Abstract
Sporotrichosis is one of the neglected tropical diseases causing subcutaneous chronic granulomatous lesion by thermally dimorphic fungi belonging to Sporothrix species. Sporothrix brasiliensis, Sporothrix mexicana and Sporothrix globosa are the common pathogenic species. In Asian countries, S. globosa constitutes nearly 99.3% of all Sporothrix species. We studied 63 cases of sporotrichosis of geographically diverse origin from India and Sporothrix isolates were characterised for its growth in different media, temperatures, ability to assimilate sugars and antifungal susceptibility profile. Molecular characterization was performed by sequencing of the calmodulin (CAL), beta tubulin (BT) and translational elongation factor 1-alpha (TEF-1α) and typing by fluorescent amplified fragment length polymorphism (FAFLP). In patients who presented with fixed (49.2%), lymphocutaneous lesions (23.8%), in 26.9% the details were not known, none had systemic dissemination. All the isolates tested were Sporothrix globosa and that could grow up to 35 °C and unable to grow at and beyond 37 °C. The assimilation of sucrose, ribitol and raffinose helps in identifying S. globosa. Sequences of CAL or BT or TEF-1α can differentiate S. globosa from other species in the complex. FAFLP results exhibited low genetic diversity. No correlation was noted between genotypes and clinical presentation, or geographic distribution. Itraconazole, terbinafine and posaconazole showed good in vitro antifungal activity against S. globosa whereas fluconazole and micafungin had no activity. S. globosa of Indian origin is relatively less pathogenic than other pathogenic Sporothrix species as it does not cause systemic dissemination and in the diagnostic laboratory, incubation of the cultures below 37 °C is essential for effective isolation.
Electronic supplementary material
The online version of this article (10.1007/s42770-020-00346-6) contains supplementary material, which is available to authorized users.
Keywords: Sporotrichosis, Sporothrix globosa, Dimorphic fungi, Genetic typing, Antifungal, Calmodulin, Taxonomy
Introduction
Sporotrichosis is a chronic granulomatous subcutaneous mycoses caused by different thermally dimorphic Sporothrix species [1]. In recent years, this dimorphic fungus has received special interest due to an increased number of infections causing epidemics and outbreaks [2]. Several phylogenetically related Sporothrix species are cosmopolitan in nature and are commonly found in tropical and subtropical countries [1, 3–6]. Humans acquire the infection by accidental percutaneous inoculation of the fungus present on a variety of vegetation, decomposing wood or soil, or by zoonotic transmission [2, 7]. Rarely, the respiratory route is the mode of acquisition [8]. Occupation-dependent infections are most frequent in the Indian subcontinent, unlike epidemics in Latin American countries where zoonotic transmission is the commonest mode of transmission [9, 10]. The severity of the disease varies from fixed cutaneous plaques/ulcerative lesions to lymphocutaneous spread to dissemination in immunocompromised patients [11, 12]. In India, the first case was reported from Calcutta in 1932 [13]. In later years, cases of sporotrichosis were reported from all over the country [7, 14–18]. As per the recent survey, it has been identified that the laboratory diagnosis of sporotrichosis is the most problematic with poor sensitivity [19].
Molecular studies from different geographical regions revealed the presence of many cryptic species in Sporothrix schenckii based on the partial calmodulin gene [20–22]. Marimon et al., proposed the existence of three pathogenic species, Sporothrix brasiliensis, Sporothrix mexicana and Sporothrix globosa within the S. schenckii complex, which were geographically distinct. In Asian countries, S. globosa is endemic and constitutes nearly 99.3% of all Sporothrix species [23]. All the Indian strains tested belonged to S. globosa [20]. Besides Asian countries, S. globosa has also been reported in Europe, the Americas and Australia [6, 23]. S. globosa, has a low degree of genetic variation in contrast to S. schenckii and S. brasiliensis [23, 24]. Recently, Sporothrix mexicana, Sporothrix pallida and Sporothrix chilensis were reported to cause human infections [23, 25, 26].
S. globosa is fourth in the virulence hierarchy after S. brasiliensis, S. schenckii, S. luriei, [27–30]. Several reports suggest variations in susceptibility profiles for Sporothrix species [31–33]. Sporotrichosis is treated by saturated potassium iodide or itraconazole for cutaneous and lymphocutaneaous forms, and amphotericin B for disseminated or itraconazole in treatment failure cases [34]. With the growing reports of treatment failure in S. globosa infections [35], routine antifungal susceptibility testing of the isolates becomes necessary.
Diversity in the genome, antifungal susceptibility, virulence of the isolates along with geographical distribution has made necessary to identify Sporothrix to species level for proper patient management. The present study is, therefore, carried out to determine the distribution and genetic diversity of Sporothrix species isolated across India and also to determine the antifungal susceptibility profiles of these isolates.
Materials and methods
Strains
A total of 63 isolates preserved as Sporothrix schenckii sensu lato at the National Collection Centre for Pathogenic Fungi (NCCPF), Postgraduate Institute of Medical Education and Research, Chandigarh, India, were included in the study. All the isolates were from human clinical samples and were isolated from geographically diverse places across India. Demographic details such as age, sex, type of lesions, year of isolation and geographic origin associated with each case were noted. The isolates were revived from the lyophilised vials and sub-cultured on potato dextrose agar (PDA) slants before using for the morphological, physiological and molecular studies.
Morphological studies
Colony morphology of all the isolates was studied on PDA, cornmeal and oatmeal agar at different temperatures (25, 30 and 35 °C). For the transition of mycelial to yeast phase, the cultures were inoculated on to brain heart infusion agar with sheep blood agar (BHIBA) incubated at 35 °C for up to 21 days. The colonies on BHIBA were suspended in saline to check for yeast conversion. To study the microscopic features of the mycelial phase, the isolates were subjected to micro-slide cultures on cornmeal agar. After incubation at 30 °C for 14 or 21 days, the coverslips were removed from the slide culture, stained with lactophenol cotton blue and examined under a light microscope.
Physiological studies
To determine the growth rate at various temperatures (25, 30, 35 and 37 °C), each isolate was inoculated with a 1-mm square piece of fungal growth (on PDA) on to the duplicate fresh PDA plates. The colonies were measured after 14 days and the mean value was recorded. The capability to assimilate sucrose, ribitol and raffinose was performed in liquid nitrogen base media in 96-well microplates as per the method described by Yarrow [36].
