Abstract
BACKGROUND AND PURPOSE:
Robust, automated segmentation algorithms are required for quantitative analysis of large imaging datasets. We developed an automated method that identifies and labels brain tumor–associated pathology by using an iterative probabilistic voxel labeling using k-nearest neighbor and Gaussian mixture model classification. Our purpose was to develop a segmentation method which could be applied to a variety of imaging from The Cancer Imaging Archive.
MATERIALS AND METHODS:
Images from 2 sets of 15 randomly selected subjects with glioblastoma from The Cancer Imaging Archive were processed by using the automated algorithm. The algorithm-defined tumor volumes were compared with those segmented by trained operators by using the Dice similarity coefficient.
RESULTS:
Compared with operator volumes, algorithm-generated segmentations yielded mean Dice similarities of 0.92 ± 0.03 for contrast-enhancing volumes and 0.84 ± 0.09 for FLAIR hyperintensity volumes. These values compared favorably with the means of Dice similarity coefficients between the operator-defined segmentations: 0.92 ± 0.03 for contrast-enhancing volumes and 0.92 ± 0.05 for FLAIR hyperintensity volumes. Robust segmentations can be achieved when only postcontrast T1WI and FLAIR images are available.
CONCLUSIONS:
Iterative probabilistic voxel labeling defined tumor volumes that were highly consistent with operator-defined volumes. Application of this algorithm could facilitate quantitative assessment of neuroimaging from patients with glioblastoma for both research and clinical indications.
Glioblastoma is the most common primary brain tumor and remains one of the deadliest human cancers.1 During the past 50 years, improvement with regard to patient outcomes has been marginal.2 A major barrier in therapeutic development is attributable to the misconception that glioblastoma constitutes a single disease. Molecular profiling has revealed that glioblastoma comprises multiple subtypes characterized by distinct molecular pathways.3 To improve the clinical outcome of patients with glioblastoma, technologies must be developed to distinguish these subtypes.
There are compelling reasons that MR imaging may serve as a tool for dissecting the variability of glioblastoma. First, radiographic data are available for every patient because the clinical management of glioblastoma tumors is largely driven by the interpretation of MR images. Second, available data suggest that the radiographic appearance of glioblastoma is related to its physiologic state.4,5 To better define this relationship, imaging archives with corresponding genomic profiling, such as The Cancer Imaging Archive (TCIA), have been launched (http://cancerimagingarchive.net/).
Much of the early work correlating MR imaging appearances of glioblastoma tumors with genomic profiling was performed by using manually delineated tumor volumes or qualitative assessments provided by trained clinicians.4,5 These approaches are limited by the inherent variability of subjective interpretation, and significant interrater discrepancies have been reported.6,7 Additionally, manual segmentation is time-consuming for large datasets. This limitation is particularly apparent when multiple radiographic features require segmentation. To address these deficiencies, effort has been devoted to developing automated algorithms for segmenting tumor volumes.8–12 These algorithms include clustering,13,14 discriminative strategies,15 and generative approaches.11,16,17 The success of these methods has been limited by widely differing MR imaging protocols for image acquisition and quality18 and the significant overlap between the radiographic appearance of glioblastoma tumors and normal cerebrum on MR imaging. Although many of these methods can generate high-quality volumes from a training set, segmentation algorithms may fail when applied to images acquired by using different protocols.
We hypothesized that a probabilistic approach by using subject-specific classifiers would reliably discriminate glioblastoma from the surrounding cerebrum. In this algorithm, termed iterative probabilistic voxel labeling (IPVL), sparse, high-specificity, preliminary volumes were created for each subject by using a combination of region-growing and K-means-based tissue segmentation. Sampling of these preliminary volumes trained k-nearest neighbor (KNN) and Gaussian mixture model (GMM) classifiers by using voxel intensity and spatial coordinates. Voxel labels are assigned probabilistically by iteratively trained classifiers. Finally, each voxel is labeled as contrast-enhancing tumor volume (CEV), FLAIR hyperintensity volume (FHV), gray matter, white matter, CSF, and blood vessel (BV). Most important, our algorithm reliably segments images from the TCIA that were acquired by a variety of scanners and protocols.
Materials and Methods
The Cancer Imaging Archive
MR images of glioblastoma tumors from The Cancer Imaging Archive were downloaded in June 2013. We identified subjects who underwent MR imaging before surgery and had a full complement of imaging, including the following: T1-weighted imaging, T1-weighted imaging with contrast enhancement (T1wCE), T2-weighted imaging, and FLAIR. Subjects were excluded when images contained a prohibitive amount of motion or distortion artifacts. Our algorithm was developed in a “pilot” set of 10 subjects from the TCIA. The algorithm was tested in 2 sets of 15 subjects selected from the TCIA that were not used during development. TCIA MR images were acquired from a number of institutions whose scanners differed by manufacturer and model and whose images varied by sequence, quality, and spatial resolution. (On-line Tables 1 and 2).
