Abstract
The geographical distribution and ecological niche of the two circulating species of the Sporothrix genus in Venezuela was established. For this, 68 isolates of Sporothrix spp. from patients of different regions of the country were analyzed. A molecular taxonomy analysis was conducted using a fragment of the calmodulin gene (CAL), and ITS regions, confirming the presence of S. schenckii (62%) and S. globosa (38%). Computational models of ecological niche for each species were obtained by the maximum entropy method using the MaxEnt software, which predicted the best environmental conditions for the presence of the two species. These models predict that the main variables influencing the presence of S. schenckii were altitude and annual mean temperature, while for S. globosa, the more influent variable was the land use, with 82% of S. globosa located at urban areas vs 56% for S. schenckii. The results here presented could contribute to understand the specific environmental factors that might modulate the occurrence of Sporothrix spp. as well as its transmission. To our knowledge, our analyses show for the first time Sporothrix spp.–specific ecological niche data, a valuable tool to promote evidence-based public health policymaking within endemic areas of sporotrichosis.
Electronic supplementary material
The online version of this article (10.1007/s42770-020-00306-0) contains supplementary material, which is available to authorized users.
Keywords: Ecological niche modeling, Sporotrichosis, MaxEnt, Sporothrix globosa, Sporothrix schenckii
Introduction
Sporotrichosis, a subcutaneous mycosis of humans and other mammals, is known to be caused by the pathogenic clade of the Sporothrix genus, of which S. brasiliensis, S. schenckii, and S. globosa are the three species of major clinical importance [1]. The etiological agents of the disease are thermodimorphic fungi, which once into a warm-blooded host can transform from a mycelial sporulating-saprophytic form to a pathogenic yeast. The infection is characterized by cutaneous and subcutaneous lesions with regional lymphocutaneous dissemination, although pulmonary and systemic manifestations have been reported [2]. The cutaneous disease begins with traumatic inoculation of the fungus by contaminated soil or plant debris, or through bites and scratches from cats bearing the fungus. Multiple infections might arise from a single source, which can lead to outbreaks [3]. The disease has a worldwide distribution, mostly in tropical and subtropical countries [4]. A previous report showed that both S. schenckii and S. globosa are circulating in Venezuela; S. globosa represented up to 30% of the clinical isolates analyzed, revealing its highest frequency reported for this species in the Americas [5].
For over a decade, the ecological niche modeling (ENM) has been proposed as a powerful tool to summarize spatial patterns of disease transmission. Its value relies on bringing together the analytical methods and conceptual bases of ecology along with the huge data compilation of epidemiology to understand the geography of a disease (pathogens, vectors, reservoirs, and host). The use of ENM on disease transmission allows then the generation of maps of risk for founded decision-making policies that could help to improve human and animal health [6, 7]. Hence, the method has been applied to different health-related issues (see [7] for a complete review on the subject), including human fungal infections [8–10].
The ENM methodology proposed by Peterson for the study of the spatial patterns of disease transmission [6] is based on the ecological niche concept [11]. Its modern definition, proposed by Soberón and Peterson [12], refers to the environmental conditions in which a species can sustain populations in the long term without need of immigration. However, the species may not use their whole niche due to biological or dispersal restrictions. To explain this modern concept, Soberón and Peterson [12] proposed the BAM framework to redefine the ecological niche of a species: biotic factors such as species-species interaction that shape the distribution of the species under study; abiotic conditions limiting the survival of the species, such as temperature or annual rain; and movement, noting geographical accessibility of the species which might be limited by geographical barriers such as mountains or oceans [12].
When applying the BAM framework to pathogenic fungi such as the Sporothrix species, the determination of the biotic factor arises as extra difficulty. The fungus presents two morphologies depending on whether it is in its pathogenic yeast form (fungus-host interaction) or its mycelial saprophytic form (fungus-vegetation interaction). In the present work, we are inferring the presence of S. schenckii and S. globosa in different regions of Venezuela by the manifested disease in the human host. By doing this analysis, we are proposing the ENM framework as a potential tool for epidemiologists in predicting likely distributions of these pathological agents and the most probable area of risk for acquiring the infection within the country.
Materials and methods
Fungal isolates used
Sixty-eight (68) clinical isolates from different Venezuelan regions were examined in this study (Supplementary Table TS1). All isolates were previously identified as Sporothrix spp. and were isolated as part of routine examination of patients at the Mycology Laboratory of Instituto de Biomedicina, Caracas, Venezuela, a national reference center for skin diseases, and no ethical approval was required for their use. They are part of the laboratory fungal collection which have been isolated from clinical samples over a period of 40 years, and carefully maintained and preserved in distilled water (Castellani’s method). Fungal isolates were recovered by growth on Sabouraud dextrose broth (SD) complemented with 150 μg/ml of chloramphenicol at 23 °C with shaking at 120 rpm for 7 days.
