Abstract
Evidence regarding protein structure and function manifest the imperative role that dynamics play in proteins, underlining reconsideration of the unanimated sequence-to-structure-to-function paradigm. Structural dynamics portray a heterogeneous energy landscape described by conformational ensembles where each structural representation can be responsible for unique functions or enable macromolecular assemblies. Using the human p27/Cdk2/Cyclin A ternary complex as an example, we highlight the vital role of intramolecular and intermolecular dynamics for target recognition, binding, and inhibition as a critical modulator of cell division. Rapidly sampling configurations is critical for the population of different conformational ensembles encoding functional roles. To garner this knowledge, we present how the integration of (sub)ensemble and single-molecule fluorescence spectroscopy with molecular dynamic simulations can characterize structural dynamics linking the heterogeneous ensembles to function. The incorporation of dynamics into the sequence-to-structure-to-function paradigm promises to assist in tackling various challenges, including understanding the formation and regulation of mesoscale assemblies inside cells.
Introduction
From sequence to ensemble(s) and function(s)
Proteins have a widespread and crucial role in maintaining the cell’s metabolism, impacting almost every metabolic process to ensure survival and evolution. Since the first three-dimensional description of a protein at atomic resolution obtained by Perutz in 1960 [1], the unanimated vision of a protein has led to the well-known sequence-to-structure-to-function paradigm. However, the classical textbook paradigm largely disregards the relevance of dynamical processes between these conformations; due to their inherent thermal fluctuations and chain flexibility, proteins continuously explore different configurations, reaching the accumulation of structural conformations that define the whole structural ensemble (Figure 1a). Then, dynamics not only allow reproducing what we observe as the native state, but also characterizes the proteins’ function(s), properties, and regulation.
Figure 1.
Relationship between sequence, structure, dynamics, and function. (a) As the original paradigm stated, the linear sequence-structure-function relationship that explained the proteins’ properties and functions obscure the relevance of the structural dynamics. Several meta-stable configurations are grouped into more stable conformations, defining the structural ensemble and what we observe as the native state. (b) Depending on the energy barriers between different structural transitions, different dynamic processes can occur between nanoseconds to milliseconds timescales depending on the energy barrier to pass. These processes conform to both intra or intermolecular ensembles. For intrinsically-disordered proteins (IDPs) (red line), the high structural heterogeneity leads to energy frustration. This heterogeneity can be decreased by employing contacts with small ligands or even other macromolecules, adopting intra- or inter-molecular structural ensembles (dashed red lines).
Using his seminal experimental findings, Anfinsen indirectly described the first evidence of inherent dynamics in the folding of a protein [2]. Building on that view, Levinthal [3], and later Wolynes [4], suggested that specific topological constraints from the amino acid chain must guide folding to satisfy the timescales typically observed in vitro and in vivo. The minimally-frustrated nature of proteins allows them to rapidly explore several short-lived configurations with high structural entropy and low transition energy barriers. Increasing the energetic barriers between configurations causes the adoption of a native/functional ensemble, highlighting the delicate balance between dynamics, structure, and function.
However, the discovery of proteins that show complex folding pathways leading to intricate functions has suggested a revision of this sequence-to-structure-to-function paradigm. Such is the case, for example, of proteins that dimerize via three-dimensional domain swapping (3D-DS) [5••]. These proteins contain local intrinsically disordered regions (IDRs), causing them to lack a well-defined, stable, and minimally-frustrated native ensemble. Moreover, several others are entirely disordered (IDPs) [6-10], showing highly-dynamical competing configurations (Figure 1b). While well-folded proteins show slower transitions as they jump over high energy barriers between distinct states, IDPs must be analyzed at shorter timescales to sample their different configurations due to their faster configurational dynamics.
Interestingly, for most locally or completely disordered proteins [11, 12, 13•], binding offers a mechanism for folding [14, 15], adding a regulatory layer. For binding reactions in proteins and other macromolecules (i.e. nucleic acids) [16, 17•,18,19], dynamics can exhibit dominant effects on association and/or dissociation rates by performing a pivotal role in specificity/promiscuity [20-22], thus affecting the lifetime of those complexes. In these cases, binding reactions allow switching between ensembles (Figure 1b). Additionally, binding, (un)folding, and dynamics can modulate micro and mesoscale molecular assemblies, such as membrane-less organelles [23,24•] and liquid phase condensates [25-27,28•], critical components in compartmentalization and other intricate functions within cells (Figure 1b). This new understanding of proteins fills a clear gap in the sequence-to-structure-to-function paradigm to explain numerous biological phenomena where the structure itself is insufficient.
Because solution nuclear magnetic resonance (NMR) can study molecules at the atomic level with a high temporal and spatial resolution [29], it is currently the gold standard ensemble approach to describe local and global structural changes of proteins in folding, binding, and function [30•,31]. As such, NMR gives experimental descriptions of the intra- and intermolecular changes between pico- to milliseconds regimes [32] and dynamic behavior between micro- to milliseconds (and beyond), allowing extensive studies into their involvement in folding and binding [11,33,34] (Figure 2a). However, NMR and other classical ensemble methodologies, although possessing high temporal resolution, struggle to characterize the short-lived configurations of highly-flexible proteins due to the need for high data throughput and ensemble averaging. For IDPs in particular, defined ensembles link to specific functions by integrating and processing signals when folded into stable structures upon binding to cellular regulatory partners, emphasizing the complexity of the (un)folding and function relationship.
Figure 2.
Conformational dynamics and experimental approaches to study. (a) Temporal and size scales covered by the combination of experimental and bioinformatic approaches. Main fluorescent methods in freely diffusing conditions (Fluorescence Correlation Spectroscopy -FCS- and Förster Resonance Energy Transfer -FRET-) are used by employing Time-Correlated Single Photon Counting (TCSPC), filtered FCS (fFCS) and burst analysis. These approaches are combined with Molecular Dynamics simulations (MD) (all-atom and coarse-grained) to cover from nano- to milliseconds in temporal resolution and from nano- to micrometers in size scale. For slower temporal scales, microscopy approaches focused on analyzing fixed molecules are ideal. (b) Flow chart to study structural dynamics using single-molecule multiparameter fluorescence spectroscopy (smMFS) toolbox. (Sub)ensemble (TCSPC and fFCS) and single-molecule (FRET and anisotropy) approaches, combined with MD can describe local structural changes at high temporal resolution. Each technique provides complementary information to each other, painting a complete picture across the accessible timescales: TCSPC: distribution of conformations present in a specified condition (monitored by FRET) that are stable on the nanoseconds timescale (> fluorophore lifetime); fFCS: solving of different relaxation times accounting for structural changes across time; smFA: high sensitivity to local flexibility changes; smFRET: quantification of different distance changes spanning a protein or protein complex via High FRET (HF) or Low FRET (LF) states, distributions of these distances, and the kinetic forward (kf) and backward (kf) rates of exchange; MD: refinement of structural models generated by the experimental considerations.
