Brain tumor location has prognostic implications so these authors explored whether location also correlated with p53 status in 182 patients. Tumors occurring in the left medial temporal and right anterior temporal lobes were associated with high expression of mutant p53 and had significantly worse progression-free survival.
Abstract
BACKGROUND AND PURPOSE:
Brain tumor location has proved to be a prognostic factor that may be associated with features of neoplastic origin. Mutation of p53 is an atypical genetic change that occurs during tumorigenesis. Thus, a potential correlation may exist between tumor location and p53 status. The purpose of the current study was to identify anatomic characteristics of mutant p53 expression by using quantitative neuroimaging analyses.
MATERIALS AND METHODS:
Preoperative MR images from 182 patients with histologically confirmed low-grade gliomas were retrospectively analyzed. All tumors were manually marked and registered to the standard space. Using a voxel-based lesion-symptom mapping analysis, we located brain regions associated with a high occurrence of p53 mutation and corrected them by using a permutation test. The acquired clusters were further included as a factor in survival analyses.
RESULTS:
Statistical analysis demonstrated that the left medial temporal lobe and right anterior temporal lobe were specifically associated with high expression of mutant p53. Kaplan-Meier curves showed that tumors located in these regions were associated with significantly worse progression-free survival compared with tumors occurring elsewhere.
CONCLUSIONS:
Our voxel-level imaging analysis provides new evidence that genetic changes during cancer may have anatomic specificity. Additionally, the current study suggests that tumor location identified on structural MR images could potentially be used for customized presurgical outcome prediction.
Gliomas originate in glial cells in the brain and are the most common type of primary brain tumor.1,2 Low-grade gliomas (LGGs) constitute 10%–20% of primary brain tumors and mainly affect young adults.3 For patients with LGGs, histologic grading remains the most important factor influencing prognosis.4 Molecular markers involved in low-grade glioma pathways also play a critical role in determining prognosis.5,6
The TP53 mutation is an early event in genetic pathogenesis that occurs in all types of cancer. The incidence of TP53 mutation in LGGs is approximately 53% in diffuse astrocytoma, 44% in oligoastrocytoma, and 13% in oligodendroglioma.5,7 Mutant p53 protein is commonly regarded as a surrogate marker for a mutation in TP53.8 Furthermore, p53 mutations acquire oncogenic properties that enable them to promote proliferation, invasion, metastasis, and cell immortalization.9,10 Although it has been widely accepted that the loss of p53 function plays an important role in glioma tumorigenesis,11,12 the prognostic role of p53 mutations has remained controversial and no consistent relationship with response to therapy or overall outcome has been reported.8,13
Associations have been made that link tumor location and genetic features in glioma. For example, anatomic characteristics have been demonstrated for several tumor-related molecular biomarkers, such as 1p and/or 19q,14 isocitrate dehydrogenase 1,15 O6-methylguanine DNA methyltransferase (MGMT),16 and TP53.17 Because location may be related to the genetic profile of the origin of a tumor cell,18–20 tumor location is one of the most important characteristics for predicting the development of a glioma and the prognosis of a patient.14,15,18,21 However, identifying brain tumor precursor cells and their distinct genetic profiles to increase survival via customized treatment paradigms is a major challenge in neuro-oncologic studies.22,23
Moreover, this issue is far from clear because anatomic characteristics of tumor-related gene expression are rarely investigated. Dichotomous statistics of tumor location based on which lobe is involved have ignored important information such as tumor size. With the goal of providing a more reliable and precise brain atlas to demonstrate the statistical correlation between p53 mutations and tumor intensity, the current study used a voxel-based lesion-symptom mapping (VLSM) approach.24 One hundred eighty-two patients with primarily diagnosed LGG were included in this study. VLSM-defined regions associated with high expression of the p53 mutant were located mainly in the left medial and right anterior temporal lobes. Notably, patients with tumors located in VLSM-defined regions had shorter progression-free survival times (PFSs) compared with those whose tumors occurred elsewhere.
