Abstract
Background
Schizophrenia (SCZ) and bipolar disorder (BIP) are debilitating neuropsychiatric disorders, collectively affecting 2% of the world’s population. Recognizing the major impact of these psychiatric disorders on the psychosocial function of more than 200 000 US Veterans, the Department of Veterans Affairs (VA) recently completed genotyping of more than 8000 veterans with SCZ and BIP in the Cooperative Studies Program (CSP) #572.
Methods
We performed genome-wide association studies (GWAS) in CSP #572 and benchmarked the predictive value of polygenic risk scores (PRS) constructed from published findings. We combined our results with available summary statistics from several recent GWAS, realizing the largest and most diverse studies of these disorders to date.
Results
Our primary GWAS uncovered new associations between CHD7 variants and SCZ, and novel BIP associations with variants in Sortilin Related VPS10 Domain Containing Receptor 3 (SORCS3) and downstream of PCDH11X. Combining our results with published summary statistics for SCZ yielded 39 novel susceptibility loci including CRHR1, and we identified 10 additional findings for BIP (28 326 cases and 90 570 controls). PRS trained on published GWAS were significantly associated with case-control status among European American (P < 10–30) and African American (P < .0005) participants in CSP #572.
Conclusions
We have demonstrated that published findings for SCZ and BIP are robustly generalizable to a diverse cohort of US veterans. Leveraging available summary statistics from GWAS of global populations, we report 52 new susceptibility loci and improved fine-mapping resolution for dozens of previously reported associations.
Keywords: schizophrenia, bipolar disorder, genome-wide association studies (GWAS), US veterans
Introduction
In the United States, the combined societal costs of treatment and loss of productivity associated with schizophrenia (SCZ) were estimated to be $62.7 billion in 20021 and more than $155 billion in 2013.2 For bipolar disorder (BIP), which is phenomenologically related and has a partially shared genetic basis,3,4 these costs exceeded $200 billion in 2015.5 Even during psychosis-free periods, cognitive deficits are thought to contribute to deteriorating functional skills in the areas of self-care, social and occupational function, and medication adherence, as well as other aspects of illness management, all needed for independent living.6 Recognizing the major impact of these disorders on the psychosocial function of more than 200 000 affected veterans, the US Department of Veterans Affairs (VA) funded and recently completed recruitment, assessment, and genotyping of 3690 veterans with SCZ and 5095 with BIP as part of the Cooperative Studies Program (CSP) #572, Genetics of Functional Disability in Schizophrenia and Bipolar Illness.7 This study included in-person assessment of a range of phenotypic variables, including extensive assessments for neurocognitive function and disability, and diagnoses were confirmed by structured clinical interviews.
Over the last decade, the genome-wide association study (GWAS) has emerged as a powerful research paradigm,8 and its success as applied to neuropsychiatric disorders exemplified by the trajectory of discovery in SCZ.3,9–12 Since the landmark Psychiatric Genomics Consortium (PGC) study that identified 108 distinct loci associated with SCZ,12 meta-analyses with new data have seen the number of susceptibility loci increase to 128,13 145,14 and 176.15 Progress for BIP risk has been comparatively modest16–18 likely owing to its clinical heterogeneity and smaller sample sizes. A critical realization is that common genetic risk variants have small effects and likely number in the thousands.19 These hard-won advances underscore the importance of ever-increasing sample sizes, well-phenotyped case-control cohorts, and the widespread accessibility of GWAS summary statistics.
We describe primary GWAS of SCZ and BIP in CSP #572 cases and screened controls from the Million Veteran Program (MVP)20 and employ polygenic risk scores (PRS) to benchmark the generalizability of published findings to our VA patient population. Noting the appreciable representation of African ancestry (AA) participants in CSP #572 and drawing upon genotype-level data from the Genomic Psychiatry Cohort (GPC),13,21 Molecular Genetics of Schizophrenia (MGS) study,9 Consortium on the Genetics of Schizophrenia (COGS),22 and Bipolar Genome Study (BiGS),23 these represent the largest such studies in AA populations to date. We combine our results with available summary statistics from several recent GWAS,13–15,18 yielding additional findings and highlighting challenges and opportunities for studies of these complex disorders.
Methods
Ascertainment and Diagnosis
All patients were required to meet lifetime (DSM-IV)24 criteria for SCZ (any subtype) or bipolar I disorder (any current phase). Participants received the Structured Clinical Interview for the DSM-IV (SCID),25 administered in-person by a trained rater. If needed, information from medical charts, patients’ clinicians, and other informants were utilized to confirm diagnoses. A diagnosis of schizoaffective disorder (SAD) based on the SCID was an exclusion criterion, in that, we did not anticipate being able to recruit a comparable number of participants with confirmed diagnoses of SAD. Patients with bipolar II disorder were not selected for participation given concerns about the reliability of this diagnosis.
Other exclusion criteria included major neurologic illnesses or systemic medical illnesses that could interfere with the central nervous system (CNS) function and test performance. We did not exclude patients with a diagnosis of substance abuse, given common co-occurrence (ie, comorbidity) and issues of representativeness: such patients were not enrolled if they appeared to be intoxicated, but could be reassessed at a later date.
Table 1 displays the demographic and clinical characteristics of CSP #572 participants with confirmed diagnoses of SCZ or BIP.
Table 1.
Demographic and Clinical Characteristics of the CSP #572 Participants
| Category | Factor | Schizophrenia | Bipolar I |
|---|---|---|---|
| N (%) | N (%) | ||
| Demographic | Age (mean ± SD years) | 55.1 ± 10.1 | 52.6 ± 11.5 |
| Female | 286 (7.3%) | 1004 (18.6%) | |
| Ancestry: | |||
| European American | 1553 (39.8%) | 3786 (70.2%) | |
| African American | 2122 (54.3%) | 1320 (24.5%) | |
| Other | 231 (5.9%) | 285 (5.3%) | |
| Marital status: | |||
| Never married | 1569 (40.0%) | 980 (18.1%) | |
| Currently married | 768 (19.5%) | 1795 (33.2%) | |
| Previously married | 1586 (40.2%) | 2628 (48.5%) | |
| Education: | |||
| Less than high school | 1709 (43.5%) | 1333 (24.7%) | |
| High school | 1833 (46.7%) | 2886 (53.4%) | |
| More than high school | 384 (9.8%) | 1184 (21.9%) | |
| Annual income: | |||
| ≤$20 000 | 1766 (51.2%) | 2071 (40.8%) | |
| >$20 000 | 1682 (48.8%) | 3006 (59.2%) | |
| Clinical | Psychiatric comorbidity: | ||
| Single | 1213 (30.8%) | 1555 (28.7%) | |
| Multiple | 1259 (31.9%) | 2799 (51.7%) | |
| Medical comorbidity | 2026 (51.4%) | 2809 (51.9%) | |
| Lifetime suicide status: | |||
| Ideation | 941 (23.9%) | 1504 (27.8%) | |
| Attempt or behavior | 1816 (46.1%) | 2953 (54.5%) | |
| Current PTSD | 867 (22.0%) | 1787 (33.0%) | |
| Lifetime major depression | 1577 (40.0%) | ||
| Current negative symptoms | 1380 (35.0%) | ||
| Impairment: | |||
| Everyday functioning UCSD Performance-based Skills Assessment (UPSA) | 1533 (38.9%) | 838 (15.5%) | |
| Any neuropsychological test | 3683 (93.4%) | 4750 (87.7%) | |
| Electronic health record (EHR): | |||
| Inpatient treatment | 2978 (75.3%) | 2978 (75.3%) | |
| Medications: | |||
| Antipsychotics | 3784 (95.7%) | 4793 (88.4%) | |
| Mood-stabilizers | 2567 (64.9%) | 5104 (94.1%) | |
| Antidepressants | 2928 (74.1%) | 4529 (83.5%) |
We selected control participants from among MVP enrollees by leveraging the VA electronic health record (EHR). We excluded individuals with any history of psychotic or affective disorders or past treatment with neuroleptic, mood-stabilizing, or antidepressant medications (see supplementary note).
