Abstract
Non-aureus staphylococci are commonly found on dairy farms. Two rarely investigated species are Staphylococcus (S.) cohnii and S. urealyticus. Since multidrug-resistant S. cohnii and S. urealyticus are known, they may serve as an antimicrobial resistance (AMR) gene reservoir for harmful staphylococcal species. In our study, nine S. cohnii and six S. urealyticus isolates from German dairy farms were analyzed by whole-genome sequencing and AMR testing. The isolates harbored various AMR genes (aadD1, str, mecA, dfrC/K, tetK/L, ermC, lnuA, fexA, fusF, fosB6, qacG/H) and exhibited non-wildtype phenotypes (resistances) against chloramphenicol, clindamycin, erythromycin, fusidic acid, rifampicin, streptomycin, tetracycline, tiamulin and trimethoprim. Although 14/15 isolates lacked the blaZ, mecA and mecC genes, they showed reduced susceptibility to a number of beta-lactam antibiotics including cefoxitin (MIC 4–8 mg/L) and penicillin (MIC 0.25–0.5 mg/L). The specificity of cefoxitin susceptibility testing for mecA or mecC gene prediction in S. cohnii and S. urealyticus seems to be low. A comparison with penicillin-binding protein (PBP) amino acid sequences of S. aureus showed identities of only 70–80% with regard to PBP1, PBP2 and PBP3. In conclusion, S. cohnii and S. urealyticus from selected German dairy farms show multiple resistances to antimicrobial substances and may carry unknown antimicrobial resistance determinants.
Subject terms: Antimicrobial resistance, Bacterial genes
Introduction
Non-aureus staphylococci (NAS) are frequently found on dairy farms and occasionally cause intramammary infections1–3. Among others, Staphylococcus (S.) cohnii was found on farms; however, it has been regarded a commensal bacterium, and not involved in severe animal infections such as bovine mastitis2,4. S. cohnii has been divided into the subspecies S. cohnii subsp. cohnii and S. cohnii subsp. urealyticus. However, recent phylogenetic studies suggested that the subspecies S. cohnii subsp. urealyticus should be regarded as an individual species S. urealyticus5. With respect to human health, S. cohnii has been reported to be an opportunistic pathogen6,7. Multidrug-resistant S. cohnii isolates were recently detected on frequently touched surfaces in a London hospital8. Accordingly, S. cohnii as well as S. urealyticus isolates of animal origin were shown to carry multiple antimicrobial resistance (AMR) genes and express phenotypic resistance to various classes of antimicrobials9–11. Even resistance to last resort antibiotics such as linezolid was demonstrated12. Thus far, only five complete genomes of S. cohnii or S. urealyticus are available in public databases such as NCBI Genome, complicating comparison of alterations in gene function with regard to AMR. Novel resistance determinants such as the fusidic acid resistance gene fusF have been discovered in S. cohnii isolates13. The most prominent resistance gene in staphylococci is mecA, which encodes a variant penicillin-binding protein (PBP) 2a. A PBP is a transpeptidase, which is crucial for bacterial cell wall synthesis. Beta-lactam antibiotics bind to the PBP and thus inhibit the transpeptidase reaction. Differences in the structure of PBP2a compared to other PBPs result in a lower binding affinity for beta-lactam antibiotics, causing resistance14,15. The mecA gene is located on the staphylococcal cassette chromosome (SCC) mec element, which has been categorized into 13 types16,17. It was shown that NAS may harbor high diversity and unknown SCCmec elements8,18 and new SCCmec elements have indeed been described for S. cohnii6,19. Determination of beta-lactam antibiotic resistance in S. aureus, a well investigated staphylococcal species, and the identification of methicillin-resistant S. aureus (MRSA) is routinely performed by cefoxitin resistance testing and detection of the mecA or mecC gene. However, several studies have reported that the determination of beta-lactam antibiotic resistance in NAS using cefoxitin susceptibility testing might be misleading and the use of oxacillin should be favored20–22.
The aim of our study was to evaluate the antimicrobial resistance potential of S. cohnii and S. urealyticus isolates from German dairy farms. The isolates were analyzed by whole-genome sequencing (WGS) and AMR testing. AMR gene prediction was linked to the respective phenotypic resistance pattern. A special focus was set on beta-lactam antibiotic resistance and related AMR genes, since beta-lactams are commonly used to treat infections such as mastitis in livestock. Since NAS might serve as an AMR gene reservoir for S. aureus, which is a major animal and human pathogenic species, monitoring of multidrug-resistant NAS is of high importance.
Results
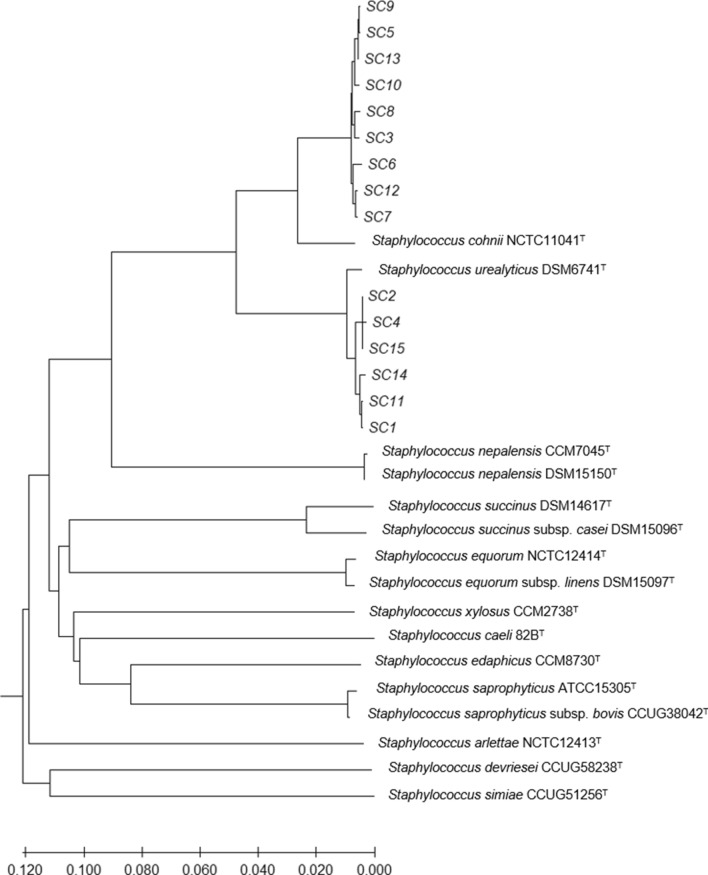
Phylogenetic analysis
Fifteen staphylococcal isolates from German dairy farms were presumptively identified as S. cohnii by MALDI-TOF-MS analysis. According to the phylogenetic analysis using the type genome server (TYGS), the isolates SC3, SC5, SC6, SC7, SC8, SC9, SC10, SC12 and SC13 were assigned to a S. cohnii type strain, while SC1, SC2, SC4, SC11, SC14 and SC15 clustered close to the type strain S. cohnii subsp. urealyticus DSM 6718 (Fig. 1). Recent phylogenetic analyses suggest the promotion of S. cohnii subsp. urealyticus to the novel species S. urealyticus5. Moreover, the genotypical differences between S. cohnii isolates within one farm were determined by comparing two different isolates from the same farm. Isolates SC7/8 and SC9/10 were therefore assigned to the same cluster, respectively.
