Abstract
Iron (Fe) is one of the essential micronutrients required by both plants and animals. In humans, Fe deficiency causes anemia, the most prevalent nutritional disorder. Most people rely on plant-based foods as their major Fe source, but plants are a poor source of dietary Fe. Therefore, there is a critical need to better understand the mechanisms involved in the uptake and trafficking of Fe and how plants adapt to Fe deficiency. Fe participates in key cellular functions such as photosynthesis and respiration. Perturbations of Fe uptake, transport, or storage affect plant growth as well as crop yield and plant product quality. Excess Fe has toxic effects due to its high redox activity. Plants, therefore, tightly regulate Fe uptake, distribution, and allocation. Here, we review the regulatory mechanisms involved at the transcriptional and post-translational levels that are critical to prevent Fe uptake except when plants experience Fe deficiency. We discuss the key regulatory network of basic helix–loop–helix (bHLH) transcription factors, including FIT, subgroup Ib, subgroup IVc, and URI (bHLH121), crucial for regulating Fe uptake in Arabidopsis thaliana. Furthermore, we describe the regulators of these transcription factors that either activate or inhibit their function, ensuring optimal Fe uptake that is essential for plant growth.
Keywords: Arabidopsis, bHLH transcription factors, E3 ligases, iron deficiency, iron homeostasis, iron uptake
A network of bHLH transcription factors and the proteins that regulate them ensures iron uptake when iron is limiting and prevents accumulation of excess iron when supplies are adequate.
Introduction
Iron (Fe) is an essential micronutrient for both plants and animals, and acts as a critical cofactor in many enzymes due to its ability to cycle between its two oxidation states: Fe2+ and Fe3+. This allows Fe to participate in key cellular processes requiring redox reactions such as photosynthesis and respiration. Perturbations of Fe uptake, transport, or storage affect plant growth as well as crop yield (Connorton et al., 2017). Although Fe is one of the most abundant elements found on earth, its availability is limited in soil since it exists mainly as insoluble ferric hydroxides and thus is bio-unavailable (Colombo et al., 2014). Fe bioavailability is further reduced in alkaline soil, which comprises one-third of the world’s arable land (Guerinot and Yi, 1994). Fe limitation to plants is a critical issue for human health as most people rely on plants for their primary Fe source. Therefore, understanding the physiological and molecular mechanisms governing plant Fe uptake is critical. Although plants often face Fe deficiency, Fe availability in the rhizosphere may not remain constant. Excess Fe can be toxic to plants due to its ability to generate reactive oxygen species (ROS) by the Fenton reaction (Halliwell and Gutteridge, 1992). Therefore, it is crucial for plants to tightly regulate Fe uptake to maintain Fe homeostasis in order to prevent both Fe deficiency and Fe toxicity (Connorton et al., 2017).
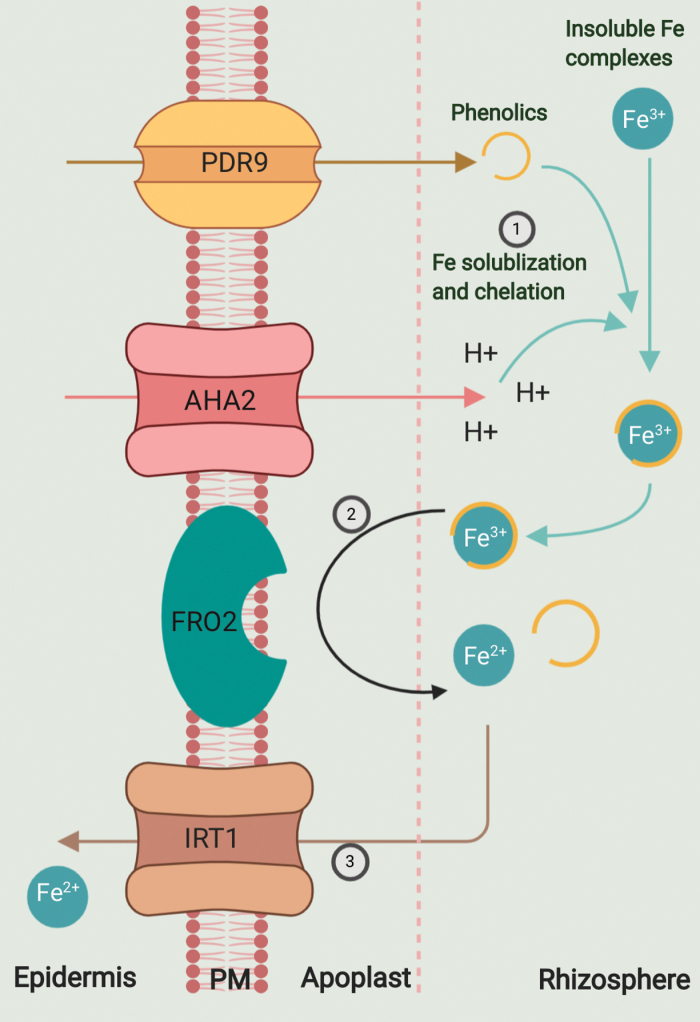
To mediate Fe uptake, plants employ reduction- and chelation-based strategies (Hindt and Guerinot, 2012). In non-grasses such as Arabidopsis thaliana, the plant uses a reduction-based strategy (Fig. 1) where insoluble Fe3+ is solubilized by acidification of the rhizosphere due to protons pumped by AHA2, a proton ATPase (Santi and Schmidt, 2009). Fe deficiency also induces secretion of phenolic compounds, particularly coumarins, to improve Fe mobilization and reduction (Connorton et al., 2017; Stringlis et al., 2019). Solubilized Fe3+ is then reduced to Fe2+ by membrane-bound ferric chelate reductase FRO2 (Robinson et al., 1999), and consequently transported into the root epidermis by a high-affinity Fe transporter IRT1 (Eide et al., 1996). The three main players, AHA2, FRO2, and IRT1, have recently been shown to assemble into a complex on the cell surface, presumably to optimize Fe uptake (Martín-Barranco et