Abstract
BACKGROUND AND PURPOSE:
The durations of acute ischemic stroke patients' CT or MR perfusion scans may be too short to fully sample the passage of the injected contrast agent through the brain. We tested the potential magnitude of hidden errors related to the truncation of data by short perfusion scans.
MATERIALS AND METHODS:
Fifty-seven patients with acute ischemic stroke underwent perfusion MR imaging within 12 hours of symptom onset, using a relatively long scan duration (110 seconds). Shorter scan durations (39.5–108.5 seconds) were simulated by progressively deleting the last-acquired images. CBV, CBF, MTT, and time to response function maximum (Tmax) were measured within DWI-identified acute infarcts, with commonly used postprocessing algorithms. All measurements except Tmax were normalized by dividing by the contralateral hemisphere values. The effects of the scan duration on these hemodynamic measurements and on the volumes of lesions with Tmax of >6 seconds were tested using regression.
RESULTS:
Decreasing scan duration from 110 seconds to 40 seconds falsely reduced perfusion estimates by 47.6%–64.2% of normal for CBV, 1.96%–4.10% for CBF, 133%–205% for MTT, and 6.2–8.0 seconds for Tmax, depending on the postprocessing method. This truncation falsely reduced estimated Tmax lesion volume by 71.5 or 93.8 mL, depending on the deconvolution method. “Lesion reversal” (ie, change from above-normal to apparently normal, or from >6 seconds to ≤6 seconds for the time to response function maximum) with increasing truncation occurred in 37%–46% of lesions for CBV, 2%–4% for CBF, 28%–54% for MTT, and 42%–44% for Tmax, depending on the postprocessing method.
CONCLUSIONS:
Hidden truncation-related errors in perfusion images may be large enough to alter patient management or affect outcomes of clinical trials.
CT and MR perfusion (MRP) imaging are used frequently in acute ischemic stroke care and are increasingly used to determine treatment eligibility in therapeutic trials.1–4 In both techniques, maps of regional hemodynamic parameters like CBF or CBV are derived from dynamic images that quantify the concentrations of an intravenously injected contrast agent as it passes through the brain. Clinical studies often omit consideration of the postprocessing algorithms that convert concentration measurements to perfusion maps, implicitly accepting their accuracy. However, postprocessing algorithms can produce significant errors,5 which typically are visually undetectable during interpretation.
One type of error results from temporal truncation of concentration measurements by scan durations that are too short to adequately sample the passage of the contrast agent bolus, which occurs most often under ischemic conditions. Bolus injections typically take at least 5 seconds, and injecting the larger volumes of contrast used for CTP may take 10 seconds or longer.6,7 After injection, the bolus travels to ischemic brain tissue via stenotic arteries and/or circuitous collateral pathways, potentially lengthening the average arm-to-brain transit time to 35 seconds or more.8 Dispersion may cause some components of the bolus to arrive far later.9 Once each part of the bolus arrives, the average time that it spends in ischemic tissue is commonly 10–20 seconds and sometimes considerably longer,10 because autoregulatory vasodilation slows blood velocity. In comparison, CTP and MRP scans may be as short as 40 seconds.6,11 Therefore, the parametric perfusion maps that are used for clinical interpretation may be derived from severely truncated concentration data.
A previous study showed that data truncation may result in underestimation of CBV in ischemic tissue.12 We hypothesized that other common perfusion measurements are also prone to truncation-related errors. We retrospectively analyzed the MRP images of patients with acute stroke, which can be acquired with longer scan durations than CTP scans, without concern for ionizing radiation exposure. We simulated progressively shorter scan durations by discarding the images acquired at the final time points in each perfusion scan. We tested whether scan duration altered calculated perfusion parameters within acute infarcts, by placing regions of interest near the center of each patient's DWI lesion, and then assessing whether perfusion measurements obtained within these ROIs were altered by increasing degrees of scan truncation. We investigated perfusion within acute infarcts because especially severe ischemia might be expected to exist within infarcts, and because some studies have substituted perfusion imaging for DWI in identifying irreversibly injured tissue.1–4,13,14 We also tested whether scan duration altered the volumes of lesions in thresholded maps of the time-to-maximum of the deconvolved residue function (Tmax), because these volumes have been used to determine eligibility for recanalization therapy in clinical trials.3,15
Materials and Methods
Patient Selection
This study was approved by our institutional review board, which waived its informed consent requirement because only retrospective data analysis was performed. We reviewed hospital records and selected all patients from a 14-month period who satisfied the following criteria: 1) clinical diagnosis of new ischemic stroke; 2) MR imaging examinations, including DWI and MRP, completed within 12 hours of the time when the patient was last seen at neurologic baseline; 3) discovery of new symptoms within 15 minutes of that time; and 4) DWI-positive anterior circulation ischemic lesion. Patients were excluded if their DWI or MRP data were too motion-degraded to permit satisfactory processing, if motion correction resulted in the sections containing the DWI lesion being no longer visualized, or if the DWI lesion was too small to accommodate a region of interest measuring 5.2 × 5.2 mm.
Image Acquisition
Images were acquired on a 1.5T MR imaging scanner (Signa; GE Healthcare, Milwaukee, Wisconsin). DWI used a balanced spin-echo echo-planar pulse sequence, incorporating two 180° radiofrequency pulses to reduce eddy current–related artifacts.16 The FOV was 22 cm, with a 128 × 128 matrix, zero-filled in k-space to produce 256 × 256 pixel images. TR was 5000 ms, and TE was as short as possible. There were 25 diffusion-encoding directions, with b=1000 s/mm2 and 3 volumes with b=0 s/mm2.
