Abstract
High-risk human papillomaviruses (hr-HPVs) are the key risk factors implicated in the development of a significant proportion of head and neck squamous cell carcinomas (HNSCCs). We aimed to investigate the distribution of hr-HPV types and HPV16 lineages in a sample of patients with HNSCC and the possible association between HPV status and the expression of P16INK4A and NF-κB in Iranian HNSCC patients. We examined 108 formalin-fixed, paraffin-embedded (FFPE) histologically confirmed primary SCC tissue specimens of different head and neck anatomical sites. HPV types and HPV16 lineages were determined by nested PCR and overlapping nested PCR assays, respectively, followed by gene sequencing and phylogenetic analysis. The expression of p16INK4a and NF-κB was evaluated by immunohistochemistry. Twenty-five (23.1%) HNSCC tissue specimens were tested positive for HPV infection. The most prevalent HPV type was HPV-16, followed by HPV18 and HPV11. HPV16 variants belonged to the lineage A and lineage D which were further sorted into sublineages A1, A2, and D2. A significant association between HPV status and p16INK4a immunoreactivity was observed in more than 76% of the HPV-related HNSCCs (P < 0.0001). The overexpression of p16INK4a and cytoplasmic NF-κB was more common in low-grade HNSCC tumors. Our data highlights that HPV16, in particular the A2 sublineage, followed by A1 and D2 sublineages are the major agents associated with HNSCCs in Iran. Based on HPV16 predominance and its lineage distribution pattern, it seems that the prophylactic vaccines developed for cervical cancer prevention could also be applicable for the prevention of HPV-related HNSCCs in our population.
Keywords: HPV types, HPV16 lineages, p16INK4a, NF-κB, HNSCC, HPV16 sublineages
Introduction
As the seventh most common cause of cancer worldwide [1], squamous cell carcinoma (SCC) of the head and neck (HNSCC) approximately accounts for 90% of head and neck cancers [2]. It is an epithelial malignancy involving mucosal anatomic sites of the larynx (glottis, subglottis, supraglottis), hypopharynx (post-cricoid region, posterior wall), oropharynx (palatine tonsil, the base of tongue), oral cavity (buccal mucosa, tongue), and nasal cavity [3]. Although tobacco smoking and alcohol consumption are the risk factors for cancers of the head and neck [1], a recent review of the literature concludes that human papillomaviruses (HPV) are the most important established risk factors for a subset of HNSCC, such as oropharyngeal SCC, where up to 60% of tumors are related to HPV infection [4]. The most common HPV type found in HNSCC is 16; but types 6 and 11 are also detectable in some head and neck cancers [5].
Based upon phylogenetic analysis of E6 and long control region (LCR) gene regions, HPV16 variants have been classified into four major lineages: lineage A, including the sublineages A1–A3 (European, E) and A4 (Asian, As); lineage B, including the sublineages B1 and B2 (African-1; Afr1a and African-1; Afr1b, respectively); lineage C, including the sublineage C1 (African-2, Afr2); and lineage D, including the sublineages D1–D3 (North American (NA)1, Asian-American (AA)2, and Asian-American (AA)1, respectively) [6]. It is suggested that when compared to HPV16 European lineage, non-European (NE) lineages have elevated risks for high-grade cervical intraepithelial neoplasia (CIN3) and cervical cancer [7].
Protein p16INK4A, the product of cyclin-dependent kinase inhibitor 2A (CDKN2A) as a tumor suppressor gene, is an important regulator in cell cycle control and cancer [8]. The overexpression of p16INK4A via the dysfunction of the retinoblastoma (Rb) protein is frequently observed in tumors as the result of HPV infection [9]. The expression of p16INK4A in HNSCCs is associated with HPV status and better prognosis. Immunostaining of p16INK4A combined with HPV-DNA PCR assay is suggested to generate sufficient sensitivity and specificity as a valuable alternative to HPV E6/E7 mRNA RT-PCR detection for evaluation of the HPV status in formalin-fixed, paraffin-embedded (FFPE) tumors. It specifies transient or inactive HPV infection and determines the elevated p16INK4A expression by non-HPV-related alterations [10].
Nuclear factor kappa B (NF-κB) is known to be modulated by several HPV oncoproteins and plays many important roles in different cellular pathways such as cell proliferation and apoptosis which are associated with the development of cancer [11]. The activation of NF-κB leads to an increased risk of metastasis resulting in a poor prognosis in a wide variety of cancers such as oral SCC [12]. Although studies revealed that NF-κB is persistently expressed in HNSCC [12, 13], little data are available about the association of NF-κB gene expression with HPV-related and unrelated HNSCC.
Human papillomaviruses diverge geographically, affecting vaccination programs, and importantly, are of critical prognostic value altering therapeutic management strategies for patients with HNSCC tumors [14]. In this study, we aimed to investigate the prevalence of high-risk HPV infection in a subset of head and neck SCC patients and the possible association between HPV status and the expression of P16INK4A and NF-κB cellular biomarkers by examining associated clinical findings. To the extent of our knowledge, this study provides the first data on the lineages and sublineages of HPV 16 isolated from HNSCC patients in Iran.
Materials and methods
Study subjects
This retrospective study included 126 formalin-fixed, paraffin-embedded (FFPE) tissue specimens histologically confirmed primary HNSCC cancers. All samples were collected from the Department of Oral and Maxillofacial Pathology, School of Dentistry, Shiraz University of Medical Sciences between January 2016 and December 2018. There were no restrictions on ethnicity, gender, and age or cancer stage at recruitment. By reviewing the hematoxylin-eosin (H&E)-stained slides, two pathologists independently graded the tumors as moderate, poor, or undifferentiated. Histological grades of HNSCC specimens were further converted into “low-grade” which means well to moderately differentiated and “high-grade” which means poorly differentiated or undifferentiated cancer cells.
DNA extraction and integrity analysis
DNA was extracted from 8 μm serial sections of FFPE tissue specimens using QIAamp Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturers’ instructions. Total genomic DNA was eluted with 50 μl of the buffer and stored at − 70 °C until analyzed. To test the adequacy of the samples and to demonstrate the absence of PCR inhibitors in the extracted DNA, 500 ng of extracted DNA was analyzed by ß-globin gene PCR amplification, resulting in a 110 bp PCO3/ PCO4 PCR product [15]. The quantity and quality of isolated DNA were measured using a UV-Vis spectrophotometer (WPA Lightwave II). Extracted DNA from the human erythroleukemia cell line (K562) available from the previous study [16] and sterile water were included as positive and negative controls, respectively.
HPV prevalence and genotyping
To screen the presence of 23 mucosotropic HPV types in HNSCC, a nested PCR assay using MY09/MY11 degenerated primer set followed by GP5+/GP6+ consensus primers was performed as previously described [17, 18]. To ensure consistency and reliability of the results, the specimens tested positive for the presence of HPV were analyzed in duplicate. In each round of amplification, 500 ng of DNA extracted from HeLa cells available from the previous study (16) and sterile water were included as positive and negative controls, respectively. Amplified PCR products were directly Sanger sequenced (Sequetech Corp., Mountain View, CA, USA) in the presence of GP5+ and GP6+ as both direction sequencing primers after purification using GF-1 PCR Clean-Up Kit (Vivantis, Malaysia). The obtained sequences were confirmed and genotyped by a BLAST similarity search using the NCBI BLAST software program (http://www.blast.ncbi.nlm.nih.gov/blast/html).
Detection of HPV16
To prevent missing this important type due to the presence of overlapping signals in sequencing chromatograms occurring in samples with multiple HPV infections, a type-specific nested PCR assay was performed on all samples for HPV16 detection. Targeting the HPV16 E6 gene region, the primer sets used for the first round of assay were 16E6F1/16E6R1 followed by 16E6F2/16E6R1 primers as the inner primers for the secondary PCR. Sequences of the primers used here were described elsewhere [16]. As a negative control in all nested PCR assays, the product of the negative control in the first step of the reaction was used as a template in the second round of PCR amplification. A plasmid containing HPV-16 DNA cloned in pBluescript (Manassas, VA) which was available from the previous study [16] was used as a positive control, and to determine the sensitivity of the assay.
