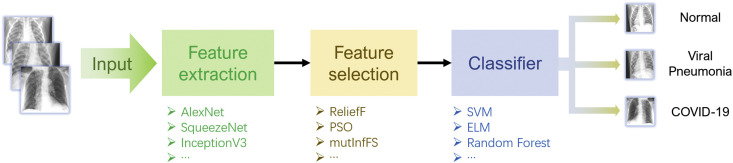
Abstract
Background
Chest X-ray radiography (CXR) has been widely considered as an accessible, feasible, and convenient method to evaluate suspected patients’ lung involvement during the COVID-19 pandemic. However, with the escalating number of suspected cases, traditional diagnosis via CXR fails to deliver results within a short period of time. Therefore, it is crucial to employ artificial intelligence (AI) to enhance CXRs for obtaining quick and accurate diagnoses. Previous studies have reported the feasibility of utilizing deep learning methods to screen for COVID-19 using CXR and CT results. However, these models only use a single deep learning network for chest radiograph detection; the accuracy of this approach required further improvement.
Methods
In this study, we propose a three-step hybrid ensemble model, including a feature extractor, a feature selector, and a classifier. First, a pre-trained AlexNet with an improved structure extracts the original image features. Then, the ReliefF algorithm is adopted to sort the extracted features, and a trial-and-error approach is used to select the n most important features to reduce the feature dimension. Finally, an SVM classifier provides classification results based on the n selected features.
Results
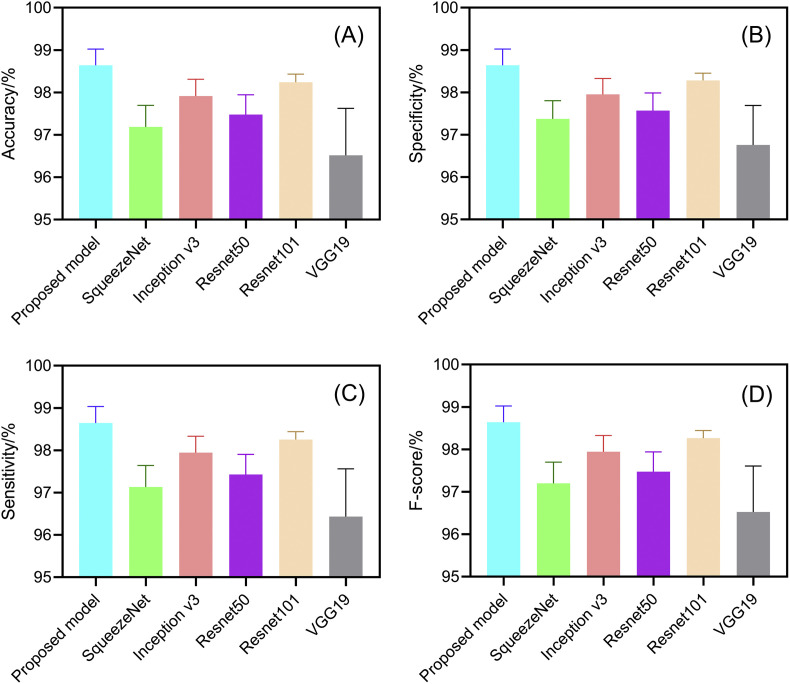
Compared to five existing models (InceptionV3: 97.916 ± 0.408%; SqueezeNet: 97.189 ± 0.526%; VGG19: 96.520 ± 1.220%; ResNet50: 97.476 ± 0.513%; ResNet101: 98.241 ± 0.209%), the proposed model demonstrated the best performance in terms of overall accuracy rate (98.642 ± 0.398%). Additionally, compared to the existing models, the proposed model demonstrates a considerable improvement in classification time efficiency (SqueezeNet: 6.602 ± 0.001s; InceptionV3: 12.376 ± 0.002s; ResNet50: 10.952 ± 0.001s; ResNet101: 18.040 ± 0.002s; VGG19: 16.632 ± 0.002s; proposed model: 5.917 ± 0.001s).
Conclusion
The model proposed in this article is practical and effective, and can provide high-precision COVID-19 CXR detection. We demonstrated its suitability to aid medical professionals in distinguishing normal CXRs, viral pneumonia CXRs and COVID-19 CXRs efficiently on small sample sizes.
Keywords: COVID-19, X-ray imaging, Transfer learning, Dimension reduction
Graphical abstract
Abbreviations
- COVID-19
Corona Virus Disease 2019
- CXR
Chest X-ray
- CT
Computed Tomography
- SVM
Support Vector Machine
- SGDM
Stochastic Gradient Descent with Momentum
- TP
True Positive
- TN
True Negative
- FP
False Positive
- FN
False Negative
- VGG
Visual Geometry Group
- RBF
Radial Basis Function
- DNN
Deep Neural Networks
- CNN
Convolutional Neural Networks
1. Introduction
1.1. Background
The COVID-19 pandemic has presented a huge challenge to global health since February 2020. It is extremely important to screen and isolate all patients with suspected COVID-19 at their first point of contact to break the chain of transmission. Chest imaging plays an essential role in the early diagnosis of patients with suspected COVID-19 chest infections because the chest X-ray radiology (CXR) can evaluate their lung abnormality and is readily available in community physician offices, urgent care clinics and hospital emergency departments [1]. In the case of COVID-19, radiological appearance obtained in CXRs is related to RT-PCR examination and patient outcome [2]. Vancheri et al. confirmed the effectiveness of employing CXR as a first-line imaging modality in the diagnostic workflow of patients with suspected COVID-19 pneumonia. Their results substantiated that chest radiography showed lung abnormalities in 75% of patients with confirmed SARS-CoV-2 infection, ranging from 63.3 to 83.9%, at 0–2 days and >9 days from the onset of symptoms [3].
Nevertheless, the rapidly accelerating number of suspected COVID-19 cases still leads to depletion of diagnostic resources due to the lack of physicians. Consequently, at present, it is imperative to utilize artificial intelligence (AI) in CXR, which can offer physicians quick and accurate diagnostic assistance, and therefore alleviate the shortage of medical resources and promote medical efficiency.