This study was approved by the VA Central Institutional Review Board, and all participants provided written informed consent; no one who required the permission of a guardian to participate was enrolled in this study.
Genotyping, Imputation, and Ancestry Assignment
Genomic data processing was performed together for 8021 CSP #572 and >350 000 MVP participants (release 2.1; www.research.va.gov/mvp). At participating VA sites, blood samples were collected on the same day as in-person assessments. The VA Central Biorepository (VACB) provided the necessary supplies directly to the sites and designed the processes by which biospecimens were collected, packaged, shipped, and logged into a computerized tracking system and by which DNA was extracted. Staff were trained at the study “kickoff” and retrained periodically.
Genotyping was performed using a custom Affymetrix Axiom Biobank array with probes for 723 305 single-nucleotide polymorphisms (SNPs). Details of quality control procedures are described elsewhere.26,27 We excluded duplicates and samples with >2.5% missing genotype calls, excess heterozygosity, or discordance between genetic sex and phenotypic gender. We excluded SNPs with missingness >5% or minor allele frequency (MAF) that deviated by >10% from the 1000 Genomes Project Phase 3 (KGP3) data.28 Cleaned genotype data were imputed using Minimac329 and the KGP3 data, giving predicted genotype dosages for 49 134 253 SNPs and insertions/deletions.
Pairwise genetic relatedness was estimated using KING.30 Individuals demonstrating excess relatedness were excluded. We removed one individual at random from each pair of first-degree relatives, preferentially retaining cases from case-control pairs.
Genome-wide average proportions of GBR (UK Caucasians), PEL (Peruvians), YRI (Yoruba from Nigeria), CHB (Han Chinese), and LWK (Luhya from Kenya) ancestries were obtained from admixture31 analysis, and we assigned individuals to broadly defined European ancestry (EA) (GBR > .8) and AA (YRI > .6) groups.27 Among CSP #572 participants, 1021 (488 SCZ and 533 BIP) did not receive an ancestry assignment.
We used flashpca232 to extract ancestry principal components (PCs) from 22 766 independent autosomal SNPs after excluding regions harboring long-range linkage disequilibrium (LD)33 (supplementary figures S1 and S2).
Following these checks, corrective actions, and assignments, 2 883 SCZ and 4 117 BIP cases and 50 436 controls were available for analysis.
GWAS and Meta-Analyses With Published Findings
We tested for association between imputed genotype dosages and case-control status by logistic regression using PLINK,34,35 including the first 10 ancestry PCs as covariates, and retaining variants with imputation quality (INFO) ≥.9 and MAF ≥.01. We combined AA and EA results using Han and Eskin’s36 random effects (RE2) model implemented in METASOFT, which is optimized to detect associations in the presence of heterogeneity. We used this method to combine sex-specific X-chromosome results.
We defined statistically independent genome-wide significant (GWS) by LD-based “clumping” of SNPs with P < 10–4 and LD r2 ≥ .1 with the lead variant within a 3 megabase (Mb) window.12 We annealed clumped intervals within 250 kilobases (kb) into physically distinct loci.
We obtained genotype-level data for the GPC, including MGS and COGS samples, and for the BiGS study23,37,38 via the database of Genotypes and Phenotypes (dbGaP; phs000017.v3.p1). We reanalyzed these data as previously described,13 applying analogous ancestry assignments. We classified as Latino ancestry (LA) any participant with ≥.25 admixed American (AMR) ancestry and <.25 AA, East Asian (EAS), or South Asian ancestry.
We combined SCZ results for CSP #572, GPC, and the meta-analysis of PGC-SCZ2 and CLOZUK (named for a series of UK-based cases registered for clozapine treatment)14,39 (https://walters.psycm.cf.ac.uk/). We also considered a recent meta-analysis of PGC-SCZ2 and multiple EAS cohorts15 (https://www.med.unc.edu/pgc/) but did not combine EAS-only results with CLOZUK + PGC because of sample overlap with 3 EAS cohorts in PGC-SCZ2,12 and because CLOZUK-only summary statistics were not available to us.
For BIP, we built on the most recent PGC GWAS (PGC-BIP),18 incorporating results for multiple ancestries from CSP #572, GPC, and BiGS.
Sample sizes by diagnosis and ancestry are displayed in Table 2.
Table 2.
Sample Sizes by Study Cohort, Assigned Ancestry, and Diagnosis
| Diagnosis | ||||
|---|---|---|---|---|
| Study | Ancestry | SCZ | BIP | Controlsa |
| CSP #572 | AA | 1683 | 1037 | 4669 |
| EA | 1200 | 3,080 | 45 767 | |
| GPC | AA | 3486 | 1641 | 2195 (2470) |
| GPC/COGS | AA | 616 | 362 | |
| GPC/MGS | AA | 1724 | 1111 | |
| GPC | EA | 2919 | 835 | 2367 |
| GPC | LA | 1234 | 1032 | 3090 |
| BiGS | AA | 349 | 958 | |
| Genotyped totals | 12 862 | 7974 | 59 561 (59 836) | |
| CLOZUK + PGC | EAb | 40 675 | 64 643 | |
| PGC-BIP | EA | 20 352 | 31 358 | |
| Meta-analysis totals (primary) | 53 537 | 28 326 | 124 204 (91 194) | |
| PGC + EAS | EA | 33 640 | 43 456 | |
| EAS | 22 778 | 35 362 | ||
| Meta-analysis totals (secondary) | 69 280 | 138 379 | ||
Note: CSP, Cooperative Studies Program; GPC, Genomic Psychiatry Cohort; COGS, Consortium on the Genetics of Schizophrenia; MGS, Molecular Genetics of Schizophrenia; BiGS, Bipolar Genome Study; CLOZUK, a series of UK-based cases registered for clozapine treatment; PGC, Psychiatric Genomics Consortium; BIP, bipolar disorder; EAS, East Asian; EA, European ancestry; AA, African ancestry; LA, Latino ancestry, SCZ, schizophrenia.
