Graphical abstract
Abbreviations: FRGs, folate-related genes; DNMs, De novo mutations; ldDNMs, likely damaging DNMs; lndDNMs, likely non-damaging DNMs; ASD, autism spectrum disorder; ID, intellectual disability; EE, epileptic encephalopathy; CHD, congenital heart disease; UDD, undiagnosed developmental disorder; ADD, all five developmental disorders; SNPs, single nucleotide polymorphisms; PTV, protein-truncating variants; Dmis, deleterious missense variants; Tmis, tolerant missense variants; TADA, Transmitted And De novo Association; pLI, probability of loss-of-function intolerance; RVIS, residual variation intolerance scores; PPI, Protein–protein interaction
Keywords: Developmental disorders, Folate-related gene, De novo mutation, Candidate disease-associated genes, Expression patterns
Abstract
Folate deficiency is an environmental risk factor for several developmental disorders. De novo mutations (DNMs) also play important etiological roles in various developmental disorders. However, it remains unclear whether DNMs in folate-related genes (FRGs) contribute to developmental disorders. We obtained a list of 1,821 FRGs from folate metabolism pathways and the Comparative Toxicogenomics Database, along with data concerning DNMs in 15,404 cases and 3,391 controls from the Gene4Denovo database. We used a TADA-Denovo model to prioritize candidate disease-associated FRGs, and characterized these genes in terms of genic intolerance, functional networks, and expression patterns. Compared with the controls, FRGs were significantly enriched in likely damaging DNMs (ldDNMs) in patients with developmental disorders (1.54 ≤ odds ratio ≤ 3.39, Padj ≤ 0.0075). Furthermore, FRGs with ldDNMs rather than with likely non-damaging DNMs (lndDNMs) overlapped significantly among the five developmental disorders included in the datasets. The TADA-Denovo model prioritized 96 candidate disease-associated FRGs, which were intolerant to genetic variants. Their functional networks mainly involved pathways associated with chromatin modification, organ development, and signal transduction pathways. DNMT3A, KMT2B, KMT2C, and YY1 emerged as hub FRGs from the protein–protein interaction network. These candidate disease-associated FRGs are preferentially expressed in the excitatory neurones during embryonic development, and in the cortex, cerebellum, striatum, and amygdala during foetal development. Overall, these findings show that DNMs in FRGs are associated with the risk of developmental disorders. Further research on these DNMs may facilitate the discovery of developmental disorder biomarkers and therapeutic targets, enabling detailed, personalized, and precise folate treatment plan.
1. Introduction
Developmental disorders, such as autism spectrum disorder (ASD), intellectual disability (ID), epileptic encephalopathy (EE), congenital heart disease (CHD), and undiagnosed developmental disorder (UDD), are a group of complex conditions that affect several aspects of development, and manifest between the embryonic period and childhood [1]. The severe symptoms of patients with developmental disorders can continue into adulthood, affecting the long-term quality of life, and placing considerable financial burdens on patients’ families and society in general. Developmental disorders exhibit some overlap, with symptoms found in one developmental disorder potentially being present in others. For example, the clinical phenotype associated with ASD is quite broad, with patients often exhibiting ID, developmental delays, or epilepsy [2]. Intriguingly, it has been reported that patients with CHD are at an increased risk for the emergence of neurodevelopmental disorders (NDDs), with approximately half of patients with CHD developing NDDs as their conditions progress [3].
Folate, a water-soluble vitamin B, plays an important role in the de novo synthesis of purines and pyrimidines, which are the components of DNA and RNA [4]. Purine and pyrimidine deficiencies can result in DNA replication errors, which can lead to mutations and chromosomal aberrations, and affect the proliferation and division of cells [5], [6]. Thus, folate is vital during the period of pregnancy, and its deficiency may affect the formation of embryos, and the growth and development of children. Several epidemiological studies have suggested that maternal folate deficiency during early pregnancy may increase the risk of children developing ASD, CHD, developmental delays, and seizures [7], [8], [9], [10], [11]. Patients with ASD and CHD reportedly have abnormally low levels of 5-methyltetrahydrofolate (5-MTHF) [12], [13], and an animal study found that folate deficiency during development caused deficits in memory and cognition [14]. More importantly, a case-control cohort of 45,300 Israelis subjects showed a preventive effect of maternal folate supplementation in the reduction of ASD [9], and a population-based study of 2,401 mothers and their children found that periconceptional folic acid supplementation reduced the risk of CHD [13]. Treatment with folinic acid could improve the clinical features of patients with developmental delays, autistic features, and seizures associated with cerebral folate deficiency [15]. These findings meet four of Hill’s criteria [16] for inferring causality (i.e., consistency, biological gradient, plausibility, experiment), and thus support the hypothesis that folate deficiency contributes to developmental disorders.
Genetic disorders of folate metabolism and transport can also affect folate status. Patients harboring loss-of-function mutations in the FOLR1 gene develop movement disorders and epileptic seizures [17], and a de novo loss-of-function mutation in the CIC gene contribute to cerebral folate deficiency by downregulating FOLR1 expression [18]. Single nucleotide polymorphisms (SNPs) in folate-related genes (FRGs) (e.g., 677C > T and 1298A > C SNPs in MTHFR, 2756A > G SNP in MTR, and 66A > G SNP in MTRR) may cause folate pathway abnormalities that increase the risks of ASD, CHD, and EE [19], [20], [21], [22]. Although the effect of each SNP is quite weak, a combination of several SNPs may account for most of the narrow-sense heritabilities. Several de novo mutations (DNMs) with relatively stronger pathogenic effects DNMs and have been shown to play vital roles in various developmental disorders, including ASD [23], UDD [24], CHD [25], EE [26], and ID [27]. For example, by comparing patients with ASD and their unaffected siblings, Iossifov et al. [23] showed that 13% of missense DNMs and 43% of functional DNMs (i.e., nonsense, frameshift, and splice-site DNMs) contributed to 12% and 9% of ASD diagnoses, respectively. Approximately 26.9%, 40%, and 42%, of DNMs are estimated to contribute to a diagnosis of EE [28], ID [29], and UDD [24], respectively. Although these findings provide important insights, our understanding of the genetic architecture of developmental disorders is still in its infancy, and there are likely additional relevant genes and genetic etiologies that remain undiscovered.
Therefore, in this study, we performed exploratory integrative genomic analyses (Fig. 1) to (1) investigate the contribution of DNMs in FRGs to the risk of developmental disorders, (2) prioritize candidate disease-associated FRGs and hub FRGs in developmental disorders, and (3) characterize the functional pathways, functional networks, and expression patterns of candidate disease-associated FRGs in the human brain.
Fig. 1.
Study overview of the Analysis Pipeline. Using whole-genome and whole-exome sequencing data, we assessed the burden of likely damaging de novo mutations (ldDNMs) in folate-related genes (FRGs) of patients with developmental disorders. The FRGs with ldDNMs were then analyzed by the Transmission And De novo Association (TADA) model to prioritize candidate disease-associated FRGs and perform exploratory integrative genomic analyses.