Figure 1.
Phylogenetic analysis by TYGS of 15 presumptive S. cohnii isolates from German dairy farms reveals two main clusters of relationship. Clustering of isolates SC1, SC2, SC4, SC11, SC14 and SC15 together with S. urealyticus DSM6718 indicates assignment of these isolates to the species.
AMR genes
The isolates harbored various AMR genes with a variable distribution among the isolates. Resistance to the antibiotic classes aminoglycosides (aadD1, str), beta-lactams (mecA), trimethoprim (dfrC/K), tetracyclines (tetK/L), macrolides (ermC), pleuromutilin–lincosamide–streptogramin (lnuA), phenicols (fexA), fusidic acid (fusF), fosfomycin (fosB6) and to quaternary ammonium compounds (qacG/H) was predicted. The number of detected AMR genes per isolate varied between eight (1/15), seven (4/15), five (1/15), four (3/15) and three (6/15).
The fusF and dfrC genes were detected in all S. cohnii and S. urealyticus isolates (Table 1). The lnuA (2/9 S. cohnii isolates), aadD1 (2/6 S. urealyticus isolates), ermC (1/9 S. cohnii isolates; 1/6 S. urealyticus isolates), tetL (2/6 S. urealyticus isolates), mphC (4/9 S. cohnii isolates), str (3/9 S. cohnii isolates; 1/6 S. urealyticus isolates), tetK (4/9 S. cohnii isolates; 1/6 S. urealyticus isolates), qacH (6/6 S. urealyticus isolates) and msrA (9/9 S. cohnii isolates) genes were only detected in some isolates or in only one of the two species. In contrast, the mecA (SC7, S. cohnii), fexA (SC4, S. urealyticus), dfrK (SC4, S. urealyticus), fosB6 (SC9, S. cohnii) and qacG (SC15, S. urealyticus) genes were each present in only one isolate. The mecA gene of isolate SC7 was located on a SCCmec type V.
Table 1.
Predicted antimicrobial resistance genes compared to results of minimum inhibitory concentration of various antimicrobial substances for the S. cohnii and S. urealyticus isolates SC1-15 from German dairy farms.
| Strain | Species | Farm | AMR1 genes | MIC2,3 (mg/L) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CHL | CIP | CLI | ERY | FUS | GEN | KAN | LZD | MUP | RIF | SMX | STR | SYN | TET | TIA | TMP | VAN | ||||
| SC1 | S. urealyticus | 1 | dfrC; fusF; qacH | 8 | 1 | 1 | ≤ 0.25 | > 4 | ≤ 1 | ≤ 4 | ≤ 1 | ≤ 0.5 | 0.06 | ≤ 64 | ≤ 4 | ≤ 0.5 | 1 | > 4 | ≤ 2 | 2 |
| SC2 | S. urealyticus | 2 | dfrC; fusF; qacH | ≤ 4 | 0.5 | 0.25 | ≤ 0.25 | > 4 | ≤ 1 | ≤ 4 | ≤ 1 | ≤ 0.5 | 0.03 | ≤ 64 | ≤ 4 | ≤ 0.5 | ≤ 0.5 | 4 | ≤ 2 | ≤ 1 |
| SC3 | S. cohnii | 3 | dfrC; fusF; msr(A) | 8 | 0.5 | 0.5 | > 8 | > 4 | ≤ 1 | ≤ 4 | ≤ 1 | ≤ 0.5 | 0.5 | 128 | ≤ 4 | ≤ 0.5 | 1 | > 4 | 4 | 2 |
| SC4 | S. urealyticus | 4 | aadD1; dfrC; dfrK; fexA; fusF; qacH; str; tet(L) | 64 | ≤ 0.25 | 0.5 | ≤ 0.25 | > 4 | ≤ 1 | 8 | ≤ 1 | ≤ 0.5 | 0.03 | ≤ 64 | > 32 | ≤ 0.5 | > 16 | 4 | > 32 | ≤ 1 |
| SC5 | S. cohnii | 5 | dfrC; fusF; msr(A) | 8 | 0.5 | ≤ 0.12 | 2 | > 4 | ≤ 1 | ≤ 4 | ≤ 1 | ≤ 0.5 | 0.12 | ≤ 64 | ≤ 4 | ≤ 0.5 | ≤ 0.5 | 1 | ≤ 2 | ≤ 1 |
| SC6 | S. cohnii | 6 | dfrC; fusF; mph(C); msr(A) | ≤ 4 | 0.5 | 0.25 | 4 | > 4 | ≤ 1 | ≤ 4 | ≤ 1 | ≤ 0.5 | 0.25 | ≤ 64 | ≤ 4 | 1 | ≤ 0.5 | 2 | ≤ 2 | 2 |
| SC7 | S. cohnii | 7 | dfrC; fusF; mecA; mph(C); msr(A); str; tet(K) | 16 | ≤ 0.25 | 0.25 | 2 | > 4 | ≤ 1 | ≤ 4 | ≤ 1 | ≤ 0.5 | 0.5 | ≤ 64 | > 32 | 1 | > 16 | > 4 | ≤ 2 | 2 |
| SC8 | S. cohnii | 7 | dfrC; fusF; msr(A) | 16 | 1 | 0.5 | > 8 | > 4 | ≤ 1 | ≤ 4 | ≤ 1 | ≤ 0.5 | 0.5 | ≤ 64 | ≤ 4 | ≤ 0.5 | 1 | > 4 | 4 | ≤ 1 |
| SC9 | S. cohnii | 8 | dfrC; fosB6; fusF; lnu(A); msr(A) | ≤ 4 | ≤ 0.25 | 0.5 | 2 | > 4 | ≤ 1 | ≤ 4 | ≤ 1 | ≤ 0.5 | 0.25 | ≤ 64 | ≤ 4 | ≤ 0.5 | ≤ 0.5 | > 4 | ≤ 2 | ≤ 1 |
| SC10 | S. cohnii | 8 | dfrC; fusF; lnu(A); mph(C); msr(A); str; tet(K) | 8 | 0.5 | 1 | 2 | > 4 | ≤ 1 | ≤ 4 | ≤ 1 | ≤ 0.5 | 0.03 | ≤ 64 | > 32 | ≤ 0.5 | > 16 | > 4 | 8 | ≤ 1 |
| SC11 | S. urealyticus | 9 | aadD1; dfrC; erm(C); fusF; qacH; tet(K); tet(L) | 8 | 0.5 | > 4 | > 8 | > 4 | ≤ 1 | 8 | ≤ 1 | ≤ 0.5 | 0.06 | ≤ 64 | ≤ 4 | 1 | > 16 | > 4 | ≤ 2 | ≤ 1 |
| SC12 | S. cohnii | 9 | dfrC; erm(C); fusF; mph(C); msr(A); str; tet(K) | ≤ 4 | ≤ 0.25 | > 4 | > 8 | 1 | ≤ 1 | ≤ 4 | ≤ 1 | ≤ 0.5 | 0.12 | ≤ 64 | ≤ 4 | ≤ 0.5 | 16 | ≤ 0.5 | ≤ 2 | ≤ 1 |
| SC13 | S. cohnii | 10 | dfrC; fusF; msr(A); tet(K) | 8 | 0.5 | ≤ 0.12 | 4 | > 4 | ≤ 1 | ≤ 4 | 2 | ≤ 0.5 | 0.25 | ≤ 64 | ≤ 4 | ≤ 0.5 | > 16 | > 4 | 4 | ≤ 1 |
| SC14 | S. urealyticus | 11 | dfrC; fusF; qacH | ≤ 4 | 0.5 | 0.25 | ≤ 0.25 | > 4 | ≤ 1 | ≤ 4 | ≤ 1 | ≤ 0.5 | 0.03 | ≤ 64 | ≤ 4 | ≤ 0.5 | ≤ 0.5 | 4 | ≤ 2 | ≤ 1 |