MRP used a gradient-echo echo-planar pulse sequence with a 22-cm FOV and a 128 × 128 matrix. TR and TE were 1500 and 35 ms, respectively, and the flip angle was 60°. Eighty volumes were acquired, resulting in a scan time of 2 minutes. Ten seconds after scan initiation, 20 mL of gadopentetate dimeglumine (Magnevist; Bayer HealthCare Pharmaceuticals, Wayne, New Jersey) was power-injected intravenously at 5 mL/s, followed by a similar volume of normal saline, injected at the same rate.
For both DWI and MRP, section thickness and spacing were 5 and 1 mm, respectively. DWI sections were prescribed to completely cover the brain. MRP coverage was limited to 14 or 15 sections, prescribed as a subset of DWI section locations.
Image Review
Without referring to perfusion maps, an experienced neuroradiologist placed a 5.2 × 5.2 mm region of interest in the geographic center of each patient's DWI lesion. A second, identically sized region of interest was placed in an anatomically similar location on the opposite side of the brain, to allow normalization of perfusion values in the lesion via comparison with presumably normal values. These ROIs were transferred to the corresponding anatomic locations on the raw perfusion images (Fig 1). Before generation of any postprocessed perfusion maps, the neuroradiologist reviewed the ROIs to ensure that neither included a macroscopically visible blood vessel whose “blooming” could unduly influence perfusion measurements. When this occurred, the region of interest was moved slightly. Whenever possible, each patient's ROIs were placed exclusively in gray matter or exclusively in white matter. In the few cases in which this placement was impossible, the lesion and contralateral ROIs were chosen to include similar proportions of gray and white matter.
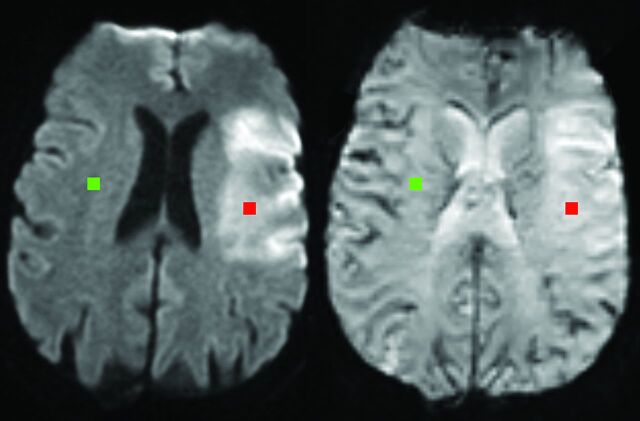
Fig 1.
Region-of-interest placement. Lesion (red) and contralateral (green) regions of interest from 1 patient are superimposed on the diffusion-weighted image that was used for their placement (left) and the perfusion source image to which they were subsequently transferred (right).
Following the completion of region-of-interest placement, the same neuroradiologist manually measured the volumes of DWI-hyperintense lesions that were thought to reflect acute infarction, for each patient.
Image Processing
From MRP data, we calculated 4 commonly studied regional hemodynamic parameters: CBV, CBF, MTT, and the time at which the deconvolved tissue response function R(t) reached its maximum (Tmax). To assess whether different computation methods have varying vulnerabilities to truncation-related artifacts, we calculated each of these 4 parameters using several alternative algorithms, so that 11 perfusion maps were created for each patient.
Generation of 10 of the 11 perfusion maps required deriving R(t) from the tissue concentration function C(t), by using deconvolution with an arterial input function. Deconvolution was performed by in-house, fully automated software, using the following steps: First, the number of baseline images (ie, before arrival of the contrast bolus) was derived by spatially smoothing each image with a Hanning filter, averaging the signal intensity of all tissue pixels at each time point, and selecting the time point before the one exhibiting the largest incremental signal change as the final baseline image. Motion correction was performed by coregistering17 each volume to the first baseline image, and the images were then converted to concentration-of-contrast-agent versus time curves, C(t).18
Arterial input function voxels were selected from the section with the largest number of high-blood-volume voxels. Candidate voxels were identified with a combination of previously described criteria,19–23 including high CBV, early contrast agent arrival time, short relative MTT, and narrow full width at half maximum concentration values. K-means cluster analysis was then performed on the candidate voxels, and the cluster with the narrowest full width at half maximum was selected as the arterial input function. The same arterial input function was used for every simulated scan duration, though the arterial input function was truncated to the same length as the C(t).
Deconvolution was performed by using 2 alternative algorithms: standard singular value decomposition (sSVD)24 and oscillation-index regulated singular value decomposition with a block circulant matrix (oSVD), a refined version of SVD that is insensitive to bolus arrival delay–related artifacts.25 Hereafter, hemodynamic measurements derived from standard singular value decomposition are labeled with the prefix “s,” whereas those derived from oSVD are labeled with the prefix “o.”
