The authors evaluated and validated a semiautomated software platform to facilitate detection of new lesions and improved MS lesions. Two neuroradiologists retrospectively assessed 161 MR imaging comparison study pairs acquired between 2009 and 2011. More comparison study pairs with new lesions and improved lesions were recorded by using the software compared with original radiology reports.
Abstract
BACKGROUND AND PURPOSE:
Treating MS with disease-modifying drugs relies on accurate MR imaging follow-up to determine the treatment effect. We aimed to develop and validate a semiautomated software platform to facilitate detection of new lesions and improved lesions.
MATERIALS AND METHODS:
We developed VisTarsier to assist manual comparison of volumetric FLAIR sequences by using interstudy registration, resectioning, and color-map overlays that highlight new lesions and improved lesions. Using the software, 2 neuroradiologists retrospectively assessed MR imaging MS comparison study pairs acquired between 2009 and 2011 (161 comparison study pairs met the study inclusion criteria). Lesion detection and reading times were recorded. We tested inter- and intraobserver agreement and comparison with original clinical reports. Feedback was obtained from referring neurologists to assess the potential clinical impact.
RESULTS:
More comparison study pairs with new lesions (reader 1, n = 60; reader 2, n = 62) and improved lesions (reader 1, n = 28; reader 2, n = 39) were recorded by using the software compared with original radiology reports (new lesions, n = 20; improved lesions, n = 5); the difference reached statistical significance (P < .001). Interobserver lesion number agreement was substantial (≥1 new lesion: κ = 0.87; 95% CI, 0.79–0.95; ≥1 improved lesion: κ = 0.72; 95% CI, 0.59–0.85), and overall interobserver lesion number correlation was good (Spearman ρ: new lesion = 0.910, improved lesion = 0.774). Intraobserver agreement was very good (new lesion: κ = 1.0, improved lesion: κ = 0.94; 95% CI, 0.82–1.00). Mean reporting times were <3 minutes. Neurologists indicated retrospective management alterations in 79% of comparative study pairs with newly detected lesion changes.
CONCLUSIONS:
Using software that highlights changes between study pairs can improve lesion detection. Neurologist feedback indicated a likely impact on management.
Multiple sclerosis affects approximately 2 million people worldwide, predominantly young adults.1 During the past decade, a number of novel disease-modifying drugs have emerged that are effective during the early phases of the disease; reducing the frequency of relapses, potentially halting disease progression, and even reversing early neurologic deficits.2 This choice in therapeutic options allows treating neurologists to alter management strategies when progression is detected.2
Because most demyelinating events are asymptomatic, MR imaging has been the primary biomarker for disease progression, and both physical disability and cognitive function have been shown to have a nonplateauing association with white matter demyelinating lesion burden, as seen on FLAIR and T2-weighted sequences.2–6
Recent advances in imaging, including 3T 3D volumetric T2 FLAIR sequences, allow better resolution of small demyelinating lesions, resulting in better clinicoradiologic correlation.7,8 Despite advances in imaging techniques, conventional side-by-side comparison (CSSC) is often subject to a reader's expertise.9 The sensitivity of detecting new lesions is also likely to be reduced when the section number is increased and scan planes are unmatched; however, to our knowledge, this reduction has not yet been investigated. In an attempt to facilitate accurate lesion-load and lesion-volume detection, much research has been devoted to fully automated computational approaches with unsatisfactory results. Robust lesion segmentation has been identified as a critical obstacle to widespread clinical adoption for several reasons: difficulties specific to MS, problems inherent to segmentation, and data variability.10
A review of fully automated MS segmentation techniques concluded that basic data-driven methods are inherently inaccurate; supervised learning methods (such as artificial neural networks) require costly and extensive training on representative data; deformable models are better; and statistical models are most promising, though these also require training on representative data.3
An alternative to total automation is to assist manual reporting with partial automation. A few semiautomated lesion-subtraction strategies have been used in the research setting on small patient populations with good lesion detection and interreader correlation.11,12
Semiautomation without segmentation is inherently easier, more robust, and less affected by data variability because the lesion count is judged manually. The software can present a number of false-positives without a negative impact on accuracy.
Our aim was to design a nonsegmentation semiautomated assistive software platform that can be integrated into vendor-agnostic PACS and validated by application to a large number of existing routine clinical scans in patients with an established diagnosis of multiples sclerosis. The approach is to merely draw the attention of the radiologist to potentially new or improved lesions rather than automate the entire process, thus preserving the expertise of neuroradiologists in determining whether a finding is real.
