Abstract
Tumor immunity represents a new avenue for cancer therapy. Immune checkpoint inhibitors have successfully improved outcomes in several tumor types. In addition, currently, immune cell-based therapy is also attracting significant attention. However, the clinical efficacy of these treatments requires further improvement. The mechanisms through which cancer cells escape the immune response must be identified and clarified. Cancer stem cells (CSCs) play a central role in multiple aspects of malignant tumors. CSCs can initiate tumors in partially immunocompromised mice, whereas non-CSCs fail to form tumors, suggesting that tumor initiation is a definitive function of CSCs. However, the fact that non-CSCs also initiate tumors in more highly immunocompromised mice suggests that the immune evasion property may be a more fundamental feature of CSCs rather than a tumor-initiating property. In this review, we summarize studies that have elucidated how CSCs evade tumor immunity and create an immunosuppressive milieu with a focus on CSC-specific characteristics and functions. These profound mechanisms provide important clues for the development of novel tumor immunotherapies.
Keywords: Cancer stem cells, Immune evasion, T cells, NK cells, Immune checkpoints
Abbreviations: ADCC, antibody-dependent cell mediated cytotoxicity; ALDH, alcohol dehydrogenase; AML, acute myeloid leukemia; ARID3B, AT-rich interaction domain-containing protein 3B; CCR7, C–C motif chemokine receptor 7; CMV, cytomegalovirus; CIK, cytokine-induced killer cell; CSC, cancer stem cell; CTL, cytotoxic T lymphocytes; CTLA-4, cytotoxic T-cell-associated antigen-4; DC, dendritic cell; DNMT, DNA methyltransferase; EMT, epithelial–mesenchymal transition; EV, extracellular vesicle; ETO, fat mass and obesity associated protein; HNSCC, head and neck squamous cell carcinoma; KDM4, lysine-specific demethylase 4C; KIR, killer immunoglobulin-like receptor; LAG3, lymphocyte activation gene 3; LILR, leukocyte immunoglobulin-like receptor; LMP, low molecular weight protein; LOX, lysyl oxidase; MDSC, myeloid-derived suppressor cell; MHC, major histocompatibility complex; MIC, MHC class I polypeptide-related sequence; NGF, nerve growth factor; NK, natural killer; NOD, nonobese diabetic; NSG, NOD/SCID IL-2 receptor gamma chain null; OCT4, octamer-binding transcription factor 4; PD-1, programmed death receptor-1; PD-L1/2, ligands 1/2; PI9, protease inhibitor 9; PSME3, proteasome activator subunit 3; SCID, severe combined immunodeficient; SOX2, sex determining region Y-box 2; TAM, tumor-associated macrophage; TAP, transporter associated with antigen processing; TCR, T cell receptor; Treg, regulatory T cell; ULBP, UL16 binding protein; uPAR, urokinase-type plasminogen activator receptor
Highlights
-
•
Cancer stem cells (CSCs) play a central role in multiple aspects of malignant tumors.
-
•
Immune evasion is a fundamental feature of CSCs.
-
•
Immune evasion mechanisms must be precisely clarified to improve tumor immunotherapy.
-
•
CSCs are promising targets for tumor immunotherapy.
1. Introduction
Tumor tissues consist of various types of noncancerous cells, including vascular and immune cells. Drugs targeting the VEGF/VEGFR signaling pathway, such as bevacizumab, ramucirumab, aflibercept, and regorafenib, suppress the proliferation of endothelial cells in tumor tissues, thereby inhibiting neovascularization in the tumor microenvironment [1]. The infiltration of immune cells, including CD4+ and CD8+ T cells, dendritic cells (DCs), and natural killer (NK) cells, into tumors is associated with the prognosis of patients with cancer. These immune cells are frequently suppressed by immune checkpoint molecules, such as programmed death receptor-1 (PD-1), its ligands (PD-L1/PD-L2), and cytotoxic T-cell-associated antigen-4 (CTLA-4), which may be restored by checkpoint inhibitors [2]. Within tumors, noncancerous cells with tumor-specific functions, including cancer-associated fibroblasts (CAFs) [3], tumor-associated macrophages (TAMs) [4], tissue-associated neutrophils [5], myeloid-derived suppressor cells (MDSCs) [6], and pericytes [7], also exist. These cells can promote tumor growth, recurrence, and metastasis through direct and indirect interactions with cancer cells [8]. Recently, it has been revealed that bacteria cells may reside within cancer and immune cells. These intratumoral bacteria affect tumor malignancy and response to immunotherapy [9]. Because of their important role in tumor growth and progression, these noncancerous cells have attracted significant interest as therapeutic targets for the development of novel cancer medicines.
Besides noncancerous cells, cancer cells exhibit genetically and nongenetically heterogeneous populations within tumor tissue. The genetic heterogeneity of cancer cells may occur as a consequence of genetic and chromosomal instabilities [10,11] and may cause significant difficulties to the overall effectiveness of precision medicine. In contrast, the nongenetic heterogeneous populations, in particular, cancer stem cells (CSCs) and non-CSCs, were found in tumor tissue. CSCs have distinct properties compared with non-CSCs, including self-renewal, differentiation into non-CSCs, and tumor initiation ability. Interestingly, non-CSCs can revert to CSCs without alteration of genomic sequences [12,13], indicating that their relation is, in part, different from that of normal stem/progenitor cells compared with their descendant cells. Because of their malignant characteristics, CSCs are also involved in recurrence, metastasis, and resistance to chemotherapy and radiotherapy. Therefore, CSCs are attracting attention as novel target for prevention and treatment of cancer.
Immune cells actively survey and eliminate cells undergoing malignant transformation. However, some of the transformed cells can evade immune surveillance and eventually form tumor. This suggests that immune evasion may also be a malignant feature of CSCs. Interestingly, immunomodulating properties have also been observed in normal stem cells [14]. Likewise, CSCs can create an immunosuppressive milieu by cooperating with other noncancerous cells in the tumor microenvironment. This function is not only intrinsic to CSCs but also extrinsic from noncancerous cells. Therefore, clarifying this mechanism and the immunological features of CSCs will increase our understanding of tumor development and recurrence and provide novel therapeutic targets for cancer immunotherapy. In this review, we summarize the current literature regarding the relation between CSCs and tumor immunity, and the manner in which CSCs evade and protect tumors from immune surveillance and destruction.
2. Tumorigenicity in immune-compromised/competent mice
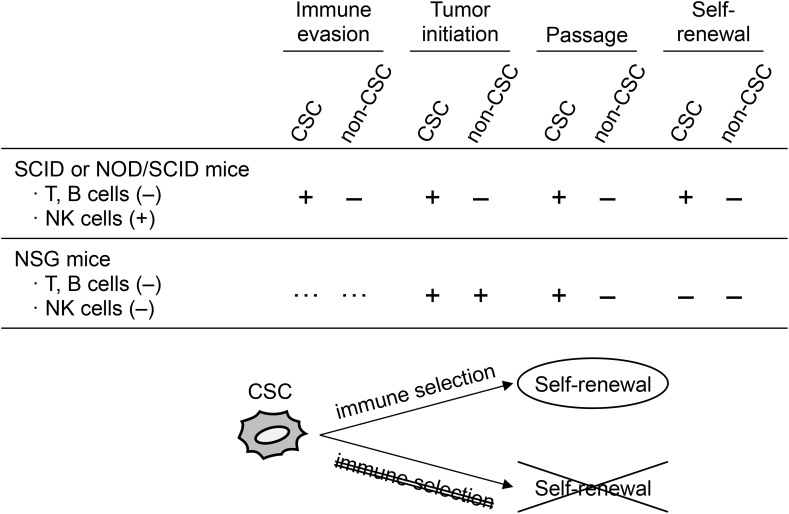
CSCs were first identified from leukemia cells using severe combined immunodeficient (SCID) mice [15], in which mature T and B cells are depleted, but the innate immune cells, including NK cells and macrophages, are retained. Thereafter, CSCs in solid tumors were identified using nonobese diabetic (NOD)/SCID mice [16], in which innate immunity activity is lower compared with SCID mice but still present. These experiments using partially immunocompromised mice demonstrated that a small subset of cancer cells can initiate tumor development. However, Quintana et al. [17] reported that tumor-initiating cells in human melanoma were not a rare population when transplanted into more highly immunocompromised NOD/SCID IL-2 receptor gamma chain null (NSG) mice, in which NK, T, and B cells are deficient. They showed that the frequency of melanoma-initiating cells determined by NOD/SCID mice was 1/106–1/105, whereas that of NSG mice was 1/9–1/4 [17]. Consistently, xenotransplantation of non-CSCs into NSG mice also resulted in tumor formation (Fig. 1), and tumor initiation was not associated with the expression of known melanoma CSC markers (CD271 and ATP-binding cassette B5), which are established indicators of tumor initiation in NOD/SCID mice [17,18]. However, in tumor tissues developed from CD271-negative melanoma cells transplanted into NSG mice, the expressions of melanoma markers, such as melanocyte-inducing transcription factor (MITF) and SP100, were no longer retained [19], suggesting the failure of self-renewal (Fig. 1). Moreover, these CD271-negative cells also failed to passage in NSG mice, whereas CD271-positive CSCs were successively propagated (Fig. 1) [19]. A similar observation was reported for CD133-positive and CD133-negative hepatocellular carcinoma cells in a syngeneic transplantation model [20]. Interestingly, after transplanting CD271-positive melanoma CSCs into NSG mice, the tumor cells did not show a similar expression pattern of melanoma markers compared with that in parental tumor tissues [19], suggesting that immune selection may be required for the maintenance of the self-renewal property (Fig. 1). The differences observed between NOD/SCID and NSG mice indicate that the tumor initiation property is a more frequent and ubiquitous feature and that the rarity of cancer cells exhibiting tumor-initiating properties may not be so important for CSC definition. Rather, tumor immune evasion may be a more important characteristic of CSCs [21,22]. Because immune evasion is also observed in normal stem cells, it may represent a fundamental stemness property endowed on CSCs [14,23,24].
Fig. 1.
Tumorigenicity in immune-compromised/competent mice. Details are discussed in the main text.
As described above, immune selection, which is a phenomenon that immune cells preferentially kill non-CSCs, and consequently promote the propagation of CSCs, may play a role in CSC maintenance and induction (Fig. 1). In other words, this phenomenon indicates that CSCs have an immune evasion property, and are capable to grow even under the surveillance of the immune system. A human hepatoma cell line, HepG2 cells, exposed to mouse macrophages or B cells exhibited increased CSC properties, concomitant with the upregulation of CSC markers [25]. In mice, cytotoxic CD8+ T lymphocytes (CTL) enriched CD24−/low/CD44+ breast CSCs by inducing the epithelial–mesenchymal transition (EMT) [26]. Trastuzumab, a specific antibody against HER2, kills HER2-positive breast cancer cells by NK-cell-mediated antibody-dependent cytotoxicity (ADCC). After ADCC, surviving breast cancer cells formed spheroids exhibiting a CD44high/CD24low CSC-like phenotype and lower HER2 expression compared with parental cells [27]. The immune selection of malignant rhabdoid tumor cells by passaging in immunocompetent mice enriched cells with increased CSC properties and decreased expression of major histocompatibility complex (MHC) molecules and costimulatory molecules [28]. The enriched CSCs also showed increased expression of lysyl oxidase (LOX) and a CSC marker, alcohol dehydrogenase (ALDH), whereas a LOX inhibitor suppressed the growth of the tumors passaged in immunocompetent mice [28]. Consistently, tumor tissues exposed to an immune challenge in vivo exhibited a CSC-like gene expression profile with increased CSC properties, including tumor initiation ability [29]. These results suggest that tumor immunity preferentially kills non-CSCs, leading to an increase in CSCs, although it is also worth investigating whether tumor immunity induces CSC properties in non-CSCs.
The immune evasion property, the tumor initiation ability in the presence of immune surveillance, is an additional characteristic of CSCs [30]. Thus, tumor-initiating cells investigated in highly immunocompromised animals would underestimate the true frequency of CSCs present in a tumor tissue [31]. As such, studies examining the in vivo function of CSCs should be performed with partially immunocompromised, syngeneic, or congenic immunocompetent animals [32]. For example, transplantation of hematopoietic stem cells transduced with the ovalbumin and MLL/AF9 fusion genes, in which the latter causes acute myeloid leukemia (AML), into wild-type mice resulted in the development of leukemia although ovalbumin-specific CTLs had expanded in the transplanted mice [33]. Interestingly, this syngeneic transplantation model revealed that the cancer cells exhibited high expression of immune checkpoint molecules, such as PD-1 and lymphocyte activation gene 3 (LAG3) (Table 1) [33]. These results indicate that an immunocompetent mouse represents a good model for the investigation of the immunological features of CSCs.
Table 1.
Regulation of effector T cells by molecules on tumor cells.
| Tumor cells | Effector T cells | Effector function | Function in CSCs |
|---|---|---|---|
| MHC class I & II | TCR | activation | [[37], [38], [39], [40], [41],43,44,[48], [49], [50],132,133] |
| CD80/B7-1 | CD28/TP44 | activation | [45] |
| CD86/B7-2 | CD152/CTLA-4 PD-L1 |
inhibition | |
| CD274/PD-L1/B7–H1 | CD279/PD-1 | inhibition | [130,141,143,[151], [152], [153], [154], [155], [156], [157], [158], [159], [160], [161], [162], [163],166,167,169] |
| CD273/PD-L2/B7-DC | |||
| CD275/ICOS-L/B7–H2 | CD278/ICOS/AILIM | activation | ? |
| CD276/B7–H3 | ? | ? | ? |
| VTCN1/B7–S1/B7–H4 | ? | ? | ? |
| VISTA/GI24/B7–H5 | CD28H/IGPR1/TMIGD2 | inhibition? | ? |
| HHLA2/B7–H7 | |||
| NCR3LG1/B7–H6 | CD337/NKp30/NCR3a | activation | [[78], [79], [80]] |
| MHC class II | CD223/LAG3 | activation | [33,186,187] |
| FGL1, galectin-3, LSECtin | inhibition | ||
| CEACAM1, galectin-9, HMGB1 | TIM3 | inhibition | ? |
| CD200 | CD200R | inhibition | [[179], [180], [181], [182]] |
AILIM, activation-inducible lymphocyte immunomodulatory molecule; CEACAM1, carcinoembryonic antigen-related cell adhesion molecule 1; CTLA-4, cytotoxic T-cell-associated antigen-4; FGL1, fibrinogen-like protein 1; HHLA2, human endogenous retrovirus-H long terminal repeat-associating protein 2; HMGB1, high-mobility group box-1; ICOS, inducible T-cell co-stimulator; ICOS-L, ICOS ligand; IGPR1, immunoglobulin and proline-rich receptor-1; LAG3, lymphocyte activation gene 3; LSECtin, liver and lymph node sinusoidal endothelial cell C-type lectin; MHC, major histocompatibility complex; NCR3, natural cytotoxicity triggering receptor 3; NCR3LG1, NCR3 ligand 1; PD-1, programmed cell death-1; PD-L1/2, PD-1 ligand 1/2; TCR, T-cell receptor; TIM3, T-cell immunoglobulin and mucin domain 3; TMIGD2, transmembrane and immunoglobulin domain containing 2; VISTA, V-domain immunoglobulin suppressor of T-cell activation; VTCN1, V-set domain-containing T-cell activation inhibitor 1.