Determination of HP16 lineages and phylogenetic analysis
To increase the sensitivity of HPV16 lineage identification, and to overcome DNA fragmentation caused by formalin fixation process, semi-nested PCR assays were performed to amplify fragments of less than 350 base-pairs. Using DNA from samples tested positive for HPV16 in the previous experiments, the primer sets used for amplification of the full-length HPV16 E6 gene were 16E6-1F/16E6-2R and 16E6-2F/16E6-3R as outer primers for the primary round of semi-nested PCR assays followed by 16E6-1F/16E6-1R, 16E6-2F/16E6-2R, and 16E6-3F/16E6-3R primers as inner primers for the secondary PCR assays, as described in Table 1. For the amplification of the long control region (LCR) of HPV16, the outer primer sets were 16LCR-1F/16LCR-2R and 16LCR-3F/16LCR-4R followed by 16LCR-1F/16LCR-1R, 16LCR-2F/16LCR-2R, 16LCR-3F/16LCR-3R, and 16LCR-4F/16LCR-4R used as inner primers for the secondary semi-nested PCR assays (Table 1). All the PCR protocols used here for DNA amplification were carried out according to the methods previously described [19, 20]. After visualization of generated amplicons on 1.5% agarose gel, sterile excised bands were purified using GF-1 PCR Clean-Up Kit (Vivantis, Malaysia) and were subjected to sequencing (Sequetech Corp., Mountain View, CA, USA) in both directions. A plasmid containing full-length HPV-16 DNA was used as a positive control in each reaction as mentioned in the previous section.
Table 1.
The sequence of primers used for amplification of HPV16 E6 gene and LCR [20]
| Primer name | Sequences (5′ to 3′) | Nucleotide positions* | Product size (bp) |
|---|---|---|---|
| 16E6-1F-outer | TTGAACCGAAACCGGTTAGT | 46–65 | 393 |
| 16E6-2R-outer | GGACACAGTGGCTTTTGACA | 419–438 | |
| 16E6-1F-inner | TTGAACCGAAACCGGTTAGT | 46–65 | 211 |
| 16E6-1R-inner | GCATAAATCCCGAAAAGCAA | 237–256 | |
| 16E6-2F-inner | GCAACAGTTACTGCGACGTG | 205–244 | 234 |
| 16E6-2R-inner | GGACACAGTGGCTTTTGACA | 419–438 | |
| 16E6-2F-outer | GCAACAGTTACTGCGACGTG | 205–224 | 386 |
| 16E6-3R-outer | TCATGCAATGTAGGTGTATCTCC | 568–590 | |
| 16E6-3F-inner | CAGCAATACAACAAACCGTTG | 371–391 | 220 |
| 16E6-3R-inner | TCATGCAATGTAGGTGTATCTCC | 568–590 | |
| 16LCR-1F-outer | GAAAACGAAAAGCTACACCCA | 7084–7104 | 497 |
| 16LCR-2R-outer | GTGCAGGTCAGGAAAACAG | 7562–7580 | |
| 16LCR-1F-inner | GAAAACGAAAAGCTACACCCA | 7084–7104 | 285 |
| 16LCR-1R-inner | CAATGAATAACCACAACACAATTA | 7345–7368 | |
| 16LCR-2F-inner | GCTTGTGTAACTATTGTGTCATG | 7289–7311 | 292 |
| 16LCR-2R- inner | GTGCAGGTCAGGAAAACAG | 7562–7580 | |
| 16LCR-3F-outer | ACTTGTACGTTTCCTGCTTG | 7525–7544 | 483 |
| 16LCR-4R-outer | TGCAGTTCTCTTTTGGTGC | 85–103 | |
| 16LCR-3F-inner | ACTTGTACGTTTCCTGCTTG | 7525–7544 | 350 |
| 16LCR-3R-inner | GTGTAACCCAAAATCGGTTTGC | 7853–7874 | |
| 16LCR-4F-inner | GTCACCCTAGTTCATACATGA | 7777–7797 | 231 |
| 16LCR-4R-inner | TGCAGTTCTCTTTTGGTGC | 85–103 |
*The numbers refer to the position of the nucleotides according to the reference sequence (GenBank accession number K02718)
The obtained sequences are available at http://www.ncbi.nlm.nih.gov/ with GenBank accession numbers MT771959 through MT771984. All sequences obtained in this study were analyzed using the BLAST software program (http://www.blast.ncbi.nlm.nih.gov/blast/html) and classified in comparison to the prototype reference sequences given in the Papillomavirus Episteme database (http://pave.niaid.nih.gov) into lineages and sublineages according to Burk et al. [6]. Phylogenetic trees were constructed using the maximum-likelihood method Mega software version 6 [21]. The reference HPV-16 E6 and LCR sequences that were used to construct the phylogenetic branches were collected from the GenBank sequence database and included K02718 (A1), AF536179 (A2), HQ644236 (A3), AF534061 (A4), AF536180 (B1), HQ644298 (B2), AF472509 (C1), HQ644257 (D1), AY686579 (D2), and AF402678 (D3). The robustness of the phylogenetic trees was assessed using 1000 bootstrap repetitions.
p16INK4a and NF-κB immunohistochemistry
The immunohistochemical staining was performed on 5-μm-thick FFPE tissue sections. Xylene was used to deparaffinize the slides twice for 30 min and rehydration was achieved through the gradual addition of ethanol solution to Tris-buffered saline (TBS). Epitope retrieval was carried out by heat-induced microwave treatment in the 10 mM citrate buffer (pH 6.0). Endogenous peroxidase activity was quenched by 10% H2O2. Protein block agent (Dako, Glostrup, Denmark) was used to block non-specific binding sites. Slides were incubated with monoclonal antibodies anti-human p16INK4a (1:50; BD Pharmingen™; BD Biosciences, Franklin Lakes, NJ, USA) and anti-human NF-κB (p65) (1:400; Santa Cruz Biotechnology, Santa Cruz, CA, USA) in a humidified chamber for 1 h at room temperature, followed by incubation with EnVision+ Dual Link System-HRP solution (Dako, Glostrup, Denmark) for 30 min. In the presence of hydrogen peroxide, 3, 30-diaminobenzidine (DAB) was used as the chromogen. Nuclei were counterstained using Meyer’s hematoxylin. The sections without primary antibody served as the negative control in each run. Formalin-fixed HeLa cell block sections known positive for p16INK4a and NF-κB immunostaining were used as the positive control of the assays.
Interpretation of p16INK4a and NF-κB immunostaining findings
A 4-tier scale from 0 to 3+ was recorded to semiquantitatively score the intensity of p16INK4a staining according to Duncan et al. [22]. Evaluation of each staining pattern was recorded as follows: 0, no staining in any tumor cells; 1+, staining less than 10% of cells without nuclear staining; 2+, mild cytoplasmic staining with or without nuclear staining in 10 to 50% of cells; 3+, strong diffuse cytoplasmic and nuclear staining in more than 50% of cells. For statistical analysis of p16INK4a immunohistochemistry (IHC) findings, the 4-level variable was converted into bipartite variables, with 0 and 1+ scores being considered as negative and 2+ and 3+ being denoted as positive results.
For NF-κB protein expression, the calculation of results based on the percentage of positive cells was considered as follows: 0, 0 to 5% of tumor cells counted as positive; 1, 5 to 25% of cells counted as positive; 2, 26 to 50% of cells counted as positive; 3, 51 to 75% of cells counted as positive; 4, more than 76% of cells counted as positive. In respect of staining intensity, 0, no staining in any tumor cells; 1, straw yellow; 2, brown; 3, tan, according to Zhou et al. [23]. For statistical analysis of NF-κB immunostaining results, the scoring index was calculated based on the sum of the items above, and the cutoff value was selected as three so that < 3 was considered as low expression, whereas ≥ 3 was defined as high expression. To study the NF-κB/relA signaling pathway in cancerous cells which can be interpreted through p65 translocation from cytoplasm to the nucleus [24], the nuclear expression of NF-κB p65 was evaluated separately. Cells were counted and examined per field of microscope at × 200 magnification.
Statistical analysis
Data were analyzed using SPSS version 21.0 (SPSS Institute, Chicago, IL, USA). Chi-square test or two-sided Fisher’s exact test were used to analyze the association between HPV PCR results, frequencies of HPV16 lineages among different anatomical sites, IHC staining index, age, gender, and other categorical factors, where appropriate. Statistical significance was assumed at a P < 0.05 level.