Materials and Methods
Patients
One hundred eighty-two patients with newly diagnosed LGGs treated at our institute between September 2006 and December 2011 were enrolled in the current retrospective study. Clinical information of these patients was collected from the Chinese Glioma Genome Atlas data base (http://www.cgga.org.cn). Inclusion criteria were the following: 1) histologic confirmation of the LGG with no previous diagnosis of any type of brain tumor, 2) no history of radiation therapy or chemotherapy, 3) no history of biopsy, 4) high-resolution presurgical T2-weighted images, and 5) tissue available for testing p53 expression. Histologic diagnoses were re-evaluated by 2 neuropathologists for all cases according to the new World Health Organization classification of tumors of the nervous system. Surgical resection was used to establish histopathologic diagnoses for all patients. Corresponding frozen tissues were reviewed to ensure a minimum of 80% tumor nuclei and a maximum of 50% necrosis.25 Additional patient characteristics are summarized in the Table. This study was approved by the institutional review board, and written informed consent was obtained from all patients.
Clinical characteristics of patients
| Characteristics | Status of p53 |
P Value | |
|---|---|---|---|
| Mutated (%) (n = 116) | Wild-Type (%) (n = 66) | ||
| Age | |||
| 40 yr and older | 41 (55) | 33 (45) | .028 |
| Younger than 40 yr | 75 (69) | 33 (31) | |
| Sex | |||
| Male | 70 (65) | 37 (35) | .469 |
| Female | 46 (61) | 29 (39) | |
| History of seizures | |||
| Yes | 70 (62) | 42 (38) | .378 |
| No | 46 (66) | 24 (34) | |
| Preoperative KPS | |||
| ≥80 | 109 (65) | 58 (35) | .499 |
| <80 | 7 (47) | 8 (53) | |
| Pathology | |||
| Oligodendroglioma | 10 (36) | 18 (64) | .003 |
| Astrocytoma | 48 (76) | 15 (24) | |
| Oligoastrocytoma | 58 (64) | 33 (36) | |
Note:—KPS indicates Karnofsky Performance Status Scale.
MR Imaging and Tumor Registration
MR imaging for most patients (n = 129) was performed on a Magnetom Trio 3T (Siemens, Erlangen, Germany) scanner. The T2-weighted image parameters included the following: TR = 5800 ms; TE = 110 ms; flip angle = 150°; FOV = 240 × 188 mm2; and voxel size = 0.6 × 0.6 × 5 mm3. A minority of clinical structural images were acquired on a Magnetom Verio 3T (Siemens) (n = 32) or Signa HD 1.5T (GE Healthcare, Milwaukee, Wisconsin) scanner (n = 21). Tumors shown on T2-weighted images were delineated by 2 senior neurosurgeons (J.W. and Z. Z., with 16 and 18 years of experience, respectively, in diagnosis by using brain MR imaging) who were blind to patient information. All lesion maps were re-evaluated by a neuroradiologist (S.W.L., with 24 years of experience). Tumor masks were combined as a new tumor mask in the analysis when <5% discrepancy was found. If ≥5% discrepancy was found, the neuroradiologist determined the tumor border. Images of each patient were then registered to a high-resolution (1.0-mm isotropic) brain atlas (Montreal Neurological Institute 152 [MNI152,]) by using a normalizing algorithm provided by SPM8 (http://www.fil.ion.ucl.ac.uk/spm/software/spm8).
Voxel-Based Lesion-Symptom Mapping
We first examined the overlap of the normalized tumor masked regions for the entire group and 2 subgroups of patients. Consequently, we performed VLSM analysis24 for the entire cohort of patients to determine brain regions that had a significant expression of the p53 mutant. A regression analysis relating p53 mutation to tumor intensity was fit at each voxel. Patients were then scored by the expression level of p53 mutants in their tumors. With age, sex, and pathology being regressed out, the voxels showing significant correlations of p53 mutants were identified, and the results were further corrected for multiple comparisons by using a permutation test (n = 500).26,27 The t value of the voxel that was greater than the t value in >95% of permutations was kept in the VLSM results (with an α set at .05; power, >.8).27
p53 Immunohistochemistry
Immunoperoxidase staining for p53 mutants was performed on formalin-fixed, paraffin-embedded tissue sections following the standard procedure introduced in our previous study.28 Briefly, 5-mm tissue sections were deparaffinized, rehydrated, and incubated with 1.0% hydrogen peroxide in tris-buffered saline to block endogenous peroxidase activity. Antigen unmasking was performed by heating samples in sodium citrate buffer (10-mmol/L sodium citrate, pH = 6.0) for 10 minutes at 100°C. Nonspecific protein binding was blocked by incubation in 5% horse serum in phosphate-buffered saline, and rabbit polyclonal p53 immune globulin G was then applied to the sections for 60 minutes at room temperature. Tumor sections were incubated in secondary antibody for 30 minutes at room temperature following washes in phosphate-buffered saline. Sections were washed and treated with Elite ABC (VECTASTAIN; Vector Laboratories, Burlingame, California), washed again, and then developed with 3,3′-diaminobenzidine tetrahydrochloride (50 mg 3,3′-diaminobenzidine in 100 mL phosphate-buffered saline and 150 mL of 3% hydrogen peroxide). After being rinsed in phosphate-buffered saline, slides were dehydrated in graded alcohols, cleared in xylene, and permanently covered. The immunohistochemical expressions of p53 protein were independently reviewed by 2 experienced pathologists and were then classified as the following: −, negative; +, isolated positive cells; ++, clusters of positive cells; and +++, mostly positive cells. The scales of positive cells were then used to score mutant p53 expression levels that were documented from 0 (−) to 3 (+++).