Our hypothesis was that CSSCs of volumetric FLAIR studies in patients with MS were prone to false-negative errors in the perception of both new and improved lesions and that more lesions would be identified by using the assistive software with improved inter- and intrareader reliability. Secondly, we hypothesized that presenting this information to clinicians would likely have changed patient management.
Materials and Methods
Software Development
Detecting lesion change in studies obtained at 2 time points, “old” and “new,” requires numerous steps including the following: 1) brain-surface extraction and masking (to remove skull and soft tissues of the head and neck), 2) coregistration and resectioning (to accurately align the 2 scans in all axes), 3) normalization of the FLAIR signal intensity (to remove global signal differences), and 4) calculating the difference in signal intensity between the old and new study at each point (to identify new T2 bright plaques and previously abnormal areas that have regained normal white matter signal). Changes between scans were presented as a color map superimposed on conventional FLAIR sequences (Fig 1). This was accomplished by bespoke code (given the trademark VisTarsier, henceforth VTS or “the software”) with the inclusion of a number of open-source components (Fig 2).
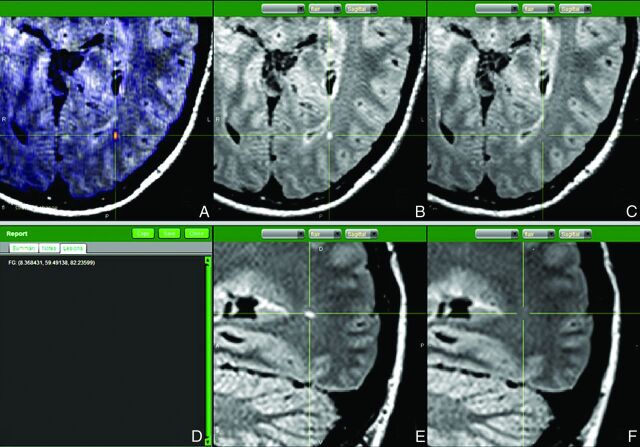
Fig 1.
Annotated capture of the software reporting screen. A, Axial FLAIR with superimposed change map shows the new occipital white matter lesion in orange. Coregistered and resectioned FLAIR sequences comparing axial of new study (B) with axial of old study (C); and sagittal of new study (E) with sagittal old study (F)—thus confirming that the lesion is real and consistent with a new demyelinating plaque. D, Each lesion is marked with 3D coordinates.
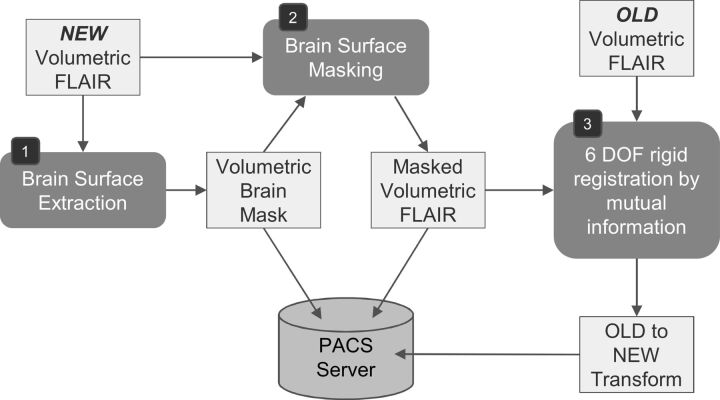
Fig 2.
Preprocessing for change-detection on receipt of a new study. A pair of old and new studies are required, each containing a volumetric series used for change detection. In our case, this series uses the FLAIR protocol. Due to significant deformation in soft tissues outside the cranium, it is preferable to register the studies by using only the brain tissue. To this end, a brain-surface extraction tool (BrainSuite from the University of Southern California)13 is fitted (1) and then used to mask the brain in the new study (2). Next, the equivalent series in the old study is retrieved and coregistered to the new study (3) by using the Mutual Information algorithm. The recovered transformation is stored in the PACS data base. Note that it is only necessary to mask the new study during registration and that rigid registration yielded sufficient accuracy after exclusion of the masked areas. DOF indicates degrees of freedom.
Step 1: Brain surface was identified by conforming a reference model (by using BrainSuite from the University of Southern California, http://brainsuite.org/)13 to the FLAIR images. This brain surface was then used to mask out the skull and extracranial soft tissues.
