Abstract
The mammalian visual system is composed of circuitry connecting sensory input from the retina to the processing core of the visual cortex. The two main retinorecipient brain targets, the superior colliculus (SC) and dorsal lateral geniculate nucleus (dLGN), bridge retinal input and visual output. The primary cilium is a conserved organelle increasingly viewed as a critical sensor for the regulation of developmental and homeostatic pathways in most mammalian cell types. Moreover, cilia have been described as crucial for neurogenesis, neuronal maturation, and survival in the cortex and retina. However, cilia in the visual relay center remain to be fully described. In this study, we characterized the ciliation profile of the SC and dLGN and found that the overall number of ciliated cells declined during development. Interestingly, shorter ciliated cells in both regions were identified as neurons, whose numbers remained stable over time, suggesting that cilia retention is a critical feature for optimal neuronal function in SC and dLGN. Our study suggests that primary cilia are important for neuronal maturation and function in cells of the SC and dLGN.
Keywords: Primary Cilia, Superior Colliculus, Dorsal Lateral Geniculate Nucleus, Retinofugal Pathway, RRID:IMSR_JAX:027967, RRID:IMSR_JAX:007677, RRID:IMSR_JAX:000664, RRID:AB_2572219, RRID:AB_630839, RRID:AB_2298772, RRID:AB_477329, RRID:AB_309787, RRID:SCR_001775, RRID:SCR_017348, RRID:SCR_002798
Graphical Abstract
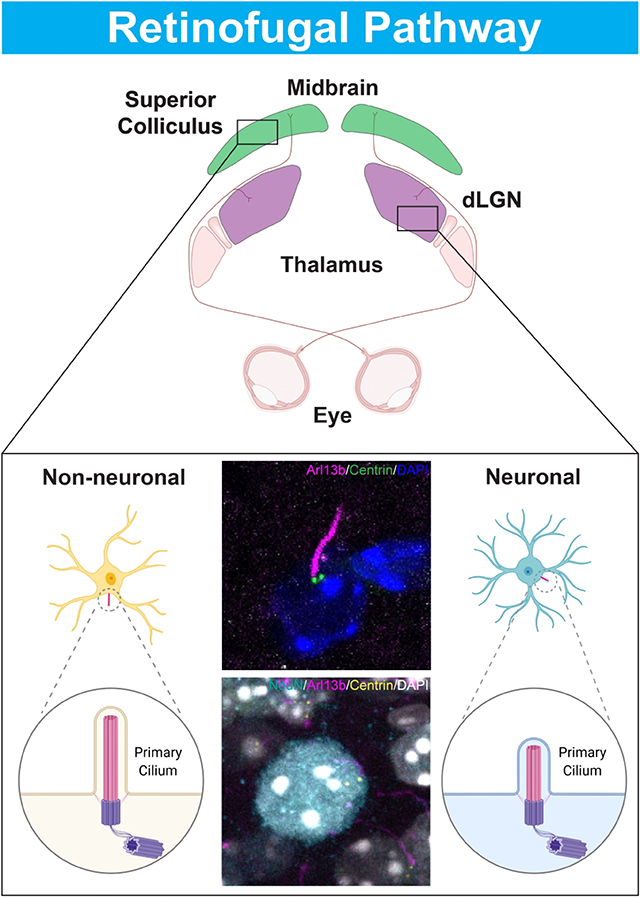
In this study we show that both the number of primary cilia and the number of ciliated neurons declines with age in major retinorecipient targets. Our data suggests that the maturation timing of neuronal and non-neural cilia differs with maturation of neuronal cilia occurring over a longer postnatal period.
INTRODUCTION
The superior colliculus (SC) and dorsal lateral geniculate nucleus (dLGN) comprise the two main retinorecipient targets of the mammalian brain, receiving input from different retinal ganglion cell (RGC) types and transmitting that signal to the primary visual cortex (V1) (Ellis et al., 2016; Martersteck et al., 2017). Located in the midbrain, the SC is involved in directing head and eye movements and discriminating moving objects, while the dLGN, which is in the thalamus, acts as the primary source of innervation for V1 (Dhande & Huberman, 2014; Felleman & Van Essen, 1991). While both the SC and dLGN receive direct input from the retina, the superficial-most SC layers project first to the dLGN, where thalamocortical relay neurons transmit signals from both collicular neurons and RGC axons to the visual cortex to be processed as conscious perception (Beltramo & Scanziani, 2019; Evangelio et al., 2018; Kerschesteiner & Guido, 2017, Piscopo et al., 2013). Retinogeniculate and retinocollicular maps are defined and segregated by the time of natural eye opening, followed by refinement after visual experience in combination with spontaneous retinal wave activity (Huberman et al., 2008; Guido 2018). Thus, developmental profile studies of SC and dLGN have contributed important insights into the formation of visual maps, receptive fields, thalamic and midbrain circuitry, and the effects of retinal spontaneous activity and visual experience in the retinofugal pathway.
The primary cilium, an antenna-like sensory organelle found in most mammalian cells, has been identified as an essential component in mechano-transduction, signaling pathways, and maintaining homeostasis in many organ systems. For instance, evidence of mechanosensory function for primary cilia has been found in bone cells, kidneys, and eyes (Malone et al., 2007; Kottgen et al., 2008; Luo et al., 2014). Moreover, the role of primary cilia in Sonic hedgehog (Shh) and Wnt signaling has been thoroughly studied, including binding of Shh ligand to Patched on the ciliary membrane and inversin protein regulation of the Wnt pathway during renal development (Huangfu et al., 2003; Simons et al., 2005; Lee et al., 2010). In fact, defects in primary cilia formation and function resulting from mutations of ciliary proteins lead to serious ramifications throughout the human body, such as impaired sight, kidney failure, and cognitive dysfunction. Lowe syndrome, an X-linked disease characterized by congenital cataracts, glaucoma, developmental delay, and renal dysfunction, is caused by a mutation in an inositol polyphosphate-5-phosphatase, OCRL, that localizes to primary cilia; fibroblasts derived from Lowe syndrome patients exhibit abnormal cilia, in addition to imbalances in phosphoinositide levels (Luo et al., 2012). Some of these genetic disorders are classified as “ciliopathies,” where a mutation in a single cilia-related gene causes a cascade of devastating health defects (Guemez-Gamboa et al., 2014; Youn et al., 2017). For instance, some mutations in ARL13B, a small GTPase localized to the primary cilium, manifest as a ciliopathy called Joubert syndrome; patients exhibit the canonical “molar tooth” sign where MRI scans reveal abnormal midbrain-hindbrain development: long, thick superior cerebellar peduncles, deep interpeduncular fossa, and vermis hypoplasia/aplasia (Cantagrel et al., 2008; Doherty 2009). ARL13b-mutant variants of Joubert syndrome have been found to disrupt the migratory rate of interneurons in the developing cortex, implicating primary cilia in development of migratory patterns in the brain (Higginbotham et al., 2012). The eye and brain abnormalities found in these disorders suggest that primary cilia play a critical role in preserving tissue homeostasis, emphasizing the need to characterize primary cilia in these organs.
Past research has focused on characterizing primary cilia within the retina and brain. Primary cilia have been found in specific retinal layers, such as the retinal pigment epithelium (RPE), which requires functional primary cilia for complete maturation (May-Simera et al., 2018). Additionally, photoreceptors, which detect light stimulation coming from the cornea, have a modified primary cilium that connects the outer segment and inner segment (Khanna et al., 2015). Furthermore, primary cilia have been identified in different regions of the brain, such as the hypothalamus and hippocampus (Green et al., 2014; Wang et al., 2011). The development and distribution of primary cilia in the neocortex have been characterized (Arellano et al., 2011). However, the distribution of cilia within SC and dLGN remains unknown.
In this study, we aimed to describe primary cilia over various stages of development in the SC and dLGN, where cilia distribution and length were measured by immunofluorescence and confocal microscopy. Furthermore, because loss of primary cilia has been found to disrupt migration patterns of inhibitory neurons as well as extension of dendrites and axons in excitatory neurons, we assessed the presence and length of primary cilia in neurons versus non-neuronal cells (Higginbotham et al., 2012; Sarkisian et al., 2014). Thus, we characterized the ciliation profile in the retinofugal pathway, including different neuronal cell types, in order to expand our base knowledge of primary cilia in visual circuitry as a whole.