Molecular characterization
For species identification, molecular characterization was performed on the 68 isolates by partial amplification of the calmodulin (CAL) locus and ITS regions, as described in [5]. Following PCR, amplicons were purified with the CONCERT™ Rapid PCR purification system (Life Technologies, USA) and sent for sequencing on both strands to Macrogen Inc. (Seoul, Korea). Assembly of the sequences was done with the Contig program within the Vector NTI suite (Vector NTI, InforMax, Inc., USA). Homology searches were performed on the GenBank database using BLAST [13]. The sequences generated in this study were deposited in the GenBank/EMBL/DDBJ database under accession numbers listed in Supplementary Table TS1.
Phylogenetic analysis
For phylogenetic analysis using the CAL sequence, 150 Sporothrix-calmodulin-related sequences were used as follows: 68 sequences generated in this study (Supplementary Table TS1), 30 sequences from Camacho et al. [5], and 52 sequences retrieved from GenBank (Supplementary Table TS4). As an outgroup, the saprophytic fungus Grosmannia serpens (Ophiostomataceae) [14] was included (Supplementary Fig. S1). For the ITS phylogenetic analyses, 146 Sporothrix-ITS-related sequences were used as follows: 68 sequences generated in the present study (Supplementary Table TS1), 32 sequences from Camacho et al. [5], and 46 sequences retrieved from GenBank (Supplementary Table TS5, Fig. S2). The multiple nucleotide sequence alignment was performed using the Clustal algorithm implemented in the MEGA7 software [15]. Phylogenetic analyses were done by the maximum likelihood and neighbor-joining methods. Evolutionary analyses were also conducted using MEGA7 with the Kimura 2-parameter distance, and a bootstrap of 1000 replicates.
Genetic variation analysis
Genetic variation was assessed by using the CAL locus as nuclear marker as previously reported [5]. Nucleotide (π) and haplotype (Hd) diversities were estimated using DnaSP software, version 5.10.01 [16]. Sites containing gaps and missing data were not considered for the analysis.
Statistical analysis
To determine differences in the epidemiological analysis between S. schenckii and S. globosa data, the Mann-Whitney non-parametric test for independent samples was used.
Geographical distribution and ecological niche modeling
To model the geographical distribution and perform the comparison of the ecological niche models for S. schenckii and S. globosa in Venezuela, the bioclimatic variables to be used were downloaded from the database WorlCLIM (www.worldclim.org) in raster format. The following bioclimate variables were used in the present study: BIO1 (annual mean temperature), BIO4 (temperature seasonality), BIO12 (annual precipitation), BIO16 (precipitation of wettest quarter), BIO17 (precipitation of driest quarter), BIO18 (precipitation of warmest quarter), BIO19 (precipitation of coldest quarter). Also, type of soil, land use (Soilgrids.org), and elevation (Global Digital Elevation Model, ASTER NASA) were included. All the variables previously mentioned were incorporated in the analysis as environmental variables, in raster format, and will be referred as so from now on.
For the niche estimation analysis of the Venezuelan Sporothrix species, the presence of clinical cases of S. schenckii and S. globosa were previously georeferenced in terms of latitude and longitude coordinates expressed in degrees, minutes, and seconds (Supplementary Tables TS2 and TS3). Only cases with enough data for georeferencing were used. Forty-three S. schenckii registers (35 from the present study and 8 from Camacho et al. [5]) and twenty-seven S. globosa registers (23 from the present study and 4 from Camacho et al. [5]) were used for the analysis (Supplementary Tables TS2 and TS3). Niche estimation analysis for each species was done by correlating the occurrence point of each species to the previously defined environmental variables. The correlation was obtained using the models generated for each variable by the maximum entropy method using the MaxEnt program, version 3.3.3.k [17]. To evaluate the relationships for the analyzed variables, a Spearman correlation was employed. A jack-knife analysis was conducted to evaluate the importance of the environmental variables in the distribution of the two species under analysis [18]. The images in raster format generated as an output by the MaxEnt program were edited with the help of the ArcGIS software suite, version 10.0 (Environmental Systems Research Institute (ESRI), Redlands, CA, USA) to generate the distribution map for each Sporothrix species (Fig. 1, panels a and b). As threshold, the values of maximum training and higher specificity were used.