CBV was calculated in 3 ways. One CBV measurement, hereafter called CBVc, was produced by integrating the C(t) in each image pixel. This simplest of CBV algorithms has been used in numerous MRP studies and does not require deconvolution.20,26 The other 2 CBV measurements were produced by integrating the R(t), and will be designated sCBVr and oCBVr, to indicate which deconvolution algorithm was used. This technique is equivalent theoretically to that used by at least 1 commercially available CTP software package.27
CBF was calculated as the maximum value of the deconvolved response function (sCBF or oCBF), and Tmax was the time at which that maximum was reached (sTmax or oTmax). MTT was calculated as the quotient of CBVc divided by sCBF or oCBF (yielding sMTTc or oMTTc, respectively), sCBVr divided by sCBF (yielding sMTTr), or oCBVr divided by oCBF (yielding oMTTr).
To assess the effects of scan duration, we created temporally truncated datasets, simulating the data that would have been acquired if shorter scans had been performed. For example, a 90-second scan duration was simulated by discarding the images from the final 30 seconds of the full 120-second scan. Sample perfusion maps produced from a single patient's data, truncated to simulate 2 different scan durations, are shown in Fig 2.
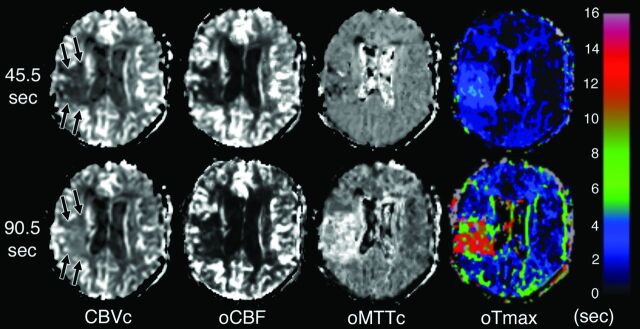
Fig 2.
Visual appearance of truncation artifacts resulting from scans of different durations. Sample perfusion maps produced from a single patient's perfusion data, which have been truncated to simulate perfusion scans of 2 different durations that are similar to those in common clinical use: 45.5 seconds (top row) and 90.5 seconds (bottom row) following contrast injection. The CBV map from the shorter 45.5-second scan shows a low-CBV lesion in the right frontal lobe (arrows, top row). However, the more accurate CBV map produced from the longer scan shows elevated rather than reduced CBV (arrows, bottom row). CBF maps produced from the 2 scan durations show little appreciable difference. The MTT map from the truncated scan shows almost no lesion, but there is a sizeable lesion with prolonged MTT in the map produced from the longer scan. The Tmax maps produced from the 2 scan durations show subjectively appreciable lesions of similar sizes. However, the color legend at the right of the figure shows that the abnormal Tmax values produced by the shorter scan are <6 seconds, so this tissue would not be classified as “at risk” by studies that use that threshold, whereas the longer scan produced a large lesion with Tmax values above the 6-second threshold.
Our MRP protocol included a 10-second delay between the beginning of image acquisition and initiation of contrast injection. To facilitate comparison with previously published results (which may not use the same preinjection delay) and to facilitate future protocol design, we will hereafter refer to our simulated scan durations in terms of their relationship to the time at which contrast injection began (eg, the duration of the full 120-second scan was 110 seconds postinjection).
Statistical Analysis
For each patient, for every simulated scan duration, normalized CBV, CBF, and MTT measurements were obtained, by dividing the mean of the lesion region-of-interest values by the mean value in the contralateral region of interest. For the 2 Tmax measurements, sTmax and oTmax, the mean absolute regional Tmax value recorded in the lesion region of interest was recorded, without normalization.
For every simulated scan duration ranging from 39.5 to 110 seconds postinjection, the mean of each hemodynamic measurement across all patients was calculated. This minimum duration was chosen for 2 reasons: First, 39.5 seconds was the shortest scan duration for which perfusion measurements incorporated at least 9 data points for all patients, thereby avoiding noisier measurements obtained from only a few data points. Second, perfusion studies have used scan durations as short as 40 seconds postinjection,6,11 and, with our TR of 1.5 seconds, the available scan duration closest to 40 seconds was 39.5 seconds.
Linear regression analyses were performed between each of these 11 hemodynamic measurements and scan duration, and against the base-10 logarithm of scan duration. To assess the magnitude of truncation-related errors that might be expected for an individual patient, we fit longitudinal fixed-effects models for each hemodynamic parameter, with patient-specific slopes and intercepts, incorporating observations from all durations, from all patients. For these fixed-effects regressions, measurements were included only if they were produced from at least 9 postarrival data points.
For each of the 11 measurements, we counted the number of patients for whom worsening truncation by shorter scan durations resulted in “lesion reversal,” which we defined as either the false creation of an apparent lesion when no lesion was truly present, or the false disappearance of a lesion when a lesion was truly present. Our goal was to reserve the classification of “lesion reversal” for only those cases in which worsening truncation caused a sustained change from above-normal ROI values to below-normal values and to avoid labeling as “lesion reversal” cases in which image noise caused perfusion values to fluctuate randomly above and below normal levels. Therefore, to determine that lesion reversal had occurred for a particular patient and any of the 9 normalized measurements, we required that decreasing scan durations (ie, worsening truncation) resulted in above-normal measurements for at least 3 consecutive scan durations, followed by below-normal measurements for at least 3 consecutive durations, and were not followed by a subsequent return to 3 above-normal measurements. For sTmax and oTmax, which were not normalized, we defined lesion reversal as a sustained transition from values of >6 seconds to ≤6 seconds. Several recent studies have proposed 6 seconds as the Tmax threshold that best identifies tissue at risk of infarction.3,15 For each of the 11 perfusion measurements, we tallied the proportion of patients for which lesion reversal occurred, and used logistic regression to assess whether the likelihood of lesion reversal was related to DWI lesion size.
