Abstract
The efficacy of targeted therapy in non-small-cell lung cancer (NSCLC) has been impeded by various mechanisms of resistance. Besides the mutations in targeted oncogenes, reversible lineage plasticity has recently considered to play a role in the development of tyrosine kinase inhibitors (TKI) resistance in NSCLC. Lineage plasticity enables cells to transfer from one committed developmental pathway to another, and has been a trigger of tumor adaptation to adverse microenvironment conditions including exposure to various therapies. More importantly, besides somatic mutation, lineage plasticity has also been proposed as another source of intratumoural heterogeneity. Lineage plasticity can drive NSCLC cells to a new cell identity which no longer depends on the drug-targeted pathway. Histological transformation and epithelial–mesenchymal transition are two well-known pathways of lineage plasticity-mediated TKI resistance in NSCLC. In the last decade, increased re-biopsy practice upon disease recurrence has increased the recognition of lineage plasticity induced resistance in NSCLC and has improved our understanding of the underlying biology. Long non-coding RNAs (lncRNAs), the dark matter of the genome, are capable of regulating variant malignant processes of NSCLC like the invisible hands. Recent evidence suggests that lncRNAs are involved in TKI resistance in NSCLC, particularly in lineage plasticity-mediated resistance. In this review, we summarize the mechanisms of lncRNAs in regulating lineage plasticity and TKI resistance in NSCLC. We also discuss how understanding these themes can alter therapeutic strategies, including combination therapy approaches to overcome TKI resistance.
Keywords: Non-small-cell lung cancer, Tyrosine kinase inhibitors, Long non-coding RNAs, Lineage plasticity
Introduction
Lung cancer is the leading cause of cancer-associated mortality worldwide, with non-small-cell lung cancer (NSCLC) as the main histological subtype with a poor 5 year survival [1]. Improved understanding of the molecular classification of lung cancer has revolutionized the treatment of NSCLC. Almost two-thirds of patients with NSCLC are oncogene addicted, approximately half of whom are exquisitely sensitive to targeted therapies [2, 3]. These include activating mutations or fusions in epidermal growth factor receptor (EGFR), serine/threonine-protein kinase b-raf (BRAF), anaplastic lymphoma kinase (ALK), and ROS1 receptor tyrosine kinase [3].
Although agents that target the tyrosine kinase domain of these oncogenes improve clinical outcomes of patients with NSCLC, responses to these drugs are generally temporary and limited by emergence of resistance [4]. Resistance to tyrosine kinase inhibitors (TKI) are generally associated with acquired somatic mutations, including genetic alterations that enable bypassing target inhibition through ineffective binding of the drugs, as well as activation of collateral or alternative survival pathways [3, 5, 6]. In addition to these genetic mechanisms of drug resistance, non-mutational mechanisms termed lineage plasticity are also associated with TKI resistance. Lineage plasticity refers to as the ability of cells transferring from one committed developmental pathway to another. It enables the adaptation and survival of tumors under adverse conditions including hypoxia and targeted therapies. Therefore, it is proposed as a mechanism of tumor cells escape from targeted dependency. Lineage plasticity can be both dependent on and a driver of intratumoral heterogeneity [7, 8]. In contrast to genetic mechanisms of TKI resistance, lineage plasticity-associated resistance primarily relies on phenotype switching, with one single genotype gives rise to different phenotypes upon drug treatment. It enables cancer cells to reversibly convert to new or hybrid lineages that is independent of TKI-targeted pathway [9].
Data from single-cell profiling and lineage tracing technologies suggest that a single cell can give rise to multiple states. Mechanistically, epigenetic modifications and transcriptomic transitions may mediate the emergence of new cell states. Long non-coding RNAs (lncRNAs) the dark matter of the genome, are capable of regulating variant malignant processes of NSCLC through epigenetic modification [10]. Importantly, lncRNAs often offer distinct advantages over proteins for some forms of epigenetic regulation [11]. Recent studies show the transcriptional heterogeneity regulated by lncRNAs might drive the phenotypic switch from one histological category to another, contributing to the lineage plasticity-mediated TKI resistance.
Targeting lineage plasticity provides a new opportunity to prevent the emergence of drug-tolerant cell states and enables TKI to achieve deeper responses. In this review, we performed a literature review covering the publication of the last 15 years on the topic of “long non-coding RNAs regulated lineage plasticity in lung cancer”. The keywords used for searching were “cell plasticity”, “cancer” and “non-coding RNAs”. We summarize current understanding of lineage plasticity as a mechanism of TKI resistance in NSCLC and discuss the implications of lncRNAs in lineage plasticity. Finally, we also discuss potential therapeutic strategies to circumvent lineage plasticity in NSCLC.
Lineage plasticity in response to targeted therapies
Lineage plasticity, the ability of cells to reversibly transfer from a certain developmental pathway to another, has been regarded as a source of intratumoral heterogeneity and of tumor adaptation to adverse tumor microenvironment such as hypoxia and exposure to targeted therapies [7]. Histological transformation of adenocarcinomas to neuroendocrine and epithelial–mesenchymal transition are two well-known phenotypes of lineage plasticity in NSCLC upon TKI treatment [7, 12]. NSCLC cells has been reported to hijack developmental process involved in epigenetic modification and transcriptional reprogramming to become phenotypically plastic, and reversibly transform into one or several drug-tolerant cell identities [13, 14]. Interestingly, the emergence of slow-cycling tumors cells is primarily considered as the first state before histological transformation and further reprogramming into a drug-resistant state [15, 16]. Adenocarcinomas with concomitant inactivation of RB1 and TP53 are more likely to transdifferentiate into neuroendocrine tumors in NSCLC [17–20]. Furthermore, loss of lung lineage-specifying transcription factor gene Nkx2-1, overexpression of lineage-specific transcription factors such as SOX genes, and dysregulation of epigenetic regulators such as histone methyltransferase, changes of key signaling pathways (IL-6-STAT3 or RXR, etc.), as well as interactions with tumor microenvironment (TME) have been associated with intratumoural heterogeneity and lineage plasticity-mediated drug resistance [7, 14] (Fig. 1).
Fig. 1.
Lineage plasticity lead to TKI resistance in NSCLC. Upon targeted therapies, various molecular events can promote lineage plasticity, thereby driving intratumoural heterogeneity and drug resistance. RB1 and TP53 mutation or loss NSCLC cells are more likely to transdifferentiate into small cell tumors. Increased histone-modifying enzymes, such as enhancer of zeste homologue 2 (EZH2) and lineage-associated transcription factors, such as SOX family genes mediates the reprogramming of NSCLC into slow-cycling, drug-tolerant cell states. These slow-cycling, drug-tolerant cells generally present neuroendocrine differentiation and epithelial-to-mesenchymal transition (EMT). Alterations of key signaling pathways and crosstalk with the tumor microenvironment also control lineage plasticity. Collectively, the plasticity-permissive molecular environment under the pressure of targeted therapies trigger the intratumoural clones presenting an alternative histology to that initially diagnosed, which might become the predominant cell type and exhibit drug resistance. Blue font: lncRNA-mediated molecular events that promote lineage plasticity
Histological transformation
Histological transformation also referred to as transdifferentiation, which presents the process that cells convert from one lineage to another [21]. Targeted therapy-induced neuroendocrine transdifferentiation has been particularly described in NSCLC and prostate cancer [12, 22]. The transformation of EGFR-mutant adenocarcinoma to a SCLC histology was first reported in a 45-year-old woman with EGFR-mutant adenocarcinoma who underwent erlotinib treatment for 18 months before relapse with the features of SCLC, and positive immunostaining of neural cell adhesion molecule 1 (NCAM1), chromogranin A and synaptophysin [23]. In rare cases, NSCLC could adopt to other neuroendocrine histology, for instance, large cell neuroendocrine carcinoma and small and large cell carcinoma [19]. Transformation to sarcomatoid carcinoma has also been observed in ALK-rearranged NSCLC in resistant to crizotinib. Importantly, the transformed SCLC tumor samples maintained their original mutation [23], which indicates these tumors were emerging from the original adenocarcinoma, rather than de novo cancers. Genetically, NSCLC cells with RB1 and TP53 loss are more likely to transdifferentiate towards a neuroendocrine identity upon TKI treatment [17, 18]. Similar observations were made in castration-resistant prostate cancer (CRPC), with approximately one-quarter of CRPC acquire androgen receptor (AR) independent resistant through phenotypic switching [22]. These AR-independent CRPC tumors are referred to as neuroendocrine prostate cancer (NEPC) [22].
Epithelial–mesenchymal transition
Epithelial–mesenchymal transition (EMT) is another type of tumor cell plasticity related to TKI resistance [12, 24], which tumor cells lose their epithelial features and acquire cellular alterations favoring more invasive, mesenchymal properties. Mesenchymal characteristics were observed in vitro and in vivo EGFR-mutant lung cancer models that acquired resistance to first-generation EGFR-TKI with no genetic alterations [25, 26]. Moreover, tumor biopsy samples from EGFR-TKI resistance patients presented increased vimentin expression and downregulated E-cadherin expression compared with tumor tissues taken before TKI treatment. Importantly, tumor cells undergo epithelial–mesenchymal plasticity retain their original mutation spectrum, indicating EMT as a mechanism of TKI resistance [12, 27]. The precise mechanism of epithelial–mesenchymal plasticity remains to be elucidated. Increased levels of pleiotropic signaling factors, such as transforming growth factor-β (TGF-β), epidermal growth factor (EGF), hepatocyte growth factor (HGF), NOTCH, fibroblast growth factor (FGF) and WNT ligands can initiate a signaling cascade resulting the expression of EMT transcription factors [13]. Furthermore, reduced expression of proapoptotic proteins such as PUMA [28] and increased drug efflux [29] have also been reported to associate with EMT process upon TKI treatment.
Deregulated lncRNAs involved in lineage plasticity upon targeted therapies
Neuroendocrine transdifferentiation-associated lncRNAs
Clonal analysis showed that complete loss of both RB1 and TP53 may predict neuroendocrine transdifferentiation in NSCLC upon EGFR-TKI treatment [18]. However, cell lines and mouse model studies of lung and prostate cancer suggested that inactivation of RB1 or TP53 alone is not sufficient for their histological transformation or effect their sensitivity to EGFR-TKI [30–32]. Additionally, epigenetic states, specially, lncRNAs are involved in cell-fate determination [33]. Crea et al. identified the first NEPC-associated lncRNA-MIAT, which contributed to the neuroendocrine transdifferentiation of CRPC [34]. Moreover, Ramnarine et al. identified lncRNAs FENDRR, H19, LINC00514, LINC00617, and SSTR5-AS1 to be implicated in the development of NEPC [35]. Altogether, these data suggest lncRNAs can be strong candidate for clinical biomarkers and therapeutic targets in preventing neuroendocrine transdifferentiation in CRPC. Nevertheless, lncRNAs associated with neuroendocrine transformation in NSCLC in response to TKI treatment warrant further investigation. (Table 1).
Table 1.
