Abstract
Serratia marcescens are gram-negative bacteria found in several environmental niches, including the plant rhizosphere and patients in hospitals. Here, we present the genome of Serratia marcescens strain N4–5 (=NRRL B-65519), which has a size of 5,074,473 bp (664-fold coverage) and contains 4840 protein coding genes, 21 RNA genes, and an average G + C content of 59.7%. N4–5 harbours a plasmid of 11,089 bp and 43.5% G + C content that encodes six unique CDS repeated 2.5× times totalling 13 CDS. Our genome assembly and manual curation uncovered the insertion of two extra copies of the 5S rRNA gene in the assembled sequence, which was confirmed by PCR and Sanger sequencing to be a misassembly. This artefact was subsequently removed from the final assembly. The occurrence of extra copies of the 5S rRNA gene was also observed in most complete genomes of Serratia spp. deposited in public databases in our comparative analysis. These elements, which also occur naturally, can easily be confused with true genetic variation. Efforts to discover and correct assembly artefacts should be made in order to generate genome sequences that represent the biological truth underlying the studied organism. We present the genome of N4–5 and discuss genes potentially involved in biological control activity against plant pathogens and also the possible mechanisms responsible for the artefact we observed in our initial assembly. This report raises awareness about the extra copies of the 5S rRNA gene in sequenced bacterial genomes as they may represent misassemblies and therefore should be verified experimentally.
Electronic supplementary material
The online version of this article (10.1007/s42770-020-00382-2) contains supplementary material, which is available to authorized users.
Keywords: Serratia marcescens, Artefacts, 5S rRNA, Biological control, Complete genome, Plant-associated bacteria
Introduction
Soil-borne plant pathogens cause diseases that result in major reductions in crop yields [1]. These diseases are typically controlled in conventional crop production systems with strategies that include chemical pesticides [2]. Biologically based methods, such as the use of microbial biological control agents, are being developed to control these soil-borne pathogens due to problems associated with the availability and effectiveness of chemical pesticides and concerns regarding the impact of these chemicals on the environment and human health [3]. Microbes control plant diseases via several mechanisms, including predation, where the biological control agent produces an assortment of enzymes such as chitinases, proteases, and glucanases that degrade pathogen cell wall and other cellular components [4, 5]. Biological control agents can also produce antibiotics and other inhibitory molecules that kill or slow growth of the pathogen and can compete with the pathogen for resources such as nutrients and space. Finally, certain biological control agents have been shown to associate with plants and induce defence responses that protect the plant from diseases [1].
The bacterium Serratia marcescens is ubiquitous in the environment and has been detected in association with plants [6] and animals [7] including humans in hospital settings [8], soil [9], water [10], and air [11]. Live cells and cell-free extracts of S. marcescens strains isolated from the environment have been shown to be effective in controlling certain soil-borne plant pathogens [12–14].
Here, we present the genome of S. marcescens N4–5, a strain isolated from soil and studied since 1996 [12] due to its effectiveness against multiple plant pathogens, including Magnaporthe poae, Pythium ultimum, and Rhizoctonia solani and its antimicrobial properties [12–15]. Strain N4–5 and its natural products, applied as seed treatments in biocontrol strategies, control seed, and seedling disease of cucurbits caused by the soil-borne plant pathogen P. ultimum [13, 15]. Among all the S. marcescens complete genomes available, only a few are from plant beneficial strains. Therefore, the addition of the N4–5 complete genome sequence into public databases will allow comparative analysis to better understand the mechanisms by which S. marcescens associates with plants and controls plant diseases, as well as the variety of lifestyles presented by this genus. Furthermore, we uncovered an assembly artefact in the genome of strain N4–5.