1.2. Related work
Recently, several researchers have proposed various models for the AI-assisted imaging diagnosis of COVID-19 and obtained some significant results. Varela-Santos et al. proposed an initial experiment using image texture feature descriptors, as well as feed-forward and convolutional neural networks on several created databases with COVID-19 images. Their work verified the effectiveness of the supervised learning model in the AI-assisted differential diagnosis between COVID-19 and other lung diseases [4]. Ozturk et al. proposed a model with classification accuracy of 98.08% for binary classification (normal versus COVID-19) and 87.02% for multi-class classification tasks (normal versus viral pneumonia versus COVID-19), which still needs improvement [5]. Zhang Yudong et al. introduced stochastic pooling to replace average pooling and max pooling with the traditional deep convolutional neural network (DCNN) model, which achieved an accuracy of 93.64% ± 1.42% in distinguishing COVID-19 cases from normal subjects [6]. Matteo et al. proposed a light convolutional neural network (CNN) design, based on the SqueezeNet, for efficient discrimination of COVID-19 CT images with respect to other community-acquired pneumonia and/or healthy CT images. Their architecture allows an accuracy of 85.03%, with fewer parameters and higher efficiency compared to that of the classical SqueezeNet [7]. Yan et al. designed an AI system to diagnose COVID-19 using multi-scale convolutional neural networks (MSCNNs), which can assess CT scan results [8]. Benbrahim et al. adopted a deep learning method using the Inceptionv3 model and the ResNet-50 model, and successfully realized classification of COVID-19 in chest X-ray images (the accuracies of those models were 99.01% and 98.03%, respectively) [9]. Shayan established two methods, deep neural network (DNN) for image fractal features and convolutional neural network (CNN) for lung images, to identify new coronary chest radiograph images. The classification results demonstrated that the CNN architecture is better than the DNN model with an accuracy of 93.2% and a sensitivity of 96.1% [10]. Shervin et al. trained four commonly used convolutional neural networks, including ResNet-18, ResNet-50, SqueezeNet, and DenseNet-121, to classify suspected COVID-19 images [11]. Among them, SqueezeNet demonstrated the best performance, reaching a sensitivity of 98% and a specificity of 92%. Toraman et al. proposed a novel artificial neural network, Convolutional CapsNet, which processed chest X-ray images with capsule networks [12]; fast and accurate diagnostics for COVID-19 were attained via two different classifications: binary classification (COVID-19 and No-Findings) and multi-class classification (COVID-19, No-Findings, and Pneumonia); this method achieved accuracies of 97.24% and 84.22% for binary class and multi-class, respectively [12]. These previous studies to screen COVID-19 are based on deep learning methods processing CT images and X-ray radiographs; these precedents affirmed the feasibility of introducing AI in COVID-19 diagnosis. Linda Wang et al. proposed COVID-Net, a deep convolutional neural network design tailored for the detection of COVID-19 cases from chest X-ray (CXR) images [13]. Lee Ki-Sun et al. fine-tuned the structures of VGG16 and VGG19 convolutional neural networks. Their experimental results showed a highest value for area under the receiver operating characteristic (ROC) curve (AUC) of 0.950 for COVID-19 classification in an experimental group fine-tuned with only 2/5 blocks of the VGG16 backbone network [14]. Chaimae et al. proposed CVDNet, a deep convolutional neural network (CNN) model to classify COVID-19 infection from normal and other pneumonia cases using chest X-ray images [15]. The proposed architecture is based on a residual neural network and is constructed using two parallel levels with different kernel sizes to capture local and global features of the inputs. Motamed et al. adopted a confrontation network (RANDGAN), which can detect images of unknown categories (COVID-19) and labeled categories (normal and viral pneumonia) from known networks without labeling and training data, but the effect is limited (the area under the ROC curve can only reach 0.77) [16]. However, these models use a single deep learning network whose processing efficacy and accuracy remain to be further improved. According to Soumya Ranjan Nayak et al.‘s comprehensive study [17], the further development of effective deep CNN models for a more accurate diagnosis of COVID-19 infection is still in urgent need because the maximum accuracy value of single CNNs did not exceed 98.33% for binary classification (COVID-19 versus normal).
Single neural network models usually need expanding structures to further improve the accuracy of the model, which complicates the model and prolongs training time. Researchers have proposed several hybrid structures to improve the accuracy and efficacy of machine learning models. Özkaya et al. used convolutional networks and an SVM on the classification task, but did not perform feature selection, and only distinguished normal chest radiographs from COVID-19 chest radiographs, which has limited application scenarios [18]. Yu Xiang et al. combined three components including feature extraction, graph-based feature reconstruction, and classification to complete the binary classification task of COVID-19 and normal CXRs [19]. Their model achieved the best accuracy of 0.9872. However, these models use a single deep learning network, whose processing efficacy and accuracy remain to be further improved.
1.3. Our work
An advanced hybrid model with feature extraction, feature selection and classification components may help solve the accuracy and efficiency issues in the differential diagnosis of COVID-19, common pneumonia and normal CXRs. To solve this problem, this paper proposes a three-step hybrid ensemble model comprising a feature extractor, feature selector, and classifier. First, an improved AlexNet serves as a feature extractor to extract the original image features. Subsequently, the ReliefF algorithm ranks the extracted features according to their importance. The best number of input features (n) is determined through the trial-and-error method, and the first n features are be input to the SVM classifier. Finally, the SVM classifier gives the classification results according to n selected features. Although the components of this hybrid model are individually relatively simple compared to the aforementioned single deep neural network, this model still manages to attain high accuracy under the synergistic effect of each component and thus excels the traditional single deep neural network in COVID-19 CXR detection.
In this article, we introduce the dataset used in model training and validation, and elaborate the model architecture and its components (AlexNet, ReliefF and SVM). In the experiment section, the concrete procedures of model training will be explained based on three aspects: feature extraction, feature selection and classification. Then, the results of self-contrast and comparative studies are presented to illustrate the superiority of the proposed model. Finally, a discussion is given on the proposed model.
2. Materials and methods
2.1. Dataset
To meet the input requirements of the AlexNet, the sizes of the images were converted to 227 × 227 × 3, before they were input to the model. The normal CXRs and viral pneumonia CXRs were obtained from the NIH Chest X-ray database [20], and the COVID-19 CXRs were collected from https://github.com/tawsifur/COVID-19-Chest-X-ray-Detection [21], and https://github.com/ieee8023/covid-chestxray-dataset [22], (shown in Table 1 ). To ensure the fairness of training, each category of pictures was randomly selected from these databases. Fig. 1 shows examples of three types of samples in the dataset used in this study.
Table 1.
Dataset used in this study.
| Class | Training set | Test set | Total |
|---|---|---|---|
| 1 COVID-19 | 380 | 163 | 543 |
| 2 Viral pneumonia | 420 | 180 | 600 |
| 3 Normal | 420 | 180 | 600 |
| Total | 1220 | 523 | 1743 |
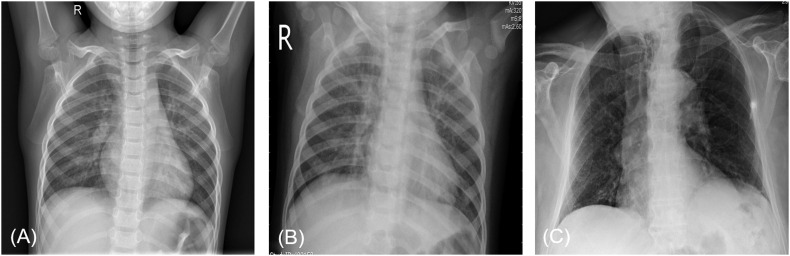
Fig. 1.
Examples of three types of samples in the dataset: (A) Normal; (B) Viral pneumonia; (C) COVID-19.