al., 2020). In graminaceous plants such as rice, barley, and maize, Fe is acquired through a chelation-based strategy which consists of synthesis and secretion of phytosiderophores belonging to the mugineic acid family (Takagi, 1976; Nozoye et al., 2011). These phytosiderophores are effluxed into the rhizosphere via TOM1 transporters (Nozoye et al., 2011), and the chelated complex is subsequently transported into the roots by yellow stripe like 1 (YSL1) transporters (Curie et al., 2001). For a detailed overview of the chelation strategy, we refer readers to other excellent reviews (Hindt and Guerinot, 2012; Connorton et al., 2017; Wang et al., 2020).
Fig. 1.
Fe uptake mechanism in Arabidopsis thaliana. Arabidopsis employs a three-step reduction-based strategy to facilitate Fe uptake from the environment. First, insoluble Fe3+ is solubilized by acidification of the rhizosphere by AHA2, a proton ATPase. Pleiotropic drug resistance 9 (PDR9) protein exports phenolics which chelate Fe3+. The second step involves the reduction of solubilized chelated Fe3+ to Fe2+ by the ferric chelate reductase FRO2. The last step involves transport of Fe2+ into the plant by IRT1, a high-affinity Fe2+ transporter.
Although the tendency has been to distinguish the grasses from the non-grasses when it comes to Fe uptake strategies, as we learn more it is becoming clear that the two uptake strategies are not that different after all (Wang et al., 2020). For example, rice can employ either a reduction- or a chelation-based strategy depending on growth conditions (Ishimaru et al., 2006; Cheng et al., 2007). Moreover, the transcription factor cascade that regulates Fe uptake in grasses may be quite similar to that described for Arabidopsis. Very recently, a transcription factor, OsbHLH156, was identified, which physically interacts with the chelation-based strategy master regulator IRO2 and regulates Fe uptake in rice (Wang et al., 2020). OsbHLH156 is an ortholog of FIT (Fer-like iron deficiency-induced transcription factor) involved in regulating the reduction-based Fe uptake in Arabidopsis. The authors discuss how the mechanism by which OsbHLH156 regulates Fe uptake via its interaction with the subgroup Ib basic helix–loop–helix (bHLH) transcription factor, IRO2, is similar to that of FIT interacting with members of the bHLH subgroup Ib transcription factors (FIT is discussed in detail in the next section). These observations suggest that although plants might employ different Fe uptake strategies, one of the key regulators, either OsbHLH156 in rice or FIT in Arabidopsis, regulates Fe deficiency responses by forming heterodimers with member of subgroup Ib bHLH transcription factors. Additionally, Wang et al. (2020) observed that OsbHLH156 is required for facilitating nuclear localization of IRO2, a phenomenon that was also recently shown to occur in Arabidopsis where FIT is required for the nuclear localization of bHLH39 (Trofimov et al., 2019). All of this provides interesting insights into the evolution of Fe uptake strategies in plants.
This review will mainly explore regulation of the Fe deficiency response by members of the bHLH transcription factor family in Arabidopsis. First a brief description of regulation of FIT in response to Fe deficiency will be presented (Fig. 2), followed by the mechanisms involved in transcriptional and post-translational regulation of specific bHLH transcription factors which tightly regulate Fe homeostasis to avoid Fe deficiency and Fe toxicity (Fig. 3). Various studies have explored how Fe is sensed, how root to shoot signaling of Fe status occurs, and how Fe is stored, but these research topics are beyond the scope of this review. Readers interested in these areas should consult reviews covering specific aspects of Fe homeostasis such as local and systematic signaling of Fe status (Gayomba et al., 2015), Fe uptake and translocation (Brumbarova et al., 2015), Fe sensors (Kobayashi and Nishizawa, 2014), and Fe storage (Briat et al., 2010; Bashir et al., 2016; Connorton et al., 2017).