Material and methods
Genome sequencing and assembly
Strain N4–5 was obtained from New Jersey (USA) soil samples [12]. Genomic DNA was extracted with the QIAGEN Blood & Tissue genomic DNA isolation kit, using the manufacturer’s protocol. The indexed library was constructed using Nextera® XT Index Kit v2 Set A, and the sequence data was generated in an Illumina NextSeq-500 using the run kit Illumina NextSeq® 500/550 High Output Kit v2. Sequencing resulted in 22,789,104 reads, with length varying from 32 to 151 bases, with an average length of 148 bp, where 94% of the reads contained 148–151 bases. The sequenced reads comprised a total of 3,369,822,757 bases and represented 664-fold genome coverage. The quality was checked with the programme FastQC v0.11.5 [16]. The paired-end library genome (2 × 149) was assembled using the SPAdes assembler available in PATRIC (Pathosystems Resource Integration Center) [17]. The 1634 contigs generated were united into 19 scaffolds using the CONTIGuator [18] with S. marcescens strain B3R3 (accession number CP013046.2) as the reference genome. Finally, gaps were closed with FGAP [19], NCBI’s BLASTn [20], and read mapping in CLC Genomics Workbench 7.
The plasmid was assembled with reads that did not map against the final N4–5 complete nucleotide sequence using the programme plasmidSPAdes. The plasmid contigs were scaffolded using the S. marcescens strain A4Y201 plasmid pG5A4Y201 (accession number KJ541069.1) as reference. The plasmid sequence was finalized using the aforementioned FGAP, NCBI’s BLASTn, and CLC Genomics Workbench. The structural and functional annotation was conducted as described above.
Genome annotation, manual curation, and analyses
The N4–5 genome was annotated using the RASTtk annotation service in PATRIC [21]. Manual curation was conducted through Artemis 16.0.0 software [22]. Translated protein sequences were confirmed with BLASTp against the UniProt database [23]. Clusters of Orthologous Groups (COGs) were inferred with the eggNOG v. 4.5.1 database [24]. Circular maps were generated using GCView Comparison Tool [25].
Manual curation revealed three copies of the 5S rRNA gene. To determine the veracity of this feature, PCR using primers designed on the regions flanking the 5S genes, MetA1F (5′- ACC GCA GGT AAC TCA TCA GG −3′) and 23S1R (5′- GAC GTT GAT AGG CTG GGT GT- 3′), followed by sequencing with the Sanger method were performed as previously described [15]. The 100 bp DNA ladder (New England BioLabs) was used to visualize the band in the gel.
Phylogenomics and chemotaxonomic classification
Digital DNA-DNA Hybridization (dDDH) and Average Nucleotide/Amino Acid Identity (ANI/AAI) comparisons were calculated using GGDC [26], JspeciesWS [27], and Kostas Lab [28].
For FAME analyses, isolate N4–5 was grown for 24 h at 28 °C on trypticase soy broth agar (TSBA), and the composition of cellular fatty acid was determined by gas chromatography. Extraction and analyses were performed according to the recommendations of the MIDI (Microbial Identification) system.
Results and discussion
Genomic features
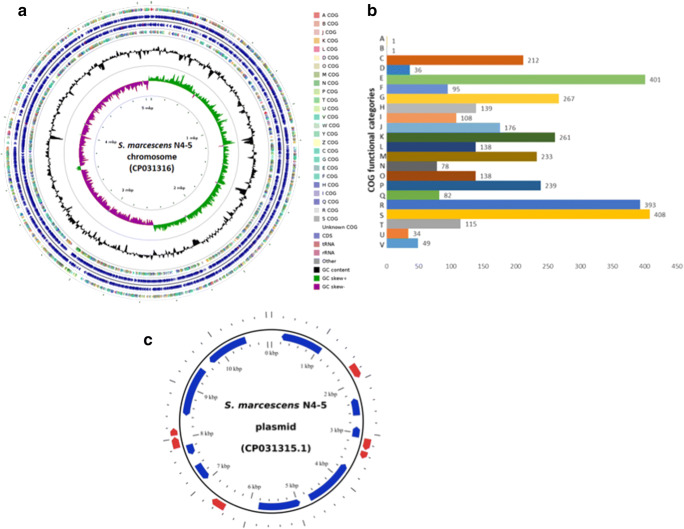
The S. marcescens N4–5 genome comprised a single chromosome of 5,074,473 bp, with 59.7% G + C content and a naturally occurring plasmid (Fig. 1). The chromosome had 4884 protein-coding genes, of which 4020 genes were functionally assigned, while the remaining genes were annotated as hypothetical proteins (Table 1). The N4–5 genomic nucleotide sequence contained 2747 transcription units and 992 operons. From the 4884 genes, 3604 (73.8%) were classified in 22 functional COG categories. The most numerous COGs contained genes with general prediction only (408 genes), no function prediction (393 genes), and 401 genes involved in amino acid transport and metabolism, whereas the COG categories with the least number of genes contained one gene for RNA processing and modification and one for chromatin function and dynamics (Fig. 1b).