Expressed on NK cells.
3. Immunosuppressive properties endowed on CSCs
It has been shown that the expression of CSC makers in human tumor tissues is correlated with the number of tumor-infiltrating immune cells [34,35], suggesting that CSCs exhibit a close relationship with the tumor immune environment. Accumulating evidence suggests that the immune response to CSCs is frequently compromised because of their immune evasion properties. For example, CSCs may secrete immunosuppressive factors and recruit immunosuppressive noncancerous cells. Subsequently, the immune suppressive cells, in turn, induce and maintain CSCs. However, the dynamics of the tumor microenvironment are complicated, and mechanisms that create the immunosuppressive milieu can vary depending on tumor development. In this section, we summarize the immunosuppressive mechanisms harnessed by CSCs.
3.1. Impaired antigen presentation of CSCs
The interaction between the T-cell receptor (TCR) on CTLs with MHC-I antigens is a prerequisite for a proper T-cell response against tumor cells. Antigens must be sufficiently degraded by proteasomes, transported to the endoplasmic reticulum by transporter proteins, and presented by MHC-I molecules. Otherwise, T cells fail to recognize transformed cells. It has been suggested that these antigen processing and presenting steps are impaired in CSCs.
Cancer cells cultured in an ultralow attachment dish form spheroids enriched with CSCs because CSCs exhibit an anchorage-independent growth [36]. Melanoma spheroids also show increased CSC properties compared with adherent cells [37]. MHC molecules are shown to be downregulated in melanoma spheroid cells, leading to the inhibition of the allogenic immune response of T cells [37]. Likewise, defects in MHC-I-mediated antigen presentation are often observed in CSCs and render the cells resistant to the cytotoxic effects of CTL [38]. A similar observation has also been reported in normal hematopoietic cells and Lgr5+ normal epithelial stem cells [39,40], suggesting that impaired antigen presentation may be a common feature of stem cells.
Neurospheres produced from primary glioblastoma were also enriched in glioblastoma CSCs, which exhibited low MHC-I expression levels and lacked MHC-II expression. Moreover, neurosphere cells also showed downregulation of antigen-processing molecules, including low-molecular-weight protein (LMP), transporter associated with antigen processing (TAP), and beta-macroglobulin [41], suggesting that not only antigen presentation but also antigen processing is impaired in glioblastoma CSCs. Consistently, T-cell activation and proliferation was more significantly impaired by these CSCs compared with non-CSCs [41,42]. It was shown that IFNs, which are potent immune modulatory cytokines, induced the expression of MHC-I molecules in glioblastoma CSCs. However, the expression level of MHC-I molecules in CSCs treated with IFNs was still lower compared with that in non-CSCs [41]. This suggests that an IFN response suppression mechanism plays a role in the immune evasion property of glioblastoma CSCs.
It was demonstrated that CSCs derived from head and neck squamous cell carcinoma (HNSCC) also exhibit downregulation of MHC molecules [43,44]. CD44+ HNSCC cells exhibited CSC properties and downregulated HLA-A2, HLA class II, and TAP2, suggesting the impairment of antigen presentation and processing such as glioblastoma CSCs [43]. Moreover, in addition to the suppression of the Th1 response, CD44+ HNSCC cells also induced regulatory T (Treg) cells and MDSCs concomitant with high expression of immune modulatory cytokines, including IL-8, granulocyte colony-stimulating factor, and TGFβ [43]. HNSCC spheroid cells, as observed in CD44+ HNSCC cells, also exhibited low expression of MHC-I and resistance to CTLs [44]. However, IFNγ efficiently induced MHC-I expression in spheroid cells and sensitized the cells to CTL-mediated cytolysis [44]. Interestingly, ALDHlow HNSCC spheroid cells were more susceptible to CTLs in the presence of IFNγ than ALDHlow spheroid cells [44].
In contrast, mouse breast ALDH+ CSCs efficiently formed tumors in immunocompetent mice, whereas the growth of bulk cells containing CSCs and non-CSCs was limited [45]. The ALDH+ CSCs downregulated the expression of TAP genes and a costimulatory molecule, CD80 (B7-1), by DNA methylation (Table 1) [45]. However, CD44+/CD24− CSCs in murine breast cancer did not show decreased TAP and CD80 expression and upregulated C-X-C motif chemokine receptor 4, which is involved in the induction of the CSC and EMT phenotypes in breast cancer [45,46]. It is not surprising that the mechanisms underlying the immune modulatory functions may vary among CSCs expressing different CSC markers, because some of these markers exhibit distinct functions in regulating CSCs [47].
Spheroid cells derived from TC-1 cells, a lung cancer cell line transduced with viral oncogene E6/E7, showed lower MHC-I expression compared with adherent cells [48]. IFNγ sensitized spheroid cells to E6/E7-specific immune attack by inducing MHC-I expression [48]. In contrast, the spheroid cells derived from 12 solid tumor cell lines (colon, pancreas, melanoma, and breast cancer) exhibited higher expression of antigen processing and presentation molecules, including LMP2/LMP7, MECL1, and TAP1/TAP2, whereas the expression levels of HLA-I and HLA-II were lower compared with those in adherent cells [49], suggesting that T-cell activation by the spheroid cells may be impaired. However, IFNγ failed to induce the expression of HLA molecules in spheroid cells [49]. Therefore, the sensitivity of CSCs to IFNγ differs among cell lines. In addition, mechanisms responsible for the regulation of antigen presentation and antigen-processing molecules in CSCs may also differ. The expression of MHC-I molecules was found to be transcriptionally downregulated in glioblastoma [50], whereas, in peripheral blood mononuclear cells, a serine protease, cathepsin G, degraded MHC-I molecules in the endosome and lysosome [51]. Although active cathepsin G is not present in glioblastoma cells [51], CSCs may utilize multiple pathways to downregulate MHC-I expression at the transcriptional and posttranscriptional levels.
3.2. Downregulation of tumor-associated antigens in CSCs
As observed in NOD/SCID and NSG mice, the melanoma initiation property depends on host immunity, and only CSCs can evade immune surveillance to form tumors in NOD/SCID mice [17,52]. In addition to the downregulation of MHC-I and MHC-II molecules, melanoma CSCs evade tumor immune surveillance through several mechanisms, one of which involves the downregulation of melanoma-associated antigens [53,54]. Consequently, T cells failed to efficiently recognize melanoma CSCs; however, CSCs highly express the marker proteins specific for CSCs, which are necessary for CSC induction and maintenance [47]. Therefore, CSC-specific proteins are desirable targets as tumor-associated antigens for cancer immune therapy. Consistently, it was demonstrated that activating the immune response to octamer-binding transcription factor 4 (OCT4), which is a well-known stemness transcription factor, suppressed the growth of teratoma cells in mice [55]. Intriguingly, it was reported that naturally occurring T cells specific for OCT4 were found in the ascites and peripheral blood of patients with ovarian cancer, although they lost the ability to eliminate OCT4+ CSCs [56,57]. However, these OCT4-specific T cells were activated by DCs primed with an OCT4-derived peptide [57].
Antibodies against another stemness transcription factor, sex-determining region Y- box 2 (SOX2), were also found in the serum of patients with lung cancer, prostate cancer, and glioblastoma [58,59]. The presence of SOX2-reactive antibodies predicted a good prognosis in patients with lung cancer [58]. SOX2-reactive T cells were also found in patients with monoclonal gammopathy, a benign disease, that sporadically progresses to multiple myeloma [60]. However, the SOX2-reactive T cells disappeared in patients with myeloma [60], suggesting that the collapse of anti-SOX2 immunity may be a mechanism of progression to multiple myeloma.
It has been clinically and experimentally demonstrated that immunotherapy targeting CSC markers, such as CD133 [61,62], CD44 [63,64], CD24 [65], and EPCAM [66,67], can suppress tumor growth. However, these CSC markers are also expressed in normal cells. Therefore, embryonic stemness markers, such as OCT4 and SOX2, may be preferential targets for CSC-specific immunotherapy [68].
3.3. Modulation of NK-cell activity by CSCs
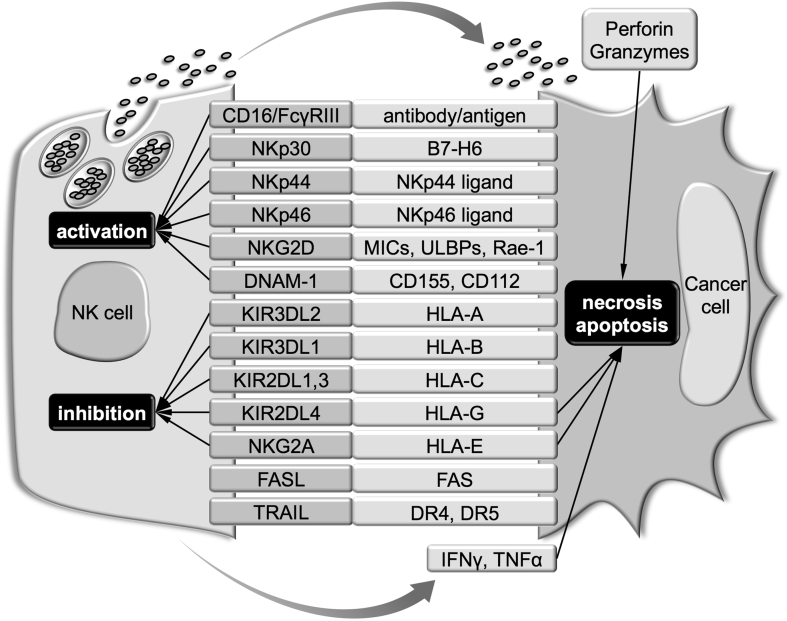
In general, it is known that NK cells attack “missing self” cells exhibiting a lack or low expression of MHC-I. This antigen-presenting molecule suppresses NK activity by interacting with NK-cell inhibitory receptors on the surface of NK cells (Fig. 2) [69]. The preferential targeting of NK cells suggests that, although CSCs could escape from MHC-restricted T-cell recognition, they would be eventually eliminated by NK cells. Nonetheless, the sensitivity of CSCs to NK-cell-mediated cell lysis remains under debate because CSCs, not non-CSCs, can form tumors in NOD/SCID mice [16]. This indicates that the tumor microenvironment containing immunosuppressive noncancerous cells should also be considered to increase our understanding of the relationship between CSCs and NK cells [70]. However, because of space limitations, only the intrinsic properties of CSCs are discussed in the following sections.
Fig. 2.
Pathways for the recognition and killing of cancer cells by NK cells. Details are discussed in the main text.
3.4. NK cell-sensitive phenotype of CSCs
It has been demonstrated that NK-cell-mediated cell lysis activity is enhanced by coculture with CSCs [71,72]. Consistently, NK cells preferentially killed CD24+/CD44+, CD133+, and ALDHhigh CSC lines and primary tumors in vitro [73]. In addition to downregulating MHC-I, NK cells may preferentially kill CSCs because of the upregulation of NK-cell-activating receptor ligands in CSCs (Fig. 2). Furthermore, it was revealed that CSCs exhibited high expression of MHC-I polypeptide-related sequence A/B (MICA/MICB; ligands for a NK-cell-activating receptor, NKG2D) and were killed by NK cells in a manner depending on NKG2D [73].
In addition, NK cells utilize not only the secretory granule-mediated cell lysis pathway involving perforin and granzymes but also the FAS/FASL and DR4/DR5/TRAIL death receptor pathways to kill tumor cells [74]. CSCs from a colon cancer cell line showed c-MYC-mediated expression of DR4 and were susceptible to TRAIL-induced apoptosis [75]. Breast CD44+/CD24−/low CSCs isolated from a spontaneous breast carcinogenesis model (B6 PyMT-MMTV transgenic mouse) expressed FAS and DR5 and were more susceptible to FASL- and TRAIL-induced apoptosis compared with non-CSCs [76]. Although these CSCs exhibited lower expression of MHC-I and higher expression of Rae-1 (NKG2D ligand) and CD155/PVR (ligand for a NK-cell-activating receptor, DNAM-1) compared with non-CSCs, the sensitivity to NK-cell-mediated cell lysis was not significantly different between the CSCs and non-CSCs [76]. Therefore, these results suggest that the sensitivity of CSCs to NK cells may not be determined by these cell surface molecules. It is noted that cancer cells are prone to be resistant against the death receptor pathways by modulating the expression of proapoptotic and antiapoptotic regulators [77].
Nonetheless, it has been suggested that various ligands for NK-cell-activating receptors are involved in the sensitization of CSCs to NK cells. Colon CSCs displayed reduced MHC-I expression and upregulation of CSC markers (CD133, CD44, Lgr5, and CD166) and ligands for the NK-cell-activating receptors, NKp30 and NKp44. As a result, they were more sensitive to allogenic NK-cell-mediated cell lysis compared with non-CSCs [78]. Ovarian CD24+ CSCs also exhibited downregulation of MHC-I molecules and upregulation of NKG2D ligands compared with non-CSCs [71]. Similarly, ovarian CSCs were more susceptible to NK-cell-mediated lysis compared with non-CSCs [71]. In patient-derived melanoma cell lines, it was found that a subpopulation of C–C motif chemokine receptor 7 (CCR7)-positive cells exhibited certain CSC characteristics, including sphere formation, tumorigenesis, metastatic potential, and CSC marker expression [79]. These CCR7-positive melanoma CSCs also exhibited reduced MHC-I molecule and CD155/PVR expression, but upregulation of ligands for NKp30 and NKp46 [79]. Consistently, allogenic NK cells more efficiently lysed CCR7-positive CSCs compared with non-CSCs, although CCR7-positive CSCs also displayed higher expression of the two immune checkpoint ligands PD-L1 and galectin-9 [79]. As a result of NKp30 ligand expression (NKp46, DNAM-1, and NKG2D), medulloblastoma cells were sensitive to NK-cell-mediated cell lysis, regardless of their CD133 expression status [80]. Further investigation on the expression of NK-cell-activating receptor ligands in CSCs may benefit the development of NK-cell-based immunotherapy.
