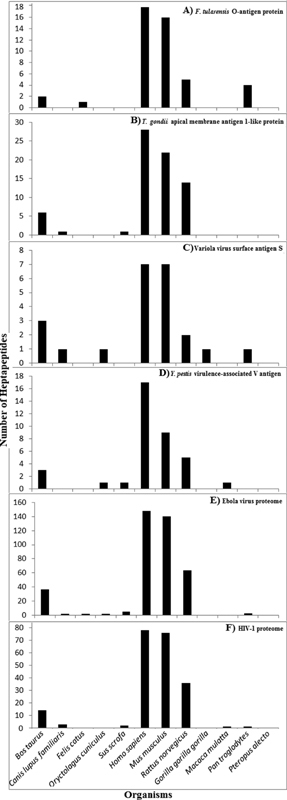
Heptapeptide sharing between mammalian proteomes and: ( A ) F. tularensis membrane protein/O-antigen protein, ( B ) T. gondii apical membrane antigen 1-like protein, ( C ) variola virus surface antigen S, ( D ) Y. pestis virulence-associated V antigen, ( E ) Ebola virus proteome, and ( F ) human immunodeficiency virus (HIV)-1 proteome.
As a preliminary observation, it is noteworthy, as already underscored elsewhere, 8 9 10 that the peptide sharing shown in Fig. 1 is highly improbable from a mathematical point of view. Indeed, the expected number of times that one heptapeptide from a protein will occur simultaneously in a second protein is given by the formula mn /N, where m is the number of heptapeptides present in the first protein, n is the number of heptapeptides present in the second protein, and N is 20 7 = 1,280,000,000, that is, the number of heptapeptides that can be composed using the 20 aa. For values of m and n << N, the probability of sharing only one heptapeptide is 0.00000000078125, that is infinitesimal.
Then, Fig. 1 shows that the peptide sharing is not stochastic, that is, the pathogen-derived heptapeptides are not distributed at random among the analyzed mammalian proteomes. Indeed, all of the analyzed pathogen proteins/proteomes, independently of their being bacterial or viral or protozoan, and independently of their aa length, share heptapeptide sequences almost exclusively with the human, murine, and rat proteomes. Zero or a low number of pathogen-derived heptapeptides are present in the proteomes from cat, dog, cow, pig, rabbit, and the three primates. As a logical consequence, Fig. 1 shows that pathologic cross-reactivity following pathogen infection/immunization might be revealed only by using mice as animal models in preclinical tests.
On the whole, the data exposed in Fig. 1 might explain the differences between humans and primates in the incidence or severity of medical conditions. In fact, communicable and noncommunicable diseases that are common in humans are practically absent or very rare in great apes. 42 43 44 Taking HIV-1 infection as an example, the progression to AIDS—common in humans and rare in great apes 44 —can be explained by the vast peptide sharing between HIV-1 and human proteins that—when altered, mutated, deficient or improperly functioning—associate with AIDS disorders, that is, immunosuppression, neurological disturbances, muscle diseases, malignancies, lipodystrophies, diarrhea, bone loss, corneal alterations, kidney disease, and hypertension, among others, which most possibly associate with molecular mimicry. 45 Instead, cross-reactions and autoimmune pathologies cannot occur in animals that do not share peptides with HIV-1. In these animals, HIV-1 infection/active immunization will be well tolerated with no adverse events as routinely reported in preclinical tests conducted in Rhesus macaques. 46
Thus, Fig. 1F offers a scientific explanation of the numerous HIV vaccine failures. Indeed, as early as 2009, Thomas 47 wrote “ to say that efficacy trials of HIV vaccines and microbicides have, to date, been disappointing is something of an understatement .” Today, in 2020, Thomas' observation is still valid and the promised effective vaccine that had to cure HIV has not yet been found, 48 and most possibly will not be found within the next decades if correct trials and correct animal models are not adopted.
Discussion
Recently, it was shown that heptapeptides from poliovirus, measles virus, dengue virus, and SARS-CoV-2 occur in the human proteome, but not in proteomes from primates and domestic animals. 29 30 The data appeared to be of relevance, since they might indicate that, starting from the very beginning of the mass polio vaccination program in 1962 49 until the current project of mass vaccination against SARS-CoV-2, 35 36 the human population has been vaccinated and revaccinated,, and it is intended to be vaccinated on the basis of protocols that used and use animal models unable to highlight adverse autoimmune pathologic consequences.