The unique advantage of single-molecule methodologies is in their ability to unravel structural heterogeneity, in most cases, without ensemble averaging. Experimental results based on fluorescence are widely exploited due to their excellent structural and temporal resolution [35-37]. Taking into advantage the different approaches and experimental corrections derived from fluorescence, single-molecule multiparameter fluorescence spectroscopy (smMFS) is a robust methodology to accurately monitor and quantify local and global dynamic changes [38-40,41•,42•]. When combined with (sub)ensemble approximations, such as Fluorescence Correlation Spectroscopy (FCS) and Time-Correlated Single-Photon Counting (TCSPC), smMFS allow the monitoring of structural changes in a broad time scale from nano-to-milliseconds [38-40,41•,42•]. For slower processes, approaches focused on fixed molecules are ideal, monitoring real-time structural changes [41•]. Specifically, single-molecule fluorescence anisotropy (smFA) allows the monitoring of local changes that reflect side-chain dynamics. Also, single-molecule Förster Resonance Energy Transfer (smFRET), when used with time-resolved fluorescence spectroscopy (TCSPC), probes distance changes and population heterogeneity with nanoseconds resolution. When coupled to burst analysis, smFRET is sensitive to dynamics over broad time scales, from milliseconds to seconds depending on instrumentation [38-40,41•,42•]. Finally, filtered FCS (fFCS) becomes ideal for following exchange processes between FRET states to quantify the structural dynamics between (sub)micro- to milliseconds (Figure 2b).
Moreover, due to the comparable timescales covered by smMFS and molecular dynamics (MD) simulations (Figure 2a), the combination of experimental results with MD lead to more accurate structural dynamics models to fully understand protein dynamics at the atomic scale [43••]. In particular, coarse-grain models, by requiring less computational resources than all-atom models to manage intra- and intermolecular interactions [44-46], is preferred when modeling larger, complex, multi-protein structures, such as quinary protein structures [47,48]. As such, coarse-grain models have become instrumental in recent modeling [49-53]. By coupling the smMFS with computational approaches, the smMFS toolbox is built (Figure 2b). This toolbox allows us to monitor several aspects of protein function, including folding [36,54], super tertiary [55•,56-59] and quaternary communications [5••,13•], and enzyme catalysis [60••,61], emphasizing how those processes create more extensive, dynamic, three-dimensional systems responsible for life.
Uncovering the role of dynamics in the sequence-to-structure-to-function paradigm: conformational heterogeneity as pivotal for proteins’ functions
One hallmark model highlighting the relevance of dynamics in protein function at high resolution is the human p27. This disordered protein causes cell cycle arrest when binding in a ternary complex with cyclin-dependent kinase (Cdk2) and cyclins (e.g. Cdk2/Cyclin A) [62,63] (Figure 3a). A recent integrative and collaborative work between multiple laboratories revealed how p27 morphs lead to the formation of the p27/ Cdk2/Cyclin A complex. Different constructs of p27 were studied using stopped-flow kinetics and the smMFS toolbox (Figure 2b) to identify the critical events that led to the initiation complex. An intricate combination of intra- and intermolecular dynamics seems to modulate this protein’s biological function (Figure 3).
Figure 3.
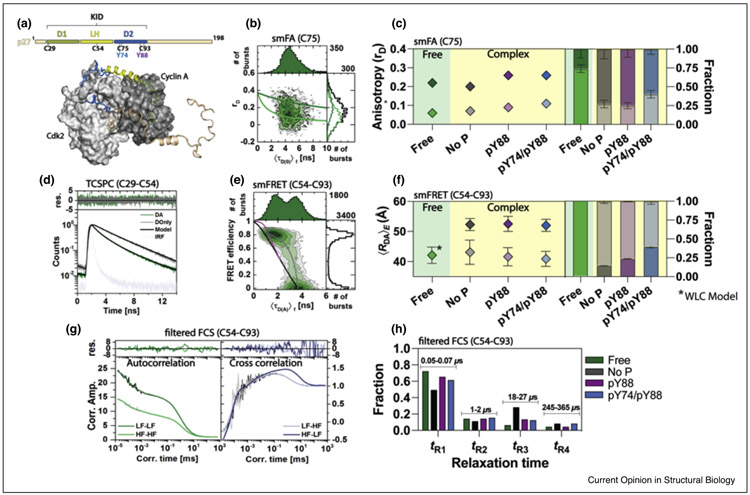
smMFS toolbox to study intra- and intermolecular dynamics of p27. (a) the topology of human p27 showing the relevant regions (D1, LH, and D2) of its Kinase Inhibitory Domain (KID). C29, C54, C75, and C93 are cysteine residues used to attach the different fluorophores, whereas Y74 and Y88 are tyrosine residues that can be phosphorylated. The ternary complex p27/Cdk2/Cyclin 2 is in cartoon. (b) smFA plot for free p27 monitoring C75 attached with Bodipy Fl, showing the two anisotropy population of the D2 region. (c) Quantitative analysis of free p27 and the ternary complex with its different phosphorylation modifications (No P, pY88, and pY74/p88), showing anisotropy values and their fraction in all conditions. (d) TCSPC plot showing fluorescence decay of free p27 monitoring C29-C54 attached with donor and acceptor of FRET (DA), donor only labeled p27 (DOnly), and the instrument response function (IRF). Differences between DA and DOnly serves as a baseline for comparison and FRET efficiency determination. (e) smFRET distribution monitoring distance changes in free p27 labeled, as mentioned in (D). The black line corresponds to the static FRET line, gray line corresponds to the dynamic FRET between DOnly and high FRET, and in pink line, a worm-like chain (WLC) model considering an equilibrium between a disordered and folded protein. (f) Quantitative analysis of (E) shows distances and fractions in free p27 in the same conditions as mentioned in (C). For free p27, FRET distance was determined using a WLC model. (g) fFCS plots show both auto- and cross-correlation between low and high FRET. (h) Quantitative analysis from data obtained in (F) for p27 in all before mentioned conditions. Data fitting found four different exchanging times (tR) for all conditions, showing the specific fraction for each one.