MATERIALS AND METHODS
Animals
Experimental procedures were in accordance with NIH guidelines and approved by the Institutional Animal Care and Use Committees at Stanford University School of Medicine. Arl13b-mCherry; Centrin2-GFP double transgenic mice were obtained from Jackson Laboratory (RRID:IMSR_JAX:027967); they have been previously reported (Higginbotham et al., 2004; Bangs et al., 2015). GAD67-GFP mice were also previously reported (Chattopadhyaya et al., 2004; RRID:IMSR_JAX:007677). Wild-type C57Bl/6J mice were obtained from Jackson Laboratory (RRID:IMSR_JAX:000664). Macaque monkey (Macaca mulatta) samples were obtained as previously described (Dhande et al., 2019). A female macaque monkey, DOB 1/28/09, was sacrificed and tissue gathered about 12/16/15.
Antibodies
Adenylate Cyclase III (AC3) was localized using a rabbit polyclonal antibody Adenylate Cyclase III against AC3 (EnCor Biotechnology, Cat# RPCA-ACIII, RRID:AB_2572219), and a rabbit polyclonal antibody A cyclase III (C-20) against AC3 (Santa Cruz Biotechnology, Cat# sc-588, RRID:AB_630839). Neurons were identified using a mouse monoclonal antibody NeuN (Millipore, Cat# MAB377, RRID:AB_2298772). GABAergic neurons expressing parvalbumin (PV+) were identified using mouse monoclonal antibody Parvalbumin (Sigma-Aldrich, Cat# P3088, RRID:AB_477329), and excitatory neurons using a monoclonal mouse antibody Ca2+/calmodulin-dependent protein kinase II alpha subunit (CaMKIIa) (Millipore, Cat# 05–532, RRID:AB_309787). Species-specific secondary antibodies conjugated to Alexa Fluor 488, 594, or 647 (Invitrogen) were used. Rabbit anti-AC3 (Santa Cruz) was used at a dilution of 1:2000. All other primary and secondary antibodies were used at a dilution of 1:1000.
Histology & Immunohistochemistry
Brains from Arl13b-mCherry; Centrin2-GFP double transgenic mice were collected at P4, P15, P33/38, and P77 to reflect relevant mouse visual development timepoints: P4 for early postnatal events, P15 for eye opening, P33 for synaptic pruning that occurs 2–3 weeks after eye opening, and P77 for adult developed mouse. GAD67-GFP mice were studied at P64, and wild-type C57Bl/6J mice at P38. Brains were perfusion fixed, harvested, and processed for immunohistochemistry as previously described (Dhande et al., 2013; Huberman et al., 2008). Briefly, mice were transcardially perfused with 1x PBS (phosphate buffered saline) followed by 4% paraformaldehyde (PFA). Brains were harvested and postfixed overnight (~12–18hrs) in 4% PFA at 4°C, then immersed in 30% sucrose for 2–3 days at 4°C, and sectioned in the coronal plane at 45μm on a sliding microtome. Tissue was incubated overnight at 4°C in primary antibodies and incubated for 2hrs at room temperature with appropriate secondary antibodies. After washing four times with 1× PBS, sections were mounted with Vectashield antifade mounting medium with DAPI (Vector laboratories).
The macaque monkey was transcardially perfused with 4% PFA, then 4% PFA with 10% sucrose, followed by 4% PFA with 20% sucrose. The brain was removed and immersed in 30% sucrose at 4°C for 1–2 weeks and sectioned at 40 μm on a sliding microtome. Slices were cleared with 5% (wt/vol) SDS in 0.1M PBS (pH 7.5) at room temperature for 2hrs, with gentle rotational shaking (Xu et al., 2017). After clearing of brain slices, the SDS was removed by washing in PBS followed by permeabilization, blocking and antibody application as for mouse samples, using NeuN and AC3 (EnCor Biotechnology) antibodies.
Imaging & Quantification
Brain sections were imaged using a Zeiss LSM 880 scanning confocal microscope at 40x with 1.6x optical zoom. Z-stacks were taken at 1 μm steps. Two to three animals of either sex were used for all analyses (at least two brains/age and two sections/brain). Only one LGN from one hemisphere was analyzed per section, chosen as consecutive sections of the same LGN. Central dLGN and SC sections were used because they include the largest extent of dLGN and SC. Ventral-medial LGN and central SC were used for counting cells. The SC area was within the SGS. Three dLGN sections per animal were used for quantification of AC3 overlap with GAD67-GFP-expressing cells. For all other analyses, at least 2 dLGN and 2 SC sections per animal were used. Three dLGN sections were used for the macaque sample quantification.
Cell counts and overlap were manually determined from the imaged stacks using Neurolucida software (www.mbfbioscience.com/neurolucida, RRID:SCR_001775)). For preliminary data analysis a volume of 100×100×30 μm3 was used, and for all subsequent analyses a volume of 50×50×15μm3 was used. About 50 cells/section were counted and a total of 2821 cilia were identified in mouse samples; ~50 cells/section were counted in the parvocellular layers of the macaque dLGN. For computing percent cells with cilia in SC and dLGN, Arl13b-mCherry signal was counted as a cilium if one end of the signal was directly adjacent to the Centrin2-GFP signal. To control for age-dependent cell density changes the total number of ciliated cells (Arl13b-labeled) was normalized to total number of DAPI stained cells within the measured area. Cilia length was computed from 3D reconstructions of cilia generated using Neurolucida Explorer software (www.mbfbioscience.com/neurolucida-explorer, RRID:SCR_017348).
Statistical analysis
All statistical analysis was performed using Graphpad8 (Prism) software (www.graphpad.com, RRID:SCR_002798). Results for total cell, neuron, and cilia counts are expressed as mean values ± SEM. Statistical analysis was performed using nested one-way analysis of variance (ANOVA) followed by Tukey’s post-hoc test for comparison of multiple age timepoints. Comparisons of cilia length between neuronal and non-neuronal cells and between SC and dLGN were analyzed by two-tailed nested t-tests. A p-value of less than 0.05 was considered statistically significant. To compare developmental trends between SC and dLGN, the mean values of total cilia, fraction of ciliated cells that are neurons, fraction of neurons that extend cilia, and cilia length were normalized to P77. To compare average cilia length of neuronal and non-neurons cells, values were normalized to NeuN+.
RESULTS
Ciliation generally decreased and ciliary length altered in the superior colliculus of the maturing mouse brain.
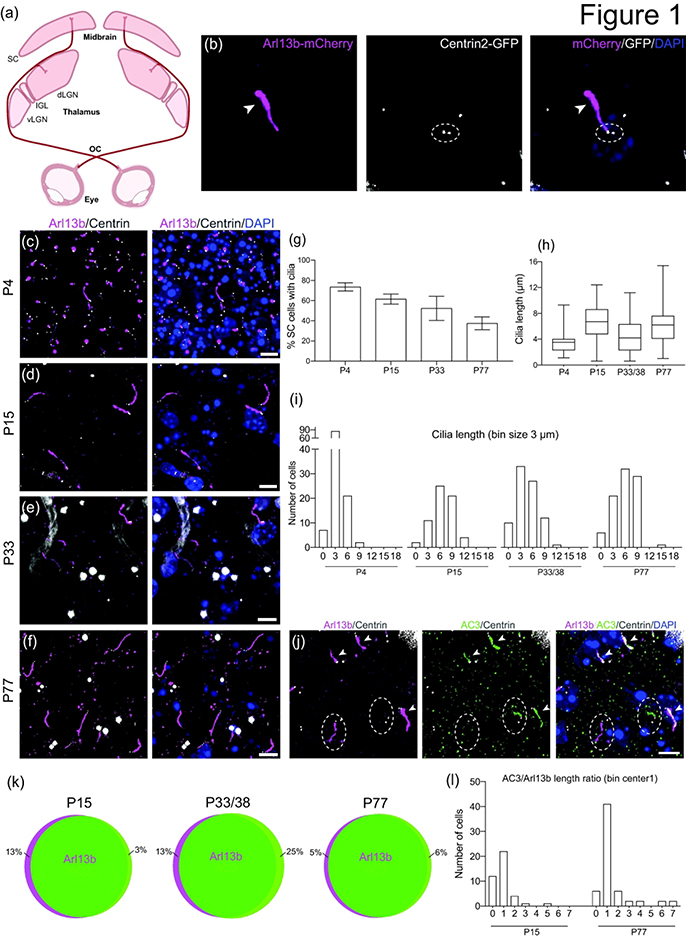
The two main retinorecipient brain targets in mammals are the superior colliculus (SC) and the dorsal lateral geniculate nucleus (dLGN) (Figure 1a) (Martersteck et al., 2017; Seabrook et al., 2017). Because Arl13b and Centrin2 are localized to primary cilia and basal bodies, respectively, they have served as surrogate markers for those ciliary structures (Kasahara et al., 2014; Paoletti et al., 1996). In mice, retinogeniculate and retinocollicular maps are defined within the first postnatal week and completely segregated by the time of natural eye opening (P12-P14) (Godement et al., 1984; Jaubert-Miazza et al., 2005; Huberman et al., 2008). Furthermore, synaptic pruning occurs over 3 weeks after eye-specific segregation, reducing the number of RGC inputs to a single dLGN neuron from more than 10 to 1–3 (Hong et al., 2011). Therefore, to investigate the full developmental profile of primary cilia in the SC and dLGN, brain sections from Arl13b-mCherry; Centrin2-GFP double transgenic mice at ages P4, P15, P33, and P77 were taken and analyzed under confocal microscopy. Primary cilia in close proximity to Centrin2-labeled basal bodies were detected in the SC at all timepoints (Figure 1b–f). Total number of ciliated cells gradually decreased with age, from 73.6 ± 4.0% SEM at P4 to 37.5 ± 6.3% SEM at P77 (p = 0.0992) (n = 381 P4, n = 146 P15, n = 372 P33, n = 647 P77 total cells counted) (Figure 1g). Throughout our analysis, we noticed that some primary cilia were shorter at different stages of development. Therefore, we subsequently quantified cilia length to examine the variance between each timepoint. We found a general shift upward in the distribution of collicular cilia length from P4 to older timepoints: a greater number of short cilia (<6μm) was observed in the SC of younger P4 mice with lower average length (P4 = 3.4 ± 0.1μm, P15 = 6.7 ± 0.3μm, P33/38 = 4.5 ± 0.3μm, P77 = 5.8 ± 0.3μm [mean ± SEM]; p = 0.0722) (n = 115 P4, n = 64 P15, n = 83 P33/38, n = 89 P77 total cilia measured) (Figure 1h,i, Figure 5d).