DNA extraction, amplification and sequencing
Sequencing reaction/molecular identification using primers to amplify CAL, BT and TEF1-α was carried out for all the 63 isolates. Degenerate primers CL1 (5′-GARTWCAAGGAGGCCTTCTC-3′) and CL2A (51-TTTTTGCATCATGAGTTGGAC-3′) were used to amplify the CAL locus region as described by O’Donnell [37]. Bt2-F [5′GG[CT]AACCA(AG)AT(ATC)GGTGC(CT)GC(CT)3′] and Bt2-R [5′ACCCTC(AG)GTGTAGTGACCCTTGGC3′] were used to amplify beta tubulin, and EF1-F (5′-CTGAGGCTCGTTACCAGGAG-3′) and EF1-R (5′-CGACTTGATGACACCGACAG-3′) primers were used to amplify TEF1-α region. Briefly, isolates were cultured on to PDA and incubated at 25 °C for 1 week, and DNA extraction and purification was carried out using a phenol chloroform extraction method. DNA was quantified using NanoDrop 2000 spectrophotometer (Thermo Fischer Scientific, Wilmington, DE, USA). The amplification of all the three gene (CAL, BT and TEF1-α) fragments was carried out directly from genomic DNA. For a reaction volume of 25 μl, 2.5 μl of the 1:10 diluted genomic DNA and 0.5 mM of each primer were added. PCR products were amplified with the following temperature profiles: denaturation for 5 min at 94 °C, followed by 35 cycles of 30 s at 95 °C, 1 min at the annealing temperature of 60 °C (CAL and BT) and 57 °C (TEF1-α) and 1 min at 72 °C, followed by final extension for 5 min at 72 °C. Sequencing PCR was carried out using Big Dye Terminator Cycle Sequencing Kit Version 3.1 (Applied Biosystems, Foster City, CA, USA) as per the kit protocol. For each gene fragment, both the positive strand and the negative strand were sequenced using both forward and reverse primer respectively. The sequence analysis was carried out with ABI 3130 genetic analyser (Applied Biosystems). The sequences of all the strains were deposited in GenBank under the accession numbers as mentioned in supplementary table – 1.
All the three gene sequences of the isolates were verified by BLASTN (http://www.ncbi.nlm.nih.gov/blast) search analysis. Furthermore, forward and reverse sequences of respective strains and genes were assembled into single contig via Lasergene’s Seqman software. Mis-paired base pairs were manually edited in the consensus sequence.
Phylogenetic analysis
Sequences of CAL, BT and TEF1-α of S. schenckii species of clinical importance belonging to the different clades were retrieved from GenBank. Multiple sequence comparison was performed using Clustal X2 algorithm and implemented in MEGA 6 software. Grosmannia serpens (CBS 141.36) [38], a saprophytic fungus belonging to Ophiostomataceae, was used as an outgroup for CAL analysis [39]. For the BT and TEF1-α analysis Ophiostoma piliferum (CBS 158.74) was used as outgroup [40].
Phylogenetic analyses were carried out using neighbour-joining method. Neighbour-joining trees were constructed using MEGA 6 software [41] and 1000 bootstrap replicates were used to estimate confidence values for individual clades [42]. The evolutionary distances were computed using the Kimura 2-parameter method [43]. Genetic diversity and haplotyping of CAL, BT and TEF1-α genes were estimated using DNAsp v5.10 [44]. Gaps and missing data were excluded in the calculation and the rate variation among sites was modelled with a gamma distribution (shape parameter = 1).
Antifungal susceptibility testing
Minimum inhibitory concentrations (MIC) were determined for antifungal agents’ fluconazole, itraconazole, voriconazole, posaconazole, amphotericin B, terbinafine and micafungin (Sigma-Aldrich, India) against 63 Sporothrix isolates. Testing was carried out by the micro-broth dilution technique according to the Clinical and Laboratory Standards Institute’s M38-A2 method [45]. The final drug concentration was adjusted from 0.03 to 16 μg/ml for fluconazole, itraconazole, voriconazole, posaconazole, amphotericin B and terbinafine, 0.03 to 4 μg/ml for micafungin. The Sporothrix isolates were freshly grown on PDA for 5–7 days at 25 °C; the conidia were harvested and used to prepare inoculum. Inoculum size was established at an optical density ranging 0.09 to 0.13 in a spectrophotometer at 530 nm. Inoculated plates were incubated at 35 °C and results were observed visually after 3 days. For quality control, Candida parapsilosis ATCC 22019 and Candida krusei ATCC 6258 strains were used.
Fluorescent amplified fragment length polymorphism genotyping
Sporothrix isolates were subjected to genotyping by fluorescent amplified fragment length polymorphism technique as previously described by Chakrabarti et al. [46]. Briefly combined restriction and ligation step was carried out using EcoRI and MseI enzymes and T4 DNA ligase (New England Biolabs, Ipswich, MA, USA). Amplification with pre-selective primers EcoRI (5′GACTGCGTACCAATTC-3′) and MseI (5′GATGAGTCCTGAGTAA-3′) was performed. EcoRI with one selective residue (5′-Flu-GACTGCGTACCAATTCAC-3′) and MseI primer with two selective residues were used (5′-GATGAGTCCTGAGTAACG-3′) for selective amplification. One of the selective primers labelled with 6-FAM. LIZ-500 was used as a standard marker. Capillary electrophoresis of the amplified products was carried out in ABI-automated DNA sequencer 3130 (Applied Biosystems, Foster City, CA, USA). Typing data were imported to BIONUMERICS v7.6 software (Applied Maths, Ghent, Belgium). Fingerprint curves were converted into bands and correct bands of each lane were assigned using the band position of the reference dye (LIZ500). The similarity coefficients were determined by the Pearson correlation with negative similarities clip to zero. Cluster analysis was performed using the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) using BIONUMERICS software.
Results
Among the 63 culture-proven sporotrichosis cases evaluated, 31 patients had a history of fixed cutaneous type and 15 patients had a lymphocutaneous type of infection whereas clinical details of 17 patients were not available. The majority of the patients were in the age group of 32 to 60 (mean age—46.7 years). The oldest patient was an 85-year-old female and the youngest patient was a 15-year-old male. Of the 44 cases with available demographic details, 21 were males and 23 females. The 63 clinical isolates were received from different geographical areas of India. Forty-seven isolates were from patients of northern India (24 from Himachal Pradesh, 21—Chandigarh and 2—Delhi), 6 isolates were from eastern India (3—Kolkata, 3—Assam) and 10 isolates were from southern India (8—Belgaum and 2—Bengaluru) (Table 1).
Table 1.
