Abstract
Objective
The endoplasmic reticulum (ER)-resident E3 ligase HRD1 and its co-activator Sel1L are major components of ER-associated degradation (ERAD) machinery. Here, we investigated the molecular mechanism and functional significance underlying the circadian regulation of HRD1/Sel1L-mediated protein degradation program in hepatic energy metabolism.
Methods
Genetically engineered animal models as well as gain- and loss-of-function studies were employed to address the circadian regulatory mechanism and functional significance. Gene expression, transcriptional activation, protein–protein interaction, and animal metabolic phenotyping analyses were performed to dissect the molecular network and metabolic pathways.
Results
Hepatic HRD1 and Sel1L expression exhibits circadian rhythmicity that is controlled by the ER-tethered transcriptional activator CREBH, the nuclear receptor peroxisome proliferator-activated receptor α (PPARα), and the core clock oscillator BMAL1 in mouse livers. HRD1/Sel1L mediates polyubiquitination and degradation of the CREBH protein across the circadian cycle to modulate rhythmic expression of the genes encoding the rate-limiting enzymes or regulators in fatty acid (FA) oxidation, triglyceride (TG) lipolysis, lipophagy, and gluconeogenesis. HRD1 liver-specific knockout (LKO) mice displayed increased expression of the genes involved in lipid and glucose metabolism and impaired circadian profiles of circulating TG, FA, and glucose due to overproduction of CREBH. The circadian metabolic activities of HRD1 LKO mice were inversely correlated with those of CREBH KO mice. Suppressing CREBH overproduction in the livers of HRD1 LKO mice restored the diurnal levels of circulating TG and FA of HRD1 LKO mice.
Conclusion
Our work revealed a key circadian-regulated molecular network through which the E3 ubiquitin ligase HRD1 and its co-activator Sel1L regulate hepatic circadian metabolism.
Keywords: Endoplasmic reticulum, ER-associated degradation, Lipid metabolism, Circadian metabolism, Transcriptional regulation
Abbreviations: ER, endoplasmic reticulum; ERAD, ER-associated degradation; HRD1, HMG-CoA reductase degradation 1 homolog; Sel1L, SEL1L adaptor subunit of ERAD E3 ubiquitin ligase; CREBH, cAMP-responsive element-binding protein, hepatic-specific; PPARα, peroxisome proliferator–activated receptor α; BMAL1, aryl hydrocarbon receptor nuclear translocator-like protein 1; UPR, unfolded protein response; LC3, microtubule-associated protein 1A/1B-light chain 3; PPARα, peroxisome proliferator-activated receptor α; ATG7, autophagy-related protein 7; BMAL1, brain and muscle ARNT-Like 1; PCK1, phosphoenolpyruvate carboxykinase 1; G6PC, glucose-6-phosphatase catalytic subunit; FA, fatty acids; TG, triglyceride; BOH, beta-hydroxybutyrate; KO, knockout; CTL, control.
Graphical abstract
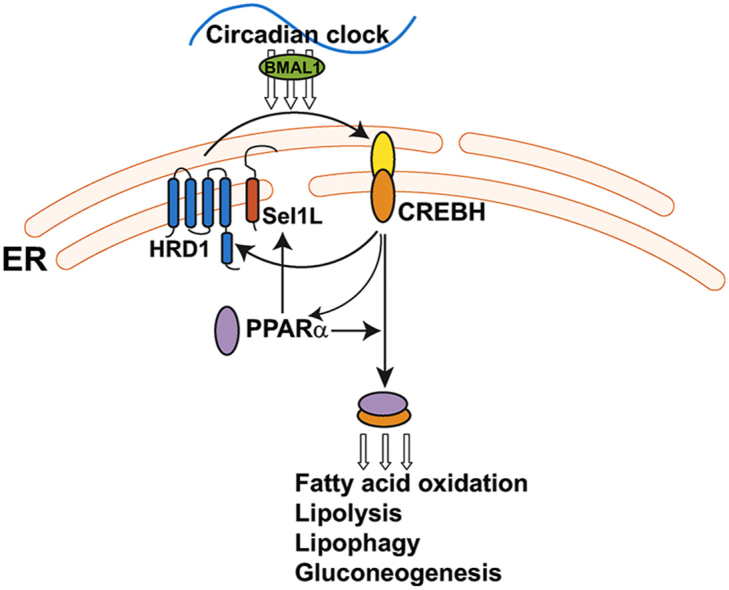
Highlights
-
•
The E3 ubiquitin ligase HRD1 and its co-factor Sel1L are regulated by the circadian clock.
-
•
HRD1-mediated protein degradation program integrates hepatic energy metabolism with circadian rhythm.
-
•
HRD1/Sel1L mediates circadian degradation of CREBH to regulate hepatic lipid and glucose homeostasis.
-
•
BMAL1, CREBH, and PPARα coordinate to control rhythmic expression of HRD1/Sel1L.
1. Introduction
Circadian rhythms are natural and internal processes that exhibit endogenous oscillations and are entrainable by light and food as well as stress signals [1]. At the molecular level, circadian oscillations are generated by a network of clock-regulated genes forming a transcriptional auto-regulatory feedback loop, which is under the control of the core circadian transcripion factors circadian locomotor output cycles kaput (CLOCK)/aryl hydrocarbon receptor nuclear translocator-like protein 1 (BMAL1) heterodimer [2]. In mammals, circadian integration of metabolic programs optimizes energy homeostasis across the circadian cycle [3]. In particular, the liver circadian clock orchestrates energy storage and utilization to maintain whole-body energy homeostasis. Disruption of the liver circadian clock leads to metabolic alterations and contributes to the development of metabolic disease [4,5]. Although significant progress has been made in understanding the relationships between circadian clock and metabolism, the major machineries that control liver energy metabolism over the circadian oscillation and their impacts in rhythmic physiology and behavior remain to be further elucidated.
Recently, we discovered that circadian clock regulates the activation of an endoplasmic reticulum (ER)-tethered, liver-enriched transcription factor, called CREBH (cAMP-responsive element binding protein, hepatocyte specific), which functions as a major circadian metabolic regulator [6]. Proteolytic activation of CREBH in the liver exhibits typical circadian rhythmicity controlled by the core clock oscillator BMAL1 and AKT/glycogen synthase kinase 3β (GSK3β) signaling pathway [6]. Functionally, CREBH regulates circadian rhythmic expression of the key genes involved in triglyceride (TG) lipolysis, fatty acid (FA) oxidation, lipophagy, gluconeogenesis, and glucogenolysis [[6], [7], [8]]. CREBH knockout (KO) mice developed profound non-alcoholic steatohepatitis (NASH) and hypertriglyceridemia under an atherogenic high-fat diet [9]. In human, patients with hypertriglyceridemia exhibit high-rate functional mutations of the CREBH gene [10,11]. At the molecular level, CREBH and peroxisome proliferator-activated receptor α (PPARα) function as binary transcriptional activators to regulate lipid and glucose homeostasis in response to metabolic alterations or circadian cues [7,8,12]. CREBH regulates expression of PPARα in the liver in response to energy demands, and activated CREBH protein interacts with PPARα to form a transcriptional complex through which both factors function in synergy to activate expression of the major metabolic regulators, including fibroblast growth factor 21 (FGF21) [7,12].
ER-associated degradation (ERAD) is the key program of ER protein quality control through which the ER directs the degradation of misfolded or unfolded proteins in order to maintain ER homeostasis under stress conditions [[13], [14], [15]]. HMG-CoA reductase degradation 1 homolog (HRD1), the major ER-resident E3 ligase, and its co-factor subunit of ERAD E3 ubiquitin ligase (Sel1L) form the most conserved branch of mammalian ERAD machinery [[16], [17], [18]]. The HRD1/Sel1L branch of ERAD not only recognizes and degrades misfolded or unfolded proteins, but also processes correctly folded proteins which are linked to pathophysiological processes. Recently, we demonstrated that HRD1 and Sel1L catalyze proteasomal degradation of the metabolic transcription factor CREBH in the liver. HRD1 KO mice displayed growth retardation, female infertility, and impaired diurnal circadian behavior partially due to hyperproduction of the hepatokine FGF21, a target of CREBH-mediated transcriptional program in the liver [12,[19], [20], [21]].
In this study, we demonstrated that the ER-resident E3 ubiquitin ligase HRD1 and its co-factor Sel1L are regulated by the circadian clock. HRD1/Sel1L and the ER-associated circadian transcriptional regulator CREBH form an interdependent regulatory loop, in which CREBH couples with the nuclear receptor PPARα, to promote the Hrd1 and Sel1L gene transcription, while HRD1 mediates degradation of CREBH protein across the circadian cycle. Functionally, our studies revealed that the HRD1/Sel1L-CREBH regulatory axis in the liver functions as a major circadian control of lipid mobilization and energy homeostasis.
2. Material and methods
2.1. Reagents
Synthetic oligonucleotides were purchased from Integrated DNA Technologies, Inc. (Coralville, IA). The commercially available antibodies were used to detect endogenous protein levels of HRD1 (Sigma), Sel1L (Invitrogen), BMAL1 (Novus Biologicals), PPARα (Millipore), SIRT1 (ABcam), and GAPDH (Sigma), respectively, in mouse liver or primary hepatocyte lysates by Western blot or Immunopreciptation (IP)-Western blot analysis. The affinity-purified rabbit polyclonal anti-CREBH antibody was previously developed in our laboratory [12]. The recombinant adenovirus expressing CREBH short hairpin RNA (shRNA) was prepared by VectorBuilder, Inc. (Chicago, IL). The adenovirus expressing mouse Bmal1 shRNA was previously described [22]. Kits for measuring TG, FA, ketone body, and glucose were purchased from BioAssay System (Hayward, CA).