Figure 1.
Developmental profile of cilia in the superior colliculus.
(a) Schematic of two major retinorecipient brain targets, the SC and the dLGN.
(b) Photomicrograph of primary cilia visualized using the Arl13b-mCherry; Centrin2-GFP double transgenic mouse. Arl13b (magenta, arrowhead at distal end) is expressed by primary cilia, and Centrin2 (white, dotted ellipse) labels centrioles (basal bodies). Nuclei are stained with DAPI (blue).
(c-f) Representative photomicrographs of cilia in the SC at P4 (c), P15 (d), P33 (e), and P77 (f).
(g) Bar graph shows the average ± SEM percent of ciliated cells in the SC at the indicated ages. n = 381 P4, n = 146 P15, n = 372 P33, n = 647 P77 total cells counted
(h) Quantification of cilia length of SC cells. n = 115 P4, n = 64 P15, n = 83 P33/38, n = 89 P77 total cilia measured
(i) Binned distribution of cilia length in the SC. The x-axis represents bin centers.
(j) Photomicrographs of SC cells immunostained for AC3 (green) and Arl13b (mCherry, magenta). Arrowheads indicate co-expression of AC3 and Ar13b, and dotted ellipse indicates lack of overlap.
(k) Schematic representation showing percent co-expression of Arl13b and AC3 in the SC. n = 344 Arl13b, n = 348 AC3 total cilia counted
(l) Binned distribution of AC3 to Arl13b ratio on a cell-by-cell basis in the SC. n = 40 P15, n = 61 P77 cells compared
Scale bar: All: 5 μm.
OC: optic chiasm; vLGN: ventral lateral geniculate nucleus; IGL: intergeniculate nucleus; dLGN: dorsal lateral geniculate nucleus; SC: superior colliculus
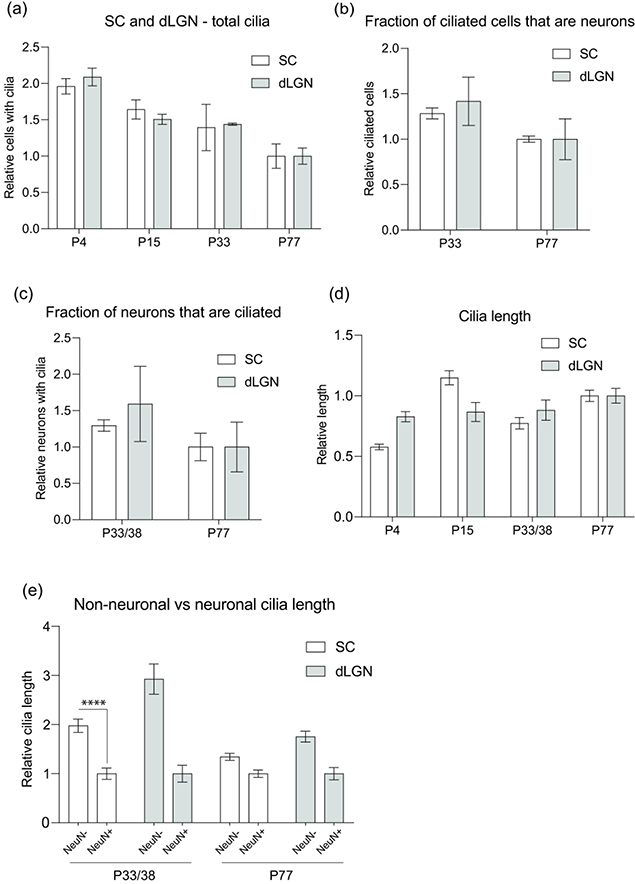
Figure 5.
Comparative analysis of the development of cilia in SC and dLGN.
(a-d) Normalized mean values (normalized to P77 of SC or dLGN, respectively) of total cilia in SC and dLGN (a), ciliated cells in dLGN and SC that are neurons (b), dLGN and SC neurons that extend cilia (c), and cilia length of dLGN and SC cells (d).
(e) Normalized mean values (normalized to NeuN+ at each age of SC and dLGN, respectively) of cilia length in non-neuronal cells and neurons; **** P<0.0001; Nested t-test (two-tailed).
Because the presence of cilia and variance in length depend on the strength of the Arl13b-mCherry signal, it was important to confirm that the fluorescence indeed corresponds with the full length of primary cilia. In addition to Arl13b, type III adenylyl cyclase (AC3) localizes to cilia throughout the mouse brain and is thus used as prominent marker for primary cilia (Bishop et al., 2007). Brain sections from Arl13b-mCherry; Centrin2-GFP mice were immunostained for AC3, and the Arl13b-mCherry signal was compared to the AC3 staining (Figure 1j). We found that Arl13b-mCherry generally overlapped with AC3 staining at all developmental stages of the SC (n = 344 Arl13b, n = 348 AC3 total cilia counted) (Figure 1k). It should be noted that co-expression was smallest at P33/38, when 25% of cilia expressing AC3 did not express Arl13b, and that this percentage dropped to a low of 6% upon reaching adulthood. Likewise, the percentage of cilia that expressed only Arl13b fell from 13% to 5% by P77. Additionally, cilia lengths identified by Arl13b and AC3 signals were also compared: an AC3 to Arl13b ratio less than one would indicate that Arl13b extended further along the axoneme than AC3 within the same cilia; a ratio approaching 1 would represent an almost complete overlap between AC3 and Arl13b along the entire length of the ciliary axoneme; and a ratio of 2 or greater would indicate cells in which AC3 extended further than Arl13b. We found that the AC3 to Arl13b cilia length ratio approached 1 as the mouse matured from P15 to P77: from 55% to 67% (n = 40 P15, n = 61 P77 cells compared) (Figure 1l). P4 staining was not compared because AC3 signal was inconclusive at this timepoint.