Boldface indicates a summation of counts, which is implicit from “totals”.
aBIP control counts are displayed parenthetically if different from SCZ analysis.
bPGC-SCZ2 included 1866 cases and 3418 controls of EAS ancestry.
PRS Profiling
We constructed PRS from published and current GWAS results (the “training” datasets), testing individual-level scores for association with case-control status in CSP #572 (the “target” dataset). Variants that met quality control requirements in both studies were clumped in the appropriate KGP3 population (r2 > .1; 500 kb window). For varying P-value thresholds in the training dataset, scores were constructed by summing the number of tested alleles by their effect estimate. We tested for case-control differences by logistic regression using the same GWAS covariates. Predictive value is reported in terms of Nagelkerke’s pseudo-R2 (fmsb package in R)40 and R2-adjusted for the proportion of cases (ie, the liability scale) assuming a 1% population prevalence for each disorder.41,42
Heritability and Genetic Correlations
We applied genome-based restricted maximum likelihood (GREML), as implemented in the genome-wide complex trait analysis (GCTA) software,43 to estimate the variance explained by the aggregate effects of genome-wide common variants (SNP-h2), and its bivariate extension to obtain the estimates of genetic correlation (rg).41 We included the same GWAS covariates and used the [--grm-cutoff .05] flag to restrict analyses to approximately unrelated individuals. Trans-ancestry rg was estimated from variants with frequency ≥.05 in both populations.44
We applied LD score regression (LDSC)45 to estimate rg between CSP #572 and CLOZUK + PGC and PGC-BIP. We extended these analyses to 597 traits from UK Biobank (UKB) and other published studies using LD Hub (http://ldsc.broadinstitute.org/ldhub/).46Given the requirement of an LD reference panel and concerns about the reliability of reference LD measures for admixed populations, we did not perform LDSC for AA samples.45
Trans-Ancestry Fine-Mapping
For independent GWS associations in each meta-analysis, we constructed credible SNP sets from variants in LD with the lead variant (r2 ≥ .1 in the relevant KGP3 populations),13–15 combining their ranked posterior probabilities until the sum exceeded 0.99.47 We compared credible sets between each discovery GWAS (eg, CLOZUK + PGC) and our expanded meta-analysis on the basis of the number of credible SNPs, the length of the corresponding genomic interval, and the smallest observed P-value. We considered a region to be fine-mapped if the interval for the reduced credible set was smaller than the interval corresponding to SNPs with LD r2 ≥ .6 with the lead variant (in KGP3 EA samples).13
Functional Annotations and Gene-Set Enrichment
We used the Functional Mapping and Annotation (FUMA)48 platform (http://fuma.ctglab.nl/) for follow-up bioinformatic analyses. For each GWS locus, variants were mapped to expression quantitative trait loci (eQTLs) within 1 Mb of a gene’s transcription start sites (ie, cis-eQTLs); we considered eQTLs in 13 CNS tissues from the Genotype-Tissue Expression (GTEx version 8) database that survived a 5% false discovery rate (FDR) correction. Gene-set enrichment analyses were performed with MAGMA using curated gene-sets and gene ontology (GO) categories from MsigDB v6.2. The major histocompatibility complex was excluded from these analyses.
We utilized the S-PrediXcan software49,50 to predict gene expression differences from GWAS summary statistics, using elastic net prediction models for the same tissues (http://predictdb.hakyimlab.org/). We applied 5% FDR corrections within and across tissues.
Results
Primary GWAS
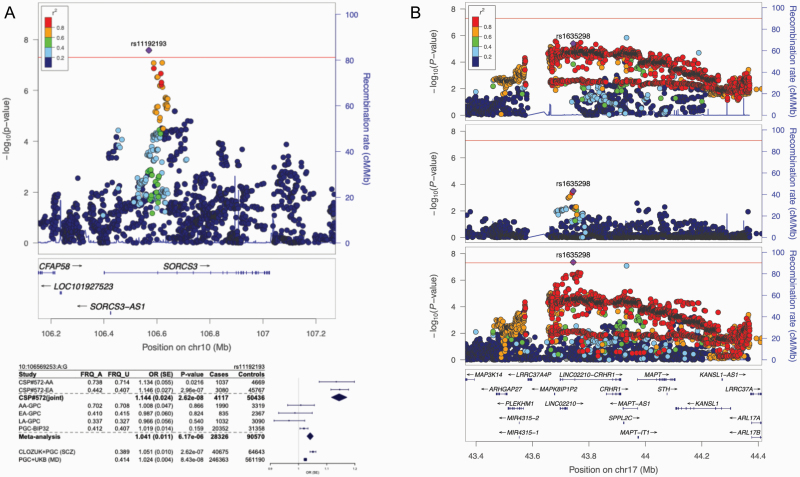
Our primary GWAS in CSP #572 uncovered an SCZ association with CHD7 variants at 8q12.2 and BIP associations with SORCS3 variants at 10q25.1 (figure 1) and downstream of PCDH11X at Xq21.32 (supplementary tables S1–S3; supplementary figures S3–S7).
Fig. 1.
Regional associations of SORCS3 in bipolar disorder (BIP) and CRHR1 in schizophrenia (SCZ). (A) LocusZoom51 plot displaying strength of linkage disequilibrium (LD) with the lead variant at SORCS3 and forest plot52 of effect sizes in genome-wide association studies (GWAS) of BIP, SCZ (based on a meta-analysis of the CLOZUK and PGC studies, as described in citation 14), and major depression (MD) (PGC + UKB).53 (B) Association of CRHR1 variants in European ancestry (EA), African ancestry (AA), and trans-ancestry meta-analyses.
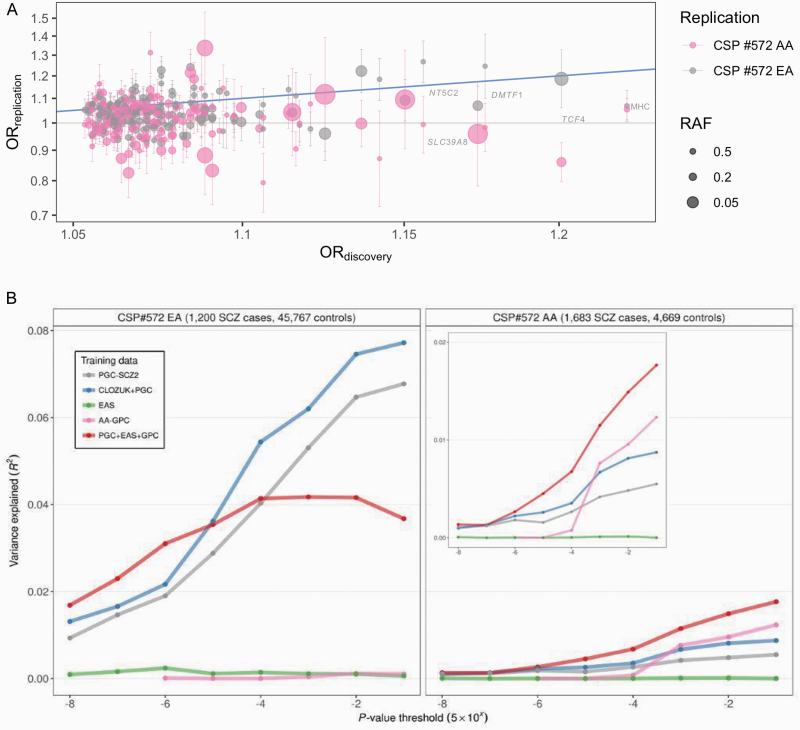
The fraction of SNPs with identical directions of allelic effects in both ancestries was significantly greater than chance (P < .001) (supplementary table S4). Comparisons with published GWAS revealed a similar pattern, with greater concordance at more inclusive P-value thresholds (supplementary table S5; figure 2A).