It was demonstrated that glioblastoma CSCs were more sensitive to NK-cell-mediated cytotoxicity compared with non-CSCs because of the higher expression of NKG2D ligands including UL16 binding protein 1 (ULBP1) and ULBP3 [81]. In contrast, another report [82] showed that glioblastoma CSCs were resistant to NK cells. However, when NK cells were activated by lectin and IL-2, these CSCs were more efficiently killed by the NK cells compared with non-CSCs [82]. Primary cancer cells isolated from nine patients with glioblastoma expressed neuronal stem cell markers and exhibited a multi-lineage differentiation potential when cultured in a stem cell medium [83]. Glioblastoma CSCs derived from all patients expressed HLA-I molecules (A, B, C) and HLA-E, which suppressed the activation and function of NK cells through killer immunoglobulin-like receptors (KIRs) and the NKG2A receptor, respectively [83,84]. However, these CSCs concomitantly expressed DNAM-1 ligands, CD155/PVR and CD112/Nectin-2, and showed low, but detectable, expression of one of the NKG2D ligands (MICA/MICB and ULBPs) [83]. Consistently, allogenic and autologous NK cells activated by IL-2 or IL-15 efficiently lysed these glioma CSCs [83].
As described in this section, NK-cell-based immunotherapy is expected to efficiently eliminate CSCs [85]. These NK cells need to be activated by cytokines to exhibit complete NK activity. Inevitably, cytokine-induced killer cells (CIKs) attract attention for immune cell therapy targeting CSCs. CIKs are induced from peripheral blood mononuclear cells following treatment with specific cytokines, such as IL-2 and IFNγ [86]. Accordingly, CIKs are a mixture of CD3−/CD56+ NK cells, CD3+/CD56− T cells, and CD3+/CD56+ bona fide CIKs [86]. CD3+/CD56+ CIKs express the NK-activating receptor, NKG2D, and exhibit MHC-unrestricted cytotoxicity against a wide variety of malignant cells, possibly through NKG2D, but not toward normal cells [86]. CIKs were shown to kill osteosarcoma CSCs without enriching CSCs [87]. CIKs were also shown to directly lyse nasopharyngeal CSCs in an NKG2D-dependent manner and prolong survival in patients with nasopharyngeal carcinoma treated with gemcitabine and cisplatin [88,89]. Moreover, CIKs combined with DCs also demonstrated significant therapeutic effects on patients with metastatic breast cancer, advanced colorectal cancer, and metastatic renal cell carcinoma [[90], [91], [92]]. CIK-mediated cytotoxicity against CSCs was also experimentally demonstrated in lymphoma [93], melanoma [94,95], and hepatocellular carcinoma [96].
3.5. NK-cell-resistant phenotype of CSCs
As described above, many CSCs exhibit downregulation of MHC-I molecules and upregulation of NK-cell-activating receptor ligands. They are susceptible to NK-cell-mediated cell lysis and, in some cases, more compared with non-CSCs. However, the fact that CSCs, not non-CSCs, can initiate tumor formation in NOD/SCID mice in which NK activity is low but detectable [16] indicates that tumor-initiating cells have an immune evasion ability, in particular, from NK cells. Hereafter, we present studies demonstrating the ability of CSCs to evade NK-cell surveillance.
It has been shown that adult T-cell leukemia downregulates NKG2D ligands, concomitant with increased expression of CD44 and the EMT marker, vimentin, and acquires an apoptosis-resistant phenotype [97]. Glioblastoma CSCs also downregulate NK-cell-activating receptor ligands, including MICA/MICB and ULBPs, and are resistant to NK-cell-mediated cytotoxicity [41]. Downregulation of NK-cell-activating receptor ligands was also observed in colon cancer cells with activated STAT3, a transcription factor that regulates colon CSCs [98,99]. STAT3 suppression in colon cancer cells induced MICA expression, which enhanced NK-cell activation [99], suggesting that STAT3 confers not only CSC properties but also NK-cell resistance.
CSCs may upregulate MHC-I molecules to acquire NK-cell resistance. It was shown that EPCAM+/CD45+ ascitic cells in patients with epithelial ovarian carcinoma possessed CSC properties, including drug resistance, high metastatic potential, and high CSC marker expression [100]. These cells also upregulated MHC-I molecules and exhibited resistance to NK-cell-mediated cell lysis by suppressing the activation of CD3−/CD56+ NK cells, whereas EPCAM single-positive cells were efficiently lysed by NK cells [100]. Another study reported that spheroid cells derived from a renal carcinoma cell line exhibited higher tumorigenicity and increased expression of stemness factors, including OCT4, NANOG, and BMI, compared with monolayer cells [101]. In these spheroid cells, MHC-I molecules were also significantly expressed, whereas NK-cell-activating receptor ligands were downregulated compared with monolayer cells. This suggests that these renal CSCs are less sensitive to NK-cell-mediated cell lysis [101].
HLA-G and HLA-E are nonclassical MHC-I molecules that may be involved in the underlying immune evasion mechanism of cancer cells from NK-cell surveillance [84,102,103]. KIR2DL4 and NKG2A on the surface of NK cells represent HLA-G- and HLA-E-specific receptors, respectively (Fig. 2), and provide an inhibitory signal to NK cells upon ligand binding [104]. In colorectal carcinoma tissues, the expression of HLA-G, HLA-E, and CSC markers was significantly higher compared with noncancerous colorectal tissues [105]. This suggests that HLA-G and HLA-E are involved in immune evasion of colorectal CSCs from NK-cell surveillance. Another study demonstrated that HLA-E was expressed in CSCs from patient-derived glioblastoma cell lines, although the expression levels were different among individual cell lines [106]. In contrast, under differentiation culture conditions, HLA-E expression was downregulated in some glioblastoma cell lines [106]. Consistently, the differentiated glioblastoma cells exhibited increased sensitivity to NK-cell-mediated cell lysis. Knockdown of HLA-E sensitized the glioblastoma cells to NK cells regardless of CD133 status [106], suggesting that HLA-E contributes to immune evasion in glioblastoma. Notably, stimulation with IFNγ induced HLA-E expression in all glioblastoma CSCs [106]. However, it has also been suggested that IFNγ enhances the immunogenicity of CSCs by upregulating MHC-I molecules [48]. Therefore, these results suggest that HLA-E is an important factor for glioblastoma CSCs to survive immune selection. Interestingly, HLA-G regulates the cell surface localization of HLA-E and thereby indirectly inhibits NK-cell activation [107]. HLA-G also has additional effects against DCs. CD105-positive renal CSCs exhibited increased expression of HLA-G, which was delivered to DCs by extracellular vesicles (EVs) [108]. Consistently, the EVs secreted by these CSCs inhibited the maturation of DCs more significantly compared with those secreted by CD105-negative non-CSCs [108]. In contrast to human HLA-G, the murine homolog, Qa-2, has been reported to be an immunogenic molecule and contribute to tumor rejection [109]. In murine CD44+/CD24− breast CSCs, Qa-2 expression was abrogated, possibly by the Src signaling pathway, whereas Src inhibitor treatment suppressed the CSC properties of breast cancer cells and restored Qa-2 expression [110]. Moreover, Qa-2 overexpression suppressed breast cancer growth and metastasis [110]. Therefore, caution is needed when interpreting the Qa-2 and HLA-G data obtained from mouse studies.
3.6. Inhibition of cytolytic granules by CSCs
Upon recognition of target cells, CTL and NK cells release cytolytic granules that contain perforin and granzymes (Fig. 2). Perforin oligomerizes and forms pores in the membranes of target cells. At high concentrations, perforin induces necrosis because of a loss of membrane integrity, whereas perforin at low concentrations induces apoptosis in a granzyme-dependent manner, which are delivered to the cytosol through perforin-formed pores or by perforin-triggered endocytosis [111]. Granzymes are serine proteases that activate pro-caspases and cleave various nuclear and mitochondrial proteins [112]. CSCs deploy several strategies to inhibit these cytolytic molecules. In spheroids of an estrogen receptor-α-positive breast cancer cell line, protease inhibitor 9 (PI9), a potent granzyme B inhibitor, was shown to be upregulated [113]. Estrogen enhanced spheroid formation and increased PI9 expression in the cells [113]. These results suggest that breast CSCs acquire resistance to CTL- and NK-cell-mediated cell lysis by PI9.
Serglycin, a proteoglycan with high affinity for granzyme B, contributes to the stabilization of granzyme B in lytic granules and delivery to target cells. Internalization of granzyme B into target cells requires an electrostatic exchange from serglycin to cell surface proteoglycans [114]. Mast cells infiltrated into glioma tissues induced serglycin expression, as well as CD44 and ZEB1, CSC and EMT markers, respectively [115]. Moreover, serglycin expression in glioma tissues was significantly associated with poor prognosis [115]. These results suggest that mast cell-induced serglycin contributes to the immune evasion of glioma CSCs from CTL and NK cells. However, multiple functions of serglycin [116] suggest that other mechanisms may also be involved in the malignant function of serglycin.
Watanabe et al. found that calreticulin was upregulated on the cell surface of pancreatic CSCs and that the expression of calreticulin was significantly associated with poor prognosis in patients with pancreatic cancer [117]. Calreticulin is a chaperone protein present in cytolytic granules of CTL and NK cells to prevent the unnecessary activation of perforin [118,119]. Accordingly, pancreatic CSCs may escape perforin-mediated cytolysis by expressing calreticulin. However, calreticulin also appears on the cell surface of apoptotic cells by binding to phosphatidylserine and promotes phagocytosis as an “eat-me signal” [120,121]. Therefore, further studies are necessary to clarify the precise role of calcineurin in CSC.
3.7. Humoral factors modulating immune response to CSCs
A number of humoral factors are involved in the regulation of immune response, which is also the case with immune evasion by CSCs. Such immunomodulatory factors are secreted not only by CSCs but also by tumor-associated noncancerous cells. In this section, we nonetheless summarized the factors that are secreted by CSCs or directly influence the immune evasion property of CSCs.
TGFβ is a multifunctional cytokine that induces CSCs and immune suppression, as well as EMT [122,123]. CD44+/CD24− breast CSCs were found to secrete a higher level of TGFβ compared with non-CSCs. In these CSCs, activation of the TGFβ signaling pathway was observed [124,125]. Consequently, TGFβ induced an EMT phenotype and enhanced stemness in not only breast cancer cells but also in normal mammary epithelial cells [[126], [127], [128]]. In contrast, metformin suppressed the TGFβ-induced EMT phenotype, self-renewal, and proliferation of trastuzumab-resistant breast CSCs [127,128]. In breast CSCs, the Sca-1 stem cell marker was suggested to be involved in TGFβ signaling [129]. It was shown that the human Sca-1 homologs Ly6K and Ly6E play an important role in TGFβ-induced SMAD2/SMAD3 activation, induced immune evasion, and eventually were associated with poor prognosis in patients with breast cancer [130]. In glioma CSCs, another CSC marker, CD133, was suggested to be involved in TGFβ production since CD133+ glioma CSCs exhibited higher expression of TGFβ compared with CD133− glioma CSCs [129]. Cytomegalovirus (CMV), with which glioma cells are frequently infected, is also suggested to be involved in TGFβ production in glioma. Interestingly, CMV-infected glioma CSCs secreted CMV IL-10 encoded by the CMV genome, induced immunosuppressive M2 macrophages, and enhanced the production of TGFβ by monocytes [131]. The CSC- or monocyte-induced TGFβ, in turn, downregulated NKG2D and MHC-II molecules in glioblastoma cells [132,133]. In addition, TGFβ secreted from glioblastoma CSCs induced the differentiation of Tregs from naïve T cells [42], enabling the escape of glioblastoma CSCs from immune recognition. Meanwhile, retinoic acid-inducible gene I (RIG-I) exhibits suppressive effects on virus infection and CSC induction. Consistently, RIG-I knockdown increased CSC marker expression in hepatocellular carcinoma [134]. These hepatic CSCs secreted TGFβ to enhance hepatic CSC properties through the SMAD2/AKT pathway in an autocrine manner [134]. TGFβ further inhibited the differentiation and maturation of DCs and recruited immature DCs with decreased expression of costimulatory molecules to hepatocellular carcinoma tissues [134]. Another mechanism involving urokinase-type plasminogen activator receptor (uPAR) was proposed for TGFβ production in CSCs. In breast cancer, uPAR increased the CD44+/CD24− CSC population with upregulation of a breast CSC marker, integrin α1β6, and enhanced tumor initiation [135]. The expression of uPAR was found in breast cancer, pancreatic cancer, and glioblastoma and was associated with the secretion of TGFβ and IL-4 [136]. These immunosuppressive cytokines induced the differentiation of macrophages into an arginase 1-positive M2 phenotype and thereby created an immunosuppressive tumor microenvironment [136]. These results suggest that TGFβ is not only a direct target of cancer therapy but also enhances tumor immunity. Other TGFβ family members, such as activin A and nodal, secreted by CSCs in pancreatic adenocarcinoma promoted cathelicidin-18 secretion from TAMs [137]. Cathelicidin-18, in turn, enhanced CSC function by binding to the formyl peptide receptor 2 and P2X purinoceptor 7 purinergic receptor [137]. The fact that TGFβ and nodal/activins share SMAD2/SMAD3 as downstream effectors renders these signaling molecules attractive targets for cancer therapy.
