Abstract
Pseudomonas aeruginosa and Staphylococcus aureus are the two most prevalent bacteria species in the lungs of cystic fibrosis (CF) patients and are associated with poor clinical outcomes. Co-infection by the two species is a frequent situation that promotes their interaction. The ability of P. aeruginosa to outperform S. aureus has been widely described, and this competitive interaction was, for a long time, the only one considered. More recently, several studies have described that the two species are able to coexist. This change in relationship is linked to the evolution of bacterial strains in the lungs. This review attempts to decipher how bacterial adaptation to the CF environment can induce a change in the type of interaction and promote coexisting interaction between P. aeruginosa and S. aureus. The impact of coexistence on the establishment and maintenance of a chronic infection will also be presented, by considering the latest research on the subject.
Keywords: P. aeruginosa, S. aureus, interaction, evolution, cystic fibrosis
Introduction
Cystic fibrosis (CF) patients suffer from severe pulmonary infections. Pseudomonas aeruginosa is one of the most prevalent bacteria within CF lungs and is the main factor responsible for poor clinical outcomes due to the difficulty of eradicating it. Indeed, this bacterium has been shown to rapidly adapt to the CF lung environment and persist efficiently despite host immune responses and antibiotic treatments. This adaptation is mainly due to the accumulation of genetic mutations that alter the expression profiles and phenotypes of P. aeruginosa (Smith et al., 2006; Marvig et al., 2015a; Winstanley et al., 2016; La Rosa et al., 2019).
Another important feature of P. aeruginosa is its ability to interact with other CF microorganisms such as Aspergillus fumigatus, Candida albicans, and Staphylococcus aureus. S. aureus is also one of the most predominant pathogens in CF lungs and is detected simultaneously with P. aeruginosa in 20 to 50% of patients (Limoli et al., 2016; Briaud et al., 2020). P. aeruginosa can exhibit particularly aggressive behavior toward S. aureus (Michelsen et al., 2016). This antagonistic interaction was the only one observed between the two species for a long time. However, P. aeruginosa strains with more tolerant behavior toward S. aureus have recently been isolated from chronic CF infections (Baldan et al., 2014; Michelsen et al., 2016; Briaud et al., 2019, 2020; Pallett et al., 2019; Camus et al., 2020). The establishment of such coexisting interaction between the two species seems to arise from their evolution in the lung ecosystem.
In this review, we aim to clarify the relationship between the adaptation of P. aeruginosa and S. aureus to the CF environment and the evolution of their interactions. We will present the genetic and phenotypic evolutions of the two partners and their impacts. The characterization of this coexisting interaction status and how it can contribute to the establishment and maintenance of a chronic infection will also be assessed.
Adaptation to the CF Environment Reduces P. aeruginosa’s Anti-Staphylococcal Behavior
P. aeruginosa Evolves During the Establishment of Chronic Infection
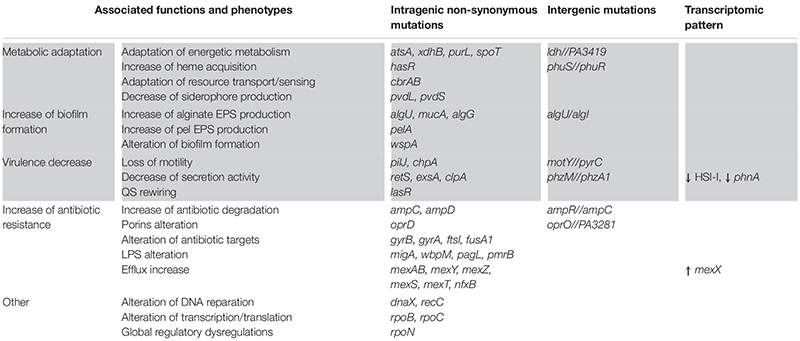
Lungs of CF patients constitute a stressful environment for colonizing microorganisms, especially due to toxic, osmotic, or oxidative stresses induced by the host immune system and recurring antibiotic treatments. Anoxia or micro-anaerobia, acidity and the nutritional characteristics of the CF environment can also hamper bacterial growth and persistence (Yang et al., 2011; La Rosa et al., 2019). Nevertheless, P. aeruginosa was shown to adapt in response to these different selective pressures. Numerous sequencing studies of longitudinal isolates of P. aeruginosa revealed that the bacterium accumulates a significant number of small mutations (SNP and small insertions and deletions) during its evolution in CF lungs. So, some genomic modification were found to be positively selected due to their beneficial impacts on P. aeruginosa fitness in the stressful CF environment (Smith et al., 2006; Marvig et al.,2015b,c; Klockgether et al., 2018; Khademi et al., 2019). There were called pathoadaptive modifications as they promote the pathogen survival and persistence at the infectious site. As a result, CF-adapted isolates of P. aeruginosa present convergent expression profiles and phenotypes, conferring advantageous features (Folkesson et al., 2012; Winstanley et al., 2016; La Rosa et al., 2019). Table 1 gathers the main pathoadaptive genomic regions of P. aeruginosa, as well as the associated phenotypes (Table 1).
TABLE 1.
Main genetic and phenotypic adaptations of Pseudomonas aeruginosa during its evolution in the CF lung environment.
|
Gray cells indicate adaptations that can impact microbial interactions of P. aeruginosa.
Firstly, CF-adapted strains of P. aeruginosa adjust their energetic metabolism to the particular nutritional composition of the CF environment (Palmer et al., 2007; La Rosa et al., 2018, 2019). This is mainly reflected by a restriction of the catabolic repertory and the slowed growth of clinical isolates in comparison to non-adapted strains (Marvig et al., 2015a; La Rosa et al., 2018, 2019). In response to iron sequestration by the host, P. aeruginosa also adapts its iron uptake mechanisms for the benefit of heme-based assimilation. Alteration of pvd genes in CF isolates was indeed shown to decrease the production of the pyoverdine siderophore and thus ferric iron acquisition. In contrast, mutations in the phuS//phuR intergenic region increase the expression of the heme receptor PhuR and thus acquisition of host-sequestered iron (Table 1; Marvig et al., 2014; Minandri et al., 2016). Iron and nutrient acquisition nonetheless remains difficult in CF lung, hampering the costly production of many iron-dependent virulence factors. P. aeruginosa virulence is also reduced by genetic alterations and/or transcriptomic dysregulations of genomic regions involved in motility (pilJ and chpA), secretion (phz and Hcp genes) and regulation of virulence (lasR, retS, exsA, and rpoN) (Marvig et al., 2015a; Winstanley et al., 2016). Most of these factors being immunogenic, the down-regulation of their production allows a better escape of P. aeruginosa from the host immune system. Such escape and resistance to immune responses is also promoted by the increased biofilm production of CF-adapted P. aeruginosa. In particular, mutations in mucA and alg genes induce the overproduction of alginate, an exopolysaccharide composing the biofilm matrix and responsible for the frequent mucoid phenotype (Marvig et al., 2015b; Winstanley et al., 2016). Finally, adapted strains of P. aeruginosa exhibit increased antibiotic resistance related to genic alterations of antibiotic targets (for instance the gyrases gyrA, gyrB), efflux pumps (mex genes), lactamases (amp genes), as well as lipopolysaccharide (LPS) and porin synthesis (pagL, pmrB, and opr genes) (Table 1; Folkesson et al., 2012; Marvig et al., 2015b; Winstanley et al., 2016). Altogether, these adaptations induce a weakly virulent but highly resistant state of P. aeruginosa, promoting its persistence within CF lungs.
Impact of P. aeruginosa Adaptation on Its Anti-staphylococcal Behavior
In addition to limiting the effectiveness of the immune response and antibiotic treatments, adaptations of P. aeruginosa can also modify its interactions with other microbial species, in particular with S. aureus. The anti-staphylococcal behavior of P. aeruginosa relies on mechanisms of bacterial lysis and growth suppression, as well as metabolic alterations, virulence and biofilm formation (Figure 1A; Hotterbeekx et al., 2017). However, several studies demonstrated that such behavior was not conserved in CF-adapted strains of P. aeruginosa, allowing the establishment of a coexisting interaction with S. aureus (Baldan et al., 2014; Michelsen et al., 2014, 2016; Limoli et al., 2017; Briaud et al., 2019). It thus appears that the relationship between P. aeruginosa and S. aureus evolves from competition to coexistence, a process resulting from the adaptation of P. aeruginosa.
FIGURE 1.
Evolution of bacterial interactions with Staphylococcus aureus related to Pseudomonas aeruginosa adaptation to CF environment. (A) Reference, environmental and acute infection P. aeruginosa isolates produce high amounts of virulence factors (LasA, HQNO, pyocyanin) or QS signals (3OC12-HSL). These factors allow P. aeruginosa to outcompete S. aureus by lysis mechanisms, growth, metabolism and virulence inhibition. Mixed-species biofilms can be formed through SpA-Psl binding, but can be disrupted by rhamnolipids and cis-2-decenoic acid. (B) P. aeruginosa isolates evolved in CF context coexist with S. aureus due to genetic and transcriptomic adaptations reducing the production of virulence and anti-staphylococcal factors. P. aeruginosa QS networks are rewired and regulated through the C4-HSL signal, that does not affect S. aureus physiology. CF-adapted isolates present a slowed and adjusted metabolism to the carbon sources of the CF environment and/or produced by S. aureus. Matrix of mixed-species biofilm are dominated by the alginate exopolysaccharide. (C) P. aeruginosa adaptation and its interaction with S. aureus are influenced by environmental factors that characterize in vivo conditions. Among them, antibiotic and oxidative stresses, depletion in oxygen, zinc and manganese, and high availability of nutrients and iron were shown to promote a coexisting interaction between the two pathogens. QS, quorum-sensing; SCV, small-colony variants; HQNO, 2-heptyl-4-hydroxyquinoline N-oxide; 3OC12-HSL, N-3-oxo-dodecanoyl homoserine lactone; C4-HSL, butyryl homoserine lactone.