In some clinical trials, patient eligibility for recanalization therapy was based on the existence of a sufficient volume of brain tissue with a Tmax of >6 seconds. To determine how scan duration might artifactually influence this measured volume, we measured the volumes of tissue with a Tmax of >6 seconds for each patient at each simulated duration. We assessed the effect of scan duration on lesion volume by using additional regression analyses similar to those described above.
Results
The study included 57 patients, 25 women, whose ages ranged from 30.4 to 93.3 years, with a mean ± standard deviation of 68.2 ±16.8 years. DWI lesion volumes ranged from 0.29 to 178.07 mL, with a mean ± standard deviation of 31.40 ± 42.65 mL.
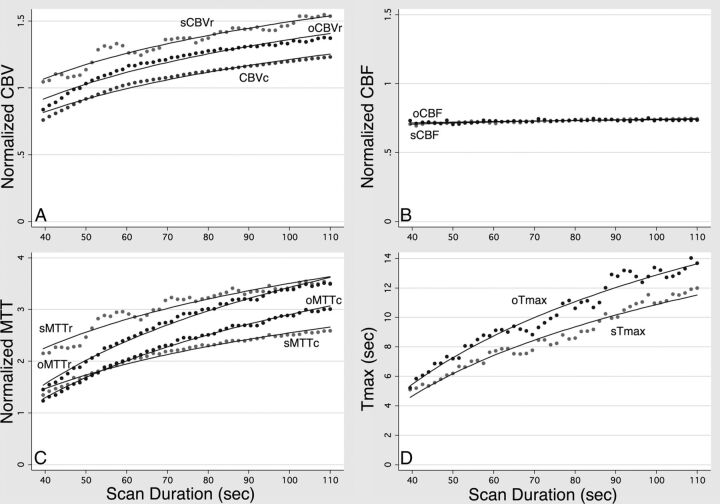
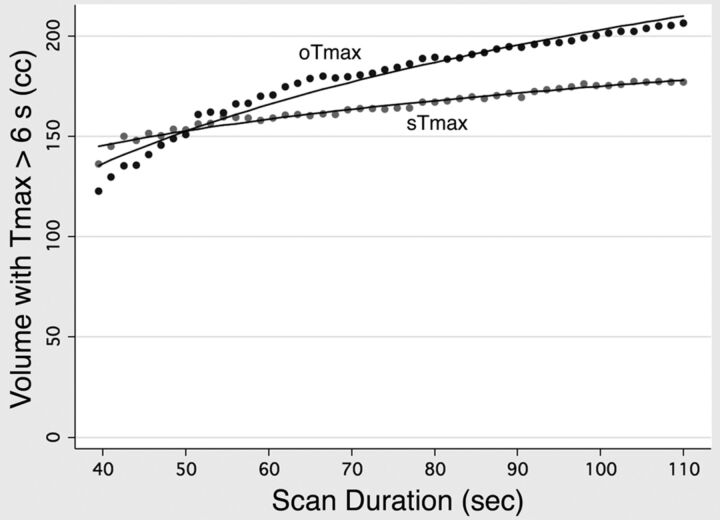
For all hemodynamic parameters, the logarithmic model produced higher R2 goodness-of-fit statistics. Therefore, only the results of the logarithmic model will be discussed. The means of all 11 parameters decreased significantly (P < .001) as truncation increased, as shown in Fig 3. However, the calculated slopes for sCBF and oCBF were much smaller than those of the other parameters, reflecting the much smaller effect that truncation had on CBF. The calculated slopes for each parameter were the following: CBVc, 0.99/log10(s); sCBVr, 1.07/log10(s); oCBVr, 1.11/log10(s); sCBF, 0.09/log10(s); oCBF, 0.06/log10(s); sMTTc, 2.70/log10(s); sMTTr, 3.16/log10(s); oMTTc, 4.04/log10(s); oMTTr, 4.69/log10(s); sTmax, 15.6 s/log10(s); and oTmax, 18.6 s/log10(s). Aggregated lesion volumes of tissue with a Tmax of >6 seconds also decreased significantly with decreasing scan duration (P < .001), with slopes of 73.8 mL/log10(s) and 167.6 mL/log10(s) for sTmax and oTmax, respectively (Fig 4).
Fig 3.
Quantitative measurement of truncation artifacts caused by scans of different durations. Mean values of the 3 different CBV measurements (A), the 2 different CBF measurements (B), the 4 different MTT measurements (C), and the 2 different Tmax measurements (D), averaged across all patients, for scan durations ranging from 39.5 to 110 seconds after contrast injection. The curves in each graph were fitted by logarithmic regression, which showed a significant (P < .001) decrease in every calculated parameter with decreasing scan time. However, the magnitude of this reduction was much smaller for CBF than for the other parameters.
Fig 4.
Artifactual effect of truncation on Tmax lesion volumes. Volumes of tissue with calculated Tmax values of >6 seconds averaged across all patients, for scan durations ranging from 39.5 to 110 seconds after contrast injection. The curves in each graph were fitted by logarithmic regression, which showed a significant (P < .001) reduction in both sTmax and oTmax with decreasing scan duration.