Demographics, clinical type, isolation place and year of Sporothrix species included in the study
| Serial. No | NCCPF No | Age/sex | Lesion type | Year | Place of origin |
|---|---|---|---|---|---|
| 1 | 220007 | 50/F | Lymphocutaneous | 1994 | Chandigarh |
| 2 | 220010 | 30/M | Fixed | 1995 | Chandigarh |
| 3 | 220011 | 22/M | Fixed | 1995 | Chandigarh |
| 4 | 220012 | 35/M | Lymphocutaneous | 1996 | Belgaum |
| 5 | 220026 | 36/F | Fixed | 1996 | Chandigarh |
| 6 | 7220027 | 85/F | Fixed | 1996 | Chandigarh |
| 7 | 220028 | 37/M | Fixed | 1996 | Belgaum |
| 8 | 220029 | 45/F | Fixed | 1996 | Kolkata |
| 9 | 220030 | 60/F | Lymphocutaneous | 1996 | Chandigarh |
| 10 | 220032 | 65/F | Fixed | 1997 | Chandigarh |
| 11 | 220037 | NA | NA | 1997 | Kolkata |
| 12 | 220038 | 48/M | Lymphocutaneous | 1997 | Belgaum |
| 13 | 220040 | 53/F | Fixed | 1997 | Chandigarh |
| 14 | 220041 | 65/M | Lymphocutaneous | 1997 | Chandigarh |
| 15 | 220045 | 35/F | Fixed | 1997 | Kolkata |
| 16 | 220047 | 46/M | Fixed | 1997 | Chandigarh |
| 17 | 220048 | 55/M | Lymphocutaneous | 1996 | Belgaum |
| 18 | 220049 | 55/F | Lymphocutaneous | 1997 | Himachal Pradesh |
| 19 | 220071 | 38/F | Fixed | 2000 | Chandigarh |
| 20 | 220072 | NA | NA | 2001 | Chandigarh |
| 21 | 220073 | NA | NA | 2001 | Chandigarh |
| 22 | 220076 | 35/M | Lymphocutaneous | 2001 | Belgaum |
| 23 | 220078 | 72/M | Lymphocutaneous | 2002 | Chandigarh |
| 24 | 220079 | 44/F | Fixed | 2002 | Himachal Pradesh |
| 25 | 220080 | 32/M | Fixed | 2003 | Chandigarh |
| 26 | 220082 | 36/F | Lymphocutaneous | 2003 | Belgaum |
| 27 | 220087 | 44/M | Lymphocutaneous | 2005 | Chandigarh |
| 28 | 220088 | 15/M | Lymphocutaneous | 2006 | Chandigarh |
| 29 | 220089 | NA | NA | 2006 | Chandigarh |
| 30 | 220090 | 65/F | Fixed | 2006 | Chandigarh |
| 31 | 220091 | 72/M | Lymphocutaneous | 2006 | Himachal |
| 32 | 220093 | 58/F | Fixed | 2007 | Chandigarh |
| 33 | 220094 | 42/M | Lymphocutaneous | 2008 | Himachal Pradesh |
| 34 | 220095 | 36/M | Fixed | 2008 | Bangalore |
| 35 | 220101 | NA | Fixed | 2010 | Delhi |
| 36 | 220103 | NA | Fixed | 2010 | Delhi |
| 37 | 220105 | NA | NA | 2010 | Bangalore |
| 38 | 220111 | NA | Lymphocutaneous | 2011 | Himachal Pradesh |
| 39 | 220112 | NA | NA | 2011 | Himachal Pradesh |
| 40 | 220113 | NA | NA | 2011 | Himachal Pradesh |
| 41 | 220114 | NA | NA | 2011 | Himachal Pradesh |
| 42 | 220116 | NA | NA | 2012 | Himachal Pradesh |
| 43 | 220118 | NA | NA | 2012 | Himachal Pradesh |
| 44 | 220119 | NA | NA | 2012 | Himachal Pradesh |
| 45 | 220120 | NA | NA | 2012 | Himachal Pradesh |
| 46 | 220122 | NA | NA | 2012 | Himachal Pradesh |
| 47 | 220124 | NA | NA | 2012 | Himachal Pradesh |
| 48 | 220125 | 25/F | Fixed | 2012 | Himachal Pradesh |
| 49 | 220126 | 45/F | Fixed | 2013 | Belgaum |
| 50 | 220127 | 45/F | Fixed | 2013 | Belgaum |
| 51 | 220129 | 57/F | Fixed | 2015 | Chandigarh |
| 52 | 220135 | NA | Fixed | 2015 | Assam |
| 53 | 220136 | NA | NA | 2015 | Assam |
| 54 | 220137 | NA | NA | 2015 | Assam |
| 55 | 220138 | 56/M | Fixed | 2015 | Himachal Pradesh |
| 56 | 220139 | 30/M | Fixed | 2015 | Himachal Pradesh |
| 57 | 220240 | 40/F | Fixed | 2015 | Himachal Pradesh |
| 58 | 220142 | 26/F | Fixed | 2015 | Himachal Pradesh |
| 59 | 220143 | 63/M | Fixed | 2015 | Himachal Pradesh |
| 60 | 220144 | 55/F | Fixed | 2015 | Himachal Pradesh |
| 61 | 220145 | 62/M | Fixed | 2015 | Himachal Pradesh |
| 62 | 220146 | 48/F | Fixed | 2015 | Himachal Pradesh |
| 63 | 220149 | NA | NA | 2015 | Himachal Pradesh |
Colony characteristics on three different media tested were similar with no significant variation. After 21 days of incubation at 30 °C, the colonies were grey which later produced brownish to dark brown–coloured colonies. The colony diameter measured between 18 and 26 mm (mean diameter = 21.8 mm) on plates incubated at 25 and 30 °C, whereas colony diameter measured from 6 to 24 mm (mean diameter = 14.2 mm) on plates incubated at 35 °C. On CMA and oatmeal agar, colonies of 63 isolates were brown to dark brown at similar incubation conditions. Colonies on BHIBA appeared white to light brown yeast-like growth after 10–12 days of incubation. Failure to convert to the yeast phase was noted in approximately half of the isolates tested (15/29). Microscopically, the yeasts are elongated or cigar-shaped measuring about 5–7 μm × 1–3 μm.
The isolates produced conidia in clusters either terminal or intercalary on well-differentiated conidiophores. Conidiophores were swollen and produced sympodial conidia on the denticles. The conidia were hyaline to subhyaline, obovoidal measuring about 3–5 μm × 1–3 μm. In addition, the second type of conidia described as sessile conidia was produced within 14 days in most of the isolates and by day 21 in all isolates. The sessile conidia were brown to dark brown, thick walled, globose to sub-globose. All the isolates assimilated sucrose and ribitol whereas raffinose was negative for all isolates.
The sequences 733, 395 and 708 nucleotides of CAL, BT and TEF-1α respectively were amplified and all the isolates were identified as S. globosa. The phylogenetic tree constructed based on CAL, BT and TEF-1α is presented in supplementary figure 1-3 respectively. The combined dataset (CAL + TEF-1α + BT) yielded a sequence alignment of 1836 positions, including 1807 invariable characters, 27 (1.5%) variable parsimony informative sites and 10 singleton variable sites. The concatenated sequences of CAL + BT + TEF-1α yielded 23 haplotypes on haplotyping analysis. Haplotype diversity (Hd) and nucleotide diversity (Pi) were 0.906 and 0.00245 respectively.