Intramolecular dynamics: structural heterogeneity of proteins as functional limiting events
Unbound or free p27 is mostly disordered while maintaining some residual alpha-helical structure in the LH subdomain consistent with prior studies [64-67]. As shown in Tsytlonok et al. [68••], free p27 adopts a compact conformation, impeding the acquisition of the ternary complex with Cdk2/Cyclin A. Hence, p27 must expand to expose its 12 residues recognition site in the D1 subdomain [69], being crucial in the association kinetics to Cyclin A by undergoing conformational rearrangement before initial binding [68••]. Similarly, the D2 region must exchange conformations for the recognition of the Cdk2 binding site. Local dynamics monitored by smFA of free p27 (Figure 3b and c) showed that free p27 shows two anisotropy values (Figure 3b and c) that reflects the flexible (low rD) and rigid (high rD) conformations. However, in the absence of target complexes, this protein is preferentially compacted (Figure 3c).
Additionally, by analyzing time-resolved fluorescence (Figure 3d) and single-molecule FRET (Figure 3e) probing various regions of p27, it was found that p27 must expand to create the ternary complex. For example, the FRET variant monitoring dynamics of regions LH and D2 (cysteines for labeling at locations C54 and C93, Figure 3a) shows a dynamic system by which p27 behaves as an intrinsically disordered protein (Figure 3e) following a worm-like-chain (WLC) model with an averaged donor-acceptor distance of 41.8 ± 2.3 Å (magenta dynamic line in Figure 3f). This result is consistent with NMR measurements, MD of the full-length p27, analytical ultracentrifugation, and small-angle- X-ray scattering of the p27/Cdk2/Cyclin A complex [70,71••]. The disordered nature of p27 permits jumps over low energy barriers and rapidly sample multiple configurations that can, over longer timescales, transition between distinct conformations or eventually accessing different structural ensembles, referred as ensemble switching [72,73]. fFCS can efficiently identify all these structural changes over a broad temporal domain (nano-to-milliseconds), corroborating that most of dynamical exchange occurs in the nanoseconds regime (Figure 3g).
Finally, the information derived from discrete MD (DMD) simulations, which samples the heterogeneous landscape, was used as an integrative element in the smMFS toolbox [74-80]. By using radius of gyration (Rg) and α-helical content on the same regions monitored by experimental observations, authors could compare interdye distances, local flexibility and polymeric behavior (like the persistence length). Thus, DMD and smMFS help each other as independent and complementary approaches without imposing physical constrains that biased either simulations of experimental observables into the attained results.
Although very useful for IDP models, this smMFS toolbox is not restricted to highly flexible proteins, but has identified transient conformations even in well-folded and minimally-frustrated models. Using the smMFS toolbox, Sanabria et al. [60••] determined the conformations of the lysozyme of bacteriophage T4 (T4L) in the catalytic cycle progression. Three major conformations that are present in the free (E), enzyme-substrate complex (ES), and enzyme-product (EP) bound states. These conformations exchange at few microseconds and hundreds of microseconds, extending the Michalis-Menten mechanism and highlighting that specific conformations favor the progression of the enzymatic reaction. In contrast, for free p27, the transitions observed imply high conformational heterogeneity and flexibility according to its disordered nature (Figure 3h), which suggests that, although disordered, p27 must overcome an expansion to bind with Cdk2/Cyclin A. These examples highlight the relevance of using smMFS toolbox to temporally characterize the structural dynamics of diverse proteins.
Intermolecular dynamics: structural dynamics in multi-step binding and partial dissociation as function modulator
Once defined that p27 must extend to bind the Cdk2/Cyclin A complex, authors studied the main changes involved in forming the ternary complex. Using smFA (Figure 3c), NMR, and X-ray crystallography (X-ray), Tsytlonok et al. [71••] discovered that p27 mostly adopts the extended conformation when it is bound to Cdk2/Cyclin A complex. Additionally, by analyzing different donor-acceptor combinations, two limiting states were obtained for p27 in complex. For example, using the FRET variant C54-C93 (covering the LH-D2 regions), authors found two distances with ÅRDAeE,exp that go from 43.1 ± 0.1 to 52.3 ± 0.1 Å via smFRET (Figure 3f), showing a good agreement with the crystallographic structure (PDBID 1JSU). When modeling the accessible volume (AV) of the dyes in such configuration using coarse-grained simulations, results showed experimental-simulations differences within ~3 Å. The anterior indicates expansion from a more compact conformation to a conformation that exposes the D2 region and adds robust stabilization in the structural dynamics, as observed in fFCS (Figure 3h) by the accumulation of transitions fraction in the mid-microseconds regime. In summary, a fully formed, fuzzy ternary complex built with a simultaneous extension of p27 was identified [68••,71••].
Furthermore, once p27 is bound to Cdk2/Cyclin A and causes cell cycle arrest, this ternary complex is finely regulated via phosphorylation of two occluded tyrosine residues by tyrosine kinases Bcr-Abl and Src for Y88 and Y74 (Figure 3a), respectively [81,82]. For these residues to be phosphorylated through dynamic anticipation, p27 exchanges between different conformations in the bound complex allow the sequential exposure of Y88, followed by Y74 anticipating phosphorylation [71••]. Each of these phosphorylation conditions allow the accessibility of different conformational ensembles. The process was observed by using the smMFS toolbox (Figure 2b) and integrating other biochemical and biophysical methods, including NMR, isothermal titration calorimetry (ITC), and X-ray crystallography.
To start, smFA (Figure 3c) showed the release of Y88 followed by Y74, supported by the increase in a population with low anisotropy values, which indicates a more freely rotating fluorophore and in agreement with chemical shift assignments of the D2 domain (e.g., C75 and C93). Next, using the same FRET variant, C54-93, smFRET showed a redistribution of states occurs, shifting the population to a more extended partially released state, thus exposing the phosphorylated Y88 (Figure 3f). In this new state, Y74 is anticipated to be sequentially phosphorylated, evidenced by the release of C75 in smFA after Y88 and Y74 are phosphorylated (Figure 3c). With fFCS, a redistribution towards the accumulation of nanoseconds fraction exchange is described, suggesting that phosphorylation allows the adoption of a highly dynamic p27 is formed [71••].
