Abstract
The abundance and diversity of anaerobic ammonium oxidation (anammox) bacteria were assessed in 152 groundwater samples in the Kathmandu Valley, Nepal. Anammox bacterial 16S rRNA genes were detected in 54% (37/68) of samples collected in the dry season at 1.6×105–8.8×106 copies L–1, and in 60% (50/84) of samples collected in the wet season at 4.3×104–1.2×107 copies L–1. The 16S rRNA genes of “Candidatus Brocadia”, “Candidatus Anammoxoglobus”, and five new deduced anammox bacterial phylotypes were detected in the shallow groundwater samples. Diverse anammox bacteria were broadly distributed in the shallow groundwater aquifer of the Kathmandu Valley.
Keywords: anammox bacteria, distribution, groundwater, Kathmandu Valley
Anaerobic ammonium oxidation (anammox) is a key microbe-driven nitrogen (N) cycle process, in which ammonium (NH4+) is oxidized with nitrite (NO2–) to produce nitrogen gas (N2) under anoxic conditions (Jetten, 2008). Anammox has been observed in various natural environments, including marine (Babbin et al., 2014), estuarine (Hou et al., 2013), freshwater (Yang et al., 2017), soil (Shen et al., 2013), and freshwater sediment (Osaka et al., 2012).
Groundwater is an important resource for domestic and drinking water; however, the contamination of groundwater by NH4+-N is a serious issue in some parts of the world (Nguyen et al., 2009; Groeschke et al., 2017). Anammox bacteria play an important role in the N cycle and N removal in freshwater aquifers (Moore et al., 2011). Nevertheless, limited information is currently available on the distribution and diversity of anammox bacteria in groundwater and, to the best of our knowledge, their distribution and diversity in the groundwater basin has not yet been examined.
We selected the Kathmandu Valley (area 664 km2; approximately 25 km long from north to south and 30 km wide) in Nepal as a case study location because its groundwaters are widely contaminated by NH4+-N (Shakya et al., 2019). The objective of the present study was to assess the distribution and diversity of anammox bacteria in order to obtain a more detailed understanding of the contribution of anammox to N removal in the groundwater basin. The shallow groundwater samples and groundwater-extracted DNA samples used in the present study were the same as those previously prepared and reported by Malla et al. (2018). In the dry season (February and March), groundwater samples (n=68) were collected from shallow dug wells in the gravel subsurface (n=24) and clay subsurface (n=44). In the wet season (August and September), shallow groundwater samples (n=84) were collected in the gravel subsurface (n=29) and clay subsurface (n=55) (Fig. S1, Table S1 and S2). To quantify the 16S rRNA genes of anammox bacteria and total bacteria, qPCR was performed using the primer sets AMX-368F (Schmid et al., 2003) and Amx-667R (van der Star et al., 2007) for the former and the primer set Eub-515F and Eub-806R (Caporaso et al., 2011) for the latter. The thermal conditions of PCR are shown in Table S3. DNA samples extracted from groundwater at Khumaltar (Table S4), a clay subsurface area, and Nayachok (Budhanilkantha) (Table S4), a gravel subsurface area, were used in the phylogenetic analysis. Anammox bacterial 16S rRNA genes were amplified using the Pla46F (Neef et al., 1998) and Amx820R primers (Schmid et al., 2000). The Pla46F and Amx 820R primers were previously reported to be an effective primer set that widely detected anammox bacteria (Nakajima et al., 2008). Amplified DNA fragments were cloned into Escherichia coli strain DH5α using a pMD19 T-vector system (Takara Bio). A PCR-Restriction Fragment Length Polymorphism (RFLP) analysis and clone library analysis were performed as previously reported (Tanaka et al., 2012). The phylogenetic tree was constructed using Clustal X (http://www.clustal.org/clustal2/). The DDBJ/EMBL/GenBank accession numbers of the 16S rRNA gene sequences of clones are LC553643 to LC553674.