Step 2: The “new” study was coregistered with the “old” study by performing a 6 df (axis of movement) rigid-body transformation, recovered by using mutual information as the distance metric.14 Both the resulting transformation and the brain surface mask were stored in a separate PACS data base (by using DCM4CHE, http://sourceforge.net/projects/dcm4che/).15 Both the old and new volumetric FLAIR datasets were resectioned into orthogonal axial, sagittal, and coronal planes, allowing exact comparison of any individual pixel regardless of the orientations at which the original scans were obtained. Trilinear interpolation (by using the ImageJ library; National Institutes of Health, Bethesda, Maryland)16 was used to preserve image quality and minimize artifacts during these transformations.
Step 3: Image signal intensity was normalized by using histogram equalization to eliminate global differences.
Step 4: Using the now masked, coregistered, and normalized volumetric new and old FLAIR sequences, we computed a volumetric image containing signed pixel differences. We used both color and transparency to encode the changes between the 2 studies, transparency to encode the magnitude of change, and color to indicate the type of change, with red indicating new lesions (NLs) and green indicating improved lesions (ILs).
The resultant images were then viewed in a bespoke DICOM viewer, with the reader able to view all 3 planes for both old and new studies, and each point could be correlated in all view panes (Fig 1). The total processing time for steps 1–4 is approximately 1 minute on a typical desktop computer (1.6 GHz), including data retrieval and storage time. Rendering of the 3D and 2D perspectives requires approximately 10 ms per viewpoint, allowing rapid scrolling through the data. Most rendering time consists of trilinear interpolation.
Validation
Institutional ethics approval was obtained for this study. The hospital PACS was queried for MR imaging brain demyelination-protocol studies performed on a single 3T magnet (Tim Trio, 12-channel head coil; Siemens, Erlangen, German) between 2009 and 2011 inclusive, for patients who had ≥2 studies during that period, yielding 367 studies. Eligibility criteria were the following: consecutive studies in patients with a confirmed diagnosis of multiple sclerosis (based on information provided on requisition forms) and availability of a diagnostic-quality MR imaging volumetric FLAIR sequence (FOV = 250, 160 sections, section thickness = 0.98 mm, matrix =258 × 258, TR = 5000 ms, TE = 350 ms, TI = 1800 ms, 72 sel inversion recovery magnetic preparation). One hundred sixty-six comparison pairs (332 studies) met the above inclusion criteria. Of these, 5 comparative study pairs (CSP) had to be excluded due to a lack of exact lesion quantification in the issued radiology reports. A final total of 161 CSP (median time between scans, 343 ± 174 days) of 153 individual patients (women = 116, men = 37; median age, 41.5 ±10.2 years) with accompanying reports were thus included in the study. MR imaging–trained radiologists at our institution reported all studies. Of the 161 CSP, 43 had initial clinical reports by 1 of the authors (reader 1).
To assess interobserver characteristics and validate the detection ability of the software, 2 fellowship-trained neuroradiologists (readers 1 and 2) with 6 and 3 years' clinical experience, respectively, retrospectively assessed all CSP by using the software. The readers were blinded to each other's findings and to the existing radiology reports (median time between clinical report being issued and assessment with the software was 449 ± 159.7 days). The time required to read a study by using the software was assessed in real-time, by using a digital stopwatch.
The CSP initially clinically read by reader 1 were reread using the PACS a second time, 12 months later, to assess intraobserver characteristics. These same CSP were also again read by reader 1 three months later, still using the software, with all images left-right reversed to reduce the risk of recalling individual lesions. The time taken to reread the studies by reader 1 was also recorded in real-time by using a digital stopwatch.
Lesion Assessment
NLs were defined as those with new focal regions of increased T2 FLAIR signal in previously normal white matter. Due to the time interval between studies, no concentrically enlarging (worsening) plaques were identified.
ILs were defined as those with either concentric reduction in lesion size or global reduction in abnormal T2 FLAIR signal.
When using the software to assess NLs, the reader scrolled through axial colored change maps, with areas of increased FLAIR signal highlighted in red. Each time a candidate lesion was identified, the area was correlated to coregistered resectioned but otherwise conventional source FLAIR images in all 3 orthogonal planes of both new and old studies. The reader assessed the lesion as one would during conventional reporting (without the aid of any assistive software), judging whether the lesion represented a new demyelinating lesion, other pathology, or artifact. When the reader was satisfied that the lesion was indeed a true finding and represented a new demyelinating plaque, it was marked with 3D Cartesian coordinates.
This process was repeated for all lesions and was similarly repeated when examining the decreased FLAIR signal maps to identify ILs (highlighted in green).