Results
Clinicopathological characteristics, HPV prevalence, and type distribution
Eleven of the 126 tissue specimens were excluded from the study because of an inadequate amount of tissue material. Of the remaining 115 cases, seven were excluded due to failure to amplify the b-globin gene region. The study included 38 tongue, 34 oral cavity, 13 larynx, 9 hypopharynx, 8 nasopharynx, and 6 tonsil SCC tissue specimens. The mean age of patients was 56 years (range 20–88 years, SD 14.63 years) and 58.3% of the patients were males. The clinicopathological data collected from medical records are described in Table 2. There were no data available regarding alcohol consumption and tobacco use by the patients. Thirteen (12%) HNSCC tumor specimens were tested positive for HPV-DNA when MY/GP+ primer sets were used in a nested PCR assay. BLASTing the obtained DNA sequences with available sequences in GenBank revealed that HPV-16 was the most commonly detected HPV type found in SCC tumors of the oral cavity (4 cases), tongue (3 cases), and tonsil (1 case). HPV-18 was also detected in the tumor tissue specimens of the larynx (2 cases), and tonsil (2 cases). Moreover, HPV-11 was detected in two patients with SCC of the hard palate and base of the tongue. All samples were further analyzed using a type-specific nested PCR assay for the detection of HPV16 and results revealed that 17 tissue specimens (15.7%) harbored HPV16 DNA. Type-specific primers could not detect HPV16 in 3 samples which previously tested positive for the presence of this type using MY/GP+ consensus primers. Multiple-type infection was only detected in a moderately differentiated SCC tissue specimen of tonsil from a female patient infected with HPV types 16 and 18. The sensitivities of the DNA amplification assays were determined, and the results showed a limit of detection of 256 and approximately 2 copies per reaction of HPV16 DNA target using MY/GP+ and HPV16 type-specific nested assays, respectively. Combining the results obtained from MY/GP+ and HPV16 type-specific nested PCR assays, total 25 (23.1%) HNSCC tissue specimens were tested positive for HPV infection. HPV16 was the most prevalent type identified in 18.5% of all HNSCC examined tissue specimens. Based on different anatomical sites, HPV-DNA was found in 3 tonsil (50%), 4 larynx (30.8%), 2 nasopharynx (25%), 2 hypopharynx (22%), 8 oral cavity (23.5%), and 6 tongue (15.8%) SCC tumors. The prevalence of HPV infection was significantly higher among younger age groups of HNSCC patients (< 56 years) than in the older age group (31% vs 14%, P = 0.036). In addition, HPV-DNA was found significantly more often in HNSCC tissue specimens from men than those from women (30.2% vs 13.3%, P = 0.041). Further sorting of the histological grade into low-grade and high-grade showed that 87.4% of HPV-positive specimens belonged to the patients with low-grade tumors. However, we found no evidence that HPV status was associated with the grade of malignancy (P = 0.700).
Table 2.
Associations between demographic, tumor characteristics, and HPV status among patients with head and neck squamous cell carcinoma
| Characteristics | All Patients (%) (N = 108) | HPV positive (%) (N = 25) | HPV negative (%) (N = 83) | P value | |
|---|---|---|---|---|---|
| Age (years) | < 56 | 58 (53.7) | 18 (72.0) | 40 (48.2) | 0.036 |
| ≥ 56 | 50 (46.3) | 7 (28.0) | 43 (51.8) | ||
| Gender | Male | 63 (58.3) | 19 (76.0) | 44 (53.0) | 0.041 |
| Female | 45 (41.7) | 6 (24.0) | 39 (47.0) | ||
| Anatomic site | Tongue | 38 (35.2) | 6 (24.0) | 32 (38.6) | 0.590 |
| Oral cavity | 34 (31.5) | 8 (32.0) | 26 (31.3) | ||
| Larynx | 13 (12.0) | 4 (16.0) | 9 (10.8) | ||
| Hypopharynx | 9 (8.3) | 2 (8.0) | 7 (8.4) | ||
| Nasopharynx | 8 (7.4) | 2 (8.0) | 6 (7.2) | ||
| Tonsil | 6 (5.6) | 3 (12.0) | 3 (3.6) | ||
| Grade (differentiation) | Well | 61 (56.5) | 17 (68.0) | 44 (53.0) | 0.383 |
| Moderate | 33 (30.6) | 5 (20.0) | 28 (33.7) | ||
| Poor | 10 (9.3) | 3 (12.0) | 7 (8.4) | ||
| Undifferentiated | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | ||
| Not recorded | 4 (3.7) | 0.0 (0.0) | 4 (4.8) | ||
| p16INK4a expression | Positive | 32 (29.6) | 19 (76.0) | 13 (15.6) | 0.0001 |
| Negative | 65 (60.2) | 6 (24.0) | 59 (71.1) | ||
| Not done | 11 (10.2) | 0.0 (0.0) | 11(13.3) | ||
| Cytoplasmic NF-κB expression | Low | 28 (25.9) | 6 (24.0) | 22 (26.5) | 0.807 |
| High | 73 (67.6) | 18 (72.0) | 55 (66.3) | ||
| Not done | 7 (6.5) | 1 (4.0) | 6 (7.2) | ||
| Nuclear NF-κB expression | Low | 77 (71.3) | 19 (76.0) | 58 (69.9) | 0.797 |
| High | 24 (22.2) | 5 (20.0) | 19 (22.9) | ||
| Not done | 7 (6.5) | 1 (4.0) | 6 (7.2) | ||
HPV 16 lineage identification
Among the 17 HNSCC tissue specimens tested positive for HPV16 using HPV16 overlapping PCR assays, 13 isolates were sequenced successfully across the E6 and/or LCR regions. In total, sixty-one E6 and forty-six LCR partial DNA fragments were compared with the HPV16 prototype of each HPV16 lineage, and sublineage and genome information revealed nucleotide signatures that allow lineage identification according to Burk’s classification [6]. The sequencing results of 4 samples were not conclusive and were not included in the analysis. HPV16 variants belonged to the lineage A (84.6%) and lineage D (15.4%), while other lineages were not detected in this study. Isolates of lineage A were further sorted into sublineages A1 (30.7%, 4/13) and A2 (53.8%, 7/13) based on four single nucleotide polymorphisms (SNPs) in E6 and LCR regions (T178A, T350G, A7507C, and C24G). The T350G SNP which results in the amino acid change L83V was found in combination with other SNPs across the E6 and LCR. The maximum similarity difference of the E6 and/or LCR sequences between any two lineages when compared to reference sequences was not more than 0.5%. Since using A532G could not distinguish (AA)1, (AA)2, and (NA)1 sublineage in the E6 gene region, A7507G and T7743G were considered as diagnostic SNPs for differentiation of the (AA)2 and (AA)1 sublineages, respectively. The specific detection of D2 (AA2) sublineage (15.4%, 2/13) was achieved by observation of four diagnostic SNPs (T350G, A532G, A7507G, and A7894C). All the three HPV16 lineages isolated from poorly differentiated hypopharyngeal and base of the tongue carcinoma tissues belonged to the A2 sublineage. HPV-16 sublineages isolated from different anatomical sites of HNSCC patients are presented in Fig. 1 and Table 3.
Fig. 1.