Fig. 2.
Transferability of published schizophrenia (SCZ) findings to the Cooperative Studies Program (CSP) #572. (A) For genome-wide significant (GWS) findings in the meta-analysis of the CLOZUK and PGC studies, as described in citation 14, odds ratios (OR) are plotted against corresponding estimates for CSP #572 (on the log scale) with standard errors and risk allele frequencies (RAF) displayed. (B) For polygenic risk scores (PRS) based on published genome-wide association studies (GWAS), the variance explained in CSP #572 is displayed in terms of R2 on the liability scale.
PRS Profiling
Scores based on the CLOZUK + PGC meta-analysis yielded the best predictive value experiment wide, accounting for upwards of 7% of the variance on the liability scale among EA participants (P < 10–140) (figure 2B; supplementary table S6). Strikingly, as applied to AA participants, CLOZUK + PGC-trained PRS explained less variance than PRS based on the smaller AA-GPC, with scores based on GPC and PGC + EAS performing best overall.
Scores based on PGC-BIP results explained ~1.7% and 0.6% of variance in EA (P < 10–69) and AA (P < 10–5) participants (supplementary table S7). Updated scores constructed from trans-ancestry GWAS explained ~1.7% of the variance among AA participants (P < 10–14).
We were able to confirm robust polygenic associations across diagnoses, both within and across populations (supplementary tables S8 and S9).
Heritability and Genetic Correlations
Estimates of SNP-h2 were 0.288 (95% CI:[0.208, 0.367]; P < 10–13) for SCZ and 0.188 (95% CI:[0.150, 0.225]; P < 10–25) for BIP in EA participants. Among AA participants, we saw lower estimates of 0.121 (95% CI:[0.040, 0.203]; P = .00149) and 0.094 (95% CI:[−0.014, 0.202]; P = .0367). Bivariate analyses returned significant estimates of both trans-ancestry and cross-disorder rg (supplementary table S10).
Using LDSC and EA summary statistics, we estimated rg between CSP #572 and CLOZUK + PGC as 0.818 (95% CI:[0.655, 0.981]; P < 10–22) and PGC-BIP as 0.721 (95% CI:[0.549,0.894]; P < 10–15).
Extension to >600 selected traits using LD Hub recapitulated previous findings54,55 and highlighted relationships between both disorders and smoking (P < 10–7), having sought treatment for anxiety or depression (P < 10–9), and deprivation index (P < 10–5) (supplementary table S11).
Joint Analyses With Published GWAS
Joint analysis of CSP #572 and GPC with CLOZUK + PGC summary statistics yielded 195 independent GWS associations in 159 physical loci. Meta-analysis of CSP #572, GPC, and PGC + EAS yielded 223 independent associations in 187 loci. Taken together, 228 distinct loci were robustly associated with SCZ and 39 are newly reported here, including 21 loci that only attained significance in joint analyses with CSP #572 (figure 1B; supplementary tables S12 and S13; supplementary figures S8 and S10).
Combining BIP results for CSP #572, GPC, and PGC-BIP yielded 23 independent associations in 23 loci, including 10 novel findings and 9 that attained significance in the CSP #572 joint analysis (supplementary table S14; supplementary figures S9 and S10).
Fine-Mapping of New and Replicated Loci
Joint analysis of CSP #572, GPC, and CLOZUK + PGC improved fine-mapping resolution for 49 independent associations; by comparison, meta-analysis with PGC + EAS yielded an improved fine-mapping resolution of 44 independent associations (supplementary Tables S15 and S16). In both analyses, 12 associations were reduced to fewer than 10 SNPs and 1 locus was mapped to a single SNP. We fine-mapped 10 of 24 independent GWS associations for BIP, with 3 reduced to fewer than 10 SNPs and 1 mapped to a single SNP (supplementary table S17; supplementary figures S11 and S12).
Functional Annotation and Predicted Gene Expression
We mapped GWS loci from the meta-analysis of CSP #572, GPC, and CLOZUK + PGC to 1072 eQTLs (79 independent loci) influencing the expression of 284 genes in one or more CNS tissues; an additional 108 eQTLs were identified via meta-analysis with PGC + EAS, the majority being attributable to a GWS association at 17q21.31 (supplementary tables S18 and S19). Among GWS loci for BIP, we identified 142 eQTLs (14 independent loci) for 52 genes (supplementary table 20).
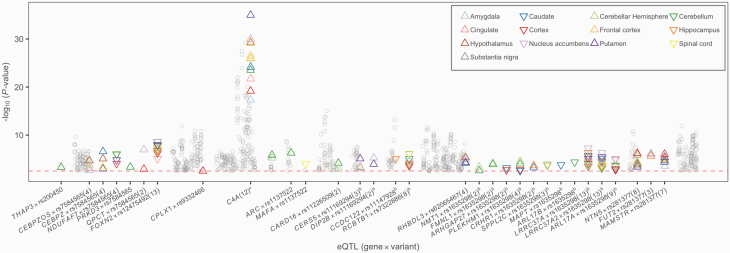
We explored these findings via transcriptome-wide association studies (TWAS) in the same tissues, imputing results directly from our summary statistics using S-PrediXcan (figure 3). Among 3152 and 416 gene-tissue pairs surviving FDR correction, the strongest findings were of predicted higher expression of complement C4A (C4A) in SCZ and X-Prolyl Aminopeptidase 3 (XPNPEP3) in BIP across multiple tissues. Intersecting these results with eQTL annotations for new associations highlighted 100 genes at 26 loci with predicted differential expression between SCZ cases and controls (supplementary tables S21–S23), including some previously reported findings.56,57
Fig. 3.
Predicted transcriptomic differences between schizophrenia (SCZ) cases and controls. Transcriptome-wide association studies (TWAS) significance (y-axis) for novel genome-wide significant (GWS) findings in the primary meta-analysis of SCZ genome-wide association studies (GWAS). Loci with significant expression quantitative trait loci (eQTL) annotations are highlighted; the number of tissues is shown parenthetically, and symbol orientation indicates higher or lower predicted expression. Additional findings at 17q21.31 are denoted with b, and replicated GWS loci appear as watermark. *C4A is highlighted for reference.