When subsequently analyzing these marked lesions, the recorded coordinates for each read were automatically compared to ensure that the same lesions were being identified. Lesions with coordinates <2 mm apart were considered as 1 lesion. For lesions with coordinates >2 mm apart, both readers performed manual review of each lesion to determine whether both coordinates belonged to 1 large lesion marked in different locations or to 2 separately detected but adjacent lesions.
Statistical Analysis
The Cohen κ interrater reliability was used to measure and compare the agreement between the 2 readers and between the readers and the originally issued radiology reports. The Spearman correlation coefficient was also used to assess interreader correlation of overall lesion load. Three sets of binary subgroups were considered (≥1 lesion, ≥2 lesions, and ≥3 lesions) when assessing interreader κ agreement. Univariate χ2 and 2-group proportion analyses were conducted to compare the number of new or improved lesions identified by the clinical report and by the readers when using the assistive software. The time taken to complete the assessment was recorded for each reader and reported as averages. The Mann-Whitney rank sum test was used to compare the time taken to read the scan data when using the side-by-side comparison with that when using the software. For all statistical tests, a 2-sided α value of .05 was used to indicate significance. Data were analyzed with STATA (Version 12.1; StataCorp, College Station, Texas).
Potential Clinical Impact
Questionnaires were sent to the referring neurologists concerning CSP if there was a change in lesion load when comparing the originally issued radiology report and the report of the readers using the software. Neurologists were asked to indicate whether their management strategies would have been changed retrospectively in regard to medication regimens, clinical follow-up interval, or MR imaging follow-up interval.
Results
Using the software, readers were able to detect changes in a greater proportion of CSP than had been detected with conventional assessment for both NL (reader 1, 37%; reader 2, 39%; CSSC, 12%) and IL (reader 1, 17%; reader 2, 24%; CSSC, 3%). In both instances, statistical significance was reached (NL, P < .001; IL, P < .001) (Table 1). Lesions were located widely throughout the brain (On-line Fig 1).
Table 1:
Demonstrating the number of study pairs showing a change in lesion load as identified using conventional side-by-side comparison and the software
| Change in Lesion Load | Issued Radiology Report | Reader 1 | Reader 2 |
|---|---|---|---|
| Study pairs with new lesions (No.) | 20 | 60 (P < .001) | 62 (P < .001) |
| Study pairs with improved lesions (No.) | 5 | 28 (P < .001) | 39 (P < .001) |
To substratify the comparison pairs with NL and IL, we considered 3 sets of dichotomized subgroups (CSP with ≥1 lesion, ≥2 lesions, and ≥3 lesions). For CSP with detected NL and IL, κ statistics indicating substantial interreader agreement were observed (Table 2). These κ values were reduced slightly due to reader 2 identifying slightly higher numbers of both NLs and ILs in each subgroup, resulting from an interreader difference in the interpretation of lobulated lesions as either 2 confluent lesions or 1 irregular lesion. The Spearman correlation coefficient demonstrated good overall interreader correlation (Spearman ρ: NL = 0.910, IL = 0.774).
Table 2:
Interreader agreement demonstrated with binary groupings of new and improved lesions when using the software
| Change in Lesion Load, Binary Grouping | κ | 95% CI |
|---|---|---|
| New lesions (0, 1+) | 0.87 | 0.79–0.95 |
| New lesions (0–1, 2+) | 0.81 | 0.71–0.91 |
| New lesions (0–2, 3+) | 0.96 | 0.90–1.00 |
| Improved lesions (0, 1+) | 0.72 | 0.59–0.85 |
| Improved lesions (0–1, 2+) | 0.79 | 0.64–0.94 |
| Improved lesions (0–2, 3+) | 0.70 | 0.49–0.91 |
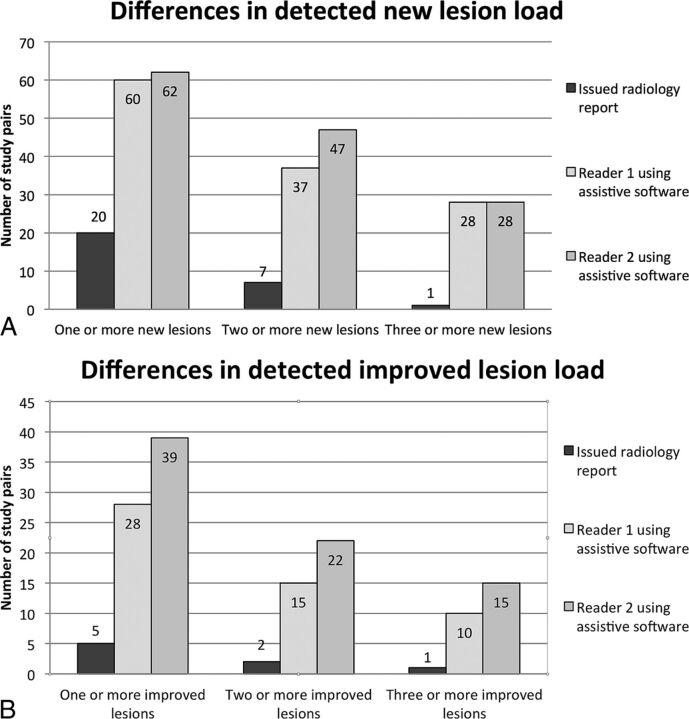
Comparing the subgroups of both NL and IL, readers detected a higher number of CSP with a changed lesion load compared to the original radiology reports (Fig 3A, -B), despite a wide variation of total background lesion load (On-line Fig 2).