Preprocessing
Images were preprocessed by using a combination of in-house and external software including the FMRIB Software Library19,20 (FSL, Version 5.0; http://fsl.fmrib.ox.ac.uk/fsl). Image distortions caused by gradient nonlinearity warping were corrected by using previous methods,21–23 followed by bias-field correction by using the FMRIB Automated Segmentation Tool (FAST),24 and were registered to the Montreal Neurological Institute-152 nonlinear sixth-generation standard brain image.25 Affine registration was performed by using the FMRIB Linear Image Registration Tool (FLIRT).26,27 To ensure removal of nonbrain tissues (eg, skull, optic nerve, and carotid arteries), we created a stringent brain mask from the T1WI by using a modified combination of the FSL Brain Extraction Tool24 and the Robust Brain Extraction tool (https://www.nitrc.org/projects/robex).28 Briefly, this method automatically compared the resultant brain with the volume derived from applying the Montreal Neurological Institute brain mask. Overestimation of >10% would adjust the fractional intensity, resulting in more restrictive brain outlines.
Preliminary Segmentation
It was crucial to generate highly specific volumes that accurately represented the range of intensity and spatial distribution for each tissue label to appropriately train our segmentation algorithm to recognize each subject's features. After skull stripping, initial tissue segmentation into preliminary WM, GM, CSF, and CEVs was performed for the available T1WI sequences by using FAST.24 The FAST-derived initial CEV consisted of both tumor-associated CEV and BV volumes. The preliminary BV volume was distinguished from the FAST-derived CEV by performing 2 morphology-based manipulations. First, CEV objects located near the cortical surface were selectively removed by using a uniform spheric 3-mm erosion of the brain mask applied to the FAST-derived CEVs. Large vessels, such as the dural veins and carotid arteries, were removed by this operation due to their proximity to the brain surface. Second, a modified region-growing algorithm was used to identify vessels that were continuous with the venous sinuses. Region-growing was seeded in the region of the torcula (confluence of the sinuses), which was identified on the template image, to which all images were registered. Voxels identified as vessels by the combination of these methods were labeled as preliminary BV volume, while the remaining CEV was assigned to preliminary tumor-associated CEV.
The FHV preliminary volume was created by first determining and applying an automatic threshold for the FLAIR image by using the Otsu method.29 FLAIR hyperintense regions on MR imaging may be tumor-associated or non-tumor-associated (eg, periventricular or pericortical). The non-tumor-associated hyperintense elements were excluded by using a spheric 3-mm erosion performed on the brain mask, while spheric 3-mm dilation was performed on the CSF volume. Together, these operations removed pericortical hyperintensities and the periventricular hyperintensities from the remainder of the preliminary FHVs.
Approximately 25% of voxels were labeled at this time. The voxels labeled were randomly sampled from regions that had the highest specificity to a particular volume of interest. For contrast enhancement, these included regions of contrast enhancement not continuous with the sagittal and transverse sinus. For FLAIR hyperintensity, these included regions above an intensity threshold >1.5 SDs above the mean intensity of the FLAIR image. The voxels assigned to each preliminary tissue label were used as the basis for training probabilistic classifiers (KNN and GMM). Voxels that were not classified into these categories during preliminary volume segmentation remained unassigned to avoid adding noise to the classifiers.
K-Nearest Neighbor and Gaussian Mixture Model Classifiers
The classifiers used to assign voxel membership were KNN and GMM. The KNN algorithm is a nonparametric method that assigns membership of a single datum on the basis of a number of neighboring training examples.30,31 GMM allows statistical clustering of data into a predefined number of Gaussian distributions, which, in this case, represent distinct imaging features. Use of these 2 probabilistic classifiers was complementary.