Nutrient Competition and Virulence
One of the main factors involved in the competitive interaction between P. aeruginosa and S. aureus is that of resources, notably iron-availability in the environment. Indeed, it has been shown that P. aeruginosa’s aggressive behavior toward the Gram positive bacterium varies according to medium richness and is even promoted during iron depletion (Mashburn et al., 2005; Filkins et al., 2015; Nguyen et al., 2015; Miller et al., 2017). In these conditions, P. aeruginosa uses S. aureus as an iron source by lysing its cells thanks to the LasA protease, whose production is regulated by the quorum-sensing (QS) regulator LasR (Toder et al., 1991; Mashburn et al., 2005; Hotterbeekx et al., 2017). As P. aeruginosa’s adaptation to CF lungs is accompanied by lasR mutations and QS network rewiring, protease activity and especially LasA production are frequently reduced in chronic clinical isolates (Table 1, Figure 1B; Manos et al., 2013; Marvig et al., 2015a; Feltner et al., 2016; van Mansfeld et al., 2016; O’Brien et al., 2017). The ability of P. aeruginosa CF strains to lyse S. aureus cells was recently assessed on a limited set of three isolates. It was nonetheless depicted that the isolate lacking LasA production and protease activity was the only one unable to outcompete and lyse S. aureus (Pallett et al., 2019). Studies including larger sets of strains with different colonization times (as longitudinal ones) would be of great interest to evaluate the relationship between P. aeruginosa’s adaption to the CF lung environment and its ability to efficiently lyse S. aureus. In addition, S. aureus lysis leads to increased concentrations of N-acetyl glucosamine (GlcNac) which induces the production of virulence factors by P. aeruginosa (Hotterbeekx et al., 2017). Thus, the absence of S. aureus lysis may contribute to the decrease in virulence of P. aeruginosa.
As a strategy to limit nutritional competition, P. aeruginosa is also able to inhibit the growth of S. aureus by altering its metabolic activities. Indeed, the secondary metabolites HQNO (2-heptyl-4-hydroxyquinoline N-oxide) and pyocyanin secreted through the PQS (Pseudomonas Quinolone Signal) system induce a fermentative metabolism in the Gram positive bacterium (Hoffman et al., 2006; Proctor et al., 2014; Hotterbeekx et al., 2017; Noto et al., 2017). This metabolic switch is also responsible for the well-described small colony variant (SCV) phenotype of S. aureus (Proctor et al., 2014; Figure 1A). Genes involved in HQNO synthesis (phn genes) are under-expressed in adapted P. aeruginosa isolates, whereas pyocyanin production through phz genes can be impacted by intergenic mutations (Table 1). As a result, adapted strains of P. aeruginosa frequently present reduced HQNO and pyocyanin production in comparison to early ones (Bianconi et al., 2015; Cullen et al., 2015; Michelsen et al., 2016; Limoli et al., 2017; O’Brien et al., 2017; Figure 1B). This is especially the case for isolates from the DK2 lineage, which is known for developing a proto-cooperative interaction with S. aureus (Michelsen et al., 2016). However, HQNO and pyocyanin production are not always reduced in CF-adapted P. aeruginosa since these molecules can still be detected at active concentrations within CF sputum of chronically infected patients (Wilson et al., 1988; Hoffman et al., 2006; Alatraktchi et al., 2020). In fact, O’Brien et al. (2017) observed considerable variances in pyocyanin production between strains from different patients, but also between lineages recovered from a single subject. These results suggest that the isolates evolve differently according to their niches within the lungs, leading to different metabolic activities and thus interactions with co-colonizing microorganisms. Therefore, growth inhibition of S. aureus would vary depending on the ability of the nearest P. aeruginosa bacterium to produce pyocyanin or HQNO.
In the same way, the QS signal N-3-oxo-dodecanoyl homoserine lactone (3OC12-HSL) was shown to inhibit S. aureus growth in a dose-dependent manner (Hotterbeekx et al., 2017; Figure 1A). Production of this long-chain acyl-homoserine lactone (AHL) is regulated by LasR and thus frequently reduced in lasR-mutated CF isolates of P. aeruginosa. Instead, these isolates generally maintain the production of the short-chain AHL butyryl-HSL (C4-HSL) whose production is regulated by the second QS system of P. aeruginosa, Rhl (Bjarnsholt et al., 2010; Feltner et al., 2016; Chen et al., 2019; Cruz et al., 2020). And interestingly, C4-HSL signals have not been shown to affect S. aureus (Figure 1B).
Mixed-Species Biofilm Formation
Another important feature of bacterial interaction is the formation of mix-biofilm. The relationship between the two pathogens within biofilms appears to be a complex situation, highly dependent on isolates and culture conditions. A first study on 63 isolates demonstrated that S. aureus biofilm formation was enhanced by the secondary metabolites HQNO and Alkyl Quinolones (AQs) secreted by P. aeruginosa. However, although the effect was clear on a specific S. aureus clinical strain used as control, it was less manifest on other clinical CF isolates (Fugère et al., 2014). Conversely, S. aureus supernatant can either stimulate or inhibit P. aeruginosa biofilm formation in a strain-dependent manner as well (Armbruster et al., 2016). These effects involve the surface protein A (SpA) of S. aureus, an immunoglobulin-binding factor responsible for immune suppression (Kobayashi and DeLeo, 2013). Besides host proteins, SpA can bind two P. aeruginosa structures important for biofilm formation: (i) type IV pili, involved in twitching motility, and (ii) Psl, one of the three main exopolysaccharides (EPS), with alginate and Pel, that form the extracellular matrix of P. aeruginosa biofilm (Leid et al., 2005; Ryder et al., 2007; Colvin et al., 2011, 2012; Armbruster et al., 2016). Chew et al. (2018) observed that Psl enables wild-type P. aeruginosa to outcompete S. aureus in early biofilm development. On the contrary, Pel is required to reduce the effective crosslinking of the matrix in late-stage biofilm development, improving super-diffusivity in microcolony regions and dual-species biofilm growth (Chew et al., 2018).
Pseudomonas aeruginosa’s adaptation to the CF environment affects both motility and exopolysaccharide production. As a result, chronical isolates frequently lack type IV pili and present a biofilm matrix dominated by alginate, especially in the mucoid phenotype (Høiby et al., 2017). Therefore, one might wonder if the modulation of mixed-species biofilm formation through Pel, Psl, and SpA occur in adapted CF isolates in the same way as for reference strains (Figure 1B).
Competitive P. aeruginosa are also able to disperse S. aureus biofilm or limit its establishment through the secretion of rhamnolipids and cis-2-decenoic acid (Hotterbeekx et al., 2017; Figure 1A). To our knowledge, the production of cis-2-decenoic acid was not assessed in CF-adapted clinical P. aeruginosa, but rhamnolipid synthesis has been studied more extensively. These molecules induce significant inflammatory host responses, promote P. aeruginosa motility, and their synthesis is favored during planktonic growth (Déziel et al., 2003; Alhede et al., 2014; Hotterbeekx et al., 2017). As these phenotypes are frequently reduced in chronic P. aeruginosa isolates, it therefore seems likely that bacterial adaptation to the CF lung is accompanied by a decrease of rhamnolipid production. Indeed, chronic isolates studied by Bjarnsholt et al., and the CF-adapted lineage DK2 were shown to produce less rhamnolipids than intermittent and reference P. aeruginosa strains (Bjarnsholt et al., 2010; Michelsen et al., 2016). Low rhamnolipid synthesis was also recently associated with alginate overproduction and the common mucoid phenotype of P. aeruginosa. The presence of alginate, either produced by mucA-mutated mucoid strains or exogenously added in the medium, induced transcriptomic and post-transcriptomic down-regulation of rhamnolipid synthesis (Limoli et al., 2017; Price et al., 2020). Alginate-producing P. aeruginosa also presented reduced ability to outcompete S. aureus (Limoli et al., 2017; Price et al., 2020). Altogether, this suggests that decreased rhamnolipid production and increased alginate synthesis in adapted P. aeruginosa isolates contribute to improve S. aureus survival during mixed-biofilm formation (Figure 1B). In connection with this, Baldan et al. (2014) showed that early and late CF P. aeruginosa isolates presented different behaviors when grown in biofilm with S. aureus. This latter exhibited better viability with the CF-adapted isolate of P. aeruginosa and was also able to alter its biofilm production (Baldan et al., 2014).
Impact of Environmental Factors on the Anti-staphylococcal Behavior of P. aeruginosa
Although bacterial features play an important role in shaping inter-species interaction, environmental factors can have an even more decisive impact on them. Environmental conditions such as antibiotic and oxidant stresses are known to shape the CF ecosystem and promote a biofilm-based lifestyle of microorganisms within lungs (Feraco et al., 2016; Riquelme et al., 2020). Biofilm conditions reducing the competitive relationship between P. aeruginosa and S. aureus, the CF environment may favor a coexistence interaction between the two pathogens independently of P. aeruginosa’s genetic adaptation. Indeed, several environmental factors inherent to CF lungs were shown to decrease the anti-staphylococcal behavior of P. aeruginosa. Pallett et al. (2019) recently observed that oxygen limitation decreased the production of P. aeruginosa virulence factors such as proteases and pyocyanin. In these conditions, not only the reference strain PAO1 but also clinical isolates thus coexisted with S. aureus despite competitive behavior under normoxia (Pallett et al., 2019). Within mixed-species biofilm formed with the PA14 reference strain, S. aureus survival was improved thanks to bacterial stratification as a function of oxygen levels (Cendra et al., 2019). In addition to oxygen, nutrient availability can also modify interaction within biofilm (Mashburn et al., 2005; Filkins et al., 2015; Nguyen et al., 2015; Hotterbeekx et al., 2017; Miller et al., 2017; Cendra et al., 2019). Miller et al. (2017) observed that S. aureus was even able to strongly outcompete P. aeruginosa within mixed-species biofilm grown under rich conditions, but not in the same diluted and thus impoverished medium. Besides nutritive richness, limited alkalization of the co-culture medium was also shown to improve S. aureus survival during interaction with the PA14 reference strain (Cendra et al., 2019). Finally, the depletion of zinc and manganese, notably observed on the edge of the biofilm architecture, represses the expression of P. aeruginosa’s virulence genes and thus S. aureus inhibition (Wakeman et al., 2016; Figure 1C).
Interestingly, all these conditions promoting coexistence interaction between the two pathogens seem to be combined in the CF environment. CF sputum is indeed considered a rich medium but it contains limited oxygen concentrations, favoring anaerobic and micro-anaerobic metabolisms (La Rosa et al., 2019). Sputa from CF patients were also shown to be relatively acidic (Bhagirath et al., 2016). Moreover, host-pathogen interfaces are known to be depleted in zinc and manganese, especially through sequestration by the host protein calprotectin (Kehl-Fie and Skaar, 2010; Hood et al., 2012; Wakeman et al., 2016). Altogether, this suggests that in vivo conditions, and especially the CF environment, may promote non-competitive interaction between P. aeruginosa and S. aureus (Figure 1C). This hypothesis is supported by several studies showing the improved survival of S. aureus during non-human in vivo co-infections with P. aeruginosa, in comparison to in vitro co-cultures. Yadav and colleagues demonstrated a higher proportion of S. aureus within an in vivo mixed-species biofilm in the presence of P. aeruginosa (Yadav et al., 2017; Millette et al., 2019). In addition, P. aeruginosa strains such as PA14 were shown to promote S. aureus colonization and maintenance in a murine lung infection model (Yadav et al., 2017; Millette et al., 2019). Similar results were obtained in the context of chronic wound co-infections (Pastar et al., 2013; DeLeon et al., 2014). Since these co-infections were maintained for a limited duration (24 h to 1 week), it is unlikely that genetic adaptation drove the establishment of a coexistence interaction between P. aeruginosa and S. aureus. This phenomenon seems to be more related to non-fixed acclimatization reducing P. aeruginosa virulence, or spatial compartmentalization isolating the two species from each other in vivo. In all cases, it appears that besides promoting P. aeruginosa genetic adaptation and then reducing anti-staphylococcal behavior, in vivo conditions are themselves favorable to non-competitive interactions with S. aureus (Figure 1C).