Genotyping analysis of the amplified fragment of three individual gene loci was performed using DNAsp software 5.10. The aligned CAL sequences were 733 bp long, including 717 invariable sites with 12 parsimony informative sites. The haplotype analysis of the CAL sequence divided the isolates into 10 Hap groups (Hd = 0.6114) (Pi = 0.00322). The aligned BT sequences were 395 bp long, including 391 invariable sites with 2 parsimony informative site and 4 singleton variable sites. The haplotype analysis of BT sequences divided the isolates into 6 Hap groups (Hd = 0.661) (pi = 0.00265). The aligned TEF1-α sequences were 708 bp long including 699 invariable sites with 3 parsimony informative sites and 6 singleton variable sites. The haplotype analysis of the TEF1-α sequence divided the isolates into 10 Hap groups. All the haplotypes (Hd = 0.7035) belonged to the same group with less nucleotide diversity (Pi = 0.00270) (Table 2).
Table 2.
Comparison of nucleotide and haplotype variations among calmodulin, beta-tubulin and Translational elongation factor 1-α genes
| Parameter | Calmodulin | Beta-tubulin | Translational elongation factor 1-α |
|---|---|---|---|
| S (number of variable sites) | 14 | 4 | 9 |
| H (total no. of mutations) | 14 | 4 | 9 |
| π (nucleotide diversity per site) | 0.00322 | 0.00265 | 0.00270 |
| h (number of haplotypes) | 10 | 6 | 10 |
| hd Haplotype diversity | 0.611 | 0.661 | 0.7035 |
| k average number of nucleotide differences | 2.344 | 1.041 | 1.111 |
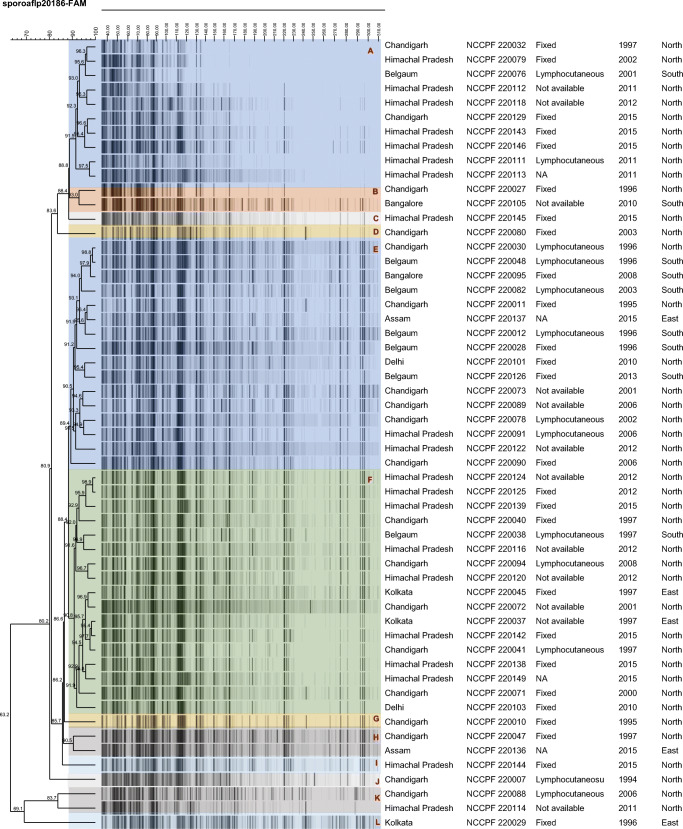
A total of 55 isolates yielding sufficient bands were included in the FAFLP (fluorescent amplified fragment length polymorphism) analysis. FAFLP profiles yielded fragments ranging from 10 to 390 base pairs, but only fragments ranging from 25 to 300 bp were included for the analysis. All Sporothrix isolates had more than 60% similarity. FAFLP profile yielded 12 clusters (Fig. 1). The majority of the isolates formed three major clusters, i.e. cluster ‘A’, ‘E’ and ‘F’. In the cluster ‘A’, all the isolates belonged to northern India except for one isolate. Cluster ‘E’ comprised most of southern India isolates. Cluster ‘C’, ‘D’, ‘G’, ‘I’, ‘J’ and ‘L’ consisted of single isolate whereas ‘B’, ‘H’ and ‘K’ had two isolates each.
Fig. 1.
Dendrogram generated by UPGMA analysis of FAFLP profiles obtained from S. globosa
The MIC, geometric mean, MIC50 and MIC90 for tested isolates against various antifungals are provided in Table 3.
Table 3.
Antifungal susceptibility profile of Sporothrix globosa isolates of Indian origin against 7 antifungal agents (values in μg/ml)
| Antifungal agent | Range | MIC50 | MIC90 | GM |
|---|---|---|---|---|
| Fluconazole | 2–16 | 16 | 16 | 13.27 |
| Itraconazole | 0.03–2 | 0.25 | 1 | 0.218 |
| Voriconazole | 0.03–16 | 0.25 | 8 | 0.454 |
| Posaconazole | 0.03–2 | 0.25 | 1 | 0.315 |
| Amphotericin B | 0.03–8 | 1 | 2 | 1.064 |
| Micafungin | 4 | 4 | 4 | 4 |
| Terbinafine | 0.03–2 | 0.03 | 0.06 | 0.043 |
Discussion
The present study describes the clinical features of sporotrichosis cases across India and characterises the isolates from those cases, both phenotypic and molecular characters, and antifungal susceptibility profile. The majority patients were males, but there was no significant difference between male and female patients similar to the report of China [47]. However, our study was only on referred cases to our centre. So no definite demographic specifics can be made from referred cases. Earlier studies from north India indicated females, who were involved in agricultural activities, were common sufferer [10, 14]. A meta-analysis by Zhang et al., in 2015 reported that S. globosa was common in the female population in different continents [23]. A study from northeast China also reported female preponderance and their involvement in agricultural activities. Sporotrichosis can affect persons regardless of age and gender. Occupational and other recreational activities influence the rate of infection in different population [48].
Phenotypic characterization of our Sporothrix isolates showed maximum growth (mean growth rate 21.8–21.9 mm) on PDA media incubated at 25 °C and 30 °C after 21 days of incubation, but stunted growth (mean 14.2 mm) at 35 °C and no growth at 37 °C. The findings are in concordance with the study by Marimon et al., in which none of the S. globosa isolates tested grew at 37 °C [49]. In contrast to our findings, S. globosa isolates of Venezuelan origin grew at 37 °C after 21 days of incubation [50] and 97% of the isolates from China exhibited stunted growth at 37 °C [51]. According to Dixon et al., Sporothrix isolates that produce pigmented conidia can grow at 37 °C and are virulent in the murine model [52]. Kwon-Chung et al., reported that the isolates obtained from lymphocutaneous lesions were able to grow at 37 °C unlike isolates from fixed cutaneous which exhibited growth at 35 °C [53]. However, our isolates irrespective of origin from lymphocutaneous or fixed cutaneous lesion produced dark brown pigmented conidia after 21 days of incubation but did not grow at 37 °C. The above phenotypic characters described by several workers were before the description of the new Sporothrix spp. It is now believed that the differences in phenotypic characters may be attributed to the different Sporothrix species implicated rather than the type of lesion and geographical location of the isolates. The report from northeast China noted S. globosa was responsible for all clinical types of sporotrichosis [54]. Generally, S. globosa isolates from all geographical regions are less virulent compared with S. schenckii sensu stricto (25,54,55).