| SC15 | S. urealyticus | 12 | dfrC; fusF; qacG; qacH | 8 | 0.5 | 0.25 | 0.5 | > 4 | ≤ 1 | ≤ 4 | ≤ 1 | ≤ 0.5 | 0.03 | ≤ 64 | ≤ 4 | ≤ 0.5 | 1 | 4 | ≤ 2 | ≤ 1 |
| EUCAST ECOFF5 | Staphylococcus aureus | ≤ 16 | ≤ 1 | ≤ 0.25 | ≤ 1 | ≤ 0.5 | ≤ 2 | ≤ 8 | ≤ 4 | ≤ 1 | ≤ 0.016 | ≤ 128 | ≤ 16 | ≤ 1 | ≤ 1 | ≤ 2 | ≤ 2 | ≤ 2 | ||
| Coagulase negative staphylococci | ≤ 16 | ≤ 1 | ≤ 0.25 | ≤ 1 | ≤ 0.5 | ND4 | ND | ≤ 2 | ND | ≤ 0.064 | ND | ND | ND | ≤ 1 | ND | ND | ≤ 4 | |||
1AMR antimicrobial resistance, 2MIC minimum inhibitory concentration, 3CHL chloramphenicol, CIP ciprofloxacin, CLI clindamycin, ERY erythromycin, FUS fusidic acid, GEN gentamycin, KAN kanamycin, LZD linezolid, MUP mupirocin, RIF rifampicin, SMX sulfamethoxazole, STR streptomycin, SYN quinupristin-dalfopristin, TET tetracycline, TIA tiamulin, TMP trimethoprim, VAN vancomycin, 4ND not defined. 5Epidemiological cut-off values as provided by EUCAST (http://www.eucast.org; accessed August 31, 2020).
Phenotypic AMR testing
The minimum inhibitory concentration (MIC) values for various antimicrobial substances are shown in Tables 1 and 2. Multidrug resistance to at least three antimicrobial classes was found in all isolates. According to the EUCAST epidemiological cut-off values (ECOFFs) for S. aureus and coagulase negative staphylococci, a non-wildtype phenotype was determined in several isolates for chloramphenicol (1/6 S. urealyticus isolates), clindamycin (5/9 S. cohnii isolates; 3/6 S. urealyticus isolates), erythromycin (9/9 S. cohnii isolates; 1/6 S. urealyticus isolates), fusidic acid (9/9 S. cohnii isolates; 6/6 S. urealyticus isolates), rifampicin (8/9 S. cohnii isolates), streptomycin (2/9 S. cohnii isolates; 1/6 S. urealyticus isolates), tetracycline (4/9 S. cohnii isolates; 2/6 S. urealyticus isolates), tiamulin (6/9 S. cohnii isolates; 6/6 S. urealyticus isolates) and trimethoprim (4/9 S. cohnii isolates; 1/6 S. urealyticus isolates) (Table 1).
Table 2.
Predicted beta-lactam antimicrobial resistance genes and minimum inhibitory concentration of S. cohnii and S. urealyticus isolates SC1-15 from twelve German dairy farms.
| Strain | Species | Farm | Beta-lactam AMR1 genes | MIC2,3 (mg/L) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FOX | OXA | PEN | AMP | FEP | ETP | IMP | MERO | FOT | FOTCLA | TAZ | TAZCLA | TEMOCI | ||||
| SC1 | S. urealyticus | 1 | – | 8 | 0.5 | 0.5 | ≤ 1 | 4 | 2 | 0.25 | 0.5 | 8 | 8 | 32 | 32 | > 128 |
| SC2 | S. urealyticus | 2 | – | 8 | 0.5 | 0.25 | ≤ 1 | 4 | 2 | ≤ 0.12 | 0.5 | 8 | 8 | 32 | 32 | > 128 |
| SC3 | S. cohnii | 3 | – | 8 | 1 | 0.5 | ≤ 1 | 4 | 2 | ≤ 0.12 | 0.5 | 16 | 8 | 64 | 32 | > 128 |
| SC4 | S. urealyticus | 4 | – | 8 | 1 | 0.5 | ≤ 1 | 8 | 2 | ≤ 0.12 | 0.5 | 16 | 8 | 64 | 32 | > 128 |
| SC5 | S. cohnii | 5 | – | 8 | 1 | 0.5 | ≤ 1 | 4 | 2 | ≤ 0.12 | 0.5 | 8 | 8 | 64 | 32 | > 128 |
| SC6 | S. cohnii | 6 | – | 4 | 1 | 0.5 | ≤ 1 | 4 | 2 | ≤ 0.12 | 0.5 | 8 | 8 | 64 | 64 | > 128 |
| SC7 | S. cohnii | 7 | mecA | > 16 | > 8 | > 2 | 4 | > 32 | > 2 | 0.25 | 2 | > 64 | 64 | 128 | > 128 | > 128 |
| SC8 | S. cohnii | 7 | – | 8 | 0.5 | 0.5 | ≤ 1 | 4 | 2 | ≤ 0.12 | 0.5 | 8 | 8 | 32 | 32 | > 128 |
| SC9 | S. cohnii | 8 | – | 4 | 0.5 | 0.25 | ≤ 1 | 2 | 2 | ≤ 0.12 | 0.25 | 4 | 4 | 32 | 16 | > 128 |
| SC10 | S. cohnii | 8 | – | 8 | 0.5 | 0.5 | ≤ 1 | 4 | 2 | ≤ 0.12 | 0.5 | 8 | 8 | 32 | 32 | > 128 |
| SC11 | S. urealyticus | 9 | – | 4 | 0.5 | 0.25 | ≤ 1 | 2 | 2 | ≤ 0.12 | 0.5 | 8 | 4 | 64 | 32 | > 128 |
| SC12 | S. cohnii | 9 | – | 8 | 0.5 | 0.25 | ≤ 1 | 2 | 2 | ≤ 0.12 | 0.5 | 4 | 4 | 32 | 16 | > 128 |
| SC13 | S. cohnii | 10 | – | 4 | 0.5 | 0.25 | ≤ 1 | 2 | 2 | ≤ 0.12 | 0.5 | 8 | 8 | 32 | 32 | > 128 |
| SC14 | S. urealyticus | 11 | – | 4 | 0.5 | 0.25 | ≤ 1 | 2 | 2 | ≤ 0.12 | 0.5 | 4 | 4 | 16 | 16 | > 128 |
| SC15 | S. urealyticus | 12 | – | 8 | 0.5 | 0.5 | ≤ 1 | 2 | 2 | ≤ 0.12 | 0.5 | 8 | 4 | 32 | 16 | > 128 |
| EUCAST ECOFF5 | Staphylococcus aureus | ≤ 4 | ≤ 2 | ≤ 0.125 | ND | ≤ 8 | ND | ≤ 0.125 | ≤ 0.5 | ≤ 4 | ND | ND | ND | ND | ||
| Coagulase negative staphylococci | ND4 | ≤ 1 | ND | ND | ND | ND | ≤ 0.125 | ≤ 0.5 | ≤ 2 | ND | ND | ND | ND | |||
aAMR antimicrobial resistance, bMIC minimum inhibitory concentration, cFOX cefoxitin, OXA oxacillin, PEN penicillin, AMP ampicillin, FEP cefepim, ETP ertapenem, IMP imipenem, MERO meropenem, FOT cefotaxime, FOTCLA cefotaxime/clavulanic acid, TAZ ceftazidime, TAZCLA ceftazidime/clavulanic acid, TEMOCI temocillin; 4ND not defined. 5Epidemiological cut-off values as provided by EUCAST (http://www.eucast.org; accessed December 7, 2020).