To showcase the role of partial dissociation and disorder in the structural dynamics, Medina et al. [5••] studied the domain-swapped dimer of the DNA-binding domain of human FoxP1. The compact and folded dimer adopted via 3D-DS exchanges with an extended dimeric, mostly disordered, intermediate ensemble adopting heterogeneous structural changes occurring between 20 μs to 5 ms. The extended intermediate is kinetically allowed due to a low average energetic barrier of ~1 kcal mol−1, resulting in the intermediate to become highly accumulated as the unfolding of the protein is promoted. This result indicates that the monomer-dimer transition overcomes the characteristic high energy barrier of three-dimensional domain swapping by containing IDRs [83,84]. Overall, the smMFS toolbox is powerful in capturing complex regulatory mechanisms from multi-step binding processes and complex folding pathways, supporting the need for updates of the current unanimated sequence-to-structure-to-function paradigm to a sequence-to-dynamics-to-function.
Perspective: from structural dynamics to function and protein assemblies
For cells to function correctly, proteins must work synergistically. Only by understanding how structural dynamics guide ensemble switching, we can understand how proteins self-assemble into multi-functional three-dimensional mesoscale architectures. Therefore, by following the relationship between dynamics and function, insights can be gained for various genetic diseases such as cancer [85-88], Huntington’s [89], autism [90], spinal muscular dystrophy [91].
The previous example of p27 binding with Cdk2/Cyclin A to form a ternary complex shows how intra- and intermolecular and intermolecular interactions must work together to regulate the cell cycle [62,63]. Binding only occurs due to the intramolecular behavior of p27, which allows a rapid sampling of multiple configurations to access the extended conformation (Figure 4). Intermolecular interactions with Cdk2/ Cyclin A impedes the cell division by arresting the cycle in phase G. This p27/Cdk2/Cyclin A association has enormous metabolic significance. Further, it is tightly regulated by specific phosphorylation modifications that trigger p27 ubiquitination followed by degradation. Degradation of p27 enables cell cycle progression [70,81]. However, as discovered, all these events inherently depend on the structural dynamics that characterize p27. The p27/Cdk2/Cyclin A complex is a clear example of where dynamics lead to a change in conformations and defined ensembles that allows the complex to adapt specific functionality. This model and others [56,92,93] have recently revealed the extreme relevance of conformational dynamics as a key functional modulator.
Figure 4.
Functional connection between structural dynamics of p27 and cell cycle. The adoption of the ternary complex between p27/Cdk2/Cyclin A depends on the conformational exchange of free p27 and different conformational ensembles. The expansion of this protein enables the binding to Cdk2/Cyclin A, leading the cell cycle to arrest in phase G, impeding the development of phase S, and therefore DNA replication. Intrinsic flexibility of p27 allows phosphorylation modifications in its tyrosine residues 74 and 88, increasing the expansion and the release from the complex, and the consequent recruitment of ubiquitination proteins that finally leads to the degradation. These events allow Cdk2/Cyclin to continue their functional role in ensuring the cell cycle progression, therefore cell division.
The next logical step is understanding high-order assemblies and their role in modulating the function of the cells. So far, there are few characterized examples by which high-order complexes communicate in relevant processes [58,94]. Such is the case of the dynamics of chromatin, where nucleosome opening/closing transitions stability can severely influence the gene expression activity inside the nucleus. A combination of single-molecule approaches with molecular dynamic simulations found that binding with external proteins severely influences nucleosome dynamics [40,41•,57,95•,96], pivotal to decipher how gene expression occurs. Dynamics are also an essential part of polyfunctional molecules, where molecular adaptors must be coordinated to ensure the appropriate function depending upon the situation [97].
Future studies are required for highly dynamic and less ordered complex systems, such as biomolecular condensates, mitotic spindles, and focal adhesions [98•]. All characterized examples focus the essential role of heterogeneity in dynamics, by which molecules may explore various conformational ensembles, each with crucial consequences in those complexes and their stability. However, although much is still left to understand micro- and mesoscale assemblies within cells, current studies are focused on applying all these high-resolution approaches inside cells to increase the understanding of structural dynamics and assemblies in a real biological context [99,100]. In the near future, we anticipate that this holistic toolbox presented will continue to unravel the sequence-to-function relationship of many mesoscale assemblies in live cells.
Acknowledgements
This work acknowledges support from Clemson University, NSF (CAREER MCB- 1749778) and US National Institutes of Health (NIH) P20GM121342. EM was partially supported by a pilot program for global engagement of Clemson University, and by Fondo Nacional de Desarrollo Científico y Tecnológico (Fondecyt grant 1170701).
Footnotes
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as
• of special interest
•• of outstanding interest
- 1.Perutz MF, Rossmann MG, Cullis AF, Muirhead H, Will G, North AC: Structure of haemoglobin: a three-dimensional Fourier synthesis at 5.5-A. Resolution, obtained by X-ray analysis. Nature 1960, 185:416–422. [DOI] [PubMed] [Google Scholar]
- 2.Anfinsen C: Principles that govern the folding of protein chains. Science 1973, 181:223–230. [DOI] [PubMed] [Google Scholar]
- 3.Levinthal C: Are there pathways for protein folding? J Chim Phys 2017, 65:44–45. [Google Scholar]
- 4.Wolynes PG, Onuchic JN, Thirumalai D: Navigating the folding routes. Science 1995, 267:1619–1620. [DOI] [PubMed] [Google Scholar]
- 5.••.Medina E, Villalobos P, Hamilton GL, Komives EA, Sanabria H, Ramirez-Sarmiento CA, Babul J: Intrinsically disordered regions of the DNA-binding domain of human FoxP1 facilitate domain swapping. J Mol Biol 2020, 432:5411–5429.In this work, authors get insights about the flexibility and dynamics of the domain-swapped dimer of FoxP1. Using a hybrid between experimental approaches such as smFRET, smFRET-FCS and hydrogen-deuterium exchange with mass spectrometry, combined with molecular dynamics simulations, they showed that dimeric FoxP1 is mostly a dynamic system containing at least two locally-extended intermediates, that facilitates the transitions between the monomer and the domain-swapped dimer.