Fig. 1.
Serratia marcescens strain N4–5 genome features. (a) Chromosome of N4–5 showing from outer circle to the centre: CDS on forward strand (coloured according to COG categories), all CDS and RNA genes on forward strand, all CDS and RNA genes on reverse strand, CDS on reverse strand (coloured according to COG categories), G + C content and GC skew. (b) COG functional classification of strain N4–5 proteome. (c) Circular map of N4–5 plasmid showing the CDSs
Table 1.
Genome features of S. marcescens strain N4–5
| Attribute | Value |
|---|---|
| Chromosome size (bp) | 5,074,473 |
| N50 | 549,421 |
| L50 | 4 |
| GC (%) | 59,69 |
| Chromosomal genes | 4884 |
| Protein coding genes | 4840 |
| Plasmid (bp) | 11,089 |
| Genes in the plasmid | 13 |
| RNA genes | 103 |
| Pseudo genes | 44 |
| Genes with function prediction | 4020 |
| CRISPR repeats | 2 |
The circular plasmid comprised 11,089 bp and had 43.5% G + C content. The size of the plasmid was confirmed by digestion with restriction enzymes followed by electrophoresis. The plasmid sequence encoded six unique CDS that were repeated 2.5× totalling 13 CDS. From the six unique CDS, four were annotated as hypothetical proteins (Fig. 1c).
Biocontrol and plant-beneficial traits
Strain N4–5 is a known producer of the broad-spectrum antimicrobial prodigiosin, which contributes to its biological control activity [13, 15]. In accordance, the genome of N4–5 harboured the 14 canonical genes for prodigiosin biosynthesis (pig cluster, pigA-N) described by [29, 30]. As seen in other bacteria [30], the N4–5 pig cluster was flanked by copA and cueR homologues; however, differently from the other studied strains, N4–5 has a putative membrane protein (41 amino acids) annotated between pigA and cueR. Sixty-nine multidrug resistance genes were found during functional annotation of the N4–5 genome, including resistance to kasugamycin, biocyclomycin, fosmidomycin, and fusaric acid, which are antibiotics produced by microorganisms. Strain N4–5 also harbours chitinase genes (chiA, chiB, chiD, and chiA1) in the genome.
Genome analysis revealed that N4–5 encodes the siderophore enterobactin gene cluster containing entA, entB, entC, entE, entF, and entH, but the vibriobactin genes were absent. Furthermore, N4–5 carried 16 tonB-dependent transporter genes, which are cellular receptors of siderophores. The production of siderophore complexes by bacteria contributes to enhance plant growth as they sequester iron from the environment and make it available for plant uptake [31]. The ability to utilize carbon sources provides a fitness advantage during microbial competition. The N4–5 genome had 267 genes responsible for carbohydrate transport and metabolism, comparable with Pseudomonas alcaliphila JAB1, a degrader of organic pollutants that had 196 genes with this functionality [32]. The surfactant serrawettin W1 was coded by one NRPS (non-ribosomal peptide synthase) gene with 3936 bp. Serrawetin W1 has antimicrobial, antitumor, and zoosporicidal activities and has potential uses in agriculture, medicine, and industry [33]. Altogether, these genome features support N4–5 as a potential biocontrol agent as well as a plant-beneficial strain.