2.2. Animal model
All animal experiments were performed with the approval of the Institutional Animal Care and Use Committees (IACUC) at the Wayne State University and the Northwestern University, in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Four-month-old wild-type and CREBH KO C57BL6 mice were housed in 12-h/12-h light/dark cycles with free access to food and water as we described previously [6]. Liver samples from 3 to 4 mice per time point per genotype group were collected every 3 or 4 h for a 24-h circadian cycle. HRD1 LKO and BMAL1 LKO mice were generated as previously described [19,23]. Measurements of circadian energy expenditure, indirect calorimetry, and food intake were performed using a comprehensive metabolic cage PhenoMaster System (TSE Systems, Inc.). For adenovirus injection experiments, recombinant adenovirus expressing mouse CREBH shRNA or scramble shRNA were injected to HRD1-LKO mice through tail-vein injection. Approximately 1 × 1010 PFU of adenovirus in 0.20 ml of phosphate-buffered saline (PBS) were intravenously injected into a mouse of approximately 20 g of body weight.
2.3. In vitro circadian synchronization of mouse primary hepatocytes
Primary hepatocytes isolated from C57BL/6 mice were infected with recombinant adenovirus expressing Bmal1 shRNA, scramble shRNA, activated CREBH (flag-tagged), or GFP at MOI (multiplicity of infection) of 100 or 150 for 24 h before being subjected to serum shock (50% horse serum) for 2 h for circadian synchronization [24]. After serum shock synchronization, the shock medium was replaced with serum-free medium. Cell lysates were collected at 8-h intervals between 24 h (circadian 0 h) and 72 h (circadian 48 h) post–serum shock for Western blot analysis.
2.4. Electrophoretic mobility shift assay (EMSA)
Liver nuclear extracts (NEs) from the mice over-expressing CREBH, PPARα, or GFP control were prepared by Subcellular Protein Fractionation kit (Thermo Scientific). EMSA was performed with Lightshift Chemiluminescent EMSA kit (Thermo Scientific). Liver NE (2.5 μg) was incubated with biotin-labeled probe (2.6 ng) in binding buffer for 30 min, and then separated in a 5% nondenaturing polyacrylamide gel. The gel was transferred to nylon membrane followed by Chemiluminescent Nucleic Acid Detection Module (Thermo Scientific).
2.5. Chromatin immunoprecipitation (ChIP) assay
ChIP assays were carried out with mouse liver tissue or Huh7 cells as previously described [8,12]. Antibodies used for immunoprecipitation were CREBH and PPARα and immunoprecipitated DNA was purified using a DNA Clean & Concentration kit (Zymo Research). The purified DNA was amplified to analyze the binding of CREBH and/or PPARα to target promoter regions and the polymerase chain reaction (PCR) products were separated by electrophoresis on a 1% agarose gel and visualized by ethidium bromide staining.
2.6. Immunoprecipitation (IP)–Western blot analyses
The endogenous protein–protein interactions between HRD1 and CREBH in mouse livers under the circadian clock were determined by IP-Western blot analysis. Approximately 200 μg of liver protein lysates were incubated with 1 μg of specific antibodies overnight at 4 °C. Protein complexes were immunoprecipitated using Dynabeads Protein G (Novex), resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred to a polyvinylidene difluoride (PVDF) membrane. The assay was followed by Western blot analysis with primary antibody against HRD1 or CREBH. The protein interaction signals were visualized using horseradish peroxidase (HRP)-conjugated Clean-Blot IP Detection Reagents (Thermo Scientific). The conjugated HRP was then developed by an enhanced chemiluminescence (ECL) detection reagent.
2.7. Statistics
The results of experiments were analyzed by several statistical methods. Unpaired Mann Whitney U testing was used for non-parametric comparisons. One-way analysis of variance (ANOVA) testing was used for parametric comparisons. Two-way ANOVA was used to distinguish the effects of genotypes from the effects of circadian time on gene expression and levels of mouse blood lipids. In all cases, a p value less than 0.05 was used to attribute statistical significance.
3. Results
3.1. Hepatic HRD1/Sel1L expression exhibits circadian rhythmicity and is regulated by CREBH and PPARα
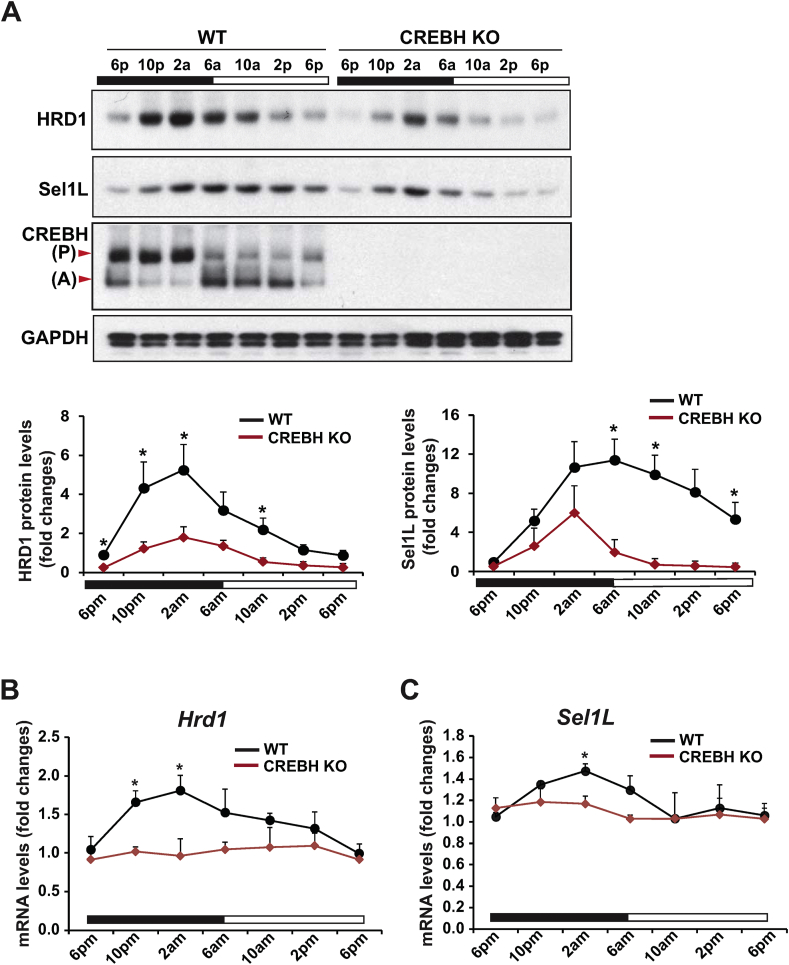
We previously demonstrated that the liver-enriched, ER-resident transcriptional factor CREBH, the target of HRD1/Sel1L degradation machinery, is regulated by the circadian clock [6,19]. As HRD1 and its co-factor Sel1L are liver-enriched protein factors [20,25], we evaluated whether the HRD1/Sel1L-mediated ERAD machinery in the liver is regulated by the circadian clock. First, we examined the 24-h rhythmic expression profile of HRD1 and Sel1L proteins in the livers of wild-type (WT) and CREBH KO mice under the circadian clock. In WT mouse livers, the levels of HRD1 protein exhibited a typical circadian rhythmic pattern, which peaked at 2 AM and troughed at 6 PM (Figure 1A). Similarly, levels of Sel1L protein displayed circadian rhythmicity with a peak at 6 AM and a trough at 6 PM. In contrast, circadian expression of HRD1 and Sel1L in the livers of CREBH KO mice was significantly reduced (Figure 1A), implicating a role of CREBH in regulating circadian expression of HRD1 and Sel1L. Next, we examined whether CRBEH, as a transcription factor, can regulate transcription of the mRNAs encoding HRD1 and Sel1L in the liver under the circadian clock. Consistent with the protein changes, Hrd1 and Sel1L transcripts also displayed circadian rhythmicity in WT mouse liver. In contrast, rhythmic expression levels of the Hrd1 and Sel1L mRNAs were repressed in the CREBH KO livers (Figure 1B–C). These results indicate that circadian rhythmic expression of HRD1 and Sel1L in the liver is regulated by CREBH.
Figure 1.
Hepatic HRD1/Sel1L expression exhibits circadian rhythmicity and is regulated by CREBH. (A) Western blot analysis of rhythmic levels of HRD1, Sel1L, CREBH, and GAPDH proteins in the livers of CREBH KO and WT mice, collected every 4 h in a 24-h circadian period. Liver protein lysates from 3 to 5 mice per time point per genotype group were used. The graphs show the quantification of the protein levels in mouse livers over the circadian cycle. The intensity of the protein signals, determined by Western blot densitometry, was normalized to that of GAPDH. Fold changes in protein levels were determined by comparison to the protein level in one of the WT control mice at 6 PM. Data are mean ± SEM (n = 3 experimental replicates). ∗P < 0.05. Black and white bars represent circadian light/dark phases. P, CREBH precursor; A, activated form of CREBH. (B–C) Rhythmic expression levels of HRD1 and Sel1L genes in the livers of CREBH KO and WT control mice across a 24-h circadian cycle. The mRNA expression levels were determined by qPCR. Fold changes in mRNA levels were determined by comparison to levels in one of the WT control mice at 6 PM. Data represent means ± SEM (n = 3–5 mice/genotype/time point). ∗P < 0.05.