Survival Analysis
Progression-free survival time was considered to be the end point of this study, measured from the date of the first surgical resection to disease progression. After subtotal resection, any volume increase of ≥25% in the MR imaging was classified as tumor progression. Any tumor regrowth after gross total resection was classified as tumor recurrence.29 Survival differences between patients with p53 mutations and wild-type p53 were evaluated by using log-rank analysis applied to Kaplan-Meier curves. Consequently, the same analysis was also performed to test the difference in outcome between tumors associated with high-expressing regions of p53 mutations and tumors not associated with these expression patterns. Additional survival analyses in subdivided patient groups, defined by tumor location and p53 status, were performed to further evaluate the effect of p53 mutations on patient outcome. Among patients having p53 mutations, tumors associated with high-expressing regions of p53 mutations and tumors not associated with these expression patterns were input into a log-rank analysis. For tumors located in VLSM-defined regions, a survival analysis was performed on 2 groups that were determined by their p53 mutation status. Last, a multivariate Cox proportional hazard analysis was performed to determine whether factors including age, sex, history of seizures, preoperative Karnofsky Performance Status score, histopathology, and p53 mutation were predictors for PFS.
Results
Demographic Characteristics
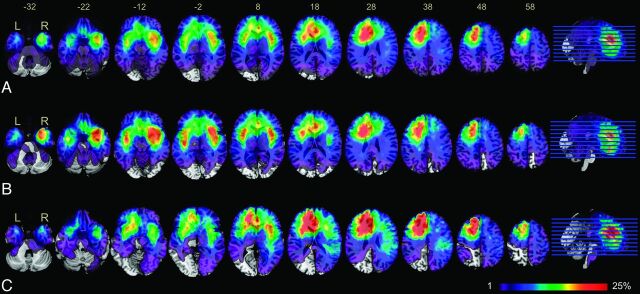
The overlap of all 182 tumors based on T2-weighted hyperintensity demonstrated 3 clusters of tumors that occurred with high frequency. Most interesting, hemispheric asymmetry was present on the overlap map (Fig 1A). Specifically, clusters in the right hemisphere were mainly located in the medial temporal lobe and insula, while most clusters in the left hemisphere were located in the frontal lobe, particularly at the subventricular zone surrounding the left anterior horn of the lateral ventricle. The asymmetric distribution of highly overlapping clusters implies that the occurrence and development of LGGs may have specific anatomic characteristics. LGGs with p53 mutations occurred more frequently in the bilateral temporal lobes and the insula, while LGGs with wild-type p53 demonstrated strong localization in the left frontal lobe (Fig 1B, -C).
Fig 1.
Overlap of tumor lesions. Overlap of all 182 patients with low-grade gliomas included in our study (A), overlap of a subgroup of 116 patients with mutant p53 gliomas (B), and overlap of a subgroup of 66 patients with wild-type p53 gliomas (C). The color range indicates the proportion of overlap of different cohorts, from violet (1 case) to red (>25% of cases overlap). Brain sections are displayed from z-coordinates −32 to +58 in the MNI space.