Examples of lncRNAs implicated in lineage plasticity in NSCLC
| lnRNA | Association with Cancer | Function | Cellular phenotypes | Refs |
|---|---|---|---|---|
| Slow cycling | ||||
| lincRNA-p21, LED | p53-induced lncRNAs | Protein interactions (interaction with hnRNPK to regulate CDKN1A; interaction with CKDN1A enhancer | Cell-cycle arrest | [61] |
| LINC-PINT, TUG1, PR-lncRNA-1, PR-lncRNA-10 | p53-induced lncRNAs | Connects p53 pathway with epigenetic silencing by PRC2 | Cell-cycle arrest | [61] |
| p15-AS1, MIR31HG, MIR100HG | Downregulated in lung cancer | Silencing pf p15 through heterochromatin formation; Protein interactions (decoy for polycomb group proteins to repress INK4A transcription) | Growth arrest and apoptosis | [65, 66] |
| MALAT1, CASC2, TINCR | Downregulated in lung cancer | Chromatin condensation by recruitment of PRC2 | Growth arrest | [68] |
| EMT | ||||
| LINC00673, CAR10, XIST, LINC81507, LINC TTN-AS1, LINC00858, H19, SOX20T, LINC00483, PRNCR1, SNHG6, ATB, PNUTS | Overexpressed in lung cancer | miRNA decoy (for miR200, miR181a) | EMT, invasion, proliferation and metastasis, drug resistance | [35] |
| ELIT-1, TBILA, LINP, HOTAIR,NKILA and LINC001186 | TGFβ-regulated lncRNAs | Interacts with various proteins; interferes with different signaling pathway | EMT, metastasis | [45–48, 56, 57] |
| MEG3 | Overexpressed in NSCLC cells | Chromatin condensation by recruitment of EZH2 to silence CDH1 and miR-200 family | EMT, metastasis | [50] |
| MALAT1 | Overexpressed in lung cancer | Chromatin condensation by recruitment of EZH2 to silence E-cadherin | EMT, metastasis | [51] |
| FEZF1-AS1 | Overexpressed in lung cancer | Chromatin condensation by recruitment of LSD1 and EZH2 to repress E-cadherin | EMT, metastasis | [52] |
| HOXA11-AS1 | Overexpressed in lung cancer | Gene methylation by recruitment of EZH2 and DNMT1 to inactivate miR200b | EMT, metastasis | [53] |
| NORAD | Overexpressed in lung cancer | Molecular scaffold for proteins (importin β1 and Smads) | EMT, metastasis | [49] |
| Epigenetic reprogramming | ||||
| HOXA11-AS | Overexpressed in NSCLC | Chromatin condensation by recruitment of PRC2 to silence miR200 | EMT, tumor progression | [53] |
| FBXL19-AS1 | Overexpressed in LUAD | miRNA Decoy (for miR-203a-3p), enhances E2F and ZEB1 | Proliferation, migration and metastasis | [76]. |
| Reprogramming transcription factors | ||||
| SOX2-OT | Overexpressed in lung cancer | Regulation the expression of SOX2 | Neural differentiation | [81, 82] |
| Key signaling pathways | ||||
| SNHG1 | Overexpressed in cisplatin-resistant NSCLC | miRNA Decoy (for miR140-5p), increases WNT/β-catenin pathway | Neural differentiation, drug resistance | [90] |
| LINC00673 | Overexpressed in LUAD | Molecular scaffold for proteins (DDX3 and CK1ε), promotes WNT/β-catenin signaling pathway | Cell migration, invasion and metastasis | [91] |
| NEAT1, FOXO2-AS1 | Overexpressed in NSCLC | Upregulation of WNT/β-catenin signaling pathway | Acquisition of cancer stem cell-like properties, proliferation | [89, 92, 93] |
| MALAT1,H19 | Overexpressed in NSCLC | miRNA decoy (for miR-124, miR-17, miR-29b-3p) to activate STAT3 | EMT, proliferation, migration and invasion | [95–97] |
| TNK2-AS1 | Overexpressed in NSCLC | Interaction with STAT3 to enhances its protein stability | Promotes angiogenesis | [98] |
| Tumor microenvironment | ||||
| GNAS-AS1 | Overexpressed in NSCLC | Promote macrophage M2 polarization | Cell migration and invasion | [112] |
| XIST | Overexpressed in NSCLC | Promote macrophage M2 polarization | Increased the progression of lung cancer and TAM-mediated drug resistance | [113] |
NSCLC non-small cell lung cancer, LUAD lung adenocarcinoma, TGFβ transforming growth factor-β, EZH2 zeste homologue 2, LSD1 lysine-specific histone demethylase 1, DNMT1 NDA methyltransferase 1, YAP1 yea-associated protein 1, SOX2 SRY-box transcription factor 2, EMT epithelial–mesenchymal transition, TAM tumor-associated macrophage
EMT-associated lncRNAs
EMT is a dynamic process which tumor cells obtain phenotypic changes through epigenetic modifications [36]. Emerging evidence highlights the involvement of lncRNAs in EMT process in NSCLC [37].
Recently, a group of lncRNAs have been shown to promote drug resistance by acting as molecular decoys to sequester miRNAs associated with EMT. For example, LINC00673, CAR10, XIST, LINC81507, TTN-AS1, LINC00858, H19, SOX20T, LINC00483, PRNCR1, SNHG6, ATB and alternative splicing-generated lncRNA-PNUTS [38] are shown to sequester miRNAs from binding EMT associated genes, such as zinc finger E-box binding homeobox 1 and 2 (ZEB1 and ZEB2), thereby initiate EMT in NSCLC [39–46]. Increased levels of transforming growth factor-β (TGFβ) are reported to facilitate EMT via interleukin-6 (IL-6) in EGFR TKI-resistant NSCLC cells [47]. Several profiling studies have identified the roles of TGFβ-regulated lncRNAs such as ELIT-1, TBILA, LINP and HOTAIR in promoting EMT in NSCLC cell lines [48–51]. Mechanistically, TGFβ-activated lncRNA ELIT-1 can bound to Smad3 and improved Smad-responsive promoter activities by recruiting Smad3 to the promoters of its target genes, which include Snail, and ELIT itself. Thus, ELIT-1 forms a positive feedback loop to promote TGFβ/Smad3 signaling pathway and, therefore, promote EMT progression [48]. NORAD, a cytoplasmic long non-coding RNA is also reported to promote TGFβ-induced EMT in NSCLC cell lines [52]. It is shown to enhance the physical interaction of importin β1 with Smads, thereby promoting the nuclear accumulation of Smad complexes induced by TGFβ. Furthermore, many lncRNAs associate with epigenetic regulators to regulate EMT. For instance, lncRNA MEG3 is able to recruit EZH2 and epigenetically silencing CDH1 and miR-200 family in NSCLC cell lines [53]. Similarly, MALAT1 can also recruit EZH2 to silence E-cadherin and, therefore, induce EMT in NSCLC [54]. LncRNA-FEZF1-AS1 was shown to epigenetically repress E-cadherin by binding with lysine-specific histone demethylase 1 (LSD1) and EZH2 in NSCLC cells [55]. HOXA11-AS1 was reported to recruit EZH2 and DNA methyltransferase 1 (DNMT1) to the promoter of miR200b and inactivate miR200b, thereby promoting EMT in NSCLC [56]. In contrast, a number of epigenetic upregulated lncRNAs can inhibit EMT through histone modification in NSCLC [57, 58]. For example, lncRNA-BANCR inhibits EMT through histone acetylation [58]. Most TGFβ-regulated lncRNA were reported to promote EMT, however, TGFβ-regulated lnRNAs NKILA and LINC001186 are reported to inhibit EMT by suppressing the expression of Snail [59, 60]. Targeting EMT-associated lncRNAs may restore the sensitivity to TKI, further studies are needed. (Table 1).
Mechanisms of lncRNA-mediated lineage plasticity
The molecular mechanisms underlying lineage plasticity-mediated TKI resistance remain to be fully established. However, tumor cells go through a slow-cycling drug-tolerant state is generally considered as the first step for lineage plasticity-mediated resistance. Moreover, epigenetic and transcription factor changes and alterations of key signaling pathways, as well as crosstalk with tumor microenvironment (TME) can favor the development of phenotypic switching and TKI resistance (Fig. 2).
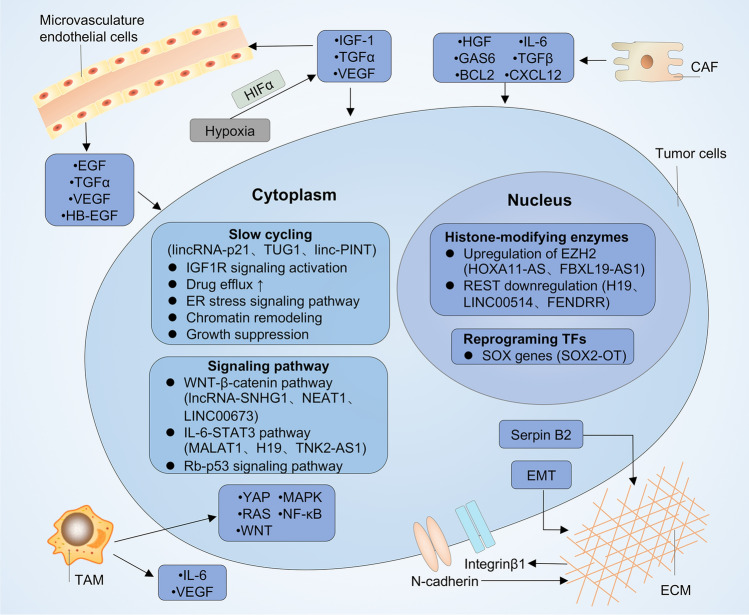
Fig. 2.
Mechanisms of lncRNA-mediated lineage transition. Transcription factors (such as SOX family), histone-modifying enzymes (such as enhancer of zeste homologue 2 (EZH2) and RE1-slienicng transcription factor (REST)) regulate the reprogramming tumor cells into slow-cycling, drug-tolerant states. LncRNAs such as lincRNA-p21, TUG1, linc-PINT, HOXA11-AS, SOX2-OT, FBXL19-AS1, LINC00514, FENDRR are implicated in the reprogramming process. Alterations in several key signaling pathways such as WNT-β-catenin pathway, IL-6/STAT3 pathway, NF-κB pathway and YAP pathway promote phenotypic switching upon TKI treatment. lncRNAs such as MALAT1, GHET1, SNHG1, NEAT1, H19 TNK2-AS1 are involved in these key signaling pathways. Crosstalk with the tumor microenvironment, through secretion of various cytokines from cancer-associated fibroblasts such as hepatocyte growth factor(HGF), growth arrest-specific protein 6 (GAS6), CXC-chemokine ligand 12 (CXCL12), interleukin-6 (IL-6) and transforming growth factorβ (TGFβ); cytokines from endothelial cells such as epidermal growth factor (EGF), transforming growth factor-α (TGFα), vascular endothelial growth factor (VEGF) and heparin-binding EGF-like growth factor (HB-EGF); Interluekin-6 (IL-6), VEGF from tumor-associated macrophage (TAM) controls tumor plasticity. The enhanced cell–cell adhesion via increased expression of integrin β1 and N-cadherin in tumor cells and increased extracellular matrix (ECM) stiffness via Serpin B2 can also promote tyrosine kinase inhibitors (TKI) resistance. LncRNAs involved in the M2 polarization of macrophage such as XIST and GNAS-AS1 played a part in TKI resistance. Lastly, low level of oxygen can active hypoxia-inducible factor 1α (HIF1α) in tumor cells, causing autocrine signaling by secretion of TGFα, VEGF and insulin-like growth factor (IGF-1), which promotes resistance to TKI therapy. TFs transcription factors, CAF cancer-associated fibroblasts
Emergence of slow-cycling cells
A drug-tolerant, slow-cycling state was first identified in bacteria [61]. In that scenario, a group of drug-tolerant slow-cycling bacteria survive in response to antibiotic treatment, and can further convert to a proliferative state and re-established drug sensitive phenotype upon drug withdrawal [61]. Similarly, reversible drug-tolerant slow-cycling persisters were reported in NSCLC. Notably, a stepwise transition may occur in NSCLC upon TKI treatment, tumor cells first reversibly enter a slow-cycling state, then regain proliferation and ultimately become drug-resistant through further epigenetic changes or via genetic modifications (such as EGFR-T790M) [6, 15]. More recently, Sanchez-Danes et al. also discovered that in basal cell carcinoma, drug-tolerant slow-cycling residuals cells can lead to relapse upon drug withdrawal [62]. Forcing these slow-cycling cells to proliferate enhanced their sensitivity to vismodegib treatment, which leaded to their elimination [62].