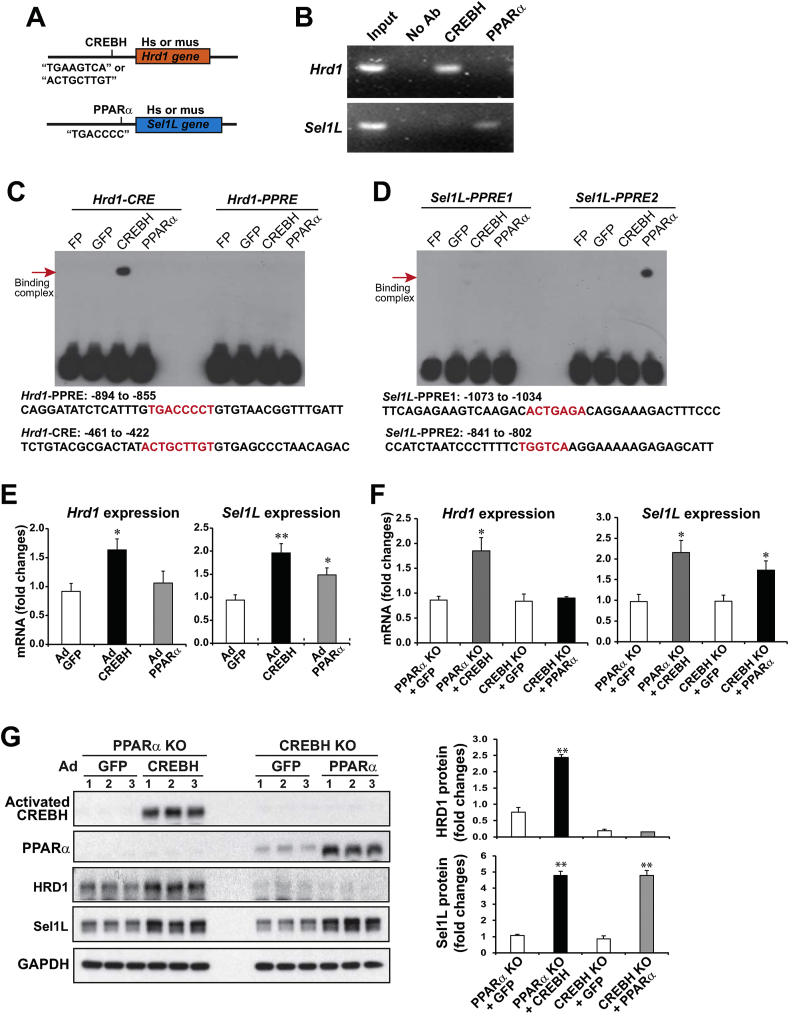
We previously demonstrated that CREBH and the nuclear receptor PPARα function as binary transcriptional activators to regulate their target genes in response to energy demands or circadian cues [8,12,26]. CREBH regulates expression of PPARα in the liver, and activated CREBH interacts with PPARα to synergistically promote expression of a number of genes encoding functions involved in lipid and glucose metabolism [12,26]. To understand the molecular basis underlying transcriptional regulation of HRD1 and Sel1L expression, we studied cis-regulatory elements in the promoter regions of human and mouse HRD1 and Sel1L genes. Highly conserved CREBH-binding elements (CREs) were identified in the promoter regions of the human and mouse HRD1 genes, while conserved PPARα-binding elements (PPREs) were found in the promoter regions of the human and mouse Sel1L genes (Figure 2A, S-Figure 1). To evaluate whether CREBH and PPARα directly regulate transcription of the HRD1 or Sel1L genes, we carried out chromatin immunoprecipitation (ChIP) analysis with WT mouse liver tissue samples to detect the binding activities of CREBH and PPARα to the Hrd1 or Sel1L gene promoter. Binding activities of CREBH, but not PPARα, to the mouse endogenous Hrd1 gene promoter region containing the CREBH-binding motif were detected in WT mouse livers (Figure 2B, S-Figure 1A, C). Consistent with the profile of PPARα-binding motif in the Sel1L gene promoter, binding activities of PPARα, but not CREBH, to the mouse endogenous Sel1L gene promoter region were detected in WT mouse livers (Figure 2B, S-Figure 1B, D). Further, we performed gel electrophoretic mobility shift assays (EMSA) with the mouse Hrd1 and Sel1L gene promoter oligonucleotides containing CREBH- and/or PPARα-binding motifs (Figure 2C–D) and mouse liver nuclear extracts (NE) expressing CREBH, PPARα, or GFP control. Binding to the Hrd1 gene promoter oligonucleotide containing the CREBH-binding element (CRE), but not the PPARα-binding element (PPRE), was detected with liver NEs from the mice expressing CREBH, but not PPARα or GFP (Figure 2C). However, binding activity to the Hrd1 promoter oligonucleotide containing PPRE was not detected with the liver NEs from the mice expressing PPARα. Further, PPARα binding activity to the mouse Sel1L gene promoter oligonucleotide containing PPRE2, but not PPRE1, was detected with the liver NEs from the mice expressing PPARα, but not CREBH or GFP (Figure 2D). These results suggested that CREBH, but not PPARα, directly bind to the Hrd1 gene promoter region containing the CREBH-binding element, while PPARα, but not CREBH, directly bind to the Sel1L gene promoter region containing the PPRE binding motif to respectively regulate their transcription.
Figure 2.
CREBH and PPARα directly activate expression of the Hrd1 and/or Sel1L genes in the liver. (A) Illustration of CREBH- or PPARα-binding sites in the promoter region of the human (hs) or mouse (ms) Hrd1 and Sel1L genes. (B) ChIP analysis of CREBH- and PPARα-binding activities to the endogenous Hrd1 and Sel1L gene promoters in the livers of WT mice. For ChIP-PCR, isolated mouse chromatins were immunoprecipitated with the antibody against CREBH, PPARα, or no antibody. Non-immunoprecipitated chromatins were included as input controls. PCR reactions were conducted using the primers amplifying the CRE or PPRE-binding motif present in the mouse Hrd1 or Sel1L gene promoter (sequences provided in S-Figure 1). The amplified PCR products were visualized on agarose gels. (C–D) Binding activities of mouse liver NEs expressing GFP, CREBH, or PPARα to the CRE- or PPRE-containing promoter regions of mouse Hrd1 or Sel1L genes. EMSA was performed using the liver NEs from WT mice infected with adenovirus expressing GFP, CREBH, or PPARα and the mouse Hrd1 or Sel1L gene probe containing the CRE- and/or PPRE-binding motifs. Free probe (FP) indicates the control reaction with the CRE or PPRE probe in the absence of NE. The sequences of mouse Hrd1 or Sel1L promoter oligonucleotides containing CRE or PPRE motif used for EMSA were indicated. The protein-DNA binding complexes were visualized by shifted signals and highlighted by arrows. (E) Expression levels of Hrd1 and Sel1L mRNAs in the livers of mice infected with adenovirus (Ad) overexpressing GFP, activated CREBH, or PPARα. mRNA expression levels were determined by qPCR. Fold changes in mRNA levels were determined. Data represent means ± SEM (n = 3). ∗P < 0.05, ∗∗P < 0.01. (F–G) PPARα-null mice were infected with recombinant adenovirus (Ad) expressing GFP or the activated form of CREBH, whereas CREBH-null mice were infected with Ad expressing GFP or PPARα via tail vein injection. Total liver RNAs and protein samples were prepared from mice under 14 h fasting. Levels of the Hrd1 and Sel1L mRNAs (F) or CREBH, PPARα, HRD1, Sel1L, and GAPDH proteins (G) in mouse livers were determined by qPCR or Western blot analysis. The graphs in panel G show the fold changes of HRD1 and Sel1L protein levels, determined by Western blot densitometry, in mouse livers. Fold change of the normalized protein levels was calculated by comparing to the level in one of PPARα KO mice expressing GFP. Data represent means ± SEM (n = 3 experimental replicates). ∗P < 0.05 and ∗∗P < 0.01 for PPARα KO + CREBH vs PPARα KO + GFP or CREBH KO + PPARα vs CREBH KO + GFP.
To evaluate the effect of CREBH or PPARα on driving expression of HRD1 and Sel1L, we over-expressed an activated form of CREBH or PPARα in mouse livers through tail vein injection of the recombinant adenovirus expressing CREBH or PPARα into WT mice. The quantitative PCR (qPCR) analyses indicated that overexpression of the activated CREBH significantly increased expression of the Hrd1 and Sel1L genes in the mouse livers (Figure 2E). In parallel, PPARα overexpression significantly increased expression of Sel1L, but not Hrd1, in the livers. These results indicate that CREBH and PPARα directly activate expression of the Hrd1 and Sel1L genes, respectively. Interestingly, activated CREBH can increase expression of the mouse Sel1L gene, although CREBH does not bind to the Sel1L gene promoter region (Figure 2B–D). This observation suggests that CREBH may promote expression of the Sel1L gene through boosting PPARα expression, as PPARα has been demonstrated to be a target of CREBH [9,12]. To further discern the roles of CREBH and PPARα in driving expression of the Hrd1 and Sel1L genes in mouse livers, we examined expression of HRD1 and Sel1L in PPARα KO mice expressing exogenous CREBH and in CREBH KO mice expressing exogenous PPARα (Figure 2F–G). Adenoviral expression of exogenous CREBH or PPARα in the liver of PPARα KO or CREBH KO mice was confirmed by immunoblotting analyses (Figure 2G). Expression of exogenous CREBH, but not GFP, in the PPARα KO mice increased expression of HRD1 and Sel1L, at both mRNA and protein levels (Figure 2F–G). However, expression of exogenous PPARα only increased expression of Sel1L, but not HRD1, in the CREBH KO mice (Figure 2F–G). These results confirmed that CREBH plays major roles in driving expression of both HRD1 and Sel1L, while PPARα activates expression of Sel1L in the liver. As CREBH-binding activity to the Sel1L gene promoter was not detected (Figure 2B), CREBH may drive expression of Sel1L by upregulating PPARα or possibly other hepatic transcriptional factors that can activate expression of the Sel1L gene [9,12].