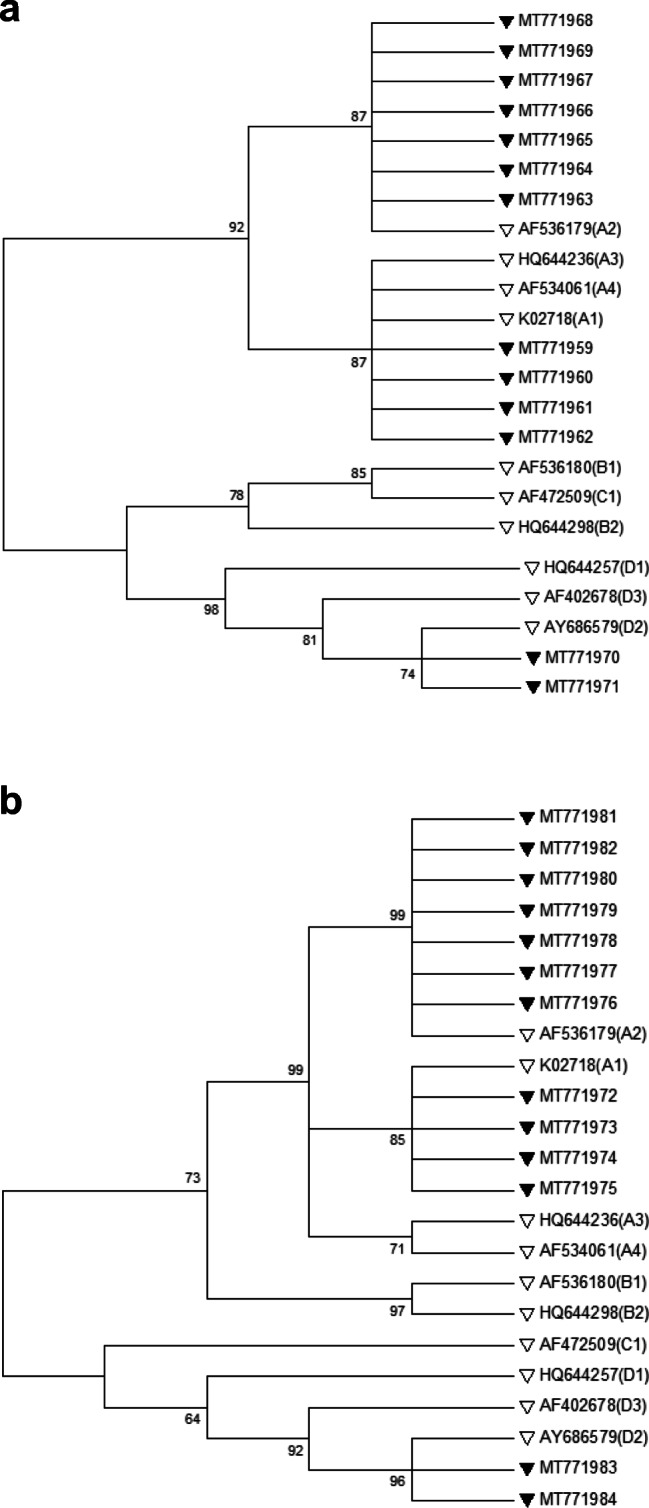
Phylogenetic analysis of HPV16 E6 (a) and LCR (b) gene regions was conducted using the maximum-likelihood method based on the Kimura 2-parameter model with bootstrap resampling (1000 replicates) by the MEGA 6 package [21]. Numbers above the branches indicate the bootstrap values. Thirteen different nucleotide patterns of studied sequences were indicated by black triangles (GenBank accession numbers MT771959 through MT771984). The accession number of reference sequences of each sublineage used for phylogenetic analysis in this study was indicated by white triangles
Table 3.
Status of HPV-16 sublineages isolated from different anatomical sites of HNSCC patients and immunoreactivity of p16INK4a and NF-κB biomarkers
| No. | HPV16* lineage | HPV16 sublineage | Anatomic site | Primary tumor grade | p16INK4a expression** | Cytoplasmic NF-κB expression*** | Nuclear NF-κB expressionC |
|---|---|---|---|---|---|---|---|
| 1 | A | A1 | Oral cavity | Well differentiated | 2+ | High | Low |
| 2 | A | A1 | Larynx | Moderately differentiated | 3+ | High | Low |
| 3 | A | A1 | Larynx | Moderately differentiated | 3+ | High | High |
| 4 | A | A1 | Oral cavity | Well differentiated | 2+ | High | Low |
| 5 | A | A2 | Oral cavity | Well differentiated | 3+ | High | Low |
| 6 | A | A2 | Larynx | Well differentiated | 2+ | High | High |
| 7 | A | A2 | Hypopharynx | Well differentiated | 3+ | Low | Low |
| 8 | A | A2 | Tonsil | Well differentiated | 3+ | High | Low |
| 9 | A | A2 | Base of the tongue | Poorly differentiated | 3+ | High | High |
| 10 | A | A2 | Base of the tongue | Poorly differentiated | 3+ | High | High |
| 11 | A | A2 | Hypopharynx | Poorly differentiated | 3+ | High | Low |
| 12 | D | D2 | Oral cavity | Well differentiated | 2+ | High | Low |
| 13 | D | D2 | Base of the tongue | Well differentiated | 3+ | High | Low |
p16INK4a and NF-κB expression status in HNSCC patients
Of 108 patients with HNSCC, 11 samples revealed unsatisfactory results for p16INK4a immunostaining and were excluded from further analysis. Thirty-two (33%) specimens tested positive for p16INK4a immunoreactivity, whereas only 19 samples tested positive for both p16INK4a and HPV-DNA detection assays. Of 97 tumor tissue specimens, 19.6% were discordant for HPV status and p16INK4a expression which 6 samples (6.2%) were HPV-DNA-positive and p16INK4a-negative, and 13 (13.4%) were HPV-DNA negative and p16INK4a-positive. However, we found a significant association between the presence of HPV-DNA and p16INK4a immunoreactivity in more than 76% of the HPV-related HNSCCs (P < 0.0001). Based on anatomical sites of HPV-positive HNSCC tumors, a consistency was observed between p16INK4a immunostaining and HPV-DNA detection across all nasopharyngeal tumor specimens (Table 4). Notably, strong p16INK4a immunoreactivity was observed in poorly differentiated tumor tissues infected with HPV16 sublineage A2 (Table 3). In addition, no association was seen between the expression of p16INK4 and the grade of tumors (P = 0.489).
Table 4.
p16INK4a and NF-κB immunoreactivity among different anatomical sites of HPV-positive and -negative HNSCC tumors
| HPV-DNA (+) | HPV-DNA (−) | HPV-DNA (+) | HPV-DNA (−) | HPV-DNA (+) | HPV-DNA (−) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p16INK4a (+) n (%) | p16INK4a (−) n (%) | p16INK4a (+) n (%) | p16INK4a (−) n (%) | High cytoplasmic NF-κB expression n (%) | Low cytoplasmic NF-κB expression n (%) | High cytoplasmic NF-κB expression n (%) | Low cytoplasmic NF-κB expression n (%) | High nuclear NF-κB expression n (%) | Low nuclear NF-κB expression n (%) | High nuclear NF-κB expression n (%) | Low nuclear NF-κB expression n (%) | |
| Tongue | 5 (83.3) | 1 (16.7) | 4 (12.5) | 28 (87.5) | 3 (50) | 3 (50) | 23 (74.2) | 8 (25.8) | 1 (16.7) | 5 (83.3) | 4 (12.9) | 27 (87.1) |
| Oral Cavity | 6 (75) | 2 (25) | 7 (36.8) | 12 (63.2) | 7 (87.5) | 1 (12.5) | 18 (75) | 6 (25) | 1 (12.5) | 7 (87.5) | 7 (29.2) | 17 (70.8) |
| Larynx | 3 (75) | 1 (25) | 1 (14.3) | 6 (85.7) | 4 (100) | 0 (0) | 5 (62.5) | 3 (37.5) | 2 (50) | 2 (50) | 2 (25) | 6 (75) |
| Hypopharynx | 0 (0) | 2 (100) | 1 (14.3) | 6 (85.7) | 1 (100) | 0 (0) | 5 (71.4) | 2 (28.6) | 1 (100) | 0 (0) | 4 (57.1) | 3 (42.9) |
| Nasopharynx | 2 (100) | 0 (0) | 0 (0) | 5 (100) | 1 (50) | 1 (50) | 3 (60) | 2 (40) | 0 (0) | 2 (100) | 1 (20) | 4 (80) |
| Tonsil | 3 (100) | 0 (0) | 0 (0) | 2 (100) | 2 (66.7) | 1 (33.3) | 1 (50) | 1 (50) | 0 (0) | 3 (100) | 1 (50) | 1 (50) |
| Total | 19 (76) | 6 (24) | 13 (18) | 59 (82) | 18 (75) | 6 (25) | 55 (71.4) | 22 (28.6) | 5 (20.8) | 19 (79.2) | 19 (24.7) | 58 (75.3) |
The NF-κB (p65) immunostaining was not applicable for 7 specimens which were consequently excluded from the analysis. Cytoplasmic expression of NF-κB was observed in 96 cases but the strong cytoplasmic expression was only revealed in 72.3% of samples. Of 101 tumor tissue specimens which were successfully examined for cytoplasmic NF-κB expression, only the pathologic grades of 99 samples were available. A significantly higher expression of cytoplasmic NF-κB was observed in low-grade HNSCC tumors as compared to high-grade cancer cells (75.3% vs 40.0%, P = 0.019). Nuclear localization of p65 was detected in 23.8% of cancerous tissues. High expression levels of cytoplasmic NF-κB (p65) protein were observed in the oral cavity (78.1%), larynx (75%), hypopharynx (75%), tongue (70.3%), tonsil (60%), and nasopharynx (57.1%) tumor tissue specimens (Table 4). Nuclear expression of NF-κB was uncommon when compared to its cytoplasmic localization and did not show an increase parallel with the increase in tumor grade. We found no evidence that HPV status was consistent with cytoplasmic or nuclear localization of p65. Tables 2 and 4 provide details regarding the association between the presence of HPV-DNA, HPV16 lineages status, p16INK4a expression, and NF-κB immunoreactivity in different anatomical sites of HNSCC tumors. Figure 2 shows representative expression patterns of p16INK4a and NF-κB in HNSCC tissues.