The present study confirms such previous data 29 30 and documents that a mathematically unexpected high molecular mimicry at the heptapeptide level occurs between high-risk pathogens, namely, F. tularensis , T. gondii , variola virus, Y. pestis , Ebola virus, and HIV-1, and the human proteome. Such high molecular mimicry is practically absent when proteomes from primates, domestic animals, and cattle are analyzed. Hence, this study might explain not only the wave of autoimmune diseases that are increasingly burdening the human population worldwide, 50 but also the repeated failures in defining immunotherapies for infectious diseases which pose a risk to public health and primary health care. 51 52 53 54 55 56 57 58 59 60
Also, it has to be considered that the present data underestimate the cross-reactivity potential by two orders of magnitude. Indeed, if one considers that a minimal immune determinant corresponds to five aa residues, 61 62 the extent of the peptide overlap of microbial versus human proteins and the consequent potential cross-reactivity risk increase exponentially. Moreover, conformational epitopes have not been considered.
In light of these additional caveats and of the consequent higher cross-reactivity risk, it appears to be mandatory to investigate the molecular mechanisms that underlie the different extents of molecular mimicry between pathogens and mammals. Possible objects of investigation might be, for example, alterations of gene transcription/translation potentially involved in the different shaping of human and primate genomes/phenomes.
In this regard, studies by Puente et al 63 already highlighted important differences in the human and chimpanzee genomes, from deletion of whole genes to small insertion/deletion events or single nucleotide changes that lead to specific gene inactivation. For example, the genes encoding Serine protease 33 (PRSS33/EOS) and Glutathione hydrolase 5 proenzyme (GGTLA1) are absent in chimpanzee, and single nucleotide changes in protease genes such as Inactive caspase-12 (CASP12) lead to functional genes in chimpanzee and pseudogenes in human. 63 Actually, although the nucleotide difference between humans and chimpanzees is surprisingly small with a value suggested to be 1 to 2%, it was reported that 80 percent of proteins are different between humans and chimpanzees. 64
In addition, segmental duplications in the genome and transposable elements are important sources of genetic/phenetic differences between humans and primates. Segmental duplications are blocks of highly homologous duplicated sequences that define hotspots of chromosomal rearrangement and act as mediators of normal variation as well as genomic diseases. 65 Studies of gene family evolution indicate that gene loss and gain are enriched within the primate lineage 66 67 and that recurrent and independent gene-containing duplications occur within the gorilla and chimpanzee, and are absent in the human lineage. 68 In particular, Blekhman et al 69 showed that not only species-specific segmental duplications are enriched with genes that are differentially expressed between species but, in addition, genes that are within species-specific segmental duplications show significantly higher absolute fold difference in expression level between human and chimpanzee compared with genes that are not associated with duplications. In this regard, it is worth mentioning that a large fraction of the KRAB-containing zinc finger (KRAB-ZF) genes—that code the largest family of transcription factors (TFs) in humans—arose from segmental duplications. 70 In primates, KRAB-ZF genes duplicate at a high rate. Due to their function as transcriptional repressors, the generation and rapid divergence of these genes may help to explain some of the transcriptome differences that have been documented between humans and our closest relatives among the apes. 71 72 73
In sum, it is not surprising that primates are not good models for many major human diseases/conditions 42 43 44 and for preclinical vaccine tests. 29 30 Literature data and the present data might explain the inefficacy and the problematics of vaccines, 51 52 53 54 55 56 57 58 59 60 thus inviting researchers and vaccinologists to study, identify, and use the correct animal models capable of revealing potential autoimmune pathogenicity connected to the peptide sharing.
Finally, as a conclusive note, it appears pertinent to recall the basic concept first stated in 2000 1 and then repeatedly illustrated ( 1 2 3 4 5 6 7 8 9 10 11 27 28 74 75 76 77 78 and additional references therein), according to which only pathogen-derived peptides, which are absent in the human proteome, that is, “non-self” peptides, can lead to safe and efficacious immunotherapies.
Funding Statement
Funding None.
Footnotes
Conflict of Interest None declared.