2.2. Model architecture
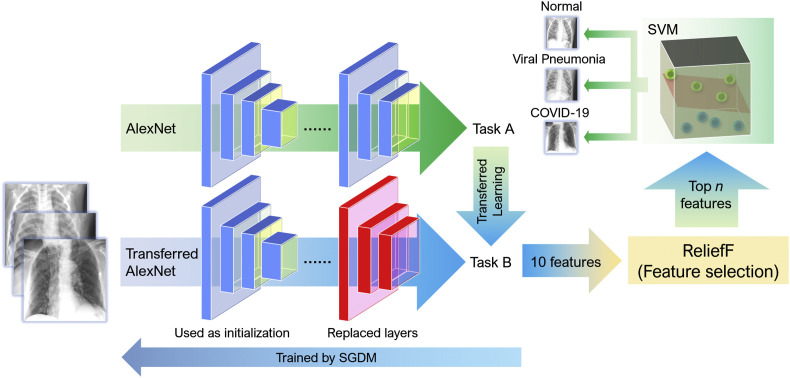
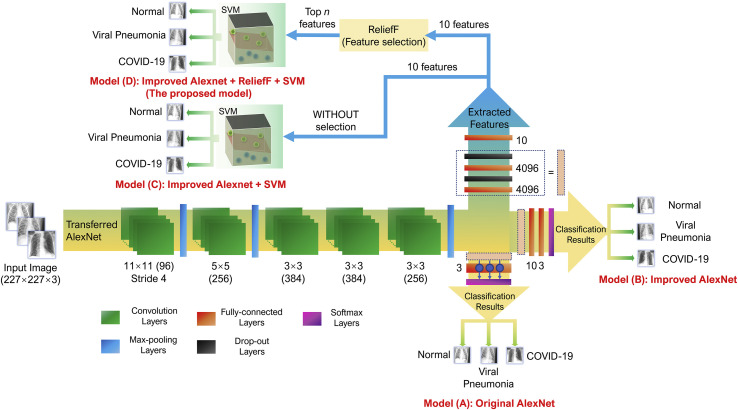
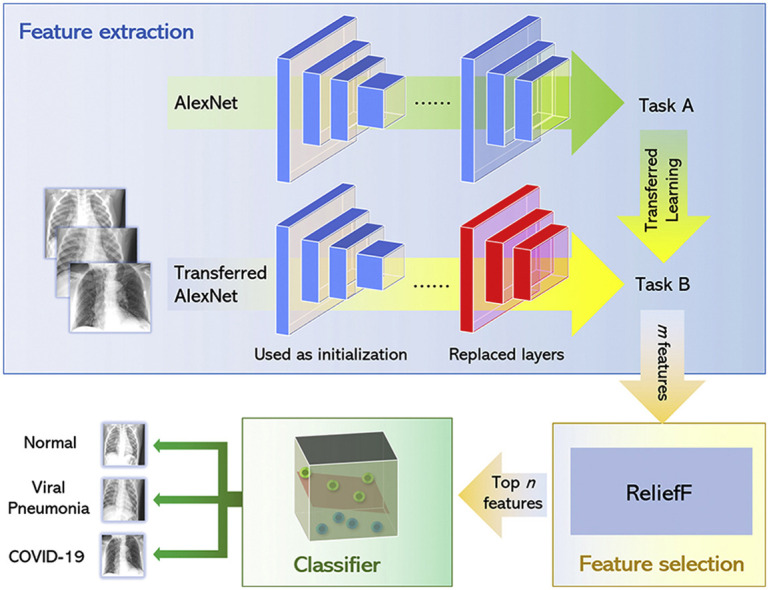
As shown in Fig. 2 , the proposed model mainly consists of three parts: feature extraction by a transferred AlexNet), feature selection with the ReliefF algorithm and SVM classifier.
Fig. 2.
An overview of model architecture: a transferred AlexNet for feature extraction, the ReliefF algorithm for feature selection and an SVM classifier.
In terms of feature extraction, all the images are input to the AlexNet, and the output of a certain layer of the network is regarded as the features of the image for classification. Because computer-aided diagnosis systems and other medical image interpretation systems are usually unable to train convolutional neural networks from scratch, common features can be migrated from trained convolutional neural networks to be used as input classifiers for imaging tasks in transfer learning.
In feature selection, the features extracted in the first step are sorted according to their importance using the ReliefF algorithm. Then the first few features that are most important for classification are selected by trial-and-error. The third part establishes the SVM model. When the features selected previously are input to the SVM model, it classifies the filtered features and obtain the final classification results.
2.3. Components of the proposed model
As mentioned in Section 2.2, the proposed model includes three parts: AlexNet, ReliefF and SVM. These three components are introduced in detail in this section.
2.3.1. AlexNet
AlexNet was originally proposed by Alex Krizhevsky et al. at the University of Toronto. It uses two GPUs for calculations, which considerably improves computational efficiency [23].
As a large network, AlexNet has 60 million parameters and 650,000 neurons, requiring a large number of labeled samples to train [23], which is a requirement that the labeled COVID-19 CXR image resources are incapable of satisfying. Under these circumstances, transfer learning is a convenient and effective method widely used to train deep neural networks when the available labeled samples are not sufficient. Employing all the parameters in a pre-trained network as an initialization step can exploit features that learned from massive datasets. These layers are mainly used for feature extraction, and the obtained parameters can help the training to converge. Furthermore, high-performance GPU and CPU are required to train deep networks, but transfer learning can be implemented on common personal computers.
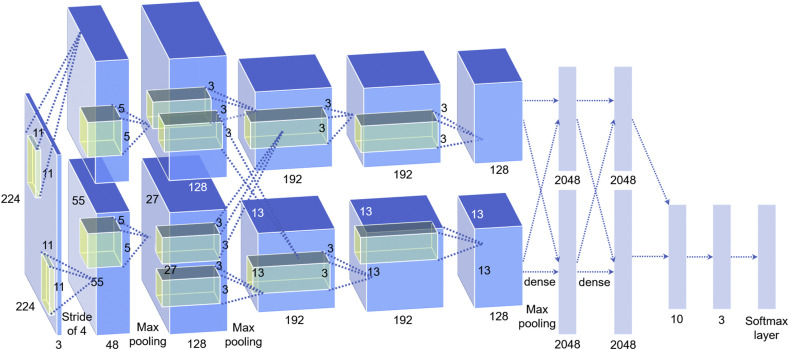
In the proposed model, we improved AlexNet by replacing the last two layers (a fully connected layer with 1000 neurons and a softmax layer) with our layers: two fully connected layers with ten and three nodes (referring to three types of categories: normal, viral pneumonia and COVID-19), respectively, and a softmax layer (shown in Fig. 3 ). The rest of the parameters of the original model were preserved and served as the initialization. Then, the entire structure is divided into two parts: the pre-trained network and the transferred network. The parameters in the pre-trained network were already trained on ImageNet with millions of images, and the extracted features have been proven effective for classification. These parameters may require marginal adjustment to adapt to the new images. The parameters in the transferred network hold a small fraction of the entire network, which is appropriate for training on a small dataset.
Fig. 3.
Structure of AlexNet used in this work.
2.3.2. ReliefF
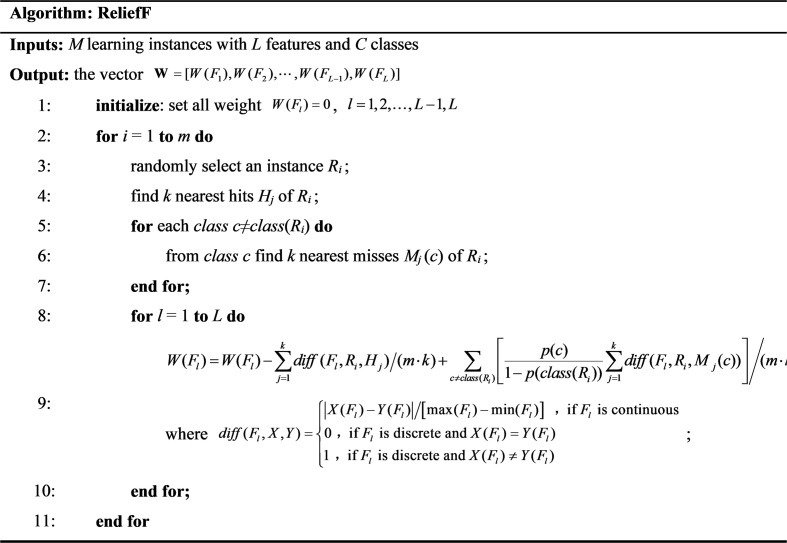
ReliefF is a dimension reduction method developed by Kira and Rendell, which can help remove unnecessary attributes from the data set and save storage space, thus reducing computational complexity and saving model training time. In 1994, the ReliefF model was improved by enhancing the noise resistance in the dataset and making it suitable for multi-class problems by ignoring missing data [24]. ReliefF aims to reveal the correlations and consistencies present in the attributes of the dataset.
The basic procedures of ReliefF are shown in Table 2 in pseudo-code [25]. In this work, the ReliefF algorithm is used to sort the extracted ten features based on their importance. The data used are the feature data of the training set, not the test set. After experimentation, we finally determined that only several of the most important features need to be used for classification to achieve the best classification speed and accuracy, which will be further illustrated in the following sections.