LncRNAs involved in regulating cell-cycling conditions in cancer are primarily defined as p53-related lncRNAs [63]. Genome-wide profiling of p53-regulated enhancer RNAs identified p53-induced lincRNA-p21, LINC-PINT, TUG1, PR-lncRNA-1, PR-lncRNA-10 and LED to support cell-cycle arrest [63]. Mechanistically, lincRNA-p21 bound to heterogeneous nuclear ribonucleoprotein K (hnRNPK) to regulate cyclin-dependent kinase inhibitor 1A (CDKN1A) is cis and arrest the cell cycle [64]. Another p53-induced lncRNA DINO was shown to interact with p53 protein and enhanced its stabilization, creating a p53 auto-amplification loop, thereby promoting damage signaling pathway and cell cycle arrest in the absence of DNA damage [65]. Moreover, alterations in cell cycle proteins, including the silencing of cyclin dependent kinase (CDK) inhibitor p16 and p15 are associated with resistance to EGFR TKI in NSCLC patients [66]. Several lncRNAs such as p15-AS-1 and MIR31HG can suppress p15 or p16 through heterochromatin formation [67, 68]. Moreover, MIR100HG encoded lncRNA interacts with HuR/ELAVL1 as well as several HuR-target genes (oncogenes) to suppress cell proliferation [69]. In addition, lncRNA TINCR, CASC2, MALAT1 were involved in FBXW7-mediated cell cycle arrest in various cancers [70]. Nevertheless, it remains to be elucidated whether these cell cycle-associated lncRNAs are implicated in the drug-tolerant refractory cells under the pressure of TKI. It is important to note that most of these studies are conducted in vitro, and should, in the future, be further validated in vivo.
Epigenetic modification
The histone methyltransferase EZH2, which is the catalytic subunit of Polycomb repressive complex 2 (PRC2), is reported to promote neuroendocrine transdifferentiation in a mouse model of lung cancer [71]. Similarly, EZH2 is also a well-established feature of NEPC [31, 72]. Pharmacological or genetic inhibition of EZH2 activity in NEPC cell lines can reverse its lineage conversion and regain drug sensitivity [30, 31, 73] RE1-slienicng transcription factor (REST), another epigenetic regulator, is found in lung and prostate cancer during neuroendocrine conversion [74, 75]. REST suppresses gene expression generally through recruiting the co-repressors such as EZH2 [75]. Elevated levels of REST are reported to promote neuroendocrine differentiation in EGFR TKI resistance NSCLC cell lines via Notch signaling pathway [76].
Many lncRNAs have been shown to scaffold and recruit epigenetic regulator to specific genome loci. For example, lncRNAs HOXA11-AS can recruit PRC2 complex to silence miR200b [56]. On the other hand, a few lncRNAs are regulated by E2F transcription factor 1 (E2F1), which is a transcription factor that induce EZH2 expression [77]. Transcription factor-binding site (TFBS) analysis identified lncRNAs FENDRR, H19, LINC00514, LINC00617 and SSTR5-AS1 include TFBS motifs for REST and E2F, and the expression of these lncRNAs were implicated in the development of NEPC [35]. Furthermore, FBXL19-AS1/miR-203a-3p axis was found to enhance E2F1 and ZEB1 in lung adenocarcinomas (LUAD) cells [78]. Therefore, lncRNAs are important players in scaffolding EZH2-dependent gene silencing and subsequent regulate EZH2-mediated phenotypic switching.
SOX family members
Several lineage-specific transcription factors are involved in lineage plasticity and drug escape both in NSCLC and prostate cancer. The SOX family is important in regulating cell fate decisions and is implicated in phenotypic conversion in various cancer models [79]. For instance, the expression of SOX2 was increased in TP53 and RB1-deficient GEMMs and xenograft models of LUAD and prostate cancers [30, 31]. Moreover, the neural lineage-specific factor BRN2, which is specifically expressed in SCLC and NEPC tumors, mediates SOX2 expression and is key for neuroendocrine transformation [80, 81]. Furthermore, insulinoma-associated-1 (INSM1), which encodes a zinc-finger transcription factor, has recently emerged as a specific neuroendocrine transcription factor and a sensitive biomarker for neuroendocrine tumors [82].
LncRNA SOX2 overlapping transcript (SOX2-OT) are functionally assumed to be associated with neuronal like differentiation and carcinogenesis [83, 84]. Concordant expression of SOX2 and SOX2-OT is found in lung and breast cancer [83]. Notably, SOX2-OT can generate six transcript variants in different cancer models [85, 86], and have been proposed to play a role in regulating the expression of SOX2 [84, 85]. Collectively, SOX reprogramming factors, together with other lineage-associated transcription factors, are key for cellular plasticity in TKI resistant NSCLC cells. LncRNA-SOX2-OT, as SOX2 overlapping transcript also makes contribution for promoting the transition toward neural crest state in NSCLC.
Key signaling pathways
Several signaling pathways play a part in phenotypic switching upon TKI treatment. The activation of WNT-β-catenin pathway is reported to promote neuroendocrine differentiation in various cancer models [87]. Meder et al. discovered NOTCH-Achaete-scure complex homolog 1(ASCL)-WNT signaling pathway could inactivate RB by phosphorylation and, therefore, promote neuroendocrine differentiation in NSCLC [88, 89]. Moreover, achaete-scute homolog 1 (ASH1) also acts as a positive regulator of WNT/β-catenin pathway, transforming NSCLC into a SCLC phenotype with neuroendocrine features both in vitro and in vivo models [90]. A number of lncRNAs have been found to active WNT/β-catenin to induce EMT and therapy resistance in NSCLC [91–93]. For instance, lncRNA-SNHG1 can sequester miR140-5p from binding WNT to active WNT/β-catenin signaling in NSCLC [92]. LINC00673 functions as a modular scaffold to strengthen the interaction between DDX3 and CK1ε, induces phosphorylation of Dvl and, therefore, promote the nuclear accumulation of β-catenin and the activation of WNT/β-catenin signaling pathway in LUAD [93]. Furthermore, the expressions of lncRNA-NEAT1, FOXO2-AS1 were positively associated with WNT/β-catenin signaling in NSCLC tissues. Knockdown of NEAT1 or FOXO2-AS1 inhibited WNT/β-catenin signaling pathway activity [91, 94, 95].
IL-6-STAT3 axis is also activated upon EGFR TKI treatment and can promote neuroendocrine differentiation in NSCLC [96]. LncRNAs such as MALAT1 and H19 are reported to, respectively, sponge miR-124, miR-17 and miR-29b-3p, subsequently activated STAT3 and promote therapy resistance [97–99]. In addition, Wang et al. showed lncRNA TNK2-AS1 was significantly upregulated in NSCLC and associated with poor survival. Mechanistically, TNK2-AS1 could interact with STAT3 to enhance its protein stability, on the other hand, STAT3 also triggers the transcription of TNK2-AS1. Thus, the positive feedback loop between TNK2-AS1 and STAT3 augmented STAT3 signaling pathway in NSCLC [100].
The tumor microenvironment
The tumor microenvironment (TME) is a multicellular system with dynamic tumor-stromal component interactions [101, 102]. Altogether, the various stromal components such as fibroblasts, endothelial cells and infiltrating immune cells influence the response to TKI therapy.
It is well established that cancer-associated fibroblasts (CAFs) can induce EMT and TKI resistance in NSCLC cells in vitro [103, 104]. The secretion of soluble factors such as hepatocyte growth factor (HGF) promoted MET or ERK activation and subsequent EGFR TKI resistance in NSCLC cells [105, 106]. CAFs can also derive the AXL ligand growth arrest-specific protein 6 (GAS6) and enhance the expression of anti-apoptotic gene BCL2, leading to TKI resistance [26, 107].
The extracellular matrix (ECM) also interacts with NSCLC cells to promote drug tolerance. A study in 3D lung cancer cell models revealed that ECM-induced ERK and PI3K/AKT signaling lead to an EGFR TKI tolerant dormant state [108]. Low levels of SerpinB2 (a serine protease inhibitor that increase ECM stiffness), is negatively associated with gefitinib resistance in vitro. And treatment with a SerpinB2-inducing agent reversed the drug-tolerant state [108]. Moreover, Elevated levels of N-cadherin and integrinβ could promote tumor cells adhesion to ECM, thereby, promoting EGFR TKI resistance [109].
Tumor-associated macrophages (TAMs) are important infiltrating immune cells with crucial role in the development of TKI resistance [110]. Patients with increased TAM infiltration within the TME had poor outcomes [111]. Computational modelling of RNA expression in a mouse model of NSCLC revealed TAMs-secreted factors can activate various signaling pathways related to EGFR TKI resistance, including the MAPK, YAP, NF-κB, PI3K, WNT and RAS pathways [112]. Furthermore, macrophage could promote EMT through the IL-6-mediated COX2/PGE2/β-catenin signaling pathway [113]. Notably, a few lncRNAs such as GNAS-AS1 and XIST are reported to promote macrophage M2 polarization in NSCLC and, therefore, play a part in TAM-induced drug tolerant [114, 115].
Finally, hypoxia and the density and distribution of vasculature are closely associated with EGFR TKI resistance. Hypoxia-inducible factor 1α (HIF1α) promotes TKI resistance in a TGFα-dependent manner and increases cancer stem phenotype via IGF1R activation [116, 117]. In addition, the secretion of vascular endothelial growth factor (VEGF) by endothelial cells under hypoxia promotes angiogenesis and drug resistance [118]. Additionally, the expression of immune checkpoint programmed cell death 1 ligand 1 (PDL1) are upregulated in some EGFR-mutated or ALK-rearranged NSCLC patients. A preliminary study has found the combination of PDL1 nanobody and gefitinib displayed effectiveness of reversing the gefitinib resistance in NSCLC [119]. However, the extent of TME contributes to TKI resistance and the involvement of lncRNAs in TME-mediated resistance warrants further investigation, particularly as novel therapies that target immune and stromal cells continue to emerge. The emerging challenge is to clarify whether there is potential for therapeutic synergy between immunotherapy and targeted therapy in preventing lineage plasticity and TKI resistance.
Targeting strategies for lineage plasticity
Targeting epigenetic regulators
Since certain epigenetic alterations are correlated with slow-cycling drug-tolerant cells, targeting the potential epigenetic regulators serves as an important way to ameliorate lineage plasticity. Notably, preventing phenotypic switching by targeting HDAC or histone demethylases KDM5A/B and KDM6A/B have promising results in early-phase studies [120]. In NSCLC, trimethylation of lysine 9 on histone (H3K9me3) represses long-interspersed repeat elements 1 (LINE-1), thus inhibiting the expression of interferon and antiviral-activated genes to promote EGFR TKI-tolerant cell survival [121, 122]. HDAC inhibitor trichostatin A or entinostat counterbalanced the drug-tolerant cells via suppressing LINE [121]. Early-phase studies of detecting HDAC inhibitors in combination with EGFR TKI are ongoing. Inhibiting demethylases KDM6 with GSK-J4 suppressed residual persister cells in glioblastoma [123, 124]. (Table 2).
Table 2.