S. aureus Adaptation to the CF Environment Can Also Impact Its Interaction With P. aeruginosa
The relationship between the two pathogens was shown to rely solely on P. aeruginosa anti-staphylococcal factors, whose production evolves during its adaptation and allows steady coexistence with S. aureus. However, although less studied, the adaptive mechanisms of S. aureus also promote its long-term persistence within CF lungs and may impact its interactions with co-colonizing P. aeruginosa strains.
S. aureus Adaptation to the CF Environment
A 21-month longitudinal study performed on 183 CF patients depicted that the clonal diversity of S. aureus strains decreased as patient aged, pointing out that some isolates tended to adapt more efficiently and dominate the airway sphere during infection history (Westphal et al., 2020). Several studies indeed highlighted that S. aureus isolates evolve in the CF lung environment and acquire features leading to better-fitted isolates (Figure 2; Chatterjee et al., 2008; Treffon et al., 2018, 2020; Herzog et al., 2019; Lennartz et al., 2019; Tan et al., 2019; Westphal et al., 2020).
FIGURE 2.
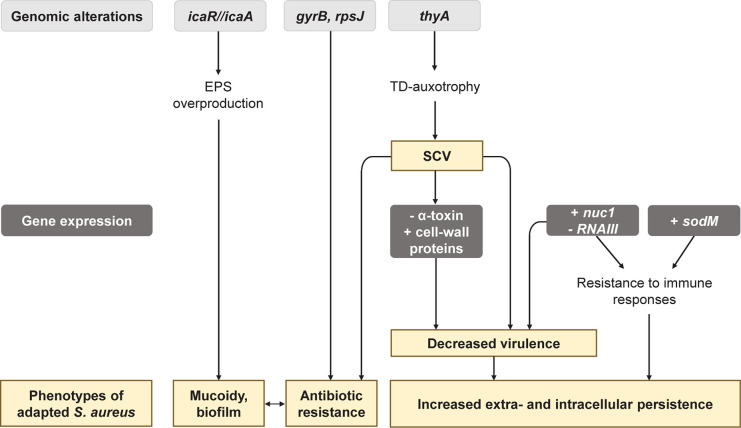
Main genetic and phenotypic adaptations of Staphylococcus aureus during its evolution in the CF lung environment. EPS, exopolysaccharide; TD, thymidine; SCV, small colony variants; NETs, Neutrophil Extracellular Traps.
Genomic, transcriptomic, and proteomic comparisons between early and late strains of S. aureus highlighted adaptations in the transport and the metabolism of carbohydrates and amino acids (Treffon et al., 2018; Tan et al., 2019). Late isolates thus often presented auxotrophies toward thymidine (TD), menadione or hemin (Kahl et al., 1998). These auxotrophies promote the formation of SCV (Chatterjee et al., 2008; Tan et al., 2019), a frequent S. aureus phenotype isolated from 4 to 50% of CF patients depending on the studies (Kahl et al., 2016). S. aureus SCVs are associated with worse lung function and patient age, in relation with their increased antibiotic resistance and persistence (Besier et al., 2007). Indeed, mutations in the thyA gene hindering TD biosynthesis were shown to facilitate resistance to trimethoprim-sulfamethoxazole (SXT) and the presence of TD-auxotrophic SCV is associated with previous antibiotic treatment with SXT (Kahl et al., 1998; Chatterjee et al., 2008; Yagci et al., 2013; Figure 2). Late isolates from antibiotic-treated patients showed other mutations in gyrB and rpsJ, that are not associated with the SCV phenotype but can explain antibiotic resistance to fluoroquinolones and cyclines (Kriegeskorte et al., 2015; Tan et al., 2019; Figure 2). Besides antibiotic resistance, SCVs are specialized for extracellular and intracellular persistence. This phenomenon is related to: (i) decreased expression of the α-toxin involved in lysis of eukaryotic cells by S. aureus, and (ii) increased expression of cell wall-associated genes which facilitate colonization to extracellular matrix proteins and internalization in eukaryotic cells (Figure 2). In line with this, S. aureus SCVs were shown to survive better intracellularly in eukaryotic cells than their isogenic parental strain (Kahl et al., 2016). CF-adapted S. aureus strains also tend to be better intracellular survivors than the corresponding early isolates (Treffon et al., 2020).
Another feature promoting the persistence of S. aureus CF isolates is the improvement of defense mechanisms against killing by polymorphonuclear leukocytes. Neutrophils usually eradicate invading pathogens by releasing reactive oxygen species (ROS) or forming Neutrophil Extracellular Trap (NETs). In comparison to their early counterparts, CF-adapted S. aureus presented an increased genic expression of the superoxide dismutase SodM involved in ROS detoxification (Treffon et al., 2018, 2020). Moreover, Herzog et al. (2019) showed that late isolates were more resistant to NETs mediated-killing through the over-expression of the nuc1 gene encoding the nuclease protein. Given the high pro-inflammatory territory in patient lungs overwhelmed by the neutrophilic attacks, the over-production of SodM and Nuc1 could be a major step in the adaptation of S. aureus in CF lungs (Figure 2).
Interestingly, S. aureus isolates overexpressing the nuc1 gene also presented a down-expression of the RNA regulator RNAIII. RNAIII is the major effector of the agr QS-system of S. aureus that positively controls the expression of many virulence factors. A decrease of RNAIII expression thus reflects reduced production of virulence factors. nuc1-overexpressing S. aureus were also shown to overexpress protein A (SpA), permease, coagulase, and adhesins (Jenul and Horswill, 2018), involved in colonization. Similarly, transcriptional analysis revealed that S. aureus SCVs have a less virulent phenotype in comparison to normal isolates (Kahl et al., 2005; Moisan et al., 2006; Seggewiss et al., 2006). Altogether, these results suggest that adaptation to the CF environment could lead toward a low-virulence but highly invasive state of S. aureus.
Finally, biofilm production appears to be increased in late S. aureus isolates. In the study of Tan et al. (2019), two out of three CF-late isolates over-produced biofilm and the same pattern was observed within late isolates of non-CF lung infections, suggesting that this characteristic was independent of the CF-lung environment. Such increased biofilm production by late isolates can be related to the development of mucoidy in S. aureus. This phenotype arises from a 5 bp deletion within the intergenic region of icaR/icaA genes, inducing an overproduction of S. aureus exopolysaccharide poly-N-acetylglucosamine (Schwartbeck et al., 2016; Figure 2). Mucoid S. aureus isolates are thus found in 2.5% of CF airways and tend to be more frequently isolated from older patients than non-mucoid isolates (Lennartz et al., 2019). These data suggest that mucoidy and strong biofilm production play a role in S. aureus adaptation to the CF environment.
Impact of S. aureus Adaptation on Its Interaction With P. aeruginosa
Some of the S. aureus adaptations to the CF environment can impact its relationship with P. aeruginosa. Among them, the SCV phenotype appears to be crucial for S. aureus survival during competitive interaction, as SCV present increased resistance to P. aeruginosa mediated-killing (Hotterbeekx et al., 2017). Once P. aeruginosa has evolved to a non-aggressive status toward S. aureus, the staphylococcus can then switch to a non-defective growth mode and coexist with its partner. The increased biofilm production of CF-adapted S. aureus isolates could also promote the formation of mixed-species biofilm with P. aeruginosa, although no direct correlation has yet been established. These hypotheses could explain (i) the high proportion of co-infected patients with S. aureus and P. aeruginosa in international cohorts (Hubert et al., 2013; Limoli et al., 2016; Briaud et al., 2020), and (ii) the high proportion of isolates presenting a non-competitive interaction (Briaud et al., 2020). Further investigations need to be conducted to confirm this model and fully understand the effects of S. aureus adaptation on its relationship with P. aeruginosa.
However, several studies suggested that S. aureus can influence the establishment of a coexisting interaction through alterations of the adaptive process of P. aeruginosa. Using an in vitro evolution assay, the adaptation of P. aeruginosa was studied in the presence or absence of S. aureus over 150 generations (Tognon et al., 2017). Mutations in the LPS biosynthesis pathway occurred only in the presence of S. aureus and increased P. aeruginosa’s fitness and resistance toward β-lactams antibiotics (Tognon et al., 2017). Repeated in vitro co-cultures with S. aureus also induced a decrease of P. aeruginosa QS regulation and may provide a departure point for a coexisting interaction (Zhao et al., 2018). It is worth recalling that modifications of LPS production, increased antibiotic resistance and down-regulation of QS are also frequently depicted in CF-adapted P. aeruginosa isolates (Folkesson et al., 2012; Marvig et al., 2015b; Winstanley et al., 2016). These results indicate that the presence of S. aureus influences P. aeruginosa’s adaptation, and that a co-evolution phenomenon could even promote a non-competitive relationship between the two pathogens. Several results obtained from CF clinical strains support these hypotheses. First, Martha et al. (2010) observed that P. aeruginosa presented a higher probability to develop a mucoid phenotype in the absence of S. aureus. Secondly, coexisting strains of S. aureus and P. aeruginosa were shown to better produce and catabolize acetoin, respectively, in comparison to competitive strains. The authors concluded that strains co-evolved to promote trophic cooperation (Camus et al., 2020). Finally, the genetic alterations of LasR that reduce P. aeruginosa anti-staphylococcal behavior are frequently observed in longitudinal studies (Marvig et al.,2015b,c). One might wonder if the presence of S. aureus in CF lungs could contribute to the selection of lasR mutants and thus non-competitive P. aeruginosa. Indeed, promoting a coexisting and even a cooperative behavior with other co-colonizing microorganisms can be a strategy to improve fitness in a stressful environment such as CF lungs. However, most of longitudinal studies focused on P. aeruginosa and no data are available concerning potential co-infections with S. aureus. It thus remains difficult to determine if S. aureus can effectively influence the fixation of lasR mutations in P. aeruginosa genome. It would be interesting to study the long-term evolution of both S. aureus and P. aeruginosa in several co-infected CF patients to reveal the bacterial adaptations leading to the establishment of coexistence.