Voxel-Based p53 Mutation Mapping
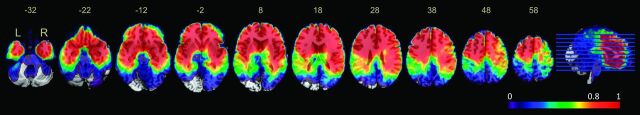
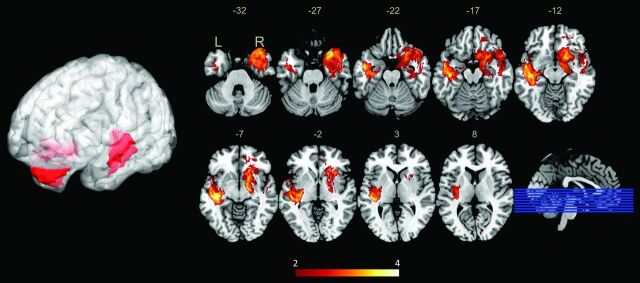
Note that overlaying the lesions yielded the distribution incidence of tumors, instead of truly specific anatomic correlates of the expression of mutant p53. To avoid this, we used VLSM analysis to show the statistically significant regions associated with specific genetic changes. Adequate voxels with high-power values (>0.8) were included in the analysis (Fig 2). Two clusters associated with high expression of mutant p53 were identified in each hemisphere following the permutation-based correction, with major expression being localized to the left medial temporal lobe and the right anterior temporal lobe (Fig 3). Overall, 78.7% (70/89) of cases were detected as having p53 mutations when their tumors involved the VLSM region; furthermore, 49.5% (46/93) of the cases showed expression of wild-type p53 when the tumor did not involve the VLSM region.
Fig 2.
Power map for voxel-based lesion-symptom mapping analysis. The color map on the MNI space shows the distribution of power, ranging from 0 (violet) to 1 (red), with α set to P < .05. Only voxels with high-power values (>0.8) were included in the VLSM analysis.
Fig 3.
Voxel-based lesion-symptom mapping–defined p53 high-mutant regions. VLSM analysis shows regions associated with high expression of mutant p53 in low-grade gliomas. The left-medial temporal lobe and right-anterior temporal lobes are significantly correlated to high mutant p53 expression. The color range indicates the level of t values from red to yellow (least to most significant). Only significant voxels are rendered on the basis of a critical threshold determined by permutation testing (n = 500, P < .05).
Survival Analysis
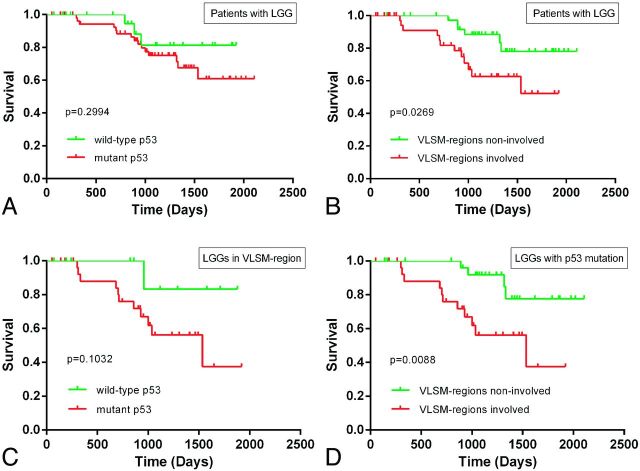
PFS information was available in 82 patients. PFS-based survival analysis showed no difference in patients with p53-mutated tumors compared with those with p53 wild-type tumors (P = .2994) (Fig 4A). In contrast, a significantly worse outcome (PFS) was observed in patients with tumors located in VLSM-defined regions compared with those with tumors outside these regions (P = .0269) (Fig 4B). To further investigate the role of p53 mutations in influencing PFS in VLSM-defined regions, we applied a survival analysis to the subdivided cohorts according to tumor location and p53 status. No significant relationship was observed among patients with tumors in VLSM-identified regions; however, a trending difference of PFS was observed among patients with wild-type and mutant p53 tumors (P = .1032) (Fig 4C). Most interesting, among p53-mutated cases, a shorter PFS was observed in patients with tumors located in VLSM-identified regions (P = .0088) compared with those with tumors outside these regions (Fig 4D). Finally, a multivariate Cox proportional hazard analysis showed that VLSM-region involvement (P = .010, hazard ratio = 3.909), age (40 years or older) (P = .015, hazard ratio = 3.400), and preoperative Karnofsky Performance Status < 80 (P = .024, hazard ratio = 9.222) were prognostic factors (On-line Table).