Selected clinical NSCLC trials of targets and compound targeting lineage plasticity
| Drug regimen | Phase | Identification | Results | Chilicaltrials.gov identifier | Refs |
|---|---|---|---|---|---|
| HDAC inhibitor | |||||
| Vorinostat + gefitinib | I/II | Relapsed/refractory advanced NSCLC | No improvement in PFS | NCT01027676 | [120] |
| Vorinostat + erlotinib | I/II | Relapsed EGFR-mutant NSCLC | No improvement in erlotinib-resistant population | NCT00503971 | [120] |
| Belinostat + Erlotinib | I | Molecularly unselected | Results pending | NCT01027676 | [120] |
| SNDX-275 + Erlotinib | II | Progression on erlotinib | Results pending | NCT00750698 | [120] |
| KDM5 and KDM6 | |||||
| YUKA1, CPI-455 (KDM5A specific), KDOAM-25 (KDM5A-D specific) + gefitinib | Preclinical | EGFR-mutant NSCLC | YUKA1 in combination with gefitinib prevents drug tolerance in EFGR-mutant NSCLC | [123, 124] | |
| CDK7/12 | |||||
| SY-1365 | I | Advanced solid tumors | Ongoing | NCT03134638 | |
| AXL inhibitor | |||||
| BGB324 + erlotinib | II | Molecularly unselected NSCLC | First efficacy end point met | NCT02424617 | [141] |
| SGI-7079 + erlotinib | Preclinical | Molecularly unselected NSCLC | Enhanced sensitivity of mesenchymal-like NSCLC cells to erlotinib | [25] | |
| TP-0903 | Ia/Ib | Advanced solid tumors | Ongoing | NCT02729298 | |
| E-cadherin | |||||
| JMF3086 | Preclinical | Molecularly unselected NSCLC | Restore sensitivity to EGFR-TKI | [130] | |
| EZH2 inhibitor | |||||
| JQEZ5 | Preclinical | EGFR or BRG1-mutant NSCLC | Decreased tumor burden in GEMM and human NSCLC models | [127, 128] | |
| GSK343 + gefitinib | Preclinical | NSCLC | Inhibited cell viability | [129] | |
NSCLC non-small-cell lung cancer, EGFR epidermal growth factor receptor, HDAC histone deacetylase, PFS progression-free survival, KDM histone demethylase, RXR retinoid X receptor, EZH2 zeste homologue 2, IL-6 interleukin-6, AXL receptor tyrosine kinase, TKI tyrosine kinase inhibitors
EZH2 is another druggable epigenetic regulator as mentioned previously [72]. Several phase I and phase II clinical trials are ongoing to inhibit EZH2 in lymphomas and multiple solid tumors [125, 126]. In a mouse model of human LUAD, using an open-source EZH2 inhibitor JQEZ5, Zhang et al. have established efficient antitumor effects [127, 128]. Another study has found that EZH2 inhibitor GSK343 synergy with gefitinib in NSCLC cells [129]. However, EZH2 inhibitor GSK2816126 failed in another early phase II trial in patients with similar indications [14]. Assessment of other epigenetic regulators, such as REST, are also ongoing. In addition, the dual HDAC and 3-hydroxy-3-methylgutaryl coenzyme A reductase inhibitor, JMF3086, which regulate the post-translational regulation of E-cadherin, can restore the sensitivity to first and third-generation EGFR-TKI in NSCLC [130]. (Fig. 3).
Fig. 3.
Principal strategies to target lineage plasticity in NSCLC. Three general approaches that target lineage plasticity are listed here: preventing lineage plasticity, targeting the emerging new cell identity and reversing the lineage plasticity. a Preventing lineage plasticity may prolong the clinical response to TKI treatment. Crucial signals and molecules that regulate the survival of slow-cycling cell, for instance, chromatin landscape remodeling modulators and cell cycle-related lncRNAs can be targeted to block tumor cellular plasticity. b The emerging drug-tolerant cell identity such as SCLC and epithelial–mesenchymal transition (EMT) feature can be eliminated through targeting neuroendocrine-related lncRNAs, AXL, TGFβ and E-cadherin. c Lineage plasticity can be reverted to resensitize NSCLC to TKI. Epigenetic regulators, such as enhancer of zeste homologue 2 (EZH2) and RE1-silencing transcription factors (REST), can be targeted for reversing lineage plasticity. NSCLC non-small-cell lung cancer, SCLC small-cell lung cancer, TKI tyrosine kinase inhibitor, KDM histone demethylase, HDAC histone deacetylase, TGFβ transforming growth factor-β
Numerous mechanistic studies support the hypothesis that targeting epigenetic regulators can synergize with TKI agents and reverse lineage plasticity in preclinical models. Nevertheless, the efficacy of epi-drugs tested in clinical trials to date has been disappointing. Overall, the epigenetic modulators exert broadening effects on cell biology and systemic physiology. Inhibitors targeting epigenetic regulators might lead to the dysregulation of cell biology [131, 132]. Therefore, novel agents are needed to target lineage plasticity more specifically. LncRNAs are excellent candidates in this respect. Several features of lncRNAs render their potential therapeutic targets in lineage plasticity-mediated TKI resistance. First, the expression of lncRNAs show strong conservation of tissue specificity [133]. Interestingly, many lncRNAs are patient and tumor specific [134]. The exclusive expression pattern of lncRNAs in specific types of tissues or cells provides an opportunity for specific regulation by lncRNA-targeting therapeutics [133, 135]. Second, chromatin modification represents an important mechanism for lncRNA, thus targeting the interaction of lncRNAs with epigenetic factors such as EZH2 can be envisioned. Third, many nuclear lncRNAs regulate neighboring gene expression in cis, thus locus-specific regulation can be achieved through lncRNA manipulation. However, the development of lncRNA therapeutics is the still in its infancy. Traditional RNAi has proven generally ineffective for lncRNA, due to their unique localization and expression [136]. Currently, antisense oligonucleotides (ASOs) and CRISPR-Cas9 are considered as promising approaches to target lncRNAs [137, 138]. Furthermore, we anticipate that future pooled CRISPR screening will be implemented to identify lineage plasticity-related lncRNAs. Nevertheless, we are beginning to understand the roles of lncRNAs in lineage plasticity. Targeting lineage plasticity associated lncRNAs in combination with TKI treatment has not been reported yet. Further translation research and clinical trials are needed.
New cell fate management
In addition to targeting epigenetic regulators involving in lineage plasticity, it is also appealing to target the emerging drug-tolerant cell identity. There are ongoing efforts to evaluate whether NSCLC that have undergone neuroendocrine differentiation share similar treatment schedules with de novo SCLC. Notably, transformed SCLC presented sensitivity to palatium-etoposide, which makes them more similar to de novo SCLC [139]. However, it is important to note that a contrasting result came from another retrospective study, in which transformed SCLC patients still displayed higher responsive to taxanes than de novo SCLC but failed response to checkpoint inhibitor therapy [19]. Therefore, treatment regimens on transformed SCLC need to be cautiously evaluated and await further functional investigation. Interestingly, our previous study demonstrated certain non-coding RNA could facilitate the therapeutic effects of EGFR-TKI in NSCLC [140]. Our unpublished data also showed that a few lncRNAs are crucial for the transformation from NSCLC to SCLC in response to TKI treatment and could be targeted to prevent lineage plasticity. Therefore, lncRNAs involving in the histological transformation will be worthwhile areas for further investigation.
EMT represents an important process determining new cell fate of cancer. The correlation between the expression of EMT signature and receptor tyrosine kinase AXL in NSCLC indicates that AXL may represent a novel target [25]. Notably, combination of AXL inhibitor SGI-7079 with EGFR TKI erlotinib increased the sensitivity of mesenchymal-like tumor cells to erlotinib in a mouse xenograft NSCLC model [25]. Preliminary findings also suggest that BGB324, the first AXL-specific-molecule inhibitor approved for clinic, can be safely administered, resulting in disease stabilization in a group of NSCLC patients [141].
Conclusion
Along with genetic alterations, lineage plasticity has recently considered to play a key part in the development of TKI resistance in NSCLC. Despite the functions of lncRNAs in cancer have been extensively studied in the past few years, we are beginning to understand the implications of lncRNA in TKI resistance in NSCLC, particularly in regulating lineage plasticity-mediated drug escape. A better understanding of the deregulated lncRNA involved in lineage plasticity will shed light on the complexity of the molecular alterations underlying TKI resistance. Recently, CRISPR-based screening has successfully identified many cancer-promoting lncRNAs. The simplicity, low cost and flexibility of pooled CRISPR screening brings transcriptome-wide screens within reach of the average molecular oncology laboratory. We anticipate that future pooled CRISPR screening will be implemented to identify lineage plasticity-related lncRNAs. The current strategies to target lineage plasticity mainly include targeting the epigenetic and transcriptional alterations. However, there remains many challenges ahead to better understand the process. First, the molecular determinants and biomarker for tumor cell phenotypic plasticity upon TKI treatment needs to be better understood. Second, the heterogeneity should be taken into account when targeting the residual tumor cells. Finally, further investigations such as CRISPR screening are required to discover lineage plasticity-related lncRNAs, and use lncRNAs as novel diagnosis and treatment modalities. Overcoming the obstacles will enable us to implement personalized medicine in the treatment of TKI resistance in NSCLC patients.
Abbreviations
- NSCLC
Non-small-cell lung cancer
- lncRNA
Long non-coding RNA
- TKI
Tyrosine kinase inhibitors
- EGFR
Epidermal growth factor receptor
- BRAF
Serine/threonine-protein kinase b-raf
- ALK
Anaplastic lymphoma kinase
- TME
Tumor microenvironment
- EZH2
Enhancer of zeste homolog 2
- ATL-1
Atractylodes macrocephula Koidz atractylenolide 1
- STAT3
Signal transducer and activator of transcription
- PI3K
Phosphatidylinositol 3-kinase
- SCLC
Small-cell lung cancer
- EMT
Epithelial-to-mesenchymal transition
- NCAM1
Neural cell adhesion molecule 1
- RB1
RB transcriptional corepressor 1
- TP53
Tumor protein p53
- CRPC
Castration-resistant prostate cancer
- AR
Androgen receptor
- NEPC
Neuroendocrine prostate cancer
- ZEB
Zinc finger E-box-binding homeobox
- TGFβ
Transforming growth factor-β
- IL-6
Interleukin 6
- LSD1
Lysine-specific demethylase 1
- DNMT1
DNA methyltransferase 1
- CDKN1A
Cyclin-dependent kinase inhibitor 1A
- YAP1
Yes-associated protein 1
- PRC2
Polycomb repressive complex 2
- REST
RE1-slienicng transcription factor
- TFBS
Transcription factor biding site
- LUAD
Lung adenocarcinoma
- E2F1
E2F transcription factor 1
- INSM1
Insulinoma-associated-1
- ASCL
Achaete-scure complex homolog 1
- ASH1
Achaete-scute homolog 1
- CAF
Cancer-associated fibroblast
- HGF
Hepatocyte growth factor
- MET
Mesenchymal–epithelial transition factor receptor
- GAS6
Growth arrest specific 6
- EGF
Epidermal growth factor
- HB-EGF
EGF-like growth factor
- ECM
Extracellular matrix
- CXCL12
CXC-chemokine ligand 12
- TAM
Tumor-associated macrophage
- MAPK
Mitogen activated kinase-like protein
- NF-κB
Nuclear factor kappa B
- RAS
Resistance to audiogenic seizures (RAS)
- COX2
Cytochrome coxidase subunit II
- PGE2
Nr5a1 enhancer region in intron 6
- HIF1α
Hypoxia-inducible factor 1α
- VEGF
Vascular endothelial growth factor
- PDL1
Programmed cell death 1 ligand 1
- HDAC
Type-2 histone deacetylase 2
- H3K9me3
Trimethylation of lysine 9 on histone
Author contributions
TYL wrote the manuscript. CC H prepared all the figures and tables. PQF, HYZ and SW W were involved in the revising of the manuscript, ZFM, QLZ and WJX contributed to the manuscript preparation. RY, JW and LX participated in the design of the manuscript and the revision. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81802277, 81872378, 81802907) and China Postdoctoral Science Foundation (2018M642198), Project of Jiangsu Provincial Medical Talent (ZDRCA2016033).
Availability of data and materials
All data generated during this study are included in this published article.
Compliance with ethical standards
conflict of interest
The authors declare no competing interests.
Consent for publication
All authors reviewed and approved the manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Tongyan Liu, Chencheng Han, Panqi Fang and Hongyu Zhu contributed equally to the paper.