IL-6 is a pleiotropic cytokine that also plays an important role in the induction of CSCs. In lung cancer, IL-6 upregulated the expression of DNA methyltransferase (DNMT) 1 through JAK2/STAT3 signaling. This subsequently silenced transcription of cell cycle regulatory factors and thereby induced lung CSCs [138]. IL-6 secreted by TAMs was shown to induce and maintain CSC properties through STAT3 in breast cancer [139]. C/EBPδ also mediated IL-6- and hypoxia-induced breast CSCs by activating the NOTCH1 pathway [140]. In prostate cancer, IL-6 induced CD44+ CSCs in an autocrine manner [141]. IL-6 and CD44 expression was significantly associated with tumor malignancy and poor prognosis [141]. IL-6 also upregulated PD-L1 expression in CD44+ CSCs and recruited MDSCs to tumors and suppressed tumor infiltration of T cells [141]. Inhibition of the IL-6/STAT3 pathway resulted in the downregulation of PD-L1 and CD44 expression, decreased CSC properties, and suppressed tumor growth [141]. STAT3 is a transcription factor required for the maintenance of pluripotency in stem cells and plays an important role in the maintenance and induction of CSCs in both IL-6-dependent and IL-6-independent manners. In bladder CSCs, histone 3 K9 trimethylation by KMT1A upregulated STAT3 expression in an IL-6-independent manner by suppressing GATA3, which suppressed STAT3 transcription [142]. Consistently, deletion of the KMT1A gene decreased CSC properties in bladder cancer [142]. CD44+ CSCs of HNSCC exhibited constitutive activation of STAT3, which upregulated PD-L1 expression [143]. These results suggest that IL-6- or STAT3-targeting therapy will benefit patients with malignant tumors.
CCL20 is also implicated in the immune evasion of CSCs. Breast cancer cells induced CSC properties by secreting CCL20, which activated NFκB via PKCζ and p38MAPK [144]. NFκB, in turn, induced CCL20 transcription by forming a positive feedback loop [144]. CCL20 and its receptor, the CCR6 axis, may recruit Tregs to tumors of hepatocellular, esophageal, and squamous cell and lung cancers to promote tumor progression, metastasis, drug resistance, and poor prognosis as a consequence of enhanced immune evasion [[145], [146], [147], [148]].
3.8. Expression of immunosuppressive molecules in CSCs
3.8.1. PD-L1
It has been demonstrated that cancer cells also employ immune checkpoint molecules while downregulating costimulatory molecules to suppress tumor immunity (Table 1) [149]. PD-L1 expression has been frequently observed in tumor cells. Upon binding to PD-1 on immune cells, PD-L1 suppresses their effector functions, and induces their exhaustion [150].
PD-L1 was found to be expressed in patient-derived Lgr5+ gastric CSCs, and its expression in gastric CSCs was associated with tumor growth [151]. In breast cancer and malignant mesothelioma, PD-L1 expression was positively correlated with CSC marker expression [152,153]. Epithelial CSCs derived from the ascites of patients with bladder cancer also exhibited PD-L1 expression as well as E-cadherin, CD24, and VEGFR2, and had more potent tumorigenicity compared with mesenchymal CSCs that exhibited constitutively active TGFβ signaling [154]. An ALDH isozyme, ALDH3A1, was shown to increase CSC properties, EMT, and the expression of pro-inflammatory factors in non-small cell lung cancer and melanoma and was significantly correlated with PD-L1 expression [155]. Moreover, ALDH3A1 overexpression also upregulated PD-L1 expression, which inhibited the proliferation of peripheral immune cells [155]. It was demonstrated that the population of CD44+/CD133+ CSCs was higher in PD-L1high pancreatic tumors compared with PD-L1low tumors [156]. However, higher infiltration of CD8+ T cells was associated with a favorable prognosis for patients even with PD-L1high pancreatic tumors [156]. These observations suggest that CSCs clinically correlate with PD-L1 expression and rationalize the activation of T cells by immune checkpoint inhibitors to overcome the PD-L1-mediated immune checkpoint pathway.
It has been suggested that several factors are involved in PD-L1 expression in CSCs, although further studies are necessary. In breast cancer cells, it was determined that Ly6K/E were required for not only TGFβ-induced SMAD activation but also IL-4-induced PD-L1 expression [130]. Consistently, PD-L1 expression was significantly correlated with Ly6K/E [130]. Notably, proteasome activator subunit 3 (PSME3) was suggested to be involved in breast CSCs [157]. PSME3 overexpression upregulated PD-L1 expression, as well as EMT and CSC markers, leading to the suppression of chemotaxis of CD8+ T cells and the induction of T-cell apoptosis [157]. Conversely, PSME3 knockdown downregulated PD-L1 expression, promoted CD8+ T-cell activation, and suppressed tumor growth [157]. On the other hand, in ALDH1+-radioresistant CSCs of oral squamous cell carcinoma, PD-L1 and DNMT3B expression was positively correlated and associated with increased MDSC infiltration into tumors [158]. Interestingly, pharmacological inhibition of DNMT3B downregulated PD-L1 expression, impaired the radioresistance of ALDH1+ cells, and suppressed MDSC infiltration [158]. In CD133+/CD44+ colorectal CSCs and drug-resistant colorectal cancer cells, increased PD-L1 expression was observed [159]. Moreover, PD-L1 increased CSC properties and expanded the CSC population in colorectal cancer cells through the PI3K/AKT and MEK/ERK pathways that were activated by direct interaction of PD-L1 with HMGA1 [159]. Another report showed that the AT-rich interaction domain-containing protein 3B (ARID3B) recruited lysine-specific demethylase 4C (KDM4C) to chromatin in colorectal cancer, which subsequently induced the expression of NOTCH-target genes, intestinal stem genes, and PD-L1 [160]. Conversely, a KDM4C inhibitor decreased ARID3B-induced CSC properties and PD-L1 expression [160]. Insulin induced PD-L1 expression through the PI3K/AKT/mTOR pathway in colon CSCs [161]. Interestingly, EGF promoted the transfer of PD-L1 protein to the cell membrane, although it did not affect PD-L1 expression in colon CSCs [161]. On the contrary, in AML, STAT3 was also suggested to play a role in PD-L1 expression since STAT3 knockdown downregulated PD-L1 expression [162]. Consistently, STAT3 knockdown in combination with toll-like receptor 9 activation resulted in the activation of tumor immunity [162]. In thyroid CSCs, acetylcholine secreted by tumor-invading neurons also increased CSC properties and PD-L1 expression through the CD133/PI3K/AKT pathway, whereas an acetylcholine receptor antagonist, 4-DAMP, suppressed CSC properties and PD-L1 expression [163]. These results suggest that the mechanisms underlying PD-L1 expression in CSCs represent novel targets to improve the therapeutic efficacy of immune checkpoint inhibitors.
3.8.2. IFNs
IFNs are potent antiviral and antineoplastic cytokines. However, in a mouse model of chronic virus infection, the blockade of the type-I IFN pathway downregulated the immunosuppressive factors IL-10 and PD-L1 and resulted in a substantial reduction in viral titers in the infected animals [164,165]. This dual role of IFNs (immunostimulative or immunosuppressive) may, in part, depend on a period of IFN exposure time (acute phase or chronic phase) [164,165]. In tumors, it was shown that PD-L1 expression was induced by IFNγ secreted from activated T and NK cells in a negative feedback manner [166]. Moreover, IFNγ also induced PD-L1 expression in CD44+ CSCs of HNSCC, possibly through STAT1 [143]. In melanoma, IFNγ also upregulated the expression of CD271/NGFR and PD-L1 and thereby increased CSC properties via NGF stimulation [167]. Interestingly, avelumab is a fully humanized anti-PD-L1 IgG1 capable of inducing ADCC in addition to inhibiting the PD-1/PD-L1 pathway [168]. The PD-L1 expression in chordoma cells was upregulated by coculture with CD8+ T cells, leading to further enhancement of avelumab-induced ADCC [168]. Therefore, the clarification of the precise mechanisms of IFNγ-induced PD-L1 expression in CSCs may provide further improvement in PD-1-/PD-L1-targeted cancer therapy.
IFNs are also involved in CSC induction and maintenance [167]. In methylcholanthrene-induced sarcoma in mice, treatment with anti-PD-L1 or anti-CTLA-4 antibodies expanded the Sca-1+/CD90− CSC population, suggesting that an alternative immune evasion mechanism may be employed by the CSCs [169]. IFNγ is involved in the expansion of the Sca-1+/CD90− CSC population in methylcholanthrene-induced sarcoma because IFNγ blockade decreased its expansion [169]. On the other hand, IFNα upregulated the expression of CSC markers and induced the EMT phenotype in pancreatic adenocarcinoma [170]. Moreover, IFNβ secreted from pancreatic adenocarcinoma cells induced TAMs to produce IFN-stimulated gene 15, which subsequently promoted the induction and maintenance of pancreatic adenocarcinoma CSCs [171]. The underlying mechanism of IFN-induced CSC properties may be dependent, at least in part, on its concentration. Thus, IFNγ at low concentrations induced CSC properties in non-small cell lung cancer through the ICAM1/PI3K/AKT/NOTCH1 pathway, whereas it induced apoptosis through the JAK1/STAT1/caspase pathway at high concentrations [172].
IFNs inhibit the tumor-initiating property and expansion of CSCs [173], and the response to IFNs was suppressed in CSCs to resist the anti-neoplastic effects. The underlying mechanisms remain to be investigated; however, they involve the suppression of autophagy [174], cyclooxygenase 2, and its product, prostaglandin E2 [175], as well as the downregulation of LCOR [176] and IFN signaling molecules [173,177]. Therefore, the two-edged functions of IFNs must be considered in IFN-based cancer therapy.
3.8.3. CD200
CD200 provides a negative signal to CD200R-expressing macrophages and DCs to induce immune tolerance [178]. The CD200/CD200R axis may be involved in immune evasion of CSCs by suppressing T-cell activation and Th1 cytokine production and inducing regulatory DCs and Tregs [179]. For example, CD200 was coexpressed with CSC markers in various types of cancers [[180], [181], [182]]. The CD200 expression in leukemia predicted poor prognosis in multiple myeloma and AML [179,183]. In B cell lymphoma, the inhibition of CD200 by a neutralizing antibody or siRNA increased the CTL response and the expression of inflammatory cytokines (IFNγ and TNFα), resulting in lymphoma cell death [184]. In HNSCC, CD200 overexpression induced the expression of BMI1 and sonic hedgehog, both of which are involved in the maintenance of CSCs [182]. Moreover, CD200 promoted chemo- and radioresistance of squamous cell carcinoma in vivo [182]. Therefore, CD200 may also be involved in the induction of CSCs.
3.8.4. Other immunosuppressive molecules
Galectin-3, a β-galactoside-binding protein of the lectin family, impairs tumor immunity through several mechanisms that remain to be defined [185]. Lung CSCs were shown to express galectin-3, which was significantly associated with CD133 and β-catenin in lung cancer tissues [186]. Consistently, the overexpression of galectin-3 induced CSC properties by activating β-catenin, which promoted the transcription of stemness-related genes [186]. Galectin-3 also increased CSC properties of prostate cancer and endowed CSCs with immune evasion and metastatic properties [187].
Inhibitory leukocyte immunoglobulin-like receptors (LILRs) on immune cells receive a negative signal from MHC-I molecules and other unidentified ligands [188]. LILRB2 and LILRB3 were shown to be expressed in AML and lung cancer and promoted their tumorigenesis and stemness [189,190]. Fat mass- and obesity-associated protein (FTO) is an RNA N6-methyladenosine demethylase that expanded the CSC population of various cancers through LILRB4 upregulation [191]. Conversely, knockdown of FTO or treatment with the FTO-specific demethylase inhibitors, CS1 and CS2, downregulated LILRB4 expression in leukemia cells, resulting in the activation of the tumor immune response and the subsequent suppression of tumor growth [191].
As summarized in Table 1, it is anticipated that CSCs employ other molecules to create an immunosuppressive tumor microenvironment. Investigation of these mechanisms will assist in the development of novel immunotherapies.
3.9. Immune modulation by CSC-intrinsic functions
Compared with non-CSCs, CSCs retain a more intact DNA repair function that reduces immunogenicity [192]. As such, CSCs employ their intrinsic functions to evade tumor immunity. Antiapoptosis proteins, such as BCL2, BCLxL, and PI3K, may protect CSCs from T-cell- and NK-cell-induced apoptosis [193,194]. Apoptosis inhibitor-5, which was upregulated in CD44high CSCs, induced CSC properties and suppressed the antigen-specific T-cell response in a fibroblast growth factor 2-dependent manner [195,196]. STAT3 in CSCs may also be involved in the expression of galectin-3 [42], suppression of phagocytosis, and secretion of IL-10 from TAMs [197]. Likewise, several CSC-regulating molecules, including NANOG [198,199], SOX2 [200], DCLK1 [201,202], and CD73 [203,204], were shown to be involved in immune evasion by CSCs.
4. Conclusions
Tumor immunity has opened new avenues for cancer therapy; however, further improvements in efficacy are needed. The mechanisms through which cancer cells evade the immune response must be precisely clarified. In this article, we summarized studies that clarified how CSCs suppress tumor immunity and create an immunosuppressive milieu, which are expected to provide important clues for the development of novel tumor immunotherapies. Moreover, in agreement with the fact that cancer cells must evade tumor immunity to form tumors, our literature survey suggests that the immune evasion property, rather than the tumor-initiating property, is a more fundamental feature of CSCs. In the present article, the underlying mechanisms of immunosuppressive properties of CSCs were summarized as: 1) the impairment of antigen processing and presenting machineries; 2) the downregulation of tumor-associated antigens; 3) the downregulation of NK-cell-activating ligands and upregulation of NK-cell inhibitory ligands; 4) the inhibition of cytolytic granules released from CTL and NK cells; 5) the secretion of immune suppressive humoral factors, such as TGFβ, IL-6, and CCL20; 6) the upregulation of immunosuppressive molecules including immune checkpoint molecules; and, 7) others. In order to suppress tumor immunity, CSCs employ these mechanisms in combination. Therefore, CSCs are expected to be a more effective target to restore tumor immunity than targeting each of the mechanisms individually. However, because of space limitations, we have reluctantly omitted immune modulatory functions of noncancerous cells in the tumor microenvironment, which also endow CSCs with the immune evasion property and vice versa. Moreover, we would also like to emphasize that additional factors, including hypoxia, cancer-specific metabolites, and EMT, remain to be discussed but must be considered for the development of immunotherapies. We expect that an integrated understanding of these factors will lead to significant improvements of current cancer immunotherapies.
Declaration of competing interest
G.S. holds more than 5% of the total shares of KanonCure Inc., and receives compensation as a member of KanonCure Inc. The other author has no competing interests.
Acknowledgments
We would like to thank Enago (www.enago.jp) for the English language review. This work was supported in part by JSPS KAKENHI Grant Number JP19K08469 (H.T.).
Footnotes
Peer review under responsibility of the Japanese Society for Regenerative Medicine.
Contributor Information
Hiroyuki Tsuchiya, Email: tsuchiyah@tottori-u.ac.jp.
Goshi Shiota, Email: gshiota@tottori-u.ac.jp.