- 6.Faust O, Grunhaus D, Shimshon O, Yavin E, Friedler A: Protein regulation by intrinsically disordered regions: a role for subdomains in the IDR of the HIV-1 rev protein. Chembiochem 2018, 19:1618–1624. [DOI] [PubMed] [Google Scholar]
- 7.Protter DSW, Rao BS, Van Treeck B, Lin Y, Mizoue L, Rosen MK, Parker R: Intrinsically disordered regions can contribute promiscuous interactions to RNP granule assembly. Cell Rep 2018, 22:1401–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright PE, Dyson HJ: Intrinsically unstructured proteins: reassessing the protein structure-function paradigm. J Mol Biol 1999, 293:321–331. [DOI] [PubMed] [Google Scholar]
- 9.Wright PE, Dyson HJ: Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol 2015, 16:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uversky VN: What does it mean to be natively unfolded? EurJ Biochem 2002, 269:2–12. [DOI] [PubMed] [Google Scholar]
- 11.Miskei M, Horvath A, Vendruscolo M, Fuxreiter M: Sequence-based prediction of fuzzy protein interactions. J Mol Biol 2020, 432:2289–2303. [DOI] [PubMed] [Google Scholar]
- 12.Tompa P, Fuxreiter M: Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci 2008, 33:2–8. [DOI] [PubMed] [Google Scholar]
- 13.•.Borgia A, Borgia MB, Bugge K, Kissling VM, Heidarsson PO, Fernandes CB, Sottini A, Soranno A, Buholzer KJ, Nettels D et al. :Extreme disorder in an ultrahigh-affinity protein complex. Nature 2018, 555:61–66.This works beautifully shows how the binding specificity and affinity do not rely exclusively in binding interfaces but also are present in disordered systems. Using a combination between ensemble, single-molecule approaches, and molecular dynamics simulations, authors found that the extremely high affinity histone H1/prothymyosin-a complex retains the intrinsics disorder that characterizes both proteins.
- 14.Zosel F, Mercadante D, Nettels D, Schuler B: A proline switch explains kinetic heterogeneity in a coupled folding and binding reaction. Nat Commun 2018, 9:3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JY, Chung HS: Diverse transition paths of coupled binding and folding of intrinsically disordered protein proved by three-color single-molecule FRET. Biophys J 2020, 118:491a–492a. [Google Scholar]
- 16.Holmstrom ED, Liu Z, Nettels D, Best RB, Schuler B: Disordered RNA chaperones can enhance nucleic acid folding via local charge screening. Nat Commun 2019, 10:2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.•.Mitra J, Ha T: Streamlining effects of extra telomeric repeat on telomeric DNA folding revealed by fluorescence-force spectroscopy. Nucleic Acids Res 2019, 47:11044–11056.In this work, authors studied the effect of G-quadruplexes (GQs)—structures observed when high-repeats of T2AG3 stack together — stability and dynamics, to understand telomeric single-stranded tails properties due their high GQs content. By combining smFRET and optical twezeers, they found that sequences having five or six T2AG3 repeats preferentially fold into GQs by the 3’ end and show extreme mechanical and structual stability.
- 18.Lee HT, Sanford S, Paul T, Choe J, Bose A, Opresko PL, Myong S: Position-dependent effect of guanine base damage and mutations on telomeric G-quadruplex and telomerase extension. Biochemistry 2020, 59:2627–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee CY, McNerney C, Ma K, Zhao W, Wang A, Myong S: R-loop induced G-quadruplex in non-template promotes transcription by successive R-loop formation. Nat Commun 2020, 11:3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dogan J, Gianni S, Jemth P: The binding mechanisms of intrinsically disordered proteins. Phys Chem Chem Phys 2014, 16:6323–6331. [DOI] [PubMed] [Google Scholar]
- 21.Mollica L, Bessa LM, Hanoulle X, Jensen MR, Blackledge M, Schneider R: Binding mechanisms of intrinsically disordered proteins: theory, simulation, and experiment. Front Mol Biosci 2016, 3:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou HX, Pang X, Lu C: Rate constants and mechanisms of intrinsically disordered proteins binding to structured targets. Phys Chem Chem Phys 2012, 14:10466–10476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banani SF, Lee HO, Hyman AA, Rosen MK: Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 2017, 18:285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.•.Dignon GL, Zheng W, Kim YC, Best RB, Mittal J : Sequence determinants of protein phase behavior from a coarse-grained model. PLoS Comput Biol 2018, 14:e1005941.This works introduces a computational framework to study intrinsically disordered proteins and protein assembly in liquid-liquid phase separation using a coarse-grained model, offering the possibility to simulate these systems without the computational resources needed when using all-atom simulations. The authors tested FUS and LAF-1 proteins, observing good correlation between experimental properties and simulations.
- 25.Brangwynne CP, Tompa P, Pappu RV: Polymer physics of intracellular phase transitions. Nat Phys 2015, 11:899–904. [Google Scholar]
- 26.Sahli L, Renard D, Sole-Jamault V, Giuliani A, Boire A: Role of protein conformation and weak interactions on gamma-gliadin liquid-liquid phase separation. Sci Rep 2019, 9:13391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin YH, Song JH, Forman-Kay JD, Chan HS: Random-phase-approximation theory for sequence-dependent, biologically functional liquid-liquid phase separation of intrinsically disordered proteins. J Mol Liquids 2017, 228:176–193. [Google Scholar]
- 28.•.Dignon GL, Zheng W, Best RB, Kim YC, Mittal J: Relation between single-molecule properties and phase behavior of intrinsically disordered proteins. Proc Natl Acad Sci U S A 2018, 115:9929–9934.Here, authors use their recently developed framework (see Ref. 24) to calculate Θ temperature (for coil-to-globule protein transition, TΘ), the Boyle temperature (TB) and the critical temperature (for phase separation, TC) of twenty proteins. They found a linear correlation between these parameters, suggesting that their correlation serve as a prediction of proteins as contributing in liquid-liquid phase separation and membrane-less organels.
- 29.Kleckner IR, Foster MP: An introduction to NMR-based approaches for measuring protein dynamics. Biochim Biophys Acta 2011, 1814:942–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.•.Adamski W, Salvi N, Maurin D, Magnat J, Milles S, Jensen MR, Abyzov A, Moreau CJ, Blackledge M: A unified description of intrinsically disordered protein dynamics under physiological conditions using NMR spectroscopy. J Am Chem Soc 2019, 141:17817–17829.An excellent and integrated description of intrinsically-disordered proteins study using NMR, highlighting the insights into the conformational changes in crowded solutions as example of complex environmental conditions, describing dynamics up to ten of nanoseconds.