Fig 4.
Kaplan-Meier curves showing the PFS for patients with low-grade gliomas. A, No significant difference was observed between patients with wild-type p53 tumors and those with mutant p53 tumors (log-rank, P = .2994). B, A statistically significant difference in PFS was observed between patients with tumors in VLSM-identified regions and patients with tumors located outside VLSM-identified regions (log-rank, P = .0269). C, Among patients with tumors located in VLSM-identified regions, a trend toward a difference in PFS was observed between patients with wild-type p53 tumors and those with mutant p53 tumors (log-rank, P = .1032). D, Among patients with mutant p53 tumors, a significantly worse PFS was observed in patients with tumors located in VLSM-identified regions compared with patients with tumors outside these regions (log-rank, P = .0088).
Discussion
The current study demonstrated the voxelwise brain risk atlas of LGG occurrence. More important, 2 regions associated with high expression of mutant p53 were identified in each hemisphere by using VLSM analysis. Furthermore, we demonstrated that patients with tumors located in VLSM-identified regions had significantly worse outcomes (PFS) compared with those with tumors occurring elsewhere in the brain.
Anatomic Characteristics of Genetic Changes in Tumors
Previous studies have demonstrated a correlation between genetic changes and tumor location. For instance, tumors with isocitrate dehydrogenase 1 mutations have frequently been found in the frontal lobe.15 Additionally, the occurrences of MGMT promoter-methylated glioblastomas have been lateralized to the left hemisphere, while MGMT-unmethylated glioblastomas have been lateralized to the right hemisphere.16 Moreover, it has been shown that phosphatase and tensin homolog loss mostly occurs in the left frontal lobe23 and that allelic loss of chromosomal arms 1p and/or 19q is more common in the frontal, parietal, and occipital lobes.14 These molecular biomarkers have been long confirmed as playing an important role in the tumorigenesis of specific regions. In the current study, overlaying a map of tumors demonstrated that LGGs with wild-type p53 mostly occurred in the frontal lobes. In comparison, LGGs with mutant p53 were predominantly located in the bilateral temporal lobes and insula. This finding is consistent with a previous report that TP53 mutations frequently arise in the temporal lobe.17
VLSM analyses were then applied to statistically identify the anatomic correlation between tumor incidence and gene expression. In this study, we found that 2 specific brain regions were associated with high-expression levels of mutant p53, which has not been documented in previous LGG studies. Because p53 mutation is an early event in the molecular pathogenesis of LGGs, the strong propensity for p53-mutation gliomas to occur in the temporal lobes and insula indicated that p53 mutations may play a particularly important role in glioma tumorigenesis in these regions.11,20 TP53 mutation and loss of 1p and/or 19q were hypothesized to be mutually exclusive on the basis of previous molecular-epidemiologic evidence.4,17 In addition, allelic loss of chromosomal arms 1p and 19q has been shown to be common in most brain regions except for the temporal lobe.14 On the basis of neuroimaging analysis, the current study showed that LGGs with mutant p53s were predominantly located in the bilateral temporal lobes. Of note, the inverse localization between brain regions highly associated with p53 mutations and those associated with the loss of 1p and/or 19q provided new radio-epidemiologic insight to the current hypothesis.
Origin of Low-Grade Gliomas
The current hypothesis on tumor origin holds that gliomas may arise from neural stem cells that are mostly isolated from the subventricular zone, which lines the lateral ventricles; the dentate gyrus within the hippocampus; and potentially along deep white matter tracts.12,20,30 Our results, in some way, support the neural stem cell hypothesis. This finding is because of the 182 LGGs examined on T2-weighted images, most of the overlay of the tumor areas was located in the subventricular zone surrounding the left lateral ventricle, anterior horn, and hippocampus. Most important, these regions have long been proposed as a possible source of human gliomas, and a number of animal models also support this theory. Specifically, animals exposed to carcinogens and cancer-causing viruses demonstrate tumor formation within the subventricular zone, and these tumors quickly migrate into deep white matter regions, masking their original source.19
Another model by using the human glial fibrillary acidic protein promoter to undertake hGFAP-cre-mediated deletion of p53 and NF1 showed that early inactivation of p53, in addition to NF1 loss, induces glioblastoma development with the earliest identifiable area of a tumor being confined to the subventricular zone.31 Although neural stem cells are the source of initiation for gliomas, their progression toward a tumorigenic state may be regulated by multiple developmental signaling pathways. Notably, gene expression involved in these pathways is asymmetric.32 For instance, the SUFU gene in the sonic hedgehog pathway is overexpressed in the left hemisphere,33 and the activation of the sonic hedgehog pathway predisposes neural progenitor cells to glioma formation,34,35 which may lead to asymmetry in tumorigenesis. The current study identified the locations associated with high p53 mutation in both hemispheres, but with asymmetric distribution; these findings may serve as a new example of the asymmetric expression associated with genetic changes in brain tumors.