References
- 1.Procaccio L., Damuzzo V., Di Sarra F., Russi A., Todino F., Dadduzio V. Safety and tolerability of anti-angiogenic protein kinase inhibitors and vascular-disrupting agents in cancer: focus on gastrointestinal malignancies. Drug Saf. 2019;42:159–179. doi: 10.1007/s40264-018-0776-6. [DOI] [PubMed] [Google Scholar]
- 2.Kudo M. Systemic therapy for hepatocellular carcinoma: latest advances. Cancers. 2018;10:412. doi: 10.3390/cancers10110412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu T., Zhou L., Li D., Andl T., Zhang Y. Cancer-associated fibroblasts build and secure the tumor microenvironment. Front Cell Dev Biol. 2018;10:E412. doi: 10.3389/fcell.2019.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chanmee T., Ontong P., Konno K., Itano N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers. 2014;6:1670–1690. doi: 10.3390/cancers6031670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mishalian I., Bayuh R., Levy L., Zolotarov L., Michaeli J., Fridlender Z.G. Tumor-associated neutrophils (TAN) develop pro-tumorigenic properties during tumor progression. Cancer Immunol Immunother. 2013;62:1745–1756. doi: 10.1007/s00262-013-1476-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schreiber R.D., Old L.J., Smyth M.J. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 7.Chantrain C.F., Henriet P., Jodele S., Emonard H., Feron O., Courtoy P.J. Mechanisms of pericyte recruitment in tumour angiogenesis: a new role for metalloproteinases. Eur J Canc. 2016;42:310–318. doi: 10.1016/j.ejca.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 8.Paolillo M., Schinelli S. Extracellular matrix alterations in metastatic processes. Int J Mol Sci. 2019;20:4947. doi: 10.3390/ijms20194947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nejman D., Livyatan I., Fuks G., Gavert N., Zwang Y., Geller L.T. The human tumor microbiome is composed of tumor type–specific intracellular bacteria. Science. 2020;368:973–980. doi: 10.1126/science.aay9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao Q., Wang Z.C., Duan M., Lin Y.H., Zhou X.Y., Worthley D.L. Cell culture system for analysis of genetic heterogeneity within hepatocellular carcinomas and response to pharmacologic agents. Gastroenterology. 2017;152:232–242.e4. doi: 10.1053/j.gastro.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Huang A., Zhao X., Yang X.R., Li F.Q., Zhou X.L., Wu K. Circumventing intratumoral heterogeneity to identify potential therapeutic targets in hepatocellular carcinoma. J Hepatol. 2017;67:293–301. doi: 10.1016/j.jhep.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 12.Zheng H., Pomyen Y., Hernandez M.O., Li C., Livak F., Tang W. Single-cell analysis reveals cancer stem cell heterogeneity in hepatocellular carcinoma. Hepatology. 2018;68:127–140. doi: 10.1002/hep.29778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W., Quan Y., Fu Q., Liu Y., Liang Y., Wu J. Dynamics between cancer cell subpopulations reveals a model coordinating with both hierarchical and stochastic concepts. PloS One. 2014;9 doi: 10.1371/journal.pone.0084654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang X.F. Immunology of stem cells and cancer stem cells. Cell Mol Immunol. 2007;4:161–171. [PubMed] [Google Scholar]
- 15.Lapidot T., Sirard C., Vormoor J., Murdoch B., Hoang T., Caceres-Cortes J. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 16.Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quintana E., Shackleton M., Sabel M.S., Fullen D.R., Johnson T.M., Morrison S.J. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quintana E., Shackleton M., Foster H.R., Fullen D.R., Sabel M.S., Johnson T.M. Phenotypic heterogeneity among tumorigenic melanoma cells from patients that is reversible and not hierarchically organized. Canc Cell. 2010;18:510–523. doi: 10.1016/j.ccr.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Civenni G., Walter A., Kobert N., Mihic-Probst D., Zipser M., Belloni B. Human CD271-positive melanoma stem cells associated with metastasis establish tumor heterogeneity and long-term growth. Canc Res. 2011;71:3098–3109. doi: 10.1158/0008-5472.CAN-10-3997. [DOI] [PubMed] [Google Scholar]
- 20.Rountree C.B., Ding W., He L., Stiles B. Expansion of CD133-expressing liver cancer stem cells in liver-specific phosphatase and tensin homolog deleted on chromosome 10-deleted mice. Stem Cell. 2009;27:290–299. doi: 10.1634/stemcells.2008-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schatton T., Frank M.H. Antitumor immunity and cancer stem cells. Ann N Y Acad Sci. 2009;1176:154–169. doi: 10.1111/j.1749-6632.2009.04568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma J., Frank M.H. Tumor initiation in human malignant melanoma and potential cancer therapies. Anticancer Agents Med Chem. 2010;10:131–136. doi: 10.2174/187152010790909254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim S.Y., Cho H.S., Yang S.H., Shin J.Y., Kim J.S., Lee S.T. Soluble mediators from human neural stem cells play a critical role in suppression of T-cell activation and proliferation. J Neurosci Res. 2009;87:2264–2272. doi: 10.1002/jnr.22050. [DOI] [PubMed] [Google Scholar]
- 24.Ljujic B., Milovanovic M., Volarevic V., Murray B., Bugarski D., Przyborski S. Human mesenchymal stem cells creating an immunosuppressive environment and promote breast cancer in mice. Sci Rep. 2013;3:2298. doi: 10.1038/srep02298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H., Yang M., Lin L., Ren H., Lin C., Lin S. HepG2 cells acquire stem cell-like characteristics after immune cell stimulation. Cell Oncol. 2016;39:35–45. doi: 10.1007/s13402-015-0249-1. [DOI] [PubMed] [Google Scholar]
- 26.Santisteban M., Reiman J.M., Asiedu M.K., Behrens M.D., Nassar A., Kalli K.R. Immune-induced epithelial to mesenchymal transition in vivo generates breast cancer stem cells. Canc Res. 2009;69:2887–2895. doi: 10.1158/0008-5472.CAN-08-3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reim F., Dombrowski Y., Ritter C., Buttmann M., Häusler S., Ossadnik M. Immunoselection of breast and ovarian cancer cells with trastuzumab and natural killer cells: selective escape of CD44high/CD24low/HER2low breast cancer stem cells. Canc Res. 2009;69:8058–8066. doi: 10.1158/0008-5472.CAN-09-0834. [DOI] [PubMed] [Google Scholar]
- 28.Golan H., Shukrun R., Caspi R., Vax E., Pode-Shakked N., Goldberg S. In Vivo expansion of cancer stemness affords novel cancer stem cell targets: malignant rhabdoid tumor as an example. Stem Cell Reports. 2018;11:795–810. doi: 10.1016/j.stemcr.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irvin D.K., Jouanneau E., Duvall G., Zhang X.X., Zhai Y., Sarayba D. T cells enhance stem-like properties and conditional malignancy in gliomas. PloS One. 2010;5 doi: 10.1371/journal.pone.0010974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bruttel V.S., Wischhusen J. Cancer stem cell immunology: key to understanding tumorigenesis and tumor immune escape? Front Immunol. 2014;5:360. doi: 10.3389/fimmu.2014.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ignatius M.S., Langenau D.M. Zebrafish as a model for cancer self-renewal. Zebrafish. 2009;6:377–387. doi: 10.1089/zeb.2009.0610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Albini A., Bruno A., Gallo C., Pajardi G., Noonan D.M., Dallaglio K. Cancer stem cells and the tumor microenvironment: interplay in tumor heterogeneity. Connect Tissue Res. 2015;56:414–425. doi: 10.3109/03008207.2015.1066780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hasegawa K., Tanaka S., Fujiki F., Morimoto S., Nakajima H., Tatsumi N. An immunocompetent mouse model for MLL/AF9 leukemia reveals the potential of spontaneous cytotoxic T-cell response to an antigen expressed in leukemia cells. PloS One. 2015;10 doi: 10.1371/journal.pone.0144594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu F., Li S., Zhang J., Wang L., Wu X., Wang J. Cancer stemness, immune cells, and epithelial-mesenchymal transition cooperatively predict prognosis in colorectal carcinoma. Clin Colorectal Canc. 2018;17:e579–e592. doi: 10.1016/j.clcc.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 35.Sang X., Wu F., Wu D., Lin S., Li J., Zhao N. Human hepatic cancer stem cells (HCSCs) markers correlated with immune infiltrates reveal prognostic significance of hepatocellular carcinoma. Front Genet. 2020;11:112. doi: 10.3389/fgene.2020.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koyama S., Tsuchiya H., Amisaki M., Sakaguchi H., Honjo S., Fujiwara Y. NEAT1 is required for the expression of the liver cancer stem cell marker CD44. Int J Mol Sci. 2020;21:1927. doi: 10.3390/ijms21061927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramgolam K., Lauriol J., Lalou C., Lauden L., Michel L., de la Grange P. Melanoma spheroids grown under neural crest cell conditions are highly plastic migratory/invasive tumor cells endowed with immunomodulator function. PloS One. 2011;6 doi: 10.1371/journal.pone.0018784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iovino F., Meraviglia S., Spina M., Orlando V., Saladino V., Dieli F. Immunotherapy targeting colon cancer stem cells. Immunotherapy. 2011;3:97–106. doi: 10.2217/imt.10.87. [DOI] [PubMed] [Google Scholar]
- 39.Le Blanc K., Tammik C., Rosendahl K., Zetterberg E., Ringdén O. HLA expression and immunologic properties of differentiated and undifferentiated mesenchymal stem cells. Exp Hematol. 2003;31:890–896. doi: 10.1016/s0301-472x(03)00110-3. [DOI] [PubMed] [Google Scholar]
- 40.Agudo J., Park E.S., Rose S.A., Alibo E., Sweeney R., Dhainaut M. Quiescent tissue stem cells evade immune surveillance. Immunity. 2018;48:271–285. doi: 10.1016/j.immuni.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Tomaso T., Mazzoleni S., Wang E., Sovena G., Clavenna D., Franzin A. Immunobiological characterization of cancer stem cells isolated from glioblastoma patients. Clin Canc Res. 2010;16:800–813. doi: 10.1158/1078-0432.CCR-09-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei J., Barr J., Kong L.Y., Wang Y., Wu A., Sharma A.K. Glioblastoma cancer-initiating cells inhibit T-cell proliferation and effector responses by the signal transducers and activators of transcription 3 pathway. Mol Canc Therapeut. 2010;9:67–78. doi: 10.1158/1535-7163.MCT-09-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chikamatsu K., Takahashi G., Sakakura K., Ferrone S., Masuyama K. Immunoregulatory properties of CD44+ cancer stem-like cells in squamous cell carcinoma of the head and neck. Head Neck. 2011;33:208–215. doi: 10.1002/hed.21420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao T., Kaufmann A.M., Qian X., Sangvatanakul V., Chen C., Kube T. Susceptibility to cytotoxic T cell lysis of cancer stem cells derived from cervical and head and neck tumor cell lines. J Canc Res Clin Oncol. 2013;139:159–170. doi: 10.1007/s00432-012-1311-2. [DOI] [PubMed] [Google Scholar]
- 45.Sultan M., Vidovic D., Paine A.S., Huynh T.T., Coyle K.M., Thomas M.L. Epigenetic silencing of TAP1 in Aldefluor+ breast cancer stem cells contributes to their enhanced immune evasion. Stem Cell. 2018;36:641–654. doi: 10.1002/stem.2780. [DOI] [PubMed] [Google Scholar]
- 46.Espinoza-Sánchez N.A., Vadillo E., Balandrán J.C., Monroy-García A., Pelayo R., Fuentes-Pananá E.M. Evidence of lateral transmission of aggressive features between different types of breast cancer cells. Int J Oncol. 2017;51:1482–1496. doi: 10.3892/ijo.2017.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsuchiya H., Shiota G. Clinical and biological implications of cancer stem cells in hepatocellular carcinoma. Yonago Acta Med. 2021;64:1–11. doi: 10.33160/yam.2021.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morrison B.J., Steel J.C., Morris J.C. Reduction of MHC-I expression limits T-lymphocyte-mediated killing of Cancer-initiating cells. BMC Canc. 2018;18:469. doi: 10.1186/s12885-018-4389-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Busse A., Letsch A., Fusi A., Nonnenmacher A., Stather D., Ochsenreither S. Characterization of small spheres derived from various solid tumor cell lines: are they suitable targets for T cells? Clin Exp Metastasis. 2013;30:781–791. doi: 10.1007/s10585-013-9578-5. [DOI] [PubMed] [Google Scholar]
- 50.Zagzag D., Salnikow K., Chiriboga L., Yee H., Lan L., Ali M.A. Downregulation of major histocompatibility complex antigens in invading glioma cells: stealth invasion of the brain. Lab Invest. 2005;85:328–341. doi: 10.1038/labinvest.3700233. [DOI] [PubMed] [Google Scholar]
- 51.Palesch D., Wagner J., Meid A. Cathepsin G-mediated proteolytic degradation of MHC class I molecules to facilitate immune detection of human glioblastoma cells. Cancer Immunol Immun. 2016;65:283–291. doi: 10.1007/s00262-016-1798-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Girouard S.D., Murphy G.F. Melanoma stem cells: not rare, but well done. Lab Invest. 2011;91:647–664. doi: 10.1038/labinvest.2011.50. [DOI] [PubMed] [Google Scholar]