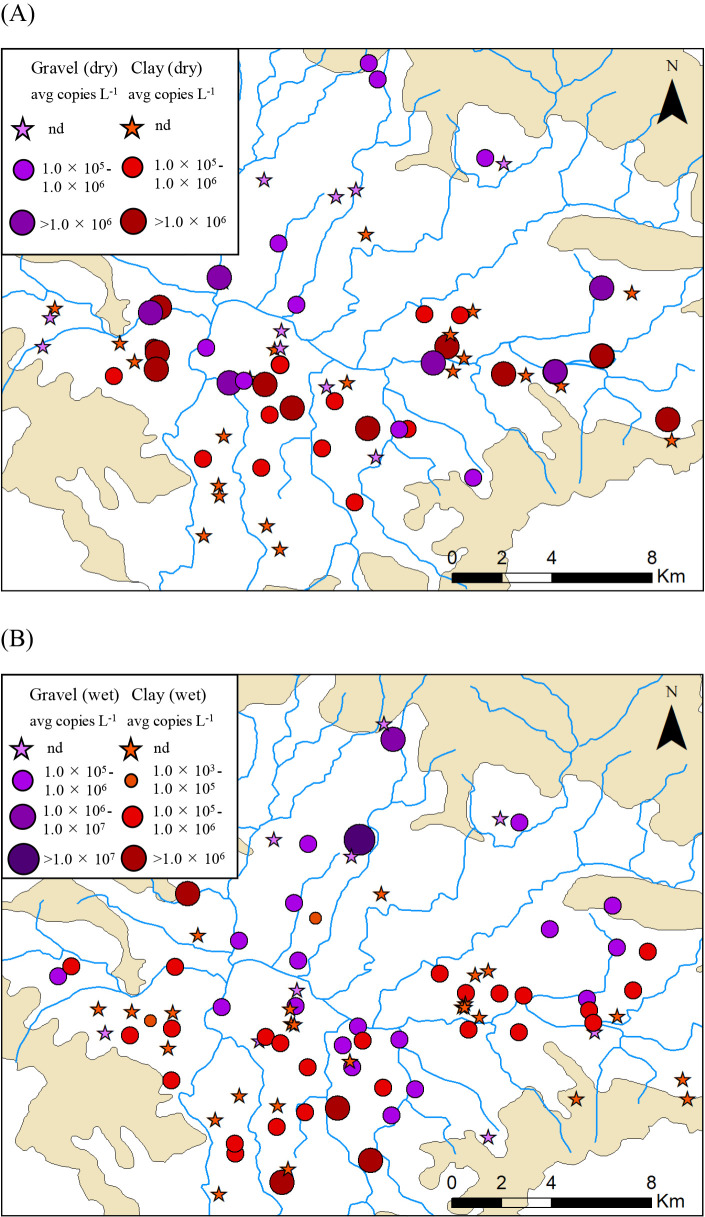
As shown in Table 1, in the dry season, the 16S rRNA genes of anammox bacteria were detected in 58% (14/24) of shallow groundwater samples in the gravel subsurface and in 52% (23/44) of those in the clay subsurface (Table 1). Anammox bacterial 16S rRNA gene abundance ranged between 1.7×105 and 7.6×106 copies L–1 in groundwater in the gravel subsurface and between 1.6×105 and 8.8×106 copies L–1 in those in the clay subsurface. In the wet season, the 16S rRNA genes of anammox bacteria were detected in 69% (20/29) of shallow groundwater samples in the gravel subsurface and in 55% (30/55) of those in the clay subsurface, with 1.2×105 to 1.2×107 copies L–1 and between 4.3×104 and 3.1×106 copies L–1 in groundwater samples in gravel and clay subsurfaces, respectively (Table 1). Anammox bacterial 16S rRNA gene abundance (copies L–1) was as high as that in other freshwater samples (Sun et al., 2014; Zhang et al., 2017). These results suggest that anammox bacteria were widely distributed throughout the Kathmandu Valley groundwater basin in the dry and wet seasons (Fig. 1).
Table 1.