2. Materials and methods
2.1. Data collection and annotation
We manually compiled a list of 1,821 FRGs (Table S1) from two different resources: (1) a set of 48 human genes from a database of the one-carbon metabolic pathway [30], and (2) a catalogue of human orthologs of 1,773 mouse genes obtained from the Comparative Toxicogenomics Database (CTD) [31], which lists specific chemical–gene relationships mentioned in published references. DNMs in patients with developmental disorders identified by whole-exome sequencing (WES) or whole-genome sequencing (WGS) were downloaded from the Gene4Denovo database (http://www.genemed.tech/gene4denovo/), which integrated data from multiple sources to allow for their convenient cataloguing, searching, downloading, browsing, and downstream analyses. Controls included an unaffected sibling and healthy individuals. The information of DNMs for developmental disorders and control subjects available in the literatures is provided in Table S2.
We annotated DNMs with ANNOVAR [32] based on RefSeq gene definitions as described in our previous studies [33], [34], [35]. Only the exonic DNMs in the FRGs (Table S3) were used for the subsequent analysis. According to the predicted functional effects, the variants were classified into four classes: protein-truncating variants (PTV, including frameshift, nonsense, and canonical splice-site mutations), deleterious missense variants (Dmis), tolerant missense variants (Tmis) and synonymous variants. Likely damaging DNMs (ldDNMs) were defined as the combination of PTVs and Dmis. Likely non-damaging DNMs (lndDNMs) were defined as the combination of Tmis and synonymous variants. We used our recently developed ReVe tool [36] to predict deleterious missense variants with scores greater than 0.7 and tolerant missense variants with scores no higher than 0.7.
2.2. Characterization of ldDNMs in FRGs
As reported previously [33], lndDNMs should be unrelated to phenotypic conditions and can therefore be used to partly remove batch effects influencing the rates of DNM detection reported in different publications. We therefore used a two-tailed Fisher’s exact test to compare the ldDNMs and lndDNMs in FRGs among patients with ASD, CHD, ID, EE, or UDD with those of the controls. Hence, we used the following general format in R (R version 4.0.2):
| fisher.test(matrix(c(x1, n1, x2, n2), alternative=“two.sided”)) |
where x1 and x2 are the number of ldDNMs in cases and controls, respectively, and n1 and n2 are the number of lndDNMs in cases and controls, respectively.
To determine whether the number of observed genes containing multiple ldDNMs was greater than the number expected from chance among FRGs, we performed one million permutations tests based on FRGs with multiple ldDNMs as described in our previous studies [33], [34]. Given that the observed numbers of FRGs with multiple ldDNMs are N1, we sampled N1 genes from all FRGs in developmental disorders with replacement based on the background DNM rate of per gene reported by Samocha et al. [37]. For each permutation, calculating the number (Li) of the same gene repeatedly arise in sampled N1 genes set. The P value was calculated as the proportion of the expected number of recurrent FRGs from the permutation is equal or greater than the observed number of recurrent FRGs. Hence, we used the following general format:
where ‘i’ denotes natural number.
We used DNENRICH software [38], which accounts for gene size, trinucleotide context, and the functional effect of the observed number of mutations, to determine whether there was significant overlap between the controls and the patients with different developmental disorders in terms of FRGs containing ldDNMs or lndDNMs. The P value was generated by the DNENRICH tool.
2.3. Prioritising candidate disease-associated FRGs
In line with previous studies [34], [35], we used the Transmitted And De novo Association (TADA) model [39] to prioritize candidate genes associated with developmental disorders, and then intersected the prioritized candidate genes with FRGs, which were then defined as candidate disease-associated FRGs. TADA-Denovo is a type of Bayesian model and requires the following parameters: the background DNM rate per-gene of mutation types, the number of risk genes of developmental disorder, the burden of mutation types, and the relative risk parameters (Table S4). We used two strategies to identify candidate disease-associated FRGs at a false discovery rate (FDR) ≤ 0.1. The first strategy involved, counting the number of ldDNMs associated with each disorder and using the TADA-Denovo method to calculate gene-specific FDR values. The second strategy involved, leveraging the power of the large cohort by pooling the ldDNMs associated with each gene across all five developmental disorders of interest to calculate gene-specific FDR values. The second strategy is viable because the five developmental disorders share genetic components [33], [40]. We classified the associations between TADA-prioritized FRGs and developmental disorder as strong (i.e., FDR ≤ 0.01), possible (0.01 < FDR ≤ 0.05), or suggestive (0.05 < FDR ≤ 0.1). We defined FRGs that only contained ldDNMs in the context of a single developmental disorder as unique FRGs, and we defined those containing ldDNMs in the contexts of multiple developmental disorders as shared genes.
To determine whether candidate disease-associated FRGs with rare variants were intolerant to variance, we used the probability of loss-of-function intolerance (pLI) score (https://gnomad.broadinstitute.org/) and the residual variation intolerance scores (RVIS) [41], which are both reliable measures of genic intolerance to ldDNMs [42]. Higher pLI and lower RVIS values indicate greater intolerance to functional and deleterious missense variants in a gene, respectively. We used the Wilcoxon rank-sum test to compare the distributional differences in pLI scores and RVIS scores across three datasets: (1) all RefSeq genes excluding the FRGs (i.e., Non-FRGs), (2) all FRGs, and (3) the candidate disease-associated FRGs.
2.4. Protein–protein interaction (PPI) network construction and analysis
To investigate the physical interactions of proteins encoded by the candidate disease-associated FRGs, we downloaded PPI datas with confidence scores of >0.4 from the STRING database (https://string-db.org/) and removed nonphysical interactions. We visualized the PPI network with Cytoscape v3.7.2 (https://cytoscape.org/). We applied a permutation test as described in our previous studies [33], [34] to evaluate whether the PPI network was more closely connected than would be expected by chance. We used the Cytoscape plugin cytoHubba to analyze the role of each gene in the PPI network based on seven topological analysis metrics: degree, closeness, radiality, maximal clique centrality, the density of the maximum neighbourhood component, the maximum neighbourhood component, and the edge percolated component. We regarded genes ranking in the top 10 for all seven metrics as being “hub genes” in the PPI network.
2.5. Gene ontology enrichment
We used Metascape [43] to perform gene ontology enrichment analysis on the candidate disease-associated FRGs within the PPI network. Only terms with a minimum count of 3, a P value < 0.01, and a minimum enrichment factor >1.5 were deemed statistically significant. We selected the most significant gene ontology term within each cluster to represent the cluster.
2.6. Expression pattern analysis of candidate disease-associated FRGs
We used the tissue-specific enrichment analysis (TSEA) tool [44] to determine the specific spatiotemporal expression patterns of candidate disease-associated FRGs in the brain. Dougherty et al. [45] provided cell-specific gene lists for different cell types and human brain regions, and/or across different developmental stages according to the specificity index probability (pSI). The user can perform enrichment analysis to estimate input candidate gene lists that significantly overlap with cell-specific gene lists, which can be identified by Fisher’s exact test with Benjamini-Hochberg correction. We calculated pSI values for input candidate gene sets in 6 different brain regions (i.e., the cortex, amygdala, hippocampus, cerebellum, striatum, and thalamus) and across 10 different developmental epochs (i.e., the early foetal period, the early mid-foetal period, the late mid-foetal period, the late foetal period, the neonatal-early infancy period, the late infancy period, the early childhood period, the middle-late childhood period, the adolescence period, and the young adulthood period) from embryonic development to adulthood. We applied Benjamini-Hochberg corrections to the P values for gene expression.