- 53.Schatton T., Schütte U., Frank N.Y., Zhan Q., Hoerning A., Robles S.C. Modulation of T-cell activation by malignant melanoma initiating cells. Canc Res. 2010;70:697–708. doi: 10.1158/0008-5472.CAN-09-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boiko A.D., Razorenova O.V., van de Rijn M., Swetter S.M., Johnson D.L., Ly D.P. Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature. 2010;466:133–137. doi: 10.1038/nature09161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen T., Liu K., Xu J., Zhan T., Liu M., Li L. Synthetic and immunological studies on the OCT4 immunodominant motif antigen-based anti-cancer vaccine. Cancer Biol Med. 2020;17:132–141. doi: 10.20892/j.issn.2095-3941.2019.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Di J., Duiveman-de Boer T., Figdor C.G., Torensma R. Aiming to immune elimination of ovarian cancer stem cells. World J Stem Cell. 2013;5:149–162. doi: 10.4252/wjsc.v5.i4.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Di J., Massuger L.F., Duiveman-de Boer T., Zusterzeel P.L., Figdor C.G., Torensma R. Functional OCT4-specific CD4+ and CD8+ T cells in healthy controls and ovarian cancer patients. OncoImmunology. 2013;2 doi: 10.4161/onci.24271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Atakan S., Bayiz H., Sak S., Poyraz A., Vural B., Yildirim A.S. Autologous anti-SOX2 antibody responses reflect intensity but not frequency of antigen expression in small cell lung cancer. BMC Clin Pathol. 2014;14:24. doi: 10.1186/1472-6890-14-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shih J., Rahman M., Luong Q.T., Lomeli S.H., Riss J., Prins R.M. Dominant B-cell epitopes from cancer/stem cell antigen SOX2 recognized by serum samples from cancer patients. Am J Clin Exp Immunol. 2014;3:84–90. [PMC free article] [PubMed] [Google Scholar]
- 60.Spisek R., Kukreja A., Chen L.C., Matthews P., Mazumder A., Vesole D. Frequent and specific immunity to the embryonal stem cell-associated antigen SOX2 in patients with monoclonal gammopathy. J Exp Med. 2007;204:831–840. doi: 10.1084/jem.20062387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kato Y., Ohishi T., Yamada S., Itai S., Furusawa Y., Sano M. Anti-CD133 monoclonal antibody CMab-43 exerts antitumor activity in a mouse xenograft model of colon cancer. Monoclon Antib Immunodiagn Immunother. 2019;38:75–78. doi: 10.1089/mab.2019.0002. [DOI] [PubMed] [Google Scholar]
- 62.Ji J., Judkowski V.A., Liu G., Wang H., Bunying A., Li Z. Identification of novel human leukocyte antigen-A∗0201-restricted, cytotoxic T lymphocyte epitopes on CD133 for cancer stem cell immunotherapy. Stem Cells Transl Med. 2014;3:356–364. doi: 10.5966/sctm.2013-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Du Y.R., Chen Y., Gao Y., Niu X.L., Li Y.J., Deng W.M. Effects and mechanisms of anti-CD44 monoclonal antibody A3D8 on proliferation and apoptosis of sphere-forming cells with stemness from human ovarian cancer. Int J Gynecol Canc. 2013;23:1367–1375. doi: 10.1097/IGC.0b013e3182a1d023. [DOI] [PubMed] [Google Scholar]
- 64.Jin L., Hope K.J., Zhai Q., Smadja-Joffe F., Dick J.E. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006;12:1167–1174. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- 65.Salnikov A.V., Bretz N.P., Perne C., Hazin J., Keller S., Fogel M. Antibody targeting of CD24 efficiently retards growth and influences cytokine milieu in experimental carcinomas. Br J Canc. 2013;108:1449–1459. doi: 10.1038/bjc.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jäger M., Schoberth A., Ruf P., Hess J., Hennig M., Schmalfeldt B. Immunomonitoring results of a phase II/III study of malignant ascites patients treated with the trifunctional antibody catumaxomab (anti-EpCAM x anti-CD3) Canc Res. 2012;72:24–32. doi: 10.1158/0008-5472.CAN-11-2235. [DOI] [PubMed] [Google Scholar]
- 67.Kubo M., Umebayashi M., Kurata K., Mori H., Kai M., Onishi H. Catumaxomab with activated T-cells efficiently lyses chemoresistant EpCAM-positive triple-negative breast cancer cell lines. Anticancer Res. 2018;38:4273–4279. doi: 10.21873/anticanres.12724. [DOI] [PubMed] [Google Scholar]
- 68.Sato N., Hirohashi Y., Tsukahara T., Kikuchi T., Sahara H., Kamiguchi K. Molecular pathological approaches to human tumor immunology. Pathol Int. 2009;59:205–217. doi: 10.1111/j.1440-1827.2009.02353.x. [DOI] [PubMed] [Google Scholar]
- 69.Tallerico R., Garofalo C., Carbone E. A new biological feature of natural killer cells: the recognition of solid tumor-derived cancer stem cells. Front Immunol. 2016;7:179. doi: 10.3389/fimmu.2016.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Klingemann H.G. Cellular therapy of cancer with natural killer cells-where do we stand? Cytotherapy. 2013;15:1185–1194. doi: 10.1016/j.jcyt.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 71.Jewett A., Tseng H.C., Arasteh A., Saadat S., Christensen R.E., Cacalano N.A. Natural killer cells preferentially target cancer stem cells; role of monocytes in protection against NK cell mediated lysis of cancer stem cells. Curr Drug Deliv. 2012;9:5–16. doi: 10.2174/156720112798375989. [DOI] [PubMed] [Google Scholar]
- 72.Koh J., Lee S.B., Park H., Lee H.J., Cho N.H., Kim J. Susceptibility of CD24(+) ovarian cancer cells to anti-cancer drugs and natural killer cells. Biochem Biophys Res Commun. 2012;427:373–378. doi: 10.1016/j.bbrc.2012.09.067. [DOI] [PubMed] [Google Scholar]
- 73.Ames E., Canter R.J., Grossenbacher S.K., Mac S., Chen M., Smith R.C. NK cells preferentially target tumor cells with a cancer stem cell phenotype. J Immunol. 2015;195:4010–4019. doi: 10.4049/jimmunol.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vrazo A.C., Hontz A.E., Figueira S.K., Butler B.L., Ferrell J.M., Binkowski B.F. Live cell evaluation of granzyme delivery and death receptor signaling in tumor cells targeted by human natural killer cells. Blood. 2015;126(8):e1–e10. doi: 10.1182/blood-2015-03-632273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sussman R.T., Ricci M.S., Hart L.S., Sun S.Y., El-Deiry W.S. Chemotherapy-resistant side-population of colon cancer cells has a higher sensitivity to TRAIL than the non-SP, a higher expression of c-Myc and TRAIL-receptor DR4. Canc Biol Ther. 2007;6:1490–1495. doi: 10.4161/cbt.6.9.4905. [DOI] [PubMed] [Google Scholar]
- 76.Li M., Knight D.A., Smyth M.J., Stewart T.J. Sensitivity of a novel model of mammary cancer stem cell-like cells to TNF-related death pathways. Cancer Immunol Immunother. 2012;61:1255–1268. doi: 10.1007/s00262-012-1200-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gras Navarro A., Björklund A.T., Chekenya M. Therapeutic potential and challenges of natural killer cells in treatment of solid tumors. Front Immunol. 2015;6:202. doi: 10.3389/fimmu.2015.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tallerico R., Todaro M., Di Franco S., Maccalli C., Garofalo C., Sottile R. Human NK cells selective targeting of colon cancer-initiating cells: a role for natural cytotoxicity receptors and MHC class I molecules. J Immunol. 2013;190:2381–2390. doi: 10.4049/jimmunol.1201542. [DOI] [PubMed] [Google Scholar]
- 79.Cristiani C.M., Turdo A., Ventura V., Apuzzo T., Capone M., Madonna G. Accumulation of circulating CCR7+ natural killer cells marks melanoma evolution and reveals a CCL19-dependent metastatic pathway. Cancer Immunol Res. 2019;7:841–852. doi: 10.1158/2326-6066.CIR-18-0651. [DOI] [PubMed] [Google Scholar]
- 80.Castriconi R., Dondero A., Negri F., Bellora F., Nozza P., Carnemolla B. Both CD133+ and CD133- medulloblastoma cell lines express ligands for triggering NK receptors and are susceptible to NK-mediated cytotoxicity. Eur J Immunol. 2007;37:3190–3196. doi: 10.1002/eji.200737546. [DOI] [PubMed] [Google Scholar]
- 81.Oh S.J., Yang J.I., Kim O., Ahn E.J., Kang W.D., Lee J.H. Human U87 glioblastoma cells with stemness features display enhanced sensitivity to natural killer cell cytotoxicity through altered expression of NKG2D ligand. Canc Cell Int. 2017;17:22. doi: 10.1186/s12935-017-0397-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Avril T., Vauleon E., Hamlat A., Saikali S., Etcheverry A., Delmas C. Human glioblastoma stem-like cells are more sensitive to allogeneic NK and T cell-mediated killing compared with serum-cultured glioblastoma cells. Brain Pathol. 2012;22:159–174. doi: 10.1111/j.1750-3639.2011.00515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Castriconi R., Daga A., Dondero A., Zona G., Poliani P.L., Melotti A. NK cells recognize and kill human glioblastoma cells with stem cell-like properties. J Immunol. 2009;182:3530–3539. doi: 10.4049/jimmunol.0802845. [DOI] [PubMed] [Google Scholar]
- 84.Iwaszko M., Bogunia-Kubik K. Clinical significance of the HLA-E and CD94/NKG2 interaction. Arch Immunol Ther Exp. 2011;59:353–367. doi: 10.1007/s00005-011-0137-y. [DOI] [PubMed] [Google Scholar]
- 85.Melaiu O., Lucarini V., Cifaldi L., Fruci D. Influence of the tumor microenvironment on NK cell function in solid tumors. Front Immunol. 2020;10:3038. doi: 10.3389/fimmu.2019.03038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Introna M. CIK as therapeutic agents against tumors. J Autoimmun. 2017;85:32–44. doi: 10.1016/j.jaut.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 87.Sangiolo D., Mesiano G., Gammaitoni L., Leuci V., Todorovic M., Giraudo L. Cytokine-induced killer cells eradicate bone and soft-tissue sarcomas. Canc Res. 2014;74:119–129. doi: 10.1158/0008-5472.CAN-13-1559. [DOI] [PubMed] [Google Scholar]
- 88.Wei F., Rong X.X., Xie R.Y., Jia L.T., Wang H.Y., Qin Y.J. Cytokine-induced killer cells efficiently kill stem-like cancer cells of nasopharyngeal carcinoma via the NKG2D-ligands recognition. Oncotarget. 2015;6:35023–35039. doi: 10.18632/oncotarget.5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li J.J., Gu M.F., Pan K., Liu L.Z., Zhang H., Shen W.X. Autologous cytokine-induced killer cell transfusion in combination with gemcitabine plus cisplatin regimen chemotherapy for metastatic nasopharyngeal carcinoma. J Immunother. 2012;35:189–195. doi: 10.1097/CJI.0b013e318241d9de. [DOI] [PubMed] [Google Scholar]
- 90.Ren J., Di L., Song G., Yu J., Jia J., Zhu Y. Selections of appropriate regimen of high-dose chemotherapy combined with adoptive cellular therapy with dendritic and cytokine-induced killer cells improved progression-free and overall survival in patients with metastatic breast cancer: reargument of such contentious therapeutic preferences. Clin Transl Oncol. 2013;15:780–788. doi: 10.1007/s12094-013-1001-9. [DOI] [PubMed] [Google Scholar]
- 91.Xie Y., Huang L., Chen L., Lin X., Chen L., Zheng Q. Effect of dendritic cell-cytokine-induced killer cells in patients with advanced colorectal cancer combined with first-line treatment. World J Surg Oncol. 2017;15:209. doi: 10.1186/s12957-017-1278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mai H.X., Mei G.H., Zhao F.L., Li B.T., Tang Y.Y., Zhang B. Retrospective analysis on the efficacy of sunitinib/sorafenib in combination with dendritic cells-cytokine-induced killer in metastasis renal cell carcinoma after radical nephrectomy. J Canc Res Therapeut. 2018;14:S427–S432. doi: 10.4103/0973-1482.180609. [DOI] [PubMed] [Google Scholar]
- 93.Contag C.H., Sikorski R., Negrin R.S., Schmidt T., Fan A.C., Bachireddy P. Definition of an enhanced immune cell therapy in mice that can target stem-like lymphoma cells. Canc Res. 2010;70:9837–9845. doi: 10.1158/0008-5472.CAN-10-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gammaitoni L., Giraudo L., Leuci V., Todorovic M., Mesiano G., Picciotto F. Effective activity of cytokine-induced killer cells against autologous metastatic melanoma including cells with stemness features. Clin Canc Res. 2013;19:4347–4358. doi: 10.1158/1078-0432.CCR-13-0061. [DOI] [PubMed] [Google Scholar]
- 95.Gammaitoni L., Giraudo L., Macagno M., Leuci V., Mesiano G., Rotolo R. Cytokine-induced killer cells kill chemo-surviving melanoma cancer stem cells. Clin Canc Res. 2017;23:2277–2288. doi: 10.1158/1078-0432.CCR-16-1524. [DOI] [PubMed] [Google Scholar]
- 96.Yang T., Zhang W., Wang L., Xiao C., Wang L., Gong Y. Co-culture of dendritic cells and cytokine-induced killer cells effectively suppresses liver cancer stem cell growth by inhibiting pathways in the immune system. BMC Canc. 2018;18:984. doi: 10.1186/s12885-018-4871-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Miyatake Y., Oliveira A.L., Jarboui M.A., Ota S., Tomaru U., Teshima T. Protective roles of epithelial cells in the survival of adult T-cell leukemia/lymphoma cells. Am J Pathol. 2013;182:1832–1842. doi: 10.1016/j.ajpath.2013.01.015. [DOI] [PubMed] [Google Scholar]
- 98.Park S.Y., Lee C.J., Choi J.H., Kim J.H., Kim J.W., Kim J.Y. The JAK2/STAT3/CCND2 Axis promotes colorectal Cancer stem cell persistence and radioresistance. J Exp Clin Canc Res. 2019;38:399. doi: 10.1186/s13046-019-1405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bedel R., Thiery-Vuillemin A., Grandclement C., Balland J., Remy-Martin J.P., Kantelip B. Novel role for STAT3 in transcriptional regulation of NK immune cell targeting receptor MICA on cancer cells. Canc Res. 2011;71:1615–1626. doi: 10.1158/0008-5472.CAN-09-4540. [DOI] [PubMed] [Google Scholar]
- 100.Akhter M.Z., Sharawat S.K., Kumar V., Kochat V., Equbal Z., Ramakrishnan M. Aggressive serous epithelial ovarian cancer is potentially propagated by EpCAM+CD45+ phenotype. Oncogene. 2018;37:2089–2103. doi: 10.1038/s41388-017-0106-y. [DOI] [PubMed] [Google Scholar]