- 31.Dyson HJ, Wright PE: Perspective: the essential role of NMR in the discovery and characterization of intrinsically disordered proteins. J Biomol NMR 2019, 73:651–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayor T, Hacker U, Stierhof YD, Nigg EA: The mechanism regulating the dissociation of the centrosomal protein C-Nap1 from mitotic spindle poles. J Cell Sci 2002, 115:3275–3284. [DOI] [PubMed] [Google Scholar]
- 33.Nolting B, Golbik R, Neira JL, Soler-Gonzalez AS, Schreiber G, Fersht AR: The folding pathway of a protein at high resolution from microseconds to seconds. Proc Natl Acad Sci U S A 1997, 94:826–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen EH, Lu TT, Hsu JC, Tseng YJ, Lim TS, Chen RP: Directly monitor protein rearrangement on a nanosecond-to-millisecond time-scale. Sci Rep 2017, 7:8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JY, Kim C, Lee NK: Real-time submillisecond single-molecule FRET dynamics of freely diffusing molecules with liposome tethering. Nat Commun 2015, 6:6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otosu T, Ishii K, Tahara T: Microsecond protein dynamics observed at the single-molecule level. Nat Commun 2015, 6:7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kondo T, Pinnola A, Chen WJ, Dall’Osto L, Bassi R, Schlau-Cohen GS: Single-molecule spectroscopy of LHCSR1 protein dynamics identifies two distinct states responsible for multitimescale photosynthetic photoprotection. Nat Chem 2017, 9:772–778. [DOI] [PubMed] [Google Scholar]
- 38.Sisamakis E, Valeri A, Kalinin S, Rothwell PJ, Seidel CAM: Accurate single-molecule FRET studies using multiparameter fluorescence detection. Single Molecule Tools, Part B:Super-Resolution, Particle Tracking, Multiparameter, and Force Based Methods. Elsevier; 2010:455–514 10.1016/s0076-6879(10)75018-7:. Methods in Enzymology. [DOI] [PubMed] [Google Scholar]
- 39.Peulen TO, Opanasyuk O, Seidel CAM: Combining graphical and analytical methods with molecular simulations to analyze time-resolved FRET measurements of labeled macromolecules accurately. J Phys Chem B 2017, 121:8211–8241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lehmann K, Felekyan S, Kuhnemuth R, Dimura M, Toth K, Seidel CAM, Langowski J: Dynamics of the nucleosomal histone H3 N-terminal tail revealed by high precision single-molecule FRET. Nucleic Acids Res 2020, 48:1551–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.•.Gansen A, Felekyan S, Kuhnemuth R, Lehmann K, Toth K, Seidel CAM, Langowski J: High precision FRET studies reveal reversible transitions in nucleosomes between microseconds and minutes. Nat Commun 2018, 9:4628.A beautiful description of structural dynamics in a complex system such as nucleosomes at single-molecule level. Using different FRET variants, the authors quantified destabilization dynamics of mononucleosomes by increasing the ionic stength, finding first opening events in the microseconds timescale by which DNA can breath, until the complete nucleosome dissasembly that is slower than 10 000 seconds.
- 42.•.Craggs TD, Sustarsic M, Plochowietz A, Mosayebi M, Kaju H, Cuthbert A, Hohlbein J, Domicevica L, Biggin PC, Doye JPK et al. : Substrate conformational dynamics facilitate structure-specific recognition of gapped DNA by DNA polymerase. Nucleic Acids Res 2019, 47:10788–10800.This work elegantly describes the structural changes of DNA required to be recognized by the DNA polymarase I, combining smFRET data from 73 variants and molecular dynamics simulations. The authors found that single-nucleotide-gapped DNA bending and fraying are necessary for the polymerase recognition and binding.
- 43.••.Dimura M, Penlen TO, Sanabria H, Rodnin D, Hemmen K, Hanke CA, Seidel CAM, Gohlke H: Automated and optimally FRET-assisted structural modeling. Nat Commun 2020. 10.1038/s41467-020-19023-1.A powerful combination of smFRET and molecular dynamics simulations is used to create a computational suite to allow the modelling of complex biomolecular systems. By using smFRET, a set of distances acts as a structural guide to perform specific molecular dynamics simulations and therefore obtain a refined model that considers the inherent dynamics.
- 44.Noid WG: Perspective: coarse-grained models for biomolecular systems. J Chem Phys 2013, 139 090901. [DOI] [PubMed] [Google Scholar]
- 45.Devane R, Shinoda W, Moore PB, Klein ML: A transferable coarse grain non-bonded interaction model for amino acids. J Chem Theory Comput 2009, 5:2115–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tozzini V: Coarse-grained models for proteins. Curr Opin Struct Biol 2005, 15:144–150. [DOI] [PubMed] [Google Scholar]
- 47.Cohen RD, Pielak GJ: Electrostatic contributions to protein Quinary structure. J Am Chem Soc 2016, 138:13139–13142. [DOI] [PubMed] [Google Scholar]
- 48.Wirth AJ, Gruebele M: Quinary protein structure and the consequences of crowding in living cells: leaving the test-tube behind. Bioessays 2013, 35:984–993. [DOI] [PubMed] [Google Scholar]
- 49.Martin EW, Holehouse AS, Peran I, Farag M, Incicco JJ, Bremer A, Grace CR, Soranno A, Pappu RV, Mittag T: Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 2020, 367:694–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harmon TS, Holehouse AS, Rosen MK, Pappu RV: Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. eLife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guin D, Gruebele M: Weak chemical interactions that drive protein evolution: crowding, sticking, and quinary structure in folding and function. Chem Rev 2019, 119:10691–10717. [DOI] [PubMed] [Google Scholar]
- 52.Chowdhury A, Bollinger JA, Dear BJ, Cheung JK, Johnston KP, Truskett TM: Coarse-grained molecular dynamics simulations for understanding the impact of short-range anisotropic attractions on structure and viscosity of concentrated monoclonal antibody solutions. Mol Pharm 2020, 17:1748–1756. [DOI] [PubMed] [Google Scholar]
- 53.Kozyrev SV, Volovich IV: Quinary lattice model of secondary structures of polymers. Phys A-Stat Mech Appl 2014, 393:86–95. [Google Scholar]
- 54.Yoo J, Louis JM, Gopich IV, Chung HS: Three-color single-molecule FRET and fluorescence lifetime analysis of fast protein folding. J Phys Chem B 2018, 122:11702–11720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.•.Yanez Orozco IS, Mindlin FA, Ma J, Wang B, Levesque B, Spencer M, Rezaei Adariani S, Hamilton G, Ding F, Bowen ME et al. : Identifying weak interdomain interactions that stabilize the supertertiary structure of the N-terminal tandem PDZ domains of PSD-95. Nat Commun 2018, 9:3724.This integrative and collaborative work uses smFRET and molecular dynamics simulations to solve structural interdomain dynamics of the PSD-95, finding that specific conformations described as open and closed that match with previous structure determination. These conformations are exchanging with a low energy barrier between them and are stabilized by specific side-chain interactions.