Furthermore, The single-cell RNA-sequencing expression data of the human mid-gestational embryonic cerebral cortex were derived from 4,000 normal individual cells of 22 brain regions, which were used to examine the expression features of candidate disease-associated FRGs in eight clusters of GABAergic inhibitory neurons (n = 968) and four clusters of glutamatergic excitatory neurones (n = 1,625) [46]. The expression value, quantified as transcripts per million (TPM), of each gene was normalized according to log2(TPM/10 + 1). We used Wilcoxon rank-sum tests to compare GABAergic and glutamatergic neurones in terms of the average expression levels of candidate disease-associated FRGs.
3. Results
3.1. Increased burden of FRGs ldDNMs in patients with developmental disorders
By cross-referencing our list of FRGs and our DNMs data, no enrichment of ldDNMs was found in the patients with developmental disorders using a set of 48 FRGs (Table S5). Owing to the small number of 48 FRGs with an even smaller number of ldDNMs, this dataset may have had insufficient power to obtain statistically significant results when comparing cases and controls. However, most of the odds ratios (ORs) were greater than 1, suggesting that there is a tendency of enrichment of ldDNMs in the 48 FRGs of patients. Significant enrichment of ldDNMs in the patients with developmental disorders was found using the set of 1,773 FRGs (Table S5). We then integrated the two gene sets for burden analysis. Patients with developmental disorders had a greater burden of ldDNMs in all FRGs than the controls (Table 1; 1.54 ≤ OR ≤ 3.39; Padj ≤ 0.0075), and for the combination of all five developmental disorders (i.e., ADD), showing a significant enrichment of ldDNMs was found ADD (OR = 1.89; Padj = 6.06 × 10−8). This analysis was replicated using the largest available exome sequencing cothorts, demonstrating a significantly increased rate of ldDNMs in developmental disorder cases, confirming our results (OR = 2.09, P = 1.81 × 10−11; Table S6).
Table 1.
Burden of ldDNMs in FRGs in five developmental disorders.
| ASD (6,511) | UDD (4,293) | EE (933) | ID (1,331) | CHD (2,645) | ADD (15,713) | Control (3,391) | |
|---|---|---|---|---|---|---|---|
| ldDNMs | 406 | 471 | 88 | 137 | 133 | 1,235 | 120 |
| lndDNMs | 577 | 532 | 66 | 90 | 193 | 1,458 | 268 |
| OR | 1.57 | 1.98 | 2.97 | 3.39 | 1.54 | 1.89 | – |
| 95% CI | 1.22–2.04 | 1.53–2.56 | 1.99–4.46 | 2.38–4.86 | 1.12–2.12 | 1.50–2.40 | – |
| P value | 0.00035 | 4.83 × 10−8 | 3.03 × 10−8 | 1.30 × 10−12 | 0.0075 | 2.12 × 10−8 | – |
| Adjusted P | 0.00042 | 7.25 × 10−8 | 6.06 × 10−8 | 7.80 × 10−12 | 0.0075 | 6.06 × 10−8 | – |
Abbreviations: ASD, autism spectrum disorder; UDD, undiagnosed developmental disorder; EE, epileptic encephalopathy; ID, intellectual disability; CHD, congenital heart disease; ADD, all five developmental disorders; FRGs, folate-related genes; DNMs, de novo mutations; OR, odds ratio; CI, confidence interval. ldDNMs were defined as the combination of protein-truncating variants (i.e. nonsense, frameshift and canonical splice site mutations) and deleterious missense mutations, which were predicted by ReVe; lndDNMs were defined as the combination of tolerant missense mutations and synonymous mutations. We performed a Fisher exact test to estimate the burden of ldDNMs in FRGs in disorders versus controls. P-values were adjusted for multiple testing using the Benjamini-Hochberg correction, and a false discovery rate (FDR) < 0.05 was considered significant.
We also observed FRGs with multiple ldDNMs in ASD, CHD, EE, ID, UDD, and controls, respectively. Interestingly, the number of FRGs with multiple ldDNMs in all five developmental disorders (P < 0.05) were significantly higher than random expectation by the permutation test, which was not the case for controls (P = 0.19) (Fig. S1). In addition, the number of FRGs with multiple ldDNMs was significantly higher than random expectation for ADD cases (P = 0.011, Fig.S1). Taken together, these results suggest that ldDNMs in FRGs may contribute to developmental disorder risk.
3.2. FRGS with ldDNMs exhibit significant overlap between different developmental disorder subtype
We observed a significantly higher degree of overlap between the ID and UDD groups, the EE and ID groups, and the EE and UDD groups, with the ratio of observed overlap to expected overlap (O/E) ranging from 5.38 to 7.38 (P ≤ 1.00 × 10−5; Table 2) in terms of FRGs harboring ldDNMs. We also found a lower degree of overlap for the ASD and ID groups, the ASD and UDD groups, and the CHD and ID groups (2.24 ≤ O/E ≤ 2.47, P ≤ 3.40 × 10−3; Table 2). The ASD and CHD groups, the ASD and EE groups, and the CHD and UDD groups showed weaker significant degree of overlap (Table 2). There was no significant degree of overlap between the control group and any of the specific developmental disorder groups in terms of FRGs with ldDNMs or with lndDNMs. There was no significant degree of overlap between any pair of specific developmental disorder groups in terms of FRGs with lndDNMs. These findings suggested that ldDNMs in FRGs may constitute a shared genetic aetiology for various developmental disorders.
Table 2.
Overlap between FRGs with ldDNMs or lndDNMs across developmental disorders and controls.
| Groups | ldDNMs in shared FRGs |
lndDNMs in shared FRGs |
||||||
|---|---|---|---|---|---|---|---|---|
| Obs | Exp | O/E | P value | Obs | Exp | O/E | P value | |
| ASD and CHD | 95 | 69.82 | 1.36 | 0.038 | 136 | 142.92 | 0.95 | 0.66 |
| ASD and EE | 83 | 52.81 | 1.57 | 0.047 | 37 | 48.13 | 0.77 | 0.88 |
| ASD and ID | 193 | 79.41 | 2.43 | 2.80 × 10−4 | 49 | 60.00 | 0.82 | 0.86 |
| ASD and UDD | 603 | 269.17 | 2.24 | 6.10 × 10−4 | 311 | 335.56 | 0.93 | 0.84 |
| ASD and Control | 56 | 54.02 | 1.04 | 0.42 | 166 | 176.28 | 0.94 | 0.73 |
| CHD and EE | 33 | 17.23 | 1.92 | 0.038 | 6 | 16.77 | 0.36 | 1.00 |
| CHD and ID | 64 | 25.91 | 2.47 | 3.40 × 10−3 | 17 | 20.98 | 0.81 | 0.79 |
| CHD and UDD | 138 | 87.90 | 1.57 | 0.054 | 108 | 117.14 | 0.92 | 0.77 |
| CHD and Control | 13 | 17.67 | 0.74 | 0.86 | 53 | 61.49 | 0.86 | 0.85 |
| EE and ID | 101 | 17.44 | 5.79 | 1.10 × 10−4 | 6 | 6.75 | 0.89 | 0.63 |
| EE and UDD | 318 | 59.15 | 5.38 | 1.70 × 10−4 | 54 | 37.64 | 1.43 | 0.019 |
| EE and Control | 8 | 11.84 | 0.68 | 0.78 | 24 | 19.80 | 1.21 | 0.22 |
| ID and UDD | 673 | 91.21 | 7.38 | 1.00 × 10−5 | 45 | 50.92 | 0.88 | 0.78 |
| ID and Control | 15 | 18.35 | 0.82 | 0.67 | 25 | 26.78 | 0.93 | 0.65 |
| UDD and Control | 51 | 63.18 | 0.81 | 0.74 | 147 | 158.71 | 0.93 | 0.78 |
Abbreviations: ASD, autism spectrum disorder; UDD, undiagnosed developmental disorder; EE, epileptic encephalopathy; ID, intellectual disability; CHD, congenital heart disease; FRGs, folate-related genes; DNMs, de novo mutations; ldDNMs were defined as the combination of protein-truncating variants (i.e., nonsense, frameshift and canonical splice site mutations) and deleterious missense mutations, which were predicted by ReVe; lndDNMs were defined as the combination of tolerant missense mutations and synonymous mutations.