- 101.Zhong Y., Guan K., Guo S., Zhou C., Wang D., Ma W. Spheres derived from the human SK-RC-42 renal cell carcinoma cell line are enriched in cancer stem cells. Canc Lett. 2010;299:150–160. doi: 10.1016/j.canlet.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 102.Amiot L., Ferrone S., Grosse-Wilde H., Seliger B. Biology of HLA-G in cancer: a candidate molecule for therapeutic intervention? Cell Mol Life Sci. 2011;68:417–431. doi: 10.1007/s00018-010-0583-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kochan G., Escors D., Breckpot K., Guerrero-Setas D. Role of non-classical MHC class I molecules in cancer immunosuppression. OncoImmunology. 2013;2 doi: 10.4161/onci.26491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pende D., Falco M., Vitale M., Cantoni C., Vitale C., Munari E. Killer ig-like receptors (KIRs): their role in NK cell modulation and developments leading to their clinical exploitation. Front Immunol. 2019;10:1179. doi: 10.3389/fimmu.2019.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Özgül Özdemir R.B., Özdemir A.T., Oltulu F., Kurt K., Yiğittürk G., Kırmaz C. A comparison of cancer stem cell markers and nonclassical major histocompatibility complex antigens in colorectal tumor and noncancerous tissues. Ann Diagn Pathol. 2016;25:60–63. doi: 10.1016/j.anndiagpath.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 106.Wolpert F., Roth P., Lamszus K., Tabatabai G., Weller M., Eisele G. HLA-E contributes to an immune-inhibitory phenotype of glioblastoma stem-like cells. J Neuroimmunol. 2012;250:27–34. doi: 10.1016/j.jneuroim.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 107.Teklemariam T., Zhao L., Hantash B.M. Full-length HLA-G1 and truncated HLA-G3 differentially increase HLA-E surface localization. Hum Immunol. 2012;73:898–905. doi: 10.1016/j.humimm.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 108.Grange C., Tapparo M., Tritta S., Deregibus M.C., Battaglia A., Gontero P. Role of HLA-G and extracellular vesicles in renal cancer stem cell-induced inhibition of dendritic cell differentiation. BMC Canc. 2015;15:1009. doi: 10.1186/s12885-015-2025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.da Silva I.L., Veloso E.S., Gonçalves I.N.N., Braga A.D., Lopes M.T.P., Cassali G.D. Qa-2 expression levels is related with tumor-infiltrating lymphocytes profile during solid Ehrlich tumor development. Biomed Pharmacother. 2017;92:750–756. doi: 10.1016/j.biopha.2017.05.135. [DOI] [PubMed] [Google Scholar]
- 110.Henrich-Noack P., Sergeeva E.G., Eber T., You Q., Voigt N., Köhler J. Electrical brain stimulation induces dendritic stripping but improves survival of silent neurons after optic nerve damage. Sci Rep. 2017;7:627. doi: 10.1038/s41598-017-00487-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Keefe D., Shi L., Feske S., Massol R., Navarro F., Kirchhausen T. Perforin triggers a plasma membrane-repair response that facilitates CTL induction of apoptosis. Immunity. 2005;23:249–262. doi: 10.1016/j.immuni.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 112.Chowdhury D., Lieberman J. Death by a thousand cuts: granzyme pathways of programmed cell death. Annu Rev Immunol. 2008;26:389–420. doi: 10.1146/annurev.immunol.26.021607.090404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lauricella M., Carlisi D., Giuliano M., Calvaruso G., Cernigliaro C., Vento R. The analysis of estrogen receptor-α positive breast cancer stem-like cells unveils a high expression of the serpin proteinase inhibitor PI-9: possible regulatory mechanisms. Int J Oncol. 2016;49:352–360. doi: 10.3892/ijo.2016.3495. [DOI] [PubMed] [Google Scholar]
- 114.Raja S.M., Metkar S.S., Höning S., Wang B., Russin W.A., Pipalia N.H. A novel mechanism for protein delivery: granzyme B undergoes electrostatic exchange from serglycin to target cells. J Biol Chem. 2005;280:20752–20761. doi: 10.1074/jbc.M501181200. [DOI] [PubMed] [Google Scholar]
- 115.Roy A., Attarha S., Weishaupt H., Edqvist P.H., Swartling F.J., Bergqvist M. Serglycin as a potential biomarker for glioma: association of serglycin expression, extent of mast cell recruitment and glioblastoma progression. Oncotarget. 2017;8:24815–24827. doi: 10.18632/oncotarget.15820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ahrens T.D., Bang-Christensen S.R., Jørgensen A.M., Løppke C., Spliid C.B., Sand N.T. The role of proteoglycans in cancer metastasis and circulating tumor cell analysis. Front Cell Dev Biol. 2020;8:749. doi: 10.3389/fcell.2020.00749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Matsukuma S., Yoshimura K., Ueno T., Oga A., Inoue M., Watanabe Y. Calreticulin is highly expressed in pancreatic cancer stem-like cells. Canc Sci. 2016;107:1599–1609. doi: 10.1111/cas.13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Andrin C., Pinkoski M.J., Burns K., Atkinson E.A., Krahenbuhl O., Hudig D. Interaction between a Ca2+-binding protein calreticulin and perforin, a component of the cytotoxic T-cell granules. Biochemistry. 1998;37:10386–10394. doi: 10.1021/bi980595z. [DOI] [PubMed] [Google Scholar]
- 119.Fraser S.A., Karimi R., Michalak M., Hudig D. Perforin lytic activity is controlled by calreticulin. J Immunol. 2000;164:4150–4155. doi: 10.4049/jimmunol.164.8.4150. [DOI] [PubMed] [Google Scholar]
- 120.Wijeyesakere S.J., Bedi S.K., Huynh D., Raghavan M. The C-terminal acidic region of calreticulin mediates phosphatidylserine binding and apoptotic cell phagocytosis. J Immunol. 2016;196:3896–3909. doi: 10.4049/jimmunol.1502122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Feng M., Marjon K.D., Zhu F., Weissman-Tsukamoto R., Levett A., Sullivan K. Programmed cell removal by calreticulin in tissue homeostasis and cancer. Nat Commun. 2018;9:3194. doi: 10.1038/s41467-018-05211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tan A.R., Alexe G., Reiss M. Transforming growth factor-beta signaling: emerging stem cell target in metastatic breast cancer? Breast Canc Res Treat. 2009;115:453–495. doi: 10.1007/s10549-008-0184-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Padua D., Massagué J. Roles of TGFbeta in metastasis. Cell Res. 2009;19:89–102. doi: 10.1038/cr.2008.316. WšSSS n. [DOI] [PubMed] [Google Scholar]
- 124.Shipitsin M., Campbell L.L., Argani P., Weremowicz S., Bloushtain-Qimron N., Yao J. Molecular definition of breast tumor heterogeneity. Canc Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 125.Chng Z., Teo A., Pedersen R.A., Vallier L. SIP1 mediates cell-fate decisions between neuroectoderm and mesendoderm in human pluripotent stem cells. Cell Stem Cell. 2010;6:59–70. doi: 10.1016/j.stem.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 126.Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cufí S., Vazquez-Martin A., Oliveras-Ferraros C., Martin-Castillo B., Joven J., Menendez J.A. Metformin against TGFβ-induced epithelial-to-mesenchymal transition (EMT): from cancer stem cells to aging-associated fibrosis. Cell Cycle. 2010;9:4461–4468. doi: 10.4161/cc.9.22.14048. [DOI] [PubMed] [Google Scholar]
- 128.Vazquez-Martin A., Oliveras-Ferraros C., Cufí S., Del Barco S., Martin-Castillo B., Menendez J.A. Metformin regulates breast cancer stem cell ontogeny by transcriptional regulation of the epithelial-mesenchymal transition (EMT) status. Cell Cycle. 2010;9:3807–3814. [PubMed] [Google Scholar]
- 129.Lottaz C., Beier D., Meyer K., Kumar P., Hermann A., Schwarz J. Transcriptional profiles of CD133+ and CD133- glioblastoma-derived cancer stem cell lines suggest different cells of origin. Canc Res. 2010;70:2030–2040. doi: 10.1158/0008-5472.CAN-09-1707. [DOI] [PubMed] [Google Scholar]
- 130.AlHossiny M., Luo L., Frazier W.R., Steiner N., Gusev Y., Kallakury B. Ly6E/K signaling to TGFβ promotes breast cancer progression, immune escape, and drug resistance. Canc Res. 2016;76:3376–3386. doi: 10.1158/0008-5472.CAN-15-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dziurzynski K., Wei J., Qiao W., Hatiboglu M.A., Kong L.Y., Wu A. Glioma-associated cytomegalovirus mediates subversion of the monocyte lineage to a tumor propagating phenotype. Clin Canc Res. 2011;17:4642–4649. doi: 10.1158/1078-0432.CCR-11-0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lee J.C., Lee K.M., Kim D.W., Heo D.S. Elevated TGF-beta1 secretion and down-modulation of NKG2D underlies impaired NK cytotoxicity in cancer patients. J Immunol. 2004;172:7335–7340. doi: 10.4049/jimmunol.172.12.7335. [DOI] [PubMed] [Google Scholar]
- 133.Facoetti A., Nano R., Zelini P., Morbini P., Benericetti E., Ceroni M. Human leukocyte antigen and antigen processing machinery component defects in astrocytic tumors. Clin Canc Res. 2005;11:8304–8311. doi: 10.1158/1078-0432.CCR-04-2588. [DOI] [PubMed] [Google Scholar]
- 134.Zhong M., Zhong C., Cui W., Wang G., Zheng G., Li L. Induction of tolerogenic dendritic cells by activated TGF-β/Akt/Smad2 signaling in RIG-I-deficient stemness-high human liver cancer cells. BMC Canc. 2019;19:439. doi: 10.1186/s12885-019-5670-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jo M., Eastman B.M., Webb D.L., Stoletov K., Klemke R., Gonias S.L. Cell signaling by urokinase-type plasminogen activator receptor induces stem cell-like properties in breast cancer cells. Canc Res. 2010;70:8948–8958. doi: 10.1158/0008-5472.CAN-10-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hu J., Jo M., Eastman B.M., Gilder A.S., Bui J.D., Gonias S.L. uPAR induces expression of transforming growth factor β and interleukin-4 in cancer cells to promote tumor-permissive conditioning of macrophages. Am J Pathol. 2014;184:3384–3393. doi: 10.1016/j.ajpath.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sainz B., Jr., Alcala S., Garcia E., Sanchez-Ripoll Y., Azevedo M.M., Cioffi M. Microenvironmental hCAP-18/LL-37 promotes pancreatic ductal adenocarcinoma by activating its cancer stem cell compartment. Gut. 2015;64:1921–1935. doi: 10.1136/gutjnl-2014-308935. [DOI] [PubMed] [Google Scholar]
- 138.Liu C.C., Lin J.H., Hsu T.W., Su K., Li A.F., Hsu H.S. IL-6 enriched lung cancer stem-like cell population by inhibition of cell cycle regulators via DNMT1 upregulation. Int J Canc. 2015;136:547–559. doi: 10.1002/ijc.29033. [DOI] [PubMed] [Google Scholar]
- 139.Korkaya H., Liu S., Wicha M.S. Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J Clin Invest. 2011;121:3804–3809. doi: 10.1172/JCI57099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Balamurugan K., Mendoza-Villanueva D., Sharan S., Summers G.H., Dobrolecki L.E., Lewis M.T. C/EBPδ links IL-6 and HIF-1 signaling to promote breast cancer stem cell-associated phenotypes. Oncogene. 2019;38:3765–3780. doi: 10.1038/s41388-018-0516-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wu C.T., Huang Y.C., Chen W.C., Chen M.F. Effect of tumor burden on tumor aggressiveness and immune modulation in prostate cancer: association with IL-6 signaling. Cancers. 2019;11:992. doi: 10.3390/cancers11070992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yang Z., He L., Lin K., Zhang Y., Deng A., Liang Y. The KMT1A-GATA3-STAT3 circuit is a novel self-renewal signaling of human bladder cancer stem cells. Clin Canc Res. 2017;23:6673–6685. doi: 10.1158/1078-0432.CCR-17-0882. [DOI] [PubMed] [Google Scholar]
- 143.Lee Y., Shin J.H., Longmire M., Wang H., Kohrt H.E., Chang H.Y. CD44+ cells in head and neck squamous cell carcinoma suppress T-cell-mediated immunity by selective constitutive and inducible expression of PD-L1. Clin Canc Res. 2016;22:3571–3581. doi: 10.1158/1078-0432.CCR-15-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chen W., Qin Y., Wang D., Zhou L., Liu Y., Chen S. CCL20 triggered by chemotherapy hinders the therapeutic efficacy of breast cancer. PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2005869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chen K.J., Lin S.Z., Zhou L., Xie H.Y., Zhou W.H., Taki-Eldin A. Selective recruitment of regulatory T cell through CCR6-CCL20 in hepatocellular carcinoma fosters tumor progression and predicts poor prognosis. PloS One. 2011;6 doi: 10.1371/journal.pone.0024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chen B., Zhang D., Zhou J., Li Q., Zhou L., Li S.M. High CCR6/CCR7 expression and Foxp3+ Treg cell number are positively related to the progression of laryngeal squamous cell carcinoma. Oncol Rep. 2013;30:1380–1390. doi: 10.3892/or.2013.2603. [DOI] [PubMed] [Google Scholar]
- 147.Zhang C.Y., Qi Y., Li X.N., Yang Y., Liu D.L., Zhao J. The role of CCL20/CCR6 axis in recruiting Treg cells to tumor sites of NSCLC patients. Biomed Pharmacother. 2015;69:242–248. doi: 10.1016/j.biopha.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 148.Lian J., Liu S., Yue Y., Yang Q., Zhang Z., Yang S. Eomes promotes esophageal carcinoma progression by recruiting Treg cells through the CCL20-CCR6 pathway. Canc Sci. 2020 Oct 28 doi: 10.1111/cas.14712. Epub ahead of print. PMID: 33113266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Maccalli C., Volontè A., Cimminiello C., Parmiani G. Immunology of cancer stem cells in solid tumours. A review. Eur J Canc. 2014;50:649–655. doi: 10.1016/j.ejca.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 150.Parsa A.T., Waldron J.S., Panner A., Crane C.A., Parney I.F., Barry J.J. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat Med. 2007;13:84–88. doi: 10.1038/nm1517. [DOI] [PubMed] [Google Scholar]
- 151.Yang Y., Wu K.E., Zhao E., Li W., Shi L., Xie G. B7-H1 enhances proliferation ability of gastric cancer stem-like cells as a receptor. Oncol Lett. 2015;9:1833–1838. doi: 10.3892/ol.2015.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Polónia A., Pinto R., Cameselle-Teijeiro J.F., Schmitt F.C., Paredes J. Prognostic value of stromal tumour infiltrating lymphocytes and programmed cell death-ligand 1 expression in breast cancer. J Clin Pathol. 2017;70:860–867. doi: 10.1136/jclinpath-2016-203990. [DOI] [PubMed] [Google Scholar]