Morphologically, Sporothrix species varies minimally and it is difficult even for the experienced mycologist to differentiate different species within the genus Sporothrix. Marimon et al., after testing a large panel of sugars, reported that assimilation of sucrose, ribitol and raffinose can help in the identification of S. globosa [20]. Hence, sugar assimilation can be successfully used in the resource constrained laboratory for the preliminary identification of S. globosa. In the present study, all isolates were ribitol and sucrose assimilation positive and raffinose negative. In the study of Marimon et al., 91% of S. globosa isolates assimilated ribitol. Possibly, such studies with more isolates from the different geographical region are required for confident identification of Sporothrix species phenotypically.
Though S. globosa is considered as a cosmopolitan species, it exhibits geographical preferences [55] with prevalence rate highest in Asia (56%) and least in Africa (5%) whereas in Europe and Americas, its prevalence is 28% and 11% respectively [55]. In this study using multigene sequencing, we confirmed that all the Sporothrix isolates obtained from various parts of India are S. globosa. Based on phylogenetic analysis, Indian isolates along with isolates of China, Japan, Spain, Italy and the USA were in the clade III clinical group (S. globosa) [20]. The majority of the Sporothrix species prevailing in Asia belong to S. globosa (99.3%) [23]. Instead of three genes, the six species of clinically important Sporothrix species can be differentiated by the CAL sequence only [20]. In this study, the calmodulin gene also identified all the Indian isolates as S. globosa (Supplementary Fig. 1) and it is the only causative species of sporotrichosis in India. Until the recent report of the presence of S. schenckii in the central part of China, it was thought that only S. globosa existed in that country [47, 54, 56]. According to Yu et al., S. globosa isolates from the Northeastern part of China differentiated into two subclades. Majority of the isolates in our study grouped under subclade I and only six isolates grouped under subclade II along with Italian isolate [54, 57]. In the present study, 16 isolates grouped along with the representative strain from subclade II of Chinese and Italian isolates. Phylogenetic analysis revealed that our isolates are genetically similar to environmental isolates of China and clinical isolates of Spain, Japan, Columbia and the USA (Supplementary Figure 1). This finding corroborates with other reports where S. globosa isolates originating from different continents displayed identical profile or low genetic diversity [23, 24, 58]. Gong et al., recently reported the utility of microsatellite typing in identifying genetically diverse S. globosa, similar to a multigene approach (CAL, TEF1-α or BT genes) [59]. However, whole genome sequencing results from Australian S. globosa isolates revealed low genome diversity [6].
Haplotyping analyses placed all our isolates into 10 haplotypes whereas S. globosa isolates belonging to Venezuela had only five haplotypes [50]. High genetic variation was observed in S. brasiliensis and S. schenckii sensu stricto [58]. Similar findings were also reported by Moussa et al., in which the S. globosa population had low diversity (9 haplotypes) compared with S. schenckii (42 haplotypes). The meta-analysis by Zhang et al., also showed that in defined endemic areas, a single molecular type of S. globosa is preponderant (> 80%) [23].
BT and TEF1-α were also used as markers to assess the genetic diversity within the species. Phylogenetically BT sequence can also differentiate S. globosa from other Sporothrix species [55]. This is in contrast with the study from Marimon et al. [20], where the BT sequences from representative Sporothrix species (S. brasiliensis AM116946, S. globosa AF116966 and S. mexicana AM498344) were in very low homology. Analysis of the BT sequence grouped our isolates into three clades (Supplementary Figure 2). But all our isolates are grouped together along with the S. globosa standard strain (AM116966). Intraspecific genetic variation is higher in the TEF1-α gene among S. globosa isolates compared with S. brasiliensis, where haplotyping revealed only three haplogroups [2]. However, the TEF1-α gene was able to differentiate two genotypes responsible for causing an epidemic in Brazil at the same time period. Haplotyping analysis of TEF1-α gene of our isolates showed 10 different haplotypes, with 31 isolates in type III, 13 isolates in type I, 7 isolates in type IX, 6 isolates in type VII and one isolate each of remaining types.
FAFLP has been shown to be a useful tool to study the molecular epidemiology of S. globosa. In a study by Gong et al., microsatellite marker–based typing has shown similar potential as FAFLP in discriminating geographic variation among S. globosa [59]. We used FAFLP in the present study as it is relatively cheaper and less cumbersome than microsatellite-based typing. FAFLP analysis divided our strains into 12 different clusters. FAFLP genotypes have not been found to be related to pathology, disease profile or geography whereas FAFLP profiles of S. globosa isolates of China exhibited regional difference [51]. The probable reason for multiple genotypes originating from a single region may be due to frequent migration of population across the country. FAFLP analysis by Zhang et al., also showed a low degree of variation among S. globosa isolates from different regions compared with other species in the Sporothrix species. This is in concordance with the study from Neyra et al., in which all the Peruvian isolates clustered into two groups irrespective of their pathobiology and geography within the area [57]. Low genetic variability among S. globosa from different continents having large geographic distance may be due to the emergence of rapid vectors that have a critical role in the dispersal of S. globosa across continent. Rangel-Gamboa et al., hypothesise that organic products and the exchange of foods via the international market could be the reason for its worldwide transmission [24]. Similar to previous reports, we also did not find any specific association between the FAFLP genotypes and susceptibility profile [51].
Antifungal susceptibility testing for Sporothrix species is performed according to the Clinical Laboratory Standard Institute (CLSI) methods [29, 60–62]. There are no defined breakpoints or epidemiological cut-off values (ECV) available for S. globosa [63]. Even reports on the antifungal susceptibility profile of S. globosa are scarce. Our results showed that itraconazole is an effective drug with MIC 90 of 1 μg/ml, whereas S. globosa isolates from Japan had poor activity against itraconazole compared with isolates tested in this study [64]. Voriconazole exhibited varied in vitro antifungal activity. While the majority (87.3%, 55 isolates) had lower MICs, only eight isolates (12.7%) exhibited higher MICs ranging between 4 and 16 μg/ml. Like other reports, our reports also confirm that fluconazole and micafungin have no activity on S. globosa [29, 60, 65]. Terbinafine showed good activity with low MIC values which is consistent with other studies on S. schenckii and S. globosa [29, 30, 51, 66–68]. But the clinical efficacy of terbinafine for the treatment of sporotrichosis due to S. globosa is not known and worth evaluating.