3.3. Prioritization of candidate disease-associated FRGs
The TADA-Denovo model prioritized 16, 3, 10, 20, and 52 candidate disease-associated FRGs in the ASD, CHD, EE, ID, and UDD groups, respectively. Given the aforementioned overlaps between these five developmental disorders in terms of FRGs with ldDNMs, we also performed a pooled TADA-Denovo analysis combining the ldDNMs associated with all five developmental disorder types, which prioritized 75 candidate disease-associated FRGs, including 21 candidate disease-associated FRGs that did not reach the significance threshold (FDR ≤ 0.1) in analyses of specific developmental disorder subtypes. After eliminating redundancies between the results of the two strategies, we obtained a list of 96 prioritized candidate disease-associated FRGs (Table 3). There was significant overlap between candidate disease-associated FRGs harboring ldDNMs with developmental disorder and FMRP targets[47], chromatin genes, essential genes, and genes encoding postsynaptic density proteins (2.25 ≤ OR ≤ 7.68, P ≤ 7.63 × 10−16, Fisher’s exact test; Table S7).
Table 3.
Prioritized candidate disease-associated FRGs with FDR less than or equal to 0.1 by TADA-Denovo.
| Rank | FDR ≤ 0.01 (n = 52) | 0.01 < FDR ≤ 0.05 (n = 27) | 0.05 < FDR ≤ 0.1 (n = 17) |
|---|---|---|---|
| Unique genes(n = 28) | BRD7a, GATA6c, EBF2e, KCNA2e, CAMTA1u, GFOD2u, SLC12A2u, SOX4u | ANKRD27a, ATP1B1a, KDM2Bi, KDM5Cu, RAD51u, IL1RAPL2u, YY1u, ASIC2u, MSI1u, OTX1u, PDX1u, GOLPH3u, SIAH1u, | CELF4a, KDM6Ba, PGAM5i, CHMP2AiKMT2Bu, PHF8u, U2AF2u |
| Shared genes in two disorders (n = 28) | RNF146, SIN3A, VEZF1, SOX11, PRPF8, WDR26, PPP2CA, TAOK1, CSNK2B, KIF1A | NGFR, TFAP4, RAB14, RARA, EPHB1, LARP4B, COL2A1, OTX2, PAX3, PSMD3 | PRPF4B, ATIC, ZNF248, PMM2, DDX50, NAT8L, NR3C2, TRPC4AP |
| Shared genes in three disorders (n = 22) | ELL2, ERI1, FOXP2, SYNCRIP, NR4A2, ASXL1, TRIO, HK1, GABBR2, BCL11A, TRIP12, LAMB1, PPM1D, CLTC, ATP1A3, SMARCA2, TCF7L2, KAT6B | PSMC4, ARID1A, | CUL1, CPD |
| Shared genes in at least four disorders (n = 18) | TAF1, SMAD6, SPRED2, AGO1, KCNQ3, RYR2, KMT2C, FGF12, POGZ, DNMT3A, ARID1B, KCNQ2, PTEN, CACNA1A, CREBBP, SCN8A | NF1, GRIN1 |
Candidate disease-associated FRGs are split into four parts based on the number of disorders and ranked into three tiers based on the strength of FDR (false discovery rate). The tier of unique genes with superscript letters including a, c, e, i, and u represents genes only carrying only ldDNMs in ASD, CHD, EE, ID, and UDD, respectively. Based on the TADA-Denovo analysis of exonic ldDNMs from five developmental disorders, we prioritized 75 candidate disease-associated FRGs, including 21 new these FRGs (bold Italic) that were not prioritized in the first strategy (see method).
Of 96 prioritized candidate disease-associated FRGs, 52 had strong associations with developmental disorders, 27 had possible associations, and 17 had a suggestive association. Furthermore, 28 were unique, 28 were shared between two developmental disorders, 22 were shared among three developmental disorders, and 18 FRGs were shared among at least four developmental disorders (Table 3). The fact that most of the prioritized candidate disease-associated FRGs were shared suggests that these five developmental disorders have substantially similar genetic mechanisms.
3.4. Genic intolerance and rare frequency of candidate disease-associated FRGs
The pLI (in which a higher score indicates more intolerant functional variants for each gene) and the RVIS [41] (in which a lower score indicates more intolerant deleterious missense variants for each gene) have been validated as reliable metrics [42] of genic intolerance to ldDNMs. The minor allele frequency of variants in gnomAD has been hypothesized to significantly contribute to complex diseases. Compared with non-FRGs, the FRGs had higher pLI scores (P < 2.22 × 10−16; Fig. S2A), lower RVIS scores (P < 2.22 × 10−16; Fig. S2B), and an extremely rare variants frequency (P = 0.0057; Fig. S2C). Furthermore, the candidate disease-associated FRGs had significantly higher pLI scores (P < 2.22 × 10−16; Fig. S2A), lower RVIS scores (P = 1.80 × 10−10; Fig. S2B) and an extremely rare variants frequency (P = 0.0011; Fig. S2C) compared with those of the other FRGs. These results suggested that FRGs harboring damaging rare mutations are more likely to exhibit genic intolerance to ldDNMs than other genes are.
3.5. PPI network of candidate disease-associated FRGs and hub FRGs
The completed PPI network for the 96 candidate disease-associated FRGs featured 78 interacting genes and 221 connections (Fig. S3A). The number of interacting genes and connections was higher than would be expected from chance drawn from the entire FRGs set (P < 1.00 × 10−6; Fig. S3B), which suggests that the candidate disease-associated FRGs within the tight network are biologically related. The PPI network showed enrichment of the candidate disease-associated FRGs within pathways related to transcriptional regulation, chromatin modification, organ development, and signal transduction pathways (Table S8). Our analyses of topological metrics showed that the PPI network contained four hub FRGs (Table S9). These were DNMT3A, KMT2B, KMT2C, and YY1, which carried 11, 6, 11, and 2 ldDNMs, respectively. The hub FRGs exhibited high degrees of connectivity within the PPI network, which suggests that they play key roles in the etiology of developmental disorders.