Detection and abundance of 16S rRNA genes of anammox bacteria and total bacteria in groundwater samples from shallow dug wells
| Season | Sample type | Samples with the gene relative to all samples (detection %) | Anammox bacterial 16S rRNA gene abundance (copies L–1) | Total bacterial 16S rRNA gene abundance (copies L–1) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Min | Max | Avea | Min | Max | Avea | ||||
| Dry season | Gravel subsurface | 14/24 (58%) | 1.7×105 | 7.6×106 | 1.4×106±1.9×106 | 6.3×106 | 1.7×1010 | 1.9×109±3.9×1010 | |
| Clay subsurface | 23/44 (52%) | 1.6×105 | 8.8×106 | 1.5×106±1.8×106 | 1.2×107 | 7.3×1010 | 3.3×109±1.1×1010 | ||
| Wet season | Gravel subsurface | 20/29 (69%) | 1.2×105 | 1.2×107 | 9.4×105±2.5×106 | 3.6×106 | 1.0×1011 | 5.0×109±1.9×1010 | |
| Clay subsurface | 30/55 (55%) | 4.3×104 | 3.1×106 | 5.3×105±7.3×105 | 3.3×106 | 3.0×1010 | 3.6×109±4.9×109 b | ||
| Total | 87/152 (57%) | 4.3×104 | 1.2×107 | 3.3×106 | 1.0×1011 | ||||
a Values are means±SD.
b Significantly higher (Student’s t-test, P<0.05) than total bacterial 16S rRNA gene abundance in dry season (gravel subsurface and clay subsurface) samples and wet season (gravel subsurface) samples.
Fig. 1.
Spatial distribution and abundance of anammox bacteria in the groundwater of shallow dug wells in the Kathmandu Valley during dry (A) and wet (B) seasons.
Water temperature, pH, and NH4+-N concentrations have been suggested to affect the abundance of anammox bacteria in freshwater environments (Sun et al., 2013; Sun et al., 2014; Yang et al., 2017). However, in the present study, anammox bacteria were widely detected in the shallow groundwater samples with a temperature range of 15.5–32.0°C, pH range of 5.7–7.3, and NH4+-N concentration range of 0–5.6 mg L–1, and their abundance did not correlate with any of these water properties (R statistical software version 3.6.0, P<0.05, Table S5). The dissolved oxygen (DO) concentration was an important environmental parameter. However, the relationship between the anammox bacterial 16S rRNA gene and DO was not investigated because the DO value was not assessed (Shakya et al., 2019). Although the underlying reasons were not elucidated in the present study, the shallow groundwater basin in the Kathmandu Valley appears to be a consistent habitat for anammox bacteria.
Based on the results of the PCR-RFLP and clone library analyses for the anammox bacterial 16S rRNA gene, 143 clones were divided into 32 RFLP groups (Table S6). A phylogenetic analysis identified eight groups: Brocadia, Anammoxoglobus, Clusters 1, 2, 3, 4, 5, and 6 (Fig. S2). The Brocadia group comprised “Candidatus Brocadia” and two RFLP groups (three clones), while the Anammoxoglobus group consisted of “Candidatus Anammoxoglobus” and RFLP18 (five clones). Cluster 1 comprised 16 RFLP groups including 66 clones. Clusters 2 and 3 each consisted of a single RFLP group, RFLP23 (46 clones) and RFLP24 (1 clone), respectively. Cluster 4 comprised three RFLP groups (13 clones), Cluster 5 consisted of six RFLP groups (seven clones), and Cluster 6 had two RFLP groups (two clones). “Ca. Brocadia” is recognized as a major anammox group in non-saline environments, such as freshwater sediments, freshwater, soils, and groundwater, while “Ca. Anammoxoglobus” has also been frequently detected in non-saline environments, including soil, freshwater, estuaries, and groundwater (Moore et al., 2011; Sonthiphand et al., 2014). In the present study, “Ca. Brocadia” and “Ca. Anammoxoglobus” were also present, but not dominant, in the shallow groundwater samples at Nayachok and Khumaltar. Clusters 1 and 2 were the dominant groups in groundwater at both Nayachok and Khumaltar in the dry and wet seasons (Fig. S3). These results suggest that diverse anammox bacteria are present in the shallow groundwater of the Kathmandu Valley. Anammox bacteria in Clusters 1 and 2 may exhibit better adaptability in the groundwater. Further studies are needed to elucidate the relationships between the anammox bacterial community and environmental factors (e.g., NH4+-N, NO2–-N, temperature, pH, DO, minerals, metals, and organic carbons) in groundwater in more detail.