Fig 3.
A, Comparative graphic representation of the number of study pairs with new lesions detected by both readers when using the software compared to the issued radiology report. B, Comparing the number of study pairs improved with demyelinating lesions detected by both readers when using the newly developed assistive software to the issued radiology report.
Three false-negatives occurred by using the software; 2 NLs and 1 IL were described in 3 respective radiology reports, not detected by the readers.
Assessment of lesion location accuracy was calculated by using the total agreed base lesion load (defined as the lowest number of NLs or ILs that both readers agreed on per CSP) and showed good interreader location accuracy (NL location accuracy = 94%, 313/333; IL location accuracy = 96%, 70/73).
Despite identifying more NLs and ILs in a greater proportion of CSP when using the software, intraobserver agreement between the first and second read of the “reader 1 subgroup” applying the software was very good and better than that with CSSC, though this did not reach statistical significance with the sample size limited to 43 CSP (Table 3 and On-line Table 1).
Table 3:
Intrareader agreement demonstrated with binary groupings of new and improved lesions using both conventional side-by-side comparison and the softwarea
| New Lesions (κ) (95% CI) | Improved Lesions (κ) (95% CI) | |
|---|---|---|
| One or more lesions | ||
| VTS 1st vs VTS 2nd read | 1.000 | 0.937 (0.815–1.000) |
| CSSC 1st vs CSSC 2nd read | 0.941 (0.826–1.000) | 0.462 (0.039–0.886) |
| Two or more lesions | ||
| VTS 1st vs VTS 2nd read | 1.000 | 0.731 (0.448–1.000) |
| CSSC 1st vs CSSC 2nd read | 0.846 (0.640–1.000) | 0.482 (−0.118–1.000) |
| Three or more lesions | ||
| VTS 1st vs VTS 2nd read | 1.000 | 0.774 (0.472–1.000) |
| CSSC 1st vs CSSC 2nd read | 0.724 (0.361–1.000) | 0.482 (−0.118–1.000) |
Correlations demonstrated substantial intrareader agreement. The software generally outperformed conventional side-by-side comparison without, however, reaching statistical significance.
Mean reporting times per CSP were <3 minutes (reader 1 = 2 minutes 15 seconds ± 1 minute 5 seconds and reader 2 = 2 minutes 45 seconds ± 1 minute 44 seconds), and there was an overall reduction in study reading times as the readers became more familiar with the software (mean read time of the first 25 studies: reader 1 = 3 minutes 9 seconds, reader 2 = 4 minutes 30 seconds versus a mean read time of the last 25 studies: reader 1 = 1 minute 36 seconds, reader 2 = 1 minute 49 seconds).
When we compared VTS with CSSC, there was a significant difference in read times (median interquartile range): VTS = 1 minute 58 seconds (range, 1 minute 37 seconds to 2 minutes 52 seconds) compared with CSSC = 3 minutes 41 seconds (range, 51 seconds to 4 minutes 12 seconds; P < .001).
Feedback forms were drafted for the 60 CSP that showed interval lesion load change when comparing the originally issued radiology reports and the lesion load detected by using VTS. A total of 47/60 completed feedback forms were returned (respondent rate of 78%). In 79% (37/47) of cases, neurologists reported that they would have been likely to change management strategies if the altered lesion load had been known at the time, prompting a change in either MR imaging follow-up interval, clinical follow-up interval, or therapeutic management (On-line Table 2).