To expedite processing and improve accuracy, we used a weighted random sampling of the preliminary volume voxels to train both KNN and GMM classifiers. The weights for sampling reflected the relative distribution of voxels assigned to tissue labels from preliminary segmentation. Weighting was performed to avoid biasing the classifiers toward any particular tissue label caused by overrepresentation attributed to sampling error. Training was performed on a subject-by-subject basis, meaning that each patient was segmented according to his or her own subject-specific classifier by using both intensity and spatial data from each voxel to define labels. After training, all voxels, including those that were unassigned during preliminary volume creation, were classified independently by both KNN and GMM probabilistically to the 6 tissue labels: CEV, FHV, CSF, GM, WM, and BV. The probability of membership for each voxel was determined by a distance metric from classifier training. For each voxel, the greatest tissue label probability determined voxel labeling. Classifier consensus was resampled and used to re-train another iteration of KNN classification at a higher voxel sampling rate. This step had the benefit of reducing noise introduced during the creation of preliminary volumes, improving both the smoothness of the final volumes and the accuracy of the tissue labels.
Final Segmentation
Voxel label probabilities from all classifiers were summed, including the iterative KNN classification, for each tissue label, and a final segmentation volume was created by assigning voxels according to the highest probability membership to each tissue label. At this time, all voxels were probabilistically assigned. A voxel continuity filter was applied that removed discontinuous clusters of limited connectivity (fewer than 150 contiguous voxels). To address voxels that had equal probabilities of belonging to ≥2 tissue labels, we set priority in the following manner from greatest to least: CEV > FHV > BV to ensure that individual voxel tissue labels were mutually exclusive. This order was determined by the confidence of labeling each feature.
Segmentation Evaluation
To assess segmentation quality, we drew CEVs and FHVs manually for 2 sets of 15 subjects selected randomly from the available pool downloaded from the TCIA. These volumes were completed by 2 independent trained operators under the supervision of a neuroradiologist (N.F.) and a neurosurgeon (C.C.C.). Manual delineation of tumor-associated volumes was performed by using the software program AMIRA (http://www.vsg3d.com/amira/overview), using threshold-assisted segmentation on whole-brain T1wCE and FLAIR images that were registered to the Montreal Neurological Institute template. Operator-derived volumes were compared with IPVL-derived volumes by using the Dice similarity coefficient (DICE). This coefficient assesses the similarity between 2 volumes by dividing twice the sum of the intersection by the sum of both volumes.32 Interoperator similarity was also compared by using this metric. A DICE equal to 1 would imply perfect similarity and overlap of 2 volumes.
Minimum Image Requirement for Adequate Segmentation
To assess the performance of our method when fewer imaging sequences were available for input, we implemented IPVL on a group of 15 subjects multiple times while removing ≥1 image, recapitulating common image combinations seen within the TCIA subjects. The segmentations that resulted from these image combinations were compared with operator volumes to determine their DICE similarities. FHVs were not segmented in image combinations that lacked FLAIR sequences.
Results
Overview of the Automated Segmentation Algorithm
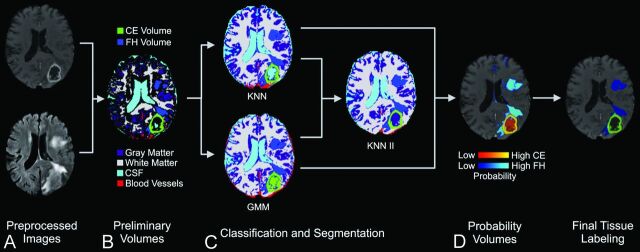
The steps of our segmentation protocol, applied to a representative case, are illustrated in Fig 1. Generally, our segmentation work flow was divided into 5 stages: preprocessing, preliminary segmentation, classification, probability labeling, and final segmentation. In preprocessing, images were loaded. Bias field correction, skull stripping, and registration to the template were then performed. The results of preprocessing created images in standard space to provide input for preliminary segmentation. Preliminary segmentation assigned voxel labels to CEV, FHV, BV, CSF, GM, WM, and unassigned (for voxels with ambiguous membership) by using k-means-based tissue segmentation and a region-growing algorithm. During classification, voxels sampled from these preliminary labels were used to train the GMM and KNN classifiers. All voxels were then classified to independent labels to identify CEV, FHV, BV, CSF, GM, and WM volumes. During probability labeling, each voxel was assigned a probability of membership to each tissue label. In the last step, final segmentation, voxels were labeled according to their greatest probability, and a voxel continuity filter was applied to eliminate clusters of <150 continuous voxels. The average time required to complete segmentation was 11.12 ± 5.63 minutes.
Fig 1.
Work flow for iterative probabilistic voxel labeling. A, Downloaded TCIA images were preprocessed. B, Preliminary segmentation was performed to generate conservative yet highly specific preliminary volumes. C, In the classification step, these volumes were used to train the GMM and KNN probabilistic classifiers. The consensus of KNN and GMM classification was resampled and used to train a new classifier (KNN II), which assigned voxel tissue labels. The classifiers integrated their respective outputs to generate tissue-specific probability volumes. D, The voxels were assigned on the basis of their greatest probability of membership to a tissue label, and a voxel continuity filter was applied to eliminate clusters of less than 150 continuous voxels.