In conclusion, various anammox bacteria were widely distributed throughout the shallow groundwater basin in the Kathmandu Valley during the wet and dry seasons. Therefore, anammox bacteria may contribute to N removal in this groundwater basin.
Citation
Nakano, M., Kamei, T., Man Shakya, B., Nakamura, T., Tanaka, Y., Haramoto, E., et al. (2021) Distribution and Community Composition of Anammox Bacteria in Shallow Groundwater of the Kathmandu Valley, Nepal. Microbes Environ 36: ME20143.
https://doi.org/10.1264/jsme2.ME20143
Supplementary Material
Acknowledgements
The authors acknowledge Dr. Bikash Malla, Dr. Rajani Ghaju Shrestha (University of Yamanashi, Japan), and Dr. Jeevan B. Sherchand (Tribhuvan University, Nepal) for their support with water sampling and DNA extraction. This research was conducted as part of the “Hydro-Microbiological Approach for Water Security in Kathmandu Valley, Nepal” project with the support of the SATREPS program of the Japan International Cooperation Agency (JICA) and the Japan Science and Technology Agency (JST).
References
- Babbin, A.R., Keil, R.G., Devol, A.H., and Ward, B.B. (2014) Organic matter stoichiometry, flux, and oxygen control nitrogen loss in the ocean. Science 344: 406–408. [DOI] [PubMed] [Google Scholar]
- Caporaso, J.G., Lauber, C.L., Walters, W.A., Berg-Lyons, D., Lozupone, C.A., Turnbaugh, P.J., et al. (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108: 4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groeschke, M., Frommen, T., Winkler, A., and Schneider, M. (2017) Sewage-borne ammonium at a river bank filtration site in central Delhi, India: Simplified flow and reactive transport modeling to support decision-making about water management strategies. Geosciences (Basel, Switz) 7: 48. [Google Scholar]
- Hou, L., Zheng, Y., Liu, M., Gong, J., Zhang, X., Yin, G., and You, L. (2013) Anaerobic ammonium oxidation (anammox) bacterial diversity, abundance, and activity in marsh sediments of the Yangtze Estuary. J Geophys Res 118: 1237–1246. [Google Scholar]
- Jetten, M.S.M. (2008) The microbial nitrogen cycle. Environ Microbiol 10: 2903–2909. [DOI] [PubMed] [Google Scholar]
- Malla, B., Shrestha, R.G., Tandukar, S., Bhandari, D., Inoue, D., Sei, K., et al. (2018) Identification of human and animal fecal contamination in drinking water source in the Kathmandu Valley, Nepal, Using Host-Associated Bacteroidales Quantitative PCR Assays. Water (Basel, Switz) 10: 1796. [Google Scholar]
- Moore, T.A., Xing, Y., Lazenby, B., Lynch, M.D. J., Schiff, S., Robertson, W.D., et al. (2011) Prevalence of anaerobic ammonium-oxidizing bacteria in contaminated groundwater. Environ Sci Technol 45: 7217–7225. [DOI] [PubMed] [Google Scholar]
- Nakajima, J., Sakka, M., Kimura, T., and Sakka, K. (2008) Detection of anaerobic ammonium oxidizing bacteria in Ago bay sediments. Biosci Biotechnol Biochem 72: 2195–2198. [DOI] [PubMed] [Google Scholar]
- Neef, A., Amann, R., Schlesner, H., and Schleifer, K.H. (1998) Monitoring a widespread bacterial group: in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiology 144: 3257–3266. [DOI] [PubMed] [Google Scholar]