3.6. Distinct expression patterns for candidate disease-associated FRGs
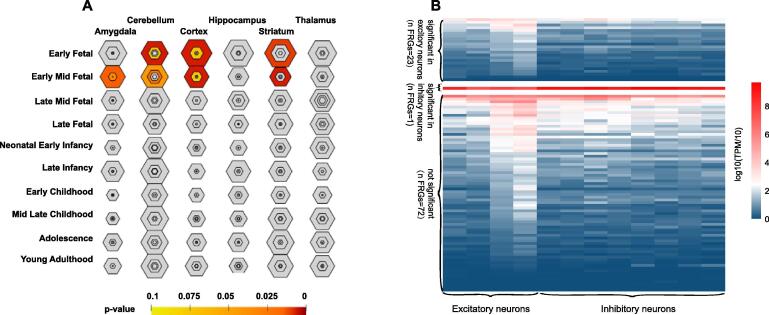
Our tissue-specific enrichment analysis showed enriched expression of the candidate disease-associated FRGs during the early-foetal periods and early-mid-foetal periods within the cortex [Benjamini–Hochberg (BH)-corrected, P value (PBH) = 0.0070 and 0.0070, respectively; Fig. 2A], cerebellum (PBH = 0.0070 and PBH = 0.043, respectively; Fig. 2A), and striatum (PBH = 0.01 and PBH = 0.0060, respectively; Fig. 2A). Expression of the candidate disease-associated FRGs was also significantly enriched in the early foetal amygdala during the early foetal period (PBH = 0.014; Fig. 2A). These findings suggested that candidate disease-associated FRGs display a prenatal bias and may play important roles in regional brain development.
Fig. 2.
Expression pattern analysis of candidate disease-associated folate-related genes (FRGs). (A) Tissue-specific expression analyses (TSEA) of the candidate disease-associated FRGs during the early foetal and mid-early foetal stages of development. The size and color of each hexagon represents the FRG’s specificity index probability, and the hexagon’s color represents the Benjamini-Hochberg–corrected P value. Positions closer to the large central hexagon indicate higher tissue specificity levels and darker colors indicate the more significant P values. (B) Heatmap of normalized expression levels of the candidate disease-associated FRGs in the inhibitory and excitatory neuron types. In the heatmap, gene expression levels are scaled as a function of [log2(TPM/10 + 1)], in which “TPM” stands for transcripts per million. Each row represents the average expression level of each gene, and the columns represent four clusters of the excitatory neurones and eight clusters of the inhibitory neurons. P values were calculated using the Wilcoxon rank sum test.
Our analyses of the expression patterns of candidate disease-associated FRGs in glutamatergic excitatory neurones and GABAergic inhibitory neurones from the human embryonic cerebral cortex [46] showed that 23 candidate disease-associated FRGs were expressed at higher levels in excitatory neurones than in inhibitory neurones, with one candidate disease-associated FRGs was expressed at a higher level in inhibitory neurones than in excitatory neurones (Fig. 2B). The 72 candidate disease-associated FRGs were expressed at similar significant levels in the excitatory and inhibitory neurones (Fig. 2B). The high number of the candidate disease-associated FRGs that were preferentially expressed in excitatory neurones suggests that such FRGs play critical roles in the cortical circuitry.
4. Discussion
The rapid development of high-throughput sequencing methods has accelerated research on the genetic architecture of developmental disorders [48]. DNMs with strong disruptive effects have been shown to be the main genetic causes of developmental disorders [49], and several environmental factors, including folate deficiency and FRG polymorphisms, can act as risk factors for developmental disorders [15], [50], [51], [52]. Furthermore, folate deficiency can result in aberrant gene expression, mutations, and chromosomal instability [53]. These lines of evidence, which point to intricate interrelationships among folate deficiency, accumulated evidence confirmed the intricate interrelationships between folate deficiency, genetic variations, and developmental disorder aetiologies, motivated us to explore the contributions of FRGs DNMs to developmental disorders.
To the best of our knowledge, this is the first study to use exome data to show that FRGs carry high DNM burdens in patients with developmental disorders. In addition, our analyses also indicated that the number of FRGs with recurrent ldDNMs in patients with developmental disorders was unlikely to be due to chance. In analyses of between-group similarities in the FRGs with ldDNMs, we observed high degrees of overlap between the ID and UDD groups, the EE and UDD groups, and the EE and ID groups. This observation is consistent with the high degree of comorbid occurrence observed for these developmental disorders. We also observed moderate degrees of overlap between the ID and ASD groups, the UDD and ASD groups, and the CHD and ID groups and weaker degrees of overlap for the CHD and ASD groups, the EE and ASD groups, and the CHD and UDD groups. These overlaps suggest that these developmental disorders should not be considered as isolated disorders. A potential future research direction would be to investigate the genetic overlaps between various developmental disorders and epilepsy, as the prevalence of epilepsy can range from 45% to 82% in patients with severe ID [2] and from 5% to 15% in patients with ASD[54].
Our TADA-Denovo model prioritised 96 candidate disease-associated FRGs, including PAX3. Lemay et al.[55] reported that a nonsense DNM in PAX3 could be responsible for cases of neural tube defects when the mothers did not take folic acid or multivitamins periconceptionally. Most of these FRGs exhibited high levels of haploinsufficiency and intolerance to ldDNMs. These results provide a useful list of developmental disorder related FRGs for future studies to focus on. We found that 68 of the candidate disease-associated FRGs carried ldDNMs in the contexts of at least two developmental disorders, which suggests that ldDNMs in certain FRGs can have multiple functional effects and contribute to multiple causes of developmental disorders. The remaining 28 candidate disease-associated FRGs, which only had ldDNMs in the context of a single developmental disorder, may be useful foci for research on molecular endophenotypes [56]. Such research could facilitate efforts to understand the genetic aetiologies of individual cases and formulate individualised folate treatment plans.
Our PPI network analyses yielded preliminary evidence that the candidate disease-associated FRGs are highly interconnected and that DNMT3A, KMT2C, KMT2B, and YY1 serve as hub FRGs. DNMT3A, KMT2C, and KMT2B are methylation-related genes, since folate deficiency affects methylation, thereby affecting the normal biological functions of these genes will be impacted. DNMT3A encodes de novo DNA methyltransferase[57], a member of the class I methyltransferase family, which is essential for DNA methylation during early embryonic development [58]. Another study showed that DNMT3A mutations are associated with overgrowth and ID [59]. Folate can be a limiting factor and methyl donor source in folate and methionine cycles, including DNMT3A methylation; in turn, a damaging mutation in DNMT3A affects the binding of the methyl donor. KMT2B and KMT2C are members of the mammalian H3K4 methyltransferase family, and mutations in these genes have been associated with developmental disorders [60], [61]. Animal studies have also shown that deletion of the KMT2B enzyme in fertilized eggs leads to the death of mouse embryos [62]. Proton-coupled folate transporter (PCFT) plays an important role in intestinal folate absorption. YY1, a trans-activating transcription factor [63], [64], resulted in a ~50% decrease in transcriptional activation of PCFT [65]. Barnard et al. [66] further demonstrated that the developmental disorder associated gene CHD8 sites were enriched in YY1 transcription factor motifs.
The candidate disease-associated FRGs are preferentially specifically expressed in the cortex, cerebellum, striatum, and amygdala during the early-foetal and early mid-foetal periods of development. This observation suggests that these early stages of brain development are particularly sensitive to ldDNMs in FRGs. Multiple lines of evidence indicate that the molecular pathologies of ID and ASD involves abnormalities in the cerebral cortex [67], [68], [69], [70] and striatum [71], [72], [73], [74]. Intriguingly, a quarter of the candidate disease-associated FRGs were preferentially expressed in the excitatory neurones of the embryonic cerebral cortex, which suggests that these FRGs play critical roles in the excitatory neuronal circuity. This idea is consistent with previous research on neurodevelopmental disorders [42].