Results of the fixed regression models with patient-specific slopes and intercepts are presented in Table 1 for region of interest–based hemodynamic measurements and lesion reversal frequencies, and in Table 2 for volumes of lesions with a Tmax of >6 seconds. All 11 parameters decreased significantly as truncation increased (P < .001). Again, the calculated slopes for CBF were much smaller than those of the other parameters. To facilitate meaningful interpretation of the magnitudes of scan duration–dependent artifacts, Table 1 includes calculations of “potential truncation effect,” which we define as the decrease in the calculated value of that parameter that would be expected for an individual patient if perfusion data were truncated by reducing the scan duration from 110 seconds to 40 seconds postinjection. Decreasing the scan duration from 110 to 40 seconds falsely reduced perfusion estimates by 47.6%–64.2% of normal for CBV, 1.96%–4.10% for CBF, 133%–205% for MTT, and 6.19–8.00 seconds for Tmax.
Table 1:
Regression-derived effects of scan duration on various hemodynamic measurements
| Hemodynamic Measurement | Deconvolution Algorithm | Function Used for CBV Integration | t-statistic | Slopea | Potential Truncation Effectb | Lesion Reversal Frequency |
|---|---|---|---|---|---|---|
| CBVc | None | Concentration | 41.53 | 1.08/log10(s) | 47.6% | 37% (21/57) |
| sCBVr | sSVD | Response | 17.99 | 1.46/log10(s) | 64.2% | 35% (20/57) |
| oCBVr | oSVD | Response | 16.72 | 1.17/log10(s) | 51.4% | 46% (26/57) |
| sCBF | sSVD | NA | 9.62 | 0.09/log10(s) | 4.10% | 4% (2/57) |
| oCBF | oSVD | NA | 3.58 | 0.04/log10(s) | 1.96% | 2% (1/57) |
| sMTTc | sSVD | Concentration | 34.49 | 3.03/log10(s) | 133% | 47% (27/57) |
| sMTTr | sSVD | Response | 35.57 | 3.73/log10(s) | 164% | 28% (16/57) |
| oMTTc | oSVD | Concentration | 42.50 | 3.79/log10(s) | 166% | 54% (31/57) |
| oMTTr | oSVD | Response | 51.15 | 4.67/log10(s) | 205% | 39% (22/57) |
| sTmax | sSVD | NA | 25.91 | 14.10/log10(s) | 6.19 seconds | 44% (25/57) |
| oTmax | oSVD | NA | 28.62 | 18.2/log10(s) | 8.00 seconds | 42% (24/57) |
Note:—NA indicates not applicable.
The slopes of all 11 hemodynamic parameters with respect to the logarithm of scan duration were significantly greater than zero (P < .001). Therefore, t-statistics rather than P values are reported.
“Potential truncation effect” refers to the expected reduction in the calculated parameter value that would result from decreasing the scan duration from 110 seconds to 40 seconds postinjection. For example, if the CBVc value derived from a 110-second scan were 107.6% of normal, the expected CBVc using a 40-second scan would be 60.0% of normal.
Table 2:
Regression-derived effects of scan duration on the volume of brain tissue with Tmax greater than 6 seconds
| Hemodynamic Measurement | Deconvolution Algorithm | t-statistic | Lesion Volume Slopea | Potential Truncation Effectb |
|---|---|---|---|---|
| sTmax | sSVD | 44.21 | 162.7 mL/log10(s) | 71.5 mL |
| oTmax | oSVD | 47.94 | 213.4 mL/log10(s) | 93.8 mL |
The slopes of sTmax and oTmax lesion volumes with respect to the logarithm of scan duration were both significantly greater than zero with P < .001. Therefore, t-statistics rather than P values are reported.
“Potential truncation effect” reflects the expected decrease in the lesion volume that would result from decreasing the scan duration from 110 seconds to 40 seconds postinjection. For example, if the volume of an oTmax lesion were measured to be 150 mL using a 110-second scan, the expected lesion volume derived from a 40-second scan would be 56.2 mL.
Lesion reversal frequencies for the 3 methods of computing CBV ranged between 37% and 46%, indicating that for more than one-third of patients, truncation of data by a short scan could result in the appearance of a low-CBV lesion when no such lesion was truly present. Lesion reversal frequencies for the 4 MTT calculations ranged between 28% and 54%, indicating that truncation errors could obscure high-MTT lesions for many patients. Lesion reversal frequencies for sTmax and oTmax were 42% and 44%, respectively, again showing that truncation errors could often prevent detection of tissue considered at risk of infarction. Lesion reversal was much rarer for oCBF and sCBF, with lesion reversal frequencies of 2% and 4%, respectively.
Logistic regression analysis showed no statistically significant relationship between lesion reversal frequency and DWI lesion size for any of the 11 perfusion parameters, except for sMTTc (odds ratio = 1.021/mL, P = .03) and sMTTr (odds ratio = 1.018/mL, P = .02).
Significant associations (P < .001) were also found between scan duration and the volume of tissue with a Tmax of >6 seconds. Truncating scan durations from 110 to 40 seconds falsely reduced the estimated Tmax lesion volume by 71.5 mL for sTmax, and 93.8 mL for oTmax (Table 2). An example of the effect of scan duration on Tmax lesion volume is shown in Fig 5.