Ciliation decreased and ciliary length altered in the dorsal lateral geniculate nucleus of the maturing mouse brain
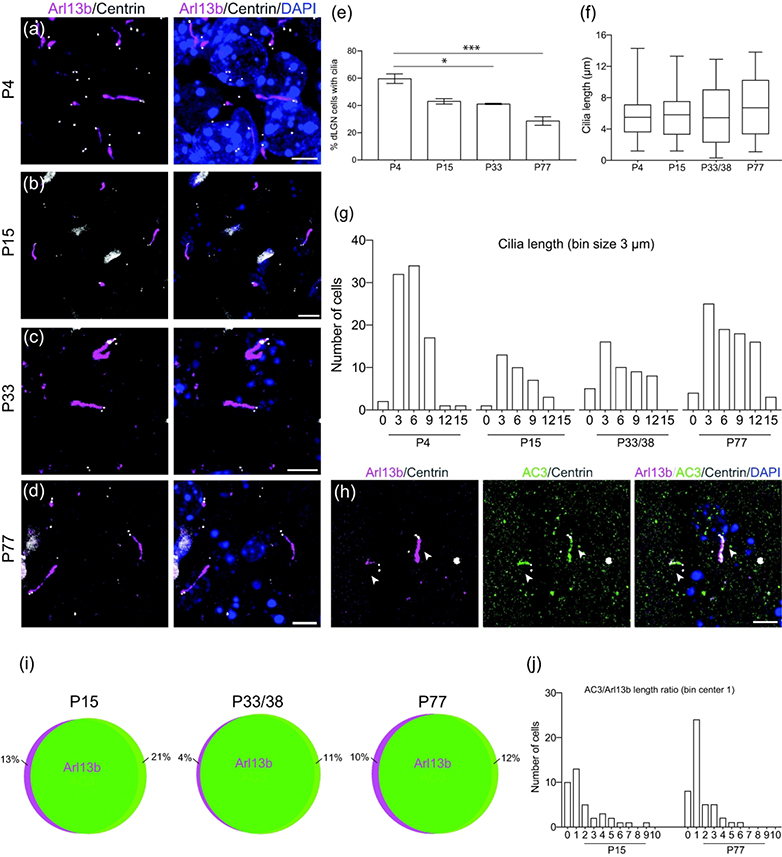
In addition to the SC, sections of the dLGN from Arl13b-mCherry; Centrin2-GFP double transgenic mice were also examined for ciliation at ages P4, P15, P33, and P77 (Figure 2a–d). Total ciliation decreased as maturation increased: from 59.6 ± 0.1% SEM at P4 to 28.5 ± 3.2% at P77 (p < 0.05) (n = 382 P4, n = 87 P15, n = 211 P33, n = 574 P77 total cells counted) (Figure 2e). Furthermore, the distribution of cilia length became wider in the dLGN of older mice: the length of most cilia remained less than 9μm at P4 but became more widely distributed between 0 and 15μm at P15, P33, and P77 (Figure 2f,g). Difference in average cilia length between timepoints was not significant (p = 0.6842) (n = 87 P4, n = 34 P15, n = 48 P33/38, n = 85 P77 total cilia measured) (Figure 5d). AC3 and Arl13b signals were also compared in the dLGN of these mice (Figure 2h). AC3 and Arl13b overlapped most of the time at all developmental stages (n = 169 Arl13b, n = 180 AC3 total cilia counted) (Figure 2i). Co-expression was lowest at P15, when 21% of cilia expressing AC3 did not express Arl13b, and decreased to a more consistent level of 11–12% as the mouse matured to P33 and P77. Moreover, comparison of ciliary lengths demonstrated by AC3 and Arl13b revealed that more cells exhibited an AC3-Arl13b ciliary length ratio approaching 1 at P77 while the ratio was more widely distributed at P15 (n = 38 P15, n = 61 P46 cells compared) (Figure 2j).
Figure 2.
Developmental profile of cilia in the dorsal lateral geniculate nucleus.
(a-d) Representative photomicrographs of cilia (Arl13b-mCherry, magenta) in the dLGN at P4 (a), P15 (b), P33 (c), and P77 (d). Centrioles labeled with GFP (white) and nuclei labeled with DAPI (blue).
(e) Bar graph shows the average ± SEM percent of ciliated cells in the SC at the indicated ages; * P<0.05, *** P<0.001; Nested ANOVA, Tukey’s multiple comparison test. n = 382 P4, n = 87 P15, n = 211 P33, n = 574 P77 total cells counted
(f) Quantification of cilia length of dLGN cells. n = 87 P4, n = 34 P15, n = 48 P33/38, n = 85 P77 total cilia measured
(g) Binned distribution of cilia length in the dLGN. The x-axis represents bin centers.
(h) Photomicrographs showing dLGN cells immunostained for AC3 (green) and Arl13b (mCherry, magenta). Arrowheads indicate co-expression of AC3 and Ar13b.
(i) Schematic representation showing percent co-expression of Arl13b and AC3 in the dLGN. n = 169 Arl13b, n = 180 AC3 total cilia counted
(j) Binned distribution of AC3 to Arl13b ratio on a cell-by-cell basis in the dLGN. n = 38 P15, n = 61 P46 cells compared
Scale bar: All: 5 μm.
Diverse types of neurons in the SC express primary cilia
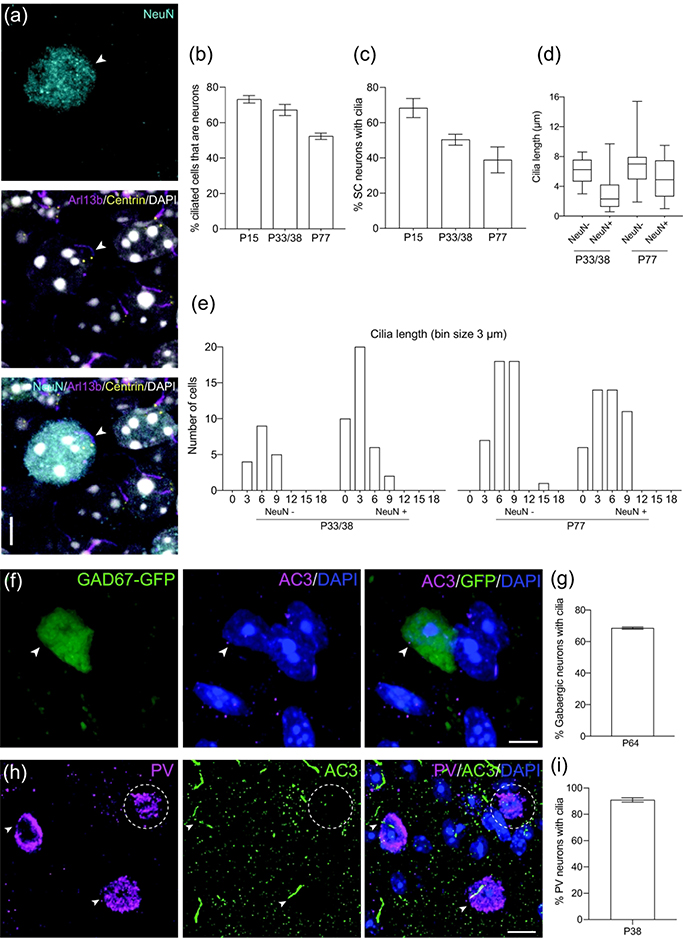
To further characterize the expression profile of primary cilia in the SC, we aimed to distinguish between neurons and non-neuronal cells associated with ciliation. Because expression of NeuN protein has been found in most types of neurons throughout the nervous system of vertebrates, including the SC of developing mice, NeuN has been used as a specific marker for neurons (Mullen et al., 1992; Arellano et al., 2011). Brain sections from the SC of Arl13b-mCherry; Centrin2-GFP mice were immunostained with NeuN at P15, P33/38, and P77 and analyzed by confocal microscopy (Figure 3a). More than half of cells with primary cilia were identified as neurons at all timepoints; the number decreased gradually with age (P15 = 73.2 ± 2.1%, P33/38 = 67.2 ± 3.1%, P77 = 52.4 ± 1.8% [mean ± SEM]; p = 0.1051) (n = 91 P15, n = 374 P33/38, n = 218 P77 total cilia counted) (Figure 3b). In fact, the average number of ciliated neurons declined dramatically from 68.4 ± 5.4% SEM at P15 to 38.9 ± 7.3% SEM at P77 (p = 0.0700) (n = 96 P15, n = 75 P33/38, n = 323 P77 total neurons counted) (Figure 3c). P4 was not analyzed because NeuN expression is weak in immature neurons (Arellano et al., 2011).
Figure 3.
Different types of neurons in the SC have cilia.
(a) Example of a P15 SC neuron (NeuN, cyan) extending a cilium (Arl13b, magenta). Centrioles labeled with GFP (yellow) and nuclei labeled with DAPI (white). Arrowhead indicates neuronal primary cilia.
(b) Quantification of ciliated SC cells that express the neuronal marker NeuN. Mean ± SEM. n = 91 P15, n = 374 P33/38, n = 218 P77 total cilia counted
(c) Quantification of SC neurons that have cilia. Mean ± SEM. n = 96 P15, n = 75 P33/38, n = 323 P77 total neurons counted
(d) Quantification of cilia length in non-neuronal cells and neurons in SC. n = 18 NeuN−, n = 38 NeuN+ P33/38 cilia; n = 44 NeuN−, n = 45 NeuN+ P77 total cilia measured
(e) Binned distribution of cilia length in non-neuronal cells and neurons in SC. The x-axis represents bin centers.
(f) Representative photomicrograph showing a cilium (AC3, magenta) extending from a P64 gabaergic SC neuron (GFP, green). Nuclei labeled with DAPI (blue). Arrowhead indicates co-expression of GFP and AC3.