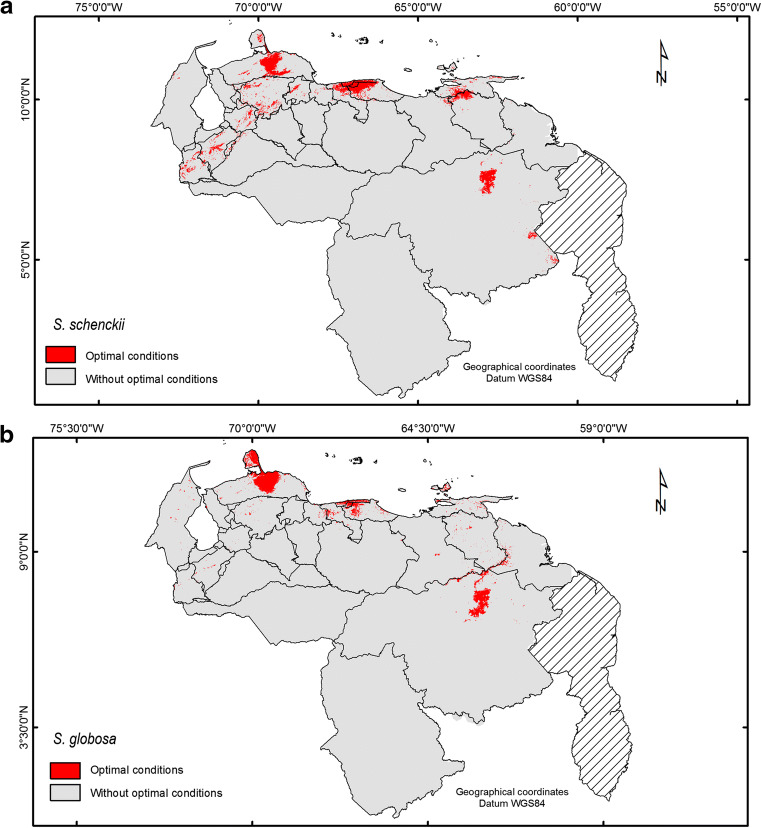
Fig. 1.
Geographical distributions predicted for Sporothrix spp. in Venezuela. The geographical distribution of the Sporothrix species is based on the optimal environmental conditions predicted by the ENM. The images generated as an output by the MaxEnt program were edited with the ArcGIS software suite, version 10.0. Panel a, geographic distribution predicted for S. schenckii. Panel b, geographical distribution predicted for S. globosa
The overlapping of S. schenckii and S. globosa niches was measured by using the framework proposed by Broennimann et al. [19], which applies kernel smoothers to densities of species incidence in a gridded environmental space to estimate metrics of niche overlap and test hypothesis of niche conservatism [19]. This framework can be used as a comparative method since it allows to estimate the climatic niches expressed on each region based on a principal component analysis (PCA). All the tests were performed with the R program, using the functions proposed by Broennimann et al. [19]. The environmental grids were produced by randomly generating 10,000 pseudo-absence locations within the area of study using Hawth’s tools in the ArcGIS software suite, version 10.00. The generated pseudo-absence locations were intersected with the layer of each environmental variable to extract the environmental values corresponding to each pseudo-absence location. In this way, a representative sampling of the environmental gradient on each area was obtained [20]. With the environmental data for the pseudo-absences and those data associated to the species occurrence points within the area of study, a PCA was done to produce the basis of the environmental grid. After the PCA was obtained, a 100 × 100 cell environmental grid was generated for each species under analysis (Fig. 2, panels a and b). Each cell represents a single vector of environmental conditions based on the boundaries of the first two principal components (PC) of the PCA, which can explain over 60% of data variance. Finally, for each cell of the grid, the niche and species occupation were calculated [19]. The indexes of niche conservatism were determined according to Petitpierre et al. [21] by overlapping the environmental grid of the ancestral species (here assumed as S. schenckii, the species with higher genetic variability, larger niche, and higher number of reports) with the grid of the recent species (here assumed as S. globosa, the species with lower genetic variability, smaller niche, and lower number of registers) (Supplementary Table TS3 and Fig. 2, panel c). The indexes of niche conservatism are represented by O, overlap index, measured by the environmental cells occupied by both species; U, unfilled index, measured by cells occupied by S. schenckii with environmental conditions suitable for, but not occupied by S. globosa; and E, expansion index, represented by the proportion of cells occupied by S. globosa but not by S. schenckii (Table 1).
Fig. 2.