Manual Segmentation Comparison
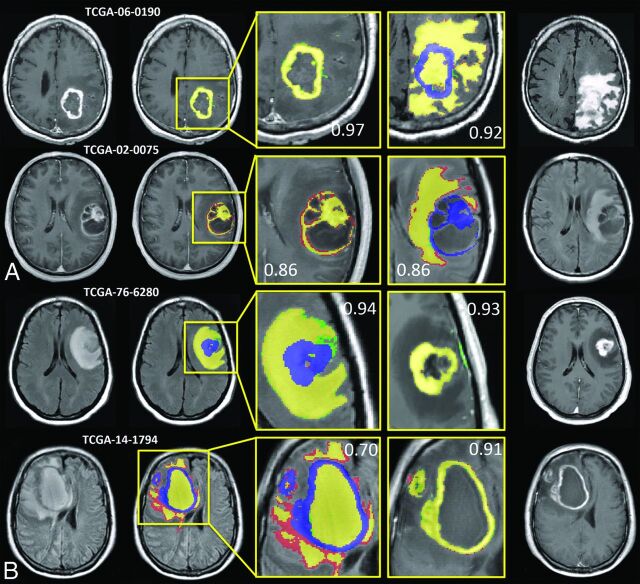
Examples from 4 subjects that represented the CEVs and FHVs with the highest and lowest DICE scores relative to operator 1 are shown in Fig 2. Corresponding FHV segmentations for CEV and CEV segmentations for FHV are included to show that segmentation success for 1 feature is not necessarily correlated with segmentation success for corresponding features. Analysis showed no statistical difference among operator-derived volumes, so operator 1 was selected as the basis for image comparison (P = .72 for CEV interoperator, and P = .39 for FHV interoperator). Figure 2 demonstrates that the algorithm generates highly analogous CEVs and FHVs relative to those derived manually.
Fig 2.
IPVL segment volumes that are highly analogous to operator-defined volumes. Results from 4 subjects representing the highest and lowest Dice similarity coefficient scores for CEV and FHV segmentations are shown. A, The highest DICE (top) and lowest DICE (bottom) examples of IPVL-segmented CEVs relative to operator-defined volumes are shown. The corresponding FHV segmentation results are shown (right) to demonstrate that CEV segmentations are independent of FHV segmentations. B, The highest DICE (top) and lowest DICE (bottom) examples of IPVL-segmented FHV relative to operator-defined volumes are shown. The corresponding CEV segmentation results are shown as well (right) to demonstrate that FHV segmentations are independent of CEV segmentations. Yellow indicates regions of intersection between operator and IPVL-defined volumes; red, operator-defined volume only; green, IPVL-defined volume only. Corresponding CEV segmentations are overlaid in blue on FLAIR images for clarity.
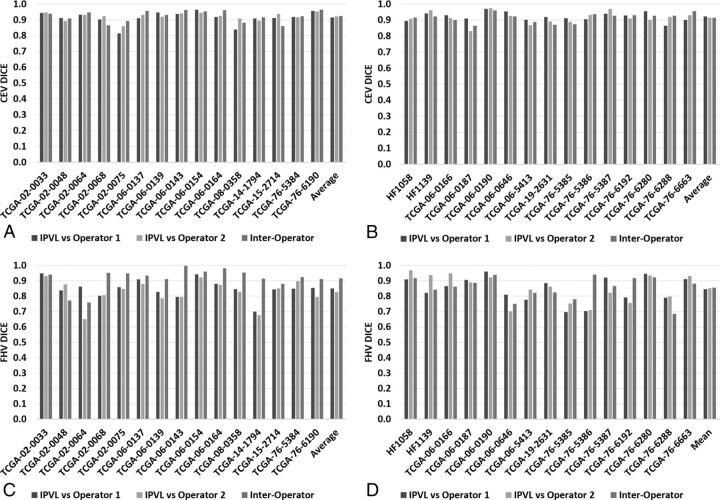
IPVL CEVs were statistically indistinguishable from volumes generated by expert operators across all subjects (P = .93). DICE scores, for automated CEVs, relative to operators 1 and 2, averaged 0.923 and 0.921, respectively. These DICE scores were highly comparable with those obtained from interoperator analysis (average of 0.923, Fig 3A). For automated FHVs, the DICE scores relative to operators 1 and 2 averaged 0.851 and 0.827, respectively. DICE scores obtained from interoperator analysis averaged 0.905 (Fig 3B). Analysis revealed that FHVs were slightly lower than interoperator comparison (P = .04). We observed that FHV DICE scores were poorer than CEV DICE scores for both the interoperator and the operator-algorithm comparisons. Overall, the DICE scores for both CEV and FHV achieved through our algorithm were improved or similar relative to those previously reported.14–20
Fig 3.