Gene Set Enrichment
Our primary SCZ results were significantly enriched for a single GO term, positive regulation of oligodendrocyte differentiation (supplementary tables S24 and S25). Across meta-analyses, GWAS of SCZ and BIP highlighted enrichment in 22 and 3 gene-sets (supplementary tables S26–S28), respectively. The enriched terms were related to the aspects of neuronal structure, dendrite growth, and synaptic transmission.
Discussion
We describe primary genomic analyses of SCZ and BIP in CSP #572, offering new insights into the etiologies of these disorders and demonstrating that published findings are largely transferable to this diverse cohort of US veterans.58 We combined our results with summary statistics from recent GWAS,13–15,18 realizing the largest and most inclusive GWAS of these disorders to date, and explore the functional biological relevance of 52 newly identified susceptibility loci.
Our primary GWAS of SCZ yielded a novel association in chromodomain helicase DNA-binding protein 7 (CHD7), a critical player in the epigenetic regulation of neuronal differentiation,59,60 disruptions of which are believed to be associated with SCZ and autism.61,62 Mutations in CHD7 are observed in developmental disorders,63–66 and common variant associations have been reported with a range of human traits.67–70
We report a novel BIP association in SORCS3, encoding sortilin-related VPS10 domain-containing receptor 3, which plays key roles in glutamate homeostasis and establishing long-term depression in hippocampal neurons.71,72SORCS3 variants have demonstrated pleiotropic effects on SCZ, major depression (MD), anorexia nervosa, attention hyperactivity disorder, Tourette’s syndrome,73 and cognition.74 This finding was not replicated in other BIP cohorts, likely reflecting heterogeneity among participants and across studies.
We observed another GWS association downstream of Protocadherin 11 X-linked (PCDH11X), one of a human-specific X/Y gene pair associated with the regulation of dendritic branching75 and neuronal differentiation and proliferation,76 and which has postulated evolutionary relevance to the neural correlates of language.77
Combining results for CSP #572 with published findings, we detected an additional 39 susceptibility loci for SCZ and 10 for BIP and fine-mapped 84 associations. New loci were enriched in eQTLs for CNS-expressed genes, including a novel BIP association in Long Intergenic Non-Protein Coding RNA 1470 (LINC01470) spanning eQTLs for neuromedin U receptor 2 (NMUR2), a neuropeptide receptor that is presynaptically expressed in GABAergic neurons and enriched in the nucleus accumbens shell and has been implicated in mediating food reward and behavioral responses to cocaine.78 Taken together, the NMUR2 and SORCS3 findings support the role of glutamatergic dysregulation in SCZ52,79,80 and perhaps BIP.81–83
We observed a novel association between SCZ and variants upstream of CRHR1 (figure 1B), encoding corticotropin-releasing hormone receptor 1 and was recently found to be associated with reexperiencing symptoms of post-traumatic stress disorder (PTSD) in the MVP.27 Nearby SNPs are GWS for neuroticism,84,85 cognitive performance,86,87 subcortical88 and intracranial volume,89 Parkinson’s disease90–93 and corticobasal degeneration,94 sleep duration,95 alcohol use disorder,96,97 various risky behaviors,98 and a number of other traits.67,99–104 The 17q21.31 locus harbors a ~900-kb inversion that has undergone positive selection in Europe but is rare among AA and EAS individuals105; the resultant pattern of long-range LD poses challenges to statistical fine-mapping and may complicate the interpretation of eQTL effects; however, we achieved a small reduction in causal credible SNPs via trans-ancestry analysis. As noted by Gelernter and colleagues,27 a weaker signal in a smaller AA sample, nonetheless, lends meaningful insight into the likely underlying association at this locus.
We observed improved fine-mapping resolution at 70 independent SCZ loci, with 21 overlapping between meta-analyses of CSP #572 and GPC with PGC + CLOZUK or PGC + EAS. Integration of diverse ancestry data yielded more fine-mapped loci in the PGC + CLOZUK meta-analysis, with slightly larger reductions in the median number of credible SNPs and length of the corresponding genomic interval. Our results are comparable to those reported in the original PGC + EAS study15 and based on analogous methods and criteria; in both studies, 44 independent associations had improved significance and smaller credible SNP sets following trans-ancestry meta-analysis, with only 3 loci overlapping across studies. This supports distinct contributions of the AA and LA data featured herein, reflecting local LD structures that are divergent from EA and EAS populations.106 In particular, the inclusion of AA study data is expected to yield the greatest improvements in fine-mapping, given a higher degree of genetic diversity and shorter haplotype blocks on average compared with out-of-Africa populations.106–108
Consistent with expectations, PRS constructed from published GWAS results were robustly associated with case-control status in CSP #572 but explained a small fraction of the overall variability in liability. Among AA participants, we saw markedly improved performance for scores constructed from multi-ancestry GWAS findings. We observed strong associations of PRS across populations and diagnoses, highlighting the importance of studying diverse populations.13,109
For both SCZ and BIP, a substantive fraction of the variance was attributable to genome-wide SNPs. Estimates of SNP-h2 for BIP were significantly lower than previously reported (P < .005),4 and this was also true for SCZ among AA participants in CSP #572.4,110
It should be noted that SNP-h2 represents a lower bound on the “narrow-sense” h2111; estimates of the total h2 of SCZ range from .31112 to .67.113
Interpretations and Conclusions
Our findings are demonstrative of a shared genetic basis of SCZ and BIP between this highly selected and severely affected veteran cohort and previous case-control studies of both disorders. We expect that the comprehensive assessment and validity of the clinical assessments in CSP #572 represent a “gold standard” for the curation of serious mental illness (SMI) phenotypes in the US veteran population. Taken together with the completeness and scale of the VA EHR, forthcoming studies of CSP #572 and the MVP hold considerable promise to advance our understanding of how genetic and environmental/experiential factors (G × E) influence disease susceptibility, neurocognitive function, treatment response, and patient outcomes. Future analyses of this dataset will explore genomic determinants of suicidal ideation and behavior, comparative risk for PTSD with non-SMI populations, and multifactorial influences on lifetime MD, negative symptoms, and other salient symptomatic domains.
Supplementary Material
Acknowledgments
The contents do not represent the views of the US Department of Veterans Affairs or the US Government. Complete acknowledgments for CSP #572, MVP, GPC, and COGS are presented in the Supplementary Materials. Dr. Harvey has served as a consultant to multiple pharmaceutical companies and device manufacturers on phase 2 or 3 development; this consulting work has been determined to be unrelated to the content of the article. No other authors report any relevant conflicts of interest.