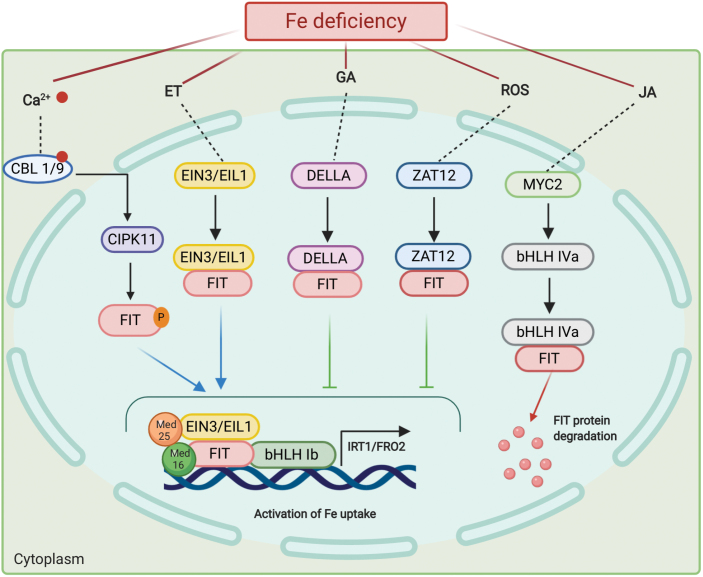
Fig. 2.
Regulation of the Fe deficiency response via FIT. The model depicts proteins that directly interact with FIT via protein–protein interactions to regulate Fe uptake (indicated by solid black lines). Binding of FIT with subgroup Ib bHLH transcription factors, MED16 and EIN3/EIL (MED25 interacts with MED16 and EIN3/EIL1), and phosphorylation by CIPK11 leads to positive regulation of the Fe deficiency response and activation of Fe uptake genes such as FRO2 and IRT1 during Fe-deficient conditions (indicated by blue arrows). Binding of FIT with DELLA and ZAT12 prevents the binding of subgroup Ib bHLHs to FIT and prevents transcription of Fe uptake genes (indicated by the green arrow), whereas FIT interactions with subgroup IVa bHLH transcription factors lead to proteasomal degradation of FIT (indicated by red arrows). Abbreviations used are Fe, iron; Ca2+, calcium; ET, ethylene; GA, gibberellic acid; ROS, reactive oxygen species; JA, jasmonic acid; red circle, calcium ion; P, phosphorylation. Red faded lines, signaling pathway; rounded rectangles, proteins; dashed circle, nucleus.
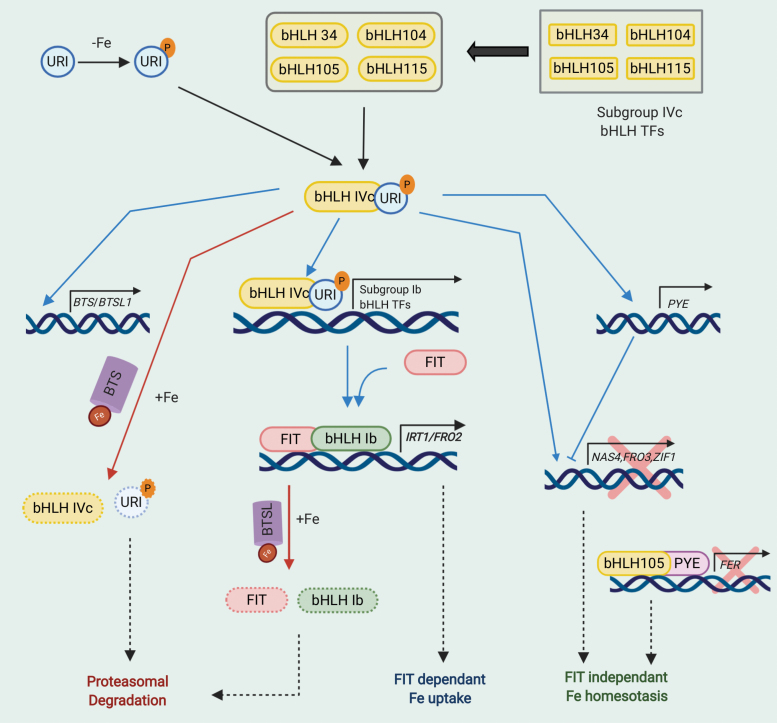
Fig. 3.