3.2. Circadian rhythmic expression of HRD1/Sel1L is under the control of BMAL1
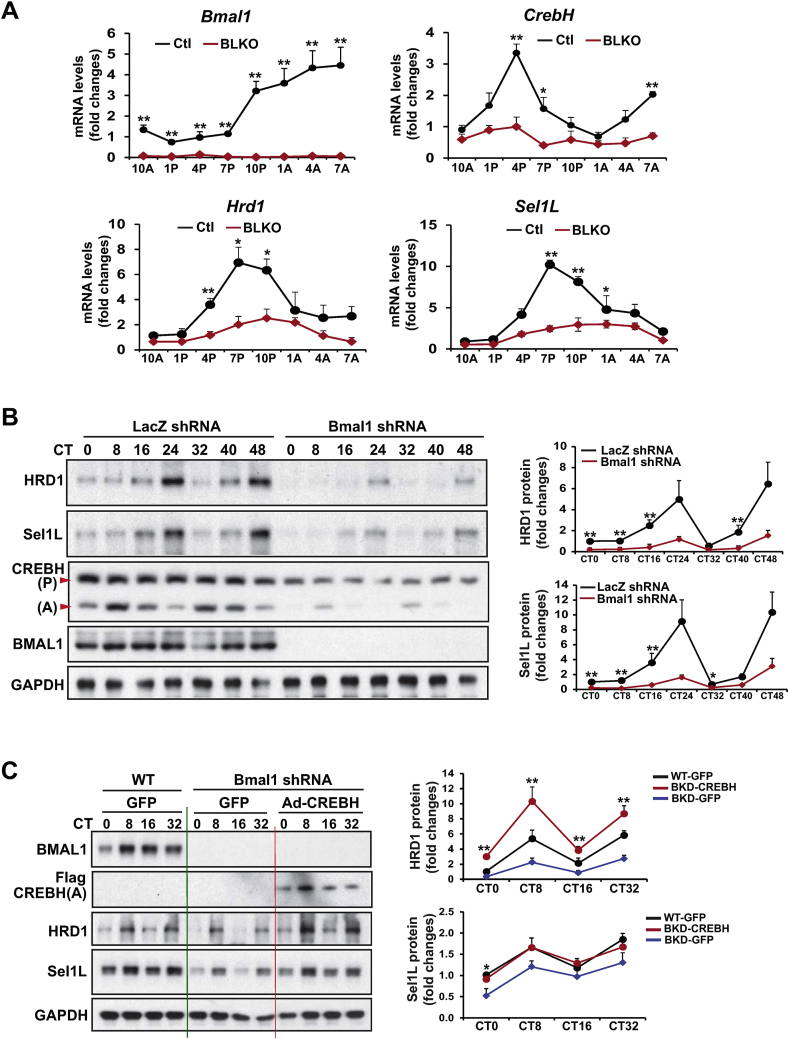
BMAL1, the core molecular circadian oscillator, plays a central role in regulating expression or activities of circadian output regulators [1]. To determine whether BMAL1 regulates circadian rhythmic expression of HRD1 and Sel1L, we examined the expression levels of Hrd1, Sel1L, and CrebH mRNAs in liver-specific Bmal1 conditional knockout (Bmal1 LKO) and control mouse livers collected every 3 h during a 24-h circadian period. The qPCR analysis showed that rhythmic expression levels and amplitudes of the Hrd1, Sel1L, and CrebH mRNAs were reduced in the livers of Bmal1 LKO mice across the circadian cycle, compared to those in the control littermates (Figure 3A). To verify the cell-autonomous function of BMAL1 in regulating HRD1 and Sel1L expression in hepatocytes, we knocked down BMAL1 in mouse primary hepatocytes using adenoviral-based expression of Bmal1 short hairpin RNA (shRNA), and then subjected the Bmal1-knockdown and control primary hepatocytes to horse serum shock for circadian synchronization [24]. Across a 48-h circadian period, both HRD1 and Sel1L expression in primary hepatocytes exhibits typical circadian rhythmicity (Figure 3B). However, the amplitudes of HRD1 and Sel1L expression in hepatocytes across the circadian cycles were significantly repressed by knockdown of BMAL1 (Figure 3B). These data confirmed that circadian rhythmic expression of HRD1 and Sel1L in primary hepatocytes is under the control of the autonomous circadian oscillator BMAL1.
Figure 3.
BMAL1 controls circadian rhythmic expression of HRD1/Sel1L in the liver through regulating CREBH. (A) Rhythmic expression levels of Bmal1, CrebH, Hrd1, and Sel1L genes in the livers of Bmal1 LKO and fl/fl control mice across a 24-h circadian cycle. The liver total RNAs were prepared from the mice collected every 3 h during a 24-h circadian period. The mRNA expression levels were determined by qPCR. Fold changes in mRNA levels were determined by comparison to levels in one of the control mice at 10 AM. Data represent means ± SEM (n = 3–4 mice/genotype/time point). ∗P < 0.05, ∗∗P < 0.01. (B) Western blot analysis of HRD1, Sel1L, CREBH, BMAL1, and GAPDH protein levels in Bmal1 knockdown and control primary hepatocytes during a 48-h circadian cycle. Mouse primary hepatocytes were infected with adenovirus expressing Bmal1 shRNA or control LacZ shRNA for 24 h before being subjected to horse serum shock for circadian synchronization. Cell lysates were collected at 8-h intervals during a 48-h circadian cycle for Western blot analysis to determine levels of HRD1, Sel1L, BMAL1, and GAPDH. The graphs show the rhythmic fold changes of HRD1 and Sel1L proteins in Bmal1 knockdown and control primary hepatocytes during the circadian cycle. Protein signals, determined by Western blot densitometry, were normalized to that of GAPDH. Fold change of the normalized HRD1 or Sel1L protein levels at each circadian time point was calculated by comparing to the level in control cells at the starting circadian time. Data are mean ± SEM (n = 3 experimental replicates). ∗P < 0.05, ∗∗P < 0.01. CT, circadian time. (C) Western blot analyses of BMAL1, CREBH, HRD1, Sel1L, and GAPDH in Bmal1-knockdown and WT control mouse primary hepatocytes expressing the activated CREBH or GFP control under the circadian clock. Mouse primary hepatocytes were infected with Ad expressing Bmal1 shRNA, the activated form of CREBH (Flag-tagged), or GFP before being subjected to horse serum shock for circadian synchronization. Primary hepatocytes from WT mice were infected with Ad expressing GFP (WT-GFP) were included as a control. Cell lysates were collected throughout a 32-h circadian cycle for Western blot analyses. The graphs show the rhythmic fold changes of HRD1 and Sel1L protein levels, determined by Western blot densitometry, in Bmal1-knockdown and WT control hepatocytes expressing CREBH or GFP control. Fold change of normalized protein levels at each circadian time point was calculated by comparing to the level in control cells at the starting circadian time. Data are mean ± SEM (n = 3 experimental replicates). ∗P < 0.05, ∗∗P < 0.01.
To evaluate whether BMAL1 controls circadian rhythmic expression of HRD1 and Sel1L in the liver through regulating CREBH, we performed reconstitution experiments by overexpressing the activated form of CRBEH or GFP control in BMAL1-knockdown primary hepatocytes across the circadian cycle. As shown by the WT and Bmal1-knockdown mouse primary hepatocytes expressing GFP control, the amplitudes of the cleaved/activated CREBH protein in the primary hepatocytes across a 32-h circadian period were significantly repressed by knockdown of BMAL1 (Figure 3C). In parallel with the reduced levels of the activated CREBH protein, rhythmic expression levels of HRD1 and Sel1L were decreased in Bmal1-knockdown primary hepatocytes during the circadian oscillation. However, when the activated form of CREBH was expressed in the BMAL1-knockdown mouse primary hepatocytes, rhythmic expression levels of HRD1 and Sel1L in the BMAL1-knockdown hepatocytes was restored, or even increased for the case of HRD1, compared to those in the WT mouse primary hepatocytes expressing GFP control (Figure 3C). These results supported the conclusion that CREBH is controlled by BMAL1 in regulating rhythmic expression of hepatic HRD1 and Sel1L under the circadian clock.