Discussion
Management strategy considerations for MS are based on clinical, biochemical, and imaging findings and are aimed at treating acute attacks, preventing relapses and progression, managing symptoms, and rehabilitation.2,17
In recent years, a number of new agents have become available, targeting various multiple sclerosis disease pathways.1,2 MR imaging plays an important role in detecting not only the total demyelinating lesion load but also, possibly more important, interval change in the number of demyelinating lesions, reflecting disease activity and potentially resulting in changes to treatment.2
Conventional comparative image assessment is subjective, dependent on the skill and consistency of the reviewer.9 To facilitate time-efficient, reproducible, and accurate lesion-load detection, many algorithms have been proposed for fully automated computer-assistive solutions.3,18 These methods use different principles, including intensity-gradient features,19 intensity thresholding,20 intensity-histogram modeling of expected tissue classes,21–23 fuzzy connectedness,24 identification of nearest neighbors in a feature space,25,26 or a combination of these. Methods such as Bayesian inference, expectation maximization, support-vector machines, k-nearest neighbor majority voting, and artificial neural networks are algorithmic approaches used to optimize segmentation.18 All of these approaches tend to show promising results; however, the results are usually on small samples, often nonreproducible and unreliable, and have not entered into routine clinical use.18
In smaller study populations, semiautomated assistive approaches have been investigated with promising results by using both MR imaging subtraction techniques and coregistered comparative volumetric FLAIR color-map overlays.11,12
Our semiautomated radiology assistive platform is computationally fast and robust, successfully processing all 322 included studies. The software allows color maps superimposed on anatomic FLAIR sequences and direct comparison between old and new studies in exactly aligned axes as well as accurate localization of any given point in all 3 planes.
In our study population, the largest reported of its kind (161 CSP; 322 individual studies), a statistically significant number of increased CSP with NLs and ILs were detected when using the assistive software compared with the originally issued MR imaging reports generated with CSSC. On the basis of responses by referring neurologists, at least 79% of CSP with changes in detected lesion loads (VTS versus CSSC) were likely to have undergone a change in management if the altered lesion load had been appreciated at the time. This represents 22% (37/161) of the whole cohort; studies reported as “stable” that actually had sufficient change in disease burden to potentially alter management.
In addition to detecting NLs, our approach demonstrates a statistically significant improvement in detecting ILs. There is, however, a larger disparity between readers when assessing ILs. After we reviewed the discrepant lesions, this does not appear to stem from software failure but from intrinsic heterogeneity in lesions that appear to be reducing in size or signal intensity. Some lesions demonstrated an unequivocal concentric reduction in size. Many lesions, however, demonstrated diffuse or ill-defined signal normalization. It was these lesions that resulted in most interreader discordance. This difficulty in clearly defining the nature of improving demyelinating lesions is echoed in studies correlating the MR imaging appearances of demyelinating lesions with lesion pathology, highlighting the heterogeneity that also exists in radiologic-pathologic correlation.27 Although the significance of new demyelinating lesions on MR imaging has been well established in the literature,1,2,4–7 the clinical significance of “improved” demyelinating lesions is less clear.
Limitations
Our study has a number of limitations. One is the single-scanner/single-sequence nature of the dataset. The software platform has been designed to be vendor-agnostic and should be able to accept any volumetric FLAIR sequence; however, this has not yet been tested, and likely, performance will vary depending on the quality of the source data. Indeed, vascular flow-induced artifacts through the anterior pons and inferior temporal lobes seen in our FLAIR sequence resulted in some difficulty in interpreting signal change in these regions, and improved sequence design may allow even greater lesion detection. Additionally, there is likely to be decreased performance if the 2 volumetric FLAIR sequences compared are from different MR imaging scanners or differ in their specifications, though again transformation, coregistration, and normalization are not dependent on identical sequences. Although not tested in this study, other volumetric sequences such as double inversion recovery should fulfill the criteria to be used with the software.
Other limitations of our study include the inability to comment on interobserver agreement on the original radiology reports because these were single-read by various MR imaging–trained radiologists in our department and a reread of all CSP by both readers was beyond the scope of this study.
Although we can also not comment on the time it took to read the original MR imaging studies in clinical practice (at our institution, we do not routinely record reporting times), we tried to address this, in part, by measuring the time taken to perform conventional interpretation by using CSSC on the PACS during the second reread by reader 1. Having done so, we nonetheless acknowledge that applying the software clinically may result in unforeseen program-related and user-related time delays. We are hoping to minimize these by incorporating the platform directly into a PACS workflow and by familiarizing MR imaging readers with the program; these changes will be explored in future work.