(g) Percent of gabaergic neurons with cilia at P64. n = 162 total GAD67+ cells counted
(h) Representative photomicrograph showing cilia (AC3, green) extending from P38 parvalbumin (PV) neurons (magenta). Nuclei labeled with DAPI (blue). Arrowheads indicate co-expression of PV and AC3, and dotted ellipse indicates lack of overlap.
(i) Percent of parvalbumin (PV) neurons in the SC with cilia at P38. n = 108 total PV+ cells counted
Scale bar: (a,f): 5μm; (h): 10μm
Because we previously noted shorter cilia in the total number of ciliated cells in the SC of younger mice, we quantified cilia length with respect to NeuN to assess if a similar trend was apparent in NeuN+ cells. The distribution of cilia length in ciliated NeuN+ cells was lower at P33/38 (1.28<x<4.2μm) than at P77 (2.65<x<7.45μm), whereas non-neuronal ciliary length distribution remained relatively consistent (P33/38 = 4.68<x<7.55μm; P77 = 4.95<x<7.9μm) (n = 18 NeuN−, n = 38 NeuN+ P33/38 cilia; n = 44 NeuN−, n = 45 NeuN+ P77 total cilia measured) (Figure 3d). From another perspective, neuronal ciliary lengths at P33/38 tended to concentrate over bins 0 and 3, representing cilia length less than 6μm, while that distribution shifted upward to between 3 and 9μm in P77 mice (Figure 3e). On the other hand, non-neuronal cilia length tended to concentrate between 3 and 9μm at both stages of development. Neuronal and non-neuronal cilia length differed at P33/38 (NeuN+ = 3.1 ± 0.4μm, NeuN− = 6.1 ± 0.4μm [mean ± SEM]; p < 0.0001) (Figure 5e).
Previous studies have demonstrated a role for primary cilia in migration and postmigratory differentiation of inhibitory and excitatory neurons in the neocortex (Sarkisian et al., 2014). The SC comprises several types of neurons that form a network for sensory input from the retina and signal transduction to deeper layers of the SC, dLGN, and the neocortex (Byun et al., 2016). Therefore, characterizing the different types of ciliated neurons in the SC would further delineate the developmental profile of primary cilia in collicular neurons. GABAergic neurons, for instance, have been shown to play a critical role in cortical plasticity in mammals and constitute approximately 30% of the neuronal population (Jones 1993; Chattopadhyaya et al., 2007). Thus, we used GAD67, a glutamic acid decarboxylase that synthesizes GABA, to identify GABAergic neurons. Quantitative analysis of brain sections from GAD67-GFP mice at P64 immunostained with AC3 revealed that 68.6 ± 0.5% SEM of GABAergic neurons expressed primary cilia (n = 162 total GAD67+ cells counted) (Figure 3f,g). However, GAD67 identifies only a portion of inhibitory neurons; GABAergic neurons expressing parvalbumin (PV), a calcium-binding protein, also form inhibitory circuits within the SC (Villalobos 2018). Sections from the SC of P38 wildtype mice stained for PV and AC3 revealed that 90.9 ± 1.7% SEM of collicular PV+ neurons were positive for primary cilia (n = 108 total PV+ cells counted) (Figure 3h,i).
Diverse types of neurons in the dLGN express primary cilia
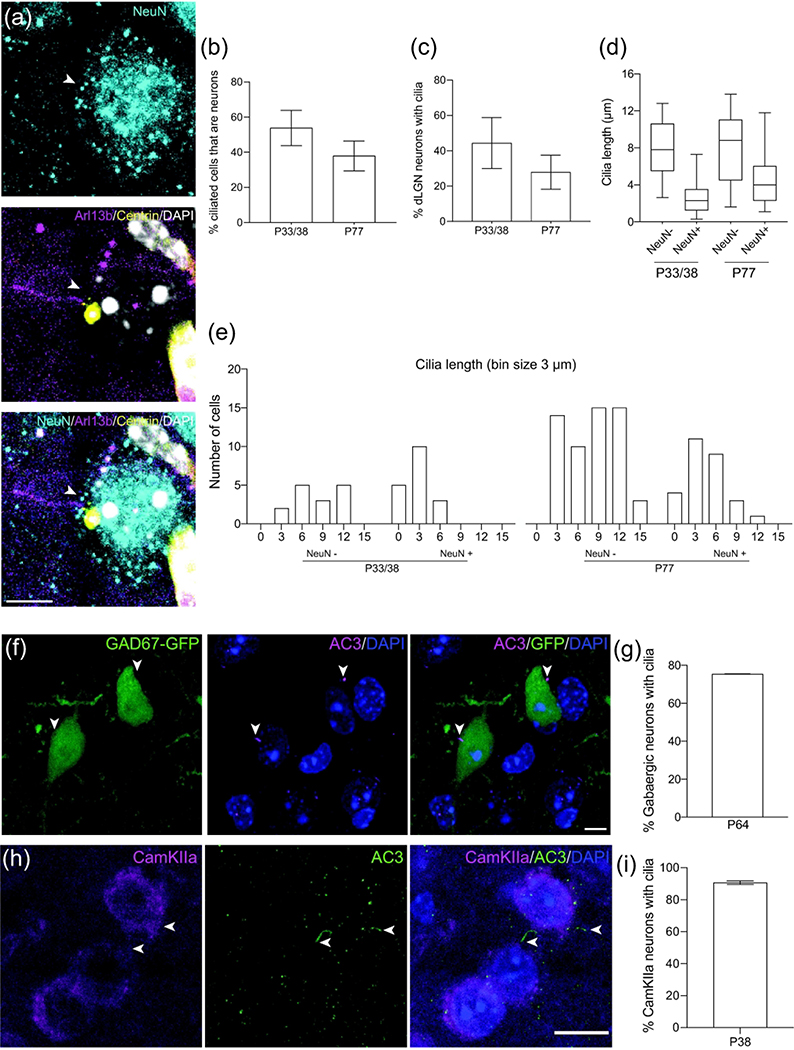
In addition to studying the SC, we investigated the types of ciliated cells in the dLGN of maturing mice. Using NeuN as a marker to stain neuronal cells in sections from Arl13b-mCherry; Centrin2-GFP double transgenic mice, 53.8 ± 10.1% SEM of all ciliated cells were identified as neurons at P33/38; the number subsequently declined to roughly 37.9 ± 8.5% SEM at P77 (n = 86 P33/38, n = 148 P77 total cilia counted) (Figure 4a,b). Furthermore, these ciliated neuronal cells constituted 44.5 ± 14.5% SEM and 27.9 ± 9.6% of all NeuN+ cells at P33/38 and P77, respectively (n = 43 P33/38, n = 248 P77 total neurons counted) (Figure 4c). Comparison of the ciliary length of neuronal versus non-neuronal cells in the dLGN showed a similar trend as in the SC: NeuN+ cilia had a smaller length distribution in younger mice (P33/38 = 1.25<x<3.53μm; P77 = 2.3<x<6.03μm) while non-neuronal cilia remained consistent at each developmental stage (P33/38 = 5.5<x<10.6μm; P77 = 4.5<x<11.05μm) (n = 15 NeuN−, n = 18 NeuN+ P33/38 cilia; n = 57 NeuN−, n = 28 NeuN+ P77 total cilia measured) (Figure 4d). Neuronal ciliary length tended to be less than 6μm at P33/38 and increased to between 3 and 9μm at P77, whereas non-neuronal cilia length was distributed over 3 and 15μm at both stages of development (Figure 4e). Average neuronal cilia length was less than non-neuronal at both P33/38 (NeuN+ = 2.7 ± 0.5μm SEM, NeuN− = 8.0 ± 0.8μm; p = 0.1377) and P77 (NeuN+ = 4.5 ± 0.6μm SEM, NeuN− = 7.9 ± 0.5μm; p = 0.0561) (Figure 5e).
Figure 4.
Different types of neurons in the dLGN extend cilia.
(a) Example of a dLGN neuron (NeuN, cyan) extending a cilium (Arl13b, magenta). Centrioles labeled with GFP (yellow) and nuclei labeled with DAPI (white). Arrowhead indicates neuronal primary cilia.
(b) Quantification of ciliated dLGN cells that express the neuronal marker NeuN. Mean ± SEM. n = 86 P33/38, n = 148 P77 total cilia counted
(c) Quantification of dLGN neurons that have cilia. Mean ± SEM. n = 43 P33/38, n = 248 P77 total neurons counted
(d) Quantification of cilia length in non-neuronal and neuronal dLGN. n = 15 NeuN−, n = 18 NeuN+ P33/38 cilia; n = 57 NeuN−, n = 28 NeuN+ P77 total cilia measured
(e) Binned distribution of cilia length in non-neuronal cells and neurons in SC. The x-axis represents bin centers.