Coexistence Promotes Cooperative Behaviors Between P. aeruginosa and S. aureus
Characterization of the Coexistence Interaction Status
Co-existence interaction leads to increased S. aureus survival in comparison to a common competitive interaction with P. aeruginosa. Bacterial enumerations performed during co-culture kinetics can thus easily reveal and quantify this phenomenon (Miller et al., 2017; Cendra et al., 2019; Pallett et al., 2019; Figure 3A). Less cumbersome procedures were nonetheless developed to assess interaction status between clinical CF isolates in large set of strains (Figure 3B; Baldan et al., 2014; Michelsen et al., 2014, 2016; Limoli et al., 2017; Briaud et al., 2019, 2020; Camus et al., 2020). Baldan and colleagues thus performed competition tests on plates during which a spot of P. aeruginosa culture is deposited on a lawn of S. aureus. Four categories of interaction status were established (no, weak, strong, and very strong inhibition) according to the size of the inhibition halo of S. aureus induced by the 24 P. aeruginosa strains tested (Baldan et al., 2014). In other studies, growth inhibition of S. aureus was observed through cross-streak assays and visual evaluation of each bacterium population after the streak intersection (Michelsen et al., 2014, 2016; Limoli et al., 2017; Figure 3C).
FIGURE 3.

Identification methods of the coexisting interaction between Pseudomonas aeruginosa and Staphylococcus aureus. (A) P. aeruginosa and S. aureus are cultivated in liquid mono- and co-culture in a rich medium during 8-h and S. aureus cells are counted in each condition. Competition is characterized by a rapid elimination of S. aureus in co-culture in comparison to the monoculture, whereas S. aureus growth is not affected during the whole kinetic in the case of a coexisting interaction (Briaud et al., 2019; Camus et al., 2020). (B) P. aeruginosa and S. aureus suspensions are prepared from overnight precultures in a rich medium and S. aureus lawn is uniformly plated on a rich agar medium. A P. aeruginosa spot is deposited in the center of the lawn and the plate is incubated 24 h. Competition is characterized by an inhibition halo of S. aureus growth in contact with the spot of P. aeruginosa, whereas S. aureus growth is not affected by P. aeruginosa during the coexisting interaction. Size of the inhibition halo can be measured (Baldan et al., 2014; Briaud et al., 2019, 2020; Camus et al., 2020). (C) P. aeruginosa streak is performed on a rich agar medium, after what a perpendicular S. aureus streak is added. The plate is incubated during 24 h. Competition is characterized by an inhibition of S. aureus growth in contact with the streak of P. aeruginosa and a low S. aureus proportion in the post-streak. Coexistence is characterized by a S. aureus growth in contact with the streak of P. aeruginosa and a high S. aureus proportion in the post-streak. Both of these parameters are visually determined (Michelsen et al., 2016; Minandri et al., 2016; Hotterbeekx et al., 2017). Arrows indicate when or where the competitive (red arrow) or coexisting (blue arrow) interaction is observed.
Despite the different methods and media employed, several studies highlighted that the interaction pattern with S. aureus was related to the adaptation of P. aeruginosa to the CF environment. Reduced anti-staphylococcal behavior was indeed observed for late isolates of P. aeruginosa, which were recovered several years after their clonal ancestor and presented common pathoadaptive traits. This evolution was thus shown in the DK2 lineage as well as in mucoid isolates (Michelsen et al., 2014, 2016; Limoli et al., 2017). However, it is noteworthy that the adaptation of P. aeruginosa in the CF lung is driven by adaptive radiation and spatial isolation mechanisms, inducing a high genotypic and phenotypic heterogeneity in the evolving population (Winstanley et al., 2016). It is therefore likely that clonally related isolates simultaneously recovered from the same patient can present different interaction statuses with S. aureus due to such diversification processes. On the other hand, it is also conceivable that the interaction statuses of competition and coexistence are dynamic and transitional within a single P. aeruginosa population. Assessing the interaction status of numerous P. aeruginosa isolates, collected longitudinally or recovered from different pulmonary niches, would lead to better understanding of this phenomenon.
In all cases, coexistence could be defined as a neutral interaction during which both bacteria are not affected by each other. However, increasing evidence shows that cooperative behaviors can positively affect P. aeruginosa and S. aureus during coexistence (Figure 4).
FIGURE 4.
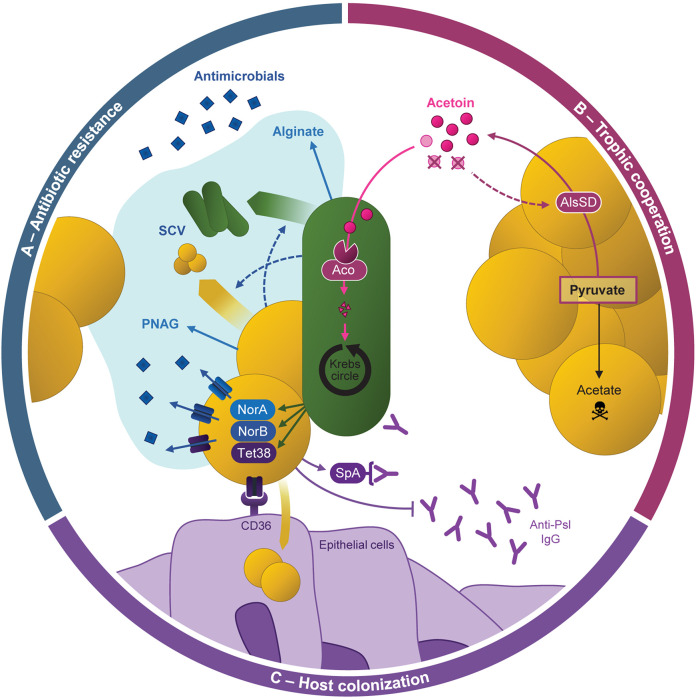
Cooperative behaviors between Pseudomonas aeruginosa and Staphylococcus aureus during coexistence. (A) Antibiotic resistance of both pathogens can be increased by the formation of mixed-species biofilm, especially through the production of alginate and poly-N-acetylglucosamine (PNAG) by P. aeruginosa and S. aureus, respectively. P. aeruginosa and S. aureus formation of small-colony variants (SCV) is also promoted during their interaction. P. aeruginosa presence was also shown to induce the over-expression of efflux pumps from the Nor family in S. aureus, enhancing its antibiotic resistance. (B) S. aureus produces acetoin from pyruvate thanks to AlsSD. P. aeruginosa catabolizes the acetoin produced by S. aureus and uses it as an alternative carbon source thanks to the aco system to feed the Krebs cycle. It improves its growth during co-culture with S. aureus. This catabolism also increases S. aureus survival potentially through a feed-back on acetoin production: the medium depletion in acetoin would promote AlsSD activity, limiting acetate production from pyruvate and thus cell acidification. (C) Resistance to immune system. P. aeruginosa-induced overexpression of tet38 improves S. aureus internalization in epithelial cells, allowing to hide from the immune system. S. aureus can limit P. aeruginosa-induced immune responses, notably through the binding of S. aureus protein A (SpA) to anti-P. aeruginosa antibodies (Anti-Psl IgG).
Increased Resistance to Antimicrobial Compounds
Several in vitro studies focusing on CF isolates highlighted that coexistence between P. aeruginosa and S. aureus can improve the antibiotic resistance of both pathogens (Figure 4A). S. aureus was recently shown to overexpress genes coding efflux pumps from the Nor family during coexisting interaction with P. aeruginosa. As a result, clinical S. aureus isolates presented significantly increased resistance to tetracycline and ciprofloxacin in the presence of P. aeruginosa, in comparison to monocultures (Briaud et al., 2019). While direct contact between bacterial cells was presumed to mediate this effect, other studies identified P. aeruginosa secreted products affecting the resistance of S. aureus to antimicrobial compounds. In particular, secondary metabolites from the 4-hydroxy-2-alkylquinolines (HAQ) family such as HQNO increase the resistance of S. aureus to aminoglycosides notably through the induction of the SCV phenotype i (Hoffman et al., 2006; Mitchell et al., 2010; DeLeon et al., 2014; Orazi and O’Toole, 2017; Orazi et al., 2019, 2020). Interestingly, late P. aeruginosa isolates from CF infections often lack HQNO production but are still able to protect S. aureus from tobramycin. The implication of other molecules than HQNO was confirmed by an atypical HAQ profile developed during adaptation to the CF environment (Michelsen et al., 2016). It thus appears that both competitive (i.e., HQNO-producing) and coexisting P. aeruginosa can protect S. aureus from antibiotic effects but through different mechanisms. Alginate produced by CF-adapted P. aeruginosa may also protect S. aureus from antimicrobial compounds since this exopolysaccharide can impact HAQ synthesis and mixed-species biofilm formation (Limoli et al., 2017; Price et al., 2020).
Conversely, interaction with S. aureus can also trigger resistance mechanisms in P. aeruginosa (Figure 4A). Co-evolution with S. aureus was shown to promote the alteration of genes from the LPS biosynthesis pathway in PAO1, leading to greater resistance to β-lactam antibiotics (Tognon et al., 2017). Michelsen and colleagues demonstrated that S. aureus cells and their supernatant can also induce the SCV phenotype and antibiotic tolerance in CF-adapted strains of P. aeruginosa (Fugère et al., 2014; Michelsen et al., 2014). Similar results were obtained using persistent CF isolates of P. aeruginosa grown in biofilm, as they presented improved tobramycin resistance upon exposure to S. aureus supernatant. In this case, the phenomenon was attributed to the formation of P. aeruginosa aggregates within the biofilm architecture (Beaudoin et al., 2017).