Environmental niche grids generated for each Sporothrix species after principal component analysis, and niche grid overlapping of the ancestral species (S. schenckii) with the recent species (S. globosa). Panels a and b show the occurrence densities per environmental cell for S. schenckii (a) and S. globosa (b) on specific environmental combinations. The solid and dashed contour lines illustrate, respectively, 100% and 50% of the available environment. The correlation circle (panel c) indicates the weight of each bioclimatic variable on the niche space defined by the first two principal components axes, PC1 (39.22%) and PC2 (23.28). Panel d shows the niche grid overlapping of the ancestral species (S. schenckii) with the niche grid of the recent species (S. globosa). Green represents the environmental conditions occupied by S. schenckii and not by S. globosa (unfilled, U). Red indicates expansion (E) and represents environmental conditions only occupied by S. globosa. Blue represents environmental conditions occupied by both species (niche overlapping)
Table 1.
Contribution of the bioclimatic variables to the principal components in each PCA
| Variable | Description | PC1 | PC2 |
|---|---|---|---|
| BIO1 | Annual mean temperature | 0.09673175 | *0.74385816 |
| BIO4 | Temperature seasonality (standard deviation *100) | 0.39218394 | 0.1903556 |
| BIO12 | Annual precipitation | *0.85930067 | 0.10212419 |
| BIO16 | Precipitation of wettest quarter | 0.62597144 | 0.18896995 |
| BIO17 | Precipitation of driest quarter | 0.6629586 | 0.00046749 |
| BIO18 | Precipitation of warmest quarter | 0.69354295 | 0.03726699 |
| BIO19 | Precipitation of coldest quarter | 0.30340937 | 0.36548879 |
| Soil | - | 0.05674086 | 0.01596698 |
| Land use | - | 0.14928295 | 0.01743091 |
| Elevation | - | 0.10542472 | 0.67938135 |
*Highest contribution values to correlation circle (Fig. 2, panel c)
Results
Phylogenetic analysis
The aligned CAL sequences were 595 bp long, including 302 invariable characters, 239 variable parsimony-informative sites (40.17%), and 43 singletons. All the 68 Venezuelan isolates clustered within the pathogenic clade [6]. Of those isolates, 42 (62%) clustered as S. schenckii, while 26 (38%) clustered as S. globosa (Fig. S1). The aligned ITS sequences were 518 bp long, including 375 invariable characters, 112 variable parsimony-informative sites (21.6%), and 25 singletons. All the Venezuelan isolates were distributed in complete agreement with the CAL-generated phylogeny, with the Venezuelan isolates of S. schenckii and S. globosa (pathogenic species), showing strong, statistically supported separation (99% bootstrap) between pathogenic and environmental clades (Fig. S2). Also, 63 of the Venezuelan S. schenckii isolates grouped within the subclade C reported by Zhou et al. [22] where isolates from the Americas and Asia are grouped, while 3 isolated grouped within subclade D.
Genetic variation
Measurement of the genetic diversity of the Venezuelan Sporothrix population was explored by using the DnaSP software [16]. A total of 7 and 2 different types were detected for S. schenckii and S. globosa, respectively. S. schenckii presented a haplotype diversity of Hd = 0.691 and nucleotide diversity of π = 0.00266. Meanwhile, S. globosa presented a haplotype diversity of Hd = 0.514, and nucleotide diversity of π = 0.00188.
Frequency of clinical forms vs Sporothrix spp.
Figure 3 shows the frequency of the sporotrichosis clinical manifestations (Supplementary Table TS1) per species. S. schenckii isolates were obtained mainly from patients with the lymphocutaneous (LC) form of disease, while S. globosa was predominantly isolated for patients with the fixed cutaneous (FC) form of the disease. This difference is statistically significant according to the Mann-Whitney non-parametric test (Fig. 3).
Fig. 3.