3.3. Hepatic HRD1 is required to maintain circadian rhythmic levels of circulating TG, FFA, and glucose
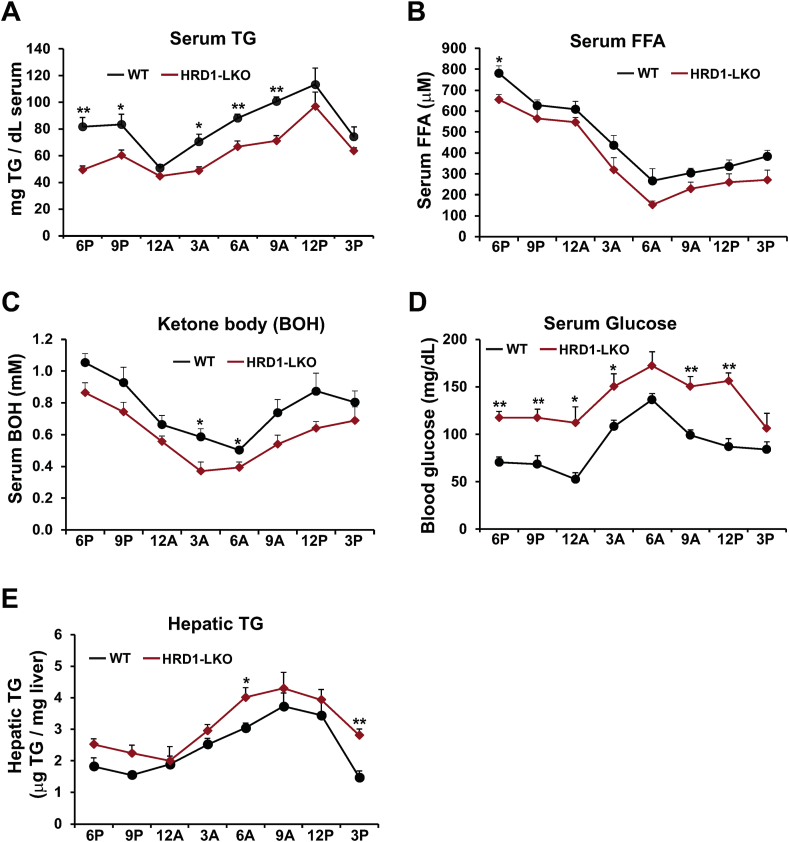
To determine whether HRD1 regulates energy homeostasis under the circadian clock, we examined rhythmic profiles of circulating lipids and glucose in the liver-specific HRD1 conditional KO (HRD1 LKO) mice and WT control mice. Rhythmic profiles of serum triglycerides (TG), free fatty acids (FFA), ketone bodies, and glucose were significantly altered in the HRD1 LKO mice under the circadian clock (Figure 4A–D). Compared to the WT control mice, HRD1 LKO mice exhibited lower levels of serum TG, FFA, and β-hydroxybutyrate (BOH), the predominant ketone body, but higher levels of glucose in the blood, over the 24-h circadian period (Figure 4A–C). The dysregulated circadian lipid and glucose profiles in HRD1 LKO mice suggested that HRD1-mediated ERAD may regulate hepatic energy homeostasis in a circadian-dependent manner. Notably, we previously showed that CREBH KO mice displayed significantly higher amplitudes of serum TG and FFA but lower amplitude of serum glucose across the circadian cycle, due to the roles of CREBH in regulating TG lipolysis, FA oxidation, and gluconeogenesis under the circadian clock [6,7]. The HRD1 LKO mice showed inverted circadian rhythmic patterns in serum lipid and glucose profiles, compared to CREBH KO mice, implicating a potential repressive relationship between HRD1 and CREBH in regulating circadian energy metabolism. HRD1 LKO mice displayed a pattern of increase in hepatic TG, compared to the control mice, during the circadian cycle, especially in the daytime period (Figure 4E). This is likely due to accumulation of CREBH protein in HRD1-deficient mouse livers, as CREBH is known to promote hepatic lipogenesis, lipid droplet growth and hepatic steatosis when it is over-expressed [9,27].
Figure 4.
HRD1 is required to maintain circadian rhythmic levels of circulating TG, FFA, BOH, and glucose in mice. (A–D) Levels of serum TG, FFA, BOH, and glucose in HRD1 LKO and WT mice under the circadian clock. Blood samples were collected every 3 h across a 24-h circadian cycle. (E) Levels of hepatic TG of HRD1 LKO and WT mice under the circadian clock. The liver tissue samples from HRD1 LKO and WT control mice were collected every 3 h across a 24-h circadian cycle. Data are mean ± SEM (n = 3 mice/genotype/time point). ∗P < 0.05, ∗∗P < 0.01. TG, triglycerides; FFA, free fatty acids; BOH, β-Hydroxybutyrate.
3.4. HRD1 regulates rhythmic expression of the gene encoding functions involved in hepatic FA oxidation, TG lipolysis, lipophagy, and gluconeogenesis
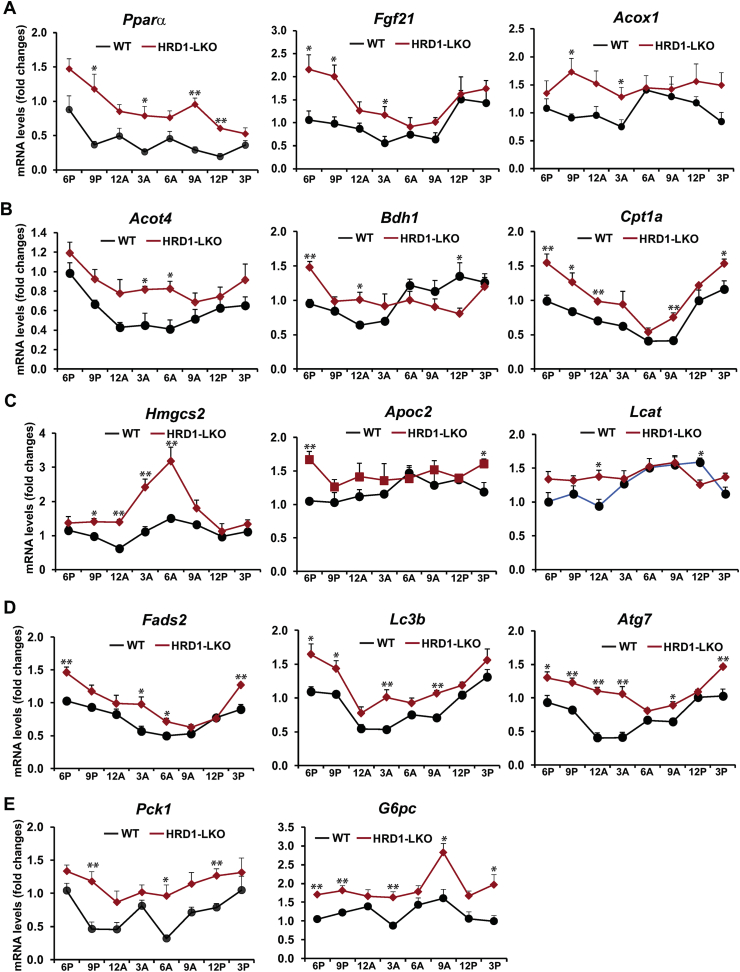
To understand the molecular basis underlying the metabolic phenotype of HRD1 LKO mice under the circadian clock, we examined circadian rhythmic expression profiles of the genes involved in hepatic lipid and glucose metabolism in the livers of HRD1 LKO and WT control mice. The qPCR analysis indicated that circadian rhythmic expression of the following genes were increased in HRD1 LKO mice (Figure 5A–E): 1) the genes encoding the enzymes or regulators in FA oxidation, including PPARα, FGF21, Acyl-CoA oxidase 1 (Acox1), Acyl-CoA thioesterase 4 (Acot4), 3-hydroxybutyrate dehydrogenase 1 (BDH1), carnitine palmitoyltransferase 1A (CPT1α), and 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 (HMGCS2); 2) the genes encoding the co-activators or enzymes in TG lipolysis, including apolipoprotein C2 (ApoC2), lecithin-cholesterol acyltransferase (Lcat), and FA desaturase 2 (Fads2); 3) the genes encoding the key component or regulator in lipophagy, including microtubule-associated protein 1 light chain 3 Beta (LC3b) and autophagy-related 7 (ATG7); and 4) the genes coding the rate-limiting enzymes in gluconeogenesis, including phosphoenolpyruvate carboxykinase 1 (PCK1) and glucose-6-phosphatase catalytic subunit (G6PC). Hepatic FA oxidation, TG lipolysis, and lipophagy, the autophagic degradation of intracellular lipid droplets [28], are all major lipid mobilization pathways in response to energy demands. As described above, the increased circadian expression of the key enzymes or regulators in FA oxidation, TG lipolysis, lipophagy, and gluconeogenesis in HRD1 LKO mouse livers is consistent with the decreased rhythmic levels of serum TG, FA, and ketone bodies but increased glucose levels in HRD1 LKO mice. Together, these results support that HRD1 functions as a metabolic regulator of circadian energy homeostasis by regulating expression of the key enzymes or regulators involved in hepatic FA oxidation, TG lipolysis, lipophagy, and gluconeogenesis.
Figure 5.
HRD1 regulates rhythmic expression of the gene involved in hepatic FA oxidation, TG lipolysis, lipophagy, and gluconeogenesis. (A–E) Rhythmic expression of the genes encoding the key enzymes or regulators involved in FA oxidation, TG lipolysis, lipophagy, and gluconeogenesis in HRD1 LKO and WT mouse livers across the circadian cycle. Levels of mRNA were determined by qPCR. Fold changes of mRNA levels were calculated by comparing to the level in one of the WT mice at 6 PM. Data are mean ± SEM (n = 3 mice/genotype/time point). ∗P < 0.05, ∗∗P < 0.01.