Funding
This research was supported by the Department of Veterans Affairs Cooperative Studies Program (CSP #572) and the Million Veteran Program (MVP-000). The MVP is supported by the Office of Research and Development, US Department of Veterans Affairs. Dr. Bigdeli is the recipient of a 2019 NARSAD Young Investigator Grant from the Brain and Behavior Research Foundation. Other relevant funding information for GPC, COGS, MGS, and BiGS is presented in the Supplementary Materials.
References
- 1. Whiteford HA, Degenhardt L, Rehm J, et al. Global burden of disease attributable to mental and substance use disorders: findings from the Global Burden of Disease Study 2010. Lancet. 2013;382(9904):1575–1586. [DOI] [PubMed] [Google Scholar]
- 2. Cloutier M, Aigbogun MS, Guerin A, et al. The economic burden of schizophrenia in the United States in 2013. J Clin Psychiatry. 2016;77(6):764–771. [DOI] [PubMed] [Google Scholar]
- 3. Purcell SM, Wray NR, Stone JL, et al. ; International Schizophrenia, Consortium . Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009;460:748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee SH, Ripke S, Neale BM, et al. ; Cross-Disorder Group of the Psychiatric Genomics Consortium . Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cloutier M, Greene M, Guerin A, Touya M, Wu E. The economic burden of bipolar I disorder in the United States in 2015. J Affect Disord. 2018;226:45–51. [DOI] [PubMed] [Google Scholar]
- 6. Reichenberg A, Harvey PD, Bowie CR, et al. Neuropsychological function and dysfunction in schizophrenia and psychotic affective disorders. Schizophr Bull. 2009;35(5):1022–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harvey PD, Siever LJ, Huang GD, et al. The genetics of functional disability in schizophrenia and bipolar illness: methods and initial results for VA Cooperative Study #572. Am J Med Genet B Neuropsychiatr Genet. 2014;165B(4):381–389. [DOI] [PubMed] [Google Scholar]
- 8. Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shi J, Levinson DF, Duan J, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature 2009;460(7256):753–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43;969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ripke S, O’Dushlaine C, Chambert K, et al. ; Multicenter Genetic Studies of Schizophrenia Consortium ; Psychosis Endophenotypes International Consortium; Wellcome Trust Case Control Consortium 2. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45(10):1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schizophrenia Working Group of the Psychiatric Genomics, Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014;511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bigdeli TB, Genovese G, Georgakopoulos P, et al. Contributions of common genetic variants to risk of schizophrenia among individuals of African and Latino ancestry. Mol Psychiatry. 2019. doi: 10.1038/s41380-019-0517-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pardiñas AF, Holmans P, Pocklington AJ, et al. ; GERAD1 Consortium ; CRESTAR Consortium. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50(3):381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lam M, Chen CY, Li Z, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium ; Indonesia Schizophrenia Consortium; Genetic REsearch on schizophreniA neTwork-China and the Netherlands (GREAT-CN). Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat Genet. 2019;51(12):1670–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mühleisen TW, Leber M, Schulze TG, et al. Genome-wide association study reveals two new risk loci for bipolar disorder. Nat Commun. 2014;5:3339. [DOI] [PubMed] [Google Scholar]
- 18. Stahl EA, Breen G, Forstner AJ, et al. ; eQTLGen Consortium ; BIOS Consortium; Bipolar Disorder Working Group of the Psychiatric Genomics Consortium. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51(5):793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. O’Connor LJ, Schoech AP, Hormozdiari F, Gazal S, Patterson N, Price AL. Extreme polygenicity of complex traits is explained by negative selection. Am J Hum Genet. 2019;105(3):456–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gaziano JM, Concato J, Brophy M, et al. Million Veteran Program: a mega-biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214–223. [DOI] [PubMed] [Google Scholar]
- 21. Pato MT, Sobell JL, Medeiros H, et al. ; Genomic Psychiatry Cohort Consortium . The Genomic Psychiatry Cohort: partners in discovery. Am J Med Genet B Neuropsychiatr Genet. 2013;162B(4):306–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Calkins ME, Dobie DJ, Cadenhead KS, et al. The Consortium on the Genetics of Endophenotypes in Schizophrenia: model recruitment, assessment, and endophenotyping methods for a multisite collaboration. Schizophr Bull. 2007;33(1):33–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Smith EN, Bloss CS, Badner JA, et al. Genome-wide association study of bipolar disorder in European American and African American individuals. Mol Psychiatry. 2009;14(8):755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. American Psychiatric Association. Diagnostic and statistical manual of mental disorders: DSM-IV-TR. Washington, DC: American Psychiatric Association; 2000. [Google Scholar]
- 25. First MB, Spitzer RL, Gibbon M, Williams JBW. Structured Clinical Interview for DSM-IV-TR Axis I Disorders: Patient Edition. New York, NY: Biometrics Research Department, Columbia University; 2005. [Google Scholar]
- 26. Klarin D, Damrauer SM, Cho K, et al. ; Global Lipids Genetics Consortium ; Myocardial Infarction Genetics (MIGen) Consortium; Geisinger-Regeneron DiscovEHR Collaboration; VA Million Veteran Program. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nat Genet. 2018;50(11):1514–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gelernter J, Sun N, Polimanti R, et al. ; Department of Veterans Affairs Cooperative Studies Program (#575B) and Million Veteran Program . Genome-wide association study of post-traumatic stress disorder reexperiencing symptoms in >165,000 US veterans. Nat Neurosci. 2019;22(9):1394–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Auton A, Brooks LD, Durbin RM, et al. ; 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Das S, Forer L, Schönherr S, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics 2010;26(22):2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Abraham G, Qiu Y, Inouye M. FlashPCA2: principal component analysis of Biobank-scale genotype datasets. Bioinformatics 2017;33(17):2776–2778. [DOI] [PubMed] [Google Scholar]
- 33. Price AL, Weale ME, Patterson N, et al. Long-range LD can confound genome scans in admixed populations. Am J Hum Genet. 2008;83(1):132–135; author reply 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Han B, Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am J Hum Genet. 2011;88(5):586–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dick DM, Foroud T, Flury L, et al. Genomewide linkage analyses of bipolar disorder: a new sample of 250 pedigrees from the National Institute of Mental Health Genetics Initiative. Am J Hum Genet. 2003;73(1):107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McInnis MG, Dick DM, Willour VL, et al. Genome-wide scan and conditional analysis in bipolar disorder: evidence for genomic interaction in the National Institute of Mental Health genetics initiative bipolar pedigrees. Biol Psychiatry. 2003;54(11):1265–1273. [DOI] [PubMed] [Google Scholar]
- 39. Hamshere ML, Walters JT, Smith R, et al. ; Schizophrenia Psychiatric Genome-wide Association Study Consortium ; Wellcome Trust Case Control Consortium+; Wellcome Trust Case Control Consortium 2. Genome-wide significant associations in schizophrenia to ITIH3/4, CACNA1C and SDCCAG8, and extensive replication of associations reported by the Schizophrenia PGC. Mol Psychiatry. 2013;18(6):708–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nakazawa M. Practices of medical and health data analysis using R. Japan: Pearson Education; 2007. [Google Scholar]