Supplementary Material
References
- 1.Natale C, Giannini T, Lucchese A, Kanduc D. Computer-assisted analysis of molecular mimicry between human papillomavirus 16 E7 oncoprotein and human protein sequences. Immunol Cell Biol. 2000;78(06):580–585. doi: 10.1046/j.1440-1711.2000.00949.x. [DOI] [PubMed] [Google Scholar]
- 2.Kanduc D, Stufano A, Lucchese G, Kusalik A. Massive peptide sharing between viral and human proteomes. Peptides. 2008;29(10):1755–1766. doi: 10.1016/j.peptides.2008.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trost B, Lucchese G, Stufano A, Bickis M, Kusalik A, Kanduc D. No human protein is exempt from bacterial motifs, not even one. Self Nonself. 2010;1(04):328–334. doi: 10.4161/self.1.4.13315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lucchese G, Kanduc D. Zika virus and autoimmunity: from microcephaly to Guillain-Barré syndrome, and beyond. Autoimmun Rev. 2016;15(08):801–808. doi: 10.1016/j.autrev.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 5.Kanduc D, Shoenfeld Y. Inter-pathogen peptide sharing and the original antigenic sin: Solving a paradox. Open Immunol J. 2018;8:11–27. [Google Scholar]
- 6.Lucchese G. Confronting JC virus and Homo sapiens biological signatures. Front Biosci. 2013;18:716–724. doi: 10.2741/4133. [DOI] [PubMed] [Google Scholar]
- 7.Kanduc D. HCV: Written in our DNA. Self Nonself. 2011;2(02):108–113. doi: 10.4161/self.2.2.15795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanduc D. Describing the hexapeptide identity platform between the influenza A H5N1 and Homo sapiens proteomes. Biologics. 2010;4:245–261. doi: 10.2147/btt.s12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucchese A, Serpico R, Crincoli V, Shoenfeld Y, Kanduc D. Sequence uniqueness as a molecular signature of HIV-1-derived B-cell epitopes. Int J Immunopathol Pharmacol. 2009;22(03):639–646. doi: 10.1177/039463200902200309. [DOI] [PubMed] [Google Scholar]
- 10.Kanduc D. From anti-SARS-CoV-2 immune responses to COVID-19 via molecular mimicry. Antibodies (Basel) 2020;9(03):E33. doi: 10.3390/antib9030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanduc D. Peptide cross-reactivity: the original sin of vaccines. Front Biosci (Schol Ed) 2012;4:1393–1401. doi: 10.2741/s341. [DOI] [PubMed] [Google Scholar]
- 12.Kanduc D. From influenza infection to anti-ADAMTS13 autoantibodies via cross-reactivity. Infect Int. 2019;7:113–120. [Google Scholar]
- 13.Kanduc D. Describing the potential crossreactome between mumps virus and spermatogenesis-associated proteins. Endocr Metab Immune Disord Drug Targets. 2014;14(03):218–225. doi: 10.2174/1871530314666140715115257. [DOI] [PubMed] [Google Scholar]
- 14.Lucchese G, Capone G, Kanduc D. Peptide sharing between influenza A H1N1 hemagglutinin and human axon guidance proteins. Schizophr Bull. 2014;40(02):362–375. doi: 10.1093/schbul/sbs197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lucchese G. Understanding neuropsychiatric diseases, analyzing the peptide sharing between infectious agents and the language-associated NMDA 2A protein. Front Psychiatry. 2016;7:60. doi: 10.3389/fpsyt.2016.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lucchese G. From toxoplasmosis to schizophrenia via NMDA dysfunction: peptide overlap between Toxoplasma gondii and N-Methyl-d-Aspartate Receptors as a potential mechanistic link . Front Psychiatry. 2017;8:37. doi: 10.3389/fpsyt.2017.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lucchese G, Kanduc D. Single amino acid repeats connect viruses to neurodegeneration. Curr Drug Discov Technol. 2014;11(03):214–219. doi: 10.2174/1570163811666140212112300. [DOI] [PubMed] [Google Scholar]
- 18.Kanduc D, Shoenfeld Y. From anti-EBV immune responses to the EBV diseasome via cross-reactivity. Glob Med Genet. 2020;7(02):51–63. doi: 10.1055/s-0040-1715641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanduc D. Potential cross-reactivity between HPV16 L1 protein and sudden death-associated antigens. J Exp Ther Oncol. 2011;9(02):159–165. [PubMed] [Google Scholar]