Table 2.
Pseudo-code of the ReliefF algorithm [25].
2.3.3. SVM
Support vector machines (SVMs) are supervised learning methods developed by Vapnik based on statistical learning theory [24]. SVM performs the learning process with the dataset divided into training and test sets. It achieves data classification by determining a decision function and detecting the hyperplane that could distinguish the data.
At present, SVMs have widespread applications in various disciplines for classification tasks such as text classification, facial recognition, handwritten character recognition, bioinformatics, and other fields. In solving multi-classification problems, SVMs divide the original classification problem into two classification problems. Hence, when applied to multi-classification, the difficulty and complexity of training accordingly increase in parallel with increasing number of sample categories. Reducing the amount of calculation and computational complexity is a known problem for SVMs, requiring new research solutions [24]. Herein, we propose to utilize the ReliefF algorithm to reduce the dimensionality of the sample data.
3. Experiment
3.1. Feature extraction
The model was fine-tuned using the transfer learning method and the pre-trained AlexNet provided by MATLAB. The specific fine-tuning method involves adding a fully connected layer between layers 22 and 23 (that is, between fc8 and drop7), with 10 neurons in the added layer (the outputs of this layer are features extracted for subsequent selection). The original fc8 layer has 1000 neurons, corresponding to the classification of 1000 types of pictures. Considering that our classification results only included three types (normal, COVID-19, and ordinary viral pneumonia), the number of neurons in this layer was set to 3.
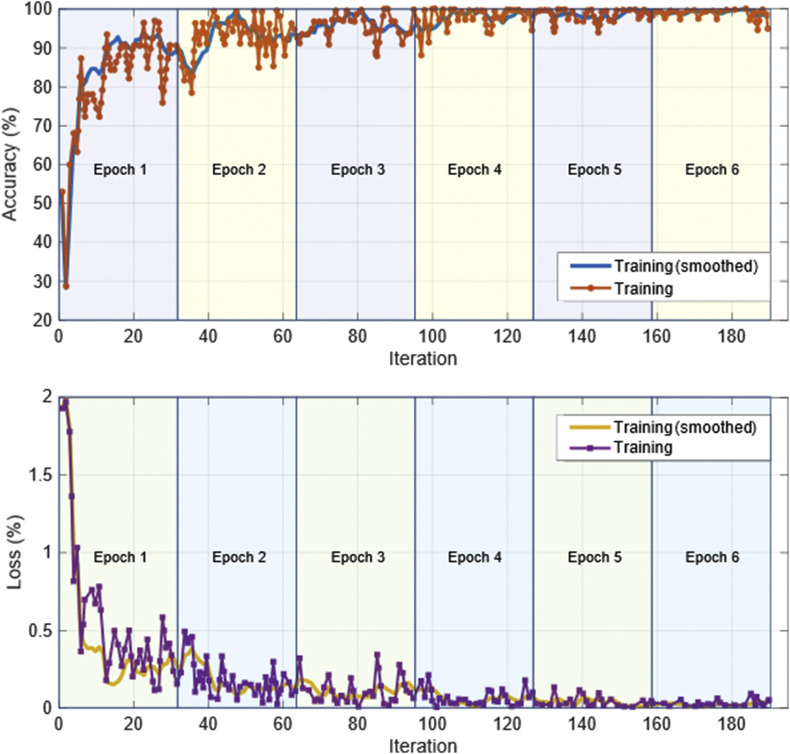
To adapt the model to be more suitable for the classification goals of this study, after the model structure is adjusted, the training set is used to train the model to fine-tune the weights. After the model was trained, all the data were input to the model to obtain 10 features of a total of 1743 pictures including the test set (521 pictures) and the training set (1222 pictures). The transferred AlexNet was trained by stochastic gradient descent with momentum (SGDM). The parameters used in training AlexNet are given in Table 3 . The training curve of the model is shown in Fig. 4 .
Table 3.
Parameters used in AlexNet training.
| Parameters | Value |
|---|---|
| Initial learn rate | 5 × 10−4 |
| Learn rate drop factor | 0.1 |
| L2 regularization | 1 × 10−4 |
| Max epochs | 6 |
| Mini batch size | 32 |
Fig. 4.
The training curve of the model.
3.2. Feature selection and classification
Taking a specific experiment as an example, we explain how the best n features are determined. This study uses SVM to classify the data after feature screening, and the division of the training and test sets is consistent with Section 3.1. The kernel function used by SVM is the RBF kernel.
As mentioned above, our proposed approach adopts an SVM to classify the previously extracted features. The classification accuracy of the SVM model is related to the number of input features. Inadequate features will lead to lower classification accuracy, while redundant features will result in a significant increase in model training time. Therefore, the ReliefF algorithm is used to sort the 10 features previously extracted by AlexNet, so that they are ranked from high to low in order of importance, and the trial-and-error method is adopted by inputting the first n features into the SVM model in turn to determine the optimal number of model input features. The division of the training and test sets is consistent with Section 3.1. We used the RBF kernel in SVM.
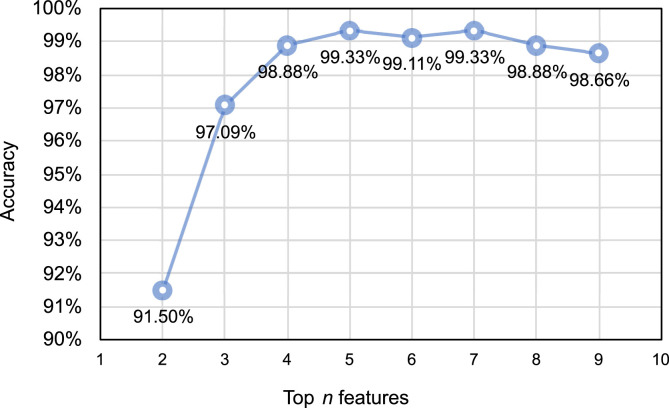
In Fig. 5 , it is shown that when the input of the SVM classifier is the top five important features, the accuracy of the classification results can reach 99.33%, which is the highest value compared with other numbers of input features. Although the accuracy can reach the same level when the first seven features are input, an increased number of input features means a longer model training time. Thus, we determined that the top five features given by the AlexNet and ReliefF algorithm were optimal for this application.
Fig. 5.
Classification accuracy of SVM classifier with different numbers of input features.
However, the process presented here is only for a certain experiment. In the accuracy comparison in the following sections, we conducted several independent repeated experiments to determine the strength of the proposed model in terms of accuracy, specificity and sensitivity compared with some existing models. In each independent repeat experiment, the optimal feature number n is determined according to the specific experimental results, and the value of n is not always 5. However, in the application of the model, we only need to determine the value of the optimal feature number n once in the model training process, which will not affect the generality of the model.
In the following self-contrast study mentioned in Section 4.2, four models (A), (B), (C) and (D), shown in Fig. 6 , are compared. The proposed model (Model (D)) uses improved AlexNet, ReliefF and SVM, and the models (A) original AlexNet, (B) improved AlexNet, (C) improved AlexNet + SVM were also built to verify the effectiveness of the model components proposed in this article.
Fig. 6.
Structures of models used in self-contrast study: (A) original AlexNet, (B) improved AlexNet, (C) improved AlexNet + SVM, (D) improved AlexNet + ReliefF + SVM.