With regard to resistance to beta-lactam antibiotics, reduced susceptibilities were detected for penicillin (9/9 S. cohnii isolates; 6/6 S. urealyticus isolates), temocillin (9/9 S. cohnii isolates; 6/6 S. urealyticus isolates), cefoxitin (6/9 S. cohnii isolates; 4/6 S. urealyticus isolates), ceftazidime (9/9 S. cohnii isolates; 6/6 S. urealyticus isolates) and cefotaxime (7/9 S. cohnii isolates; 5/6 S. urealyticus isolates) (Table 2). The combination of cefotaxime or ceftazidime with the beta-lactamase inhibitor clavulanic acid only slightly lowered the MICs in four and seven isolates, respectively. With regard to cefotaxime, the MIC was lowered from 16 to 8 mg/L in one S. cohnii isolate and one S. urealyticus isolate, and from 8 to 4 mg/L in two more S. urealyticus isolates. Regarding ceftazidime, after addition of clavulanic acid a MIC reduction from 64 to 32 mg/L was detected in two S. cohnii and two S. urealyticus isolates, as well as from 32 to 16 mg/L in two S. cohnii isolates and one S. urealyticus isolate (Table 2). In contrast, all mecA/mecC negative isolates were susceptible to oxacillin, ampicillin, cefepime, meropenem, ertapenem and imipenem. The mecA gene harboring S. cohnii isolate SC7 exhibited a non-wildtype phenotype to all beta-lactam antibiotics.
Beta-lactam AMR genes
With regard to beta-lactam antibiotic resistance, several genes of interest were analyzed. With the exception of S. cohnii isolate SC7, not only the mecA and mecC genes, but also mecB and mecD were absent from all genomes. In addition, there were no femB, femC, femD, mgrA or mprF genes which might be associated with beta-lactam resistance23. Only the femA and gdpP genes were detected.
Resistance to beta-lactam antibiotics might be mediated by the BlaZ beta-lactamase, which is encoded by the blaZ gene. However, the blaZ gene was not detected in any S. cohnii isolate in our study either using the NCBI amrfinder database or NCBI BLAST against known blaZ gene sequences. According to reference S. cohnii PBP gene sequences, all isolates from our study carried one PBP1, PBP2 and PBP3 gene each. Between isolates, the amino acid sequences of each respective PBP showed high identities, whereby the amino acid sequence identities between PBP1, 2 and 3 varied as also shown for the PBPs of S. aureus (Fig. 2). Interestingly, PBPs 1, 2 and 3 shared only approximately 70–80% amino acid sequence identity to the known PBP1, PBP2 and PBP3 of S. aureus (Fig. 2).
Figure 2.
Neighbor-joining phylogenetic tree with reference S. aureus penicillin-binding protein (PBP) 1, PBP2 and PBP3 amino acid sequences in comparison to PBPs of the investigated S. cohnii and S. urealyticus isolates from German dairy farms. PBP1-3 genes were detected in each S. cohnii and S. urealyticus isolate. Amino acid sequence identity of PBPs from S. aureus and S. cohnii/S. urealyticus was only 70–80%.
Discussion
In our study, the phylogenetic relationship and antimicrobial resistance potential of nine S. cohnii and six S. urealyticus isolates from German dairy farms were analyzed. The staphylococcal species S. cohnii has been reported as a prevalent commensal bacterium on dairy farms that can be found on teat apices and in other milking parlor-related extramammary niches4,24. Previous studies suggested that S. cohnii is largely uninvolved in severe infections such as mastitis in dairy cows2,4. However, NAS may harbor a wide range of AMR genes and serve as a resistance gene reservoir for pathogenic species9,10.
The phylogenetic analysis revealed a clear genomic distinction from other NAS species. It is likely that isolates SC1, SC2, SC4, SC11, SC14 and SC15 belong to the subspecies S. cohnii subsp. urealyticus, which was recently declared to be a separate species S. urealyticus5, since these isolates clustered together with the respective type strain. Multidrug-resistant S. cohnii subsp. urealyticus (now S. urealyticus5) strains were also reported from a goat´s nasal cavity sample from Tanzania11 and from an ear swab of a healthy dog25.
S. cohnii isolates SC7 and SC8 as well as SC9 and SC10, each pair originating from one farm, grouped closely together, indicating the high clonality of these isolates. Thus, the spread of one S. cohnii clone within one farm seems likely. The transmission of staphylococci within dairy farms may be due to interaction of animals, farm workers and/or insufficient hygiene procedures during the milking process26.
The phenotypic resistance to fusidic acid in all S. cohnii and S. urealyticus isolates was in accordance with the detection of the fusF gene indicating its contribution to the resistant phenotype. Our study implies a wide distribution of this gene in S. cohnii and S. urealyticus isolates across the investigated German dairy farms. The fusF gene was originally detected in S. cohnii subsp. urealyticus (now S. urealyticus5) isolates and was suggested to be an intrinsic factor in this species13. Chloramphenicol resistance with a MIC value of 64 mg/L was determined for only one S. urealyticus isolate, SC4. This was the only isolate that harbored the fexA gene. This gene encodes a chloramphenicol/florfenicol efflux MFS transporter and was likely involved in the manifested resistance. In accordance with the low frequency of detection of the fexA gene in our study, a low fexA gene prevalence was also shown in NAS from Canadian dairy herds10 and low resistance rates to chloramphenicol were observed in MRSA from German dairy farms27,28.
S. cohnii isolate SC11 and S. urealyticus isolate SC12 showed increased MIC values for the lincosamide clindamycin (MIC > 4 mg/L) and the macrolide erythromycin (MIC > 8 mg/L). Both isolates carried the ermC gene, which most likely led to the resistance phenotype. The ermC gene is the most widespread erm gene in staphylococci and it is often located on plasmids29. A cross-resistance to macrolides and lincosamides in staphylococci mediated by constitutive expression of the ermC gene has been described in several studies30,31, and the mode of action of these antibiotics is very similar. Erythromycin resistance according to EUCAST ECOFFs was also detected in S. cohnii isolates SC3, SC5, SC6, SC7, SC8, SC9, SC10 and SC13. These isolates harbored the msrA gene, which encodes an ATP-binding protein that mediates erythromycin resistance32 and this AMR gene may have played a role in erythromycin resistance in our isolates. The msrA gene was also associated with erythromycin resistance in NAS isolates from dairy cattle10.
