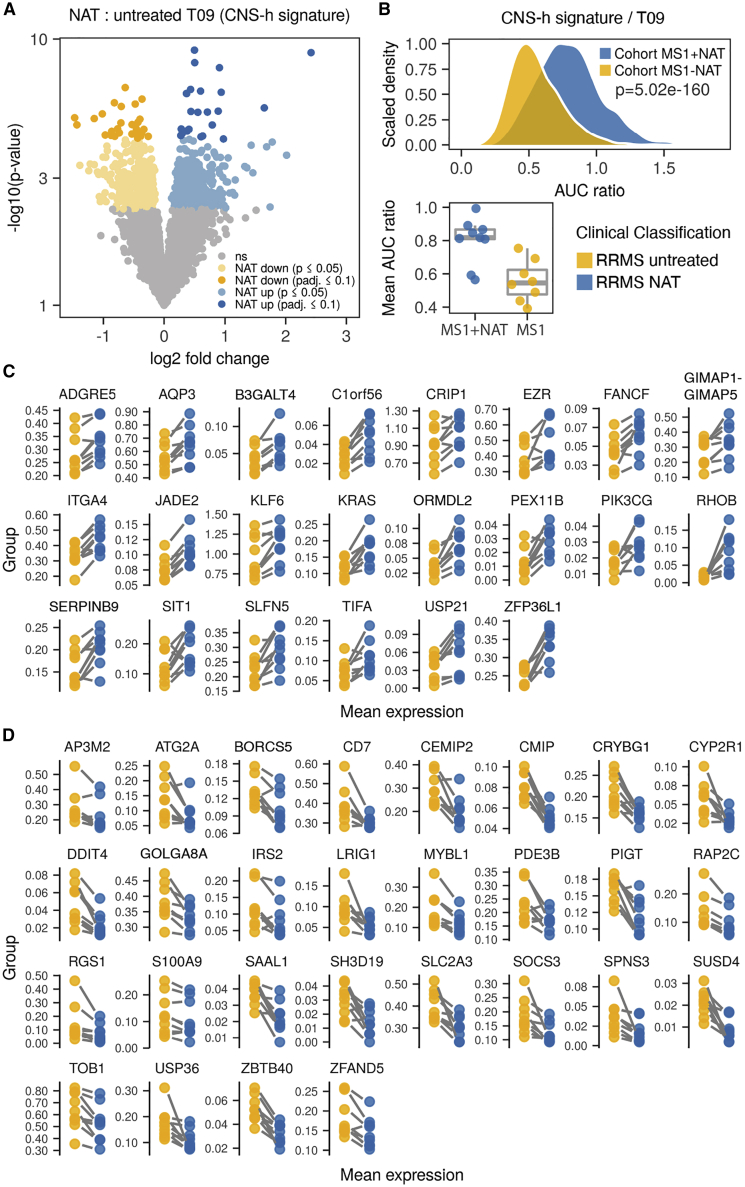
Figure 4.
Differential gene expression analysis between T09 cells with and without natalizumab treatment identifies their CNS-homing signature
(A) Differentially expressed genes in cluster T09 between natalizumab-treated and untreated time points (cohort MS1), termed CNS-homing signature (CNS-h).
(B) Comparison of single-cell gene set enrichment of the CNS-h signature in cluster T09 for n = 150 cells per sample and mean gene set enrichment averaged for each sample (lower panel). n = 9 samples from natalizumab-treated RRMS patients and n = 8 samples from untreated time points of the same patients (cohort MS1) are shown.
(C and D) Mean mRNA expression in cluster T09 of indicated genes for n = 9 RRMS patients with and without natalizumab treatment (cohort MS1).
(C) Significantly upregulated genes with natalizumab treatment (adjusted p value of DEseq2 analysis ≤ 0.1).
(D) Significantly downregulated genes with natalizumab treatment (adjusted p value of DEseq2 analysis ≤ 0.1).
p values in (A, C, and D) are derived from differential gene expression analysis with DEseq2. False discovery rate (FDR)-adjusted paired two-tailed t tests were used in (B).