Quantitative comparison between IPVL-defined volumes and operator-derived volumes compared with interoperator comparisons. A and B, DICE comparisons for 2 sets of IPVL-defined and operator-defined CEVs are shown. DICE scores were calculated comparing CEVs generated by IPVL, operator 1, and operator 2. C and D, DICE score comparisons for IPVL-defined and operator-defined FHVs. DICE scores were calculated comparing FHVs generated by IPVL, operator 1, and operator 2.
To ensure that these results were generalizable, we randomly selected 15 additional subjects for analysis. The results from this analysis are highly comparable with those reported above. DICE scores for automated CEVs relative to operators 1 and 2 averaged 0.921 and 0.901, respectively. These DICE scores were highly comparable with those obtained from interoperator analysis (DICE = 0.905). For automated FHVs, the DICE scores relative to operators 1 and 2 averaged 0.846 and 0.823, respectively. DICE scores obtained from interoperator analysis averaged 0.812.
It was possible that difficult cases, including tumors with multifocal patterns, or tumors with attachment to large vessels or the brain surface, may cause errors in automatic segmentation. Of the images analyzed, 2 glioblastomas (The Cancer Genome Atlas [TCGA]-06–0139, TCGA-06–0166) were multifocal. For these subjects, IPVL-defined CEV and FHV showed mean DICE scores of 0.94 (range, 0.92–0.95) and 0.92 (range, 0.91–0.93) relative to expert defined volumes, respectively. Seven tumors (TCGA-02–0048, TCGA-06–0164, TCGA-08–0358, TCGA-76–6280, TCGA-76–6192, TCGA-76–5386, and HF1139) were located on the surface of the cerebrum. For these tumors, IPVL-defined CEV and FHV showed mean DICE scores of 0.92 (range, 0.84–0.96) and 0.84 (range, 0.70–0.95). One tumor (TCGA-76–5385) was attached to a major vessel (the MCA). For this tumor, IPVL-defined CEV and FHV showed mean DICE scores of 0.90 (range, 0.89–0.91) and 0.72 (range, 0.70–0.75). These results suggest that our algorithm performs adequately in anatomic locations and in difficult cases that are historically challenging to previously published algorithms.
Minimal Image Requirement for Adequate Segmentation
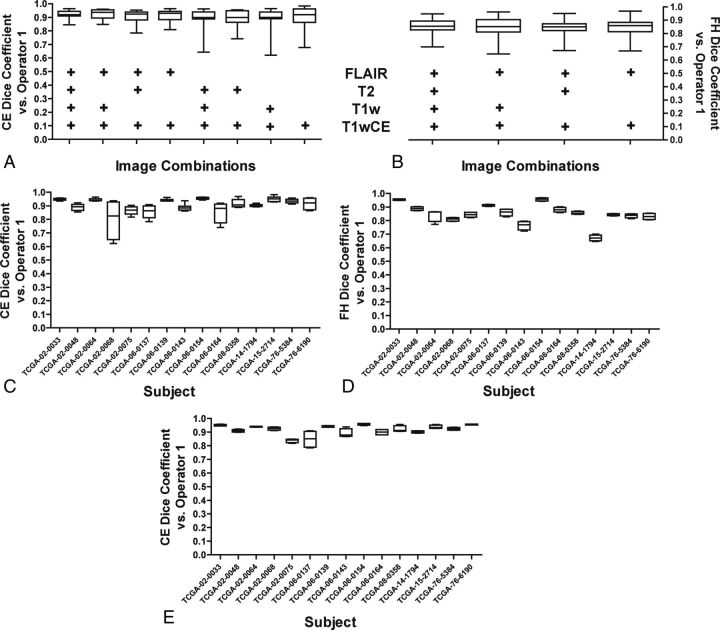
The TCIA and other image databases include many subjects who do not have the full complement of T1WI, T1wCE, T2WI, and FLAIR images. In the TCIA, this full set of imaging was available in only 52% of subjects. Therefore, it was of interest to determine how our algorithm would perform when limited imaging modalities were available. To this end, we examined how the sequential removal of the various image sequences impacted segmentation performance. DICE scores were determined for a subject's segmentations by using each combination of images relative to operator-defined volumes. These DICE scores were plotted compared with the DICE scores derived from segmentations by using all 4 imaging sequences.