References
- 1.Hanahan D. Rethinking the war on cancer. Lancet. 2014;383(9916):558–563. doi: 10.1016/S0140-6736(13)62226-6. [DOI] [PubMed] [Google Scholar]
- 2.Altorki NK, Markowitz GJ, Gao D, Port JL, Saxena A, Stiles B, McGraw T, Mittal V. The lung microenvironment: an important regulator of tumour growth and metastasis. Nat Rev Cancer. 2019;19(1):9–31. doi: 10.1038/s41568-018-0081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rotow J, Bivona TG. Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer. 2017;17(11):637–658. doi: 10.1038/nrc.2017.84. [DOI] [PubMed] [Google Scholar]
- 4.Mayekar MK, Bivona TG. Current landscape of targeted therapy in lung cancer. Clin Pharmacol Ther. 2017;102(5):757–764. doi: 10.1002/cpt.810. [DOI] [PubMed] [Google Scholar]
- 5.Roesch A. Tumor heterogeneity and plasticity as elusive drivers for resistance to MAPK pathway inhibition in melanoma. Oncogene. 2015;34(23):2951–2957. doi: 10.1038/onc.2014.249. [DOI] [PubMed] [Google Scholar]
- 6.Hata AN, Niederst MJ, Archibald HL, Gomez-Caraballo M, Siddiqui FM, Mulvey HE, Maruvka YE, Ji F, Bhang HE, Krishnamurthy Radhakrishna V, Siravegna G, Hu H, Raoof S, Lockerman E, Kalsy A, Lee D, Keating CL, Ruddy DA, Damon LJ, Crystal AS, Costa C, Piotrowska Z, Bardelli A, Iafrate AJ, Sadreyev RI, Stegmeier F, Getz G, Sequist LV, Faber AC, Engelman JA. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med. 2016;22(3):262–269. doi: 10.1038/nm.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quintanal-Villalonga A, Chan JM, Yu HA, Pe’er D, Sawyers CL, Sen T, Rudin CM. Lineage plasticity in cancer: a shared pathway of therapeutic resistance. Nat Rev Clin Oncol. 2020 doi: 10.1038/s41571-020-0340-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barkley D, Yanai I. Plasticity and clonality of cancer cell states. Trends Cancer. 2019;5(11):655–656. doi: 10.1016/j.trecan.2019.09.002. [DOI] [PubMed] [Google Scholar]
- 9.Ramirez M, Rajaram S, Steininger RJ, Osipchuk D, Roth MA, Morinishi LS, Evans L, Ji W, Hsu CH, Thurley K, Wei S, Zhou A, Koduru PR, Posner BA, Wu LF, Altschuler SJ. Diverse drug-resistance mechanisms can emerge from drug-tolerant cancer persister cells. Nat Commun. 2016;7:10690. doi: 10.1038/ncomms10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, Yang B, Zhang M, Guo W, Wu Z, Wang Y, Jia L, Li S, Cancer Genome Atlas Research N, Xie W, Yang D lncRNA Epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell. 2018;33(4):706–720. doi: 10.1016/j.ccell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338(6113):1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 12.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger S, Cosper AK, Akhavanfard S, Heist RS, Temel J, Christensen JG, Wain JC, Lynch TJ, Vernovsky K, Mark EJ, Lanuti M, Iafrate AJ, Mino-Kenudson M, Engelman JA. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3(75):75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan S, Norgard RJ, Stanger BZ. Cellular plasticity in cancer. Cancer Discov. 2019;9(7):837–851. doi: 10.1158/2159-8290.CD-19-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boumahdi S, de Sauvage FJ. The great escape: tumour cell plasticity in resistance to targeted therapy. Nat Rev Drug Discov. 2019 doi: 10.1038/s41573-019-0044-1. [DOI] [PubMed] [Google Scholar]
- 15.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, Wong KK, Brandstetter K, Wittner B, Ramaswamy S, Classon M, Settleman J. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141(1):69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menon DR, Das S, Krepler C, Vultur A, Rinner B, Schauer S, Kashofer K, Wagner K, Zhang G, Rad EB, Haass NK, Soyer HP, Gabrielli B, Somasundaram R, Hoefler G, Herlyn M, Schaider H. A stress-induced early innate response causes multidrug tolerance in melanoma. Oncogene. 2015;34(34):4545. doi: 10.1038/onc.2014.432. [DOI] [PubMed] [Google Scholar]
- 17.Oser MG, Niederst MJ, Sequist LV, Engelman JA. Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol. 2015;16(4):e165–172. doi: 10.1016/S1470-2045(14)71180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JK, Lee J, Kim S, Kim S, Youk J, Park S, An Y, Keam B, Kim DW, Heo DS, Kim YT, Kim JS, Kim SH, Lee JS, Lee SH, Park K, Ku JL, Jeon YK, Chung DH, Park PJ, Kim J, Kim TM, Ju YS. Clonal history and genetic predictors of transformation into small-cell carcinomas from lung adenocarcinomas. J Clin Oncol. 2017;35(26):3065–3074. doi: 10.1200/JCO.2016.71.9096. [DOI] [PubMed] [Google Scholar]
- 19.Marcoux N, Gettinger SN, O’Kane G, Arbour KC, Neal JW, Husain H, Evans TL, Brahmer JR, Muzikansky A, Bonomi PD, Del Prete S, Wurtz A, Farago AF, Dias-Santagata D, Mino-Kenudson M, Reckamp KL, Yu HA, Wakelee HA, Shepherd FA, Piotrowska Z, Sequist LV. EGFR-Mutant adenocarcinomas that transform to small-cell lung cancer and other neuroendocrine carcinomas: clinical outcomes. J Clin Oncol. 2019;37(4):278–285. doi: 10.1200/JCO.18.01585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niederst MJ, Sequist LV, Poirier JT, Mermel CH, Lockerman EL, Garcia AR, Katayama R, Costa C, Ross KN, Moran T, Howe E, Fulton LE, Mulvey HE, Bernardo LA, Mohamoud F, Miyoshi N, VanderLaan PA, Costa DB, Janne PA, Borger DR, Ramaswamy S, Shioda T, Iafrate AJ, Getz G, Rudin CM, Mino-Kenudson M, Engelman JA. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat Commun. 2015;6:6377. doi: 10.1038/ncomms7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tata PR, Rajagopal J. Cellular plasticity: 1712 to the present day. Curr Opin Cell Biol. 2016;43:46–54. doi: 10.1016/j.ceb.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davies AH, Beltran H, Zoubeidi A. Cellular plasticity and the neuroendocrine phenotype in prostate cancer. Nat Rev Urol. 2018;15(5):271–286. doi: 10.1038/nrurol.2018.22. [DOI] [PubMed] [Google Scholar]
- 23.Zakowski MF, Ladanyi M, Kris MG, Memorial Sloan-Kettering Cancer Center Lung Cancer OncoGenome G EGFR mutations in small-cell lung cancers in patients who have never smoked. N Engl J Med. 2006;355(2):213–215. doi: 10.1056/NEJMc053610. [DOI] [PubMed] [Google Scholar]
- 24.Gower A, Hsu WH, Hsu ST, Wang Y, Giaccone G. EMT is associated with, but does not drive resistance to ALK inhibitors among EML4-ALK non-small cell lung cancer. Mol Oncol. 2016;10(4):601–609. doi: 10.1016/j.molonc.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z, Lee JC, Lin L, Olivas V, Au V, LaFramboise T, Abdel-Rahman M, Wang X, Levine AD, Rho JK, Choi YJ, Choi CM, Kim SW, Jang SJ, Park YS, Kim WS, Lee DH, Lee JS, Miller VA, Arcila M, Ladanyi M, Moonsamy P, Sawyers C, Boggon TJ, Ma PC, Costa C, Taron M, Rosell R, Halmos B, Bivona TG. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44(8):852–860. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Byers LA, Diao L, Wang J, Saintigny P, Girard L, Peyton M, Shen L, Fan Y, Giri U, Tumula PK, Nilsson MB, Gudikote J, Tran H, Cardnell RJ, Bearss DJ, Warner SL, Foulks JM, Kanner SB, Gandhi V, Krett N, Rosen ST, Kim ES, Herbst RS, Blumenschein GR, Lee JJ, Lippman SM, Ang KK, Mills GB, Hong WK, Weinstein JN, Wistuba II, Coombes KR, Minna JD, Heymach JV. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res. 2013;19(1):279–290. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung JH, Rho JK, Xu X, Lee JS, Yoon HI, Lee CT, Choi YJ, Kim HR, Kim CH, Lee JC. Clinical and molecular evidences of epithelial to mesenchymal transition in acquired resistance to EGFR-TKIs. Lung Cancer. 2011;73(2):176–182. doi: 10.1016/j.lungcan.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 28.Wu WS, Heinrichs S, Xu D, Garrison SP, Zambetti GP, Adams JM, Look AT. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell. 2005;123(4):641–653. doi: 10.1016/j.cell.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 29.Saxena M, Stephens MA, Pathak H, Rangarajan A. Transcription factors that mediate epithelial-mesenchymal transition lead to multidrug resistance by upregulating ABC transporters. Cell Death Dis. 2011;2:e179. doi: 10.1038/cddis.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mu P, Zhang Z, Benelli M, Karthaus WR, Hoover E, Chen CC, Wongvipat J, Ku SY, Gao D, Cao Z, Shah N, Adams EJ, Abida W, Watson PA, Prandi D, Huang CH, de Stanchina E, Lowe SW, Ellis L, Beltran H, Rubin MA, Goodrich DW, Demichelis F, Sawyers CL. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science. 2017;355(6320):84–88. doi: 10.1126/science.aah4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ku SY, Rosario S, Wang Y, Mu P, Seshadri M, Goodrich ZW, Goodrich MM, Labbe DP, Gomez EC, Wang J, Long HW, Xu B, Brown M, Loda M, Sawyers CL, Ellis L, Goodrich DW. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science. 2017;355(6320):78–83. doi: 10.1126/science.aah4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niederst MJ, Engelman JA. Bypass mechanisms of resistance to receptor tyrosine kinase inhibition in lung cancer. Sci Signal. 2013;6(294):re6. doi: 10.1126/scisignal.2004652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li M, Izpisua Belmonte JC. Roles for noncoding RNAs in cell-fate determination and regeneration. Nat Struct Mol Biol. 2015;22(1):2–4. doi: 10.1038/nsmb.2946. [DOI] [PubMed] [Google Scholar]
- 34.Crea F, Venalainen E, Ci X, Cheng H, Pikor L, Parolia A, Xue H, Nur Saidy NR, Lin D, Lam W, Collins C, Wang Y. The role of epigenetics and long noncoding RNA MIAT in neuroendocrine prostate cancer. Epigenomics. 2016;8(5):721–731. doi: 10.2217/epi.16.6. [DOI] [PubMed] [Google Scholar]