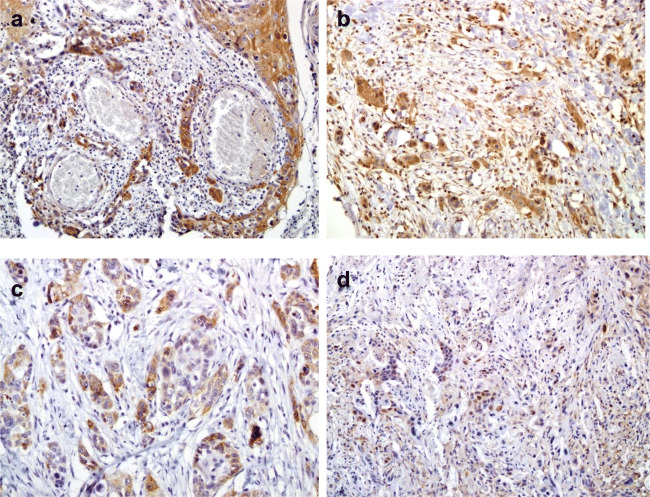
Fig. 2.
Immunohistochemical analysis of HNSCC cancerous tissues from HPV-positive cases with hematoxylin counterstain; magnification, × 200. Cytoplasmic NF-κB (p65) immunostaining in moderately (a) and poorly (b) differentiated HNSCC. c Cytoplasmic p16INK4a immunohistochemical staining in moderately differentiated HNSCC. d Nuclear p16INK4a immunostaining in poorly differentiated HNSCC
Discussion
The association of HPV with a subset of nasopharyngeal carcinomas and other HNSCC cancers including oral cavity cancers is now well established [25–29]. The goal of this study was to investigate the prevalence of high-risk HPV infection in a number of HNSCC patients to evaluate the incidence of HPV infection in the tumors and to determine the possible correlation between specific HPV16 lineages and expression of P16INK4A and NF-κB cellular biomarkers by examining associated clinical findings. This study recorded a prevalence of 23.1% for HPV infection in 108 HNSCC cases. Based on different reports, rates of HPV infection in HNSCC cancers vary considerably from 0 to 70% worldwide [4, 30–32]. A systematic assessment of the HPV burden in different cancers in Iran estimated a prevalence of 32.4% HPV infection in HNSCC Iranian patients [33]. This variation is certainly due to differences in laboratory methods, tumor site selection, the geographical distribution of HPV types, circulating variants, heterogeneity in patient populations, certain sexual behaviors, and poor oral hygiene [34]. For example, in this study, MY/GP+ consensus primers and HPV16 type-specific primers were employed in nested PCR assays to detect HPV infections. Results demonstrated suboptimal sensitivity of MY/GP+ primers in detecting HPV16 DNA when compared to type-specific primers. But, on the other hand, type-specific primers could not detect HPV16 DNA in 3 tissue specimens which were tested positive for the same type using MY/GP+ primers. Since in comparison with the HPV-negative HNSCC cancers, a favorable prognosis and better patients’ overall survival is associated with HPV-positive HNSCC cancers [35], recruitment of highly sensitive HPV detection methods such as a combination of various consensus and type-specific nested PCR assays is necessary to provide more accurate information about HPV status for clinical decision-making and management of HNSCC cancer patients.
HPV16 was the most common type detected in this study, a finding that is consistent with many other studies [25, 31, 34, 36]. Although some studies demonstrated a strong association between HPV16 infection and tonsillar and base of tongue SCCs [36], however, our findings on the prevalence of HPV16 showed no large differences in various anatomical locations of the HNSCC cancers. Such a disagreement can be explained by the large number of tongue and oral cavity SCC cases in our study which can generally lead to observed differences in frequencies of viral infection. The second most prevalent type was HPV18 detected in 3.7% of samples. Among different anatomic sites, HPV18 DNA was only detected in 33.3% of the oropharynx (palatine tonsils) and 15.4% of larynx (glottis) SCC cancers. Michaud et al. reported a significant association between HPV18 infection and oropharyngeal cancers and Mineta et al. found HPV18 in 15% of oropharyngeal and 0% of oral cavity SCC samples [37, 38]. Although some studies suggest HPV18 infection is more common in oral SCCs than in oropharyngeal cancers [14], it appears that the predominance of HPV18 infection in oropharyngeal cancers is a more common finding compared to other anatomical subsites of HNSCC. Furthermore, we also detected HPV11 DNA in a minority of cases (1.85%). The low HPV11 prevalence reported in this study was comparable to other findings published in the literature [14, 35]. The presence of low-risk HPV11 infection in SCC tumors of hard palate and base of tongue indicated that it might not be an entirely benign type while infecting oropharyngeal and oral cavity sites, as previously suggested by other studies [14, 39]. In addition, patients’ characteristic data showed that the incidence of HPV-related HNSCC cancers is higher among men than women. This finding is consistent with previous studies suggesting that men are at higher risk of developing HPV-related HNSCC cancers than women [40–42]. Due to the predominance of HPV-related HNSCC male patients even when compared to their female counterparts for acquiring HPV infection, this finding highlights the importance of implementing preventive HPV vaccination in men.
Although carcinogenic properties and sequence variability of HPV16 in cervical specimens have been studied for decades [43], very limited data are available regarding HPV16 lineage distribution in HNSCC worldwide. To our knowledge, this is the first study reporting HPV16 variability in HNSCC tumors in Iran. Among the 13 HNSCC tissue specimens tested positive for HPV16 using overlapping PCR assays, the predominance of lineage A (84.6%) followed by lineage D (15.4%) was observed. This finding is consistent with the study conducted by Gillison et al. which reported the presence of European and Asian lineages in 75% and 17% of HPV16-infected HNSCC tumors, respectively [25]. It is also comparable to a previously published report on HPV16 lineage analysis describing HPV16 lineages A and D are more prevalent in cervical samples of Iranian women [44]. Vaezi et al. confirmed that the majority of cervical samples were infected with lineage D (70.7%) and only 29.3% belonged to lineage A [44]. Our findings revealed that lineage E was more prevalent in HNSCC tumors. Although Gillison et al. found a notable similarity in the distribution of the various HPV16 lineages in cervical cancers and HNSCC in the USA [25], it appears that the distribution of HPV16 lineages in HNSCC tumor specimens of Iranian patients is not consistent with the circulating lineages previously reported for the cervical samples [44]. Most studies on HPV16 lineages in cervical samples were compared for lineage A (European-Asian, sublineages A1–A4) versus lineages B/C/D (non-European, sublineages B1, B2, C1, D1–D3) and concluded that non-European HPV16 lineages were associated with higher viral persistence and progression to cancer [45, 46]. Later studies on invasive cervical cancers revealed that even in lineage A, a higher risk-associated with sublineage A4 could be observed when compared to A1–A3 sublineages [47]. Mirabello et al. clearly showed huge differences in the carcinogenic properties of different HPV16 sublineages associated with an increased risk of cervical cancer [48]. Their findings indicated that A4, D3, and particularly D2 sublineages intensely influenced the risk of histological types of cervical lesions. In the present study, we further sorted the isolates of lineage A into sublineages A1 (30.7%) and A2 (53.8%) and linage D to sublineage D2 (15.4%). We evaluated the association between HPV16 lineages and sublineages and HNSCC primary tumor grade, anatomic site, and expression of two cellular biomarkers. However, such a relationship was not observed in our study, which may be due to the various challenges in research on HNSCC cancers in association with HPV lineages. The inadequate sample size for each anatomical site and the lack of comprehensive data for each patient in order to evaluate the risk of viral persistence and lesion progression to cancer associated with certain HPV lineages might be some of the existing challenges.