p53 Mutations, Tumor Location, and Survival Outcome
The prognostic or predictive role of p53 mutations has remained controversial. Inconsistent associations have been reported between the overexpression of mutant p53 and patient outcome or tumor response to therapy.13,29 A recent meta-analysis suggested that p53 expression is not a significant prognostic marker for human astrocytomas (P = .531, hazard ratio = 1.034).8 Most interesting, the same results were demonstrated in this study. There are 2 possible explanations for this. First, it could be that p53 status has no significant impact on survival prognosis. Second, p53 could interact with a multitude of other factors to influence prognosis.
In the current study, the shortest PFS was identified in patients with mutant p53 tumors localized to VLSM-identified regions when cohorts were subdivided according to tumor location and p53 status. This finding suggests that mutant p53 has a significant influence on the prognosis of tumors involving VLSM-identified regions rather than other regions. Survival benefits from region-specific gene expression in tumors have been reported in other molecular biomarkers. As previously discussed, tumors with a methylated MGMT promoter are associated with significantly longer survival periods when located in specific regions in the left hemisphere compared with tumors located in the right hemisphere that have an unmethylated MGMT promoter.16 Additionally, it has been shown that tumors with mutations in isocitrate dehydrogenase 1 in the frontal lobe are associated with a favorable prognosis.15 This study observed a significantly poor prognosis for patients with tumors located in VLSM-identified clusters, which suggests that the impact of p53 status on prognosis may also have regional specificity. We further infer that p53 mutations could play a major role in the tumorigenesis of gliomas arising from the temporal-insula area. Additionally, a multivariate survival analysis revealed that a preoperative Karnofsky Performance Status < 80, VLSM-region involvement, and age older than 40 years are significant prognostic factors for PFS in LGGs. On the basis of these findings, regions associated with a higher incidence of genetic changes (as can be identified by using neuroimaging) could potentially be used for customized molecular therapy and survival prediction.
In the current study, VLSM analysis24,36 was used to identify the anatomic specificity of lesion-related characteristics. Although entitled “lesion-symptom mapping,” the principle of this approach allowed us to investigate the anatomic correlates of any characteristic that could be associated with tumors (genetic changes in this case). A related method, entitled “analysis of differential involvement,” has demonstrated the practical role of voxel-based neuroimaging analysis in localizing tumor-associated molecular events. On the basis of a voxelwise Fisher exact test, analysis of differential involvement has been used to identify the anatomic correlates of tumor-specific biomarkers (such as MGMT methylation16 and isocitrate dehydrogenase 115) in the brain. The VLSM approach was used in the current study, however, because the regression model is more appropriate than the Fisher exact test when analyzing the graded data of gene expression.
Limitations
Several limitations to the current study should be considered. First, PFS was used as the end point of this study because patients with LGGs have a relatively long survival time; most of our patients are still alive. Nevertheless, patients who were not available for follow-up assessment may have influenced the integrity of our analyses. This may be the reason that no significant difference in PFS was observed between patients with mutant and wild-type p53 tumors when VLSM regions were involved. Additionally, clinical image registration of anatomically distorted brain structures into a 3D standard space can be challenging. Thus, every tumor mask was determined and manually checked by 2 independent neurosurgeons to minimize inaccuracy.
Conclusions
This analysis allowed us to identify anatomic correlations among genetic lesions on a voxel basis and levels of mutant p53 expression in a large cohort of patients with LGGs. In addition, we demonstrated that tumor location could be an independent prognostic factor for tumor progression and that tumors located in VLSM-defined regions associated with high expression of mutant p53 have generally worse PFS. Our study provides an example that molecular-related brain regions identified on structural MRI could be used in preoperative surgical planning and clinical survival predictions.