- 41. Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88(3):294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee SH, Goddard ME, Wray NR, Visscher PM. A better coefficient of determination for genetic profile analysis. Genet Epidemiol. 2012;36(3):214–224. [DOI] [PubMed] [Google Scholar]
- 43. Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brown BC, Ye CJ, Price AL, Zaitlen N; Asian Genetic Epidemiology Network Type 2 Diabetes Consortium . Transethnic genetic-correlation estimates from summary statistics. Am J Hum Genet. 2016;99(1):76–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bulik-Sullivan BK, Loh PR, Finucane HK, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium . LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zheng J, Erzurumluoglu AM, Elsworth BL, et al. ; Early Genetics and Lifecourse Epidemiology (EAGLE) Eczema Consortium . LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 2017;33(2):272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huang H, Fang M, Jostins L, et al. ; International Inflammatory Bowel Disease Genetics Consortium . Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 2017;547(7662):173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gamazon ER, Wheeler HE, Shah KP, et al. ; GTEx Consortium . A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47(9):1091–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Barbeira AN, Dickinson SP, Bonazzola R, et al. ; GTEx Consortium . Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun. 2018;9(1):1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2010;26(18):2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Coyle JT, Tsai G, Goff D. Converging evidence of NMDA receptor hypofunction in the pathophysiology of schizophrenia. Ann N Y Acad Sci. 2003;1003:318–327. [DOI] [PubMed] [Google Scholar]
- 53. Howard DM, Adams MJ, Clarke TK, et al. ; 23andMe Research Team ; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22(3):343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bulik-Sullivan B, Finucane HK, Anttila V, et al. ; ReproGen Consortium ; Psychiatric Genomics Consortium; Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47(11):1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Anttila V, Bulik-Sullivan B, Finucane HK, et al. ; Brainstorm Consortium . Analysis of shared heritability in common disorders of the brain. Science 2018;360(6395):eaap8757. doi: 10.1126/science.aap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Huckins LM, Dobbyn A, Ruderfer DM, et al. ; CommonMind Consortium ; Schizophrenia Working Group of the Psychiatric Genomics Consortium; iPSYCH-GEMS Schizophrenia Working Group. Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nat Genet. 2019;51(4):659–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hall LS, Medway CW, Pain O, et al. A transcriptome-wide association study implicates specific pre- and post-synaptic abnormalities in schizophrenia. Hum Mol Genet. 2020;29(1):159–167. doi: 10.1093/hmg/ddz253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Harvey PD, Sun N, Bigdeli TB, et al. ; Consortium on the Genetics of Schizophrenia (COGS) . Genome-wide association study of cognitive performance in U.S. veterans with schizophrenia or bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 2020;183(3):181–194. [DOI] [PubMed] [Google Scholar]
- 59. Feng W, Khan MA, Bellvis P, et al. The chromatin remodeler CHD7 regulates adult neurogenesis via activation of SoxC transcription factors. Cell Stem Cell 2013;13(1):62–72. [DOI] [PubMed] [Google Scholar]
- 60. Feng W, Kawauchi D, Körkel-Qu H, et al. Chd7 is indispensable for mammalian brain development through activation of a neuronal differentiation programme. Nat Commun. 2017;8:14758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. McCarthy SE, Gillis J, Kramer M, et al. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol Psychiatry. 2014;19(6):652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liu J, Chen J, Ehrlich S, et al. Methylation patterns in whole blood correlate with symptoms in schizophrenia patients. Schizophr Bull. 2014;40(4):769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vissers LE, van Ravenswaaij CM, Admiraal R, et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36(9):955–957. [DOI] [PubMed] [Google Scholar]
- 64. Cadman SM, Kim SH, Hu Y, González-Martínez D, Bouloux PM. Molecular pathogenesis of Kallmann’s syndrome. Horm Res. 2007;67(5):231–242. [DOI] [PubMed] [Google Scholar]
- 65. O’Roak BJ, Vives L, Girirajan S, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 2012;485(7397):246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Homsy J, Zaidi S, Shen Y, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science 2015;350(6265):1262–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Astle WJ, Elding H, Jiang T, et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell 2016;167(5):1415–1429.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Teumer A, Li Y, Ghasemi S, et al. Genome-wide association meta-analyses and fine-mapping elucidate pathways influencing albuminuria. Nat Commun. 2019;10(1):4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Day FR, Ruth KS, Thompson DJ, et al. ; PRACTICAL consortium ; kConFab Investigators; AOCS Investigators; Generation Scotland; EPIC-InterAct Consortium; LifeLines Cohort Study. Large-scale genomic analyses link reproductive aging to hypothalamic signaling, breast cancer susceptibility and BRCA1-mediated DNA repair. Nat Genet. 2015;47(11):1294–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kichaev G, Bhatia G, Loh PR, et al. Leveraging polygenic functional enrichment to improve GWAS power. Am J Hum Genet. 2019;104(1):65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Willnow TE, Petersen CM, Nykjaer A. VPS10P-domain receptors — regulators of neuronal viability and function. Nat Rev Neurosci.. 2008;9:899–909. [DOI] [PubMed] [Google Scholar]
- 72. Breiderhoff T, Christiansen GB, Pallesen LT, et al. Sortilin-related receptor SORCS3 is a postsynaptic modulator of synaptic depression and fear extinction. PLoS One. 2013;8(9):e75006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cross-Disorder Group of the Psychiatric Genomics Consortium. Electronic address: plee0@mgh.harvard.edu, and Cross-Disorder Group of the Psychiatric Genomics Consortium. Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell 2019;179:1469–1482.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lam M, Hill WD, Trampush JW, et al. Pleiotropic meta-analysis of cognition, education, and schizophrenia differentiates roles of early neurodevelopmental and adult synaptic pathways. Am J Hum Genet. 2019;105(2):334–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wu C, Niu L, Yan Z, et al. Pcdh11x negatively regulates dendritic branching. J Mol Neurosci. 2015;56(4):822–828. [DOI] [PubMed] [Google Scholar]
- 76. Zhang P, Wu C, Liu N, et al. Protocadherin 11 x regulates differentiation and proliferation of neural stem cell in vitro and in vivo. J Mol Neurosci. 2014;54(2):199–210. [DOI] [PubMed] [Google Scholar]
- 77. Priddle TH, Crow TJ. The protocadherin 11X/Y (PCDH11X/Y) gene pair as determinant of cerebral asymmetry in modern Homo sapiens. Ann N Y Acad Sci. 2013;1288:36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kasper JM, McCue DL, Milton AJ, et al. Gamma-aminobutyric acidergic projections from the dorsal raphe to the nucleus accumbens are regulated by neuromedin U. Biol Psychiatry. 2016;80(11):878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Coyle JT. NMDA receptor and schizophrenia: a brief history. Schizophr Bull. 2012;38(5):920–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Goff DC. Drug development in schizophrenia: are glutamatergic targets still worth aiming at? Curr Opin Psychiatry. 2015;28(3):207–215. [DOI] [PubMed] [Google Scholar]