- 56.Dahiya V, Agam G, Lawatscheck J, Rutz DA, Lamb DC, Buchner J: Coordinated conformational processing of the tumor suppressor protein p53 by the Hsp70 and Hsp90 chaperone machineries. Mol Cell 2019, 74:816–830 e817. [DOI] [PubMed] [Google Scholar]
- 57.Kameda T, Awazu A, Togashi Y: Histone tail dynamics in partially disassembled nucleosomes during chromatin remodeling. Front Mol Biosci 2019, 6:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gudkov AT, Behlke J, Vtiurin NN, Lim VI: Tertiary and quaternary structure for ribosomal protein L7 in solution. FEBS Lett 1977, 82:125–129. [DOI] [PubMed] [Google Scholar]
- 59.Peulen T-O, Hengstenberg CS, Biehl R, Dimura M, Lorenz C, Valeri A, Ince S, Vopel T, Farago B, Gohlke H et al. : Integrative dynamic structural biology unveils conformers essential for the oligomerization of a large GTPase. arXiv: Biol Phys 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.••.Sanabria H, Rodnin D, Hemmen K, Peulen TO, Felekyan S, Fleissner MR, Dimura M, Koberling F, Kuhnemuth R, Hubbell W et al. : Resolving dynamics and function of transient states in single enzyme molecules. Nat Commun 2020, 11:1231.Using a combination of smFRET, smFA, fFCS and molecular simulations, a short-lived configurational state of lyzome T4L was found to compliment the already known open and close state. With this discovery, an extended Michaelis-Menten mechanism for enzymes is explained and supported.
- 61.Sielaff H, Singh D, Grüber G, Börsch M: Analyzing Conformational Changes in Single FRET-labeled A1 Parts of Archaeal A1AO-ATP synthase. SPIE; 2018. [Google Scholar]
- 62.Polyak K, Lee MH, Erdjument-Bromage H, Koff A, Roberts JM, Tempst P, Massague J: Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell 1994, 78:59–66. [DOI] [PubMed] [Google Scholar]
- 63.Toyoshima H, Hunter T: p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell 1994, 78:67–74. [DOI] [PubMed] [Google Scholar]
- 64.Bienkiewicz EA, Adkins JN, Lumb KJ: Functional consequences of preorganized helical structure in the intrinsically disordered cell-cycle inhibitor p27(Kip1). Biochemistry 2002, 41:752–759. [DOI] [PubMed] [Google Scholar]
- 65.Otieno S, Grace CR, Kriwacki RW: The role of the LH subdomain in the function of the Cip/Kip cyclin-dependent kinase regulators. Biophys J 2011, 100:2486–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Otieno S, Kriwacki R: Probing the role of nascent helicity in p27 function as a cell cycle regulator. PLoS One 2012, 7:e47177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bowman P, Galea CA, Lacy E, Kriwacki RW: Thermodynamic characterization of interactions between p27(Kip1) and activated and non-activated Cdk2: intrinsically unstructured proteins as thermodynamic tethers. Biochim Biophys Acta 2006, 1764:182–189. [DOI] [PubMed] [Google Scholar]
- 68.••.Tsytlonok M, Hemmen K, Hamilton G, Kolimi N, Felekyan S, Seidel CAM, Tompa P, Sanabria H: Specific conformational dynamics and expansion underpin a multi-step mechanism for specific binding of p27 with Cdk2/Cyclin A. J Mol Biol 2020, 432:2998–3017.This indept study of p27 uses smFRET, smFA, fFCS, stopped-flow kinetics, and replica-exchange discrete molecular dynamics simulations to determine the multi-step mechanism used to bind p27 to the Cdk2/Cyclin A complex. It was found that the binding mechanism depends on local and long-range configurations.
- 69.Russo AA, Jeffrey PD, Patten AK, Massague J, Pavletich NP: Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature 1996, 382:325–331. [DOI] [PubMed] [Google Scholar]
- 70.Galea CA, Nourse A, Wang Y, Sivakolundu SG, Heller WT, Kriwacki RW: Role of intrinsic flexibility in signal transduction mediated by the cell cycle regulator, p27 Kip1. J Mol Biol 2008, 376:827–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.••.Tsytlonok M, Sanabria H, Wang Y, Felekyan S, Hemmen K, Phillips AH, Yun MK, Waddell MB, Park CG, Vaithiyalingam S et al. : Dynamic anticipation by Cdk2/Cyclin A-bound p27 mediates signal integration in cell cycle regulation. Nat Commun 2019, 10:1676.This study shows the importance of protein flexiblity for signaling and function by studying the phosphorylation of p27 bound to Cdk2/Clyclin A. Using NMR spectroscopy, smFA, smFRET and fFCS it was determined that Y88 is first phosphorylated which allows access for Y74 to be phosphorylated. Isothermal titration calorimetry were then used to monitor how tryosine phsophorylations allow the dissasembly of the p27/Cdk2/Cyclin A complex to promote cell division.