Phylogenomics and chemotaxonomic data
Phylogenetic trees constructed with whole genome sequences and with sequences of the 16S gene placed strain N4–5 within the S. marcescens clade (Supplementary Fig. S1 and S2). The identity of these seven copies of the 16S rRNA gene found in the genome of strain N4–5 varied from 99.7 to 100%, indicating that strain N4–5 possesses low intragenomic variation in the ribosomal genes. Further analyses, including Average Nucleotide/Amino Acid Identity (ANI/AAI) and digital DNA-DNA Hybridization (dDDH) confirmed the classification of strain N4–5 as S. marcescens. The values for ANI, AAI, and dDDH were above the cut off for species delineation, 95, 95, and 70%, respectively, when compared with other Serratia genomes (Table S1).
The major components of the S. marcescens N4–5 fatty acid profile were C16:0 (22.4%), C17:0 cyclo (12.13%), C10:0 3OH (12.07%), and C12:0 3OH (5.15%). Minor fatty acid components were identified at less than 5%, most of which being common among previously identified species within the genus Serratia: C12:0, C12:0 2OH, C14:0, C14:0, C18:0, and C19:0 cyclo w8c. Some fatty acids isolated from N4–5 co-occurred in just a few other species. For example, C10:0 3OH was only shared with S. plymuthica and S. rubidaea, whereas C12:1 3OH, C12:0 3OH, C14:0 2OH, and C17:0 were only identified in other S. marcescens-GC subgroups.
Genome assembly artefact
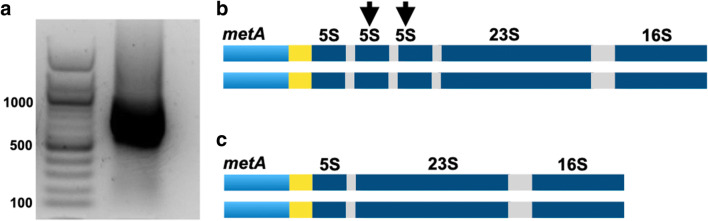
Two extra copies of the rRNA 5S gene were found in the fifth ribosomal cluster of strain N4–5 after the initial assembly performed in SPAdes. These extra copies of the 5S rRNA gene were also present in all the other four assemblies we performed with data from different sequencing lanes with coverage 160–167×. The combined assembly, with 664× coverage was chosen due to the best metrics it returned, namely, highest N50 and N75, lowest L50 and L75, largest contig size, and overall total length assembled. To verify whether it was an artefact or a natural feature in strain N4–5, PCR amplification and sequencing with the Sanger method of the rRNA 5S gene region were performed. The primers were anchored in the gene metA and in the 23S rRNA gene (Fig. 2). The 755 bp sequence obtained was mapped to the 989 bp region in the assembly and unequivocally showed that these extra copies of the 5S rRNA gene were assembly artefacts. The genome sequence was corrected accordingly, and therefore, strain N4–5 had regular ribosomal operons, i.e. one copy of the 5S, 16S, and 23S genes per cluster (Fig. 2). In strain N4–5, the assembly process was responsible for generating the artefact, but we currently do not know how the programme produces them. One possibility, although we did not test this hypothesis, is the fact that we used a genome with an extra copy of the 5S rRNA gene as the reference.
Fig. 2.