- 81. Bodkin JA, Coleman MJ, Godfrey LJ, et al. Targeted treatment of individuals with psychosis carrying a copy number variant containing a genomic triplication of the glycine decarboxylase gene. Biol Psychiatry. 2019;86(7):523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Veldic M, Millischer V, Port JD, et al. Genetic variant in SLC1A2 is associated with elevated anterior cingulate cortex glutamate and lifetime history of rapid cycling. Transl Psychiatry. 2019;9(1):149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Stahl EA, Breen G, Forstner AJ, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51(5):793–803. doi: 10.1038/s41588-019-0397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Okbay A, Baselmans BM, De Neve JE, et al. ; LifeLines Cohort Study . Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet. 2016;48(6):624–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Nagel M, Watanabe K, Stringer S, Posthuma D, van der Sluis S. Item-level analyses reveal genetic heterogeneity in neuroticism. Nat Commun. 2018;9(1):905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lam M, Trampush JW, Yu J, et al. Large-scale cognitive GWAS meta-analysis reveals tissue-specific neural expression and potential nootropic drug targets. Cell Rep. 2017;21(9):2597–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Davies G, Lam M, Harris SE, et al. Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function. Nat Commun. 2018;9(1):2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ikram MA, Fornage M, Smith AV, et al. ; Early Growth Genetics Consortium ; Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium. Common variants at 6q22 and 17q21 are associated with intracranial volume. Nat Genet. 2012;44(5):539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hibar DP, Stein JL, Renteria ME, et al. ; Alzheimer’s Disease Neuroimaging Initiative ; CHARGE Consortium; EPIGEN; IMAGEN; SYS. Common genetic variants influence human subcortical brain structures. Nature 2015;520(7546):224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chang D, Nalls MA, Hallgrímsdóttir IB, et al. ; International Parkinson’s Disease Genomics Consortium ; 23andMe Research Team. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat Genet. 2017;49(10):1511–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Nalls MA, Pankratz N, Lill CM, et al. ; International Parkinson’s Disease Genomics Consortium (IPDGC) ; Parkinson’s Study Group (PSG) Parkinson’s Research: The Organized GENetics Initiative (PROGENI); 23andMe; GenePD; NeuroGenetics Research Consortium (NGRC); Hussman Institute of Human Genomics (HIHG); Ashkenazi Jewish Dataset Investigator; Cohorts for Health and Aging Research in Genetic Epidemiology (CHARGE); North American Brain Expression Consortium (NABEC); United Kingdom Brain Expression Consortium (UKBEC); Greek Parkinson’s Disease Consortium; Alzheimer Genetic Analysis Group. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat Genet. 2014;46(9):989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Nalls MA, Plagnol V, Hernandez DG, et al. ; International Parkinson Disease Genomics Consortium . Imputation of sequence variants for identification of genetic risks for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet 2011;377:641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Simón-Sánchez J, Schulte C, Bras JM, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet. 2009;41(12):1308–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kouri N, Ross OA, Dombroski B, et al. Genome-wide association study of corticobasal degeneration identifies risk variants shared with progressive supranuclear palsy. Nat Commun. 2015;6:7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Doherty A, Smith-Byrne K, Ferreira T, et al. GWAS identifies 14 loci for device-measured physical activity and sleep duration. Nat Commun. 2018;9(1):5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Sanchez-Roige S, Palmer AA, Fontanillas P, et al. ; 23andMe Research Team, the Substance Use Disorder Working Group of the Psychiatric Genomics Consortium . Genome-wide association study meta-analysis of the alcohol use disorders identification test (AUDIT) in two population-based cohorts. Am J Psychiatry. 2019;176(2):107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Liu M, Jiang Y, Wedow R, et al. ; 23andMe Research Team ; HUNT All-In Psychiatry. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51(2):237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Karlsson Linnér R, Biroli P, Kong E, et al. ; 23and Me Research Team ; eQTLgen Consortium; International Cannabis Consortium; Social Science Genetic Association Consortium. Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet. 2019;51(2):245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Höglinger GU, Melhem NM, Dickson DW, et al. ; PSP Genetics Study Group . Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat Genet. 2011;43(7):699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Wain LV, Shrine N, Artigas MS, et al. ; Understanding Society Scientific Group ; Geisinger-Regeneron DiscovEHR Collaboration. Genome-wide association analyses for lung function and chronic obstructive pulmonary disease identify new loci and potential druggable targets. Nat Genet. 2017;49(3):416–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Phelan CM, Kuchenbaecker KB, Tyrer JP, et al. ; AOCS study group ; EMBRACE Study; GEMO Study Collaborators; HEBON Study; KConFab Investigators; OPAL study group. Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nat Genet. 2017;49(5):680–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Tachmazidou I, Hatzikotoulas K, Southam L, et al. ; arcOGEN Consortium . Identification of new therapeutic targets for osteoarthritis through genome-wide analyses of UK Biobank data. Nat Genet. 2019;51(2):230–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Pickrell JK, Berisa T, Liu JZ, Ségurel L, Tung JY, Hinds DA. Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet. 2016;48(7):709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Pirastu N, Joshi PK, de Vries PS, et al. GWAS for male-pattern baldness identifies 71 susceptibility loci explaining 38% of the risk. Nat Commun. 2017;8(1):1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Stefansson H, Helgason A, Thorleifsson G, et al. A common inversion under selection in Europeans. Nat Genet. 2005;37(2):129–137. [DOI] [PubMed] [Google Scholar]
- 106. Asimit JL, Hatzikotoulas K, McCarthy M, Morris AP, Zeggini E. Trans-ethnic study design approaches for fine-mapping. Eur J Hum Genet. 2016;24(9):1330–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Morris AP. Transethnic meta-analysis of genomewide association studies. Genet Epidemiol. 2011;35(8):809–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Gurdasani D, Carstensen T, Tekola-Ayele F, et al. The African genome variation project shapes medical genetics in Africa. Nature 2015;517(7534):327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Peterson RE, Kuchenbaecker K, Walters RK, et al. Genome-wide association studies in ancestrally diverse populations: opportunities, methods, pitfalls, and recommendations. Cell 2019;179(3):589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. de Candia TR, Lee SH, Yang J, et al. ; International Schizophrenia Consortium ; Molecular Genetics of Schizophrenia Collaboration. Additive genetic variation in schizophrenia risk is shared by populations of African and European descent. Am J Hum Genet. 2013;93(3):463–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Zaitlen N, Kraft P. Heritability in the genome-wide association era. Hum Genet. 2012;131(10):1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Light G, Greenwood TA, Swerdlow NR, et al. Comparison of the heritability of schizophrenia and endophenotypes in the COGS-1 family study. Schizophr Bull. 2014;40(6):1404–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wray NR, Gottesman II. Using summary data from the danish national registers to estimate heritabilities for schizophrenia, bipolar disorder, and major depressive disorder. Front Genet. 2012;3:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.