- 35.Ramnarine VR, Alshalalfa M, Mo F, Nabavi N, Erho N, Takhar M, Shukin R, Brahmbhatt S, Gawronski A, Kobelev M, Nouri M, Lin D, Tsai H, Lotan TL, Karnes RJ, Rubin MA, Zoubeidi A, Gleave ME, Sahinalp C, Wyatt AW, Volik SV, Beltran H, Davicioni E, Wang Y, Collins CC. The long noncoding RNA landscape of neuroendocrine prostate cancer and its clinical implications. Gigascience. 2018 doi: 10.1093/gigascience/giy050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14(10):611–629. doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gugnoni M, Ciarrocchi A. Long noncoding RNA and epithelial mesenchymal transition in cancer. Int J Mol Sci. 2019 doi: 10.3390/ijms20081924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grelet S, Link LA, Howley B, Obellianne C, Palanisamy V, Gangaraju VK, Diehl JA, Howe PH. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat Cell Biol. 2017;19(9):1105–1115. doi: 10.1038/ncb3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu W, Zhang H, Niu Y, Wu Y, Sun W, Li H, Kong J, Ding K, Shen HM, Wu H, Xia D, Wu Y. Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150-5p. Mol Cancer. 2017;16(1):118. doi: 10.1186/s12943-017-0685-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ge X, Li GY, Jiang L, Jia L, Zhang Z, Li X, Wang R, Zhou M, Zhou Y, Zeng Z, Xiang J, Li Z. Long noncoding RNA CAR10 promotes lung adenocarcinoma metastasis via miR-203/30/SNAI axis. Oncogene. 2019;38(16):3061–3076. doi: 10.1038/s41388-018-0645-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng W, He D, Shan B, Wang J, Shi W, Zhao W, Peng Z, Luo Q, Duan M, Li B, Cheng Y, Dong Y, Tang F, Zhang C, Duan C. LINC81507 act as a competing endogenous RNA of miR-199b-5p to facilitate NSCLC proliferation and metastasis via regulating the CAV1/STAT3 pathway. Cell Death Dis. 2019;10(7):533. doi: 10.1038/s41419-019-1740-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jia Y, Duan Y, Liu T, Wang X, Lv W, Wang M, Wang J, Liu L. LncRNA TTN-AS1 promotes migration, invasion, and epithelial mesenchymal transition of lung adenocarcinoma via sponging miR-142-5p to regulate CDK5. Cell Death Dis. 2019;10(8):573. doi: 10.1038/s41419-019-1811-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Q, Li X, Li X, Li X, Chen Z. LncRNA H19 promotes epithelial-mesenchymal transition (EMT) by targeting miR-484 in human lung cancer cells. J Cell Biochem. 2018;119(6):4447–4457. doi: 10.1002/jcb.26537. [DOI] [PubMed] [Google Scholar]
- 44.Yang QS, Li B, Xu G, Yang SQ, Wang P, Tang HH, Liu YY. Long noncoding RNA LINC00483/microRNA-144 regulates radiosensitivity and epithelial-mesenchymal transition in lung adenocarcinoma by interacting with HOXA10. J Cell Physiol. 2019;234(7):11805–11821. doi: 10.1002/jcp.27886. [DOI] [PubMed] [Google Scholar]
- 45.Liang R, Xiao G, Wang M, Li X, Li Y, Hui Z, Sun X, Qin S, Zhang B, Du N, Liu D, Ren H. SNHG6 functions as a competing endogenous RNA to regulate E2F7 expression by sponging miR-26a-5p in lung adenocarcinoma. Biomed Pharmacother. 2018;107:1434–1446. doi: 10.1016/j.biopha.2018.08.099. [DOI] [PubMed] [Google Scholar]
- 46.Cao Y, Luo X, Ding X, Cui S, Guo C. LncRNA ATB promotes proliferation and metastasis in A549 cells by down-regulation of microRNA-494. J Cell Biochem. 2018;119(8):6935–6942. doi: 10.1002/jcb.26894. [DOI] [PubMed] [Google Scholar]
- 47.Yao Z, Fenoglio S, Gao DC, Camiolo M, Stiles B, Lindsted T, Schlederer M, Johns C, Altorki N, Mittal V, Kenner L, Sordella R. TGF-beta IL-6 axis mediates selective and adaptive mechanisms of resistance to molecular targeted therapy in lung cancer. Proc Natl Acad Sci USA. 2010;107(35):15535–15540. doi: 10.1073/pnas.1009472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakai S, Ohhata T, Kitagawa K, Uchida C, Aoshima T, Niida H, Suzuki T, Inoue Y, Miyazawa K, Kitagawa M. Long noncoding RNA ELIT-1 acts as a Smad3 cofactor to facilitate TGFbeta/Smad signaling and promote epithelial-mesenchymal transition. Cancer Res. 2019;79(11):2821–2838. doi: 10.1158/0008-5472.CAN-18-3210. [DOI] [PubMed] [Google Scholar]
- 49.Lu Z, Li Y, Che Y, Huang J, Sun S, Mao S, Lei Y, Li N, Sun N, He J. The TGFbeta-induced lncRNA TBILA promotes non-small cell lung cancer progression in vitro and in vivo via cis-regulating HGAL and activating S100A7/JAB1 signaling. Cancer Lett. 2018;432:156–168. doi: 10.1016/j.canlet.2018.06.013. [DOI] [PubMed] [Google Scholar]
- 50.Zhang C, Hao Y, Wang Y, Xu J, Teng Y, Yang X. TGF-beta/SMAD4-regulated LncRNA-LINP1 inhibits epithelial-mesenchymal transition in lung cancer. Int J Biol Sci. 2018;14(12):1715–1723. doi: 10.7150/ijbs.27197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ren Y, Jia HH, Xu YQ, Zhou X, Zhao XH, Wang YF, Song X, Zhu ZY, Sun T, Dou Y, Tian WP, Zhao XL, Kang CS, Mei M. Paracrine and epigenetic control of CAF-induced metastasis: the role of HOTAIR stimulated by TGF-ss1 secretion. Mol Cancer. 2018;17(1):5. doi: 10.1186/s12943-018-0758-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawasaki N, Miwa T, Hokari S, Sakurai T, Ohmori K, Miyauchi K, Miyazono K, Koinuma D. Long noncoding RNA NORAD regulates transforming growth factor-beta signaling and epithelial-to-mesenchymal transition-like phenotype. Cancer Sci. 2018;109(7):2211–2220. doi: 10.1111/cas.13626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Terashima M, Tange S, Ishimura A, Suzuki T. MEG3 Long noncoding RNA contributes to the epigenetic regulation of epithelial-mesenchymal transition in lung cancer cell lines. J Biol Chem. 2017;292(1):82–99. doi: 10.1074/jbc.M116.750950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu L, Luo F, Liu Y, Liu X, Shi L, Lu X, Liu Q. Posttranscriptional silencing of the lncRNA MALAT1 by miR-217 inhibits the epithelial-mesenchymal transition via enhancer of zeste homolog 2 in the malignant transformation of HBE cells induced by cigarette smoke extract. Toxicol Appl Pharmacol. 2015;289(2):276–285. doi: 10.1016/j.taap.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 55.He R, Zhang FH, Shen N. LncRNA FEZF1-AS1 enhances epithelial-mesenchymal transition (EMT) through suppressing E-cadherin and regulating WNT pathway in non-small cell lung cancer (NSCLC) Biomed Pharmacother. 2017;95:331–338. doi: 10.1016/j.biopha.2017.08.057. [DOI] [PubMed] [Google Scholar]
- 56.Chen JH, Zhou LY, Xu S, Zheng YL, Wan YF, Hu CP. Overexpression of lncRNA HOXA11-AS promotes cell epithelial-mesenchymal transition by repressing miR-200b in non-small cell lung cancer. Cancer Cell Int. 2017;17:64. doi: 10.1186/s12935-017-0433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu Y, Lyu H, Liu H, Shi X, Song Y, Liu B. Downregulation of the long noncoding RNA GAS5-AS1 contributes to tumor metastasis in non-small cell lung cancer. Sci Rep. 2016;6:31093. doi: 10.1038/srep31093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun M, Liu XH, Wang KM, Nie FQ, Kong R, Yang JS, Xia R, Xu TP, Jin FY, Liu ZJ, Chen JF, Zhang EB, De W, Wang ZX. Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Mol Cancer. 2014;13:68. doi: 10.1186/1476-4598-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu Z, Li Y, Wang J, Che Y, Sun S, Huang J, Chen Z, He J. Long non-coding RNA NKILA inhibits migration and invasion of non-small cell lung cancer via NF-kappaB/Snail pathway. J Exp Clin Cancer Res. 2017;36(1):54. doi: 10.1186/s13046-017-0518-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hao Y, Yang X, Zhang D, Luo J, Chen R. Long noncoding RNA LINC01186, regulated by TGF-beta/SMAD3, inhibits migration and invasion through epithelial-mesenchymal-transition in lung cancer. Gene. 2017;608:1–12. doi: 10.1016/j.gene.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 61.Balaban NQ, Gerdes K, Lewis K, McKinney JD. A problem of persistence: still more questions than answers? Nat Rev Microbiol. 2013;11(8):587–591. doi: 10.1038/nrmicro3076. [DOI] [PubMed] [Google Scholar]
- 62.Sanchez-Danes A, Larsimont JC, Liagre M, Munoz-Couselo E, Lapouge G, Brisebarre A, Dubois C, Suppa M, Sukumaran V, Del Marmol V, Tabernero J, Blanpain C. A slow-cycling LGR5 tumour population mediates basal cell carcinoma relapse after therapy. Nature. 2018;562(7727):434–438. doi: 10.1038/s41586-018-0603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dimitrova N, Zamudio JR, Jong RM, Soukup D, Resnick R, Sarma K, Ward AJ, Raj A, Lee JT, Sharp PA, Jacks T. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell. 2014;54(5):777–790. doi: 10.1016/j.molcel.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schmitt AM, Garcia JT, Hung T, Flynn RA, Shen Y, Qu K, Payumo AY, Peres-da-Silva A, Broz DK, Baum R, Guo S, Chen JK, Attardi LD, Chang HY. An inducible long noncoding RNA amplifies DNA damage signaling. Nat Genet. 2016;48(11):1370–1376. doi: 10.1038/ng.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang J, Gu Y, Liu J, Wu R, Fu L, Zhao J, Guan Y. Coexistence of p16/CDKN2A homozygous deletions and activating EGFR mutations in lung adenocarcinoma patients signifies a poor response to EGFR-TKIs. Lung Cancer. 2016;102:101–107. doi: 10.1016/j.lungcan.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 67.Montes M, Nielsen MM, Maglieri G, Jacobsen A, Hojfeldt J, Agrawal-Singh S, Hansen K, Helin K, van de Werken HJG, Pedersen JS, Lund AH. The lncRNA MIR31HG regulates p16(INK4A) expression to modulate senescence. Nat Commun. 2015;6:6967. doi: 10.1038/ncomms7967. [DOI] [PubMed] [Google Scholar]
- 68.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451(7175):202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun Q, Tripathi V, Yoon JH, Singh DK, Hao Q, Min KW, Davila S, Zealy RW, Li XL, Polycarpou-Schwarz M, Lehrmann E, Zhang Y, Becker KG, Freier SM, Zhu Y, Diederichs S, Prasanth SG, Lal A, Gorospe M, Prasanth KV. MIR100 host gene-encoded lncRNAs regulate cell cycle by modulating the interaction between HuR and its target mRNAs. Nucleic Acids Res. 2018;46(19):10405–10416. doi: 10.1093/nar/gky696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yeh CH, Bellon M, Nicot C. FBXW7: a critical tumor suppressor of human cancers. Mol Cancer. 2018;17(1):115. doi: 10.1186/s12943-018-0857-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu M, Chen X, Lin K, Zeng K, Liu X, Pan B, Xu X, Xu T, Hu X, Sun L, He B, Pan Y, Sun H, Wang S. The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol Cancer. 2018;17(1):141. doi: 10.1186/s12943-018-0894-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BV, Varambally S, Tomlins SA, Nanus DM, Tagawa ST, Van Allen EM, Elemento O, Sboner A, Garraway LA, Rubin MA, Demichelis F. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22(3):298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dardenne E, Beltran H, Benelli M, Gayvert K, Berger A, Puca L, Cyrta J, Sboner A, Noorzad Z, MacDonald T, Cheung C, Yuen KS, Gao D, Chen Y, Eilers M, Mosquera JM, Robinson BD, Elemento O, Rubin MA, Demichelis F, Rickman DS. N-Myc Induces an EZH2-Mediated Transcriptional Program Driving Neuroendocrine Prostate Cancer. Cancer Cell. 2016;30(4):563–577. doi: 10.1016/j.ccell.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lim JS, Ibaseta A, Fischer MM, Cancilla B, O'Young G, Cristea S, Luca VC, Yang D, Jahchan NS, Hamard C, Antoine M, Wislez M, Kong C, Cain J, Liu YW, Kapoun AM, Garcia KC, Hoey T, Murriel CL, Sage J. Intratumoural heterogeneity generated by Notch signalling promotes small-cell lung cancer. Nature. 2017;545(7654):360–364. doi: 10.1038/nature22323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121(4):645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 76.Lim JS, Ibaseta A, Fischer MM, Cancilla B, O’Young G, Cristea S, Luca VC, Yang D, Jahchan NS, Hamard C, Antoine M, Wislez M, Kong C, Cain J. Intratumoural heterogeneity generated by Notch signalling promotes small-cell lung cancer. Nature. 2017;545(7654):360–364. doi: 10.1038/nature22323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Santos M, Martinez-Fernandez M, Duenas M, Garcia-Escudero R, Alfaya B, Villacampa F, Saiz-Ladera C, Costa C, Oteo M, Duarte J, Martinez V, Gomez-Rodriguez MJ, Martin ML, Fernandez M, Viatour P, Morcillo MA, Sage J, Castellano D, Rodriguez-Peralto JL, de la Rosa F, Paramio JM. In vivo disruption of an Rb-E2F-Ezh2 signaling loop causes bladder cancer. Cancer Res. 2014;74(22):6565–6577. doi: 10.1158/0008-5472.CAN-14-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang L, Zhang X, Liu Y, Xu S. Long noncoding RNA FBXL19-AS1 induces tumor growth and metastasis by sponging miR-203a-3p in lung adenocarcinoma. J Cell Physiol. 2019 doi: 10.1002/jcp.29251. [DOI] [PubMed] [Google Scholar]