For CEV segmentations, removal of T1WI and T2WI did not significantly affect performance. The DICE scores obtained when comparing volumes delineated by using only T1wCE and FLAIR were comparable with those obtained when all 4 imaging sequences were processed by our algorithm (Fig 4A). Similarly, FHV segmentations were minimally impacted by image reduction, and DICE scores by using all 4 imaging sequences were comparable with those obtained when using only T1wCE and FLAIR (Fig 4B).
Fig 4.
Effects of removing image sequences on IPVL segmentation. A, Select image sequences (such as T1WI) were removed before IPVL CEV segmentations for each subject. The image sequences that were available during IPVL segmentation are indicated by a plus sign. DICE scores were calculated for the resultant CEVs relative to operator 1–defined CEVs. The distribution of DICE scores across all subjects because of image sequence removal is shown as a boxplot. B, Select image sequences were removed before IPVL FHV segmentations. DICE scores were calculated for the resultant FHV segmentations relative to operator 1–defined FHVs. The distribution of DICE scores across all subjects due to image-sequence removal is shown as a boxplot. C, A boxplot demonstrates the range of DICE scores for IPVL-segmented CEVs relative to operator-defined CEVs per patient for all image combinations tested. D, A boxplot demonstrates the range of DICE scores for IPVL-segmented FHVs relative to operator-defined FHVs per patient for all image combinations tested. E, A boxplot demonstrates the range of DICE scores for IPVL-segmented CEVs relative to operator-defined CEVs per patient when only T1WI and FLAIR source images were used for IPVL segmentation.
To further characterize the impact of reducing the number of image sequences on the performance of CEV and FHV segmentations, we also plotted the range of DICE scores that resulted from removing ≥1 image series for each subject. For most subjects, removing images minimally impacted DICE scores—that is, the segmentation quality was not significantly altered (Fig 4C for CEV, Fig 4D for FHV). For FHV segmentations, removal of T2WI and T1WI did not significantly alter segmentation performance (Fig 4D for FHV).
Only 2 subjects, TCGA-02–0068 and TCGA-06–0164, had increased vessel contamination of the CEVs when FLAIR images were removed during image-reduction analysis. CEV segmentations for image combinations that contained at least a FLAIR and T1wCE image were highly comparable across all subjects (Fig 4E). While adequate CEV segmentation required only T1wCE for most cases, FLAIR images improved CEV and BV discrimination and may be required for segmentation in a subset of subjects. Most surprising, CEV segmentation for T1wCE and FLAIR alone was nearly identical to CEV segmentation results that used all available imaging series. These results suggest that our algorithm requires only T1wCE and FLAIR images for robust volume segmentation of both tumor-associated CEVs and FHVs.
Discussion
The success of any automated segmentation process hinges on the a priori definition of the features that constitute a volume of interest. Human vision can integrate visual data along with both experience and assumption to distinguish and classify independent features. This task is often challenging for a computer because the cross-section of data available to the computer is often simplified. We hypothesized that the complexity of image segmentation could be largely recapitulated by using iterating probabilistic classifiers trained on sparse subject-specific preliminary features. To test this hypothesis, we predefined the voxels that were most likely associated with the features of interest to generate preliminary volumes. We then used k-nearest neighbor and GMM probabilistic classifiers to refine the segmentation process. In our algorithm, these complementary probabilistic classifiers were integrated in an iterative manner to converge on a segmentation result for the various features of an MR image. Our results demonstrate that IPVL image segmentation is highly comparable with segmentations that were drawn manually.
Most important, the time required for segmentation per subject averaged 11.2 minutes when all 4 image sequences were used. In contrast, manual segmentation by experts required 1–3 hours depending on the size and complexity of the volume to be segmented. As such, our method presents an opportunity for high-throughput quantitative analysis of TCIA images and other imaging databases. The insensitivity of our algorithm to interinstitutional methodologic differences in MR imaging supports its utility for this application. Further supporting the utility of our algorithm, we demonstrated that only 2 image sequences (the T1wCE and FLAIR images) are needed for reliable segmentation of tumor CEVs and FHVs. Finally, using a common template space will provide a platform for future analyses, intersubject comparisons, and longitudinal studies.