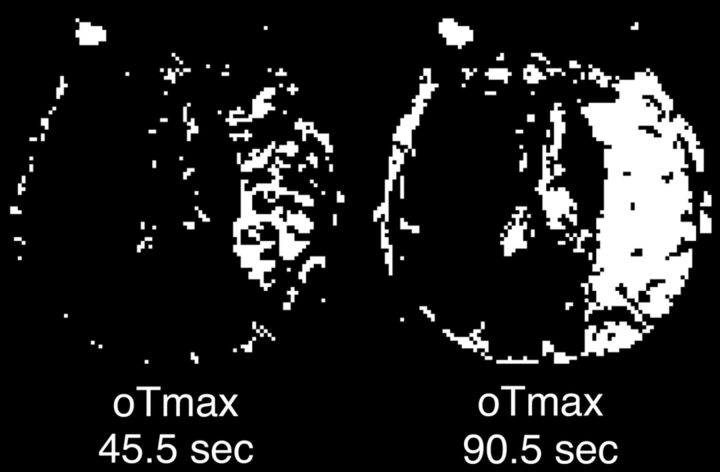
Fig 5.
Change in Tmax lesion volume resulting from truncation artifacts. Tmax maps produced from a single patient's perfusion data, which have been truncated to simulate perfusion scans of 2 different durations that are similar to those in common clinical use: 45.5 seconds (left) and 90.5 seconds (right) following contrast injection. High-contrast window settings were used to depict pixels with a Tmax of >6 seconds as white and other pixels as black. The 90.5-second scan demonstrates a large lesion that would be considered “at risk” by using the 6-second criterion and could potentially make the patient eligible for thrombolytic therapy. The 45.5-second scan shows a much smaller Tmax lesion. Tmax lesion sizes measured across all image sections for this patient were 430.2 mL for the 90.5-second scan and 79.2 mL for the 45.5-second scan.
Discussion
Perfusion imaging research studies frequently provide little or no information regarding the postprocessing algorithms used, and design details of proprietary postprocessing software typically are not revealed. However, inattention to methodology may conceal errors in perfusion images that could significantly change their implications for patient care. Our results demonstrate that common perfusion postprocessing algorithms may produce very different results when scans of different durations are performed on the same patient, resulting in varying degrees of data truncation errors.
Longer scans more completely sample the passage of the contrast bolus through the brain, and presumably provide more accurate perfusion measurements. Shorter scans may produce perfusion maps in which CBV, MTT, Tmax, and, to a much lesser degree, CBF values are all underestimated in ischemic brain tissue to varying degrees, depending on truncation severity. Because scan durations vary widely across institutions, the severity of truncation artifacts likely varies extensively as well. Perfusion measurements reported in different studies from different institutions may not only fail to reflect the true hemodynamic status of the brain but may not even be reliably comparable with each other.
The severity of truncation artifacts is presumably most severe in stroke patients, whose arterial lesions delay the arrival of the contrast agent bolus and prolong its transit through the brain. However, truncation can also be worsened for other reasons. For example, a slower contrast injection rate, a narrower intravenous catheter, longer or larger injection tubing, or an increase in patient size could all result in increased truncation artifacts. Even if these factors were constant, moment-to-moment changes in cardiac output could change the rate at which contrast material is delivered to the brain and, therefore, the degree of truncation. Therefore, even data acquired at different times at the same institution by using the same imaging protocol may be difficult to compare if the scan duration is too short.
In acute stroke imaging, perfusion maps are often used to identify brain tissue that is putatively at risk of infarction.4 Some studies have proposed defining tissue at risk as visibly appreciable MTT lesions. As shown in Fig 2, the truncation artifacts introduced by a shorter scan could make such lesions less conspicuous or erase them entirely. Other studies have proposed delineating at-risk tissue by using quantitative hemodynamic thresholds, for example, Tmax values of >6 seconds. In our study, calculated Tmax values changed from above that threshold to below it in nearly half of ischemic lesions, when scan durations were reduced. Our analysis estimates that truncation of perfusion data may reduce the measured volume of putative “at-risk” tissue by >70 mL, depending on truncation severity and the postprocessing algorithm used. These truncation-related errors in MTT and Tmax maps could result in failure to administer thrombolytic therapy to patients who could be eligible for treatment in clinical trials using Tmax inclusion criteria.
Some studies have also proposed that perfusion imaging can also be used to identify “core” brain tissue that has irreversible ischemic injury, a role usually filled by DWI, because low-CBV lesions approximate the size and location of DWI lesions.2,13 Our results show that identifying core tissue with perfusion imaging also may be challenging, because shorter scan durations could cause CBV to appear low when it is actually elevated. If a large low-CBV lesion is presumed to reflect extensive completed infarction that would make thrombolytic therapy futile or dangerous, then truncation-related errors in the CBV calculation could result in failure to treat patients who otherwise would be eligible.
Our study is limited in that only a handful of postprocessing algorithms were tested. Although these algorithms are among the most commonly used in clinical research and practice, other algorithms potentially could be more resistant to truncation-related errors. In addition, oTmax and sTmax volumes appear to diverge with increased scan duration. However, it is more likely that both will converge to different steady-state values representing the extent of tissue with abnormal hemodynamics. This outcome is likely due to the threshold used for identifying tissue at risk being based on literature values that used SVD for deconvolution. It is possible that if a higher Tmax threshold were used, similar volumetric results would be obtained. This finding of volume dependency exemplifies the need to disclose the algorithms used for calculating perfusion maps, even those as simple as Tmax.