In conclusion, all the Sporothrix isolates across different geographic regions of India are S. globosa. None of our isolates were from disseminated cases and did not grow at 37 °C. Sucrose, ribitol and raffinose could be employed for differentiating S. globosa from other Sporothrix species. CAL, BT and TEF1-α genes could be used to differentiate S. globosa from other species in the complex. The translational elongation factor gene has more genetic variability compared with other genes. FAFLP analysis yielded a significant number of genotypes but is not associated with phenotypic characters or geographic locations. Itraconazole and terbinafine exhibited good in vitro activity.
Electronic supplementary material
(DOCX 59 kb)
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.López-Romero E, del Rocio Reyes-Montes M, Pérez-Torres A, et al. Sporothrix schenckii complex and sporotrichosis, an emerging health problem. Future Microbiol. 2011;6:85–102. doi: 10.2217/fmb.10.157. [DOI] [PubMed] [Google Scholar]
- 2.Rodrigues AM, de Melo TM, de Hoog GS, et al. Phylogenetic analysis reveals a high prevalence of Sporothrix brasiliensis in feline sporotrichosis outbreaks. PLoS Negl Trop Dis. 2013;7:e2281. doi: 10.1371/journal.pntd.0002281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pappas PG, Tellez I, Deep AE, et al. Sporotrichosis in Peru: description of an area of hyperendemicity. Clin Infect Dis. 2000;30:65–70. doi: 10.1086/313607. [DOI] [PubMed] [Google Scholar]
- 4.Ghosh A, Chakrabarti A, Sharma VK, Singh K, Singh A. Sporotrichosis in Himachal Pradesh (North India) Trans R Soc Trop Med Hyg. 1999;93:41–45. doi: 10.1016/S0035-9203(99)90173-6. [DOI] [PubMed] [Google Scholar]
- 5.Schubach A, Schubach TMP, de Lima Barros MB, Wanke B. Cat-transmitted sporotrichosis, Rio de Janeiro, Brazil. Emerg Infect Dis. 2005;11:1952–1954. doi: 10.3201/eid1112.040891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.New D, Beukers AG, Kidd SE, Merritt AJ, Weeks K, van Hal SJ, Arthur I. Identification of multiple species and subpopulations among Australian clinical Sporothrix isolates using whole genome sequencing. Med Mycol. 2019;57:905–908. doi: 10.1093/mmy/myy126. [DOI] [PubMed] [Google Scholar]
- 7.Yegneswaran PP, Sripathi H, Bairy I, Lonikar V, Rao R, Prabhu S. Zoonotic sporotrichosis of lymphocutaneous type in a man acquired from a domesticated feline source: report of a first case in southern Karnataka, India. Int J Dermatol. 2009;48:1198–1200. doi: 10.1111/j.1365-4632.2008.04049.x. [DOI] [PubMed] [Google Scholar]
- 8.Aung AK, Teh BM, McGrath C, Thompson PJ. Pulmonary sporotrichosis: case series and systematic analysis of literature on clinico-radiological patterns and management outcomes. Med Mycol. 2013;51:534–544. doi: 10.3109/13693786.2012.751643. [DOI] [PubMed] [Google Scholar]
- 9.Bravo TC. New observations on the epidemiology of sporotrichosis and Sporothrix schenckii complex 1. Sources Infect Occup Risks. 2012;59:88–100. [Google Scholar]
- 10.Verma S, Verma GK, Singh G, Kanga A, Shanker V, Singh D, Gupta P, Mokta K, Sharma V. Sporotrichosis in sub-himalayan India. PLoS Negl Trop Dis. 2012;6:e1673. doi: 10.1371/journal.pntd.0001673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agarwal S, Gopal K, Umesh, Kumar B. Sporotrichosis in Uttarakhand (India): a report of nine cases. Int J Dermatol. 2008;47:367–371. doi: 10.1111/j.1365-4632.2008.03538.x. [DOI] [PubMed] [Google Scholar]
- 12.Carvalho MT, de Castro AP, Baby C, Werner B, Filus Neto JQ-TF. Disseminated cutaneous sporotrichosis in a patient with AIDS: report of a case. Rev Soc Bras Med Trop. 2002;35:655–659. doi: 10.1590/S0037-86822002000600018. [DOI] [PubMed] [Google Scholar]
- 13.Ghosh L. An unusual case of sporotrichosis. Indian Med Gaz. 1932;67:570. [PMC free article] [PubMed] [Google Scholar]
- 14.Mahajan VK, Sharma NL, Sharma RC, Gupta ML, Garg G, Kanga AK. Cutaneous sporotrichosis in Himachal Pradesh, India. Mycoses. 2005;48:25–31. doi: 10.1111/j.1439-0507.2004.01058.x. [DOI] [PubMed] [Google Scholar]
- 15.Randhawa HS, Chand R, Mussa AY, et al. Sporotrichosis in India: first case in a Delhi resident and an update. Indian J Med Microbiol. 2003;21:12–16. [PubMed] [Google Scholar]
- 16.Baruah BD, Saikia TC, Bhuyan RN. Sporotrichosis in Assam (a clinical and mycological study) J Indian Med Assoc. 1976;67:223–229. [PubMed] [Google Scholar]
- 17.Devi KRSR, Devi MU, Singh TN, Devi KS, Sharma SS, Singh LR, Singh HL, Singh NB. Emergence of sporotrichosis in Manipur. Indian J Med Microbiol. 2006;24:216–219. [PubMed] [Google Scholar]
- 18.Kamalam A, Thambiah AS. Sporotrichosis--first case report from Madras. Mykosen. 2008;25:2008. doi: 10.1111/j.1439-0507.1982.tb01923.x. [DOI] [PubMed] [Google Scholar]
- 19.Hay R, Denning DW, Bonifaz A, Queiroz-Telles F, Beer K, Bustamante B, Chakrabarti A, Chavez-Lopez MG, Chiller T, Cornet M, Estrada R, Estrada-Chavez G, Fahal A, Gomez BL, Li R, Mahabeer Y, Mosam A, Soavina Ramarozatovo L, Rakoto Andrianarivelo M, Rapelanoro Rabenja F, van de Sande W, Zijlstra EE. The diagnosis of fungal neglected tropical diseases (fungal NTDs) and the role of investigation and laboratory tests: an expert consensus report. Trop Med Infect Dis. 2019;4:122. doi: 10.3390/tropicalmed4040122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marimon R, Cano J, Gené J, et al. Sporothrix brasiliensis, S. globosa, and S. mexicana, three new Sporothrix species of clinical interest. J Clin Microbiol. 2007;45:3198–3206. doi: 10.1128/JCM.00808-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marimon R, Gené J, Cano J, Guarro J. Sporothrix luriei: a rare fungus from clinical origin. Med Mycol. 2008;46:621–625. doi: 10.1080/13693780801992837. [DOI] [PubMed] [Google Scholar]