3.5. HRD1 LKO animals contrast with CREBH KO animals in circadian rhythmic profiles of energy consumption
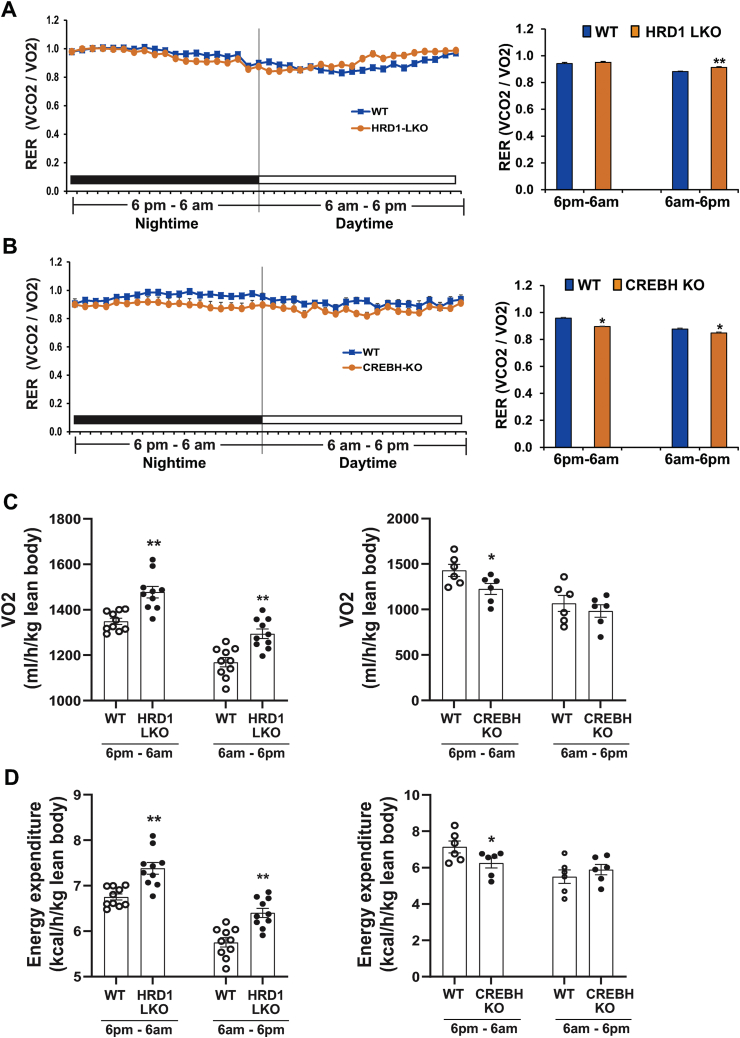
We previously demonstrated that HRD1/Sel1L promotes proteome-mediated degradation of ER-resident CREBH protein in the liver [19,20]. As shown above (Figure 4, Figure 5) and as we described previously [6], HRD1 LKO and CREBH KO mice displayed opposite regulation of circadian lipid profiles and metabolic gene expression involved in FA and TG catabolism. Accordingly, we questioned whether HRD1 LKO mice are opposite to CREBH KO mice in circadian rhythmicity of energy expenditure. To address this question, we examined circadian metabolic rates of HRD1 LKO, CREBH KO, and WT control mice. Across the circadian clock, HRD1 LKO mice showed increased metabolic rates, especially during the daytime period (6 AM–6 PM), as indicted by the respiratory exchange ratio (RER) (Figure 6A). This is in line with the increased expression of the genes encoding the key enzymes or regulators for hepatic FA oxidation, TG lipolysis, lipophagy, and gluconeogenesis in HRD1 LKO mice (Figure 5). In contrast, CREBH KO mice displayed decreased metabolic rates under the circadian clock (Figure 6B), consistent with the roles of CREBH in activating expression of the genes involved in hepatic FA oxidation, TG lipolysis, lipophagy, and gluconeogenesis [[6], [7], [8],29]. Further analysis of lean body oxygen consumption and energy expenditure indicated that HRD1 LKO mice consumed more oxygen and energy, while CREBH KO mice consumed less oxygen and energy than the control mice (Figure 6C–D). In agreement with the increased energy consumption of HRD1 LKO mice, the HRD1 LKO mice consumed more food during the daytime period (S-Figure 2), but displayed reduced body weights (S-Figure 3) compared to the control mice.
Figure 6.
HRD1 LKO mice contrast with CREBH KO mice in circadian rhythmic profiles of energy consumption. (A–B) Circadian rhythmic profiles of RER (respiratory exchange ratio) of HRD1 LKO, CREBH KO, and WT control mice under the circadian clock, determined by the comprehensive metabolic cage PhenoMaster System. The graphs show quantification of RERs of HRD1 LKO, CREBH KO, and WT control mice during the nighttime (6 PM–6 AM) and daytime (6 AM–6 PM) periods. Each bar donates mean ± SEM (n = 10 mice for HRD1 LKO vs WT or 6 mice for CREBH KO vs WT). ∗p < 0.05; ∗∗p < 0.01. (C–D) Quantification of lean body VO2 (C) and energy expenditure (D) of HRD1 LKO, CREBH KO, and WT control mice during the nighttime and daytime periods. The quantitative analyses were based on the data generated through the measurement by the comprehensive metabolic cage PhenoMaster System. Each bar denotes mean ± SEM (n = 10 mice for HRD1 LKO vs WT or 6 mice for CREBH KO vs WT). ∗p < 0.05; ∗∗p < 0.01.
3.6. HRD1 mediates circadian rhythmic polyubiquitination and degradation of CREBH protein
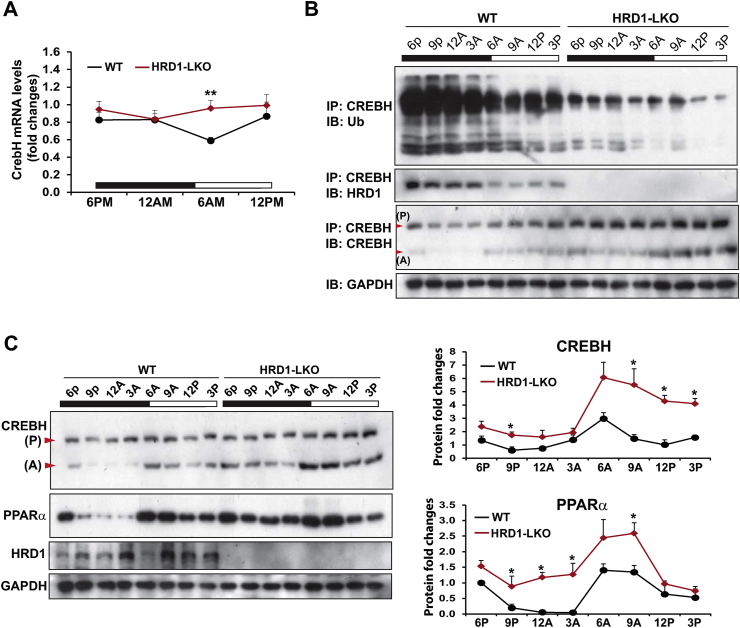
To understand the molecular basis underlying the functional connecon between HRD1 and CREBH in regulating circadian lipid metabolism, we examined whether HRD1 modulates CREBH protein stability under the circadian clock. Immunoblotting and qPCR analyses indicated that circadian expression of the CREBH protein, but not the CrebH mRNA, was significantly elevated in the HRD1 LKO mice (Figure 7A, C), suggesting that HRD1, as a major E3 ligase of ERAD machinery, may promote CREBH protein degradation under the circadian clock. Supporting this notion, polyubiquitination of CREBH protein in mouse livers exhibited circadian rhythmicity (Figure 7B, the 1st row). However, the polyubiquitination of CREBH proein was diminished in the HRD1 LKO mouse liver, indicating that HRD1 is an E3 ubiquitin ligase that mediates CREBH ubiquitination under the circadian clock. Coincident with the circadian-regulated CREBH ubiquitination, CREBH protein rhythmically interacted with HRD1 in the livers of WT, but not HRD1 LKO, mice under the circadian clock (Figure 7B, the 2nd row). Consistently, across the circadian cycle, the levels of CREBH proteins in the livers of HRD1 LKO mice were significantly higher than those of WT mice (Figure 7C). Taken together, these data indicated that HRD1 rhythmically interacts with CREBH and mediates circadian rhythmic ubiquitination and degradation of CREBH protein in the liver. Additionally, we previously demonstrated that CREBH regulates expression of the nuclear receptor PPARα to activate gene expression programs involved in hepatic lipid metabolism [12]. Correlated with the increased CREBH protein levels in the HRD1 LKO mouse livers, circadian rhythmic levels of PPARα protein were elevated in the livers of HRD1 LKO mice (Figure 7C), suggesting that HRD1 may regulate the CREBH/PPARα-mediated transcriptional program through controlling CREBH protein stability under the circadian clock.
Figure 7.
HRD1 promotes circadian rhythmic polyubiquitination and degradation of CREBH protein. (A) Circadian rhythmic expression of the CrebH mRNA in HRD1 LKO and WT mouse livers. Levels of mRNA were determined by qPCR. Fold changes of mRNA levels were calculated by comparing to the level in one of the WT mice at 6 PM. Data are mean ± SEM (n = 3 mice/genotype/time point). ∗∗P < 0.01. (B) IP-Western blot analyses of CREBH protein polyubiquitination and CREBH-HRD1 interaction in the livers of HRD1 LKO and WT control mice across the circadian cycle. The liver protein lysates were prepared from liver tissues of HRD1 LKO and WT mice collected every 3 h during a 24-h circadian period (n = 3 mice/genotype/time point). The liver protein lysates were pulled down by the rabbit polyclonal anti-CREBH antibody, followed by immunoblotting with the anti-ubiquitin (Ub), anti-HRD1, or anti-CREBH antibody. Levels of GAPDH were determined by immunoblotting as controls. (C) Western blot analyses of CREBH, PPARα, HRD1, and GAPDH protein levels in the livers of HRD1 LKO and WT control mice across the circadian cycle. The liver protein lysates were prepared from liver tissues of HRD1 LKO and WT mice collected every 3 h during a 24-h circadian period (n = 3 mice/genotype/time point). P, CREBH precursor/full-length protein; A, activated/cleaved CREBH protein. The graphs show rhythmic fold changes of CREBH and PPARα protein levels, determined by Western blot densitometry, in the livers of HRD1 LKO and control. Fold change of the normalized protein levels was determined by comparing to the level in one of the WT mice at 6 PM. Data are mean ± SEM (n = 3 experimental replicates). ∗P < 0.05.