Although intrareader correlation was shown to be very good in the subset of cases that reader 1 reread (Table 3), the accuracy of the other radiology reports may have been influenced by factors related to the daily demands of a busy radiology department, such as time pressures and interruptions (factors not simulated when testing the software). All MR imaging readers in our department are experienced; thus, radiologic expertise is unlikely to present a limitation. If anything, it is likely that the lesion-detection improvement would be larger for radiologists with less neuroradiology experience.
We also acknowledge that we did not directly assess the clinical impact of a second read without the software; however, we believe we have, in part, explored this by having reader 1 additionally reread all the studies he originally assessed (n = 43) by using CSSC. The agreement between the 2 reads was high (Table 3). More important, in only a single patient did the reads differ in categorization (On-line Table 1). The reports of a second read by using CSSC would have been, in all except 1 case, indistinguishable from the initial clinical reports; thus, no change in management would be expected. As such, retrospective changes to management reported by treating clinicians are attributable to the software, rather than to merely a second read.
Future Work
Our current development work is focused on deploying the software to the live clinical PACS workflow at our institution, which will allow us to carry out prospective research and ensure that the findings of this study are replicated in terms of improved lesion detection without the burden of false-positives. We are also hoping to make the software available to other institutions for further validation by introducing additional readers and by using a variety of FLAIR sequences. The functionality of the software can also be extended in the future by adding semiautomated segmentation for quantification of lesion volume.
Conclusions
We have developed a semiautomated software platform to assist the radiologist reading comparative MR imaging MS follow-up study pairs with accurate and timely reporting. We have shown that it significantly outperforms CSSC in the identification of NLs and ILs, while maintaining high intra- and interobserver reliability. On the basis of neurologists' feedback, it is likely that identifying new lesions would result in some management change. Given the availability and efficacy of numerous disease-modifying drugs, even a small change in prescribing practices could result in sizable effects on the long-term patient disability.
Supplementary Material
Acknowledgments
The authors acknowledge Andy Le, Suria Barelds, Irene Ngui, and Alex Stern for their assistance with data gathering and integration. The National Information and Communications Technology of Australia Research Centre of Excellence (NICTA) contribution to this work was supported by the Australian Research Council and by the NICTA Victorian Research Laboratory.
ABBREVIATIONS:
- CSP
comparative study pairs
- CSSC
conventional side-by-side comparison
- IL
improved lesion
- NL
new lesion
- VTS
VisTarsier software
Footnotes
Disclosures: Frank Gaillard—UNRELATED: Employment: Founder/CEO Radiopaedia.org, Radiopaedia Australia Pty Ltd; Grants/Grants Pending: multiple grants in progress for this and related projects, through the National Health and Medical Research Council development grants,* Business Development Initiative Victoria Government (successful first round),* and Royal Melbourne Hospital Foundation*; Payment for Lectures (including service on Speakers Bureaus): Medtronic Australia, Comments: travel and payment to give talks on MRI safety; Other: technical consultant, Medtronic Australasia Pty Ltd. David Rawlinson—RELATED: Other: Our institute, NICTA/University of Melbourne, was funded by state and federal governments, which paid our wages.* Patricia M. Desmond—UNRELATED: Grants/Grants Pending: National Health and Medical Research Council.* *Money paid to the institution.
References
- 1. Jones JL, Coles AJ. New treatment strategies in multiple sclerosis. Exp Neurol 2010;225:34–39 [DOI] [PubMed] [Google Scholar]
- 2. Weiner HL. The challenge of multiple sclerosis: how do we cure a chronic heterogeneous disease? Ann Neurol 2009;65:239–48 [DOI] [PubMed] [Google Scholar]
- 3. Mortazavi D, Kouzani AZ, Soltanian-Zadeh H. Segmentation of multiple sclerosis lesions in MR images: a review. Neuroradiology 2012;54:299–320 [DOI] [PubMed] [Google Scholar]
- 4. Caramanos Z, Francis SJ, Narayanan S, et al. Large, nonplateauing relationship between clinical disability and cerebral white matter lesion load in patients with multiple sclerosis. Arch Neurol 2012;69:89–95 [DOI] [PubMed] [Google Scholar]