Another limitation of our study is our inability to achieve exactly the same proportion of gray matter and white matter in each patient's lesion and contralateral ROIs, for those patients in whom ROIs could not be placed entirely within either gray matter or white matter structures. As a result, noise may have been introduced into the ratios that were used for normalization of CBV, CBF, and MTT values. Therefore, the lesion reversal frequencies that we calculated for each parameter are best considered approximations of the frequencies that would be computed by using a larger sample size. However, because each patient's lesion and contralateral ROIs did not change positions across different scan durations, volume averaging–related errors in normalization ratios would not invalidate our finding that increasing truncation causes artifactual decreases in calculated perfusion parameters.
The most straightforward way to avoid truncation-related errors in perfusion maps is simply to use CTP and MRP scans that are as long as is feasibly possible. Unfortunately, in the case of CTP, longer scans entail increased exposure to ionizing radiation. This increase in exposure may be mitigated by combining an increase in scan duration with a decrease in sampling frequency, for example, acquisition of CTP images every 3 seconds instead of every 1 second. However, reducing sampling frequency also presumably degrades perfusion maps, causing errors whose nature and severity are dependent on the imaging technique and postprocessing algorithm used. Such artifacts are beyond the scope of the current study but have been investigated by previous studies that used methodology similar to ours, in which the effects of various sampling frequencies were tested by acquiring a scan with a high sampling frequency and then simulating lower frequencies by selectively omitting some images.28–30
Conclusions
Our results show that truncation of data by short perfusion scans may introduce large errors in perfusion maps of patients with acute ischemic stroke. Our findings highlight the importance of considering the vulnerabilities of different algorithms to truncation errors when choosing among them and interpreting the results of published studies that have relied on perfusion imaging. Just as stroke researchers have recognized the importance of assessing postprocessing errors related to bolus arrival delay,31 our results show that truncation-related errors may be important as well. Testing perfusion postprocessing software for these and other shortcomings is more difficult when vendors of proprietary software do not disclose the details of their postprocessing algorithms. Therefore, our results also show the importance of transparency in revealing algorithms that may be used in clinical research or patient care.
ABBREVIATIONS:
- MRP
MR perfusion imaging
- SVD
singular value decomposition
- Tmax
time-to-maximum of the deconvolved tissue response function
Footnotes
Disclosures: Ona Wu—RELATED: Grant: National Institutes of Health/National Institute of Neurological Disorders and Stroke (R01NS059775)*; UNRELATED: Consultancy: Penumbra, Comments: I consulted on work unrelated to the topic of this article; Grants/Grants Pending: National Institutes of Health (P50NS051343, R01NS059775, R01NS082285);* Royalties: US Patent 7,512,435, March 31, 2009,* Comments: delay-compensated calculation of tissue blood flow. This patent has been licensed by GE Healthcare, Siemens, Imaging Biometrics, and Olea Medical. *Money paid to the institution.
This work was supported by National Institutes of Health grants P50NS051343, R01NS059775, and R01NS063925.
This research was presented in part at: “Perfusion MRI: Standardization, Beyond CBF and Everyday Clinical Applications,” conducted by the International Society for Magnetic Resonance in Medicine Scientific Workshop, October 11–14, 2012; Amsterdam, the Netherlands.
REFERENCES
- 1. Hacke W, Furlan AJ, Al-Rawi Y, et al. Intravenous desmoteplase in patients with acute ischaemic stroke selected by MRI perfusion-diffusion weighted imaging or perfusion CT (DIAS-2): a prospective, randomised, double-blind, placebo-controlled study. Lancet Neurol 2009;8:141–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Parsons M, Spratt N, Bivard A, et al. A randomized trial of tenecteplase versus alteplase for acute ischemic stroke. N Engl J Med 2012;366:1099–107 [DOI] [PubMed] [Google Scholar]
- 3. Ma H, Parsons MW, Christensen S, et al. A multicentre, randomized, double-blinded, placebo-controlled Phase III study to investigate EXtending the time for Thrombolysis in Emergency Neurological Deficits (EXTEND). Int J Stroke 2012;7:74–80 [DOI] [PubMed] [Google Scholar]
- 4. Kidwell CS, Jahan R, Gornbein J, et al. A trial of imaging selection and endovascular treatment for ischemic stroke. N Engl J Med 2013;368:914–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kudo K, Christensen S, Sasaki M, et al. Accuracy and reliability assessment of CT and MR perfusion analysis software using a digital phantom. Radiology 2013;267:201–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ho CY, Hussain S, Alam T, et al. Accuracy of CT cerebral perfusion in predicting infarct in the emergency department: lesion characterization on CT perfusion based on commercially available software. Emerg Radiol 2013;20:203–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Murphy BD, Fox AJ, Lee DH, et al. Identification of penumbra and infarct in acute ischemic stroke using computed tomography perfusion-derived blood flow and blood volume measurements. Stroke 2006;37:1771–77 [DOI] [PubMed] [Google Scholar]