Development of Trophic Cooperation
Recent transcriptomic studies suggested that trophic cooperation could establish between S. aureus and P. aeruginosa (Figure 4B). The four-carbon molecule acetoin, produced by S. aureus, was shown to induce the overexpression of the aco system in CF P. aeruginosa strains during coexistence interaction (Camus et al., 2020). In line with this, P. aeruginosa presented an enhanced ability to catabolize acetoin produced by S. aureus as an alternative carbon source, resulting in increased survival during co-culture. Acetoin catabolism was also shown to benefit S. aureus and improve its survival in co-culture. Acetoin thus appears to be the keystone of trophic cooperation between P. aeruginosa and S. aureus during which both partners are beneficiaries. Interestingly, this cooperative behavior seems to be selected during bacterial evolution in the CF environment. Acetoin production by S. aureus and catabolism by P. aeruginosa were indeed more efficient for coexisting isolates in comparison to competitive ones (Camus et al., 2020). As acetoin could be detected in CF sputa, these results go along with the trophic adaptation of P. aeruginosa strains to resources present in the CF environment (Španěl et al., 2016; La Rosa et al., 2018, 2019; Camus et al., 2020). Other studies suggest that lactate, also detected in CF sputa, may also play a role in metabolic interactions between P. aeruginosa and S. aureus (Filkins et al., 2015; Tognon et al., 2017; La Rosa et al., 2019). Co-culture with P. aeruginosa was shown to induce the overexpression of genes involved in glucose fermentation and lactate production in S. aureus, leading to lactate accumulation in the medium. In turn, P. aeruginosa overexpressed genes responsible for lactate utilization and was able to use the molecule as a carbon source (Filkins et al., 2015; Tognon et al., 2017). However, this phenomenon was observed during a competitive interaction (using PAO1 or PA14 strains) and the impacts of lactate catabolism on both pathogens remain unknown.
Modification of Host-Bacterium Interaction
Cooperative behaviors such as protection against antimicrobials and trophic cooperation probably contribute to the establishment and maintenance of P. aeruginosa and S. aureus co-infections. In addition, interactions between the two pathogens were shown to affect their abilities to colonize host and settle as chronic infections (Figure 4C). Adhesion to host components is an important feature for colonization, and S. aureus can attach more efficiently to abiotic surfaces during co-culture with P. aeruginosa (Kumar and Ting, 2015). In these conditions, S. aureus presented an up-regulation of several proteins involved in adhesion to platelets or to the extracellular matrixes of various hosts, such as serine rich glycoproteins and the Ebh protein. These results suggest that co-culture may increase S. aureus’s adhesion to host cells (Kumar and Ting, 2015). In line with this, co-culture with P. aeruginosa CF strains was shown to induce the overexpression of nine virulence factors of S. aureus. Among them, the Tet38 transporter promoted the internalization of S. aureus within epithelial pulmonary cells during coexistence with P. aeruginosa (Briaud et al., 2019). Finally, HQNO-mediated induction of the SCV phenotype can increase the intracellular survival of S. aureus (Mitchell et al., 2010). Cell internalization and the SCV phenotype, both promoted by P. aeruginosa, could thus contribute to the success of S. aureus infection.
Limiting the induction and efficiency of immune responses is a strategy already developed by P. aeruginosa during its genetic adaptation to the CF environment. Interestingly, P. aeruginosa-S. aureus co-infection induces a specific host response different from mono-infections in a rat otitis model (Yadav et al., 2017). Using bacterial supernatants, Chekabab et al. (2015) observed that molecules secreted by S. aureus decreased the hosts’ immune response induced by P. aeruginosa supernatant. Thus, the protein SpA produced by S. aureus was shown to bind either to P. aeruginosa Psl exopolysaccharide or to anti-Psl IgG antibodies, protecting P. aeruginosa from Psl recognition by neutrophils and thus phagocytosis (Armbruster et al., 2016; Figure 4C). In addition, S. aureus presence induces transcriptomic down-regulation of several antigenic factors in P. aeruginosa, such as genes involved in secretion and flagellum synthesis (Miller et al., 2017).
Consequently, several studies suggest that P. aeruginosa and S. aureus co-infection favors chronic infections. Cigana et al. (2018) indeed observed that preliminary colonization by S. aureus increased the ability of PA14 and CF-adapted strains to establish a chronic infection in a murine model. Interestingly, such infection kinetics are frequent in CF patients as S. aureus is one of the first colonizers in the lungs of young children, whereas P. aeruginosa infection occurs upon adolescence (French Cystic Fibrosis Register, 2017). On another note, S. aureus-P. aeruginosa co-infections were shown to delay chronic wound healing and thus bacterial clearing (Pastar et al., 2013). Finally, Zhao et al. (2018) observed that mice presented an improved survival when their lungs were infected by a mix of P. aeruginosa, S. aureus and Klebsiella pneumoniae, in comparison to mice infected only with P. aeruginosa. Although this effect cannot be specifically attributed to the presence of S. aureus and/or K. pneumoniae, co-infection appears to reduce the host mortality of P. aeruginosa infections and thus promote longer infections. Altogether, these studies suggest that co-infection favors the establishment of chronicity, since the concomitant presence of pathogens will improve their persistence within the infection site (Limoli and Hoffman, 2019).
P. aeruginosa and S. aureus Are Not Alone
So far, the adaptive process and interactions of P. aeruginosa and S. aureus remain those most documented, as their infections are the most prevalent and severe in the context of CF (Machan et al., 1992; Mashburn et al., 2005; Hotterbeekx et al., 2017). However, numerous other microorganisms can colonize the CF environment and are thus susceptible to interact with P. aeruginosa (French Cystic Fibrosis Register, 2017). Among them, the fungus Aspergillus fumigatus is involved in a nutritional competition with P. aeruginosa. In this context, growth, biofilm formation and hyphal structure of the fungus are altered by P. aeruginosa secreted factors such as rhamnolipids, phenazines and the QS signals 3OC12-HSL and PQS (O’Brien and Fothergill, 2017; Briard et al., 2019; Sass et al., 2019; Chatterjee et al., 2020). As P. aeruginosa’s adaptation to the CF lung environment affects the secretion of these factors, it is likely that its relationship with A. fumigatus also evolves toward a coexistence-like interaction allowing the establishment of stable P. aeruginosa-A. fumigatus co-infections. Although no direct evidence has been established yet, this hypothesis is supported by the reduced antifungal activity shown for mucoid P. aeruginosa isolates (Briard et al., 2019; Chatterjee et al., 2020). Moreover, P. aeruginosa infection rather precedes A. fumigatus colonization and can even promote aspergillosis (O’Brien and Fothergill, 2017; Briard et al., 2019; Chatterjee et al., 2020). The aggressive behavior of P. aeruginosa is also observed toward the yeast C. albicans and the bacteria from the Burkholderia spp (Fourie et al., 2016; O’Brien and Fothergill, 2017; Fourie and Pohl, 2019; Limoli et al., 2019). However, it appears that these microorganisms are not powerless in front of P. aeruginosa and can even take advantage of this interaction within mixed-species biofilm. Antimicrobial resistance and virulence of C. albicans and P. aeruginosa are thus enhanced during their interaction, particularly through an increased production of phenazines by P. aeruginosa and ethanol by C. albicans (Fourie et al., 2016; O’Brien and Fothergill, 2017; Fourie and Pohl, 2019; Alam et al., 2020; Bandara et al., 2020; Hùlková et al., 2020; Ibberson and Whiteley, 2020). In the same way, P. aeruginosa alginate can protect B. cenocepacia from the host immune system (O’Brien and Fothergill, 2017).
If microbial adaptation to the CF environment favors such win-win interactions is yet to be determined. In particular, one might wonder if the increased virulence within mixed-species biofilm with C. albicans is a conserved response in CF-adapted P. aeruginosa, as the bacterium preferentially evolves toward an avirulent lifestyle (Bianconi et al., 2015; Marvig et al., 2015a; Riquelme et al., 2020). One another note, and as described earlier for S. aureus, P. aeruginosa is not alone in evolving. Notably, modifications in LPS and siderophore production, as well as a decrease of mucoidy, virulence and biofilm formation, were demonstrated during B. cenocepacia’s adaption to the CF environment (Cullen et al., 2015; Maldonado et al., 2016). These alterations are susceptible to affect its interactions with P. aeruginosa, and with other co-colonizing microorganisms.
Concluding Remarks and Perspectives
In addition to the persistence features of microorganisms, their interactions play a key role in their survival within infectious sites. Although poorly described, this “social” aspect is particularly essential in CF lung infections as they gather significant microorganism richness and densities. P. aeruginosa’s adaptation to the CF environment appears to drastically impact its microbial interactions, allowing the development of neutralist and even cooperative behaviors with co-colonizing microorganisms such as S. aureus or C. albicans. This raises questions about the impacts of these microorganisms on P. aeruginosa’s adaptation to the CF lung environment: can their presence constitute a selective force and promote the establishment of cooperative interactions?
Such cooperation is indeed an attractive strategy to promote microbial persistence within lungs: (i) it is less costly for both partners as the production of virulence or resistance factors is no longer required, and (ii) it can provide advantageous features to both partners of the interaction. CF-adapted strains of P. aeruginosa and S. aureus can thus benefit from reciprocal protection against antibiotics and metabolic cooperation, despite strong competition in vitro and/or between non-adapted strains.
Ultimately, these cooperative interactions could contribute to the establishment of “Climax communities,” i.e., microbial communities with a steady structure within the CF ecosystem (Quinn et al., 2016). Interestingly, S. aureus and P. aeruginosa are part of one of these Climax communities in vivo, suggesting that their interaction may stabilize and maintain co-infection by these two pathogens. This phenomenon could explain the high proportion of P. aeruginosa-S. aureus co-infected patients within CF cohorts (Limoli et al., 2016; Briaud et al., 2020), and more broadly the positive or negative associations observed between different pathogens (Granchelli et al., 2018). Nevertheless, the impacts of P. aeruginosa and S. aureus co-infections on clinical outcomes remain unclear and poorly described (Limoli et al., 2016; Briaud et al., 2020). Taking into account the nature of their interaction might unveil new aspects of their pathogenesis and their ability to durably persist within CF lungs.
Author Contributions
KM and LC were primarily responsible for preparing the review. FV and PB contributed to writing the review and editing the final version. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Anne Doléans-Jordheim and Rachel Zapf for proofreading and advice.
Footnotes
Funding. This work was supported by the Fondation pour la Recherche Médicale, grant number ECO20170637499 to LC, Finovi Foundation to KM, and associations “Vaincre la Mucoviscidose” and “Gregory Lemarchal” to KM.