While our study was focused on the development of an algorithm for research use in terms of radiographic biomarker discovery, reliable volume segmentation by using our algorithm may also impact clinical practice. For example, it is often difficult to detect subtle differences in the radiographic appearance of a tumor during disease progression. As a result, changes in serial MR imaging may be underappreciated until a patient becomes symptomatic. Automated segmentation and longitudinal quantitative comparison may help facilitate the detection of subtle radiographic changes, such as tumor progression, thereby allowing clinicians to perform procedures to prevent clinical deterioration in select patients. Application of these methods may also aid the evaluation of the therapeutic response in clinical trials.
Careful study of the discrepancies between the volumes generated by IPVL and expert-defined volumes revealed a few limitations. The algorithm can fail to detect tumor contrast enhancement or FLAIR hyperintensity in regions of these volumes that fall below a single voxel (∼1 mm). This limitation, a result of voxel sampling and partial volume effect, could be mitigated with higher resolution imaging. In a few subjects (eg, TCGA-02–0068, TCGA-06–0154), reliable delineation of BV volume from tumor CEVs remained challenging when image combinations lacking FLAIR images were used for segmentation, leading us to conclude that FLAIR and T1wCE are required for our method. Segmentation of FLAIR volumes remains a challenge, but this challenge is shared by the human eye as demonstrated by the interobserver discrepancies reported previously. The use of higher order image processing, such as textural analysis, may facilitate the improvement of our algorithm in the near future.
Conclusions
We demonstrate that iterative probabilistic voxel labeling is a reliable and robust tool for automatic segmentation of MR images in the TCIA dataset. Application of this method could facilitate quantitative radiographic assessment of glioblastoma for both researchers and clinicians alike.
Supplementary Material
ABBREVIATIONS:
- BV
blood vessel
- CEV
contrast-enhancing volume
- DICE
Dice similarity coefficient
- FHV
FLAIR hyperintensity volume
- GMM
Gaussian mixture modeling
- IPVL
iterative probabilistic voxel labeling
- KNN
k-nearest neighbor
- T1wCE
T1WI with contrast enhancement
- TCIA
The Cancer Imaging Archive
- TCGA
The Cancer Genome Atlas
Footnotes
Disclosures: Anders M. Dale—RELATED: Grant: National Institutes of Health,* Department of Defense,* National Science Foundation*; UNRELATED: Grants/Grants Pending: National Institutes of Health,* Department of Defense,* National Science Foundation*; Patents (planned, pending or issued): University of California, San Diego,* CorTechs,* Labs Inc*; Stock/Stock Options: CorTechs, Labs Inc, Human Longevity Inc, Comments: I hold equity in CorTechs Labs Inc and serve on its Scientific Advisory Board. I am also a member of the Scientific Advisory Board of Human Longevity Inc; the terms of these arrangements have been reviewed and approved by the University of California, San Diego, in accordance with its conflict of interest policies; Other: GE Healthcare,* Medtronic,* Comments: I receive support from GE Healthcare and Medtronic under Research Agreements with the University of California, San Diego. *Money paid to the institution.
C.C.C. is supported by the Doris Duke Charitable Foundation Clinical Scientist Development Award, Sontag Foundation Distinguished Scientist Award, Burroughs Wellcome Fund Career Awards for Medical Scientists, Kimmel Scholar award, a Discovery Grant from the American Brain Tumor Association, and a National Cancer Institute K12 award.