4. Results and discussion
4.1. Metrics for evaluation
The four metrics used for model evaluation are accuracy, specificity, sensitivity, and F-score. They are defined as follows:
where TP (true positives) refers to the correctly predicted COVID-19 cases, FP (false positives) refers to normal or common viral pneumonia cases that were classified as COVID-19 by a model, TN (true negatives) refers to normal or common viral pneumonia cases that were classified as non-COVID-19 cases, while FN (false negatives) refers to COVID-19 cases that were classified as normal or as common viral pneumonia cases.
In this study, confusion matrices were also used in the model evaluation. The confusion matrix is an error matrix commonly used in evaluating the performance of supervised learning algorithm. In a confusion matrix, each column represents the predicted category, and the total number of each column represents the number of data predicted to be that category. Each row represents the true attribution category of the data, and the total number of data in each row represents the number of data instances of that category.
4.2. Self-contrast study
To verify the effectiveness of the model components proposed in this article, this section compares four models, namely (A) original AlexNet, (B) improved AlexNet, (C) improved AlexNet + SVM, and (D) the proposed model (improved AlexNet + ReliefF + SVM). The same dataset division and training parameters are used to train each model, and the results are shown in Fig. 7 , Fig. 8 and Table 4 .
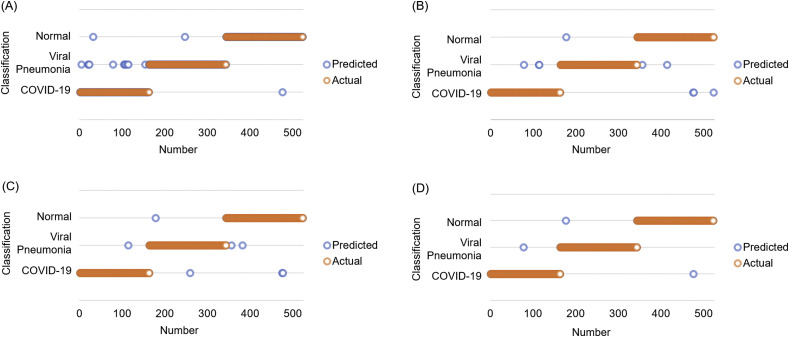
Fig. 7.
Classification results of four models: (A) Original AlexNet, (B) Improved AlexNet, (C) Improved AlexNet + SVM, and (D) Proposed model (improved AlexNet + ReliefF + SVM). The results displayed in this figure correspond to the results with the highest classification accuracy of each model.
Fig. 8.
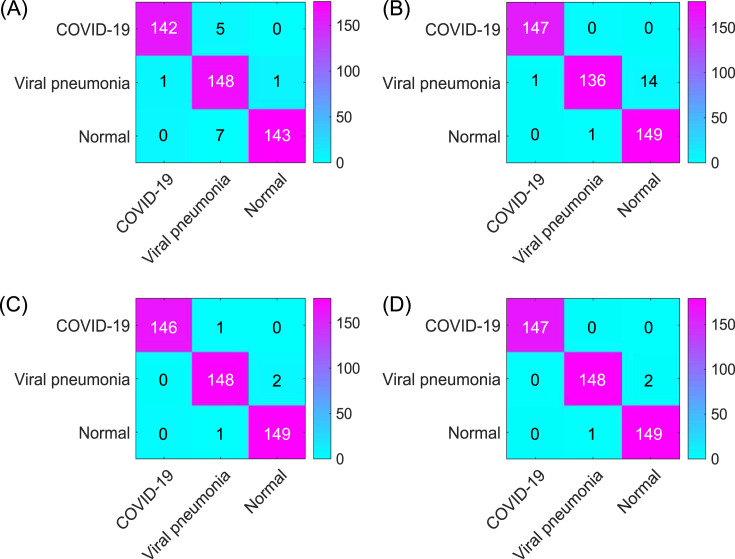
Confusion matrices of four models: (A) Original AlexNet (95.98%), (B) Improved AlexNet (98.09%), (C) Improved AlexNet + SVM (98.47%), and (D) Proposed model (improved AlexNet + ReliefF + SVM) (99.43%) The results displayed in the confusion matrix correspond to the results with the highest classification accuracy of each model.
Table 4.
Classification results.
| (A) Original AlexNet (n = 40) | ||||
|---|---|---|---|---|
| Classification | Accuracy | Specificity | Sensitivity | F-score |
| 1 COVID-19 | 96.864 ± 1.655% | 98.244 ± 1.713% | 91.656 ± 6.102% | 94.704 ± 3.078% |
| 2 Viral pneumonia | 96.272 ± 1.835% | 92.110 ± 4.613% | 97.833 ± 2.290% | 94.803 ± 2.389% |
| 3 Normal | 98.528 ± 0.675% | 98.145 ± 1.767% | 97.611 ± 1.978% | 97.854 ± 0.996% |
| Total | 95.832 ± 1.895% | 96.167 ± 1.540% | 95.700 ± 2.007% | 95.787 ± 1.953% |
| (B) Improved AlexNet (n = 40) | ||||
|---|---|---|---|---|
| Classification | Accuracy | Specificity | Sensitivity | F-score |
| 1 COVID-19 | 97.897 ± 0.941% | 98.980 ± 0.414% | 94.233 ± 3.325% | 96.516 ± 1.622% |
| 2 Viral pneumonia | 97.648 ± 1.000% | 94.656 ± 2.719% | 98.833 ± 0.960% | 96.676 ± 1.362% |
| 3 Normal | 98.872 ± 0.229% | 98.466 ± 1.208% | 98.278 ± 1.155% | 98.360 ± 0.329% |
| Total | 97.208 ± 0.955% | 97.367 ± 0.821% | 97.115 ± 1.027% | 97.184 ± 0.977% |
| (C) Improved AlexNet + SVM (n = 40) | ||||
|---|---|---|---|---|
| Classification | Accuracy | Specificity | Sensitivity | F-score |
| 1 COVID-19 | 98.834 ± 0.436% | 98.279 ± 0.555% | 97.975 ± 1.262% | 98.123 ± 0.712% |
| 2 Viral pneumonia | 98.642 ± 0.343% | 97.289 ± 1.350% | 98.833 ± 0.665% | 98.047 ± 0.478% |
| 3 Normal | 98.815 ± 0.217% | 98.934 ± 0.546% | 97.611 ± 0.644% | 98.266 ± 0.320% |
| Total | 98.145 ± 0.404% | 98.168 ± 0.384% | 98.140 ± 0.426% | 98.145 ± 0.410% |
| (D) The proposed model (Improved AlexNet + ReliefF + SVM) (n = 40) | ||||
|---|---|---|---|---|
| Classification | Accuracy | Specificity | Sensitivity | F-score |
| 1 COVID-19 | 99.082 ± 0.335% | 98.412 ± 0.577% | 98.650 ± 0.950% | 98.528 ± 0.540% |
| 2 Viral pneumonia | 99.082 ± 0.369% | 98.302 ± 1.061% | 99.056 ± 0.375% | 98.674 ± 0.528% |
| 3 Normal | 99.120 ± 0.273% | 99.218 ± 0.467% | 98.222 ± 0.861% | 98.715 ± 0.403% |
| Total | 98.642 ± 0.398% | 98.644 ± 0.388% | 98.643 ± 0.406% | 98.639 ± 0.398% |
It can be seen from the experimental results (Figs. 7 and 8, and Table 4) that all models have satisfactory accuracy. Compared with model (A), model (B) is optimized on the structure of the original AlexNet. From the results, the accuracy of the two is close, whereas model (B) (improved AlexNet) could contribute to the performance improvement of model (C) to a significant extent because if the model (C) is built on the basis of model (A) instead of model (B), it has to classify 1000 features, which makes the training time considerably longer while an accuracy improvement is not obvious.