- 79.Sarkar A, Hochedlinger K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12(1):15–30. doi: 10.1016/j.stem.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bishop JL, Thaper D, Vahid S, Davies A, Ketola K, Kuruma H, Jama R, Nip KM, Angeles A, Johnson F, Wyatt AW, Fazli L, Gleave ME, Lin D, Rubin MA, Collins CC, Wang Y, Beltran H, Zoubeidi A. The Master neural transcription factor BRN2 Is an androgen receptor-suppressed driver of neuroendocrine differentiation in prostate cancer. Cancer Discov. 2017;7(1):54–71. doi: 10.1158/2159-8290.CD-15-1263. [DOI] [PubMed] [Google Scholar]
- 81.Sakaeda M, Sato H, Ishii J, Miyata C, Kamma H, Shishido-Hara Y, Shimoyamada H, Fujiwara M, Endo T, Tanaka R, Kondo H, Goya T, Aoki I, Yazawa T. Neural lineage-specific homeoprotein BRN2 is directly involved in TTF1 expression in small-cell lung cancer. Lab Invest. 2013;93(4):408–421. doi: 10.1038/labinvest.2013.2. [DOI] [PubMed] [Google Scholar]
- 82.Chen C, NoTKIns AL, Lan MS. Insulinoma-associated-1: from neuroendocrine tumor marker to cancer therapeutics. Mol Cancer Res. 2019;17(8):1597–1604. doi: 10.1158/1541-7786.MCR-19-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ahmad A, Strohbuecker S, Tufarelli C, Sottile V. Expression of a SOX1 overlapping transcript in neural differentiation and cancer models. Cell Mol Life Sci. 2017;74(22):4245–4258. doi: 10.1007/s00018-017-2580-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Saghaeian Jazi M, Samaei NM, Ghanei M, Shadmehr MB, Mowla SJ. Identification of new SOX2OT transcript variants highly expressed in human cancer cell lines and down regulated in stem cell differentiation. Mol Biol Rep. 2016;43(2):65–72. doi: 10.1007/s11033-015-3939-x. [DOI] [PubMed] [Google Scholar]
- 85.Shahryari A, Jazi MS, Samaei NM, Mowla SJ. Long non-coding RNA SOX2OT: expression signature, splicing patterns, and emerging roles in pluripotency and tumorigenesis. Front Genet. 2015;6:196. doi: 10.3389/fgene.2015.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shahryari A, Rafiee MR, Fouani Y, Oliae NA, Samaei NM, Shafiee M, Semnani S, Vasei M, Mowla SJ. Two novel splice variants of SOX2OT, SOX2OT-S1, and SOX2OT-S2 are coupregulated with SOX2 and OCT4 in esophageal squamous cell carcinoma. Stem Cells. 2014;32(1):126–134. doi: 10.1002/stem.1542. [DOI] [PubMed] [Google Scholar]
- 87.Kim JT, Liu C, Zaytseva YY, Weiss HL, Townsend CM, Jr, Evers BM. Neurotensin, a novel target of Wnt/beta-catenin pathway, promotes growth of neuroendocrine tumor cells. Int J Cancer. 2015;136(6):1475–1481. doi: 10.1002/ijc.29123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Meder L, Konig K, Ozretic L, Schultheis AM, Ueckeroth F, Ade CP, Albus K, Boehm D, Rommerscheidt-Fuss U, Florin A, Buhl T, Hartmann W, Wolf J, Merkelbach-Bruse S, Eilers M, Perner S, Heukamp LC, Buettner R. NOTCH, ASCL1, p53 and RB alterations define an alternative pathway driving neuroendocrine and small cell lung carcinomas. Int J Cancer. 2016;138(4):927–938. doi: 10.1002/ijc.29835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tenjin Y, Kudoh S, Kubota S, Yamada T, Matsuo A, Sato Y, Ichimura T, Kohrogi H, Sashida G, Sakagami T, Ito T. Ascl1-induced Wnt11 regulates neuroendocrine differentiation, cell proliferation, and E-cadherin expression in small-cell lung cancer and Wnt11 regulates small-cell lung cancer biology. Lab Invest. 2019;99(11):1622–1635. doi: 10.1038/s41374-019-0277-y. [DOI] [PubMed] [Google Scholar]
- 90.Osada H, Tomida S, Yatabe Y, Tatematsu Y, Takeuchi T, Murakami H, Kondo Y, Sekido Y, Takahashi T. Roles of achaete-scute homologue 1 in DKK1 and E-cadherin repression and neuroendocrine differentiation in lung cancer. Cancer Res. 2008;68(6):1647–1655. doi: 10.1158/0008-5472.CAN-07-5039. [DOI] [PubMed] [Google Scholar]
- 91.Rong L, Zhao R, Lu J. Highly expressed long non-coding RNA FOXD2-AS1 promotes non-small cell lung cancer progression via Wnt/beta-catenin signaling. Biochem Biophys Res Commun. 2017;484(3):586–591. doi: 10.1016/j.bbrc.2017.01.141. [DOI] [PubMed] [Google Scholar]
- 92.Shi SL, Zhang ZH. Long non-coding RNA SNHG1 contributes to cisplatin resistance in non-small cell lung cancer by regulating miR-140-5p/Wnt/beta-catenin pathway. Neoplasma. 2019;66(5):756–765. doi: 10.4149/neo_2018_181218N980. [DOI] [PubMed] [Google Scholar]
- 93.Guan H, Zhu T, Wu S, Liu S, Liu B, Wu J, Cai J, Zhu X, Zhang X, Zeng M, Li J, Song E, Li M. Long noncoding RNA LINC00673-v4 promotes aggressiveness of lung adenocarcinoma via activating WNT/beta-catenin signaling. Proc Natl Acad Sci USA. 2019;116(28):14019–14028. doi: 10.1073/pnas.1900997116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jiang P, Xu H, Xu C, Chen A, Chen L, Zhou M, Haq IU, Wu X, Mariyam Z, Feng Q. NEAT1 contributes to the CSC-like traits of A549/CDDP cells via activating Wnt signaling pathway. Chem Biol Interact. 2018;296:154–161. doi: 10.1016/j.cbi.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 95.Sun SJ, Lin Q, Ma JX, Shi WW, Yang B, Li F. Long non-coding RNA NEAT1 acts as oncogene in NSCLC by regulating the Wnt signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21(3):504–510. [PubMed] [Google Scholar]
- 96.Chang KT, Tsai CM, Chiou YC, Chiu CH, Jeng KS, Huang CY. IL-6 induces neuroendocrine dedifferentiation and cell proliferation in non-small cell lung cancer cells. Am J Physiol Lung Cell Mol Physiol. 2005;289(3):L446–453. doi: 10.1152/ajplung.00089.2005. [DOI] [PubMed] [Google Scholar]
- 97.Liu L, Liu L, Lu S. lncRNA H19 promotes viability and epithelial-mesenchymal transition of lung adenocarcinoma cells by targeting miR-29b-3p and modifying STAT3. Int J Oncol. 2019;54(3):929–941. doi: 10.3892/ijo.2019.4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li S, Mei Z, Hu HB, Zhang X. The lncRNA MALAT1 contributes to non-small cell lung cancer development via modulating miR-124/STAT3 axis. J Cell Physiol. 2018;233(9):6679–6688. doi: 10.1002/jcp.26325. [DOI] [PubMed] [Google Scholar]
- 99.Huang Z, Lei W, Hu HB, Zhang H, Zhu Y. H19 promotes non-small-cell lung cancer (NSCLC) development through STAT3 signaling via sponging miR-17. J Cell Physiol. 2018;233(10):6768–6776. doi: 10.1002/jcp.26530. [DOI] [PubMed] [Google Scholar]
- 100.Wang Y, Han D, Pan L, Sun J. The positive feedback between lncRNA TNK2-AS1 and STAT3 enhances angiogenesis in non-small cell lung cancer. Biochem Biophys Res Commun. 2018;507(1–4):185–192. doi: 10.1016/j.bbrc.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 101.Chen X, Song E. Turning foes to friends: targeting cancer-associated fibroblasts. Nat Rev Drug Discov. 2019;18(2):99–115. doi: 10.1038/s41573-018-0004-1. [DOI] [PubMed] [Google Scholar]
- 102.Liu T, Han C, Wang S, Fang P, Ma Z, Xu L, Yin R. Cancer-associated fibroblasts: an emerging target of anti-cancer immunotherapy. J Hematol Oncol. 2019;12(1):86. doi: 10.1186/s13045-019-0770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Choe C, Shin YS, Kim C, Choi SJ, Lee J, Kim SY, Cho YB, Kim J. Crosstalk with cancer-associated fibroblasts induces resistance of non-small cell lung cancer cells to epidermal growth factor receptor tyrosine kinase inhibition. Onco Targets Ther. 2015;8:3665–3678. doi: 10.2147/OTT.S89659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yoshida T, Ishii G, Goto K, Neri S, Hashimoto H, Yoh K, Niho S, Umemura S, Matsumoto S, Ohmatsu H, Iida S, Niimi A, Nagai K, Ohe Y, Ochiai A. Podoplanin-positive cancer-associated fibroblasts in the tumor microenvironment induce primary resistance to EGFR-TKIs in lung adenocarcinoma with EGFR mutation. Clin Cancer Res. 2015;21(3):642–651. doi: 10.1158/1078-0432.CCR-14-0846. [DOI] [PubMed] [Google Scholar]
- 105.Apicella M, Giannoni E, Fiore S, Ferrari KJ, Fernandez-Perez D, Isella C, Granchi C, Minutolo F, Sottile A, Comoglio PM, Medico E, Pietrantonio F, Volante M, Pasini D, Chiarugi P, Giordano S, Corso S. Increased lactate secretion by cancer cells sustains non-cell-autonomous adaptive resistance to MET and EGFR targeted therapies. Cell Metab. 2018;28(6):848–865. doi: 10.1016/j.cmet.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 106.Suzuki E, Yamazaki S, Naito T, Hashimoto H, Okubo S, Udagawa H, Goto K, Tsuboi M, Ochiai A, Ishii G. Secretion of high amounts of hepatocyte growth factor is a characteristic feature of cancer-associated fibroblasts with EGFR-TKI resistance-promoting phenotype: A study of 18 cases of cancer-associated fibroblasts. Pathol Int. 2019;69(8):472–480. doi: 10.1111/pin.12838. [DOI] [PubMed] [Google Scholar]