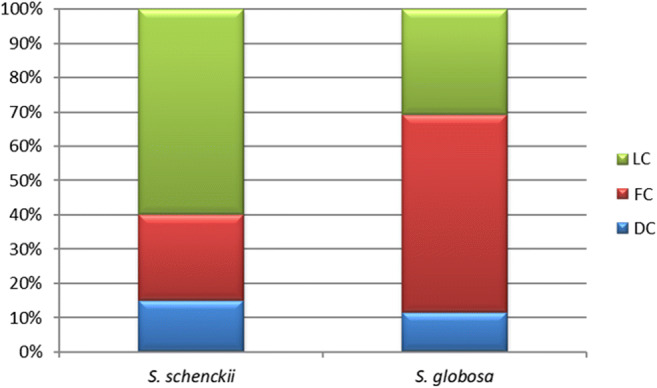
Frequency of clinical forms of sporotrichosis vs phylogenetic species. DS, disseminated; LC, lymphocutaneous; FC, fixed cutaneous. Information corresponds to clinical cases with available epidemiological data (Supplementary Table TS1). Statistical significance for the Mann-Whitney non-parametric test p < 0.05
Geographical distribution and ecological niche modeling
For each species, a maximum entropy analysis was performed, giving as result of a correlation between the location of the Sporothrix species under study and each of the ten variables considered in the present study (see Materials and methods). The jack-knife analysis for S. schenckii and each of the ten variables demonstrated that the two variables with more gain for this species were elevation and annual mean temperature, while for S. globosa, the land use was the variable with more gain (Fig. 4). Regarding the presence of cases according to elevation for the two species under study, both are mostly present over 897 mamsl, with 88% and 68% of the sporotrichosis cases corresponding to S. schenckii and S. globosa, respectively. Considering the land use, the sporotrichosis cases caused by both species are mostly reported in urban areas, 82% of them attributable to S. globosa while 56% were due to S. schenckii. The latter is also present in evergreen forests (data not shown).
Fig. 4.
Jack-knife analyses (JKA) of regularized training gain for Sporothrix spp. present in Venezuela. Panel a, JKA for S. schenckii. Panel b, JKA for S. globosa. The red bar shows the gain when using all variables. For each variable, the light blue bar shows the gain when that specific variable is excluded from analysis, and a lower gain indicates that the specific variable has more information that is not present in other variables. In contrast, the dark blue bar shows the gain when the specific variable is used in isolation. For S. schenckii, the variable with highest gain is BIO1 (average annual temperature) and elevation. For S. globosa, the variable with highest gain is the land use
Based on the optimal environmental conditions predicted by the ENM for the two species analyzed, the best conditions for the occurrence of S. schenckii are predicted North of the country, specifically to the North-Central area (Miranda, Aragua, and Vargas states), towards the West from the Andean region up to the North-Western region (Falcón and Lara states), the Eastern region (Sucre state), and down to the East-Central region (Bolívar state) (Fig. 1, panel a). For S. globosa, the best climatic conditions are predicted to the North-Central area of the country (Miranda, Aragua, and Vargas states), the North-Western region (Falcón and Lara states), and down to the East-Central region (Bolívar state) (Fig. 1, panel b).
The environmental grids showed the occurrence densities per environmental cell for each species on specific environmental combinations, allowing the modeling of the niches occupied per species (Fig. 2, panels a and b). Annual precipitation (BIO12) was the most contributive variable, followed by the annual mean temperature (BIO1) (Table 1). S. schenckii is present where annual precipitation values ranged from 835 to 1289 mm, while S. globosa is present in areas slightly more wet where annual precipitation ranged from 874 to 1289 mm. Regarding annual mean temperature, both species are present at temperatures from a minimum of 17 °C to a maximum of 26.6 °C for S. globosa or a maximum of 25.7 °C for S. schenckii.
The niche conservatism indexes were obtained by overlapping the environmental grid obtained for S. schenckii with the environmental grid obtained for S. globosa (Table 2). The unfilled index (U) indicated that 33% of the environmental conditions occupied by S. schenckii are not occupied by S. globosa, while the expansion index (E) showed that 13% of environmental conditions are exclusively occupied by S. globosa, and both species analyzed shared 54% of the environmental conditions (O) (Table 2).
Table 2.
Niche conservatism indexes obtained by overlapping the environmental grid for S. schenckii on the environmental grid for S. globosa
| Niche overlapping | Overlap index (O) | Unfilled index (U) (S. schenckii) | Expansion index (E) (S. globosa) |
|---|---|---|---|
| S. schenckii/S. globosa | 0.802 (54%) | 0.495 (33%) | 0.197 (13%) |
Discussion
The present report shows that S. schenckii and S. globosa are the most prevalent etiological agents of human sporotrichosis in Venezuela, with 62% and 38% of the clinical isolates, respectively. This corroborates our previous study to the highest report of S. globosa incidence in the Americas, highlighting the importance of this pathogen in the region [5].
Although both S. schenckii and S. globosa were isolated from patients with the three clinical manifestation of the disease (lymphocutaneous, fixed cutaneous, and cutaneous disseminated), there was a clear predominance of lymphocutaneous sporotrichosis on patients infected with S. schenckii, while the fixed cutaneous clinical form was mostly manifested in patients infected with S. globosa (Fig. 3). The differences in clinical manifestations might be related to the patient’s immunological conditions, infection route, and strain virulence. Some reports have also suggested that differences in virulence levels within the Sporothrix pathogenic clade might be species-related, with S. brasiliensis as the most virulent species, and S. globosa as the less virulent species, while variable levels of virulence within S. schenckii could be strain-related [23–25]. Further analyses of the Venezuelan S. schenckii vs S. globosa isolates regarding their virulence patterns might shed a light on this issue.