3.7. Suppression of CREBH in HRD1 LKO mouse livers rescues the phenotype of hyperactive circadian lipid mobilization caused by HRD1 deficiency
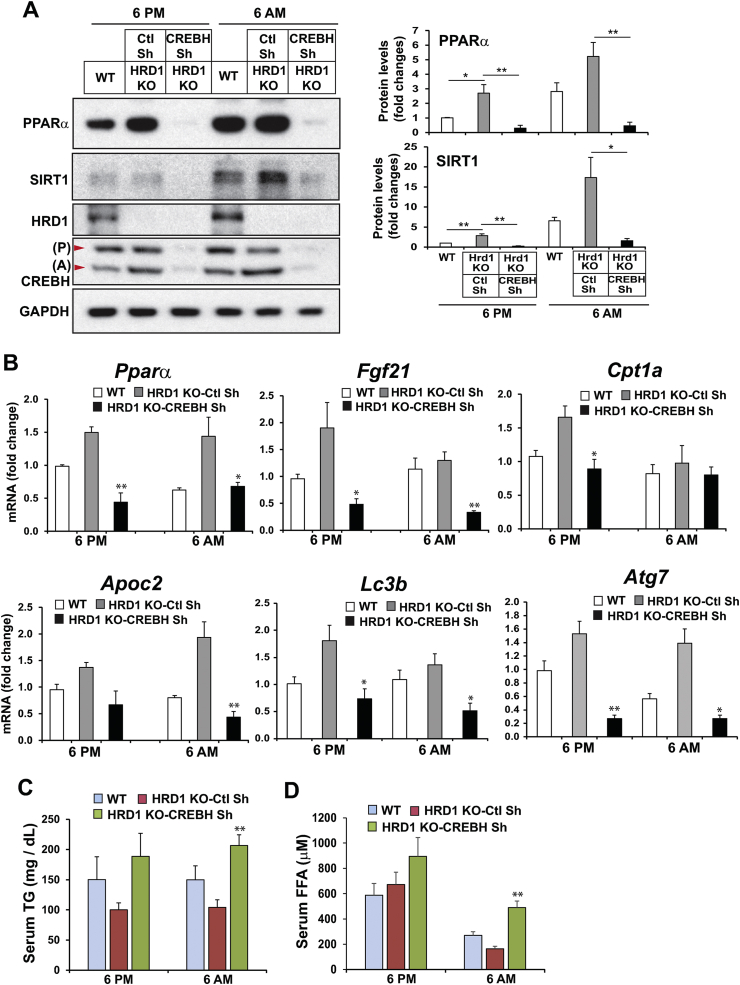
To validate whether HRD1 modulates circadian FA and TG metabolism through promoting degradation of CREBH, recombinant adenovirus (Ad) expressing CrebH shRNA was delivered into HRD1 LKO mice through tail vein infusion. We confirmed that the levels of CrebH mRNA and protein were barely detectable 5 days after injection of adenovirus expressing CrebH shRNA (Figure 8A, S-Figure 4). Consistent with the role of HRD1 in repressing hepatic FA oxidation and TG lipolysis through degrading CREBH, the levels of PPARα and SIRT1, the targets of CREBH and major transcriptional regulators of FA and TG metabolism [12,26], were significantly elevated in the livers of HRD1 LKO mice injected with Ad-scramble shRNA at the representative daytime 6 AM and the nighttime 6 PM (Figure 8A). However, when CREBH was knocked down in the livers of HRD1 LKO mice, the levels of PPARα and SIRT1 in the HRD1 LKO mouse livers were reduced (Figure 8A), thus confirming that elevation of CREBH is responsible for the upregulation of PPARα and SIRT1 in HRD1 LKO mouse livers (Figure 8A).
Figure 8.
Suppression of CREBH rescues the elevated circadian expression of the major lipid mobilization-promoting factors and the reduction of serum lipids in HRD1 LKO mice. (A) Western blot analyses of PPARα, SIRT1, HRD1, CREBH, and GAPDH in the livers of WT or HRD1 LKO mice infected with Ad expressing CREBH shRNA or control shRNA at the representative nighttime (6 PM) or daytime (6 AM) period. The liver protein lysates were prepared from liver tissues of WT or HRD1 LKO mice (n = 3 mice/genotype/time point). The graphs show the rhythmic fold changes of PPARα and SIRT1 protein levels, determined by Western blot densitometry, in WT or HRD1 LKO mouse livers expressing CREBH shRNA or control shRNA. Fold change of the normalized protein levels was calculated by comparing to the level in one of WT mice at 6 PM. The bars denote mean ± SEM (n = 3 experimental replicates). ∗P < 0.05, ∗∗P < 0.01. (B) Expression levels of the genes involved in FA oxidation, TG lipolysis, and lipophagy in the livers of WT or HRD1 LKO mice infected with Ad expressing CrebH shRNA or control shRNA at the representative nighttime (6 PM) or daytime (6 AM) period. Levels of mRNA were determined by qPCR. Fold changes of mRNA levels were calculated by comparing to the level in one of the WT mice at 6 PM. Data are mean ± SEM (n = 3 mice/genotype/time point). ∗P < 0.05 and ∗∗P < 0.01 for HRD1 KO-CrebH shRNA vs HRD1 KO-ctl shRNA. (C–D) Levels of serum TG and free FA in WT or HRD1 LKO mice infected with Ad expressing CrebH shRNA or control shRNA at the representative nighttime or daytime period. Data are mean ± SEM (n = 3 mice/genotype/time point). ∗∗P < 0.01 for HRD1 KO-CrebH shRNA vs HRD1 KO-ctl shRNA.
Further, we determined whether the elevation of the genes encoding functions in FA oxidation, TG lipolysis, and lipophagy by HRD1 deletion can be brought down by CREBH knockdown. Expression levels of the genes encoding the key enzymes or regulators in FA oxidation (PPARα, FGF21, Cpt1a), lipolysis (ApoC2), and lipophagy (LC3b, ATG7) were increased in the livers of HRD1 LKO mice injected with Ad-scramble shRNA during both the nighttime (6 PM) and daytime (6 AM) periods (Figure 8B). However, CREBH knockdown decreased the elevated expression of the genes involved in FA oxidation, TG lipolysis, and lipophagy in HRD1 LKO mouse livers (Figure 8B). These results confirmed that upregulation of the genes involved in FA and TG metabolism in the HRD1 LKO mouse livers is through upregulation of CREBH across the day–night circadian cycle.
Functionally, we measured the circulating TG and FA in the HRD1 LKO mice injected with Ad-scramble shRNA or Ad-CrebH shRNA. The levels of serum TG were decreased in the HRD1 LKO mice injected with Ad-scramble shRNA across the circadian cycle (Figure 8C–D). Knockdown of CREBH in HRD1 LKO mouse livers significantly increased the levels of serum FA and TG in HRD1 LKO mice across the day–night cycle (Figure 8C–D), thus confirming that HRD1 modulates FA and TG metabolism through promoting CREBH protein degradation in the liver.
4. Discussion
In this study, we revealed that the ER-resident E3 ubiquitin ligase HRD1 and its co-factor Sel1L are regulated by the circadian clock in the liver. The HRD1/Sel1L-CREBH/PPARα regulatory axis functions as a major metabolic regulator of lipid and glucose homeostasis under the circadian clock. Our major findings include the following: 1) expression of HRD1/Sel1L in the liver is regulated by the circadian clock in BMAL1- and CREBH-dependent manners; 2) at the protein level, HRD1/Sel1L mediates polyubiquitination and degradation of ER-resident CREBH protein under the circadian clock; 3) HRD1-CREBH regulatory axis is required to maintain circadian rhythmic levels of circulating TG, FA, and glucose; 4) HRD1 regulates rhythmic expression of the genes involved in hepatic FA oxidation, TG lipolysis, lipophagy and gluconeogenesis through suppressing CREBH; and 5) suppression of CREBH in HRD1 LKO mouse livers can rescue the phenotype of hyperactive circadian lipid mobilization caused by HRD1 deficiency. These findings revealed that the HRD1/Sel1L-CREBH/PPARα regulatory axis integrates a protein degradation program with circadian lipid and glucose metabolism in the liver, and therefore is critical to maintain whole-body energy homeostasis (S-Figure 5). The identification of the regulation of HRD1 and HRD1-mediated degradation of CREBH by the circadian clock and its functional activity in lipid and glucose homeostasis provides novel insights into the molecular basis of circadian metabolism and has important implications in the prevention and treatment of metabolic orders.
Circadian clock disruption impairs lipid and glucose metabolism in animals, as shown by hyperlipidemia, hepatic steatosis, and defective gluconeogenesis [[30], [31], [32]]. Our recent works demonstrated that CREBH is a circadian-regulated transcription factor that activates expression of the genes encoding the key enzymes or regulators in hepatic FA oxidation, TG lipolysis, lipophagy, and glucose metabolism in response to energy demands [[6], [7], [8]]. The current work reveals that circadian activity of CREBH is under the control of HRD1, the E3 ligase controlling CREBH polyubiquitination and degradation across the circadian clock. We proved that HRD1 and its co-factor Sel1L are BMAL1-regulated liver circadian metabolic regulators of FA oxidation, TG lipolysis, lipophagy, and gluconeogenesis. Circadian rhythmic levels of circulating TG, FA, ketone body, and glucose were significantly altered in HRD1 LKO mice. These findings reveal that the core circadian transcriptional oscillators may regulate hepatic energy metabolism through the ER-associated protein degradation program.
The circadian rhythmic regulation of HRD1/Sel1L expression and its roles in circadian energy homeostasis are consistent with our previous observation that HRD1/Sel1L and CREBH are activated by distinct metabolic conditions or circadian signals [6,19]. Mice exhibit limited feeding behavior during the daytime, while they take most of their food during the nighttime. As shown by this study, the HRD1-mediated regulation of the lipid-catabolizing pathways, including FA oxidation, TG lipolysis, lipophagy and gluconeogenesis, during the daytime period is coincident with downregulation of HRD1 but upregulation of CREBH (Figure 4, Figure 5, Figure 6) [6,19]. Additionally, the refeeding or over-nutrient signals may represent metabolic cues similar to that of mice during the nighttime when they consume most of their food of the day. Indeed, refeeding or high-fat feeding stimulates HRD1/Sel1L activation but suppresses CREBH activity in transcriptional activation of TG and FA mobilization programs [6,19]. This is consistent with the scenarios that HRD1/Sel1L promotes CREBH degradation and that CREBH expression and activation profiles are downregulated in mouse livers during the nighttime period, although CREBH remains a modest activity in promoting energy storage during the nighttime period [6].