- 153.Inaguma S., Lasota J., Wang Z., Czapiewski P., Langfort R., Rys J. Expression of ALCAM (CD166) and PD-L1 (CD274) independently predicts shorter survival in malignant pleural mesothelioma. Hum Pathol. 2018;71:1–7. [Google Scholar]
- 154.Jinesh G.G., Manyam G.C., Mmeje C.O., Baggerly K.A., Kamat A.M. Surface PD-L1, E-cadherin, CD24, and VEGFR2 as markers of epithelial cancer stem cells associated with rapid tumorigenesis. Sci Rep. 2017;7:9602. doi: 10.1038/s41598-017-08796-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Terzuoli E., Bellan C., Aversa S., Ciccone V., Morbidelli L., Giachetti A. ALDH3A1 overexpression in melanoma and lung tumors drives cancer stem cell expansion, impairing immune surveillance through enhanced PD-L1 output. Cancers. 2019;11:1963. doi: 10.3390/cancers11121963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hou Y.C., Chao Y.J., Hsieh M.H., Tung H.L., Wang H.C., Shan Y.S. Low CD8⁺ T cell infiltration and high PD-L1 expression are associated with level of CD44⁺/CD133⁺ cancer stem cells and predict an unfavorable prognosis in pancreatic cancer. Cancers. 2019;11:541. doi: 10.3390/cancers11040541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yi Z., Yang D., Liao X., Guo F., Wang Y., Wang X. PSME3 induces epithelial-mesenchymal transition with inducing the expression of CSC markers and immunosuppression in breast cancer. Exp Cell Res. 2017;358:87–93. doi: 10.1016/j.yexcr.2017.05.017. [DOI] [PubMed] [Google Scholar]
- 158.Tsai M.S., Chen W.C., Lai C.H., Chen Y.Y., Chen M.F. Epigenetic therapy regulates the expression of ALDH1 and immunologic response: relevance to the prognosis of oral cancer. Oral Oncol. 2017;73:88–96. doi: 10.1016/j.oraloncology.2017.08.007. [DOI] [PubMed] [Google Scholar]
- 159.Wei F., Zhang T., Deng S.C., Wei J.C., Yang P., Wang Q. PD-L1 promotes colorectal cancer stem cell expansion by activating HMGA1-dependent signaling pathways. Canc Lett. 2019;450:1–13. doi: 10.1016/j.canlet.2019.02.022. [DOI] [PubMed] [Google Scholar]
- 160.Liao T.T., Lin C.C., Jiang J.K., Yang S.H., Teng H.W., Yang M.H. Harnessing stemness and PD-L1 expression by AT-rich interaction domain-containing protein 3B in colorectal cancer. Theranostics. 2020;10:6095–6112. doi: 10.7150/thno.44147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chen M., Sharma A., Lin Y., Wu Y., He Q., Gu Y. Insluin and epithelial growth factor (EGF) promote programmed death ligand 1(PD-L1) production and transport in colon cancer stem cells. BMC Canc. 2019;19:153. doi: 10.1186/s12885-019-5364-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hossain D.M., Dos Santos C., Zhang Q., Kozlowska A., Liu H., Gao C. Leukemia cell-targeted STAT3 silencing and TLR9 triggering generate systemic antitumor immunity. Blood. 2014;123:15–25. doi: 10.1182/blood-2013-07-517987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wang Z., Liu W., Wang C., Li Y., Ai Z. Acetylcholine promotes the self-renewal and immune escape of CD133+ thyroid cancer cells through activation of CD133-Akt pathway. Canc Lett. 2020;471:116–124. doi: 10.1016/j.canlet.2019.12.009. [DOI] [PubMed] [Google Scholar]
- 164.Wilson E.B., Yamada D.H., Elsaesser H., Herskovitz J., Deng J., Cheng G. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science. 2013;340:202–207. doi: 10.1126/science.1235208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Teijaro J.R., Ng C., Lee A.M., Sullivan B.M., Sheehan K.C., Welch M. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science. 2013;340:207–211. doi: 10.1126/science.1235214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ayers M., Lunceford J., Nebozhyn M., Murphy E., Loboda A., Kaufman D.R. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127:2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Furuta J., Inozume T., Harada K., Shimada S. CD271 on melanoma cell is an IFN-γ-inducible immunosuppressive factor that mediates downregulation of melanoma antigens. J Invest Dermatol. 2014;134:1369–1377. doi: 10.1038/jid.2013.490. [DOI] [PubMed] [Google Scholar]
- 168.Fujii R., Friedman E.R., Richards J., Tsang K.Y., Heery C.R., Schlom J. Enhanced killing of chordoma cells by antibody-dependent cell-mediated cytotoxicity employing the novel anti-PD-L1 antibody avelumab. Oncotarget. 2016;7:33498–33511. doi: 10.18632/oncotarget.9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gross E.T.E., Peinado C.D., Jung Y., Han S., Liu B., Santosa E.K. Identification and editing of stem-like cells in methylcholanthrene-induced sarcomas. OncoImmunology. 2018;8 doi: 10.1080/2162402X.2017.1404212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhu Y., Karakhanova S., Huang X., Deng S.P., Werner J., Bazhin A.V. Influence of interferon-α on the expression of the cancer stem cell markers in pancreatic carcinoma cells. Exp Cell Res. 2014;324:146–156. doi: 10.1016/j.yexcr.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 171.Sainz B., Jr., Martín B., Tatari M., Heeschen C., Guerra S. ISG15 is a critical microenvironmental factor for pancreatic cancer stem cells. Canc Res. 2014;74:7309–7320. doi: 10.1158/0008-5472.CAN-14-1354. [DOI] [PubMed] [Google Scholar]
- 172.Song M., Ping Y., Zhang K., Yang L., Li F., Zhang C. Low-Dose IFNγ induces tumor cell stemness in tumor microenvironment of non-small cell lung cancer. Canc Res. 2019;79:3737–3748. doi: 10.1158/0008-5472.CAN-19-0596. [DOI] [PubMed] [Google Scholar]
- 173.Castiello L., Sestili P., Schiavoni G., Dattilo R., Monque D.M., Ciaffoni F. Disruption of IFN-I signaling promotes HER2/neu tumor progression and breast cancer stem cells. Cancer Immunol Res. 2018;6:658–670. doi: 10.1158/2326-6066.CIR-17-0675. [DOI] [PubMed] [Google Scholar]
- 174.Li J., Chen J.N., Zeng T.T., He F., Chen S.P., Ma S. CD133+ liver cancer stem cells resist interferon-gamma-induced autophagy. BMC Canc. 2016;16:15. doi: 10.1186/s12885-016-2050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zelenay S., van der Veen A.G., Böttcher J.P., Snelgrove K.J., Rogers N., Acton S.E. Cyclooxygenase-dependent tumor growth through evasion of immunity. Cell. 2015;162:1257–1270. doi: 10.1016/j.cell.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Celià-Terrassa T., Liu D.D., Choudhury A., Hang X., Wei Y., Zamalloa J. Normal and cancerous mammary stem cells evade interferon-induced constraint through the miR-199a-LCOR axis. Nat Cell Biol. 2017;19:711–723. doi: 10.1038/ncb3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhan X., Guo S., Li Y., Ran H., Huang H., Mi L. Glioma stem-like cells evade interferon suppression through MBD3/NuRD complex-mediated STAT1 downregulation. J Exp Med. 2020;217 doi: 10.1084/jem.20191340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Barclay A.N., Wright G.J., Brooke G., Brown M.H. CD200 and membrane protein interactions in the control of myeloid cells. Trends Immunol. 2002;23:285–290. doi: 10.1016/s1471-4906(02)02223-8. [DOI] [PubMed] [Google Scholar]
- 179.Kawasaki B.T., Farrar W.L. Cancer stem cells, CD200 and immunoevasion. Trends Immunol. 2008;29:464–468. doi: 10.1016/j.it.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 180.Kawasaki B.T., Mistree T., Hurt E.M., Kalathur M., Farrar W.L. Co-expression of the toleragenic glycoprotein, CD200, with markers for cancer stem cells. Biochem Biophys Res Commun. 2007;364:778–782. doi: 10.1016/j.bbrc.2007.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ho J.M., Dobson S.M., Voisin V., McLeod J., Kennedy J.A., Mitchell A. CD200 expression marks leukemia stem cells in human AML. Blood Adv. 2020;4:5402–5413. doi: 10.1182/bloodadvances.2020001802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Jung Y.S., Vermeer P.D., Vermeer D.W., Lee S.J., Goh A.R., Ahn H.J. CD200: association with cancer stem cell features and response to chemoradiation in head and neck squamous cell carcinoma. Head Neck. 2015;37:327–335. doi: 10.1002/hed.23608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tonks A., Hills R., White P., Rosie B., Mills K.I., Burnett A.K. CD200 as a prognostic factor in acute myeloid leukaemia. Leukemia. 2007;21:566–568. doi: 10.1038/sj.leu.2404559. [DOI] [PubMed] [Google Scholar]
- 184.Wong K.K., Khatri I., Shaha S., Spaner D.E., Gorczynski R.M. The role of CD200 in immunity to B cell lymphoma. J Leukoc Biol. 2010;88:361–372. doi: 10.1189/jlb.1009686. [DOI] [PubMed] [Google Scholar]
- 185.Gilson R.C., Gunasinghe S.D., Johannes L., Gaus K. Galectin-3 modulation of T-cell activation: mechanisms of membrane remodelling. Prog Lipid Res. 2019;76:101010. doi: 10.1016/j.plipres.2019.101010. [DOI] [PubMed] [Google Scholar]
- 186.Chung L.Y., Tang S.J., Wu Y.C., Sun G.H., Liu H.Y., Sun K.H. Galectin-3 augments tumor initiating property and tumorigenicity of lung cancer through interaction with β-catenin. Oncotarget. 2015;6:4936–4952. doi: 10.18632/oncotarget.3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Caputo S., Grioni M., Brambillasca C.S., Monno A., Brevi A., Freschi M. Galectin-3 in prostate cancer stem-like cells is immunosuppressive and drives early metastasis. Front Immunol. 2020;11:1820. doi: 10.3389/fimmu.2020.01820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.van der Touw W., Chen H.M., Pan P.Y., Chen S.H. LILRB receptor-mediated regulation of myeloid cell maturation and function. Cancer Immunol Immunother. 2017;66:1079–1087. doi: 10.1007/s00262-017-2023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zheng J., Umikawa M., Cui C., Li J., Chen X., Zhang C. Inhibitory receptors bind ANGPTLs and support blood stem cells and leukaemia development. Nature. 2012;485:656–660. doi: 10.1038/nature11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Liu X., Yu X., Xie J., Zhan M., Yu Z., Xie L. ANGPTL2/LILRB2 signaling promotes the propagation of lung cancer cells. Oncotarget. 2015;6:21004–21015. doi: 10.18632/oncotarget.4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Su R., Dong L., Li Y., Gao M., Han L., Wunderlich M. Targeting FTO suppresses cancer stem cell maintenance and immune evasion. Canc Cell. 2020;38:79–96. doi: 10.1016/j.ccell.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Sica A., Porta C., Amadori A., Pastò A. Tumor-associated myeloid cells as guiding forces of cancer cell stemness. Cancer Immunol Immunother. 2017;66:1025–1036. doi: 10.1007/s00262-017-1997-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Abdullah L.N., Chow E.K. Mechanisms of chemoresistance in cancer stem cells. Clin Transl Med. 2013;2:3. doi: 10.1186/2001-1326-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ni J., Cozzi P., Hao J., Duan W., Graham P., Kearsley J. Cancer stem cells in prostate cancer chemoresistance. Curr Cancer Drug Targets. 2014;14:225–240. doi: 10.2174/1568009614666140328152459. [DOI] [PubMed] [Google Scholar]
- 195.Noh K.H., Kim S.H., Kim J.H., Song K.H., Lee Y.H., Kang T.H. API5 confers tumoral immune escape through FGF2-dependent cell survival pathway. Canc Res. 2014;74:3556–3566. doi: 10.1158/0008-5472.CAN-13-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Song K.H., Cho H., Kim S., Lee H.J., Oh S.J., Woo S.R. API5 confers cancer stem cell-like properties through the FGF2-NANOG axis. Oncogenesis. 2017;6:e285. doi: 10.1038/oncsis.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wu A., Wei J., Kong L.Y., Wang Y., Priebe W., Qiao W. Glioma cancer stem cells induce immunosuppressive macrophages/microglia. Neuro Oncol. 2010;12:1113–1125. doi: 10.1093/neuonc/noq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Noh K.H., Kim B.W., Song K.H., Cho H., Lee Y.H., Kim J.H. Nanog signaling in cancer promotes stem-like phenotype and immune evasion. J Clin Invest. 2012;122:4077–4093. doi: 10.1172/JCI64057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Noh K.H., Lee Y.H., Jeon J.H., Kang T.H., Mao C.P., Wu T.C. Cancer vaccination drives Nanog-dependent evolution of tumor cells toward an immune-resistant and stem-like phenotype. Canc Res. 2012;72:1717–1727. doi: 10.1158/0008-5472.CAN-11-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chen J., Ding P., Li L., Gu H., Zhang X., Zhang L. CD59 regulation by SOX2 is required for epithelial cancer stem cells to evade complement surveillance. Stem Cell Reports. 2017;8:140–151. doi: 10.1016/j.stemcr.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wu X., Qu D., Weygant N., Peng J., Houchen C.W. Cancer stem cell marker DCLK1 correlates with tumorigenic immune infiltrates in the colon and gastric adenocarcinoma microenvironments. Cancers. 2020;12:274. doi: 10.3390/cancers12020274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Yan R., Li J., Zhou Y., Yao L., Sun R., Xu Y. Inhibition of DCLK1 down-regulates PD-L1 expression through Hippo pathway in human pancreatic cancer. Life Sci. 2020;241:117150. doi: 10.1016/j.lfs.2019.117150. [DOI] [PubMed] [Google Scholar]
- 203.Beavis P.A., Divisekera U., Paget C., Chow M.T., John L.B., Devaud C. Blockade of A2A receptors potently suppresses the metastasis of CD73+ tumors. Proc Natl Acad Sci U S A. 2013;110:14711–14716. doi: 10.1073/pnas.1308209110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Katsuta E., Tanaka S., Mogushi K., Shimada S., Akiyama Y., Aihara A. CD73 as a therapeutic target for pancreatic neuroendocrine tumor stem cells. Int J Oncol. 2016;48:657–669. doi: 10.3892/ijo.2015.3299. [DOI] [PubMed] [Google Scholar]