- Nguyen, V.N., Bang, S., Viet, P.H., and Kim, K.W. (2009) Contamination of groundwater and risk assessment for arsenic exposure in Ha Nam province, Vietnam. Environ Int 35: 466–472. [DOI] [PubMed] [Google Scholar]
- Osaka, T., Kimura, Y., Otsubo, Y., Suwa, Y., Tsuneda, S., and Isaka, K. (2012) Temperature dependence for anammox bacteria enriched from freshwater sediments. J Biosci Bioeng 114: 429–434. [DOI] [PubMed] [Google Scholar]
- Schmid, M.C., Twachtmann, U., Klein, M., Strous, M., Juretschko, S., Jetten, M., et al. (2000) Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst Appl Microbiol 23: 93–106. [DOI] [PubMed] [Google Scholar]
- Schmid, M.C., Walsh, K., Webb, R., Rijpstra, W.I., van de Pas-Schoonen, K., Verbruggen, M.J., et al. (2003) Candidatus “Scalindua brodae”, sp. nov., Candidatus “Scalindua wagneri”, sp. nov., two new species of anaerobic ammonium oxidizing bacteria. Syst Appl Microbiol 26: 529–538. [DOI] [PubMed] [Google Scholar]
- Shakya, B.M., Nakamura, T., Kamei, T., Shrestha, S.D., and Nishida, K. (2019) Seasonal groundwater quality status and nitrogen contamination in the shallow aquifer system of the Kathmandu Valley, Nepal. Water (Basel, Switz) 11: 2184. [Google Scholar]
- Shen, L., Liu, S., Lou, L., Liu, W., Xu, X., Zheng, P., and Hu, B. (2013) Broad distribution of diverse anaerobic ammonium-oxidizing bacteria in Chinese agricultural soils. Appl Environ Microbiol 79: 6167–6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonthiphand, P., Hall, M.W., and Neufeld, J.D. (2014) Biogeography of anaerobic ammonia-oxidizing (anammox) bacteria. Front Microbiol 5: 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, W., Xu, M.Y., Wu, W.M., Guo, J., Xia, C.Y., Sun, G.P., and Wang, A.J. (2013) Molecular diversity and distribution of anammox community in sediments of the Dongjiang River, a drinking water source of Hong Kong. Appl Microbiol 166: 464–476. [DOI] [PubMed] [Google Scholar]
- Sun, W., Xia, C., Xu, M., Guo, J., Wang, A., and Sun, G. (2014) Diversity and distribution of planktonic anaerobic ammonium-oxidizing bacteria in the Dongjiang River, China. Microbiol Res 169: 897–906. [DOI] [PubMed] [Google Scholar]
- Tanaka, Y., Tamaki, H., Matsuzawa, H., Nigaya, M., Mori, K., and Kamagata, Y. (2012) Microbial community analysis in the roots of aquatic plants and isolation of novel microbes including an organism of the candidate phylum OP10. Microbes Environ 27: 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Star, W.R.L., Abma, W.R., Blommers, D., Mulder, J.W., Tokutomi, T., Strous, M., et al. (2007) Startup of reactors for anoxic ammonium oxidation: experiences from the first full-scale anammox reactor in Rotterdam. Water Res 41: 4149–4163. [DOI] [PubMed] [Google Scholar]
- Yang, Y., Dai, Y., Li, N., Li, B., Xie, S., and Liu, Y (2017) Temporal and spatial dynamics of sediment anaerobic ammonium oxidation (anammox) bacteria in freshwater lakes. Microb Ecol 73: 285–295. [DOI] [PubMed] [Google Scholar]
- Zhang, S., Xia, X., Liu, T., Xia, L., Zhang, L., Jia, Z., et al. (2017) Potential roles of anaerobic ammonium oxidation (anammox) in overlying water of rivers with suspended sediments. Biogeochemistry 132: 237–249. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.