- 72.Choi UB, Sanabria H, Smirnova T, Bowen ME, Weninger KR: Spontaneous switching among conformational ensembles in intrinsically disordered proteins. Biomolecules 2019, 9:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Choi UB, McCann JJ, Weninger KR, Bowen ME: Beyond the random coil: stochastic conformational switching in intrinsically disordered proteins. Structure 2011, 19:566–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sali A, Berman HM, Schwede T, Trewhella J, Kleywegt G, Burley SK, Markley J, Nakamura H, Adams P, Bonvin AM et al. : Outcome of the first wwPDB hybrid/integrative methods task force workshop. Structure 2015, 23:1156–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matsunaga Y, Sugita Y: Linking time-series of single-molecule experiments with molecular dynamics simulations by machine learning. eLife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dimura M, Peulen TO, Hanke CA, Prakash A, Gohlke H, Seidel CA: Quantitative FRET studies and integrative modeling unravel the structure and dynamics of biomolecular systems. Curr Opin Struct Biol 2016, 40:163–185. [DOI] [PubMed] [Google Scholar]
- 77.Ding F, Tsao D, Nie H, Dokholyan NV: Ab initio folding of proteins with all-atom discrete molecular dynamics. Structure 2008, 16:1010–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shirvanyants D, Ding F, Tsao D, Ramachandran S, Dokholyan NV: Discrete molecular dynamics: an efficient and versatile simulation method for fine protein characterization. J Phys Chem B 2012, 116:8375–8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Proctor EA, Ding F, Dokholyan NV: Discrete molecular dynamics. Wiley Interdiscip Rev-Comput Mol Sci 2011, 1:80–92. [Google Scholar]
- 80.Ding F, Dokholyan NV: Discrete molecular dynamics simulation of biomolecules. In Computational Modeling of Biological Systems. Edited by Dokholyan NV. Springer US;; 2012:55–73 10.1007/978-1-4614-2146-7_3. Biological and Medical Physics, Biomedical Engineering. [DOI] [Google Scholar]
- 81.Grimmler M, Wang Y, Mund T, Cilensek Z, Keidel EM, Waddell MB, Jakel H, Kullmann M, Kriwacki RW, Hengst L: Cdk-inhibitory activity and stability of p27Kip1 are directly regulated by oncogenic tyrosine kinases. Cell 2007, 128:269–280. [DOI] [PubMed] [Google Scholar]
- 82.Chu I, Sun J, Arnaout A, Kahn H, Hanna W, Narod S, Sun P, Tan CK, Hengst L, Slingerland J: p27 phosphorylation by Src regulates inhibition of cyclin E-Cdk2. Cell 2007, 128:281–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rousseau F, Schymkowitz JW, Itzhaki LS: The unfolding story of three-dimensional domain swapping. Structure 2003, 11:243–251. [DOI] [PubMed] [Google Scholar]
- 84.Liu L, Byeon IJ, Bahar I, Gronenborn AM: Domain swapping proceeds via complete unfolding: a 19F- and 1H-NMR study of the Cyanovirin-N protein. J Am Chem Soc 2012,134:4229–4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tannapfel A, Grund D, Katalinic A, Uhlmann D, Kockerling F, Haugwitz U, Wasner M, Hauss J, Engeland K, Wittekind C: Decreased expression of p27 protein is associated with advanced tumor stage in hepatocellular carcinoma. Int J Cancer 2000, 89:350–355. [DOI] [PubMed] [Google Scholar]
- 86.Li Z, Tao Y, Wang X, Jiang P, Li J, Peng M, Zhang X, Chen K, Liu H, Zhen P et al. : Tumor-secreted exosomal miR-222 promotes tumor progression via regulating p27 expression and relocalization in pancreatic cancer. Cell Physiol Biochem 2018, 51:610–629. [DOI] [PubMed] [Google Scholar]
- 87.Patel P, Tsiperson V, Gottesman SRS, Somma J, Blain SW: Dual inhibition of CDK4 and CDK2 via targeting p27 tyrosine phosphorylation induces a potent and durable response in breast cancer cells. Mol Cancer Res 2018, 16:361–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kolsi LE, Leal AS, Yli-Kauhaluoma J, Liby KT, Moreira VM: Dehydroabietic oximes halt pancreatic cancer cell growth in the G1 phase through induction of p27 and downregulation of cyclin D1. Sci Rep 2018, 8:15923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Savas JN, Makusky A, Ottosen S, Baillat D, Then F, Krainc D, Shiekhattar R, Markey SP, Tanese N: Huntington’s disease protein contributes to RNA-mediated gene silencing through association with Argonaute and P bodies. Proc Natl Acad Sci U S A 2008, 105:10820–10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gall JG: Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol 2000, 16:273–300. [DOI] [PubMed] [Google Scholar]
- 91.Antonellis A, Ellsworth RE, Sambuughin N, Puls I, Abel A, Lee-Lin SQ, Jordanova A, Kremensky I, Christodoulou K, Middleton LT et al. : Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am J Hum Genet 2003, 72:1293–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Heiss G, Ploetz E, Voith von Voithenberg L, Viswanathan R, Glaser S, Schluesche P, Madhira S, Meisterernst M, Auble DT, Lamb DC: Conformational changes and catalytic inefficiency associated with Mot1-mediated TBP-DNA dissociation. Nucleic Acids Res 2019, 47:2793–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Barth A, Hendrix J, Fried D, Barak Y, Bayer EA, Lamb DC: Dynamic interactions of type I cohesin modules fine-tune the structure of the cellulosome of Clostridium thermocellum. Proc Natl Acad Sci U S A 2018, 115:E11274–E11283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gudkov AT: The L7/L12 ribosomal domain of the ribosome: structural and functional studies. FEBS Lett 1997, 407:253–256. [DOI] [PubMed] [Google Scholar]
- 95.•.Kilic S, Felekyan S, Doroshenko O, Boichenko I, Dimura M, Vardanyan H, Bryan LC, Arya G, Seidel CAM, Fierz B: Single-molecule FRET reveals multiscale chromatin dynamics modulated by HP1alpha. Nat Commun 2018, 9:235.This excelent work combines freely-difussing and fixed smFRET approaches to obtain chromatin dynamics at high temporal and structural resolution. Using three FRET variants, the authors could solve conformational changes from microseconds to seconds with sub nanometer precision, finding not only highly-dynamical stacking interactions between nucleosomes but also that, although compacted by HPF1-α, chromatin fibers maitains their intrinsic dynamics.
- 96.Mivelaz M, Cao AM, Kubik S, Zencir S, Hovius R, Boichenko I, Stachowicz AM, Kurat CF, Shore D, Fierz B: Chromatin fiber invasion and nucleosome displacement by the Rap1 transcription factor. Mol Cell 2020, 77:488–500 e489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fox SW: Self-sequencing of amino acids and origins of polyfunctional protocells. Orig Life 1984, 14:485–488. [DOI] [PubMed] [Google Scholar]
- 98.•.Riback JA, Zhu L, Ferrolino MC, Tolbert M, Mitrea DM, Sanders DW, Wei MT, Kriwacki RW, Brangwynne CP: Composition-dependent thermodynamics of intracellular phase separation. Nature 2020, 581 :209–214.This scientific milestone gives us a deep understanding about multicomponent liquid-liquid phase separation by analyzing intracellular condensates, using the partition coefficient concept, different protein fusions and micrsocopy. By integrating those elements, authors found not only that heterotypic interactions are relevant, but also its interplay with composition-dependent thermodynamics of condensates assemblies inside cells.
- 99.Heckmeier PJ, Agam G, Teese MG, Hoyer M, Stehle R, Lamb DC, Langosch D: Determining the stoichiometry of small protein oligomers using steady-state fluorescence anisotropy. Biophys J 2020, 119:99–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Coullomb A, Bidan CM, Qian C, Wehnekamp F, Oddou C, Albiges-Rizo C, Lamb DC, Dupont A: QuanTI-FRET: a framework for quantitative FRET measurements in living cells. Sci Rep 2020, 10:6504. [DOI] [PMC free article] [PubMed] [Google Scholar]