Assembly artefact in the genome of S. marcescens strain N4–5. (a) Gel image representing the expected band size of 755 bp for a single 5S gene and its flanking regions. Band sizes are given in bp. (b) Schematic representation of the ribosomal operon with the assembly artefact in the genome of strain N4–5. Both DNA strands are represented. The rRNA genes are represented in dark blue, the gene metA is shown in light blue, the spacers in the ribosomal operon in grey, and the intergenic region between metA and the ribosomal operon in yellow. The black arrows indicate the extra copies of the 5S gene in both DNA strands. This genomic region is not drawn to scale. (c) Ribosomal operon without the assembly artefact in strain N4–5
Extra copies of the 5S gene were also found in 72 completely sequenced genomes of Serratia strains deposited in public databases (Supplementary Table S2). The extra copies of the 5S gene were found in the complete genomes of species in other genera, including Serratia, Proteus, Vibrio, Yersinia (Supplementary Table S3), and possibly other bacterial genera. There is limited information on the biological implications of extra copies of the 5S gene, and it is certainly a subject that deserves further investigation. Many of the genomes that contain extra copies of the 5S gene were sequenced by long reads technologies such as PacBio (Supplementary Table 2), which is supposed to obtain reads that encompass the whole ribosomal region. Nevertheless, the occurrence of the extra copies of the 5S rRNA gene in these genomes should be verified with sequencing technologies with read lengths longer than 600 bp.
The number of copies of the 5S gene in bacterial genomes is thought to be identical to the number of copies of the other genes (23S and 16S) in the ribosomal operon. This assumption is supported by the fact that ribosomal genes and multigene families are homogenized by recombination through concerted evolution [34]. However, an estimate by rrnDB, a database that documents the number of rRNA and tRNA genes in bacteria and archaea, revealed that 23.6% of the bacterial genomes have unequal copies of the rRNA genes, due mainly to additional copies of the 5S rRNA gene [35]. In the genus Serratia, this unequal number of 5S rRNA genes occurs in 96% of the sequenced genomes (Supplementary Table 2). It would be interesting to verify if these numbers are real or are artefacts caused by the in silico assembly programmes. The discovery and correction of assembly errors in draft genomes is a crucial problem that persists in Eulerian assemblies and genome assemblies in general [36].
Strain N4–5 is among the three S. marcescens with complete genomes in public databases without extra copies of the 5S rRNA gene. This does not mean that genomes without extra copies of the 5S rRNA genes are correctly assembled and the other ones are misassemblies. With these results, we want to emphasize the need to verify these in silico assemblies with standard laboratory experimental procedures.
Electronic supplementary material
(PDF 1124 kb)
(DOCX 52 kb)
Acknowledgements
LCF and JTS acknowledge the financial support from CNPq, Brazil.
Footnotes
Data deposition: Trimmed sequence data and assembly are deposited in GenBank (accession numbers: NZ_CP031316.1 and NZ_CP031315.1).
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Raaijmakers JM, Paulitz TC, Steinberg C, Alabouvette C, Moënne-Loccoz Y. The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil. 2009;321:341–361. doi: 10.1007/s11104-008-9568-6. [DOI] [Google Scholar]
- 2.Chauhan MS, Yadav JPS, Gangopadhyay S. Chemical control of soilborne fungal pathogen complex of seedling cotton. Trop Pest Man. 2008;34:159–161. doi: 10.1080/09670878809371233. [DOI] [Google Scholar]