- 22.de Meyer EM, de Beer ZW, Summerbell RC, Moharram AM, de Hoog GS, Vismer HF, Wingfield MJ. Taxonomy and phylogeny of new wood- and soil-inhabiting Sporothrix species in the Ophiostoma stenoceras-Sporothrix schenckii complex. Mycologia. 2008;100:647–661. doi: 10.3852/07-157R. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Hagen F, Stielow B, Rodrigues AM, Samerpitak K, Zhou X, Feng P, Yang L, Chen M, Deng S, Li S, Liao W, Li R, Li F, Meis JF, Guarro J, Teixeira M, al-Zahrani HS, de Camargo ZP, Zhang L, de Hoog GS. Phylogeography and evolutionary patterns in Sporothrix spanning more than 14 000 human and animal case reports. Persoonia. 2015;35:1–20. doi: 10.3767/003158515X687416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rangel-Gamboa L, Martínez-Hernandez F, Maravilla P, Arenas-Guzmán R, Flisser A. Update of phylogenetic and genetic diversity of Sporothrix schenckii sensu lato. Med Mycol. 2016;54:248–255. doi: 10.1093/mmy/myv096. [DOI] [PubMed] [Google Scholar]
- 25.Morrison AS, Lockhart SR, Bromley JG, Kim JY, Burd EM. An environmental Sporothrix as a cause of corneal ulcer. Med Mycol Case Rep. 2013;2:88–90. doi: 10.1016/j.mmcr.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodrigues AM, Cruz Choappa R, Fernandes GF, de Hoog GS, de Camargo ZP. Sporothrix chilensis sp. nov. (Ascomycota: Ophiostomatales), a soil-borne agent of human sporotrichosis with mild-pathogenic potential to mammals. Fungal Biol. 2016;120:246–264. doi: 10.1016/j.funbio.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 27.Arrillaga-Moncrieff I, Capilla J, Mayayo E, Marimon R, Marine M, Genis J, Cano J, Guarro J. Different virulence levels of the species of Sporothrix in a murine model. Clin Microbiol Infect. 2009;15:651–655. doi: 10.1111/j.1469-0691.2009.02824.x. [DOI] [PubMed] [Google Scholar]
- 28.Fernández-Silva F, Capilla J, Mayayo E, Guarro J. Virulence of Sporothrix luriei in a murine model of disseminated infection. Mycopathologia. 2012;173:245–249. doi: 10.1007/s11046-011-9506-7. [DOI] [PubMed] [Google Scholar]
- 29.Marimon R, Serena C, Gené J, et al. In vitro antifungal susceptibilities of five species of Sporothrix. Antimicrob Agents Chemother. 2008;52:732–734. doi: 10.1128/AAC.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silveira CP, Torres-Rodríguez JM, Alvarado-Ramírez E, et al. MICs and minimum fungicidal concentrations of amphotericin B, itraconazole, posaconazole and terbinafine in Sporothrix schenckii. J Med Microbiol. 2009;58:1607–1610. doi: 10.1099/jmm.0.007609-0. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Huang H, Feng P, Zhang J, Zhong Y, Xue R, Xie Z, Li M, Xi L. In vitro activity of itraconazole in combination with terbinafine against clinical strains of itraconazole-insensitive Sporothrix schenckii. Eur J Dermatol. 2011;21:573–576. doi: 10.1684/ejd.2011.1400. [DOI] [PubMed] [Google Scholar]
- 32.Alvarado-Ramírez E, Torres-Rodríguez JM. In vitro susceptibility of Sporothrix schenckii to six antifungal agents determined using three different methods. Antimicrob Agents Chemother. 2007;51:2420–2423. doi: 10.1128/AAC.01176-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliveira DC, Lopes PGM, Spader TB, Mahl CD, Tronco-Alves GR, Lara VM, Santurio JM, Alves SH. Antifungal susceptibilities of Sporothrix albicans, S. brasiliensis, and S. luriei of the S. schenckii complex identified in Brazil. J Clin Microbiol. 2011;49:3047–3049. doi: 10.1128/JCM.00255-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kauffman CA, Bustamante B, Chapman SW, Pappas PG. Clinical practice guidelines for the management of sporotrichosis: 2007 update by the Infectious Diseases Society of America. Clin Infect Dis. 2007;45:1255–1265. doi: 10.1086/522765. [DOI] [PubMed] [Google Scholar]
- 35.Gompertz OF, Rodrigues AM, Fernandes GF, et al. Case report: atypical clinical presentation of sporotrichosis caused by Sporothrix globosa resistant to itraconazole. Am J Trop Med Hyg. 2016;94:1218–1222. doi: 10.4269/ajtmh.15-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.D Y (1988) Methods for the isolation, maintenance and identification of yeasts. In: Kurtzman C. and JWF (ed) The Yeasts, A Taxonomic Study, 4th ed. Elsevier, pp 77–100
- 37.O’Donnell K, Nirenberg HI, Aoki T, Cigelnik E. A multigene phylogeny of the Gibberella fujikuroi species complex: detection of additional phylogenetically distinct species. Mycoscience. 2000;41:61–78. doi: 10.1007/BF02464387. [DOI] [Google Scholar]
- 38.Duong TA, de Beer ZW, Wingfield BD, Wingfield MJ. Phylogeny and taxonomy of species in the Grosmannia serpens complex. Mycologia. 2012;104:715–732. doi: 10.3852/11-109. [DOI] [PubMed] [Google Scholar]
- 39.Rodrigues AM, de Hoog S, de Camargo ZP. Emergence of pathogenicity in Sporothrix schenckii complex. Med Mycol. 2013;51:405–412. doi: 10.3109/13693786.2012.719648. [DOI] [PubMed] [Google Scholar]
- 40.Spatafora JW, Sung G-H, Johnson D et al A five-gene phylogeny of Pezizomycotina. Mycologia 98:1018–1028 [DOI] [PubMed]
- 41.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary eistance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hillis DM, Bull JJ. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol. 1993;42:182–192. doi: 10.1093/sysbio/42.2.182. [DOI] [Google Scholar]
- 43.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 44.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 45.CLSI Reference method for broth dilution antifungal susceptibility testing of filamentous fungi; approved standard - CLSI document M38-A2. Clin Lab Stand Inst. 2008;28:52. [Google Scholar]
- 46.Chakrabarti A, Rudramurthy SM, Kale P, Hariprasath P, Dhaliwal M, Singhi S, Rao KLN. Epidemiological study of a large cluster of fungaemia cases due to Kodamaea ohmeri in an Indian tertiary care Centre. Clin Microbiol Infect. 2014;20:O83–O89. doi: 10.1111/1469-0691.12337. [DOI] [PubMed] [Google Scholar]