- 20.Capone G, Kanduc D. Peptide sharing between Bordetella pertussis proteome and human sudden death proteins: a hypothesis for a causal link. Future Microbiol. 2013;8(08):1039–1048. doi: 10.2217/fmb.13.72. [DOI] [PubMed] [Google Scholar]
- 21.Kanduc D, Shoenfeld Y.Human papillomavirus epitope mimicry and autoimmunity: the molecular truth of peptide sharing Pathobiology 201986(5-6):285–295. [DOI] [PubMed] [Google Scholar]
- 22.Kanduc D. Influenza and sudden unexpected death: the possible role of peptide cross-reactivity. Infect Int. 2019;7:121–131. [Google Scholar]
- 23.Kanduc D, Shoenfeld Y. On the molecular determinants of the SARS-CoV-2 attack. Clin Immunol. 2020;215:108426. doi: 10.1016/j.clim.2020.108426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanduc D. Measles virus hemagglutinin epitopes are potential hotspots for crossreactions with immunodeficiency-related proteins. Future Microbiol. 2015;10(04):503–515. doi: 10.2217/fmb.14.137. [DOI] [PubMed] [Google Scholar]
- 25.Kanduc D, Polito A. From viral infections to autistic neurodevelopmental disorders via cross-reactivity. J Psychiatry Brain Sci. 2018;3:14. [Google Scholar]
- 26.Kanduc D. From hepatitis C virus immunoproteomics to rheumatology via cross-reactivity in one table. Curr Opin Rheumatol. 2019;31(05):488–492. doi: 10.1097/BOR.0000000000000606. [DOI] [PubMed] [Google Scholar]
- 27.Kanduc D. Immunobiology: on the inexistence of a negative selection process. Adv Stud Biol. 2020;12:19–28. [Google Scholar]
- 28.Kanduc D. Hydrophobicity and the physico-chemical basis of immunotolerance. Pathobiology. 2020;87(04):268–276. doi: 10.1159/000508903. [DOI] [PubMed] [Google Scholar]
- 29.Kanduc D, Shoenfeld Y. Molecular mimicry between SARS-CoV-2 spike glycoprotein and mammalian proteomes: implications for the vaccine. Immunol Res. 2020;68(05):310–313. doi: 10.1007/s12026-020-09152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanduc D, Shoenfeld Y. Medical, genomic, and evolutionary aspects of the peptide sharing between pathogens, primates, and humans. Glob Med Genet. 2020;7(02):64–67. doi: 10.1055/s-0040-1716334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abee C R, Keeling M E, Mansfield K, Tardif S, Morris T. Academic Press, Elsevier; Oxford, UK: 2012. Nonhuman Primates in Biomedical Research. Vol. 2: Diseases. [Google Scholar]
- 32.Pasetti M F, Resendiz-Albor A, Ramirez K. Heterologous prime-boost strategy to immunize very young infants against measles: pre-clinical studies in rhesus macaques. Clin Pharmacol Ther. 2007;82(06):672–685. doi: 10.1038/sj.clpt.6100420. [DOI] [PubMed] [Google Scholar]
- 33.Verstrepen B E, Oostermeijer H, Fagrouch Z. Vaccine-induced protection of rhesus macaques against plasma viremia after intradermal infection with a European lineage 1 strain of West Nile virus. PLoS One. 2014;9(11):e112568. doi: 10.1371/journal.pone.0112568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stephenson K E, Wegmann F, Tomaka F. Comparison of shortened mosaic HIV-1 vaccine schedules: a randomised, double-blind, placebo-controlled phase 1 trial (IPCAVD010/HPX1002) and a preclinical study in rhesus monkeys (NHP 17-22) Lancet HIV. 2020;7(06):e410–e421. doi: 10.1016/S2352-3018(20)30001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Doremalen N, Lambe T, Spencer A.ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques Nature 2020586(7830):578–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu S, Zhao Y, Yu W. Comparison of nonhuman primates identified the suitable model for COVID-19. Signal Transduct Target Ther. 2020;5(01):157. doi: 10.1038/s41392-020-00269-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun W, Singh A K.Plague vaccine: recent progress and prospects npjVaccines 2019(e-pub ahead of print) 10.1038/s41541-019-0105-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shedlock D J, Silvestri G, Weiner D B. Monkeying around with HIV vaccines: using rhesus macaques to define ‘gatekeepers’ for clinical trials. Nat Rev Immunol. 2009;9(10):717–728. doi: 10.1038/nri2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.UniProt Consortium UniProt: a worldwide hub of protein knowledge Nucleic Acids Res 201947(D1):D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.UniProt Consortium . Chen C, Li Z, Huang H, Suzek B E, Wu C H. A fast Peptide Match service for UniProt Knowledgebase. Bioinformatics. 