In the present research, the expression of p16INK4a was determined in 90% of all studied tumors, and we observed a significant correlation between HPV-DNA status and p16INK4a expression among Iranian HNSCC patients. Based on such a correlation, p16INK4a immunoreactivity is associated with younger age (P = 0.036) but not with gender or histological grades of HNSCC tumors. In 6.2% of HPV-DNA-positive cases, p16INK4a immunoreactivity was not observed and 13.4% of p16INK4a overexpressed cases were found to be negative for the presence of HPV-DNA. The expression of p16INK4a is considered as a surrogate marker for HPV infection [10, 22, 49]. However, some studies have shown that p16INK4a is not a reliable biomarker for the detection of transcriptionally active HPV infection in HNSCC tumors [50, 51]. This discrepancy, independent of HPV status, might be explained by pointing to geographical differences and populations’ exposure to additional non-viral carcinogens, which may alter p16INK4a expression. Since many studies have associated HPV-induced overexpression of p16INK4a with better survival in HNSCC patients compared to HPV-unrelated cases [4, 52], the use of p16INK4a as a surrogate biomarker of active HPV infection to prove its pathogenic role in tumor causation needs to be considered in patients from the studied geographical region.
We examined the expression of p65, as the major and the most abundant member of the NF-κB protein family in HNSCC specimens by immunohistochemistry assay. As several studies have suggested the impact of p65 translocation from the cytoplasm to the nucleus on activation of the NF-κB/relA signaling network [53], all the specimens were evaluated for cytoplasmic and nuclear localization of p65 protein. Our results demonstrated a high expression of cytoplasmic NF-κB in approximately 57–78% of all HNSCC cancers from different anatomical sites. In addition, we found nuclear localization of p65 in 23.8% of HNSCC cases. This finding is supported by various studies demonstrating an overexpression of cytoplasmic and nuclear localization of p65 in almost 75% and 23–52% of oral squamous cell carcinomas, respectively [53–55]. The expression of cytoplasmic NF-κB depicted a statistically significant gradual increase in low-grade HNSCC cancers when compared to high-grade tumors. This result proposes that the activation of the NF-κB/relA signaling network is an early event in HNSCC cancer development. Many studies have demonstrated increased expression of NF-κB in HPV-related cervical, laryngeal, tongue, and penile cancers, suggesting a role for NF-κB in the life cycle of HPV and its carcinogenesis [54, 56–60]. In the present study, we analyzed the effect of high-risk HPV infection on the cytoplasmic and nuclear expression of NF-κB by comparing the expression levels of p65 in HPV-positive and HPV-negative HNSCC cancers. Although the upregulation of cytoplasmic NF-κB was observed in 75% of HPV-related HNSCC tumors, it was not significant when compared to cases without HPV infection. Some studies reported that deletion or mutation in cylindromatosis (CYLD) and TNF receptor-associated factor 3 (TRAF3) proteins, which leads to activation of NF-κB and disruption of the innate immune signaling network, was found in approximately 30% of HPV-related oropharyngeal cancers [61, 62]. There is also evidence that defects in other NF-κB regulators such as mitogen-activated protein kinase kinase kinase 14 (MAP3K14), baculoviral IAP repeat-containing 3 (BIRC3), TNF receptor-associated factor (TRAF2), and myeloid differentiation primary response 88 (MYD88) can induce NF-κB activity. It seems that disruption of these genes and upregulation of NF-κB along with inhibition of innate immune responses play a critical role in the survival of HPV-infected cells, delaying apoptosis and triggering tumor development in HPV-related HNSCC cancers.
Conclusions
We have shown that HPV-related HNSCC tumors account for considerable proportions of total HNSCCs in western Iran and HPV infection and p16INK4a overexpression are common in low-grade tumors and among younger HNSCC patients. Our data suggest that for optimal assessment of active HPV infection in FFPE tissue specimens, a combination of HPV nested PCR assay and p16INK4a staining is required. Our data highlights that HPV16, in particular the A2 sublineage, followed by A1 and D2 sublineages are the major causative agents associated with HNSCCs in Iran. HPV16 is the most predominant type and its lineage distribution is not similar to that of the data previously reported for cervical samples in the country. It seems that the prophylactic vaccines developed for cervical cancer prevention and control could also be applicable for the prevention of HPV-related HNSCCs in our population.
Acknowledgments
The authors would like to thank the staff of the Diagnostic Laboratory Sciences and Technology Research Center (DLSTRC) and Oral & Maxillofacial Pathology Laboratory, school of Dentistry, Shiraz University of Medical Sciences, Shiraz, Iran.
Authors’ contributions
Writing—original draft preparation, methodology, and investigation: F.P. Conceptualization, methodology, writing review and editing, supervision, and visualization: A.F. Methodology and validation: T.P. Methodology and investigation: A.A.T. Project administration, software, and formal analysis: P.A. Resources and methodology: A.B.B. and M.J.A. All the authors provided a critical revision of this manuscript.
Funding
This study was supported by a grant from Shiraz University of Medical Sciences, Shiraz, Iran, under the agreement no. 93-01-45-8241.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Ethical statement
The Ethical Committee of Shiraz University of Medical Sciences approved this study (SUMS, ethical reference No. IR.SUMS.REC.1393.8241) given the retrospective nature of the study.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wild CP, Weiderpass E, Stewart BW (2020) World cancer report: cancer research for cancer prevention. Lyon: International Agency for Research on Cancer. 2020; Chapter 5.2. ISBN 978-92-832-0448-0
- 2.Vigneswaran N, Williams MD. Epidemiologic trends in head and neck cancer and aids in diagnosis. Oral Maxillofac Surg Clin North Am. 2014;26(2):123–141. doi: 10.1016/j.coms.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ndiaye C, Mena M, Alemany L, Arbyn M, Castellsagué X, Laporte L, Bosch FX, de Sanjosé S, Trottier H. HPV DNA, E6/E7 mRNA, and p16INK4a detection in head and neck cancers: a systematic review and meta-analysis. Lancet Oncol. 2014;15(12):1319–1331. doi: 10.1016/S1470-2045(14)70471-1. [DOI] [PubMed] [Google Scholar]
- 4.Spence T, Bruce J, Yip KW, Liu FF. HPV associated head and neck cancer. Cancers. 2016;8(8):75. doi: 10.3390/cancers8080075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rautava J, Kuuskoski J, Syrjänen K, Grenman R, Syrjänen S. HPV genotypes and their prognostic significance in head and neck squamous cell carcinomas. J Clin Virol. 2012;53(2):116–120. doi: 10.1016/j.jcv.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 6.Burk RD, Harari A, Chen Z. Human papillomavirus genome variants. Virology. 2013;445(1–2):232–243. doi: 10.1016/j.virol.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freitas LB, Chen Z, Muqui EF, Boldrini NA, Miranda AE, et al. Human papillomavirus 16 non-European variants are preferentially associated with high-grade cervical lesions. PLoS One. 2014;9(7):e100746. doi: 10.1371/journal.pone.0100746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366(6456):704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 9.Romagosa C, Simonetti S, Lopez-Vicente L, Mazo A, Lleonart ME, et al. p16 Ink4a overexpression in cancer: a tumor suppressor gene associated with senescence and high-grade tumors. Oncogene. 2011;30(18):2087–2097. doi: 10.1038/onc.2010.614. [DOI] [PubMed] [Google Scholar]