- 47.Li J, Zhan P, Jiang Q et al (2019) Prevalence and antifungal susceptibility of Sporothrix species in Jiangxi, central China. Med Mycol:1–8. 10.1093/mmy/myy163 [DOI] [PubMed]
- 48.Chakrabarti A, Bonifaz A, Gutierrez-Galhardo MC, et al. Global epidemiology of sporotrichosis. Med Mycol. 2015;53:3–14. doi: 10.1093/mmy/myu062. [DOI] [PubMed] [Google Scholar]
- 49.Marimon R, Gené J, Cano J, et al. Molecular phylogeny of Sporothrix schenckii. J Clin Microbiol. 2006;44:3251–3256. doi: 10.1128/JCM.00081-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Camacho E, León-Navarro I, Rodríguez-Brito S, Mendoza M, Niño-Vega GA. Molecular epidemiology of human sporotrichosis in Venezuela reveals high frequency of Sporothrix globosa. BMC Infect Dis. 2015;15:1–10. doi: 10.1186/s12879-015-0839-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao L, Cui Y, Zhen Y, Yao L, Shi Y, Song Y, Chen R, Li S. Genetic variation of Sporothrix globosa isolates from diverse geographic and clinical origins in China. Emerg Microbes Infect. 2017;6:1–13. doi: 10.1038/emi.2017.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dixon DM, Salkin IF, Duncan RA, et al. Isolation and characterization of Sporothrix schenckii from clinical and environmental sources associated with the largest U.S. epidemic of sporotrichosis. J Clin Microbiol. 1991;29:1106–1113. doi: 10.1128/JCM.29.6.1106-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kwon-Chung KJ. Comparison of isolates of Sporothrix schenckii obtained from fixed cutaneous lesions with isolates from other types of lesions. J Infect Dis. 1979;139:424–431. doi: 10.1093/infdis/139.4.424. [DOI] [PubMed] [Google Scholar]
- 54.Yu X, Wan Z, Zhang Z, Li F, Li R, Liu X. Phenotypic and molecular identification of Sporothrix isolates of clinical origin in Northeast China. Mycopathologia. 2013;176:67–74. doi: 10.1007/s11046-013-9668-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou X, Rodrigues AM, Feng P, De Hoog GS. Global ITS diversity in the Sporothrix schenckii complex. Fungal Divers. 2014;66:153–165. doi: 10.1007/s13225-013-0220-2. [DOI] [Google Scholar]
- 56.Liu T, Zhang K, Zhou X. Molecular identification of Sporothrix clinical isolates in China. J Zhejiang Univ Sci B. 2014;15:100–108. doi: 10.1631/jzus.B1300136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neyra E, Fonteyne P, Swinne D, et al. Epidemiology of human sporotrichosis investigated by amplified fragment length polymorphism. J Clin Microbiol. 2005;43:1348–1352. doi: 10.1128/JCM.43.3.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodrigues AM, de Hoog GS, Zhang Y, de Camargo ZP. Emerging sporotrichosis is driven by clonal and recombinant Sporothrix species. Emerg Microbes Infect. 2014;3:1–10. doi: 10.1038/emi.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gong J, Zhang M, Wang Y, Li R, He L, Wan Z, Li F, Zhang J. Population structure and genetic diversity of Sporothrix globosa in China according to 10 novel microsatellite loci. J Med Microbiol. 2019;68:248–254. doi: 10.1099/jmm.0.000896. [DOI] [PubMed] [Google Scholar]
- 60.Trilles L, Fernández-torres B, Lazéra S, et al. In vitro antifungal susceptibilities of Sporothrix schenckii in two growth phases in vitro antifungal susceptibilities of Sporothrix schenckii in two growth phases. Antimicrob Agents Chemother. 2005;49:9–12. doi: 10.1128/AAC.49.9.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gutierrez-Galhardo MC, Zancopé-Oliveira RM, Monzón A, Rodriguez-Tudela JL, Cuenca-Estrella M. Antifungal susceptibility profile in vitro of Sporothrix schenckii in two growth phases and by two methods: microdilution and E-test. Mycoses. 2010;53:227–231. doi: 10.1111/j.1439-0507.2009.01701.x. [DOI] [PubMed] [Google Scholar]
- 62.Brilhante RSN, Rodrigues AM, Sidrim JJC, Rocha MFG, Pereira SA, Gremião IDF, Schubach TMP, de Camargo ZP. In vitro susceptibility of antifungal drugs against Sporothrix brasiliensis recovered from cats with sporotrichosis in Brazil. Med Mycol. 2016;54:275–279. doi: 10.1093/mmy/myv039. [DOI] [PubMed] [Google Scholar]
- 63.Espinel-Ingroff A, Abreu DPB, Almeida-Paes R et al (2017) Multicenter and international study of MIC/MEC distributions for definition of epidemiological cutoff values (ECVs) for species of Sporothrix identified by molecular methods. Antimicrob Agents Chemother:1–17. 10.1128/AAC.01057-17 [DOI] [PMC free article] [PubMed]
- 64.Suzuki R, Yikelamu A, Tanaka R, Igawa K, Yokozeki H, Yaguchi T. Studies in phylogeny, development of rapid identification methods, antifungal susceptibility, and growth rates of clinical strains of Sporothrix schenckii Complex in Japan. Med Mycol J. 2016;57:E47–E57. doi: 10.3314/mmj.16-00005. [DOI] [PubMed] [Google Scholar]
- 65.Lortholary O, Denning DW, Dupont B. Endemic mycoses: a treatment update. J Antimicrob Chemother. 1999;43:321–331. doi: 10.1093/jac/43.3.321. [DOI] [PubMed] [Google Scholar]
- 66.Mahmoudi S, Zaini F, Kordbacheh P, Safara M, Heidari M. Sporothrix schenckii complex in Iran: molecular identification and antifungal susceptibility. Med Mycol. 2016;54:593–599. doi: 10.1093/mmy/myw006. [DOI] [PubMed] [Google Scholar]
- 67.Ottonelli Stopiglia CD, Magagnin CM, Castrillón MR, Mendes SDC, Heidrich D, Valente P, Scroferneker ML. Antifungal susceptibilities and identification of species of the Sporothrix schenckii complex isolated in Brazil. Med Mycol. 2014;52:56–64. doi: 10.3109/13693786.2013.818726. [DOI] [PubMed] [Google Scholar]
- 68.Galhardo MCG, De Oliveira RMZ, Do Valle ACF, et al. Molecular epidemiology and antifungal susceptibility patterns of Sporothrix schenckii isolates from a cat-transmitted epidemic of sporotrichosis in Rio de Janeiro, Brazil. Med Mycol. 2008;46:141–151. doi: 10.1080/13693780701742399. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 59 kb)