Our study has several limitations that should be taken into consideration when interpreting the results. First, DNMs were collected based on integration of datasets from previously published studies using different cohorts, technologies, and coverages. These integrated DNMs were analyzed by the TADA-Denovo model to increase the power for detecting candidate genes, but individual heterogeneity should be taken to care. Second, further research is needed to verify the existence of specific regulatory relationships between folic acid and most of the FRGs listed in the CTD. Third, the combination of two FRG datasets of vastly different size (the 48 FRGs set and the core 1,773 FRGs set) may fall into the Simpson’s paradox so that the final results are explained mainly by the core 1,773 FRGs set. Fourth, although we divided missense mutations into Dmis and Tmis, the genes associated with hotspots of Tmis may be better understood about protein structure. Fifth, our analysis was based on bioinformatics methods to prioritize candidate disease-associated genes and cannot be used unequivocally to establish causality. Thus, functional experiments will be necessary to validate their pathophysiological importance of the identified genes. Sixth, although it is essential to understand the relationship between folate and disease phenotypic change, the existing phenotypes associated with the developmental disorders and FRG prevalence were not available. Further, detailed clinical data may aid in the understanding the clinical diagnosis and genetic etiology of developmental disorders.
In summary, our results suggest that ldDNMs in FRGs contribute to the risk of patients with developmental disorder and may provide useful information for the genetic counselling and the treatment of some patients. Furthermore, our analyses highlighted several the candidate disease-associated FRGs and hub FRGs that may be useful foci for further research into the pathophysiology of developmental disorders. Although further research is necessary to determine the optimal list of the candidate disease-associated FRGs, our results clearly implicate FRGs in perturbations of the neuronal circuity. Taken together, our findings elucidate the role of FRGs in developmental disorders, enabling further research is planned to integrate multi-omics data for identifying additional candidate disease-associated FRGs.
Author contributions
JL, KX, and QP were involved in study conception and design and participated in review of manuscript and offered valuable advices. TL and KL collected and analyzed the data. TL and KL wrote the paper. All authors contributed to the preparation of the manuscript and read and approved the final manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81801133 to J. Li; 81730036 to K. Xia), Young Elite Scientist Sponsorship Program of the China Association for Science and Technology (2018QNRC001 to J. Li), Central South University’s Innovation-Driven Project (20180033040004 to J. Li), Science and Technology Major Projectof Hunan Provincial Science and Technology Department (2018SK1030 to K. Xia ) and the Hunan Natural Science Foundation Outstanding Youth Fund (2020JJ3059 to J. Li). The funders had no role in the study design; collection, analysis, and interpretation of data; writing of the report; and in the decision to submit the article for publication.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.02.011.
Contributor Information
Qian Pan, Email: panqian@sklmg.edu.cn.
Kun Xia, Email: xiakun@sklmg.edu.cn.
Jinchen Li, Email: lijinchen@csu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Sheridan E., Wright J., Small N., Corry P.C., Oddie S., Whibley C. Risk factors for congenital anomaly in a multiethnic birth cohort: an analysis of the Born in Bradford study. Lancet. 2013;382(9901):1350–1359. doi: 10.1016/S0140-6736(13)61132-0. [DOI] [PubMed] [Google Scholar]
- 2.Shepherd C., Hosking G. Epilepsy in school children with intellectual impairments in Sheffield: the size and nature of the problem and the implications for service provision. J Ment Defic Res. 1989;33(Pt 6):511–514. doi: 10.1111/j.1365-2788.1989.tb01508.x. [DOI] [PubMed] [Google Scholar]
- 3.Marino B.S., Lipkin P.H., Newburger J.W., Peacock G., Gerdes M., Gaynor J.W. Neurodevelopmental outcomes in children with congenital heart disease: evaluation and management: a scientific statement from the American Heart Association. Circulation. 2012;126(9):1143–1172. doi: 10.1161/CIR.0b013e318265ee8a. [DOI] [PubMed] [Google Scholar]
- 4.Desai A., Sequeira J.M., Quadros E.V. The metabolic basis for developmental disorders due to defective folate transport. Biochimie. 2016;126:31–42. doi: 10.1016/j.biochi.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 5.Duthie S.J. Folic acid deficiency and cancer: mechanisms of DNA instability. Br Med Bull. 1999;55(3):578–592. doi: 10.1258/0007142991902646. [DOI] [PubMed] [Google Scholar]
- 6.Li G.-M., Presnell S.R., Gu L. Folate deficiency, mismatch repair-dependent apoptosis, and human disease. J Nutr Biochem. 2003;14(10):568–575. doi: 10.1016/s0955-2863(03)00115-3. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt RJ, Tancredi DJ, Ozonoff S, Hansen RL, Hartiala J, Allayee H, et al. Maternal periconceptional folic acid intake and risk of autism spectrum disorders and developmental delay in the CHARGE (CHildhood Autism Risks from Genetics and Environment) case-control study. Am J Clin Nutr. 2012;96(1):80-9. [DOI] [PMC free article] [PubMed]
- 8.Berry R.J., Crider K.S., Yeargin-Allsopp M. Periconceptional folic acid and risk of autism spectrum disorders. JAMA. 2013;309(6):611–613. doi: 10.1001/jama.2013.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levine S.Z., Kodesh A., Viktorin A., Smith L., Uher R., Reichenberg A. Association of maternal use of folic acid and multivitamin supplements in the periods before and during pregnancy with the risk of autism spectrum disorder in offspring. JAMA Psychiatry. 2018;75(2):176. doi: 10.1001/jamapsychiatry.2017.4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czeizel A.E., Dudas I., Vereczkey A., Banhidy F. Folate deficiency and folic acid supplementation: the prevention of neural-tube defects and congenital heart defects. Nutrients. 2013;5(11):4760–4775. doi: 10.3390/nu5114760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu S., Joseph K.S., Luo W., León J.A., Lisonkova S., Van den Hof M. Effect of Folic Acid Food Fortification in Canada on Congenital Heart Disease Subtypes. Circulation. 2016;134(9):647–655. doi: 10.1161/CIRCULATIONAHA.116.022126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frye R.E., Sequeira J.M., Quadros E.V., James S.J., Rossignol D.A. Cerebral folate receptor autoantibodies in autism spectrum disorder. Mol Psychiatry. 2013;18(3):369–381. doi: 10.1038/mp.2011.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Beynum I.M., Kapusta L., Bakker M.K., den Heijer M., Blom H.J., de Walle H.E.K. Protective effect of periconceptional folic acid supplements on the risk of congenital heart defects: a registry-based case-control study in the northern Netherlands. Eur Heart J. 2010;31(4):464–471. doi: 10.1093/eurheartj/ehp479. [DOI] [PubMed] [Google Scholar]