Similarly to other reports, where S. schenckii exhibits a higher genetic diversity while S. globosa presents a low genetic diversity within the Sporothrix pathogenic clade [5, 22, 26–28], our measurements of the genetic variation among the Venezuelan isolates show higher haplotype number (7) and haplotype diversity (Hd = 0.691) for the S. schenckii isolates, when compared with the S. globosa isolates (2 haplotypes, Hd = 0.514).
The phylogenetic analysis performed grouped all the Venezuelan isolates in the same clade. For the analysis done with the CAL locus, the Venezuelan isolates grouped into the clade IIa defined by Marimon et al. [29] (Fig. S1). On the other hand, the phylogenetic analysis using the ITS region grouped almost all of the Venezuelan S. schenckii isolates into clade C, as defined by Zhou et al. [22], whom points that clade C groups isolates from the Americas and Asia (Fig. S2), thus our results agree with this statement. Conversely, the Venezuelan S. globosa isolates grouped within the S. globosa clade for isolates from different regions of the world (Fig. S2), supporting the low genetic variability observed for this species.
As Rodrigues et al. [30] point out, numerous factors might be influencing the epidemiology of sporotrichosis: (i) biological factors (strain virulence, fitness of the host’s immune system), (ii) ecological factors, such as temperature, atmospheric humidity, geological conditions, and interaction with other organisms, and (iii) socioeconomic factors, such as poverty, hygiene, prophylactic habits of the population, and migration. In order to understand at least some of those factors and determine if they can be related to the epidemiology of the disease in Venezuela, we performed a modeling of the ecological niche and geographical distribution of the two Sporothrix species circulating in the country. From the ENM, the importance of the variables tested on the distribution of the two species under study was determined by a jack-knife analysis (Fig. 4), which indicated that the main variables influencing the presence of S. schenckii were altitude and annual mean temperature. S. schenckii was isolated at elevations ranging from 341 to 1633 mamsl, with 88% of isolates obtained at elevations from 857 mamsl and up. The optimal annual mean temperature for cases of sporotrichosis related to S. schenckii was determine in the range of 17 °C to 25.7 °C.
In the case of sporotrichosis cases related to S. globosa, the jack-knife analysis showed that the variable more influent on the presence of sporotrichosis cases related to this species was the land use (Fig. 4); 82% of S. globosa–related cases of the disease located at urban areas. For sporotrichosis cases related to S. schenckii, although more than half the cases were urban-related (56%), an important share of cases was present in evergreen forests (data not shown). These results could be relevant for epidemiological studies establishing an association between the species causing the disease and the patient’s occupation. However, in the present study, there was not enough data to conclude on this; further studies would be needed to determine if such a relation is real.
Considering the overlap of data for each of the ten variables on each georeferenced case per species, a niche modeling was generated for both, S. schenckii and S. globosa. The results demonstrate an environmental grid as described by Broennimann et al. [19], showing a gradient of densities of niche occupancy for each species (Fig. 2, panels a and b). From this analysis, it is clear that S. schenckii presents a larger niche than S. globosa. This result together with the higher genetic variability of S. schenckii suggests that S. schenckii have been present in the country for a longer time (ancestral species) than S. globosa, which could be suggested as the recent species (lower genetic diversity, smaller niche). By overlapping both niche models, we could determine that the variables that contribute the most to the correlation circle as proposed by Broennimann et al. [19] were annual precipitation and annual mean temperature (Fig. 2, panel c). Both species are then predicted to be found when the following environmental conditions are present: annual precipitation in the range of 835–1289 mm and annual mean temperature of 17 °C to 25.7 °C. Table 1 shows the results of the niche overlapping analysis, indicating that the niches for both species share over 50% of environmental conditions (O = 0.802); however, both species do not share identical niches (Fig. 2, panel d), a key feature to be taken into account when determining maps of risk of infection.
The ENM analysis also allowed to propose a geographical distribution model for each species in the country (Fig. 1 a and b), based on the variables here considered and the occurrence of cases due to each agent, S. schenckii and S. globosa. In general, the geographical distribution for both species is defined on the northern mountain chain of the country, with a major distribution for S. schenckii which is also predicted over the Andean mountain chain (Fig. 1, panel a). An important point to be made for this geographical distribution is that the model predicts that at the North-West of the country (Falcón and Lara states), all the conditions for occurrence of both species are present. However, for the Falcón state, a low record of sporotrichosis cases are reported [31], perhaps due to misdiagnosis of cases in the region mostly affected with a high annual number of chromoblastomycosis cases [31]. The present work could serve as starting point for epidemiologists to develop prophylactic measures on the occurrence of sporotrichosis, but more importantly, it might provide the fundamental basis for further research on the epidemiology of other fungal diseases in the country.