- 3.Geiger F, Bengtsson J, Berendse F, Weisser WW, Emmerson M, Morales MB, Ceryngier P, Liira J, Tscharntke T, Winqvist C, Eggers S, Bommarco R, Part T, Bretagnolle V, Plantegenest M, Clement LW, Dennis C, Palmer C, Onate JJ, Guerrero I, Hawro V, Aavik T, Thies C, Flohre A, Hanke S, Fischer C, Goedhart PW, Inchausti P. Persistent negative effects of pesticides on biodiversity and biological control potential on European farmland. Basic Appl Ecol. 2010;11:97–105. doi: 10.1016/j.baae.2009.12.001. [DOI] [Google Scholar]
- 4.Someya N, Kataoka N, Komagata T, Hirayae K, Hibi T. Potential of Serratia marcescens strain B2 for biological control of rice sheath blight. Biocontrol Sci Tech. 2005;15:105–109. doi: 10.1080/09583150400016092. [DOI] [Google Scholar]
- 5.Wang K, Yan PS, Cao LX (2014) Chitinase from a novel strain of Serratia marcescens JPP1 for biocontrol of aflatoxin: molecular characterization and production optimization using response surface methodology. Biomed Res Int 482623. 10.1155/2014/482623 [DOI] [PMC free article] [PubMed]
- 6.Afzal I, Iqrar I, Shinwari ZK, Yasmin A. Plant growth-promoting potential of endophytic bacteria isolated from roots of wild Dodonaea viscosa L. Plant Growth Regul. 2017;81:339–408. doi: 10.1007/s10725-016-0216-5. [DOI] [Google Scholar]
- 7.Vicente CS, Nascimento FX, Ikuyo Y, Cock PJ, Mota M, Hasegawa K. The genome and genetics of a high oxidative stress tolerant Serratia sp. LCN16 isolated from the plant parasitic nematode Bursaphelenchus xylophilus. BMC Genomics. 2016;17:301. doi: 10.1186/s12864-016-2626-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnin RA, Girlich D, Imanci D, Dortet L, Naas T. Draft genome sequence of the Serratia rubidaea CIP 103234T reference strain, a human-opportunistic pathogen. Genome Announc. 2015;3:e01340–e01315. doi: 10.1128/genomeA.01340-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lavania M, Nautiyal CS. Solubilization of tricalcium phosphate by temperature and salt tolerant Serratia marcescens NBRI1213 isolated from alkaline soils. Afr J Microbiol Res. 2013;7:4403–4413. doi: 10.5897/AJMR2013.5773. [DOI] [Google Scholar]
- 10.Kampfer P, Glaeser SP. Serratia aquatilis sp. nov., isolated from drinking water systems. Int J Syst Evol Microbiol. 2016;66:407–413. doi: 10.1099/ijsem.0.000731. [DOI] [PubMed] [Google Scholar]
- 11.Bencini MA, Yzermana EPF, Bruina JP, Den Boer JW. Airborne dispersion of Serratia marcescens as a model for spread of Legionella from a whirlpool. R Inst Public Health. 2008;122:962–964. doi: 10.1016/j.puhe.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi DY, El-Barrad N. Selection of bacterial antagonists using enrichment cultures for the control of summer patch disease in Kentucky bluegrass. Curr Microbiol. 1996;32:106–111. doi: 10.1007/s002849900019. [DOI] [Google Scholar]
- 13.Roberts DP, Lakshman DK, McKenna LF, Emche SE, Maul JE, Bauchan G. Seed treatment with ethanol extract of Serratia marcescens is compatible with Trichoderma isolates for control of damping-off of cucumber caused by Pythium ultimum. Plant Dis. 2016;100:1278–1287. doi: 10.1094/PDIS-09-15-1039-RE. [DOI] [PubMed] [Google Scholar]
- 14.Roberts DP, McKenna LF, Buyer JS. Consistency of control of damping-off of cucumber is improved by combining ethanol extract of Serratia marcescens with other biologically based technologies. Crop Prot. 2017;96:59–67. doi: 10.1016/j.cropro.2017.01.007. [DOI] [Google Scholar]
- 15.Roberts DP, McKenna LF, Lakshman DK, Meyer SLF, Kong H, De Souza JT, Lydon J, Baker CJ, Chung S. Suppression of damping-off of cucumber caused by Pythium ultimum with live cells and extracts of Serratia marcescens. Soil Biol Biochem. 2007;39:2275–2288. doi: 10.1016/j.soilbio.2007.03.029. [DOI] [Google Scholar]
- 16.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed October 2018