References
- Alam F., Catlow D., Di Maio A., Blair J. M. A., Hall R. A. (2020). Candida albicans enhances meropenem tolerance of Pseudomonas aeruginosa in a dual-species biofilm. J. Antimicrob. Chemother. 75 925–935. 10.1093/jac/dkz514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alatraktchi F. A., Dimaki M., Støvring N., Johansen H. K., Molin S., Svendsen W. E. (2020). Nanograss sensor for selective detection of Pseudomonas aeruginosa by pyocyanin identification in airway samples. Anal. Biochem. 593 113586. 10.1016/j.ab.2020.113586 [DOI] [PubMed] [Google Scholar]
- Alhede M., Bjarnsholt T., Givskov M., Alhede M. (2014). Pseudomonas aeruginosa biofilms: mechanisms of immune evasion. Adv. Appl. Microbiol. 86 1–40. [DOI] [PubMed] [Google Scholar]
- Armbruster C. R., Wolter D. J., Mishra M., Hayden H. S., Radey M. C., Merrihew G., et al. (2016). Staphylococcus aureus protein a mediates interspecies interactions at the cell surface of Pseudomonas aeruginosa. mBio 7:e00538-16. 10.1128/mBio.00538-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldan R., Cigana C., Testa F., Bianconi I., De Simone M., Pellin D., et al. (2014). Adaptation of Pseudomonas aeruginosa in cystic fibrosis airways influences virulence of Staphylococcus aureus in vitro and murine models of co-infection. 3. PLoS One 9:e89614. 10.1371/journal.pone.0089614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandara H. M. H. N., Wood D. L. A., Vanwonterghem I., Hugenholtz P., Cheung B. P. K., Samaranayake L. P. (2020). Fluconazole resistance in Candida albicans is induced by Pseudomonas aeruginosa quorum sensing. Sci. Rep. 10:7769. 10.1038/s41598-020-64761-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin T., Yau Y. C. W., Stapleton P. J., Gong Y., Wang P. W., Guttman D. S., et al. (2017). Staphylococcus aureus interaction with Pseudomonas aeruginosa biofilm enhances tobramycin resistance. NPJ Biofilms Microbiomes 3:25. 10.1038/s41522-017-0035-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besier S., Ludwig A., Ohlsen K., Brade V., Wichelhaus T. A. (2007). Molecular analysis of the thymidine-auxotrophic small colony variant phenotype of Staphylococcus aureus. Int. J. Med. Microbiol. 297 217–225. 10.1016/j.ijmm.2007.02.003 [DOI] [PubMed] [Google Scholar]
- Bhagirath A. Y., Li Y., Somayajula D., Dadashi M., Badr S., Duan K. (2016). Cystic fibrosis lung environment and Pseudomonas aeruginosa infection. BMC Pulm Med. 16:174. 10.1186/s12890-016-0339-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianconi I., Jeukens J., Freschi L., Alcalá-Franco B., Facchini M., Boyle B., et al. (2015). Comparative genomics and biological characterization of sequential Pseudomonas aeruginosa isolates from persistent airways infection. BMC Genom. 16:1105. 10.1186/s12864-015-2276-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnsholt T., Jensen P. Ø, Jakobsen T. H., Phipps R., Nielsen A. K., Rybtke M. T., et al. (2010). Quorum sensing and virulence of Pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PLoS One 5:e10115. 10.1371/journal.pone.0010115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briard B., Mislin G. L. A., Latgé J.-P., Beauvais A. (2019). Interactions between Aspergillus fumigatus and pulmonary bacteria: current state of the field, new data, and future perspective. J. Fungi (Basel) 5:48. 10.3390/jof5020048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briaud P., Bastien S., Camus L., Boyadijian M., Reix P., Mainguy C., et al. (2020). Impact of coexistence phenotype between Staphylococcus aureus and Pseudomonas aeruginosa isolates on clinical outcomes among Cystic Fibrosis patients. Front. Cell Infect. Microbiol. 10:266. 10.3389/fcimb.2020.00266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briaud P., Camus L., Bastien S., Doléans-Jordheim A., Vandenesch F., Moreau K. (2019). Coexistence with Pseudomonas aeruginosa alters Staphylococcus aureus transcriptome, antibiotic resistance and internalization into epithelial cells. Sci. Rep. 9:16564. 10.1038/s41598-019-52975-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus L., Briaud P., Bastien S., Elsen S., Doléans-Jordheim A., Vandenesch F., et al. (2020). Trophic cooperation promotes bacterial survival of Staphylococcus aureus and Pseudomonas aeruginosa. ISME J. 14 3093–3105. 10.1038/s41396-020-00741-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cendra M. D. M., Blanco-Cabra N., Pedraz L., Torrents E. (2019). Optimal environmental and culture conditions allow the in vitro coexistence of Pseudomonas aeruginosa and Staphylococcus aureus in stable biofilms. Sci. Rep. 9:16284. 10.1038/s41598-019-52726-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee I., Kriegeskorte A., Fischer A., Deiwick S., Theimann N., Proctor R. A., et al. (2008). In vivo mutations of thymidylate synthase (encoded by thyA) are responsible for thymidine dependency in clinical small-colony variants of Staphylococcus aureus. J. Bacteriol. 190 834–842. 10.1128/jb.00912-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee P., Sass G., Swietnicki W., Stevens D. A. (2020). Review of potential pseudomonas weaponry, relevant to the pseudomonas-aspergillus interplay, for the mycology community. J. Fungi (Basel.) 6:81. 10.3390/jof6020081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekabab S. M., Silverman R. J., Lafayette S. L., Luo Y., Rousseau S., Nguyen D. (2015). Staphylococcus aureus inhibits IL-8 responses induced by Pseudomonas aeruginosa in airway epithelial cells. PLoS One 10:e0137753. 10.1371/journal.pone.0137753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Déziel E., Groleau M.-C., Schaefer A. L., Greenberg E. P. (2019). Social cheating in a Pseudomonas aeruginosa quorum-sensing variant. Proc. Natl. Acad. Sci. U.S.A. 116 7021–7026. 10.1073/pnas.1819801116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew S. C., Yam J. K. H., Matysik A., Seng Z. J., Klebensberger J., Givskov M., et al. (2018). Matrix polysaccharides and SiaD diguanylate cyclase alter community structure and competitiveness of pseudomonas aeruginosa during dual-species biofilm development with Staphylococcus aureus. mBio 9:e00585-18. 10.1128/mBio.00585-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigana C., Bianconi I., Baldan R., De Simone M., Riva C., Sipione B., et al. (2018). Staphylococcus aureus impacts Pseudomonas aeruginosa Chronic respiratory disease in murine models. J. Infect. Dis. 217 933–942. 10.1093/infdis/jix621 [DOI] [PubMed] [Google Scholar]
- Colvin K. M., Gordon V. D., Murakami K., Borlee B. R., Wozniak D. J., Wong G. C. L., et al. (2011). The pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog 7:e1001264. 10.1371/journal.ppat.1001264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin K. M., Irie Y., Tart C. S., Urbano R., Whitney J. C., Ryder C., et al. (2012). The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 14 1913–1928. 10.1111/j.1462-2920.2011.02657.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz R. L., Asfahl K. L., Bossche SV den, Coenye T., Crabbé A., Dandekar A. A. (2020). RhlR-regulated Acyl-homoserine lactone quorum sensing in a cystic fibrosis isolate of Pseudomonas aeruginosa. mBio 11:e00532-20. 10.1128/mBio.00532-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen L., Weiser R., Olszak T., Maldonado R. F., Moreira A. S., Slachmuylders L., et al. (2015). Phenotypic characterization of an international Pseudomonas aeruginosa reference panel: strains of cystic fibrosis (CF) origin show less in vivo virulence than non-CF strains. Microbiology (Reading Engl.) 161 1961–1977. 10.1099/mic.0.000155 [DOI] [PubMed] [Google Scholar]
- DeLeon S., Clinton A., Fowler H., Everett J., Horswill A. R., Rumbaugh K. P. (2014). Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect. Immun. 82 4718–4728. 10.1128/IAI.02198-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déziel E., Lépine F., Milot S., Villemur R. (2003). rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology (Reading Engl.) 149 2005–2013. 10.1099/mic.0.26154-0 [DOI] [PubMed] [Google Scholar]
- Feltner J. B., Wolter D. J., Pope C. E., Groleau M.-C., Smalley N. E., Greenberg E. P., et al. (2016). LasR variant cystic fibrosis isolates reveal an adaptable quorum-sensing hierarchy in Pseudomonas aeruginosa. mBio 7:e01513-16. 10.1128/mBio.01513-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feraco D., Blaha M., Khan S., Green J. M., Plotkin B. J. (2016). Host environmental signals and effects on biofilm formation. Microb. Pathog. 99 253–263. 10.1016/j.micpath.2016.08.015 [DOI] [PubMed] [Google Scholar]
- Filkins L. M., Graber J. A., Olson D. G., Dolben E. L., Lynd L. R., Bhuju S., et al. (2015). Coculture of Staphylococcus aureus with Pseudomonas aeruginosa Drives S. aureus towards fermentative metabolism and reduced viability in a cystic fibrosis model. J. Bacteriol. 197 2252–2264. 10.1128/jb.00059-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkesson A., Jelsbak L., Yang L., Johansen H. K., Ciofu O., Høiby N., et al. (2012). Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. 12. Nat. Rev. Microbiol. 10 841–851. 10.1038/nrmicro2907 [DOI] [PubMed] [Google Scholar]
- Fourie R., Ells R., Swart C. W., Sebolai O. M., Albertyn J., Pohl C. H. (2016). Candida albicans and Pseudomonas aeruginosa Interaction, with Focus on the Role of Eicosanoids. Front. Physiol. 7:64. 10.3389/fphys.2016.00064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourie R., Pohl C. H. (2019). Beyond antagonism: the interaction between candida species and Pseudomonas aeruginosa. J. Fungi. (Basel) 5:34. 10.3390/jof5020034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French Cystic Fibrosis Register (2017). Available online at: https://www.vaincrelamuco.org/2019/05/17/statistiques-et-publications-204 (accessed February 18, 2021). [Google Scholar]