Resistance to the aminoglycoside streptomycin was found in S. urealyticus isolate SC4 and S. cohnii isolates SC7 and SC10. Accordingly, these isolates harbored the str gene, which mediates resistance to streptomycin. In contrast to studies which demonstrated resistance to several aminoglycosides such as gentamicin or kanamycin in MRSA isolates from dairy cows28,33 and NAS from veal calves34, the isolates in our study showed only resistance to streptomycin. However, reduced susceptibility to kanamycin (MIC 8 mg/L) was detected in S. urealyticus isolates SC4 and SC11. Both isolates carried the aadD1 gene, which may mediate resistance to aminoglycosides such as tobramycin with a MIC ≥ 128 mg/L35, but apparently only leads to a reduced susceptibility to kanamycin.
Resistance to tetracycline was detected in four S. cohnii and two S. urealyticus isolates and in all cases was most likely mediated by the tetL and/or tetK genes. The tetL or tetK genes were also prevalent in NAS isolates from Canadian dairy herds10 and from Belgian veal calves34. Tetracyclines have been extensively used on animal farms, thus promoting the survival of tetracycline resistant isolates36. However, in comparison to previous studies with MRSA isolates from dairy farms, in which the tetracycline resistance prevalence reached 95–100%28,33,37, the 44% (4/9) or 33% (2/6) tetracycline resistance frequency of the S. cohnii and S. urealyticus isolates in our study is rather low.
Resistance to tiamulin, a pleuromutilin, with a MIC value > 2 mg/L was observed in six S. cohnii and all S. urealyticus isolates. This antimicrobial is only licensed for use in pigs and poultry in Germany, but not in cattle. Pleuromutilin resistance may be mediated by the cfr or vgaA/E genes in staphylococci but these genes were not detected in the isolates of our study. Most likely, S. cohnii and S. urealyticus isolates harbor unidentified pleuromutilin resistance genes that are not covered by the NCBI amrfinder database, which is mostly based on sequence data from S. aureus. Accordingly, resistance to tiamulin could not be attributed to any predicted AMR gene in a study of NAS on dairy farms38. Moreover, rifampicin resistance according to EUCAST ECOFFs for coagulase negative staphylococci (MIC ≤ 0.064 mg/L) was detected in eight S. cohnii isolates and, with regard to the EUCAST ECOFFs for S. aureus (≤ 0.016 mg/L), in all S. cohnii and S. urealyticus isolates in our study. Rifampicin resistance may be mediated by point mutations in the rpoB gene39. Due to the limited data in public databases with respect to rpoB genes from S. cohnii or S. urealyticus as well as to antimicrobial resistant S. cohnii or S. urealyticus isolates in general, it is difficult to link mutations in the rpoB gene of the isolates from our study to the manifestation of rifampicin resistance.
In our study, resistance to trimethoprim was detected in S. urealyticus isolate SC4 with a very high MIC value (> 32 mg/L), whereas S. cohnii isolates SC3, SC8, SC10 and SC13 showed resistance with lower MIC values of 4–8 mg/L. Most likely, the high level trimethoprim resistance of S. urealyticus isolate SC4 was mediated by the dfrK gene. Since several strains harboring the dfrC gene did not show resistance to trimethoprim, this gene was apparently nonfunctional with respect to trimethoprim resistance in our isolates. The authors of one study described a physical linkage of the dfrK and tetL genes40, which might also be the case for S. urealyticus isolate SC4 in our study. However, S. urealyticus isolate SC11 harbored tetL without dfrK.
Only one S. cohnii isolate, SC9, might have been resistant to fosfomycin as predicted according to the presence of the fosB6 gene. In clinical MRSA isolates from a Chinese hospital, the fosB6 gene was located on a small plasmid and it was associated with a high level fosfomycin resistance (MIC > 256 mg/L)41.
Detection of qacG/H genes, which encode multidrug efflux pumps, was exclusively shown for the six S. urealyticus isolates. Qac efflux pumps may mediate resistance to disinfectant agents, including quaternary ammonium compounds, intercalating dyes and some antibiotics42. A widespread distribution of qac genes among staphylococci of bovine and caprine origin was shown in a study from Norway43. Moreover, another study reported qacA/C genes in NAS isolates from veal calves34. However, reports about qacG/H genes in NAS and in particular in S. cohnii or S. urealyticus isolates from livestock are rare and the first description of a qacG gene in livestock-associated MRSA sequence type 398 was not until 201544.
The prediction of AMR genes and the phenotypic resistance to several beta-lactam antibiotics disagreed in 14/15 isolates. The acquisition of a SCCmec element appears to be rare in the analyzed S. cohnii isolates and was not observed in S. urealyticus. Only one S. cohnii isolate (SC7) harbored the mecA gene, located on a SCCmec type V. The SCCmec element type V was reported to be common in MRSA from dairy farms26,33,37. In our study, the mecA gene harboring S. cohnii isolate showed resistance to all tested beta-lactam antibiotics. Interestingly, all other isolates, in which mecA or mecC genes were missing, also exhibited a reduced susceptibility to cefotaxime (MIC 4–16 mg/L), ceftazidime (MIC 16–64 mg/L), cefoxitin (MIC 4–8 mg/L), temocillin (MIC > 128 mg/L) and penicillin (MIC 0.25–0.5 mg/L). In contrast, these isolates were susceptible to the beta-lactam antibiotics oxacillin, ampicillin, cefepime, meropenem, ertapenem and imipenem. The inconsistent resistance pattern indicates a different mechanism than the mecA/mecC gene transmitted mechanism, which results in resistance to virtually all beta-lactam antibiotics, as also determined for isolate SC7 in our study. The mecA/mecC gene mediated resistance is based on a modification of PBP2. Likewise, PBPs of the isolates in our study showed divergence from the known S. aureus PBPs. The amino acid sequence identities to the reference S. aureus PBP1, PBP2 and PBP3 were only 70–80%. It is possible that the differences in the amino acid sequences are associated with a structural difference in the protein, which may have lowered the binding affinity for some of the tested beta-lactam antibiotics. Since there is very little information on S. cohnii or S. urealyticus available in public databases and only sparse data regarding antimicrobial susceptibility of S. cohnii or S. urealyticus were published thus far, the possible function of the divergent PBPs in beta-lactam resistance is currently difficult to prove. The focus of our study and the respective methodological approach was the investigation of antimicrobial resistant S. cohnii and S. urealyticus isolates from dairy farms and thus no beta-lactam susceptible isolates were obtained for the necessary comparison of the AMR sequence data. However, one previous genomic study also conducted antimicrobial susceptibility testing with a S. cohnii strain11. This strain was susceptible to cefoxitin according to VITEK2 testing and also exhibited divergent PBPs, as did the isolates from our study (data not shown). Therefore, it is questionable whether the divergent PBPs of the S. cohnii isolates in our study triggered the partial beta-lactam antibiotic resistance. A borderline oxacillin-resistance in S. aureus isolates due to modified PBPs was previously suggested in a review about methicillin resistance in staphylococci45. Moreover, studies recently reported borderline cefoxitin- and oxacillin-resistant S. aureus isolates with substituted PBP amino acid sequences23,46,47. However, in both cases, the borderline resistance was associated with the hyperproduction of the BlaZ beta-lactamase. In our study, all mecA/mecC negative S. cohnii and S. urealyticus isolates were susceptible to oxacillin and the beta-lactamase encoding blaZ gene was not detected in any of these isolates. In addition, the susceptibility to ampicillin clearly indicates a lack of beta-lactamase expression, since this would have led to a reduced susceptibility to ampicillin. Moreover, the combination of cefotaxime or ceftazidime with clavulanic acid, a beta lactamase inhibitor, only marginally affected the susceptibility to the respective cephalosporins and did not lead to a susceptible wildtype phenotype. According to the NCBI Nucleotide database, blaZ genes may be present in S. cohnii isolates. However, further in depth analysis of S. cohnii blaZ genes using NCBI BLAST revealed that the sequences only show low identities (< 80%) to any other published staphylococcal blaZ gene. Along with the mecA and mecC gene, the femA and gdpP genes were associated with beta-lactam antibiotic resistance23 and both genes were detected in the isolates of our study. In general, however, data availability regarding AMR and its respective genes in S. cohnii and S. urealyticus is extremely limited and for this reason it is unclear how reliable the annotation and function of the so far published S. cohnii blaZ genes is. Due to the lack of comparable public data from susceptible and non-susceptible strains, the association of AMR with specific gene alterations currently cannot be proven.