Supplementary Material
Acknowledgments
We would like to thank Yuling Yang for tissue-sample collection and clinical data retrieval and Drs J. Wang and Z. Zhang for their effort in tumor segmentation.
ABBREVIATIONS:
- LGG
low-grade glioma
- MGMT
O6-methylguanine DNA methyltransferase
- MNI
Montreal Neurological Institute
- PFS
progression-free survival
- VLSM
voxel-based lesion-symptom mapping
Footnotes
Disclosures: Lei Wang—UNRELATED: Grants/Grants Pending: Beijing Committee of Scientific Technology,* Comments: This grant is to support the research on biologic features and clinical treatment of gliomas. Tao Jiang—RELATED: Grants: China Ministry of Science and Technology,* Comments: This study was supported by funds from the National 973 Program (No.2011CB707804), and the National High Technology Research and Development Program (No. 2012AA02A508). *Money paid to the institution.
This work was supported by the National 973 Program No. 2011CB707804 (T.J.) and the National High Technology Research and Development Program No. 2012AA02A508 (T.J.).
References
- 1. Yang P, Wang Y, Peng X, et al. Management and survival rates in patients with glioma in China (2004–2010): a retrospective study from a single-institution. J Neurooncol 2013;113:259–66 [DOI] [PubMed] [Google Scholar]
- 2. Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009;360:765–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Youland RS, Brown PD, Giannini C, et al. Adult low-grade glioma: 19-year experience at a single institution. Am J Clin Oncol 2013;36:612–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hartmann C, Hentschel B, Tatagiba M, et al. Molecular markers in low-grade gliomas: predictive or prognostic? Clin Cancer Res 2011;17:4588–99 [DOI] [PubMed] [Google Scholar]
- 5. Weiler M, Wick W. Molecular predictors of outcome in low-grade glioma. Curr Opin Neurol 2012;25:767–73 [DOI] [PubMed] [Google Scholar]
- 6. Bourne TD, Schiff D. Update on molecular findings, management and outcome in low-grade gliomas. Nat Rev Neurol 2010;6:695–701 [DOI] [PubMed] [Google Scholar]
- 7. Okamoto Y, Di Patre PL, Burkhard C, et al. Population-based study on incidence, survival rates, and genetic alterations of low-grade diffuse astrocytomas and oligodendrogliomas. Acta Neuropathol 2004;108:49–56 [DOI] [PubMed] [Google Scholar]
- 8. Levidou G, El-Habr E, Saetta AA, et al. P53 immunoexpression as a prognostic marker for human astrocytomas: a meta-analysis and review of the literature. J Neurooncol 2010;100:363–71 [DOI] [PubMed] [Google Scholar]
- 9. Cho Y, Gorina S, Jeffrey PD, et al. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science 1994;265:346–55 [DOI] [PubMed] [Google Scholar]
- 10. England B, Huang T, Karsy M. Current understanding of the role and targeting of tumor suppressor p53 in glioblastoma multiforme. Tumour Biol 2013;34:2063–74 [DOI] [PubMed] [Google Scholar]
- 11. Muñoz DM, Tung T, Agnihotri S, et al. Loss of p53 cooperates with K-ras activation to induce glioma formation in a region-independent manner. Glia 2013;61:1862–72 [DOI] [PubMed] [Google Scholar]
- 12. Muñoz DM, Guha A. Mouse models to interrogate the implications of the differentiation status in the ontogeny of gliomas. Oncotarget 2011;2:590–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ständer M, Peraud A, Leroch B, et al. Prognostic impact of TP53 mutation status for adult patients with supratentorial World Health Organization grade II astrocytoma or oligoastrocytoma: a long-term analysis. Cancer 2004;101:1028–35 [DOI] [PubMed] [Google Scholar]
- 14. Zlatescu MC, TehraniYazdi A, Sasaki H, et al. Tumor location and growth pattern correlate with genetic signature in oligodendroglial neoplasms. Cancer Res 2001;61:6713–15 [PubMed] [Google Scholar]
- 15. Lai A, Kharbanda S, Pope WB, et al. Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin. J Clin Oncol 2011;29:4482–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ellingson BM, Cloughesy TF, Pope WB, et al. Anatomic localization of O6-methylguanine DNA methyltransferase (MGMT) promoter methylated and unmethylated tumors: a radiographic study in 358 de novo human glioblastomas. Neuroimage 2012;59:908–16 [DOI] [PubMed] [Google Scholar]