Peripheral clocks, such as in the liver, get synchronized with the central clock located in SCN through complicated regulatory networks, including neural, hormonal, behavioral, and environmental cues [33]. At the molecular level, the components of the central SCN and peripheral clocks are the same, with both identically organized in multiple transcriptional and translational feedback systems. We revealed that the circadian clock regulates rhythmic expression of the ER-resident E3 ubiquitin ligase HRD1 and its co-activator Sel1L, through a BMAL1-CREBH/PPARα regulatory axis, as evidenced by the following: 1) the core circadian oscillator BMAL1 regulates rhythmic expression of HRD1 and Sel1L through CREBH; 2) the reciprocally-regulated transcriptional factors CREBH and PPARα directly regulate expression of HRD1 and Sel1L, respectively, in a circadian-dependent manner. However, remaining circadian expression of HRD1 and Sel1L was detected in the Bmal1- or CREBH-KO or knockdown mouse livers or primary hepatocytes (Figure 1, Figure 3). It is known that many circadian-regulated targets are controlled by multiple layers of regulations [34]. When BMAL1 or CREBH is ablated, alternative regulation of the circadian targets may be triggered as a feedback or compensatory response, which likely accounts for the remaining rhythmic expression of HRD1 and Sel1L in mouse livers or primary hepatocytes. Additionally, it appears that the circadian regulation of HRD1 and Sel1L at the protein level was more pronounced than that at the mRNA level (Figure 1). It is possible that the stability of HRD1 and Sel1L proteins, regulated through post-translational modifications, is under the circadian control. This is an interesting question to be investigated in the future.
In summary, our work revealed that HRD1/Sel1L controls CREBH protein turnover while CREBH, in cooperation with PPARα, regulates HRD1 and Sel1L expression, thus forming a self-regulatory mechanism that provides an important feedback loop for HRD1/Sel1L activity under the circadian clock. The reciprocal regulation between HRD1/Sel1L and CREBH may provide an avenue by which the ER-associated protein degradation program and circadian regulators are integrated to influence the whole-body metabolism, an interesting paradigm to be further explored in the future.
Author contributions
KZ and HK designed and conducted the experiments, analyzed the data, and wrote the manuscript; JW, ZS, EM, RZ, LL, XC, JDL, and DF performed the experiments and acquired the data; LS, BPJ, JDL, and DF provided key reagents or critical comments. Drs. Kezhong Zhang and Deyu Fang are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Acknowledgment
Portions of this work were supported by National Institutes of Health (NIH) grants DK090313 and ES017829 (to KZ), AR066634 (to DF and KZ), DK120330 (to DF), DK110314 (to XC), American Heart Association Grants 0635423Z and 09GRNT2280479 (to KZ), and a Pilot and Feasibility Grant (to HK) from the Michigan Diabetes Research Center (NIH Grant P30-DK020572).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101192.
Contributor Information
Deyu Fang, Email: fangd@northwestern.edu.
Kezhong Zhang, Email: kzhang@med.wayne.edu.
Conflict of interest
All authors (HK, JW, ZS, EM, LS, RZ, LL, XC, BPJ, JDL, DF, and KZ) have no any financial, professional, or personal conflict related to this work to declare.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Lowrey P.L., Takahashi J.S. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annual Review of Genomics and Human Genetics. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reppert S.M., Weaver D.R. Molecular analysis of mammalian circadian rhythms. Annual Review of Physiology. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 3.Bass J., Takahashi J.S. Circadian integration of metabolism and energetics. Science. 2010;330(6009):1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatori M., Vollmers C., Zarrinpar A., DiTacchio L., Bushong E.A., Gill S. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metabolism. 2012;15(6):848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang X., Downes M., Yu R.T., Bookout A.L., He W., Straume M. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126(4):801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 6.Zheng Z., Kim H., Qiu Y., Chen X., Mendez R., Dandekar A. CREBH couples circadian clock with hepatic lipid metabolism. Diabetes. 2016;65(11):3369–3383. doi: 10.2337/db16-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim H., Zheng Z., Walker P.D., Kapatos G., Zhang K. CREBH maintains circadian glucose homeostasis by regulating hepatic glycogenolysis and gluconeogenesis. Molecular and Cellular Biology. 2017;37(14) doi: 10.1128/MCB.00048-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim H., Williams D., Qiu Y., Song Z., Yang Z., Kimler V. Regulation of hepatic autophagy by stress-sensing transcription factor CREBH. FASEB Journal. 2019;33(7):7896–7914. doi: 10.1096/fj.201802528R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang C., Wang G., Zheng Z., Maddipati K.R., Zhang X., Dyson G. Endoplasmic reticulum-tethered transcription factor cAMP responsive element-binding protein, hepatocyte specific, regulates hepatic lipogenesis, fatty acid oxidation, and lipolysis upon metabolic stress in mice. Hepatology. 2012;55(4):1070–1082. doi: 10.1002/hep.24783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J.H., Giannikopoulos P., Duncan S.A., Wang J., Johansen C.T., Brown J.D. The transcription factor cyclic AMP-responsive element-binding protein H regulates triglyceride metabolism. Nature Medicine. 2011;17(7):812–815. doi: 10.1038/nm.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cefalu A.B., Spina R., Noto D., Valenti V., Ingrassia V., Giammanco A. Novel CREB3L3 nonsense mutation in a family with dominant hypertriglyceridemia. Arteriosclerosis, Thrombosis, and Vascular Biology. 2015;35(12):2694–2699. doi: 10.1161/ATVBAHA.115.306170. [DOI] [PubMed] [Google Scholar]
- 12.Kim H., Mendez R., Zheng Z., Chang L., Cai J., Zhang R. Liver-enriched transcription factor CREBH interacts with peroxisome proliferator-activated receptor alpha to regulate metabolic hormone FGF21. Endocrinology. 2014;155(3):769–782. doi: 10.1210/en.2013-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Werner E.D., Brodsky J.L., McCracken A.A. Proteasome-dependent endoplasmic reticulum-associated protein degradation: an unconventional route to a familiar fate. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(24):13797–13801. doi: 10.1073/pnas.93.24.13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu X., Rapoport T.A. Mechanistic insights into ER-associated protein degradation. Current Opinion in Cell Biology. 2018;53:22–28. doi: 10.1016/j.ceb.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hetz C., Zhang K., Kaufman R.J. Mechanisms, regulation and functions of the unfolded protein response. Nature Reviews Molecular Cell Biology. 2020;21(8):421–438. doi: 10.1038/s41580-020-0250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carvalho P., Goder V., Rapoport T.A. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126(2):361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 17.Bays N.W., Gardner R.G., Seelig L.P., Joazeiro C.A., Hampton R.Y. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nature Cell Biology. 2001;3(1):24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- 18.Bordallo J., Plemper R.K., Finger A., Wolf D.H. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Molecular Biology of the Cell. 1998;9(1):209–222. doi: 10.1091/mbc.9.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei J., Chen L., Li F., Yuan Y., Wang Y., Xia W. HRD1-ERAD controls production of the hepatokine FGF21 through CREBH polyubiquitination. The EMBO Journal. 2018;37(22) doi: 10.15252/embj.201898942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhattacharya A., Sun S., Wang H., Liu M., Long Q., Yin L. Hepatic Sel1L-Hrd1 ER-associated degradation (ERAD) manages FGF21 levels and systemic metabolism via CREBH. The EMBO Journal. 2018;37(22) doi: 10.15252/embj.201899277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L., Wei J., Zhu H., Pan H., Fang D. Energy supplementation rescues growth restriction and female infertility of mice with hepatic HRD1 ablation. American Journal of Translational Research. 2020;12(5):2018–2027. [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang D., Tong X., Arthurs B., Guha A., Rui L., Kamath A. Liver clock protein BMAL1 promotes de novo lipogenesis through insulin-mTORC2-AKT signaling. Journal of Biological Chemistry. 2014;289(37):25925–25935. doi: 10.1074/jbc.M114.567628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molusky M.M., Ma D., Buelow K., Yin L., Lin J.D. Peroxisomal localization and circadian regulation of ubiquitin-specific protease 2. PLoS One. 2012;7(11) doi: 10.1371/journal.pone.0047970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balsalobre A., Damiola F., Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93(6):929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 25.Wei J., Yuan Y., Chen L., Xu Y., Zhang Y., Wang Y. ER-associated ubiquitin ligase HRD1 programs liver metabolism by targeting multiple metabolic enzymes. Nature Communications. 2018;9(1):3659. doi: 10.1038/s41467-018-06091-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim H., Mendez R., Chen X., Fang D., Zhang K. Lysine acetylation of CREBH regulates fasting-induced hepatic lipid metabolism. Molecular and Cellular Biology. 2015;35(24):4121–4134. doi: 10.1128/MCB.00665-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu X., Park J.G., So J.S., Lee A.H. Transcriptional activation of Fsp27 by the liver-enriched transcription factor CREBH promotes lipid droplet growth and hepatic steatosis. Hepatology. 2015;61(3):857–869. doi: 10.1002/hep.27371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee M.W., Chanda D., Yang J., Oh H., Kim S.S., Yoon Y.S. Regulation of hepatic gluconeogenesis by an ER-bound transcription factor, CREBH. Cell Metabolism. 2010;11(4):331–339. doi: 10.1016/j.cmet.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 30.Lamia K.A., Storch K.F., Weitz C.J. Physiological significance of a peripheral tissue circadian clock. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(39):15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turek F.W., Joshu C., Kohsaka A., Lin E., Ivanova G., McDearmon E. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308(5724):1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimba S., Ogawa T., Hitosugi S., Ichihashi Y., Nakadaira Y., Kobayashi M. Deficient of a clock gene, brain and muscle Arnt-like protein-1 (BMAL1), induces dyslipidemia and ectopic fat formation. PLoS One. 2011;6(9) doi: 10.1371/journal.pone.0025231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukherji A., Bailey S.M., Staels B., Baumert T.F. The circadian clock and liver function in health and disease. Journal of Hepatology. 2019;71(1):200–211. doi: 10.1016/j.jhep.2019.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kornmann B., Schaad O., Bujard H., Takahashi J.S., Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biology. 2007;5(2):e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.