- Fugère A., Lalonde Séguin D., Mitchell G., Déziel E., Dekimpe V., Cantin A. M., et al. (2014). Interspecific small molecule interactions between clinical isolates of Pseudomonas aeruginosa and Staphylococcus aureus from adult cystic fibrosis patients. PLoS One 9:e86705. 10.1371/journal.pone.0086705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granchelli A. M., Adler F. R., Keogh R. H., Kartsonaki C., Cox D. R., Liou T. G. (2018). Microbial interactions in the cystic fibrosis airway. J. Clin. Microbiol. 56:e00354-18. 10.1128/JCM.00354-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog S., Dach F., de Buhr N., Niemann S., Schlagowski J., Chaves-Moreno D., et al. (2019). High nuclease activity of long persisting Staphylococcus aureus isolates within the airways of cystic fibrosis patients protects against NET-mediated killing. Front. Immunol. 10:2552. 10.3389/fimmu.2019.02552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman L. R., Déziel E., D’Argenio D. A., Lépine F., Emerson J., McNamara S., et al. (2006). Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. 52. Proc. Natl. Acad. Sci. U.S.A. 103 19890–19895. 10.1073/pnas.0606756104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høiby N., Bjarnsholt T., Moser C., Jensen P. Ø, Kolpen M., Qvist T., et al. (2017). Diagnosis of biofilm infections in cystic fibrosis patients. APMIS 125 339–343. 10.1111/apm.12689 [DOI] [PubMed] [Google Scholar]
- Hood M. I., Mortensen B. L., Moore J. L., Zhang Y., Kehl-Fie T. E., Sugitani N., et al. (2012). Identification of an Acinetobacter baumannii zinc acquisition system that facilitates resistance to calprotectin-mediated zinc sequestration. PLoS Pathog. 8:e1003068. 10.1371/journal.ppat.1003068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotterbeekx A., Kumar-Singh S., Goossens H., Malhotra-Kumar S. (2017). In vivo and In vitro interactions between Pseudomonas aeruginosa and Staphylococcus spp. Front. Cell. Infect. Microbiol. 7:106. 10.3389/fcimb.2017.00106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert D., Réglier-Poupet H., Sermet-Gaudelus I., Ferroni A., Le Bourgeois M., Burgel P.-R., et al. (2013). Association between Staphylococcus aureus alone or combined with Pseudomonas aeruginosa and the clinical condition of patients with cystic fibrosis. J. Cyst. Fibros. 12 497–503. 10.1016/j.jcf.2012.12.003 [DOI] [PubMed] [Google Scholar]
- Hùlková M., Soukupová J., Carlson R. P., Maršálek B. (2020). Interspecies interactions can enhance Pseudomonas aeruginosa tolerance to surfaces functionalized with silver nanoparticles. Colloid. Surf B Biointerfac. 192:111027. 10.1016/j.colsurfb.2020.111027 [DOI] [PubMed] [Google Scholar]
- Ibberson C. B., Whiteley M. (2020). The social life of microbes in chronic infection. Curr. Opin. Microbiol. 53 44–50. 10.1016/j.mib.2020.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenul C., Horswill A. R. (2018). Regulation of Staphylococcus aureus virulence. Microbiol. Spectr. 6:1128. 10.1128/microbiolspec.GPP3-0031-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahl B., Herrmann M., Everding A. S., Koch H. G., Becker K., Harms E., et al. (1998). Persistent infection with small colony variant strains of Staphylococcus aureus in patients with cystic fibrosis. J. Infect. Dis. 177 1023–1029. 10.1086/515238 [DOI] [PubMed] [Google Scholar]
- Kahl B. C., Becker K., Löffler B. (2016). Clinical significance and pathogenesis of staphylococcal small colony variants in persistent infections. Clin. Microbiol. Rev. 29 401–427. 10.1128/cmr.00069-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahl B. C., Belling G., Becker P., Chatterjee I., Wardecki K., Hilgert K., et al. (2005). Thymidine-dependent Staphylococcus aureus small-colony variants are associated with extensive alterations in regulator and virulence gene expression profiles. Infect. Immun. 73 4119–4126. 10.1128/iai.73.7.4119-4126.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehl-Fie T. E., Skaar E. P. (2010). Nutritional immunity beyond iron: a role for manganese and zinc. Curr. Opin. Chem. Biol. 14 218–224. 10.1016/j.cbpa.2009.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khademi S. M. H., Sazinas P., Jelsbak L. (2019). Within-host adaptation mediated by intergenic evolution in Pseudomonas aeruginosa. Genome Biol. Evol. 11 1385–1397. 10.1093/gbe/evz083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klockgether J., Cramer N., Fischer S., Wiehlmann L., Tümmler B. (2018). Long-term microevolution of Pseudomonas aeruginosa differs between mildly and severely affected cystic fibrosis lungs. 2. Am. J. Respir. Cell Mol. Biol. 59 246–256. 10.1165/rcmb.2017-0356oc [DOI] [PubMed] [Google Scholar]
- Kobayashi S. D., DeLeo F. R. (2013). Staphylococcus aureus protein A promotes immune suppression. mBio 4:e00764-13. 10.1128/mBio.00764-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegeskorte A., Lorè N. I., Bragonzi A., Riva C., Kelkenberg M., Becker K., et al. (2015). Thymidine-dependent Staphylococcus aureus small-colony variants are induced by trimethoprim-sulfamethoxazole (SXT) and have increased fitness during SXT challenge. Antimicrob. Agents Chemother. 59 7265–7272. 10.1128/aac.00742-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Ting Y. P. (2015). Presence of Pseudomonas aeruginosa influences biofilm formation and surface protein expression of Staphylococcus aureus. Environ. Microbiol. 17 4459–4468. 10.1111/1462-2920.12890 [DOI] [PubMed] [Google Scholar]
- La Rosa R., Johansen H. K., Molin S. (2018). Convergent Metabolic specialization through distinct evolutionary paths in Pseudomonas aeruginosa. mBio 9:e00269-18. 10.1128/mBio.00269-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa R., Johansen H. K., Molin S. (2019). Adapting to the airways: metabolic requirements of Pseudomonas aeruginosa during the infection of cystic fibrosis patients. Metabolites 9:234. 10.3390/metabo9100234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leid J. G., Willson C. J., Shirtliff M. E., Hassett D. J., Parsek M. R., Jeffers A. K. (2005). The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J. Immunol. 175 7512–7518. 10.4049/jimmunol.175.11.7512 [DOI] [PubMed] [Google Scholar]
- Lennartz F. E., Schwartbeck B., Dübbers A., Große-Onnebrink J., Kessler C., Küster P., et al. (2019). The prevalence of Staphylococcus aureus with mucoid phenotype in the airways of patients with cystic fibrosis-A prospective study. Int. J. Med. Microbiol. 309 283–287. 10.1016/j.ijmm.2019.05.002 [DOI] [PubMed] [Google Scholar]
- Limoli D. H., Hoffman L. R. (2019). Help, hinder, hide and harm: what can we learn from the interactions between Pseudomonas aeruginosa and Staphylococcus aureus during respiratory infections? 7. Thorax 74 684–692. 10.1136/thoraxjnl-2018-212616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limoli D. H., Warren E. A., Yarrington K. D., Donegan N. P., Cheung A. L., O’Toole G. A. (2019). Interspecies interactions induce exploratory motility in Pseudomonas aeruginosa. Elife 8:e47365. 10.7554/eLife.47365.sa2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limoli D. H., Whitfield G. B., Kitao T., Ivey M. L., Davis M. R., Grahl N., et al. (2017). Pseudomonas aeruginosa alginate overproduction promotes coexistence with Staphylococcus aureus in a model of cystic fibrosis respiratory infection. mBio 8:e00186-17. 10.1128/mBio.00186-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limoli D. H., Yang J., Khansaheb M. K., Helfman B., Peng L., Stecenko A. A., et al. (2016). Staphylococcus aureus and Pseudomonas aeruginosa co-infection is associated with cystic fibrosis-related diabetes and poor clinical outcomes. Eur. J. Clin. Microbiol. Infect. Dis. 35 947–953. 10.1007/s10096-016-2621-0 [DOI] [PubMed] [Google Scholar]
- Machan Z. A., Taylor G. W., Pitt T. L., Cole P. J., Wilson R. (1992). 2-Heptyl-4-hydroxyquinoline N-oxide, an antistaphylococcal agent produced by Pseudomonas aeruginosa. J. Antimicrob. Chemother. 30 615–623. 10.1093/jac/30.5.615 [DOI] [PubMed] [Google Scholar]
- Maldonado R. F., Sá-Correia I., Valvano M. A. (2016). Lipopolysaccharide modification in Gram-negative bacteria during chronic infection. FEMS Microbiol. Rev. 40 480–493. 10.1093/femsre/fuw007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manos J., Hu H., Rose B. R., Wainwright C. E., Zablotska I. B., Cheney J., et al. (2013). Virulence factor expression patterns in Pseudomonas aeruginosa strains from infants with cystic fibrosis. Eur. J. Clin. Microbiol. Infect. Dis. 32 1583–1592. 10.1007/s10096-013-1916-7 [DOI] [PubMed] [Google Scholar]
- Martha B., Croisier D., Fanton A., Astruc K., Piroth L., Huet F., et al. (2010). Factors associated with mucoid transition of Pseudomonas aeruginosa in cystic fibrosis patients. Clin. Microbiol. Infect. 16 617–623. 10.1111/j.1469-0691.2009.02786.x [DOI] [PubMed] [Google Scholar]
- Marvig R. L., Damkiær S., Khademi S. M. H., Markussen T. M., Molin S., Jelsbak L. (2014). Within-host evolution of Pseudomonas aeruginosa reveals adaptation toward iron acquisition from hemoglobin. mBio 5:e00966-14. 10.1128/mBio.00966-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvig R. L., Dolce D., Sommer L. M., Petersen B., Ciofu O., Campana S., et al. (2015a). Within-host microevolution of Pseudomonas aeruginosa in Italian cystic fibrosis patients. BMC Microbiol. 15:218. 10.1186/s12866-015-0563-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvig R. L., Sommer L. M., Jelsbak L., Molin S., Johansen H. K. (2015b). Evolutionary insight from whole-genome sequencing of Pseudomonas aeruginosa from cystic fibrosis patients. 4. Future Microbiol. 10 599–611. 10.2217/fmb.15.3 [DOI] [PubMed] [Google Scholar]
- Marvig R. L., Sommer L. M., Molin S., Johansen H. K. (2015c). Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. 1. Nat. Genet. 47 57–64. 10.1038/ng.3148 [DOI] [PubMed] [Google Scholar]