Our study illustrates that the prediction of AMR phenotypes by genomic approaches in the case of rarely investigated staphylococcal species such as S. cohnii and S. urealyticus needs further research and improvement of established gene databases. Moreover, as already shown for other NAS species20–22, the specificity of cefoxitin testing for the detection of mecA or mecC genes and respective methicillin resistance in S. cohnii and S. urealyticus seems to be quite low. As reported for other Staphylococcus spp., oxacillin testing may be more reliable, but this needs to be confirmed on a larger strain collection.
In conclusion, our findings provide further impetus for the investigation of antimicrobial resistance in NAS, since these organisms have been thus far largely overlooked in this respect.
Methods
Ethical statement
Ethical review and approval was not required for the study because sampling of quarter milk samples was carried out in accordance with German legislation within the framework of diagnostic investigations on the dairy farms. No ethical approval from the Institutional Ethics Committee or the National Animal Experimentation Council was required. Samples were collected by a trained veterinarian with consent from the owners of the animals.
Sample collection and whole-genome sequencing
For this study, 15 presumptive S. cohnii isolates, which were retrospectively identified as nine S. cohnii and six S. urealyticus isolates5, were selected for the analysis of antimicrobial resistance potential. The isolates were obtained from quarter milk samples of cows during a sampling campaign in 2018 and 2019 on twelve dairy farms from nine German federal states. Farms had been pre-selected based on a history of MRSA detection in the dairy cow herds26. The isolation procedure was based on a two-step selection with cefoxitin-containing media (3.5 and 4 mg/L). Along with MRSA, cefoxitin-resistant NAS were also detected by the isolation procedure and identified by MALDI-TOF analysis as described previously26. For evaluation of within farm genotypical differences, two independent S. cohnii isolates were analyzed from two farms. DNA from one 1 µl inoculation loop filled with colonies was extracted using the Qiagen DNeasy Blood and Tissue Kit (Qiagen, Germany) according to the manufacturer´s protocol modified by a 10 µl lysostaphin addition to the lysis buffer. The DNA library was prepared using an Illumina Nextera DNA Flex kit (Illumina Inc., USA) and the 150 bp paired-end sequencing run was performed on an Illumina NextSeq 500 instrument.
Bioinformatic analysis
Raw Illumina reads were trimmed and de novo assembled with the in-house developed Aquamis pipeline (https://gitlab.com/bfr_bioinformatics/AQUAMIS/), which implements fastp48 for trimming and shovill (based on SPAdes) (https://github.com/tseemann/shovill) for assembly. Furthermore, it performs mash (v 2.1) for reference search49 as well as quast (v 5.0.2) for assembly quality control50. The assembled sequences were submitted to NCBI under the BioProject PRJNA641762. ANI search of NCBI revealed values of ≥ 96% for all isolates. Phylogenetic typing of isolates was performed using TYGS (https://tygs.dsmz.de/)51. Bacterial characterization was conducted with the in-house developed Bakcharak pipeline (https://gitlab.com/bfr_bioinformatics/bakcharak), which implements ABRicate (https://github.com/tseemann/abricate) for screening of antimicrobial resistance genes using the NCBI amrfinder database52. SCCmec-types were predicted using the software tool SCCmecFinder 1.2 from the Centre for Genomic Epidemiology (https://cge.cbs.dtu.dk/services/). Moreover, PBP1-3 genes of four annotated S. cohnii complete circular genomes (NCBI accession numbers UHDA01000001.1, CP027422.1, CP019597.1, CP033735.1), which were retrieved by NCBI Nucleotide search for “penicillin binding protein” and “Staphylococcus cohnii”, were extracted into an in-house database and S. cohnii/S. urealyticus assembled sequences from our study were searched by NCBI blastn against these known PBP genes. Translation of DNA to amino acid sequences and alignment to PBP1-3 of S. aureus extracted from NCBI as well as among the different PBPs of the detected S. cohnii/S. urealyticus isolates was conducted using MEGA X version 10.1.7 and ClustalW. A neighbor-joining phylogenetic tree was subsequently calculated comparing the PBP1-3 amino acid sequences from S. aureus and S. cohnii/S. urealyticus isolates. In addition, presence of the mecB (NG_047954.1), mecC (NG_047955.1), mecD (NG_054960.1), blaZ (KR270450.2), femA (AF145333.1), femB (GQ284649.1), femC (AP018376.1), femD (Y09570.1), gdpP (MF071106.1), mgrA (AP017922.1) and mprF (HM140977.1) genes was checked by individual NCBI BLAST against S. aureus or Macrococcus caseolyticus (mecB and mecD) reference genes.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed by broth microdilution according to the EUCAST and CLSI guidelines (ISO 20776-1:2006 or CLSI M31-A3). It was carried out using a standardized antibiotic panel (EUST scheme) that is recommended for use in all member states of the European Union for resistance monitoring on staphylococci from livestock and food53. Moreover, the standardized scheme was extended by a larger number of beta-lactam antibiotics as well as in combination with the beta-lactamase inhibitor clavulanic acid for analyzing resistance to beta-lactams in more detail. For interpretation of MIC of the individual isolates, the EUCAST ECOFFs for S. aureus and coagulase negative staphylococci were used, since specific MIC values for S. cohnii are not available (Tables 1, 2). For quality control of resistance testing, the S. aureus isolates ATCC 29213 and ATCC 25923 were used.
Acknowledgements
We thank all farmers, who voluntarily participated in the study. Many thanks to Thomas Peters, Ulrike Sorge, Karsten Donat, Sabine Reinhold, Oliver Claushues, Karin Eulenberger and Natalie Morgenstern for supporting the identification of study farms. Special thanks to Daniel Leeser-Boek, Pascal Witt and Ylanna Kelner-Burgos for NGS preparation and Heidi Wichmann-Schauer for help with study conception. We also thank Carlus Deneke and Simon Tausch for bioinformatics analyses. Moreover, we thank the German Federal Ministry of Education and Research (BMBF) for funding our research in the framework of the project #1HealthPREVENT (Grant No. 01KI1727C).