2013;29(21):2808–2809. doi: 10.1093/bioinformatics/btt484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.UniProt Consortium . Morgat A, Lombardot T, Coudert E. Enzyme annotation in UniProtKB using Rhea. Bioinformatics. 2020;36(06):1896–1901. doi: 10.1093/bioinformatics/btz817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Varki A. A chimpanzee genome project is a biomedical imperative. Genome Res. 2000;10(08):1065–1070. doi: 10.1101/gr.10.8.1065. [DOI] [PubMed] [Google Scholar]
- 43.Olson M V, Varki A. Sequencing the chimpanzee genome: insights into human evolution and disease. Nat Rev Genet. 2003;4(01):20–28. doi: 10.1038/nrg981. [DOI] [PubMed] [Google Scholar]
- 44.Varki A, Altheide T K. Comparing the human and chimpanzee genomes: searching for needles in a haystack. Genome Res. 2005;15(12):1746–1758. doi: 10.1101/gr.3737405. [DOI] [PubMed] [Google Scholar]
- 45.Lucchese G, Stufano A, Calabro M, Kanduc D. Charting the peptide crossreactome between HIV-1 and the human proteome. Front Biosci (Elite Ed) 2011;3:1385–1400. doi: 10.2741/e341. [DOI] [PubMed] [Google Scholar]
- 46.Barouch D H, Tomaka F L, Wegmann F.Evaluation of a mosaic HIV-1 vaccine in a multicentre, randomised, double-blind, placebo-controlled, phase 1/2a clinical trial (APPROACH) and in rhesus monkeys (NHP 13-19) Lancet 2018392(10143):232–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas C. Roadblocks in HIV research: five questions. Nat Med. 2009;15(08):855–859. doi: 10.1038/nm0809-855. [DOI] [PubMed] [Google Scholar]
- 48.Cohen J.Another HIV vaccine strategy fails in large-scale study Science 2020(e-pub ahead of print) 10.1126/science.abb1480 [DOI] [Google Scholar]
- 49.Sabin A B. Oral poliovirus vaccine: history of its development and use and current challenge to eliminate poliomyelitis from the world. J Infect Dis. 1985;151(03):420–436. doi: 10.1093/infdis/151.3.420. [DOI] [PubMed] [Google Scholar]
- 50.Lerner A, Jeremias P, Matthias T. The world incidence and prevalence of autoimmune diseases is increasing. Int J Celiac Dis. 2015;3:151–155. [Google Scholar]
- 51.Bond K A, Franklin L J, Sutton B, Firestone S M. Q-Vax Q fever vaccine failures, Victoria, Australia 1994-2013. Vaccine. 2017;35(51):7084–7087. doi: 10.1016/j.vaccine.2017.10.088. [DOI] [PubMed] [Google Scholar]
- 52.Breakwell L, Moturi E, Helgenberger L. Measles outbreak associated with vaccine failure in adults-Federated States of Micronesia, February-August 2014. MMWR Morb Mortal Wkly Rep. 2015;64(38):1088–1092. doi: 10.15585/mmwr.mm6438a7. [DOI] [PubMed] [Google Scholar]
- 53.Cherry J D. Epidemic pertussis and acellular pertussis vaccine failure in the 21st century. Pediatrics. 2015;135(06):1130–1132. doi: 10.1542/peds.2014-4118. [DOI] [PubMed] [Google Scholar]
- 54.Lopez-Lacort M, Collado S, Díez-Gandía A, Díez-Domingo J. Rotavirus, vaccine failure or diagnostic error? Vaccine. 2016;34(48):5912–5915. doi: 10.1016/j.vaccine.2016.10.032. [DOI] [PubMed] [Google Scholar]
- 55.Mahalingam S, Herring B L, Halstead S B. Call to action for dengue vaccine failure. Emerg Infect Dis. 2013;19(08):1335–1337. doi: 10.3201/eid1908.121864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moinho R, Brett A, Ferreira G, Lemos S. Pneumococcal vaccine failure: can it be a primary immunodeficiency? BMJ Case Rep. 2014;2014:bcr2014204714. doi: 10.1136/bcr-2014-204714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.PROVIDE study teams . Naylor C, Lu M, Haque R. Environmental enteropathy, oral vaccine failure and growth faltering in infants in Bangladesh. EBioMedicine. 2015;2(11):1759–1766. doi: 10.1016/j.ebiom.2015.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poland G A. Influenza vaccine failure: failure to protect or failure to understand? Expert Rev Vaccines. 2018;17(06):495–502. doi: 10.1080/14760584.2018.1484284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramsay M, Brown K. The public health implications of secondary measles vaccine failure. J Prim Health Care. 2013;5(02):92. [PubMed] [Google Scholar]