Model (C) uses AlexNet to extract the features of the original image and then establishes an SVM model to classify the extracted features. Comparing the results of models (B) and (C), it can be found that the accuracy of model (C) reaches 98.642 ± 0.398% and model (C) also has a better performance in terms of specificity, sensitivity, and F-score than model (B), which shows that model (C) has a significant improvement in performance on the basis of model (B). Model (C) is superior to model (B) because model (C) is an ensemble model that uses AlexNet as a feature extractor and SVM as a classifier.
Moreover, model (D), our proposed model, is further improved based on model (C). In model (D), the ReliefF algorithm is used to further sort the features extracted by AlexNet, and the trial-and-error method is adopted to determine the optimal number of feature inputs to improve the classification performance of the SVM, thereby improving the accuracy of the overall model. Additionally, it can be seen from Table 5 that compared to model (C), as the number of feature inputs decreases, model (D) takes less time (5.917 ± 0.001s) than model (C) (5.924 ± 0.001s) to complete the classification task while the accuracy was improved from 98.145 ± 0.404% to 98.642 ± 0.398% (P < 0.05, n = 40, for details, see Supplement 5). From the perspective of model accuracy, model (D) has an accuracy rate of 98.642 ± 0.398%, which stands out among all models (A) –(D).
Table 5.
Classification time consumed by models (C) AlexNet + SVM and (D) AlexNet + ReliefF + SVM (n = 40).
| Models | Feature extraction time/s | Time for classifier/s | Total/s |
|---|---|---|---|
| (C) AlexNet + SVM | 5.896 ± 0.001 | 0.028 ± 0.000 | 5.924 ± 0.001 |
| (D) AlexNet + ReliefF + SVM | 5.896 ± 0.001 | 0.022 ± 0.001 | 5.917 ± 0.001 |
In general, the performance of the four compared models progressively improves with each, clearly demonstrating that in the proposed model, each component has a positive contribution to the performance improvement of the ensemble model. The integrated model proposed in the present work can achieve the task of CXR COVID-19 detection both accurately and effectively.
4.3. Comparative study
To verify the effectiveness of the proposed model, we compare the performance of the five existing models (InceptionV3 [9], VGG19 [26], SqueezeNet [27], ResNet50, ResNet101) and the proposed model and compare the experimental training set and test set divisions consistent with the previous article Table 6, Table 7 and Fig. 9, Fig. 10, Fig. 11, Fig. 12 show the results of the comparative experiment.
Table 6.
Total time* consumed by models in comparative study (n = 40).
| Models | Total/s |
|---|---|
| SqueezeNet | 6.602 ± 0.001 |
| InceptionV3 | 12.376 ± 0.002 |
| ResNet50 | 10.952 ± 0.001 |
| ResNet101 | 18.040 ± 0.002 |
| VGG19 | 16.632 ± 0.002 |
| AlexNet + ReliefF + SVM | 5.917 ± 0.001 |
| * The time required for total classification in seconds. | |
Table 7.
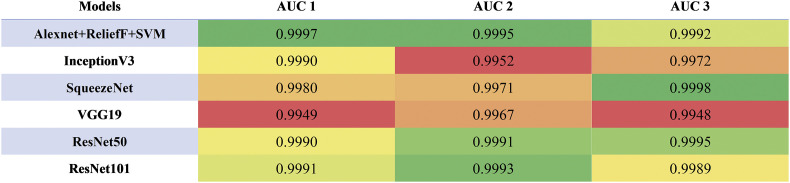
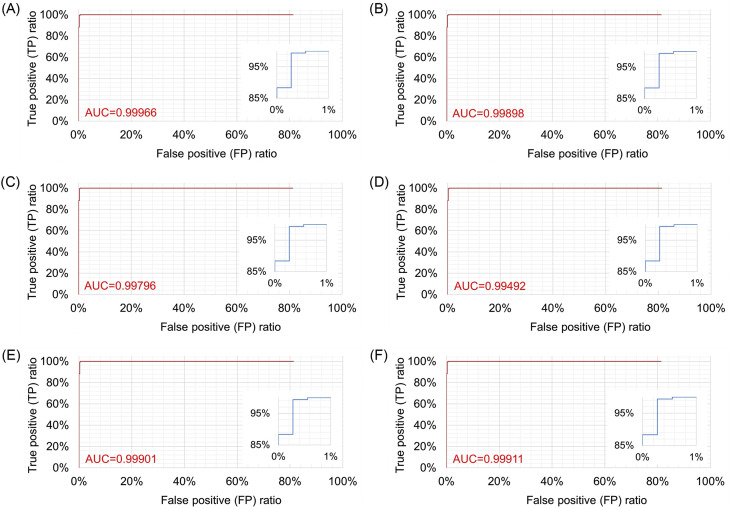
AUC (Area under ROC curves) values. (AUC 1, 2, 3 are defined when true positive results are defined as COVID-19, viral pneumonia, and normal samples are accurately recognized, respectively).
Fig. 9.
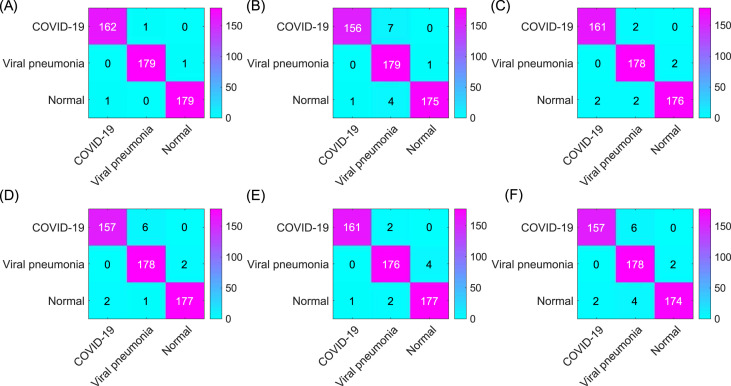
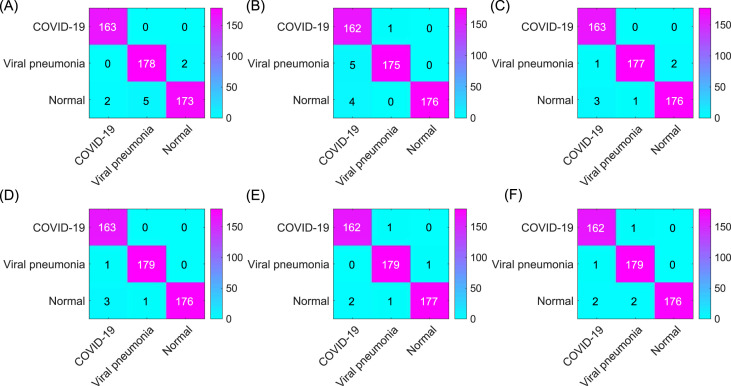
Confusion matrix of (A) The proposed model (improved AlexNet + ReliefF + SVM) (99.43%); (B) InceptionV3 (98.47%); (C) SqueezeNet (97.51%); (D) ResNet-50 (97.90%); (E) ResNet-101 (98.27%); and (F) VGG19 (97.32%). The results displayed in the confusion matrix correspond to the results with the highest classification accuracy of each model.
Fig. 10.
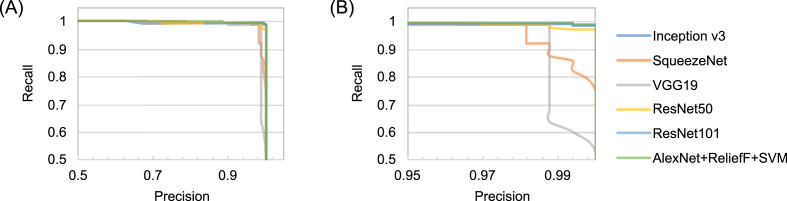
Precision-recall curves of (1) proposed model (improved AlexNet + ReliefF + SVM); (2) InceptionV3; (3) SqueezeNet; (4) ResNet50; (5) ResNet101; and (6) VGG19: (A) Overall; (B) Magnified.