- 107.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, Kosaka T, Holmes AJ, Rogers AM, Cappuzzo F, Mok T, Lee C, Johnson BE, Cantley LC, Janne PA. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316(5827):1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 108.Karachaliou N, Codony-Servat J, Teixido C, Pilotto S, Drozdowskyj A, Codony-Servat C, Gimenez-Capitan A, Molina-Vila MA, Bertran-Alamillo J, Gervais R, Massuti B, Moran T, Majem M, Felip E, Carcereny E, Garcia-Campelo R, Viteri S, Gonzalez-Cao M, Morales-Espinosa D, Verlicchi A, Crisetti E, Chaib I, Santarpia M, Luis Ramirez J, Bosch-Barrera J, Felipe Cardona A, de Marinis F, Lopez-Vivanco G, Miguel Sanchez J, Vergnenegre A, Sanchez Hernandez JJ, Sperduti I, Bria E, Rosell R. BIM and mTOR expression levels predict outcome to erlotinib in EGFR-mutant non-small-cell lung cancer. Sci Rep. 2015;5:17499. doi: 10.1038/srep17499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang Y, Zhang T, Guo L, Ren T, Yang Y. Stromal extracellular matrix is a microenvironmental cue promoting resistance to EGFR tyrosine kinase inhibitors in lung cancer cells. Int J Biochem Cell Biol. 2019;106:96–106. doi: 10.1016/j.biocel.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 110.Wang LC, Lo A, Scholler J, Sun J, Majumdar RS, Kapoor V, Antzis M, Cotner CE, Johnson LA, Durham AC, Solomides CC, June CH, Pure E, Albelda SM. Targeting fibroblast activation protein in tumor stroma with chimeric antigen receptor T cells can inhibit tumor growth and augment host immunity without severe toxicity. Cancer Immunol Res. 2014;2(2):154–166. doi: 10.1158/2326-6066.CIR-13-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang B, Zhang Y, Zhao J, Wang Z, Wu T, Ou W, Wang J, Yang B, Zhao Y, Rao Z, Gao J. M2-polarized macrophages contribute to the decreased sensitivity of EGFR-TKIs treatment in patients with advanced lung adenocarcinoma. Med Oncol. 2014;31(8):127. doi: 10.1007/s12032-014-0127-0. [DOI] [PubMed] [Google Scholar]
- 112.Choi H, Sheng J, Gao D, Li F, Durrans A, Ryu S, Lee SB, Narula N, Rafii S, Elemento O, Altorki NK, Wong ST, Mittal V. Transcriptome analysis of individual stromal cell populations identifies stroma-tumor crosstalk in mouse lung cancer model. Cell Rep. 2015;10(7):1187–1201. doi: 10.1016/j.celrep.2015.01.040. [DOI] [PubMed] [Google Scholar]
- 113.Che D, Zhang S, Jing Z, Shang L, Jin S, Liu F, Shen J, Li Y, Hu J, Meng Q, Yu Y. Macrophages induce EMT to promote invasion of lung cancer cells through the IL-6-mediated COX-2/PGE2/beta-catenin signalling pathway. Mol Immunol. 2017;90:197–210. doi: 10.1016/j.molimm.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 114.Sun Y, Xu J. TCF-4 Regulated lncRNA-XIST promotes M2 polarization of macrophages and is associated with lung cancer. Onco Targets Ther. 2019;12:8055–8062. doi: 10.2147/OTT.S210952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li Z, Feng C, Guo J, Hu X, Xie D. GNAS-AS1/miR-4319/NECAB3 axis promotes migration and invasion of non-small cell lung cancer cells by altering macrophage polarization. Funct Integr Genomics. 2019 doi: 10.1007/s10142-019-00696-x. [DOI] [PubMed] [Google Scholar]
- 116.Murakami A, Takahashi F, Nurwidya F, Kobayashi I, Minakata K, Hashimoto M, Nara T, Kato M, Tajima K, Shimada N, Iwakami S, Moriyama M, Moriyama H, Koizumi F, Takahashi K. Hypoxia increases gefitinib-resistant lung cancer stem cells through the activation of insulin-like growth factor 1 receptor. PLoS ONE. 2014;9(1):e86459. doi: 10.1371/journal.pone.0086459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Minakata K, Takahashi F, Nara T, Hashimoto M, Tajima K, Murakami A, Nurwidya F, Yae S, Koizumi F, Moriyama H, Seyama K, Nishio K, Takahashi K. Hypoxia induces gefitinib resistance in non-small-cell lung cancer with both mutant and wild-type epidermal growth factor receptors. Cancer Sci. 2012;103(11):1946–1954. doi: 10.1111/j.1349-7006.2012.02408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, Boon RA, Dimmeler S. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res. 2014;114(9):1389–1397. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- 119.Yin W, Yu X, Kang X, Zhao Y, Zhao P, Jin H, Fu X, Wan Y, Peng C, Huang Y. Remodeling tumor-associated macrophages and neovascularization overcomes EGFR(T790M)—associated drug resistance by PD-L1 nanobody-mediated codelivery. Small. 2018;14(47):e1802372. doi: 10.1002/smll.201802372. [DOI] [PubMed] [Google Scholar]
- 120.Yang GJ, Ko CN, Zhong HJ, Leung CH, Ma DL. Structure-based discovery of a selective KDM5A inhibitor that exhibits anti-cancer activity via inducing cell cycle arrest and senescence in breast cancer cell lines. Cancers (Basel) 2019 doi: 10.3390/cancers11010092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Guler GD, Tindell CA, Pitti R, Wilson C, Nichols K, KaiWai Cheung T, Kim HJ, Wongchenko M, Yan Y, Haley B, Cuellar T, Webster J, Alag N, Hegde G, Jackson E, Nance TL, Giresi PG, Chen KB, Liu J, Jhunjhunwala S, Settleman J, Stephan JP, Arnott D, Classon M. Repression of stress-induced LINE-1 expression protects cancer cell subpopulations from lethal drug exposure. Cancer Cell. 2017;32(2):221–237. doi: 10.1016/j.ccell.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 122.McKeithen D, Graham T, Chung LW, Odero-Marah V. Snail transcription factor regulates neuroendocrine differentiation in LNCaP prostate cancer cells. Prostate. 2010;70(9):982–992. doi: 10.1002/pros.21132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liau BB, Sievers C, Donohue LK, Gillespie SM, Flavahan WA, Miller TE, Venteicher AS, Hebert CH, Carey CD, Rodig SJ, Shareef SJ, Najm FJ, van Galen P, Wakimoto H, Cahill DP, Rich JN, Aster JC, Suva ML, Patel AP, Bernstein BE. Adaptive chromatin remodeling drives glioblastoma stem cell plasticity and drug tolerance. Cell Stem Cell. 2017;20(2):233–246. doi: 10.1016/j.stem.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, Bantscheff M, Bountra C, Bridges A, Diallo H, Eberhard D, Hutchinson S, Jones E, Katso R, Leveridge M, Mander PK, Mosley J, Ramirez-Molina C, Rowland P, Schofield CJ, Sheppard RJ, Smith JE, Swales C, Tanner R, Thomas P, Tumber A, Drewes G, Oppermann U, Patel DJ, Lee K, Wilson DM. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488(7411):404–408. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Knutson SK, Kawano S, Minoshima Y, Warholic NM, Huang KC, Xiao Y, Kadowaki T, Uesugi M, Kuznetsov G, Kumar N, Wigle TJ, Klaus CR, Allain CJ, Raimondi A, Waters NJ, Smith JJ, Porter-Scott M, Chesworth R, Moyer MP, Copeland RA, Richon VM, Uenaka T, Pollock RM, Kuntz KW, Yokoi A, Keilhack H. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol Cancer Ther. 2014;13(4):842–854. doi: 10.1158/1535-7163.MCT-13-0773. [DOI] [PubMed] [Google Scholar]
- 126.Vaswani RG, Gehling VS, Dakin LA, Cook AS, Nasveschuk CG, Duplessis M, Iyer P, Balasubramanian S, Zhao F, Good AC, Campbell R, Lee C, Cantone N, Cummings RT, Normant E, Bellon SF, Albrecht BK, Harmange JC, Trojer P, Audia JE, Zhang Y, Justin N, Chen S, Wilson JR, Gamblin SJ. Identification of (R)-N-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-2-methyl-1-(1-(1 -(2,2,2-trifluoroethyl)piperidin-4-yl)ethyl)-1H-indole-3-carboxamide (CPI-1205), a potent and selective inhibitor of histone methyltransferase EZH2, suitable for phase I clinical trials for B-cell lymphomas. J Med Chem. 2016;59(21):9928–9941. doi: 10.1021/acs.jmedchem.6b01315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang H, Qi J, Reyes JM, Li L, Rao PK, Li F, Lin CY, Perry JA, Lawlor MA, Federation A, De Raedt T, Li YY, Liu Y, Duarte MA, Zhang Y, Herter-Sprie GS, Kikuchi E, Carretero J, Perou CM, Reibel JB, Paulk J, Bronson RT, Watanabe H, Brainson CF, Kim CF, Hammerman PS, Brown M, Cichowski K, Long H, Bradner JE, Wong KK. Oncogenic deregulation of EZH2 as an opportunity for targeted therapy in lung cancer. Cancer Discov. 2016;6(9):1006–1021. doi: 10.1158/2159-8290.CD-16-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Frankel AE, Liu X, Minna JD. Developing EZH2-targeted therapy for lung cancer. Cancer Discov. 2016;6(9):949–952. doi: 10.1158/2159-8290.CD-16-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gong H, Yuan Y, Li Y, Zhang H, Li Y, Li W, Wang P, Shi R, Liu C, Cui L, Liu H, Chen J. Role of EZH2 inhibitor combined with gefitinib in EGFR-TKIs resistant lung cancer cells. Zhongguo Fei Ai Za Zhi. 2019;22(5):255–263. doi: 10.3779/j.issn.1009-3419.2019.05.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Weng CH, Chen LY, Lin YC, Shih JY, Lin YC, Tseng RY, Chiu AC, Yeh YH, Liu C, Lin YT, Fang JM, Chen CC. Epithelial-mesenchymal transition (EMT) beyond EGFR mutations per se is a common mechanism for acquired resistance to EGFR TKI. Oncogene. 2019;38(4):455–468. doi: 10.1038/s41388-018-0454-2. [DOI] [PubMed] [Google Scholar]
- 131.Bates SE. Epigenetic therapies for cancer. N Engl J Med. 2020;383(7):650–663. doi: 10.1056/NEJMra1805035. [DOI] [PubMed] [Google Scholar]
- 132.Mohammad HP, Barbash O, Creasy CL. Targeting epigenetic modifications in cancer therapy: erasing the roadmap to cancer. Nat Med. 2019;25(3):403–418. doi: 10.1038/s41591-019-0376-8. [DOI] [PubMed] [Google Scholar]
- 133.Washietl S, Kellis M, Garber M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res. 2014;24(4):616–628. doi: 10.1101/gr.165035.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 135.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12(11):847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gutschner T, Baas M, Diederichs S. Noncoding RNA gene silencing through genomic integration of RNA destabilizing elements using zinc finger nucleases. Genome Res. 2011;21(11):1944–1954. doi: 10.1101/gr.122358.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Esposito R, Bosch N, Lanzos A, Polidori T, Pulido-Quetglas C, Johnson R. Hacking the cancer genome: profiling therapeutically actionable long non-coding RNAs using CRISPR-Cas9 screening. Cancer Cell. 2019;35(4):545–557. doi: 10.1016/j.ccell.2019.01.019. [DOI] [PubMed] [Google Scholar]
- 138.Goyal A, Myacheva K, Gross M, Klingenberg M, Duran Arque B, Diederichs S. Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res. 2017;45(3):e12. doi: 10.1093/nar/gkw883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ferrer L, Giaj Levra M, Brevet M, Antoine M, Mazieres J, Rossi G, Chiari R, Westeel V, Poudenx M, Letreut J, Gervais R, Osman G, Girard N, Toffart AC, Novello S, Moro-Sibilot D. A brief report of transformation from NSCLC to SCLC: molecular and therapeutic characteristics. J Thorac Oncol. 2019;14(1):130–134. doi: 10.1016/j.jtho.2018.08.2028. [DOI] [PubMed] [Google Scholar]
- 140.Qiu M, Xia W, Chen R, Wang S, Xu Y, Ma Z, Xu W, Zhang E, Wang J, Fang T, Hu J, Dong G, Yin R, Wang J, Xu L. The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma. Cancer Res. 2018;78(11):2839–2851. doi: 10.1158/0008-5472.CAN-17-2808. [DOI] [PubMed] [Google Scholar]
- 141.Sheridan C. First Axl inhibitor enters clinical trials. Nat Biotechnol. 2013;31(9):775–776. doi: 10.1038/nbt0913-775a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated during this study are included in this published article.