REFERENCES
- 1. Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA 2013;310:1842–50 [DOI] [PubMed] [Google Scholar]
- 2. Minniti G, Muni R, Lanzetta G, et al. Chemotherapy for glioblastoma: current treatment and future perspectives for cytotoxic and targeted agents. Anticancer Res 2009;29:5171–84 [PubMed] [Google Scholar]
- 3. Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell 2013;155:462–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Diehn M, Nardini C, Wang DS, et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci U S A 2008;105:5213–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gutman DA, Cooper LA, Hwang SN, et al. MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology 2013;267:560–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deeley MA, Chen A, Datteri R, et al. Comparison of manual and automatic segmentation methods for brain structures in the presence of space-occupying lesions: a multi-expert study. Phys Med Boil 2011;56:4557–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weltens C, Menten J, Feron M, et al. Interobserver variations in gross tumor volume delineation of brain tumors on computed tomography and impact of magnetic resonance imaging. Radiother Oncol 2001;60:49–59 [DOI] [PubMed] [Google Scholar]
- 8. Zhu Y, Young GS, Xue Z, et al. Semi-automatic segmentation software for quantitative clinical brain glioblastoma evaluation. Acad Radiol 2012;19:977–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Diaz I, Boulanger P, Greiner R, et al. An automatic brain tumor segmentation tool. Conf Proc IEEE Eng Med Biol Soc 2013;2013:3339–42 [DOI] [PubMed] [Google Scholar]
- 10. Harati V, Khayati R, Farzan A. Fully automated tumor segmentation based on improved fuzzy connectedness algorithm in brain MR images. Comput Boil Med 2011;41:483–92 [DOI] [PubMed] [Google Scholar]
- 11. Prastawa M, Bullitt E, Moon N, et al. Automatic brain tumor segmentation by subject specific modification of atlas priors. Acad Radiol 2003;10:1341–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zikic D, Glocker B, Konukoglu E, et al. Decision forests for tissue-specific segmentation of high-grade gliomas in multi-channel MR. Med Image Comput Comput Assist Interv 2012;15:369–76 [DOI] [PubMed] [Google Scholar]
- 13. Phillips WE 2nd, Phuphanich S, Velthuizen RP, et al. Automatic magnetic resonance tissue characterization for three-dimensional magnetic resonance imaging of the brain. J Neuroimaging 1995;5:171–77 [DOI] [PubMed] [Google Scholar]
- 14. Clark MC, Hall LO, Goldgof DB, et al. Automatic tumor segmentation using knowledge-based techniques. IEEE Trans Medi Imaging 1998;17:187–201 [DOI] [PubMed] [Google Scholar]
- 15. Lee CH, Wang S, Murtha A, et al. Segmenting brain tumors using pseudo-conditional random fields. Med Image Comput Comput Assist Interv 2008;11(pt 1):359–66 [PubMed] [Google Scholar]
- 16. Prastawa M, Bullitt E, Ho S, et al. A brain tumor segmentation framework based on outlier detection. Med Image Anal 2004;8:275–83 [DOI] [PubMed] [Google Scholar]
- 17. Menze B, van Leemput K, Lashkari D, et al. A generative model for brain tumor segmentation in multi-modal images. In: Jiang T, Navab N, Pluim JW, et al., eds. Medical Image Computing and Computer-Assisted Intervention: MICCAI 2010. Berlin: Springer-Verlag Berlin; 2010:151–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gordillo N, Montseny E, Sobrevilla P. State of the art survey on MRI brain tumor segmentation. Magn Reson Imaging 2013;31:1426–38 [DOI] [PubMed] [Google Scholar]
- 19. Jenkinson M, Beckmann CF, Behrens TE, et al. FSL. Neuroimage 2012;62:782–90 [DOI] [PubMed] [Google Scholar]
- 20. Smith SM, Jenkinson M, Woolrich MW, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 2004;23(suppl 1):S208–19 [DOI] [PubMed] [Google Scholar]
- 21. Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage 1999;9:179–94 [DOI] [PubMed] [Google Scholar]
- 22. Jovicich J, Czanner S, Greve D, et al. Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. Neuroimage 2006;30:436–43 [DOI] [PubMed] [Google Scholar]
- 23. Holland D, Kuperman JM, Dale AM. Efficient correction of inhomogeneous static magnetic field-induced distortion in echo planar imaging. Neuroimage 2010;50:175–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smith SM. Fast robust automated brain extraction. Hum Brain Mapp 2002;17:143–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grabner G, Janke AL, Budge MM, et al. Symmetric atlasing and model based segmentation: an application to the hippocampus in older adults. Med Image Comput Comput Assist Interv 2006;9(pt 2):58–66 [DOI] [PubMed] [Google Scholar]
- 26. Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal 2001;5:143–56 [DOI] [PubMed] [Google Scholar]
- 27. Jenkinson M, Bannister P, Brady M, et al. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 2002;17:825–41 [DOI] [PubMed] [Google Scholar]
- 28. Iglesias JE, Liu CY, Thompson PM, et al. Robust brain extraction across datasets and comparison with publicly available methods. IEEE Trans Med Imaging 2011;30:1617–34 [DOI] [PubMed] [Google Scholar]
- 29. Otsu N. A threshold selection method from gray-level histograms. IEEE Transactions on Systems, Man, and Cybernetics 1979;9:62–66 [Google Scholar]
- 30. Steenwijk MD, Pouwels PJ, Daams M, et al. Accurate white matter lesion segmentation by k nearest neighbor classification with tissue type priors (kNN-TTPs). Neuroimage Clin 2013;3:462–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Anbeek P, Vincken KL, van Bochove GS, et al. Probabilistic segmentation of brain tissue in MR imaging. Neuroimage 2005;27:795–804 [DOI] [PubMed] [Google Scholar]
- 32. Dice LR. Measures of the amount of ecologic association between species. Ecology 1945;26:297–302 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.