Author contributions
T.L.: conceptualization and design of the study, samples collection, laboratory work, data analysis, original draft preparation. A.S.: conceptualization and design of the study; samples collection; laboratory work; critically reviewing the manuscript. J.A.H.: laboratory work; critically reviewing the manuscript. S.F.M.: data analysis, critically reviewing the manuscript; S.M.: data analysis, critically reviewing the manuscript; B.-A.T.: conceptualization and design of the study, critically reviewing the manuscript. All authors read and approved the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
The assembled sequences of all isolates in our study are deposited in NCBI under the BioProject PRJNA641762.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tobias Lienen, Email: Tobias.Lienen@bfr.bund.de.
Bernd-Alois Tenhagen, Email: Bernd-Alois.Tenhagen@bfr.bund.de.
References
- 1.Mahmmod YS, Klaas IC, Svennesen L, Pedersen K, Ingmer H. Communications of Staphylococcus aureus and non-aureus Staphylococcus species from bovine intramammary infections and teat apex colonization. J. Dairy Sci. 2018;101:7322–7333. doi: 10.3168/jds.2017-14311. [DOI] [PubMed] [Google Scholar]
- 2.Condas LAZ, et al. Distribution of non-aureus staphylococci species in udder quarters with low and high somatic cell count, and clinical mastitis. J. Dairy Sci. 2017;100:5613–5627. doi: 10.3168/jds.2016-12479. [DOI] [PubMed] [Google Scholar]
- 3.De Visscher A, Piepers S, Haesebrouck F, Supre K, De Vliegher S. Coagulase-negative Staphylococcus species in bulk milk: Prevalence, distribution, and associated subgroup- and species-specific risk factors. J. Dairy Sci. 2017;100:629–642. doi: 10.3168/jds.2016-11476. [DOI] [PubMed] [Google Scholar]
- 4.Wuytack A, De Visscher A, Piepers S, Haesebrouck F, De Vliegher S. Fecal non-aureus Staphylococci are a potential cause of bovine intramammary infection. Vet. Res. 2020;51:32. doi: 10.1186/s13567-020-00761-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madhaiyan M, Wirth JS, Saravanan VS. Phylogenomic analyses of the Staphylococcaceae family suggest the reclassification of five species within the genus Staphylococcus as heterotypic synonyms, the promotion of five subspecies to novel species, the taxonomic reassignment of five Staphylococcus species to Mammaliicoccus gen. nov., and the formal assignment of Nosocomiicoccus to the family Staphylococcaceae. Int. J. Syst. Evol. Microbiol. 2020;70:5926–5936. doi: 10.1099/ijsem.0.004498. [DOI] [PubMed] [Google Scholar]
- 6.Mendoza-Olazaran S, et al. Draft genome sequences of two opportunistic pathogenic strains of Staphylococcus cohnii isolated from human patients. Stand. Genomic Sci. 2017;12:49. doi: 10.1186/s40793-017-0263-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garza-Gonzalez E, et al. Microbiological and molecular characterization of human clinical isolates of Staphylococcus cohnii, Staphylococcus hominis, and Staphylococcus sciuri. Scand. J. Infect. Dis. 2011;43:930–936. doi: 10.3109/00365548.2011.598873. [DOI] [PubMed] [Google Scholar]
- 8.Cave R, Misra R, Chen J, Wang S, Mkrtchyan HV. Whole genome sequencing revealed new molecular characteristics in multidrug resistant staphylococci recovered from high frequency touched surfaces in London. Sci. Rep. 2019;9:9637. doi: 10.1038/s41598-019-45886-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loncaric I, et al. Prevalence of methicillin-resistant Staphylococcus sp. (MRS) in different companion animals and determination of risk factors for colonization with MRS. Antibiotics (Basel) 2019;8:1. doi: 10.3390/antibiotics8020036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nobrega DB, et al. Prevalence and genetic basis of antimicrobial resistance in non-aureus staphylococci isolated from Canadian dairy herds. Front. Microbiol. 2018;9:256. doi: 10.3389/fmicb.2018.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seni J, et al. Draft genome sequence of a multidrug-resistant caprine isolate of Staphylococcus cohnii subsp. urealyticus from Tanzania encoding ermB, tet(K), dfrG, fusF and fosD. J. Glob. Antimicrob. Resist. 2019;18:163–165. doi: 10.1016/j.jgar.2019.07.006. [DOI] [PubMed] [Google Scholar]
- 12.Cui L, et al. Cfr-mediated linezolid-resistance among methicillin-resistant coagulase-negative staphylococci from infections of humans. PLoS ONE. 2013;8:e57096. doi: 10.1371/journal.pone.0057096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen HJ, et al. A novel fusidic acid resistance determinant, fusF, in Staphylococcus cohnii. J. Antimicrob. Chemother. 2015;70:416–419. doi: 10.1093/jac/dku408. [DOI] [PubMed] [Google Scholar]
- 14.Fishovitz J, Hermoso JA, Chang M, Mobashery S. Penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. IUBMB Life. 2014;66:572–577. doi: 10.1002/iub.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peacock SJ, Paterson GK. Mechanisms of methicillin resistance in Staphylococcus aureus. Annu. Rev. Biochem. 2015;84:577–601. doi: 10.1146/annurev-biochem-060614-034516. [DOI] [PubMed] [Google Scholar]
- 16.Lakhundi S, Zhang K. Methicillin-resistant Staphylococcus aureus: Molecular characterization, evolution, and epidemiology. Clin. Microbiol. Rev. 2018 doi: 10.1128/CMR.00020-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shore AC, Coleman DC. Staphylococcal cassette chromosome mec: Recent advances and new insights. Int. J. Med. Microbiol. 2013;303:350–359. doi: 10.1016/j.ijmm.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Chen XP, et al. Extreme diversity and multiple SCCmec elements in coagulase-negative Staphylococcus found in the Clinic and Community in Beijing, China. Ann. Clin. Microbiol. Antimicrob. 2017;16:57. doi: 10.1186/s12941-017-0231-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zong Z, Lu X. Characterization of a new SCCmec element in Staphylococcus cohnii. PLoS ONE. 2010;5:e14016. doi: 10.1371/journal.pone.0014016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Humphries RM, et al. Evaluation of surrogate tests for the presence of mecA-mediated methicillin resistance in Staphylococcus haemolyticus, Staphylococcus hominis, Staphylococcus capitis and Staphylococcus warneri. J. Clin. Microbiol. 2020 doi: 10.1128/jcm.02290-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huse HK, et al. Evaluation of oxacillin and cefoxitin disk diffusion and MIC breakpoints established by the clinical and laboratory standards institute for detection of mecA-mediated oxacillin resistance in Staphylococcus schleiferi. J. Clin. Microbiol. 2018 doi: 10.1128/jcm.01653-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu MT, et al. Evaluation of oxacillin and cefoxitin disk and MIC breakpoints for prediction of methicillin resistance in human and veterinary isolates of Staphylococcus intermedius group. J. Clin. Microbiol. 2016;54:535. doi: 10.1128/JCM.02864-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scholtzek AD, et al. Molecular characterization of equine Staphylococcus aureus isolates exhibiting reduced oxacillin susceptibility. Toxins (Basel) 2019 doi: 10.3390/toxins11090535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Visscher A, et al. Further evidence for the existence of environmental and host-associated species of coagulase-negative staphylococci in dairy cattle. Vet. Microbiol. 2014;172:466–474. doi: 10.1016/j.vetmic.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 25.Bean DC, Wigmore SM, Wareham DW. Draft genome sequence of Staphylococcuscohnii subsp. urealyticus isolated from a healthy dog. Genome Announc. 2017 doi: 10.1128/genomeA.01628-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schnitt A, Lienen T, Wichmann-Schauer H, Cuny C, Tenhagen BA. The occurrence and distribution of livestock-associated methicillin-resistant Staphylococcus aureus ST398 on German dairy farms. J. Dairy Sci. 2020 doi: 10.3168/jds.2020-18958. [DOI] [PubMed] [Google Scholar]