(f) Representative photomicrograph showing a cilium (AC3, magenta) extending from a P64 gabaergic dLGN neuron (GFP, green). Nuclei labeled with DAPI (blue). Arrowheads indicate co-expression of GFP and AC3.
(g) Percent of gabaergic neurons with cilia at P64. n = 49 total GAD67+ cells counted
(h) Representative photomicrograph showing cilia (AC3, green) extending from P38 CamKIIa expressing neurons (magenta) in the dLGN. Nuclei labeled with DAPI (blue). Arrowheads indicate co-expression of CamKIIa and AC3.
(i) Percent of CamKIIa neurons in the dLGN with cilia at P38. n = 104 total CamKIIa+ cells counted
Scale bar: (a, f): 5μm; (h): 10μm
The portion of all types of neurons that GABAergic neurons constitute in the dLGN of GAD67-GFP mice has been previously described (Evangelio et al., 2018). Therefore, we used GAD67 together with AC3 to identify the number of GABAergic neurons with primary cilia in the dLGN (Figure 4f). Quantitative analysis revealed that 75.4 ± 0.1% SEM of GABAergic neurons in the dLGN expressed primary cilia (n = 49 total GAD67+ cells counted) (Figure 4g). However, identifying GABAergic neurons excludes a large population of neuronal types in the dLGN, the excitatory neurons. Calmodulin-dependent protein kinase IIα (CaMKIIa) has been described as a critical mediator of neuronal plasticity and is known to stain a subtype of dLGN neurons (Fink & Meyer 2002; Higo et al. 2000). Hence, we used anti-CaMKIIa as an excitatory neuronal marker alongside AC3 to investigate the expression of primary cilia in excitatory neurons of the dLGN (Figure 4h). A great majority of CaMKIIa+ cells (90.7 ± 1.1% SEM) expressed primary cilia at P38 (n = 104 total CamKIIa+ cells counted) (Fig. 4I).
Ciliation in the dLGN of the macaque monkey
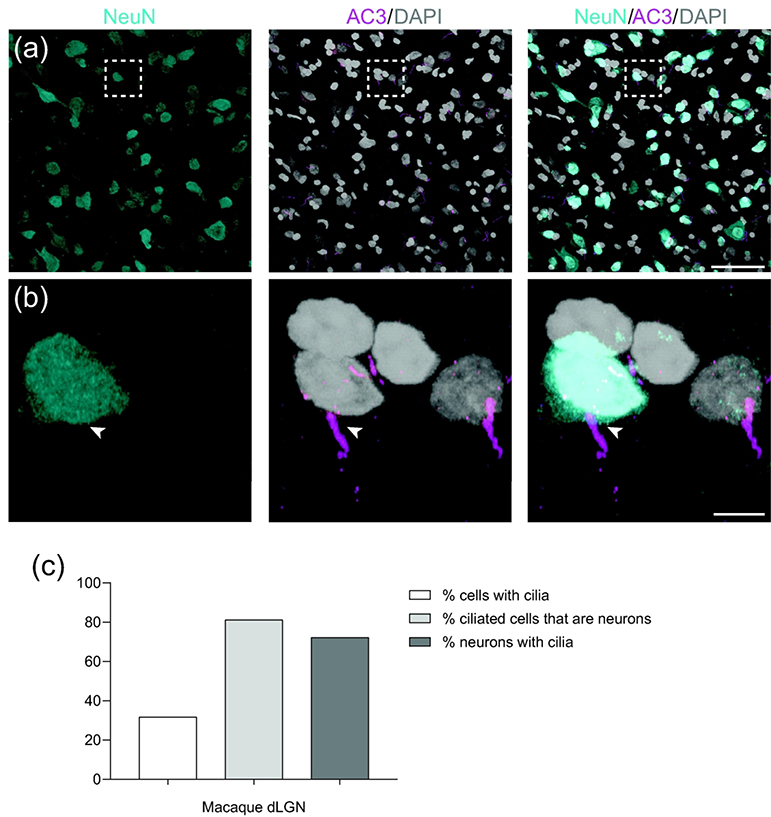
The mouse visual system has been the predominant model for mammalian visual circuitry due to its highly conserved neuronal architecture. For instance, the shell region of the mouse dLGN, which receives projections from the SC, resembles other mammalian brain regions like the C-laminae of cats and the koniocellular division of some primates (Guido 2018). However, key differences between mice and primates exist. For example, eye-specific segregation and spontaneous retinal wave activity occur prenatally in monkeys in contrast to before eye-opening in mice, indicating a species-dependent difference in axon mapping and rearrangement in the dLGN during development (Huberman et al., 2008; Hong et al., 2011). Furthermore, about 10% of RGCs project to the SC in primates compared to about 90% in mice, a striking contrast (Perry et al., 1984; Seabrook et al., 2017). Such differences in visual circuitry between species necessitate analysis of primary tissue from primates. We stained sections from a macaque monkey for AC3 and NeuN to identify primary cilia in the parvocellular layer of the dLGN (Figure 6a,b): 31.8% of all cells were ciliated, most of these ciliated cells (81.2%) were neurons, and most neurons (72.3%) were ciliated (n = 145 total cells counted) (Figure 6c).
Figure 6.
Primary cilia profile in the dorsal lateral geniculate nucleus of the macaque monkey.
(a) Overview of dLGN section marked with NeuN, AC3, and DAPI.
(b) Example of a dLGN neuron (NeuN, cyan) extending a cilium (AC3, magenta). Nuclei labeled with DAPI (white). Arrowhead indicates co-expression of NeuN and AC3.
(c) Percent of ciliated cells, ciliated cells that are neurons, and neurons that have cilia in the parvocellular layer of macaque dLGN. n = 145 total cells counted
Scale bar: (a): 50μm; (b): 5μm.
DISCUSSION
The ciliation profile of SC and dLGN cells undergoes similar alterations during brain maturation
Primary cilia are thought to play a critical role in development and tissue homeostasis in the retina and cortex of the visual system. Characterizing the primary cilia profile of sensory relay centers for this visual pathway, such as the SC and dLGN, provides a foundation for understanding cilia at this intersection. We found that the number of primary cilia significantly declined with advancing stages of development in dLGN, similar to a general decrease in SC, suggesting that maturation of primary cilia is associated with the general maturation process in these tissues (Figure 5a). We also noted that AC3/Arl13b co-expression increases as a mouse matures to P77 while the subset of primary cilia expressing neither AC3 or Arl13b declines. We hypothesize that the general decrease in ciliation levels observed in the SC and dLGN primarily affects this subset of ciliated cells lacking either AC3 or Arl13b. In this context, cilia co-expressing Arl13b and AC3 are retained and increase further into adulthood while other cilia are gradually removed, indicating that co-expression of both these proteins might be important for cilia stability.
The ciliary profile of SC and dLGN neurons diverges slightly during brain maturation
The fraction of ciliated cells that are neurons is higher in dLGN tissue at P33/38 than at P77, and the total number of ciliated neurons in the dLGN declines as development proceeds (Figure 4b, c). Similarly, the fraction of neurons that are ciliated declines with maturation in SC, but at a lower relative value than in dLGN (1.29 and 1.59) (Figure 5c). The reduction of cilia in SC and dLGN neurons may partially contribute to the general decline in primary cilia observed over the stages of maturation, but the rate of reduction is somewhat higher in dLGN, suggesting that dLGN tissue may need more time to complete maturation than SC. It is also notable that neurons constituted the greatest proportion of ciliated cells in the SC, but not the dLGN, at every timepoint (Figure 3b, Figure 4b) Taken together, the faster maturation and higher retention rate in the SC suggest that primary cilia may be critically important in neuronal development and function in the SC.