- 17.Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, Gillespie JJ, Gough R, Hix D, Kenyon R, Machi D, Mao C, Nordberg EK, Olson R, Overbeek R, Pusch GD, Shukla M, Schulman J, Stevens RL, Sullivan DE, Vonstein V, Warren A, Will R, Wilson MJ, Yoo HS, Zhang C, Zhang Y, Sobral BW. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014;42:D581–D591. doi: 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galardini M, Biondi EG, Bazzicalupo M, Mengoni A. CONTIGuator: a bacterial genomes finishing tool for structural insights on draft genomes. Source Code Biol Med. 2011;6:11. doi: 10.1186/1751-0473-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piro VC, Faoro H, Weiss VA, Steffens MB, Pedrosa FO, Souza EM, Raittz RT. FGAP: an automated gap closing tool. BMC Res Notes. 2014;7:371. doi: 10.1186/1756-0500-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1999;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, Stevens R, Vonstein V, Wattam AR, Xia F. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. 2015;5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream M, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 23.The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, Rattei T, Mende DR, Sunagawa S, Kuhn M, Jensen LJ, von Mering C, Bork P. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44(D1):D286–D293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grant JR, Arantes AS, Stothard P. Comparing thousands of circular genomes using the CGView comparison tool. BMC Genomics. 2012;13:202–1582. doi: 10.1093/bib/bbx081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Auch AF, Klenk HP, Göker M. Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand Genomic Sci. 2010;2:142–148. doi: 10.4056/sigs.541628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richter M, Rosselló-Móra R, Glöckner FO, Peplies J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2015;32(6):929–931. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez RLM, Konstantinidis KT. Bypassing cultivation to identify bacterial species. Microbe. 2014;9:111–118. doi: 10.1128/microbe.9.111.1. [DOI] [Google Scholar]
- 29.Cerdeño AM, Bibb MJ, Challis GL. Analysis of the prodiginine biosynthesis gene cluster of Streptomyces coelicolor A3(2): new mechanisms for chain initiation and termination in modular multienzymes. Chem Biol. 2001;119:1–13. doi: 10.1016/S1074-5521(01)00054-0. [DOI] [PubMed] [Google Scholar]
- 30.Harris AKP, Williamson NR, Slater H, Cox A, Abbasi S, Foulds I, Simonsen HT, Leeper FJ, Salmond GPC. The Serratia gene cluster encoding biosynthesis of the red antibiotic, prodigiosin, shows species- and strain-dependent genome context variation. Microbiology. 2004;150:3547–3560. doi: 10.1099/mic.0.27222-0. [DOI] [PubMed] [Google Scholar]
- 31.Scavino AF, Pedraza RO. The role of siderophores in plant growth-promoting bacteria. In: Maheshwari D, Saraf M, Aeron A, editors. Bacteria in agrobiology: crop productivity. Berlin, Heidelberg: Springer; 2013. [Google Scholar]
- 32.Ridl J, Suman J, Fraraccio S, Hradilova M, Strejcek M, Cajthaml T, Zubrova A, Macek T, Strnad H, Uhlik O. Complete genome sequence of Pseudomonas alcaliphila JAB1 (=DSM 26533), a versatile degrader of organic pollutants. Stand Genomic Sci. 2018;13:3. doi: 10.1186/s40793-017-0306-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thies S, Santiago-Schubel B, Kovacic F, Rosenau F, Hausmann R, Jaeger KE. Heterologous production of the lipopeptide biosurfactant serrawettin W1 in Escherichia coli. J Biotechnol. 2014;181:27–30. doi: 10.1016/j.jbiotec.2014.03.037. [DOI] [PubMed] [Google Scholar]
- 34.Hillis DM, Moritz C, Porter CA, Baker RJ. Evidence for biased gene conversion in concerted evolution of ribosomal DNA. Science. 1991;251:308–310. doi: 10.1126/science.1987647. [DOI] [PubMed] [Google Scholar]
- 35.Lee ZM-P, Bussema C, Schmidt TM. rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res. 2008;37:D489–D493. doi: 10.1093/nar/gkn689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muggli MD, Puglisi SJ, Ronen R, Boucher C. Misassembly detection using paired-end sequence reads and optical mapping data. Bioinformatics. 2015;31:i80–i88. doi: 10.1093/bioinformatics/btv262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1124 kb)
(DOCX 52 kb)