- 5. Sormani MP, Rovaris M, Comi G, et al. A reassessment of the plateauing relationship between T2 lesion load and disability in MS. Neurology 2009;73:1538–42 [DOI] [PubMed] [Google Scholar]
- 6. Fisniku LK, Brex PA, Altmann DR, et al. Disability and T2 MRI lesions: a 20-year follow-up of patients with relapse onset of multiple sclerosis. Brain 2008;131:808–17 [DOI] [PubMed] [Google Scholar]
- 7. Stankiewicz JM, Glanz BI, Healy BC, et al. Brain MRI lesion load at 1.5T and 3T versus clinical status in multiple sclerosis. J Neuroimaging 2011;21:e50–e56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bink A, Schmitt M, Gaa J, et al. Detection of lesions in multiple sclerosis by 2D FLAIR and single-slab 3D FLAIR sequences at 3.0 T: initial results. Eur Radiol 2006;16:1104–10 [DOI] [PubMed] [Google Scholar]
- 9. Abraham AG, Duncan DD, Gange SJ, et al. Computer-aided assessment of diagnostic images for epidemiological research. BMC Med Res Methodol 2009;9:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. García-Lorenzo D, Francis S, Narayanan S, et al. Review of automatic segmentation methods of multiple sclerosis white matter lesions on conventional magnetic resonance imaging. Med Image Anal 2013;17:1–18 [DOI] [PubMed] [Google Scholar]
- 11. Moraal B, Meier DS, Poppe PA, et al. Subtraction MR images in a multiple sclerosis multicenter clinical trial setting. Radiology 2009;250:506–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bilello M, Arkuszewski M, Nasrallah I, et al. Multiple sclerosis lesions in the brain: computer-assisted assessment of lesion load dynamics on 3D FLAIR MR images. Neuroradiol J 2012;25:17–21 [DOI] [PubMed] [Google Scholar]
- 13. Shattuck DW, Leahy RM. BrainSuite: an automated cortical surface identification tool. Med Image Anal 2002;6:129–42 [DOI] [PubMed] [Google Scholar]
- 14. Cover TM, Thomas JA. Elements of Information Theory. New York: John Wiley & Sons; 1991 [Google Scholar]
- 15. Warnock MJ, Toland C, Evans D, et al. Benefits of using the DCM4CHE DICOM archive. J Digit Imaging 2007;20:125–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rasband WS. ImageJ. Bethesda; National Institutes of Health; 1997–2014. http://imagej.nih.gov/ij/. Accessed April 1, 2013. [Google Scholar]
- 17. Murray TJ. Diagnosis and treatment of multiple sclerosis. BMJ 2006;332:525–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vrenken H, Jenkinson M, Horsfield MA, et al. ; MAGNIMS Study Group. Recommendations to improve imaging and analysis of brain lesion load and atrophy in longitudinal studies of multiple sclerosis. J Neurol 2013;260:2458–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pachai C, Zhu YM, Grimaud J, et al. A pyramidal approach for automatic segmentation of multiple sclerosis lesions in brain MRI. Comput Med Imaging Graph 1998;22:399–408 [DOI] [PubMed] [Google Scholar]
- 20. Goldberg-Zimring D, Achiron A, Miron S, et al. Automated detection and characterization of multiple sclerosis lesions in brain MR images. Magn Reson Imaging 1998;16:311–18 [DOI] [PubMed] [Google Scholar]
- 21. Shiee N, Bazin PL, Ozturk A, et al. A topology-preserving approach to the segmentation of brain images with multiple sclerosis lesions. Neuroimage 2010;49:1524–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Khayati R, Vafadust M, Towhidkhah F, et al. Fully automatic segmentation of multiple sclerosis lesions in brain MR FLAIR images using adaptive mixtures method and Markov random field model. Comput Biol Med 2008;38:379–90 [DOI] [PubMed] [Google Scholar]
- 23. Freifeld O, Greenspan H, Goldberger J. Multiple sclerosis lesion detection using constrained GMM and curve evolution. Int J Biomed Imaging 2009;2009:715124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Horsfield MA, Bakshi R, Rovaris M, et al. Incorporating domain knowledge into the fuzzy connectedness framework: application to brain lesion volume estimation in multiple sclerosis. IEEE Trans Med Imaging 2007;26:1670–80 [DOI] [PubMed] [Google Scholar]
- 25. Anbeek P, Vincken KL, van Osch MJ, et al. Automatic segmentation of different-sized white matter lesions by voxel probability estimation. Med Image Anal 2004;8:205–15 [DOI] [PubMed] [Google Scholar]
- 26. Mohamed FB, Vinitski S, Gonzalez CF, et al. Increased differentiation of intracranial white matter lesions by multispectral 3D-tissue segmentation: preliminary results. Magn Reson Imaging 2001;19:207–18 [DOI] [PubMed] [Google Scholar]
- 27. Filippi M, Rocca MA, Barkhof F, et al. ; Attendees of the Correlation between Pathological MRI Findings in MS workshop. Association between pathological and MRI findings in multiple sclerosis. Lancet Neurol 2012;11:349–60 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.