- 14.Berrocal-Zaragoza M.I., Sequeira J.M., Murphy M.M., Fernandez-Ballart J.D., Abdel Baki S.G., Bergold P.J. Folate deficiency in rat pups during weaning causes learning and memory deficits. Br J Nutr. 2014;112(8):1323–1332. doi: 10.1017/S0007114514002116. [DOI] [PubMed] [Google Scholar]
- 15.Moretti P., Sahoo T., Hyland K., Bottiglieri T., Peters S., del Gaudio D. Cerebral folate deficiency with developmental delay, autism, and response to folinic acid. Neurology. 2005;64(6):1088–1090. doi: 10.1212/01.WNL.0000154641.08211.B7. [DOI] [PubMed] [Google Scholar]
- 16.Hill A.B. The environment and disease: association or causation? Proc R Soc Med. 1965;58(5):295–300. doi: 10.1177/003591576505800503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steinfeld R., Grapp M., Kraetzner R., Dreha-Kulaczewski S., Helms G., Dechent P. Folate receptor alpha defect causes cerebral folate transport deficiency: a treatable neurodegenerative disorder associated with disturbed myelin metabolism. Am J Hum Genet. 2009;85(3):354–363. doi: 10.1016/j.ajhg.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao X, Wolf A, Kim SE, Cabrera RM, Wlodarczyk BJ, Zhu H, et al. CIC de novo loss of function variants contribute to cerebral folate deficiency by downregulating FOLR1 expression. J Med Genet. 2020. [DOI] [PMC free article] [PubMed]
- 19.Chen L., Liu L., Hong K., Hu J., Cheng X. Three genetic polymorphisms of homocysteine-metabolizing enzymes and risk of coronary heart disease: a meta-analysis based on 23 case-control studies. DNA Cell Biol. 2012;31(2):238–249. doi: 10.1089/dna.2011.1281. [DOI] [PubMed] [Google Scholar]
- 20.Wang W., Wang Y., Gong F., Zhu W., Fu S., Miao X.-P. MTHFR C677T polymorphism and risk of congenital heart defects: evidence from 29 case-control and TDT studies. PLoS ONE. 2013;8(3):e58041. doi: 10.1371/journal.pone.0058041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaik Mohammad N., Sai Shruti P., Bharathi V., Krishna Prasad C., Hussain T., Alrokayan S.A. Clinical utility of folate pathway genetic polymorphisms in the diagnosis of autism spectrum disorders. Psychiatr Genet. 2016;26(6):281–286. doi: 10.1097/YPG.0000000000000152. [DOI] [PubMed] [Google Scholar]
- 22.Coppede F. The genetics of folate metabolism and maternal risk of birth of a child with Down syndrome and associated congenital heart defects. Front Genet. 2015;6:223. doi: 10.3389/fgene.2015.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iossifov I., O’Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515(7526):216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deciphering Developmental Disorders S. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542(7642):433-8. [DOI] [PMC free article] [PubMed]
- 25.Jin S.C., Homsy J., Zaidi S., Lu Q., Morton S., DePalma S.R. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017;49(11):1593–1601. doi: 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heyne H.O., Singh T., Stamberger H., Abou Jamra R., Caglayan H., Craiu D. De novo variants in neurodevelopmental disorders with epilepsy. Nat Genet. 2018;50(7):1048–1053. doi: 10.1038/s41588-018-0143-7. [DOI] [PubMed] [Google Scholar]
- 27.Lelieveld S.H., Reijnders M.R.F., Pfundt R., Yntema H.G., Kamsteeg E.-J., de Vries P. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat Neurosci. 2016;19(9):1194–1196. doi: 10.1038/nn.4352. [DOI] [PubMed] [Google Scholar]
- 28.Hamdan F.F., Myers C.T., Cossette P., Lemay P., Spiegelman D., Laporte A.D. High rate of recurrent de novo mutations in developmental and epileptic encephalopathies. Am J Hum Genet. 2017;101(5):664–685. doi: 10.1016/j.ajhg.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vissers L.E.L.M., Gilissen C., Veltman J.A. Genetic studies in intellectual disability and related disorders. Nat Rev Genet. 2016;17(1):9–18. doi: 10.1038/nrg3999. [DOI] [PubMed] [Google Scholar]
- 30.Bhat M.K., Gadekar V.P., Jain A., Paul B., Rai P.S., Satyamoorthy K. 1-CMDb: A Curated Database of Genomic Variations of the One-Carbon Metabolism Pathway. Public Health Genomics. 2017;20(2):136–141. doi: 10.1159/000475805. [DOI] [PubMed] [Google Scholar]
- 31.Davis AP, Grondin CJ, Johnson RJ, Sciaky D, McMorran R, Wiegers J, et al. The Comparative Toxicogenomics Database: update 2019. Nucleic Acids Res. 2019;47(D1):D948-D54. [DOI] [PMC free article] [PubMed]
- 32.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. [DOI] [PMC free article] [PubMed]
- 33.Li J., Cai T., Jiang Y.i., Chen H., He X., Chen C. Genes with de novo mutations are shared by four neuropsychiatric disorders discovered from NPdenovo database. Mol Psychiatry. 2016;21(2):290–297. doi: 10.1038/mp.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J., Wang L., Guo H., Shi L., Zhang K., Tang M. Targeted sequencing and functional analysis reveal brain-size-related genes and their networks in autism spectrum disorders. Mol Psychiatry. 2017;22(9):1282–1290. doi: 10.1038/mp.2017.140. [DOI] [PubMed] [Google Scholar]
- 35.Zhao G, Li K, Li B, Wang Z, Fang Z, Wang X, et al. Gene4Denovo: an integrated database and analytic platform for de novo mutations in humans. Nucleic Acids Res. 2020;48(D1):D913-D26. [DOI] [PMC free article] [PubMed]
- 36.Li J, Zhao T, Zhang Y, Zhang K, Shi L, Chen Y, et al. Performance evaluation of pathogenicity-computation methods for missense variants. Nucleic Acids Res. 2018;46(15):7793-804. [DOI] [PMC free article] [PubMed]
- 37.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46(9):944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fromer M., Pocklington A.J., Kavanagh D.H., Williams H.J., Dwyer S., Gormley P. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506(7487):179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He X., Sanders S.J., Liu L.i., De Rubeis S., Lim E.T., Sutcliffe J.S. Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet. 2013;9(8):e1003671. doi: 10.1371/journal.pgen.1003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takata A., Miyake N., Tsurusaki Y., Fukai R., Miyatake S., Koshimizu E. Integrative analyses of de novo mutations provide deeper biological insights into autism spectrum disorder. Cell Rep. 2018;22(3):734–747. doi: 10.1016/j.celrep.2017.12.074. [DOI] [PubMed] [Google Scholar]