- 17. Mueller W, Hartmann C, Hoffmann A, et al. Genetic signature of oligoastrocytomas correlates with tumor location and denotes distinct molecular subsets. Am J Pathol 2002;161:313–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wechsler-Reya R, Scott MP. The developmental biology of brain tumors. Annu Rev Neurosci 2001;24:385–428 [DOI] [PubMed] [Google Scholar]
- 19. Sanai N, Alvarez-Buylla A, Berger MS. Neural stem cells and the origin of gliomas. N Engl J Med 2005;353:811–22 [DOI] [PubMed] [Google Scholar]
- 20. Jacques TS, Swales A, Brzozowski MJ, et al. Combinations of genetic mutations in the adult neural stem cell compartment determine brain tumour phenotypes. EMBO J 2010;29:222–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Simpson JR, Horton J, Scott C, et al. Influence of location and extent of surgical resection on survival of patients with glioblastoma multiforme: results of three consecutive Radiation Therapy Oncology Group (RTOG) clinical trials. Int J Radiat Oncol Biol Phys 1993;26:239–44 [DOI] [PubMed] [Google Scholar]
- 22. Barani IJ, Benedict SH, Lin PS. Neural stem cells: implications for the conventional radiotherapy of central nervous system malignancies. Int J Radiat Oncol Biol Phys 2007;68:324–33 [DOI] [PubMed] [Google Scholar]
- 23. Ellingson BM, Lai A, Harris RJ, et al. Probabilistic radiographic atlas of glioblastoma phenotypes. AJNR Am J Neuroradiol 2013;34:533–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bates E, Wilson SM, Saygin AP, et al. Voxel-based lesion-symptom mapping. Nat Neurosci 2003;6:448–50 [DOI] [PubMed] [Google Scholar]
- 25. Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455:1061–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Medina J, Kimberg DY, Chatterjee A, et al. Inappropriate usage of the Brunner-Munzel test in recent voxel-based lesion-symptom mapping studies. Neuropsychologia 2010;48:341–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kimberg DY, Coslett HB, Schwartz MF. Power in voxel-based lesion-symptom mapping. J Cogn Neurosci 2007;19:1067–80 [DOI] [PubMed] [Google Scholar]
- 28. Li S, Jiang T, Li G, et al. Impact of p53 status to response of temozolomide in low MGMT expression glioblastomas: preliminary results. Neurol Res 2008;30:567–70 [DOI] [PubMed] [Google Scholar]
- 29. Peraud A, Kreth FW, Wiestler OD, et al. Prognostic impact of TP53 mutations and P53 protein overexpression in supratentorial WHO grade II astrocytomas and oligoastrocytomas. Clin Cancer Res 2002;8:1117–24 [PubMed] [Google Scholar]
- 30. Li G, Fang L, Fernandez G, et al. The ventral hippocampus is the embryonic origin for adult neural stem cells in the dentate gyrus. Neuron 2013;78:658–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhu Y, Guignard F, Zhao D, et al. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell 2005;8:119–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sun T, Walsh CA. Molecular approaches to brain asymmetry and handedness. Nat Rev Neurosci 2006;7:655–62 [DOI] [PubMed] [Google Scholar]
- 33. Sun T, Patoine C, Abu-Khalil A, et al. Early asymmetry of gene transcription in embryonic human left and right cerebral cortex. Science 2005;308:1794–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pomeroy SL, Tamayo P, Gaasenbeek M, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature 2002;415:436–42 [DOI] [PubMed] [Google Scholar]
- 35. Hsieh A, Ellsworth R, Hsieh D. Hedgehog/GLI1 regulates IGF dependent malignant behaviors in glioma stem cells. J Cell Physiol 2011;226:1118–27 [DOI] [PubMed] [Google Scholar]
- 36. Nakagawa Y, Matsumura K, Iwasa M, et al. Single photon emission computed tomography and statistical parametric mapping analysis in cirrhotic patients with and without minimal hepatic encephalopathy. Ann Nucl Med 2004;18:123–29 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.