Ciliary length patterns remain consistent between SC and dLGN
Some genetic disorders manifest themselves in shorter cilia resulting from the lack of proper cilia formation. For instance, fibroblasts derived from Lowe syndrome patients exhibit shorter cilia than normal, and individuals with Joubert syndrome variants expressing abnormal cilia demonstrate disrupted migratory and positional potential of interneurons in the developing cortex (Guemez-Gamboa et al., 2014; Youn et al., 2017; Luo et al., 2012; Higginbotham et al., 2012). Therefore, characterizing not only the presence of cilia but also ciliary length in the maturing mouse brain is critical in determining the role of primary cilia in the SC and dLGN. We observed shorter cilia in all cells during early stages of development than at later stages, an almost 2-fold increase in average cilia length from P4 to P77 in the SC (Figure 5d). Interestingly, non-neuronal cells exhibited longer primary cilia than neuronal cells at all developmental stages in both the SC and dLGN, yet ciliary length in NeuN+ cells increased over time. Maximal extension may be a sign of final maturation, and these results suggest that neurons in these regions may need more time to terminally extend primary cilia and develop into mature neurons (Figure 5e). Similar observations were found for primary cilia in the neocortex, where cilia elongation starts at P4 and continues postnatally for up to 3 months to reach maximum average length (Arellano 2011). This observation across different parts of the visual pathway suggests that primary cilia play similar roles in neurons of the neocortex and retinofugal pathway. Because inhibition of ciliogenesis through Arl13b, IFT88, and Kif3a has been found to disrupt both dendritic and axonal outgrowth involved in excitatory neuron connectivity and the migratory patterns of inhibitory interneurons in neocortex, it would be interesting to examine how disruption in cilia elongation affects different neuronal types in the SC and dLGN (Sarkisian et al., 2014; Kerschesteiner & Guido, 2017; Piscopo et al., 2013). Furthermore, the observed decline in ciliation of non-neuronal cells could be attributed to a number of different types of cells, including glia, which have been implicated in proper brain development and function through neuronal-glial interactions (Nedergaard et al., 2003; Tasker et al., 2012).
Ciliated cells in primate dLGN identified as neurons, and primate dLGN neurons expressed primary cilia
Primate visual circuitry such as that of the macaque monkey has been thoroughly studied, from the distribution of cortical maps in the cerebral cortex to the determination of direction selectivity by the connectivity of RGCs (Felleman & Van Essen, 1991; Dhande et al., 2019). However, the retinofugal pathway remains to be further investigated. We observed that, although most cells in the parvocellular layer of the macaque dLGN were not ciliated, the ciliated cells were mostly neuronal (81.2%). Moreover, a majority of neurons exhibited primary cilia (72.3%) (Figure 6c). Thus, primary cilia appear to be important in neurons of the primate dLGN. Because primary cilia are necessary for proper brain development and neuronal differentiation, it is plausible that the large number of ciliated neurons indicates an important developmental role for primary cilia in primate dLGN, including the laminar organization of magnocellular and parvocellular layers. Furthermore, because these layers comprise the two major pathways (M and P) for relaying visual information to the cortex, primary cilia in neurons of these M and P pathways may contribute to delivering these signals for cortical processing.
Visual circuitry differs across species, and here we note some interesting distinctions between the ciliation profile of mice and primates. In both species, most cells did not have primary cilia. However, in our primate dLGN sample, most ciliated cells were neurons and most neurons had primary cilia, whereas in mouse dLGN most ciliated cells were nonneuronal and less than half of all neurons had primary cilia. This disparity in neuronal cilia suggests that primary cilia in neurons may play role in primate dLGN development and function and necessitates further investigation. Furthermore, the functional relevance of primary cilia may also extend to neurons in the SC.
Acknowledgments
This work was supported by NIH/NEI K08-EY022058 (Y.S.), R01-EY025295 (Y.S.), VA merit CX001298 (Y.S.), Ziegler Foundation for the Blind (Y.S.), R01-EY-023295 (Y.H.) and R01-EY024932 (Y.H.), Children’s Health Research Institute Award (Y.S.), Research for Prevention of Blindness Unrestricted grant (Stanford Ophthalmology), American Glaucoma Society (Y.S.), Lowe syndrome association (Y.S.), Knights Templar Eye Foundation (Y.S.) and P30 Vision Center grant to Stanford Ophthalmology department. Y.S. is a Laurie Kraus Lacob Faculty Scholar in Pediatric Translational Medicine. We are grateful for Dr. Andrew Huberman for providing space and resources for this study. We thank Dr. Xing Wei and Dr. Massimo Scanziani for generously providing GAD67-GFP transgenic mice and Dr. Tia Kowal for assistance with tissue collection. We are grateful to the Sun lab for helpful discussions and comments on the manuscript.
ABBREVIATIONS LIST
- SC
superior colliculus
- dLGN
dorsal lateral geniculate nucleus
- RGC
retinal ganglion cell
- RPE
retinal pigment epithelium
- AC3
type III adenylyl cyclase
- Shh
Sonic hedgehog
- AC3
anti-Adenylate Cyclase III
- NeuN
anti-Neuronal Nuclei
- PV
parvalbumin
- CaMKIIa
Ca2+/calmodulin-dependent protein kinase II alpha subunit
Footnotes
Conflict of interest statement: The authors declare no conflict of interest.
Data Availability Statement: The data and images that support the findings of this study are available from the corresponding authors upon request.
REFERENCES
- Arellano JI, Guadiana SM, Breunig JJ, Rakic P, & Sarkisian MR (2012). Development and distribution of neuronal cilia in mouse neocortex. Journal of Comparative Neurology, 520(4), 848–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangs FK, Schrode N, Hadjantonakis AK, & Anderson KV (2015). Lineage specificity of primary cilia in the mouse embryo. Nature Cell Biology, 17(2), 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltramo R, & Scanziani M (2019). A collicular visual cortex: Neocortical space for an ancient midbrain visual structure. Science, 363(6422), 64–69. [DOI] [PubMed] [Google Scholar]
- Bishop GA, Berbari NF, Lewis J, & Mykytyn K (2007). Type III adenylyl cyclase localizes to primary cilia throughout the adult mouse brain. Journal of Comparative Neurology, 505, 562–571. [DOI] [PubMed] [Google Scholar]
- Byun H, Kwon S, Ahn HJ, Liu H, Forrest D, Demb JB, & Kim IJ (2016). Molecular features distinguish ten neuronal types in the mouse superficial superior colliculus. Journal of Comparative Neurology, 524, 2300–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantagrel V, Silhavy JL, Bielas SL, Swistun D, Marsh SE, Bertrand JY, Audollent S, Attie-Bitach T, Holden KR, Dobyns WB, Traver D, Al-Gazali L, Ali BR, Lindner TH, Caspary T, Otto EA, Hildebrandt F, Glass IA, Logan CV, Johnson CA, Bennett C, Brancati F, Valente EM, Woods CF, & Gleeson JG (2008). Mutations in the Cilia Gene ARL13B Lead to the Classical Form of Joubert Syndrome. The American Journal of Human Genetics, 83, 170–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyaya B, Di Cristo G, Wu CZ, Knott GW, Kuhlman S, Fu Y, Palmiter RD, & Huang ZJ (2007). GAD67-mediated GABA synthesis and signaling regulate inhibitory synaptic innervation in the visual cortex. Neuron, 54, 889–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyaya B, Di Cristo G, Higashiyama H, Knott GW, Kuhlman SJ, Welker E, & Huang ZJ (2004). Experience and activity-dependent maturation of perisomatic GABAergic innervation in primary visual cortex during a postnatal critical period. Journal of Neuroscience, 24(43), 9598–9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhande OS, Bhatt S, Anishchenko A, Elstrott J, Iwasato T, Swindell EC, Xu HP, Jamrich M, Itohara S, Feller MB, & Crair MC (2012). Role of adenylate cyclase 1 in retinofugal map development. Journal of Comparative Neurology, 520(7), 1562–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhande OS, & Huberman AD (2014). Visual circuits: Mouse retina no longer a level playing field. Current Opinion in Neurobiology, 24, 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhande OS, Stafford BK, Franke K, El-Danaf R, Percival KA, Phan AH, Li P, Hansen BJ, Nguyen PL, Berens P, Taylor WR, Callaway E, Euler T, Huberman AD (2019). Molecular fingerprinting of on–off direction-selective retinal ganglion cells across species and relevance to primate visual circuits. Journal of Neuroscience, 39(1), 78–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty D (2009). Joubert Syndrome: Insights Into Brain Development, Cilium Biology, and Complex Disease. Seminars in Pediatric Neurology, 16(3), 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis EM, Gauvain G, Sivyer B, & Murphy GJ (2016). Shared and distinct retinal input to the mouse superior colliculus and dorsal lateral geniculate nucleus. Journal of Neurophysiology, 116, 602–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelio M, García-Amado M, & Clascá F (2018). Thalamocortical projection neuron and interneuron numbers in the visual thalamic nuclei of the adult C57BL/6 mouse. Frontiers in Neuroanatomy, 12, Article 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felleman DJ, & Van Essen DC (1991). Distributed hierarchical processing in the primate cerebral cortex. Cerebral Cortex, 1(1), 1–47. [DOI] [PubMed] [Google Scholar]