Fig. 11.
Comparative result: performances of proposed method (improved AlexNet + ReliefF + SVM), SqueezeNet, InceptionV3, ResNet50, ResNet101 and VGG19 (n = 40).
Fig. 12.
ROC curves of (A) improved AlexNet + ReliefF + SVM, (B) SqueezeNet, (C) InceptionV3, (D) VGG19, (E) ResNet50 and (E) ResNet101 when true positive results are defined as that COVID-19 samples are accurately recognized.
It can be seen from Fig. 11 that all three models have been fully trained and have good training accuracy. The experimental results show that the model proposed in this study has the highest accuracy rate. Compared to five existing comparison models (InceptionV3: 97.916 ± 0.408%; SqueezeNet: 97.189 ± 0.526%; VGG19: 96.520 ± 1.220%; ResNet50: 97.476 ± 0.513%; ResNet101: 98.241 ± 0.209%), the proposed model has the best performance in the overall accuracy rate (98.642 ± 0.398%) (for details about statistical significance tests, see Supplement 4), which demonstrates that the model proposed in this article is practical and effective, and can provide high-precision COVID-19 CXR detection. In addition, as shown in Table 6, compared to the existing models, the model proposed in this study has a great improvement in efficiency (taking only 5.917 ± 0.001s (n = 40) to classify the test set) because the proposed model has a simpler neural network structure. While ensuring accuracy, our model can significantly shortentraining time. Meanwhile, it can be seen from Fig. 10 (PR curves) and 12 (ROC curves) that our model shows satisfactory performance with an overall improvement in AUC values (shown in Table 7; other ROC curves for calculating AUC values 2 and 3 in the table are shown in Supplement 6). In general, the ensemble model proposed in this study can accomplish the task of COVID-19 detection with improved efficiency and accuracy than existing models.
4.4. Further study
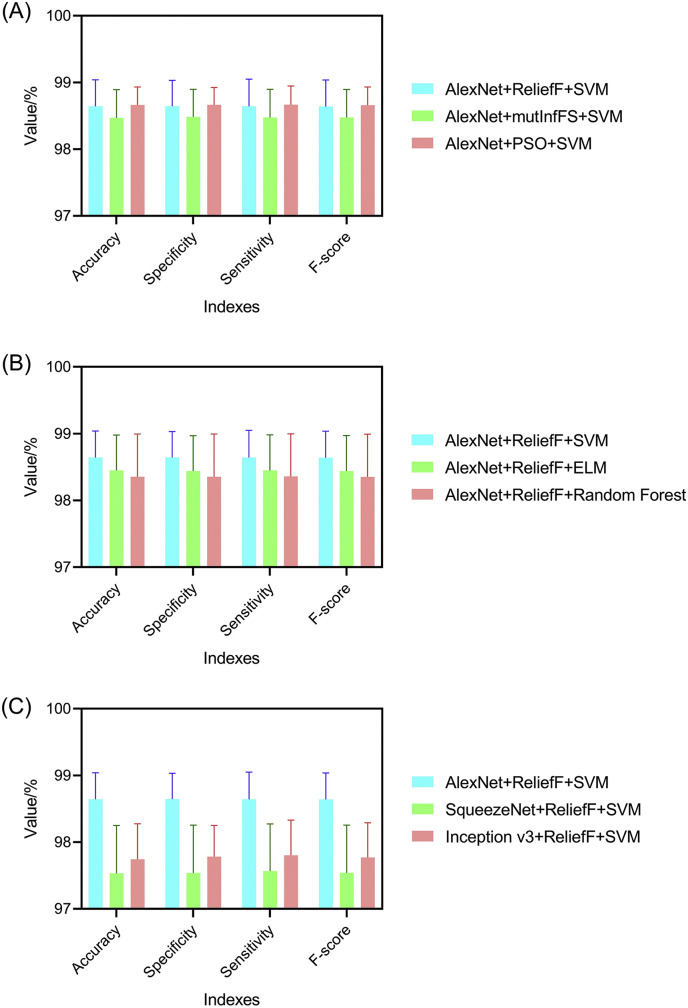
As shown in Fig. 13 , the three parts (feature extraction, feature selection and classifier) of our hybrid model can be re-modeled with different algorithms. To verify the validity and generality of the model structure with different components, we replaced one component while keeping other components unchanged and re-conducted all the classification experiments. The confusion matrices and indexes of the models are shown in Fig. 14, Fig. 15 , respectively.
Fig. 13.
Possible components in the proposed model structure.
Fig. 14.
Confusion matrix of (A) InceptionV3+ReliefF + SVM (98.27%); (B) SqueezeNet + ReliefF + SVM (98.09%); (C) AlexNet + ReliefF + Random Forest (98.66%); (D) AlexNet + ReliefF + ELM (99.04%); (E) AlexNet + PSO + SVM (99.04%); (F) AlexNet + mutInfFS + SVM (98.85%).
Fig. 15.
Classification performances of models (A) using different feature extractors; (B) using different feature selectors; (C) using different final classifiers.
It can be seen from the experimental results that the models using different components can still achieve satisfactory classification results. On the assumption that the data sets used are the same, our proposed model (improved AlexNet + ReliefF + SVM) has the highest accuracy value, which demonstrates the excellence of this approach. According to the experimental results, it can be found that the feature extraction component has the greatest impact on the accuracy of the model. As shown in the Supplement 1, when the InceptionV3, SqueezeNet, and AlexNet were used as feature extractors, the accuracy values were 97.744 ± 0.531%, 97.533 ± 0.718% and 98.642 ± 0.398%, respectively. Replacing the other two components (feature selector and classifier) had a relatively small impact on the performance of the model (see Supplements 2 and 3). Therefore, it becomes crucial to find a suitable network for feature extraction in performing this classification task.
Meanwhile, as shown in Table 8 , there are differences in the time needed for different networks to extract features. In addition to accuracy, the AlexNet we used as feature extractor has the least model running time (5.917 ± 0.001s), which demonstrates the efficiency of our proposed model (AlexNet + ReliefF + SVM) as presented in the previous section.
Table 8.
Total classification time* of models in comparative study (n = 40).
| (A) Using different feature extractors. | |
|---|---|
| Models | Total time/s |
| AlexNet + ReliefF + SVM | 5.917 ± 0.001 |
| InceptionV3 + ReliefF + SVM | 6.428 ± 0.001 |
| SqueezeNet + ReliefF + SVM | 5.918 ± 0.002 |
| (B) Using different feature selectors. | |
|---|---|
| Models | Total time/s |
| AlexNet + ReliefF + SVM | 5.917 ± 0.001 |
| AlexNet + PSO + SVM | 5.918 ± 0.001 |
| AlexNet + mutInfFS + SVM | 5.918 ± 0.001 |
| (C) Using different final classifiers. | |
|---|---|
| Models | Total time/s |
| AlexNet + ReliefF + SVM | 5.917 ± 0.001 |
| AlexNet + ReliefF + Random Forest | 7.271 ± 0.001 |
| AlexNet + ReliefF + ELM | 6.029 ± 0.001 |
(* The time required for total classification in seconds.)