Electronic supplementary material
(DOCX 18 kb)
(DOCX 16 kb)
(DOCX 14 kb)
(DOCX 19 kb)
(DOCX 20 kb)
(PNG 7038 kb)
(PNG 6465 kb)
Funding information
The data here presented were part of the M.Sc. thesis work of LB at IVIC. This work was partially supported by Research Project N° 112 to SR and GAN-V from the Instituto Venezolano de Investigaciones Cientificas (IVIC, Venezuela).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.de Beer ZW, Duong TA, Wingfield MJ. The divorce of Sporothrix and Ophiostoma: solution to a problematic relationship. Stud Mycol. 2016;83:165–191. doi: 10.1016/j.simyco.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Callens SFJ, Kitetele F, Lukun P, et al. Pulmonary Sporothrix schenckii infection in a HIV positive child. J Trop Pediatr. 2006;52:144–146. doi: 10.1093/tropej/fmi101. [DOI] [PubMed] [Google Scholar]
- 3.Chakrabarti A, Bonifaz A, Gutierrez-Galhardo MC, Mochizuki T, Li S. Global epidemiology of sporotrichosis. Med Mycol. 2014;53:3–14. doi: 10.1093/mmy/myu062. [DOI] [PubMed] [Google Scholar]
- 4.Lopes-Bezerra LM, Schubach A, Costa RO. Sporothrix schenckii and sporotricosis. An Acad Bras Cienc. 2006;78:293–308. doi: 10.1590/S0001-37652006000200009. [DOI] [PubMed] [Google Scholar]
- 5.Camacho E, León-Navarro I, Rodríguez-Brito S, Mendoza M, Niño-Vega GA. Molecular epidemiology of human sporotrichosis in Venezuela reveals high frequency of Sporothrix globosa. BMC Infect Dis. 2015;15:1–10. doi: 10.1186/s12879-015-0839-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peterson AT. Ecologic niche modeling and spatial patterns of disease transmission. Emerg Infect Dis. 2006;12:1822–1826. doi: 10.3201/eid1212.060373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Escobar LE, Craft ME. Advances and limitations of disease biogeography using ecological niche modeling. Front Microbiol. 2016;7:1–21. doi: 10.3389/fmicb.2016.01174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reed KD, Meece JK, Archer JR, Peterson AT. Ecologic niche modeling of Blastomyces dermatitidis in Wisconsin. PLoS One. 2008;3(4):e2034. doi: 10.1371/journal.pone.0002034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baptista-Rosas RC, Hinojosa A, Riquelme M. Ecological niche modeling of Coccidioides spp. in western North American deserts. Ann N Y Acad Sci. 2007;1111:35–46. doi: 10.1196/annals.1406.003. [DOI] [PubMed] [Google Scholar]
- 10.Mak S, Klinkenberg B, Bartlett K, Fyfe M. Ecological niche modeling of Cryptococcus gattii in British Columbia, Canada. Environ Health Perspect. 2010;118:653–658. doi: 10.1289/ehp.0901448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peterson AT, Soberón J, Pearson RG, Anderson RP, Martínez-Meyer E, Nakamura M, Araújo MB. Ecological niches and geographic distributions. Oxford: Princenton University Press; 2011. [Google Scholar]
- 12.Soberon J, Peterson AT. Interpretation of models of fundamental ecological niches and species’ distributional areas. Biodivers Inform. 2005;2:1–10. doi: 10.17161/bi.v2i0.4. [DOI] [Google Scholar]
- 13.Zhang J, Madden TL. PowerBLAST: a new network BLAST application for interactive or automated sequence analysis and annotation. Genome Res. 1997;7:649–656. doi: 10.1101/gr.7.6.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duong TA, de Beer ZW, Wingfield BD, Wingfield MJ. Phylogeny and taxonomy of species in the Grosmannia serpens complex. Mycologia. 2012;104:715–732. doi: 10.3852/11-109. [DOI] [PubMed] [Google Scholar]
- 15.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 17.Phillips SJ, Anderson RP, Schapire RE. Maximum entropy modeling of species geographic distributions. Econ Model. 2006;190:231–259. doi: 10.1016/j.ecolmodel.2005.03.026. [DOI] [Google Scholar]