- 41.Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B., Williams S.M. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9(8) doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coe B.P., Stessman H.A.F., Sulovari A., Geisheker M.R., Bakken T.E., Lake A.M. Neurodevelopmental disease genes implicated by de novo mutation and copy number variation morbidity. Nat Genet. 2019;51(1):106–116. doi: 10.1038/s41588-018-0288-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1) doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu X., Wells A.B., O'Brien D.R., Nehorai A., Dougherty J.D. Cell type-specific expression analysis to identify putative cellular mechanisms for neurogenetic disorders. J Neurosci. 2014;34(4):1420–1431. doi: 10.1523/JNEUROSCI.4488-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dougherty J.D., Schmidt E.F., Nakajima M., Heintz N. Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucl Acids Res. 2010;38(13):4218–4230. doi: 10.1093/nar/gkq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fan X., Dong J.i., Zhong S., Wei Y., Wu Q., Yan L. Spatial transcriptomic survey of human embryonic cerebral cortex by single-cell RNA-seq analysis. Cell Res. 2018;28(7):730–745. doi: 10.1038/s41422-018-0053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Darnell J., Van Driesche S., Zhang C., Hung K., Mele A., Fraser C. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146(2):247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lappalainen T., Scott A.J., Brandt M., Hall I.M. Genomic analysis in the age of human genome sequencing. Cell. 2019;177(1):70–84. doi: 10.1016/j.cell.2019.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Acuna-Hidalgo R., Veltman J.A., Hoischen A. New insights into the generation and role of de novo mutations in health and disease. Genome Biol. 2016;17(1):241. doi: 10.1186/s13059-016-1110-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramaekers V.T., Rothenberg S.P., Sequeira J.M., Opladen T., Blau N., Quadros E.V. Autoantibodies to folate receptors in the cerebral folate deficiency syndrome. N Engl J Med. 2005;352(19):1985–1991. doi: 10.1056/NEJMoa043160. [DOI] [PubMed] [Google Scholar]
- 51.Ramaekers V., Blau N., Sequeira J., Nassogne M.-C., Quadros E. Folate receptor autoimmunity and cerebral folate deficiency in low-functioning autism with neurological deficits. Neuropediatrics. 2007;38(6):276–281. doi: 10.1055/s-2008-1065354. [DOI] [PubMed] [Google Scholar]
- 52.Caccamo D., Condello S., Gorgone G., Crisafulli G., Belcastro V., Gennaro S. Screening for C677T and A1298C MTHFR polymorphisms in patients with epilepsy and risk of hyperhomocysteinemia. Neuromolecular Med. 2004;6(2-3):117–126. doi: 10.1385/NMM:6:2-3:117. [DOI] [PubMed] [Google Scholar]
- 53.Das K.C., Herbert V. In vitro DNA synthesis by megaloblastic bone marrow: effect of folates and cobalamins on thymidine incorporation and de novo thymidylate synthesis. Am J Hematol. 1989;31(1):11–20. doi: 10.1002/ajh.2830310103. [DOI] [PubMed] [Google Scholar]
- 54.Geschwind D.H., State M.W. Gene hunting in autism spectrum disorder: on the path to precision medicine. Lancet Neurol. 2015;14(11):1109–1120. doi: 10.1016/S1474-4422(15)00044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lemay P., Guyot M.-C., Tremblay É., Dionne-Laporte A., Spiegelman D., Henrion É. Loss-of-function de novo mutations play an important role in severe human neural tube defects. J Med Genet. 2015;52(7):493–497. doi: 10.1136/jmedgenet-2015-103027. [DOI] [PubMed] [Google Scholar]
- 56.Walters J.T.R., Owen M.J. Endophenotypes in psychiatric genetics. Mol Psychiatry. 2007;12(10):886–890. doi: 10.1038/sj.mp.4002068. [DOI] [PubMed] [Google Scholar]
- 57.Xie S., Wang Z., Okano M., Nogami M., Li Y., He W.-W. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene. 1999;236(1):87–95. doi: 10.1016/s0378-1119(99)00252-8. [DOI] [PubMed] [Google Scholar]
- 58.Okano M., Bell D.W., Haber D.A., Li E.n. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 59.Tatton-Brown K., Seal S., Ruark E., Harmer J., Ramsay E., del Vecchio Duarte S. Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat Genet. 2014;46(4):385–388. doi: 10.1038/ng.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tan Y.C., Chow V.T. Novel human HALR (MLL3) gene encodes a protein homologous to ALR and to ALL-1 involved in leukemia, and maps to chromosome 7q36 associated with leukemia and developmental defects. Cancer Detect Prev. 2001;25(5):454–469. [PubMed] [Google Scholar]
- 61.Ruault M., Brun M.E., Ventura M., Roizès G., De Sario A. MLL3, a new human member of the TRX/MLL gene family, maps to 7q36, a chromosome region frequently deleted in myeloid leukaemia. Gene. 2002;284(1-2):73–81. doi: 10.1016/s0378-1119(02)00392-x. [DOI] [PubMed] [Google Scholar]
- 62.Glaser S., Schaft J., Lubitz S., Vintersten K., van der Hoeven F., Tufteland K.R. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development. 2006;133(8):1423–1432. doi: 10.1242/dev.02302. [DOI] [PubMed] [Google Scholar]
- 63.Klenova E.M., Nicolas R.H., Paterson H.F., Carne A.F., Heath C.M., Goodwin G.H. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol Cell Biol. 1993;13(12):7612–7624. doi: 10.1128/mcb.13.12.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi Y., Seto E., Chang L.S., Shenk T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67(2):377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 65.Stark M., Gonen N., Assaraf Y.G. Functional elements in the minimal promoter of the human proton-coupled folate transporter. Biochem Biophys Res Commun. 2009;388(1):79–85. doi: 10.1016/j.bbrc.2009.07.116. [DOI] [PubMed] [Google Scholar]
- 66.Barnard R.A., Pomaville M.B., O'Roak B.J. Mutations and Modeling of the Chromatin Remodeler CHD8 Define an Emerging Autism Etiology. Front Neurosci. 2015;9:477. doi: 10.3389/fnins.2015.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rubenstein J.L. Annual Research Review: Development of the cerebral cortex: implications for neurodevelopmental disorders. J Child Psychol Psychiatry. 2011;52(4):339–355. doi: 10.1111/j.1469-7610.2010.02307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Voineagu I., Wang X., Johnston P., Lowe J.K., Tian Y., Horvath S. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474(7351):380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Satterstrom F.K., Kosmicki J.A., Wang J., Breen M.S., De Rubeis S., An J.Y. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell. 2020;180(3):568–84 e23. doi: 10.1016/j.cell.2019.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhong S., Ding W., Sun L.e., Lu Y., Dong H., Fan X. Decoding the development of the human hippocampus. Nature. 2020;577(7791):531–536. doi: 10.1038/s41586-019-1917-5. [DOI] [PubMed] [Google Scholar]
- 71.Langen M., Bos D., Noordermeer S.D.S., Nederveen H., van Engeland H., Durston S. Changes in the development of striatum are involved in repetitive behavior in autism. Biol Psychiatry. 2014;76(5):405–411. doi: 10.1016/j.biopsych.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 72.Kaya N., Alsagob M., D'Adamo M.C., Al-Bakheet A., Hasan S., Muccioli M. KCNA4 deficiency leads to a syndrome of abnormal striatum, congenital cataract and intellectual disability. J Med Genet. 2016;53(11):786–792. doi: 10.1136/jmedgenet-2015-103637. [DOI] [PubMed] [Google Scholar]
- 73.Platt R.J., Zhou Y., Slaymaker I.M., Shetty A.S., Weisbach N.R., Kim J.-A. Chd8 mutation leads to autistic-like behaviors and impaired striatal circuits. Cell Rep. 2017;19(2):335–350. doi: 10.1016/j.celrep.2017.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shohat S., Ben-David E., Shifman S. Varying intolerance of gene pathways to mutational classes explain genetic convergence across neuropsychiatric disorders. Cell Rep. 2017;18(9):2217–2227. doi: 10.1016/j.celrep.2017.02.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.