- Fink CC, & Meyer T (2002). Molecular mechanisms of CaMKII activation in neuronal plasticity. Current Opinions in Neurobiology, 12(3), 293–299. [DOI] [PubMed] [Google Scholar]
- Godement P, Salaün J, & Imbert M (1984). Prenatal and Postnatal Development of Retinogeniculate and Retinocollicular Projections in the Mouse. Journal of Comparative Neurology, 230, 552–575. [DOI] [PubMed] [Google Scholar]
- Green JA, & Mykytyn K (2014). Neuronal primary cilia: An underappreciated signaling and sensory organelle in the brain. Neuropsychopharmacology, 39(1), 244–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guemez-Gamboa A, Coufal NG, Gleeson JG (2014). Primary Cilia in the Developing and Mature Brain. Neuron, 82(3), 511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guido W (2018). Development, form, and function of the mouse visual thalamus. Journal of Neurophysiology, 120, 211–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginbotham H, Bielas S, Tanaka T, & Gleeson JG (2004). Transgenic mouse line with green-fluorescent protein-labeled Centrin 2 allows visualization of the centrosome in living cells. Transgenic Research, 13, 155–164. [DOI] [PubMed] [Google Scholar]
- Higginbotham H, Eom TY, Mariani LE, Bachleda A, Hirt J, Gukassyan V, Cusack CL, Lai C, Caspary T, & Anton ES (2012). Arl13b in Primary Cilia Regulates the Migration and Placement of Interneurons in the Developing Cerebral Cortex. Developmental Cell, 23, 925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo N, Oishi T, Yamashita A, Matsuda K & Hayashi M (2000). Expression of GAP-43 and SCG10 mRNAs in Lateral Geniculate Nucleus of Normal and Monocularly Deprived Macaque Monkeys. Journal of Neuroscience, 20(16), 6030–6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong YK, & Chen C (2011). Wiring and rewiring of the retinogeniculate synapse. Current Opinion in Neurobiology, 21(2), 228–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, & Anderson KV (2003). Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature, 426, 83–87. [DOI] [PubMed] [Google Scholar]
- Huberman AD, Feller MB, & Chapman B (2008). Mechanisms Underlying Development of Visual Maps and Receptive Fields. Annual Review of Neuroscience, 31, 479–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaubert-Miazza L, Green E, Lo F-S, Bui K, Mills J, & Guido W (2005). Structural and functional composition of the developing retinogeniculate pathway in the mouse. Visual Neuroscience, 22, 661–676. [DOI] [PubMed] [Google Scholar]
- Jones EG (1993). Gabaergic neurons and their role in cortical plasticity in primates. Cerebral Cortex, 3(5), 361–372. [DOI] [PubMed] [Google Scholar]
- Kasahara K, Kawakami Y, Kiyono T, Yonemura S, Kawamura Y, Era S, Matsuzaki F, Goshima N, & Inagaki M (2014). Ubiquitin-proteasome system controls ciliogenesis at the initial step of axoneme extension. Nature Communications, 5, Article 5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerschenstiner D, & Guido W (2017). Organization of the dorsal lateral geniculate nucleus in the mouse. Visual Neuroscience, 34, e008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna H (2015). Photoreceptor Sensory Cilium: Traversing the Ciliary Gate. Cells, 4(4), 674–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köttgen M, Buchholz B, Garcia-Gonzalez MA, Kotsis F, Fu X, Doerken M, Boehlke C, Steffl D, Tauber R, Wegierski T, Nitschke R, Suzuki M, Kramer-Zucker A, Germino GG, Watnick T, Prenen J, Nilius B, Kuehn EW, & Walz G (2008). TRPP2 and TRPV4 form a polymodal sensory channel complex. Journal of Cell Biology, 182(3), 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, & Gleeson JG (2010). The role of primary cilia in neuronal function. Neurobiology of Disease, 38(2), 167–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo N, Conwell MD, Chen X, Kettenhofen CI, Westlake CJ, Cantor LB, Wells CD, Weinreb RN, Corson TW, Spandau DF, Joos KM, Iomini C, Obukhov AG, Sun Y (2014). Primary cilia signaling mediates intraocular pressure sensation. PNAS, 111(35), 12871–12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo N, West CC, Murga-Zamalloa CA, Sun L, Anderson RM, Wells CD, Weinreb RN, Travers JB, Khanna H, & Sun Y (2012). OCRL localizes to the primary cilium: A new role for cilia in Lowe syndrome. Human Molecular Genetics, 21(15), 3333–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone AMD, Anderson CT, Tummala P, Kwon RY, Johnston TR, Stearns T, & Jacobs CR (2007). Primary cilia mediate mechanosensing in bone cells by a calcium-independent mechanism. PNAS, 104(33), 13325–13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martersteck EM, Hirokawa KE, Evarts M, Bernard A, Duan X, Li Y, Ng L, Oh SW, Ouellette B, Royall JJ, Stoecklin M, Wang Q, Zeng H, Sanes JR, & Harris JA (2017). Diverse Central Projection Patterns of Retinal Ganglion Cells. Cell Reports, 18(8), 2058–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May-Simera HL, Wan Q, Jha BS, Hartford J, Khristov V, Dejene R, Chang J, Patnaik S, Lu Q, Banerjee P, Silver J, Insinna-Kettenhofen C, Patel D, Lotfi M, Malicdan M, Hotaling N, Maminishkis A, Sridharan R, Brooks B, Miyagishima K, Gunay-Aygun M, Pal R, Westlake C, Miller S, Sharma R, & Bharti K (2018). Primary Cilium-Mediated Retinal Pigment Epithelium Maturation Is Disrupted in Ciliopathy Patient Cells. Cell Reports, 22(1), 189–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen RJ, Buck CR, Smith AM. (1992). NeuN, a neuronal specific nuclear protein in vertebrates. Development, 116, 201–211. [DOI] [PubMed] [Google Scholar]
- Nedergaard M, Ransom B, & Goldman SA (2003). New roles for astrocytes: Redefining the functional architecture of the brain. Trends in Neurosciences, 26(10), 523–530. [DOI] [PubMed] [Google Scholar]
- Paoletti A, Moudjou M, Paintrand M, Salisbury JL, & Bornens M (1996). Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. Journal of Cell Science, 109, 3089–3102. [DOI] [PubMed] [Google Scholar]
- Piscopo DM, El-Danaf RN, Huberman AD, & Niell CM (2013). Diverse Visual Features Encoded in Mouse Lateral Geniculate Nucleus. Journal of Neuroscience, 33(11), 4642–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry VH, & Cowey A (1984). Retinal ganglion cells that project to the superior colliculus and pretectum in the macaque monkey. Neuroscience 12(4), 1125–1137. [DOI] [PubMed] [Google Scholar]
- Sarkisian MR, & Guadiana SM (2015). Influences of primary cilia on cortical morphogenesis and neuronal subtype maturation. Neuroscientist 21(2), 136–151. [DOI] [PubMed] [Google Scholar]
- Seabrook TA, Burbridge TJ, Crair MC, & Huberman AD 2017a. Architecture, Function, and Assembly of the Mouse Visual System. Annual Review of Neuroscience, 40, 499–538. [DOI] [PubMed] [Google Scholar]
- Seabrook TA, Dhande OS, Ishiko N, Wooley VP, Nguyen PL, & Huberman AD (2017b). Strict Independence of Parallel and Poly-synaptic Axon-Target Matching during Visual Reflex Circuit Assembly. Cell Reports, 21(11), 3049–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M, Gloy J, Ganner A, Bullerkotte A, Bashkurov M, Krönig C, Schermer B, Benzing T, Cabello OA, Jenny A, Mlodzik M, Polok B, Driever W, Obara T, & Walz G (2005). Inversin, the gene product mutated in nephronophthisis type II, functions as a molecular switch between Wnt signaling pathways. Nature Genetics, 37(5), 537–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasker JG, Oliet SHR, Bains JS, Brown CH, Stern JE (2012). Glial Regulation of Neuronal Function: From Synapse to Systems Physiology. Journal of Neuroendocrinology, 24(4), 566–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalobos CA, Wu Q, Lee PH, May PJ, Basso MA. 2018. Parvalbumin and GABA microcircuits in the mouse superior colliculus. Front Neural Circuits 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Phan T, Storm DR. 2011. The type 3 adenylyl cyclase is required for novel object learning and extinction of contextual memory: Role of camp signaling in primary cilia. J Neurosci 31:5557–5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y 1999. Segregation and Convergence of Functionally Defined V2 Thin Stripe and Interstripe Compartment Projections to Area V4 of Macaques. Cereb Cortex 9:792–804. [DOI] [PubMed] [Google Scholar]
- Xu N, Tamadon A, Liu Y, Ma T, Leak RK, Chen J, Gao Y, Feng Y. 2017. Fast free-of acrylamide clearing tissue (FACT) - An optimized new protocol for rapid, high-resolution imaging of three-dimensional brain tissue. Sci Rep 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn YH, Han YG. 2018. Primary Cilia in Brain Development and Diseases. Am J Pathol 188:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]