- 18.Phillips SJ, Dudík M. Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography. 2008;31:161–175. doi: 10.1111/j.0906-7590.2008.5203.x. [DOI] [Google Scholar]
- 19.Broennimann O, Fitzpatrick MC, Pearman PB, Petitpierre B, Pellissier L, Yoccoz NG, Thuiller W, Fortin MJ, Randin C, Zimmermann NE, Graham CH, Guisan A. Measuring ecological niche overlap from occurrence and spatial environmental data. Glob Ecol Biogeogr. 2012;21:481–497. doi: 10.1111/j.1466-8238.2011.00698.x. [DOI] [Google Scholar]
- 20.Sillero N, Barbosa AM, Martínez-Freiría F, Real R (2010) Los modelos de nicho ecológico en la herpetología ibérica: pasado, presente y futuro. Bol Asoc Herpetol Esp 21:2–24
- 21.Petitpierre B, Kueffer C, Broennimann O, Randin C, Curtis Daehler AG. Climatic niche shifts are rare among terrestrial plant invaders. Science. 2012;335(80):1344–1348. doi: 10.1126/science.1215933. [DOI] [PubMed] [Google Scholar]
- 22.Zhou X, Rodrigues AM, Feng P, De Hoog GS. Global ITS diversity in the Sporothrix schenckii complex. Fungal Divers. 2014;66:153–165. doi: 10.1007/s13225-013-0220-2. [DOI] [Google Scholar]
- 23.Arrillaga-Moncrieff I, Capilla J, Mayayo E, Marimon R, Marine M, Genis J, Cano J, Guarro J. Different virulence levels of the species of Sporothrix in a murine model. Clin Microbiol Infect. 2009;15:651–655. doi: 10.1111/j.1469-0691.2009.02824.x. [DOI] [PubMed] [Google Scholar]
- 24.Romeo O, Scordino F, Criseo G. New insight into molecular phylogeny and epidemiology of Sporothrix schenckii species complex based on calmodulin-encoding gene analysis of Italian isolates. Mycopathologia. 2011;172:179–186. doi: 10.1007/s11046-011-9420-z. [DOI] [PubMed] [Google Scholar]
- 25.Fernandes KSS, Mathews HL, Bezerra LML. Differences in virulence of Sporothrix schenckii conidia related to culture conditions and cell-wall components. J Med Microbiol. 1999;48:195–203. doi: 10.1099/00222615-48-2-195. [DOI] [PubMed] [Google Scholar]
- 26.Rodrigues AM, De Hoog S, De Camargo ZP. Emergence of pathogenicity in the Sporothrix schenckii complex. Med Mycol. 2013;51:405–412. doi: 10.3109/13693786.2012.719648. [DOI] [PubMed] [Google Scholar]
- 27.Rodrigues AM, De Hoog G, Zhang Y, De Camargo ZP. Emerging sporotrichosis is driven by clonal and recombinant Sporothrix species. Emerg Microbes Infect. 2014;3(0):1–10. doi: 10.1038/emi.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Hagen F, Stielow B, Rodrigues AM, Samerpitak K, Zhou X, Feng P, Yang L, Chen M, Deng S, Li S, Liao W, Li R, Li F, Meis JF, Guarro J, Teixeira M, al-Zahrani HS, de Camargo ZP, Zhang L, de Hoog GS. Phylogeography and evolutionary patterns in Sporothrix spanning more than 14 000 human and animal case reports. Persoonia. 2015;35:1–20. doi: 10.3767/003158515X687416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marimon R, Cano J, Gené J, et al. Sporothrix brasiliensis, S. globosa, and S. mexicana, three new Sporothrix species of clinical interest. J Clin Microbiol. 2007;45:3198–3206. doi: 10.1128/JCM.00808-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodrigues AM, Teixeira MDM, De Hoog GS, et al. Phylogenetic analysis reveals a high prevalence of Sporothrix brasiliensis in feline sporotrichosis outbreaks. PLoS Negl Trop Dis. 2013;7(6):e2281. doi: 10.1371/journal.pntd.0002281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martínez Méndez D, Hernández Valles R, Alvarado P, Mendoza M. Las micosis en Venezuela: Casuística de los Grupos de Trabajo en Micología (1984-2010) Rev Iberoam Micol. 2013;30:39–46. doi: 10.1016/j.riam.2012.10.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 18 kb)
(DOCX 16 kb)
(DOCX 14 kb)
(DOCX 19 kb)
(DOCX 20 kb)
(PNG 7038 kb)
(PNG 6465 kb)