- 10.Wang H, Sun R, Lin H, Hu WH. P16INK4A as a surrogate biomarker for human papillomavirus-associated oropharyngeal carcinoma: consideration of some aspects. Cancer Sci. 2013;104(12):1553–1559. doi: 10.1111/cas.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vandermark ER, Deluca KA, Gardner CR, Marker DF, Schreiner CN, Strickland DA, Wilton KM, Mondal S, Woodworth CD. Human papillomavirus type 16 E6 and E 7 proteins alter NF-kB in cultured cervical epithelial cells and inhibition of NF-kB promotes cell growth and immortalization. Virology. 2012;425(1):53–60. doi: 10.1016/j.virol.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anbo N, Ogi K, Sogabe Y, Shimanishi M, Kaneko T, Dehari H, Miyazaki A, Hiratsuka H. Suppression of NF-κB/p65 inhibits the proliferation in oral squamous cancer cells. J Cancer Ther. 2013;4:891–897. doi: 10.4236/jct.2013.44100. [DOI] [Google Scholar]
- 13.Monisha J, Kishor Roy N, Bordoloi D, Kumar A, Golla R, Kotoky J, Padmavathi G, B. Kunnumakkara A. A. Nuclear factor kappa B: a potential target to persecute head and neck cancer. Curr Drug Targets. 2017;18(2):232–253. doi: 10.2174/1389450117666160201112330. [DOI] [PubMed] [Google Scholar]
- 14.Kreimer AR, Clifford GM, Boyle P, Franceschi S. Human papillomavirus types in head and neck squamous cell carcinomas worldwide: a systematic review. Cancer Epidemiol Biomark Prev. 2005;14(2):467–475. doi: 10.1158/1055-9965.EPI-04-0551. [DOI] [PubMed] [Google Scholar]
- 15.Saiki RK, Bugawan TL, Horn GT, Mullis KB, Erlich HA. Analysis of enzymatically amplified β-globin and HLA-DQα DNA with allele-specific oligonucleotide probes. Nature. 1986;324(6093):163–166. doi: 10.1038/324163a0. [DOI] [PubMed] [Google Scholar]
- 16.Farhadi A, Behzad-Behbahani A, Geramizadeh B, Sekawi Z, Rahsaz M, Sharifzadeh S. High-risk human papillomavirus infection in different histological subtypes of renal cell carcinoma. J Med Virol. 2014;86(7):1134–1144. doi: 10.1002/jmv.23945. [DOI] [PubMed] [Google Scholar]
- 17.Manos MM. The use of polymerase chain reaction amplification for the detection of genital human papillomavirus. Cancer Cells. 1989;7:209–214. [Google Scholar]
- 18.de Roda Husman AM, Walboomers JM, van den Brule AJ, Meijer CJ, Snijders PJ. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J Gen Virol. 1995;76(4):1057–1062. doi: 10.1099/0022-1317-76-4-1057. [DOI] [PubMed] [Google Scholar]
- 19.De Boer MA, Peters LA, Aziz MF, Siregar B, Cornain S, et al. Human papillomavirus type 16 E6, E7, and L1 variants in cervical cancer in Indonesia, Suriname, and The Netherlands. Gynecol Oncol. 2004;94(2):488–494. doi: 10.1016/j.ygyno.2004.05.037. [DOI] [PubMed] [Google Scholar]
- 20.Pientong C, Wongwarissara P, Ekalaksananan T, Swangphon P, Kleebkaow P, Kongyingyoes B, Siriaunkgul S, Tungsinmunkong K, Suthipintawong C. Association of human papillomavirus type 16 long control region mutation and cervical cancer. Virol J. 2013;10(1):30. doi: 10.1186/1743-422X-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duncan LD, Winkler M, Carlson ER, Heidel RE, Kang E, Webb D. p16 immunohistochemistry can be used to detect human papillomavirus in oral cavity squamous cell carcinoma. J Oral Maxillofac Surg. 2013;71(8):1367–1375. doi: 10.1016/j.joms.2013.02.019. [DOI] [PubMed] [Google Scholar]
- 23.Zhou S, Ye W, Shao Q, Qi Y, Zhang M, Liang J. Prognostic significance of XIAP and NF-κB expression in esophageal carcinoma with postoperative radiotherapy. World J Surg Oncol. 2013;11(1):288. doi: 10.1186/1477-7819-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baud V, Karin M. Is NF-κB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov. 2009;8(1):33–40. doi: 10.1038/nrd2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, Wu L, Zahurak ML, Daniel RW, Viglione M, Symer DE, Shah KV, Sidransky D. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92(9):709–720. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 26.Klussmann JP, Weissenborn SJ, Wieland U, Dries V, Kolligs J, Jungehuelsing M, Eckel HE, Dienes HP, Pfister HJ, Fuchs PG. Prevalence, distribution, and viral load of human papillomavirus 16 DNA in tonsillar carcinomas. Cancer. 2001;92(11):2875–2884. doi: 10.1002/1097-0142(20011201)92:11<2875::aid-cncr10130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 27.Singhi AD, Califano J, Westra WH. High-risk human papillomavirus in nasopharyngeal carcinoma. Head Neck. 2012;34(2):213–218. doi: 10.1002/hed.21714. [DOI] [PubMed] [Google Scholar]
- 28.Lee LA, Huang CG, Liao CT, Lee LY, Hsueh C, Chen TC, Lin CY, Fan KH, Wang HM, Huang SF, Chen IH, Kang CJ, Ng SH, Yang SL, Tsao KC, Chang YL, Yen TC Human papillomavirus-16 infection in advanced oral cavity cancer patients is related to an increased risk of distant metastases and poor survival. PLoS One 2012;7(7). 10.1371/journal.pone.0040767 [DOI] [PMC free article] [PubMed]
- 29.Duray A, Descamps G, Decaestecker C, Remmelink M, Sirtaine N, Lechien J, Ernoux-Neufcoeur P, Bletard N, Somja J, Depuydt CE, Delvenne P, Saussez S. Human papillomavirus DNA strongly correlates with a poorer prognosis in oral cavity carcinoma. Laryngoscope. 2012;122(7):1558–1565. doi: 10.1002/lary.23298. [DOI] [PubMed] [Google Scholar]
- 30.Götz C, Bischof C, Wolff KD, Kolk A. Detection of HPV infection in head and neck cancers: promise and pitfalls in the last ten years: a meta-analysis. Mol Clin Oncol. 2019;10(1):17–28. doi: 10.3892/mco.2018.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Termine N, Panzarella V, Falaschini S, Russo A, Matranga D, Lo Muzio L, Campisi G. HPV in oral squamous cell carcinoma vs head and neck squamous cell carcinoma biopsies: a meta-analysis (1988–2007) Ann Oncol. 2008;19(10):1681–1690. doi: 10.1093/annonc/mdn372. [DOI] [PubMed] [Google Scholar]
- 32.Guo L, Yang F, Yin Y, Liu S, Li P, Zhang X, Chen D, Liu Y, Wang J, Wang K, Zhu Y, Lv Q, Wang X, Sun X. Prevalence of human papillomavirus type-16 in head and neck cancer among the Chinese population: a meta-analysis. Front Oncol. 2018;8:619. doi: 10.3389/fonc.2018.00619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jalilvand S, Shoja Z, Hamkar R. Human papillomavirus burden in different cancers in Iran: a systematic assessment. Asian Pac J Cancer Prev. 2014;15(17):7029–7035. doi: 10.7314/apjcp.2014.15.17.7029. [DOI] [PubMed] [Google Scholar]
- 34.Kingma DW, Allen RA, Moore W, Caughron SK, Melby M, Gillies EM, Marlar RA, Dunn ST. HPV genotype distribution in oral and oropharyngeal squamous cell carcinoma using seven in vitro amplification assays. Anticancer Res. 2010;30(12):5099–5104. [PubMed] [Google Scholar]
- 35.Nauta IH, Rietbergen MM, van Bokhoven AA, Bloemena E, Lissenberg-Witte BI, et al. Evaluation of the eighth TNM classification on p16-positive oropharyngeal squamous cell carcinomas in the Netherlands and the importance of additional HPV DNA testing. Ann Oncol. 2018;29(5):1273–1279. doi: 10.1093/annonc/mdy060. [DOI] [PubMed] [Google Scholar]
- 36.Ni G, Huang K, Luan Y, Cao Z, Chen S, Ma B, Yuan J, Wu X, Chen G, Wang T, Li H, Walton S, Liu F, Chen B, Wang Y, Pan X, Liu X, Frazer IH (2019) Human papillomavirus infection among head and neck squamous cell carcinomas in southern China. PLoS One 14(9). 10.1371/journal.pone.0221045 [DOI] [PMC free article] [PubMed]