- Mashburn L. M., Jett A. M., Akins D. R., Whiteley M. (2005). Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. 2. J. Bacteriol. 187 554–566. 10.1128/jb.187.2.554-566.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelsen C. F., Christensen A.-M. J., Bojer M. S., Hoiby N., Ingmer H., Jelsbak L. (2014). Staphylococcus aureus alters growth activity, autolysis, and antibiotic tolerance in a human host-adapted Pseudomonas aeruginosa lineage. J. Bacteriol. 196 3903–3911. 10.1128/jb.02006-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelsen C. F., Khademi S. M. H., Johansen H. K., Ingmer H., Dorrestein P. C., Jelsbak L. (2016). Evolution of metabolic divergence in Pseudomonas aeruginosa during long-term infection facilitates a proto-cooperative interspecies interaction. ISME J. 10 1323–1336. 10.1038/ismej.2015.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. L., Van Laar T. A., Chen T., Karna S. L. R., Chen P., You T., et al. (2017). Global transcriptome responses including small RNAs during mixed-species interactions with methicillin-resistant Staphylococcus aureus and Pseudomonas aeruginosa. Microbiologyopen 6:e00427. 10.1002/mbo3.427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millette G., Langlois J.-P., Brouillette E., Frost E. H., Cantin A. M., Malouin F. (2019). Despite antagonism in vitro, Pseudomonas aeruginosa enhances Staphylococcus aureus colonization in a murine lung infection model. Front. Microbiol. 10:2880. 10.3389/fmicb.2019.02880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minandri F., Imperi F., Frangipani E., Bonchi C., Visaggio D., Facchini M., et al. (2016). Role of iron uptake systems in Pseudomonas aeruginosa virulence and airway infection. Infect. Immun. 84 2324–2335. 10.1128/iai.00098-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell G., Séguin D. L., Asselin A.-E., Déziel E., Cantin A. M., Frost E. H., et al. (2010). Staphylococcus aureus sigma B-dependent emergence of small-colony variants and biofilm production following exposure to Pseudomonas aeruginosa 4-hydroxy-2-heptylquinoline-N-oxide. BMC Microbiol. 10:33. 10.1186/1471-2180-10-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisan H., Brouillette E., Jacob C. L., Langlois-Bégin P., Michaud S., Malouin F. (2006). Transcription of virulence factors in Staphylococcus aureus small-colony variants isolated from cystic fibrosis patients is influenced by SigB. J. Bacteriol. 188 64–76. 10.1128/jb.188.1.64-76.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A. T., Jones J. W., Ruge M. A., Kane M. A., Oglesby-Sherrouse A. G. (2015). Iron depletion enhances production of antimicrobials by Pseudomonas aeruginosa. J. Bacteriol. 197 2265–2275. 10.1128/jb.00072-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noto M. J., Burns W. J., Beavers W. N., Skaar E. P. (2017). Mechanisms of pyocyanin toxicity and genetic determinants of resistance in Staphylococcus aureus. J. Bacteriol. 199:e00221-17. 10.1128/JB.00221-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien S., Fothergill J. L. (2017). The role of multispecies social interactions in shaping Pseudomonas aeruginosa pathogenicity in the cystic fibrosis lung. FEMS Microbiol. Lett. 364:fnx128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien S., Williams D., Fothergill J. L., Paterson S., Winstanley C., Brockhurst M. A. (2017). High virulence sub-populations in Pseudomonas aeruginosa long-term cystic fibrosis airway infections. BMC Microbiol. 17:30. 10.1186/s12866-017-0941-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orazi G., Jean-Pierre F., O’Toole G. A. (2020). Pseudomonas aeruginosa PA14 enhances the efficacy of norfloxacin against Staphylococcus aureus Newman biofilms. J. Bacteriol. 202:e00159-20. 10.1128/JB.00159-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orazi G., O’Toole G. A. (2017). Pseudomonas aeruginosa alters Staphylococcus aureus sensitivity to vancomycin in a biofilm model of cystic fibrosis infection. mBio 8:e00873-17. 10.1128/mBio.00873-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orazi G., Ruoff K. L., O’Toole G. A. (2019). Pseudomonas aeruginosa increases the sensitivity of biofilm-grown Staphylococcus aureus to membrane-targeting antiseptics and antibiotics. mBio 10:e01501-19. 10.1128/mBio.01501-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallett R., Leslie L. J., Lambert P. A., Milic I., Devitt A., Marshall L. J. (2019). Anaerobiosis influences virulence properties of Pseudomonas aeruginosa cystic fibrosis isolates and the interaction with Staphylococcus aureus. Sci. Rep. 9:6748. 10.1038/s41598-019-42952-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer K. L., Aye L. M., Whiteley M. (2007). Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 189 8079–8087. 10.1128/jb.01138-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastar I., Nusbaum A. G., Gil J., Patel S. B., Chen J., Valdes J., et al. (2013). Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS One 8:e56846. 10.1371/journal.pone.0056846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C. E., Brown D. G., Limoli D. H., Phelan V. V., O’Toole G. A. (2020). Exogenous alginate protects Staphylococcus aureus from killing by Pseudomonas aeruginosa. J. Bacteriol. 202:e00559-19. 10.1128/JB.00559-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor R. A., Kriegeskorte A., Kahl B. C., Becker K., Löffler B., Peters G. (2014). Staphylococcus aureus small colony variants (SCVs): a road map for the metabolic pathways involved in persistent infections. Front. Cell Infect. Microbiol. 4:99. 10.3389/fcimb.2014.00099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn R. A., Whiteson K., Lim Y. W., Zhao J., Conrad D., LiPuma J. J., et al. (2016). Ecological networking of cystic fibrosis lung infections. NPJ Biofilms Microb. 2:4. 10.1038/s41522-016-0002-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme S. A., Wong Fok, Lung T., Prince A. (2020). Pulmonary pathogens adapt to immune signaling metabolites in the airway. Front. Immunol. 11:385. 10.3389/fimmu.2020.00385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder C., Byrd M., Wozniak D. J. (2007). Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol. 10 644–648. 10.1016/j.mib.2007.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass G., Nazik H., Penner J., Shah H., Ansari S. R., Clemons K. V., et al. (2019). Aspergillus-Pseudomonas interaction, relevant to competition in airways. Med. Mycol. 57 S228–S232. 10.1093/mmy/myy087 [DOI] [PubMed] [Google Scholar]
- Schwartbeck B., Birtel J., Treffon J., Langhanki L., Mellmann A., Kale D., et al. (2016). Dynamic in vivo mutations within the ica operon during persistence of Staphylococcus aureus in the airways of cystic fibrosis patients. PLoS Pathog. 12:e1006024. 10.1371/journal.ppat.1006024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seggewiss J., Becker K., Kotte O., Eisenacher M., Yazdi M. R. K., Fischer A., et al. (2006). Reporter metabolite analysis of transcriptional profiles of a Staphylococcus aureus strain with normal phenotype and its isogenic hemB mutant displaying the small-colony-variant phenotype. J. Bacteriol. 188 7765–7777. 10.1128/jb.00774-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. E., Buckley D. G., Wu Z., Saenphimmachak C., Hoffman L. R., D’Argenio D. A., et al. (2006). Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. 22. Proc. Natl. Acad. Sci. U.S.A. 103 8487–8492. 10.1073/pnas.0602138103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Španěl P., Sovová K., Dryahina K., Doušová T., Dřevínek P., Smith D. (2016). Do linear logistic model analyses of volatile biomarkers in exhaled breath of cystic fibrosis patients reliably indicate Pseudomonas aeruginosa infection? 3. J. Breath Res. 10:036013. 10.1088/1752-7155/10/3/036013 [DOI] [PubMed] [Google Scholar]
- Tan X., Coureuil M., Ramond E., Euphrasie D., Dupuis M., Tros F., et al. (2019). Chronic Staphylococcus aureus lung infection correlates with proteogenomic and metabolic adaptations leading to an increased intracellular persistence. Clin. Infect. Dis. 69 1937–1945. 10.1093/cid/ciz106 [DOI] [PubMed] [Google Scholar]
- Toder D. S., Gambello M. J., Iglewski B. H. (1991). Pseudomonas aeruginosa LasA: a second elastase under the transcriptional control of lasR. Mol. Microbiol. 5 2003–2010. 10.1111/j.1365-2958.1991.tb00822.x [DOI] [PubMed] [Google Scholar]
- Tognon M., Köhler T., Gdaniec B. G., Hao Y., Lam J. S., Beaume M., et al. (2017). Co-evolution with Staphylococcus aureus leads to lipopolysaccharide alterations in Pseudomonas aeruginosa. ISME J. 11 2233–2243. 10.1038/ismej.2017.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treffon J., Block D., Moche M., Reiss S., Fuchs S., Engelmann S., et al. (2018). Adaptation of Staphylococcus aureus to airway environments in patients with cystic fibrosis by upregulation of superoxide dismutase M and iron-scavenging proteins. J. Infect. Dis. 217 1453–1461. 10.1093/infdis/jiy012 [DOI] [PubMed] [Google Scholar]
- Treffon J., Chaves-Moreno D., Niemann S., Pieper D. H., Vogl T., Roth J., et al. (2020). Importance of superoxide dismutases A and M for protection of Staphylococcus aureus in the oxidative stressful environment of cystic fibrosis airways. Cell Microbiol. 22:e13158. 10.1111/cmi.13158 [DOI] [PubMed] [Google Scholar]
- van Mansfeld R., de Been M., Paganelli F., Yang L., Bonten M., Willems R. (2016). Within-host evolution of the dutch high-prevalent Pseudomonas aeruginosa clone ST406 during chronic colonization of a patient with cystic fibrosis. PLoS One 11:e0158106. 10.1371/journal.pone.0158106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeman C. A., Moore J. L., Noto M. J., Zhang Y., Singleton M. D., Prentice B. M., et al. (2016). The innate immune protein calprotectin promotes Pseudomonas aeruginosa and Staphylococcus aureus interaction. Nat. Commun. 7:11951. 10.1038/ncomms11951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal C., Görlich D., Kampmeier S., Herzog S., Braun N., Hitschke C., et al. (2020). Antibiotic treatment and age are associated with Staphylococcus aureus carriage profiles during persistence in the airways of cystic fibrosis patients. Front. Microbiol. 11:230. 10.3389/fmicb.2020.00230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R., Sykes D. A., Watson D., Rutman A., Taylor G. W., Cole P. J. (1988). Measurement of Pseudomonas aeruginosa phenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infect. Immun. 56 2515–2517. 10.1128/iai.56.9.2515-2517.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winstanley C., O’Brien S., Brockhurst M. A. (2016). Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends Microbiol. 24 327–337. 10.1016/j.tim.2016.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav M. K., Chae S.-W., Go Y. Y., Im G. J., Song J.-J. (2017). In vitro Multi-species biofilms of methicillin-resistant Staphylococcus aureus and Pseudomonas aeruginosa and Their host interaction during In vivo colonization of an otitis media rat model. Front. Cell Infect. Microbiol. 7:125. 10.3389/fcimb.2017.00125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagci S., Hascelik G., Dogru D., Ozcelik U., Sener B. (2013). Prevalence and genetic diversity of Staphylococcus aureus small-colony variants in cystic fibrosis patients. Clin. Microbiol. Infect. 19 77–84. 10.1111/j.1469-0691.2011.03742.x [DOI] [PubMed] [Google Scholar]
- Yang L., Jelsbak L., Molin S. (2011). Microbial ecology and adaptation in cystic fibrosis airways. Environ. Microbiol. 13 1682–1689. 10.1111/j.1462-2920.2011.02459.x [DOI] [PubMed] [Google Scholar]
- Zhao K., Du L., Lin J., Yuan Y., Wang X., Yue B., et al. (2018). Pseudomonas aeruginosa quorum-sensing and type VI secretion system can direct interspecific coexistence during evolution. Front. Microbiol. 9:2287. 10.3389/fmicb.2018.02287 [DOI] [PMC free article] [PubMed] [Google Scholar]