- 27.Tenhagen BA, Vossenkuhl B, Käsbohrer A, Alt K, Kraushaar B, Guerra B, Schroeter A, Fetsch A. Methicillin-resistant Staphylococcus aureus in cattle food chains—Prevalence, diversity, and antimicrobial resistance in Germany. J. Anim. Sci. 2014;92:2741–2751. doi: 10.2527/jas2014-7665. [DOI] [PubMed] [Google Scholar]
- 28.Tenhagen BA, et al. Short communication: Methicillin-resistant Staphylococcus aureus in conventional and organic dairy herds in Germany. J. Dairy Sci. 2018;101:3380–3386. doi: 10.3168/jds.2017-12939. [DOI] [PubMed] [Google Scholar]
- 29.Feßler A, et al. Small antimicrobial resistance plasmids in livestock-associated methicillin-resistant Staphylococcus aureus CC398. Front. Microbiol. 2018;9:2063. doi: 10.3389/fmicb.2018.02063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leclercq R. Mechanisms of resistance to macrolides and lincosamides: Nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 2002;34:482–492. doi: 10.1086/324626. [DOI] [PubMed] [Google Scholar]
- 32.Ross JI, Eady EA, Cove JH, Baumberg S. Identification of a chromosomally encoded ABC-transport system with which the staphylococcal erythromycin exporter MsrA may interact. Gene. 1995;153:93–98. doi: 10.1016/0378-1119(94)00833-E. [DOI] [PubMed] [Google Scholar]
- 33.Feßler AT, et al. Characterization of methicillin-resistant Staphylococcus aureus CC398 obtained from humans and animals on dairy farms. Vet. Microbiol. 2012;160:77–84. doi: 10.1016/j.vetmic.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 34.Argudín MA, Vanderhaeghen W, Butaye P. Diversity of antimicrobial resistance and virulence genes in methicillin-resistant non-Staphylococcus aureus staphylococci from veal calves. Res. Vet. Sci. 2015;99:10–16. doi: 10.1016/j.rvsc.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 35.Ida T, et al. Identification of aminoglycoside-modifying enzymes by susceptibility testing: Epidemiology of methicillin-resistant Staphylococcus aureus in Japan. J. Clin. Microbiol. 2001;39:3115. doi: 10.1128/JCM.39.9.3115-3121.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Granados-Chinchilla F, Rodriguez C. Tetracyclines in food and feedingstuffs: From regulation to analytical methods, bacterial resistance, and environmental and health implications. J. Anal. Methods Chem. 2017;2017:1315497. doi: 10.1155/2017/1315497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadlec K, Entorf M, Peters T. Occurrence and characteristics of livestock-associated methicillin-resistant Staphylococcus aureus in quarter milk samples from dairy cows in Germany. Front. Microbiol. 2019;10:1295. doi: 10.3389/fmicb.2019.01295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frey Y, Rodriguez JP, Thomann A, Schwendener S, Perreten V. Genetic characterization of antimicrobial resistance in coagulase-negative staphylococci from bovine mastitis milk. J. Dairy Sci. 2013;96:2247–2257. doi: 10.3168/jds.2012-6091. [DOI] [PubMed] [Google Scholar]
- 39.Campbell EA, et al. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 2001;104:901–912. doi: 10.1016/S0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 40.Kadlec K, Feßler AT, Hauschild T, Schwarz S. Novel and uncommon antimicrobial resistance genes in livestock-associated methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 2012;18:745–755. doi: 10.1111/j.1469-0691.2012.03842.x. [DOI] [PubMed] [Google Scholar]
- 41.Fu Z, et al. Characterization of fosfomycin resistance gene, fosB, in methicillin-resistant Staphylococcus aureus isolates. PLoS ONE. 2016;11:e0154829. doi: 10.1371/journal.pone.0154829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wassenaar TM, Ussery D, Nielsen LN, Ingmer H. Review and phylogenetic analysis of qac genes that reduce susceptibility to quaternary ammonium compounds in Staphylococcus species. Eur. J. Microbiol. Immunol. (Bp) 2015;5:44–61. doi: 10.1556/EUJMI-D-14-00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bjorland J, et al. Widespread distribution of disinfectant resistance genes among staphylococci of bovine and caprine origin in Norway. J. Clin. Microbiol. 2005;43:4363–4368. doi: 10.1128/JCM.43.9.4363-4368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Couto N, Belas A, Kadlec K, Schwarz S, Pomba C. Clonal diversity, virulence patterns and antimicrobial and biocide susceptibility among human, animal and environmental MRSA in Portugal. J. Antimicrob. Chemother. 2015;70:2483–2487. doi: 10.1093/jac/dkv141. [DOI] [PubMed] [Google Scholar]
- 45.Chambers HF. Methicillin resistance in staphylococci: Molecular and biochemical basis and clinical implications. Clin. Microbiol. Rev. 1997;10:781–791. doi: 10.1128/CMR.10.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Argudín MA, et al. Genetic diversity among Staphylococcus aureus isolates showing oxacillin and/or cefoxitin resistance not linked to the presence of mec genes. Antimicrob. Agents Chemother. 2018;62:e00091–e118. doi: 10.1128/AAC.00091-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ba X, et al. Novel mutations in penicillin-binding protein genes in clinical Staphylococcus aureus isolates that are methicillin resistant on susceptibility testing, but lack the mec gene. J. Antimicrob. Chemother. 2014;69:594–597. doi: 10.1093/jac/dkt418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen S, Zhou Y, Chen Y, Gu J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ondov BD, et al. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mikheenko A, Prjibelski A, Saveliev V, Antipov D, Gurevich A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics. 2018;34:i142–i150. doi: 10.1093/bioinformatics/bty266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meier-Kolthoff JP, Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019;10:2182. doi: 10.1038/s41467-019-10210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feldgarden M, et al. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype–phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 2019 doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.European Food Safety Authority Technical specifications on the harmonised monitoring and reporting of antimicrobial resistance in methicillin-resistant Staphylococcus aureus in food-producing animals and food. EFSA J. 2012;10:1. doi: 10.2903/j.efsa.2012.2897. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The assembled sequences of all isolates in our study are deposited in NCBI under the BioProject PRJNA641762.