- 37.Michaud DS, Langevin SM, Eliot M, Nelson HH, Pawlita M, McClean MD, Kelsey KT. High-risk HPV types and head and neck cancer. Int J Cancer. 2014;135(7):1653–1661. doi: 10.1002/ijc.28811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mineta H, Ogino T, Amano HM, Ohkawa Y, Araki K, Takebayashi S, Miura K. Human papilloma virus (HPV) type 16 and 18 detected in head and neck squamous cell carcinoma. Anticancer Res. 1998;18(6B):4765–4768. [PubMed] [Google Scholar]
- 39.Syrjänen S. The role of human papillomavirus infection in head and neck cancers. Ann Oncol. 2010;21(suppl_7):vii243–vii245. doi: 10.1093/annonc/mdq454. [DOI] [PubMed] [Google Scholar]
- 40.D’Souza G, Westra WH, Wang SJ, Van Zante A, Wentz A, et al. Differences in the prevalence of human papillomavirus (HPV) in head and neck squamous cell cancers by sex, race, anatomic tumor site, and HPV detection method. JAMA Oncol. 2017;3(2):169–177. doi: 10.1001/jamaoncol.2016.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, Jiang B, Goodman MT, Sibug-Saber M, Cozen W, Liu L, Lynch CF, Wentzensen N, Jordan RC, Altekruse S, Anderson WF, Rosenberg PS, Gillison ML. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29(32):4294–4301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Field N, Lechner M. Exploring the implications of HPV infection for head and neck cancer. Sex Transm Dis. 2015;91:229–230. doi: 10.1136/sextrans-2014-051808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamada T, Wheeler CM, Halpern AL, Stewart AC, Hildesheim A, Jenison SA. Human papillomavirus type 16 variant lineages in United States populations characterized by nucleotide sequence analysis of the E6, L2, and L1 coding segments. J Virol. 1995;69(12):7743–7753. doi: 10.1128/JVI.69.12.7743-7753.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaezi T, Shoja Z, Hamkar R, Shahmahmoodi S, Nozarian Z, Marashi SM, Jalilvand S. Human papillomavirus type 16 lineage analysis based on E6 region in cervical samples of Iranian women. Infect Genet Evol. 2017;55:26–30. doi: 10.1016/j.meegid.2017.08.025. [DOI] [PubMed] [Google Scholar]
- 45.Schiffman M, Rodriguez AC, Chen Z, Wacholder S, Herrero R, Hildesheim A, Desalle R, Befano B, Yu K, Safaeian M, Sherman ME, Morales J, Guillen D, Alfaro M, Hutchinson M, Solomon D, Castle PE, Burk RD. A population-based prospective study of carcinogenic human papillomavirus variant lineages, viral persistence, and cervical neoplasia. Cancer Res. 2010;70(8):3159–3169. doi: 10.1158/0008-5472.CAN-09-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zuna RE, Moore WE, Shanesmith RP, Dunn ST, Wang SS, Schiffman M, Blakey GL, Teel T. Association of HPV16 E6 variants with diagnostic severity in cervical cytology samples of 354 women in a US population. Int J Cancer. 2009;125(11):2609–2613. doi: 10.1002/ijc.24706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicolás-Párraga S, Alemany L, De Sanjosé S, Bosch FX, Bravo IG, et al. Differential HPV16 variant distribution in squamous cell carcinoma, adenocarcinoma and adenosquamous cell carcinoma. Int J Cancer. 2017;140(9):2092–2100. doi: 10.1002/ijc.30636. [DOI] [PubMed] [Google Scholar]
- 48.Mirabello L, Yeager M, Cullen M, Boland JF, Chen Z, et al. HPV16 sublineage associations with histology-specific cancer risk using HPV whole-genome sequences in 3200 women. J Natl Cancer Inst. 2016;108(9):djw100. doi: 10.1093/jnci/djw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rietbergen MM, Snijders PJ, Beekzada D, Braakhuis BJ, Brink A, et al. Molecular characterization of p16-immunopositive but HPV DNA-negative oropharyngeal carcinomas. Int J Cancer. 2014;134(10):2366–2372. doi: 10.1002/ijc.28580. [DOI] [PubMed] [Google Scholar]
- 50.Gheit T, Anantharaman D, Holzinger D, Alemany L, Tous S, Lucas E, Prabhu PR, Pawlita M, Ridder R, Rehm S, Bogers J, Maffini F, Chiocca S, Lloveras B, Kumar RV, Somanathan T, de Sanjosé S, Castellsagué X, Arbyn M, Brennan P, Sankaranarayanan R, Pillai MR, Gangane N, Tommasino M, the HPV-AHEAD study group Role of mucosal high-risk human papillomavirus types in head and neck cancers in central India. Int J Cancer. 2017;141(1):143–151. doi: 10.1002/ijc.30712. [DOI] [PubMed] [Google Scholar]
- 51.Castellsagué X, Alemany L, Quer M, Halec G, Quirós B, et al. HPV involvement in head and neck cancers: comprehensive assessment of biomarkers in 3680 patients. J Natl Cancer Inst. 2016;108(6):djv403. doi: 10.1093/jnci/djv403. [DOI] [PubMed] [Google Scholar]
- 52.Porceddu SV, Milne R, Brown E, Bernard A, Rahbari R, Cartmill B, Foote M, McGrath M, Coward J, Panizza B (2017) Validation of the ICON-S staging for HPV-associated oropharyngeal carcinoma using a pre-defined treatment policy. Oral Oncol 66:81–86. 10.1016/j.oraloncology.2017.01.002 [DOI] [PubMed]
- 53.Yan M, Xu Q, Zhang P, Zhou XJ, Zhang ZY, Chen WT (2010) Correlation of NF-κB signal pathway with tumor metastasis of human head and neck squamous cell carcinoma. BMC Cancer 10(1):437. 10.1186/1471-2407-10-437 [DOI] [PMC free article] [PubMed]
- 54.Sawhney M, Rohatgi N, Kaur J, Shishodia S, Sethi G, Gupta SD, Deo SVS, Shukla NK, Aggarwal BB, Ralhan R (2007) Expression of NF-kappaB parallels COX-2 expression in oral precancer and cancer: association with smokeless tobacco. Int J Cancer 120:2545–2556. 10.1002/ijc.22657 [DOI] [PubMed]
- 55.Kamperos G, Nikitakis N, Sfakianou A, Avgoustidis D, Sklavounou-Andrikopoulou A (2016) Expression of NF-κB and IL-6 in oral precancerous and cancerous lesions: an immunohistochemical study. Medicina Oral Patologia Oral y Cirugia Bucal 21(1):e6. 10.4317/medoral.20570 [DOI] [PMC free article] [PubMed]
- 56.Nakahara T, Tanaka K, Ohno SI, Egawa N, Yugawa T, Kiyono T. Activation of NF-κB by human papillomavirus 16 E1 limits E1-dependent viral replication through degradation of E1. J Virol. 2015;89(9):5040–5059. doi: 10.1128/JVI.00389-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li J, Jia H, Xie L, Wang X, Wang X, He H, Lin Y, Hu L. Association of constitutive nuclear factor-κB activation with aggressive aspects and poor prognosis in cervical cancer. Int J Gynecol Cancer. 2009;19(8):1421–1426. doi: 10.1111/IGC.0b013e3181b70445. [DOI] [PubMed] [Google Scholar]
- 58.Du J, Chen GG, Vlantis AC, Xu H, Tsang RK, et al. The nuclear localization of NFκB and p53 is positively correlated with HPV16 E7 level in laryngeal squamous cell carcinoma. J Histochem Cytochem. 2003;51(4):533–539. doi: 10.1177/002215540305100415. [DOI] [PubMed] [Google Scholar]
- 59.Gupta S, Kumar P, Kaur H, Sharma N, Gupta S, et al. Constitutive activation and overexpression of NF-κB/c-Rel in conjunction with p50 contribute to aggressive tongue tumorigenesis. Oncotarget. 2018;9(68):33011. doi: 10.18632/oncotarget.26041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Senba M, Mori N, Fujita S, Jutavijittum P, Yousukh A, et al. Relationship among human papillomavirus infection, p16INK4a, p53 and NF-κB activation in penile cancer from northern Thailand. Oncol Lett. 2010;1(4):599–603. doi: 10.3892/ol_00000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hajek M, Sewell A, Kaech S, Burtness B, Yarbrough WG, Issaeva N. TRAF3/CYLD mutations identify a distinct subset of human papillomavirus associated head and neck squamous cell carcinoma. Cancer. 2017;123:1778–1790. doi: 10.1002/cncr.30570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pan C, Issaeva N, Yarbrough WG. HPV-driven oropharyngeal cancer: current knowledge of molecular biology and mechanisms of carcinogenesis. Cancers Head Neck. 2018;3(1):12. doi: 10.1186/s41199-018-0039-3. [DOI] [PMC free article] [PubMed] [Google Scholar]