- 60.Wiedermann U, Garner-Spitzer E, Wagner A. Primary vaccine failure to routine vaccines: why and what to do? Hum Vaccin Immunother. 2016;12(01):239–243. doi: 10.1080/21645515.2015.1093263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pieczenik G. Are the universes of antibodies and antigens symmetrical? Reprod Biomed Online. 2003;6(02):154–156. doi: 10.1016/s1472-6483(10)61702-6. [DOI] [PubMed] [Google Scholar]
- 62.Kanduc D. Pentapeptides as minimal functional units in cell biology and immunology. Curr Protein Pept Sci. 2013;14(02):111–120. doi: 10.2174/1389203711314020003. [DOI] [PubMed] [Google Scholar]
- 63.Puente X S, Gutiérrez-Fernández A, Ordóñez G R, Hillier L W, López-Otín C. Comparative genomic analysis of human and chimpanzee proteases. Genomics. 2005;86(06):638–647. doi: 10.1016/j.ygeno.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 64.Glazko G, Veeramachaneni V, Nei M, Makałowski W. Eighty percent of proteins are different between humans and chimpanzees. Gene. 2005;346:215–219. doi: 10.1016/j.gene.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 65.Sharp A J, Locke D P, McGrath S D. Segmental duplications and copy-number variation in the human genome. Am J Hum Genet. 2005;77(01):78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hahn M W, Demuth J P, Han S G. Accelerated rate of gene gain and loss in primates. Genetics. 2007;177(03):1941–1949. doi: 10.1534/genetics.107.080077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dumas L, Kim Y H, Karimpour-Fard A. Gene copy number variation spanning 60 million years of human and primate evolution. Genome Res. 2007;17(09):1266–1277. doi: 10.1101/gr.6557307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marques-Bonet T, Kidd J M, Ventura M.A burst of segmental duplications in the genome of the African great ape ancestor Nature 2009457(7231):877–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blekhman R, Oshlack A, Gilad Y. Segmental duplications contribute to gene expression differences between humans and chimpanzees. Genetics. 2009;182(02):627–630. doi: 10.1534/genetics.108.099960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kapopoulou A, Mathew L, Wong A, Trono D, Jensen J D. The evolution of gene expression and binding specificity of the largest transcription factor family in primates. Evolution. 2016;70(01):167–180. doi: 10.1111/evo.12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hamilton A T, Huntley S, Tran-Gyamfi M, Baggott D M, Gordon L, Stubbs L. Evolutionary expansion and divergence in the ZNF91 subfamily of primate-specific zinc finger genes. Genome Res. 2006;16(05):584–594. doi: 10.1101/gr.4843906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Khaitovich P, Hellmann I, Enard W.Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees Science 2005309(5742):1850–1854. [DOI] [PubMed] [Google Scholar]
- 73.Enard W, Khaitovich P, Klose J.Intra- and interspecific variation in primate gene expression patterns Science 2002296(5566):340–343. [DOI] [PubMed] [Google Scholar]
- 74.Kanduc D. “Self-nonself” peptides in the design of vaccines. Curr Pharm Des. 2009;15(28):3283–3289. doi: 10.2174/138161209789105135. [DOI] [PubMed] [Google Scholar]
- 75.Kanduc D. The self/nonself issue: a confrontation between proteomes. Self Nonself. 2010;1(03):255–258. doi: 10.4161/self.1.3.11897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lucchese G, Stufano A, Kanduc D. Proposing low-similarity peptide vaccines against Mycobacterium tuberculosis. J Biomed Biotechnol. 2010;2010:832341. doi: 10.1155/2010/832341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Angelini G, Bonamonte D, Lucchese A. Preliminary data on Pemphigus vulgaris treatment by a proteomics-defined peptide: a case report. J Transl Med. 2006;4:43. doi: 10.1186/1479-5876-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kanduc D. Immunogenicity, immunopathogenicity, and immunotolerance in one graph. Anticancer Agents Med Chem. 2015;15(10):1264–1268. doi: 10.2174/1871520615666150716105543. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