- 8. Chiu FY, Kao YH, Teng MM, et al. Validation and absolute quantification of MR perfusion compared with CT perfusion in patients with unilateral cerebral arterial stenosis. Eur J Radiol 2012;81:4087–93 [DOI] [PubMed] [Google Scholar]
- 9. Calamante F, Gadian DG, Connelly A. Delay and dispersion effects in dynamic susceptibility contrast MRI: simulations using singular value decomposition. Magn Reson Med 2000;44:466–73 [DOI] [PubMed] [Google Scholar]
- 10. Carrera E, Jones PS, Iglesias S, et al. The vascular mean transit time: a surrogate for the penumbra flow threshold? J Cereb Blood Flow Metab 2011;31:1027–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schramm P, Schellinger PD, Klotz E, et al. Comparison of perfusion computed tomography and computed tomography angiography source images with perfusion-weighted imaging and diffusion-weighted imaging in patients with acute stroke of less than 6 hours' duration. Stroke 2004;35:1652–58 [DOI] [PubMed] [Google Scholar]
- 12. Deipolyi AR, Wu O, Macklin EA, et al. Reliability of cerebral blood volume maps as a substitute for diffusion-weighted imaging in acute ischemic stroke. J Magn Reson Imaging 2012;36:1083–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wintermark M, Flanders AE, Velthuis B, et al. Perfusion-CT assessment of infarct core and penumbra: receiver operating characteristic curve analysis in 130 patients suspected of acute hemispheric stroke. Stroke 2006;37:979–85 [DOI] [PubMed] [Google Scholar]
- 14. Kamalian S, Kamalian S, Maas MB, et al. CT cerebral blood flow maps optimally correlate with admission diffusion-weighted imaging in acute stroke but thresholds vary by postprocessing platform. Stroke 2011;42:1923–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lansberg MG, Straka M, Kemp S, et al. MRI profile and response to endovascular reperfusion after stroke (DEFUSE 2): a prospective cohort study. Lancet Neurol 2012;11:860–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reese TG, Heid O, Weisskoff RM, et al. Reduction of eddy-current-induced distortion in diffusion MRI using a twice-refocused spin echo. Magn Reson Med 2003;49:177–82 [DOI] [PubMed] [Google Scholar]
- 17. Collins DL, Neelin P, Peters TM, et al. Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J Comput Assist Tomogr 1994;18:192–205 [PubMed] [Google Scholar]
- 18. Rosen BR, Belliveau JW, Chien D. Perfusion imaging by nuclear magnetic resonance. Magn Reson Q 1989;5:263–81 [PubMed] [Google Scholar]
- 19. Carroll TJ, Rowley HA, Haughton VM. Automatic calculation of the arterial input function for cerebral perfusion imaging with MR imaging. Radiology 2003;227:593–600 [DOI] [PubMed] [Google Scholar]
- 20. Rempp KA, Brix G, Wenz F, et al. Quantification of regional cerebral blood flow and volume with dynamic susceptibility contrast-enhanced MR imaging. Radiology 1994;193:637–41 [DOI] [PubMed] [Google Scholar]
- 21. Mouridsen K, Christensen S, Gyldensted L, et al. Automatic selection of arterial input function using cluster analysis. Magn Reson Med 2006;55:524–31 [DOI] [PubMed] [Google Scholar]
- 22. Rausch M, Scheffler K, Rudin M, et al. Analysis of input functions from different arterial branches with gamma variate functions and cluster analysis for quantitative blood volume measurements. Magn Reson Imaging 2000;18:1235–43 [DOI] [PubMed] [Google Scholar]
- 23. Mlynash M, Eyngorn I, Bammer R, et al. Automated method for generating the arterial input function on perfusion-weighted MR imaging: validation in patients with stroke. AJNR Am J Neuroradiol 2005;26:1479–86 [PMC free article] [PubMed] [Google Scholar]
- 24. Østergaard L, Weisskoff RM, Chesler DA, et al. High resolution measurement of cerebral blood flow using intravascular tracer bolus passages. Part I: mathematical approach and statistical analysis. Magn Reson Med 1996;36:715–25 [DOI] [PubMed] [Google Scholar]
- 25. Wu O, Østergaard L, Weisskoff RM, et al. Tracer arrival timing-insensitive technique for estimating flow in MR perfusion-weighted imaging using singular value decomposition with a block-circulant deconvolution matrix. Magn Reson Med 2003;50:164–74 [DOI] [PubMed] [Google Scholar]
- 26. Sorensen AG, Copen WA, Østergaard L, et al. Hyperacute stroke: simultaneous measurement of relative cerebral blood volume, relative cerebral blood flow, and mean tissue transit time. Radiology 1999;210:519–27 [DOI] [PubMed] [Google Scholar]
- 27. CT Perfusion 4D User Guide. Revision 2. Milwaukee: GE Healthcare Publications; 2011 [Google Scholar]
- 28. Wintermark M, Smith WS, Ko NU, et al. Dynamic perfusion CT: optimizing the temporal resolution and contrast volume for calculation of perfusion CT parameters in stroke patients. AJNR Am J Neuroradiol 2004;25:720–29 [PMC free article] [PubMed] [Google Scholar]
- 29. Kamena A, Streitparth F, Grieser C, et al. Dynamic perfusion CT: optimizing the temporal resolution for the calculation of perfusion CT parameters in stroke patients. Eur J Radiol 2007;64:111–18 [DOI] [PubMed] [Google Scholar]
- 30. Abels B, Klotz E, Tomandl BF, et al. CT perfusion in acute ischemic stroke: a comparison of 2-second and 1-second temporal resolution. AJNR Am J Neuroradiol 2011;32:1632–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wintermark M, Albers GW, Alexandrov AV, et al. Acute stroke imaging research roadmap. Stroke 2008;39:1621–28 [DOI] [PubMed] [Google Scholar]