5. Conclusion
A three-step hybrid ensemble model, which comprises of a feature extractor, feature selector, and classifier, is proposed in this work. First, the improved AlexNet extracts the image features, and then the ReliefF algorithm sorts the extracted features according to their importance. The optimized number of input features (n) is acquired through the trial-and-error method, and the first n features are input to the SVM classifier. Finally, the SVM classifier gives the classification results. . Compared with five existing comparison models (InceptionV3: 97.916 ± 0.408%; SqueezeNet: 97.189 ± 0.526%; VGG19: 96.520 ± 1.220%; ResNet50: 97.476 ± 0.513%; ResNet101: 98.241 ± 0.209%), the proposed model has the best performance in the overall accuracy rate (98.642 ± 0.398%), which demonstrates the feasibility and effectiveness of the proposed model.
The superiority of the proposed model can be enumerated as follows: (1) On the whole, the final classification result of the model reached 98.642 ± 0.398% proving its feasibility for COVID-19 CXR detection; (2) Compared with the direct application of neural networks for classification, the hybrid method proposed in this article demonstrates its high accuracy. In addition, the transfer learning method adopted in this work can remarkably reduce the time required for deep learning network training and the size of training sets. Despite having only 1222 images as the training set for network training, this work still achieves satisfactory classification results; (3) Compared with a model without feature selection (AlexNet + SVM), the proposed hybrid model (AlexNet + ReliefF + SVM) is more accurate and less time-consuming. The reason is that model performance is not necessarily proportional to the number of input features because the data redundancy affects the speed and accuracy of the algorithm and increases the difficulty of learning tasks; (4) The components in the hybrid model structure presented in this paper can also be replaced according to the characteristics of the data, which can maintain a good classification effectiveness.
Declaration of competing interest
The authors have no conflicts of interest to declare.
Acknowledgements
This study was funded by the National Science Foundation of China (81571629, 82071990), Project of Shanghai Science and Technology Commission (19411965200), Shanghai Chest Hospital Project of Collaborative Innovative Grant (YJT20191015) and Shanghai Municipal Medical and Health Outstanding Young Talents Training Program (GWV-10.2-YQ28).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2021.104252.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Radiology U.C.L.A. COVID-19 chest X-ray guideline. https://www.uclahealth.org/radiology/covid-19-chest-x-ray-guideline
- 2.Cozzi D., Albanesi M., Cavigli E., et al. Chest X-ray in new Coronavirus Disease 2019 (COVID-19) infection: findings and correlation with clinical outcome. Radiol. Med. 2020;125(8):730–737. doi: 10.1007/s11547-020-01232-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vancheri S.G., Savietto G., Ballati F., et al. Radiographic findings in 240 patients with COVID-19 pneumonia: time-dependence after the onset of symptoms. Eur. Radiol. 2020 doi: 10.1007/s00330-020-06967-7. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varela-Santos S., Melin P. A new approach for classifying coronavirus COVID-19 based on its manifestation on chest X-rays using texture features and neural networks. Inf. Sci. 2021;545(January):403–414. doi: 10.1016/j.ins.2020.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Rajendra Acharya U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020;121(April) doi: 10.1016/j.compbiomed.2020.103792. 103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y.-D., Satapathy S., Shuaiqi L., Li G.-R. A five-layer deep convolutional neural network with stochastic pooling for chest CT-based COVID-19 diagnosis. Mach. Vis. Appl. 2020;123 doi: 10.1007/s00138-020-01128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polsinelli M., Cinque L., Placidi G. A light CNN for detecting COVID-19 from CT scans of the chest. Pattern Recogn. Lett. 2020;140(January):95–100. doi: 10.1016/j.patrec.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan T., Wong P.K., Ren H., Wang H., Wang J., Li Y. Automatic distinction between COVID-19 and common pneumonia using multi-scale convolutional neural network on chest CT scans. Chaos, Solit. Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110153. 110153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benbrahim H., Hachimi H., Amine A. Deep transfer learning with Apache spark to detect COVID-19 in chest X-ray images. Rom. J. Inf. Sci. Technol. 2020;23(April):117–129. [Google Scholar]
- 10.Hassantabar S., Ahmadi M., Sharifi A. Diagnosis and detection of infected tissue of COVID-19 patients based on lung x-ray image using convolutional neural network approaches. Chaos, Solit. Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110170. 110170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minaee S., Kafieh R., Sonka M., Yazdani S., Jamalipour Soufi G. Deep-COVID: predicting COVID-19 from chest X-ray images using deep transfer learning. Med. Image Anal. 2020;65 doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toraman S., Alakus T.B., Turkoglu I. Convolutional capsnet: a novel artificial neural network approach to detect COVID-19 disease from X-ray images using capsule networks. Chaos, Solit. Fractals. 2020:140. doi: 10.1016/j.chaos.2020.110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L., Lin Z.Q., Wong A. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci. Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee K.-S., Kim J.Y., Jeon E.-T., Choi W.S., Kim N.H., Lee K.Y. Evaluation of scalability and degree of fine-tuning of deep convolutional neural networks for COVID-19 screening on chest X-ray images using explainable deep-learning algorithm. J. Personalized Med. 2020;10(4) doi: 10.3390/jpm10040213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ouchicha C., Ammor O., Meknassi M. CVDNet: a novel deep learning architecture for detection of coronavirus (Covid-19) from chest x-ray images. Chaos, Solit. Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Motamed S., Rogalla P., Khalvati F. RANDGAN: randomized generative adversarial network for detection of COVID-19 in chest X-ray. http://arxiv.org/abs/2010.06418 Published online 2020:1-10. [DOI] [PMC free article] [PubMed]
- 17.Nayak S.R., Nayak D.R., Sinha U., Arora V., Pachori R.B. Application of deep learning techniques for detection of COVID-19 cases using chest X-ray images: a comprehensive study. Biomed. Signal Process Contr. 2021;64(January) doi: 10.1016/j.bspc.2020.102365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Özkaya U., Öztürk Ş., Budak S., Melgani F., Polat K. vol. 19. 2020. Classification of COVID-19 in Chest CT Images Using Convolutional Support Vector Machines; pp. 1–20. December 2019. [Google Scholar]
- 19.Yu X., Wang S.H., Zhang Y.D. CGNet: a graph-knowledge embedded convolutional neural network for detection of pneumonia. Inf. Process. Manag. 2021;58(1) doi: 10.1016/j.ipm.2020.102411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.https://www.kaggle.com/nih-chest-xrays/data No title.
- 21.Wang X., Peng Y., Lu L., Lu Z., Bagheri M., Summers R.M. ChestX-ray: hospital-scale chest X-ray database and benchmarks on weakly supervised classification and localization of common thorax diseases. Adv. Comput. Vis. Pattern Recognit. 2019:369–392. doi: 10.1007/978-3-030-13969-8_18. Published online. [DOI] [Google Scholar]
- 22.Cohen J.P., Morrison P., Dao L., Roth K., Duong T.Q., Ghassemi M. COVID-19 image data collection: prospective predictions are the future. 2020. https://github.com/ieee8023/covid-chestxray-dataset Published online.
- 23.Lu S., Lu Z., Zhang Y. Pathological brain detection based on AlexNet and transfer learning. J. Comput. Sci. 2019;30:41–47. doi: 10.1016/j.jocs.2018.11.008. [DOI] [Google Scholar]
- 24.Kilicarslan S., Adem K., Celik M. Diagnosis and classi fi cation of cancer using hybrid model based on ReliefF and convolutional neural network. Med. Hypotheses. 2020;137(January) doi: 10.1016/j.mehy.2020.109577. 109577. [DOI] [PubMed] [Google Scholar]
- 25.Urbanowicz R.J., Meeker M., Cava W La, Olson R.S., Moore J.H. Relief-based feature selection : introduction and review. J. Biomed. Inf. 2018;85(June):189–203. doi: 10.1016/j.jbi.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020;43(2):635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abbas A., Abdelsamea M.M., Gaber M.M. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl. Intell. 2020 doi: 10.1007/s10489-020-01829-7. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.