bHLH-dependent transcriptional regulation of Fe deficiency in Arabidopsis thaliana. During Fe deficiency, URI (bHLH121) is phosphorylated and interacts with subgroup IVc bHLH transcription factors, bHLH34, bHLH104, bHLH115, and bHLH105. These heterodimers transcriptionally regulate expression of genes including subgroup Ib bHLH transcription factors, BTS, BTSL, and PYE (indicated by blue arrows). Heterodimerization of FIT with subgroup Ib bHLHs induces expression of Fe uptake genes IRT1 and FRO2. PYE is a known transcriptional repressor that inhibits expression of genes involved in Fe transport and sequestration. BTS and BTSL are also negative regulators of Fe deficiency whose expression is induced by Fe deficiency. During Fe-sufficient conditions, BTS and BTSL bind to available Fe and interact with members of bHLH subgroups IVc and Ib, and URI and FIT transcription factors, and cause their proteasomal degradation (indicated by red arrows). Fe-dependent turnover by BTS and BTSL is crucial for tight regulation of Fe deficiency response to prevent toxic effects of excess Fe. Abbreviations used are –Fe, Fe deficiency; +Fe, Fe sufficiency; P, phosphorylation; Fe, iron metal. Rounded rectangles and circle, proteins; rectangles, genes; solid black arrow, transcription and translation; blue arrows, transcriptional regulation; red arrows; proteasomal degradation; purple cylinder, BTS and BTSL; red X, transcriptional repression.
Transcriptional regulation of the Fe deficiency response
Great progress has been made in identifying bHLH transcription factors that regulate Fe homeostasis in the model plant A. thaliana. The bHLH proteins belong to the largest family of transcription factors found in plants, with 159 members in A. thaliana that have been grouped into 26 subfamilies (Bailey et al., 2003; Heim et al., 2003; Toledo-Ortiz et al., 2003; Carretero-Paulet et al., 2010; Pires and Dolan, 2010). To date, 17 bHLH transcription factors belonging to five different subfamilies have been implicated in the response to Fe homeostasis (Gao et al., 2019, 2020; Kim et al., 2019; Lei et al., 2020). These studies have shown how complex and interconnected the network of bHLH transcription factors is, with different clades and individual transcriptional regulators having unique roles in controlling Fe homeostasis in plants (Fig. 3). Despite the fact that several of these bHLH transcription factors have been identified along with their target genes, relatively little is known about the molecular regulation of the bHLH transcriptional cascade that controls Fe homeostasis. This review provides an overview of Fe homeostasis and current research on how the transcriptional cascade is regulated at the molecular level during Fe deficiency. Fe uptake and translocation within the plant requires coordinated expression and proper localization of these bHLH transcription factors to efficiently regulate Fe homeostasis. Gao et al. (2019) nicely not only summarized the bHLH transcriptional network but also explored expression (promoter activity) and localization of various bHLH transcription factors involved in the Fe deficiency response. The current review adds another bHLH transcription factor to the mix (URI/bHLH121) (Kim et al., 2019; Gao et al., 2020; Lei et al., 2020) and summarizes studies on interaction partners of the bHLH transcription factors involved in the regulation of Fe homeostasis. For a more global view of how various nutrient response pathways such as the Fe deficiency response are interconnected, readers are directed to Brumbarova and Ivanov (2019).
The expression of AHA2, FRO2, and IRT1 is inducible by Fe deficiency, and these genes are transcriptionally regulated by various bHLH transcription factors. In Arabidopsis, FIT is known to regulate Fe uptake in roots (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005). FIT belongs to subgroup IIIa bHLH transcription factors (Pires and Dolan, 2010) and is the functional ortholog of FER, a bHLH transcription factor required for regulation of Fe deficiency response in tomato (Ling et al., 2002). FIT is induced upon Fe deficiency (Colangelo and Guerinot, 2004) and binds to its target gene promoter regions as a heterodimer with subgroup Ib bHLH transcription factors (bHLH38, bHLH39, bHLH100, and bHLH101) to activate the transcription of FRO2 and IRT1 (Yuan et al., 2008; Wang et al., 2013). Mutant studies in Arabidopsis revealed that the members of subgroup Ib bHLH transcription factors are functionally redundant (Yuan et al., 2008; Sivitz et al., 2012), but it is possible that they might be regulating the Fe deficiency response by activating different downstream genes (Sivitz et al., 2012). The expression of bHLH38, bHLH39, bHLH100, and bHLH101 is induced during Fe deficiency (Wang et al., 2007), which suggests the presence of upstream regulators that control expression of FIT and subgroup Ib bHLH transcription factors. Recently it was demonstrated that bHLH121/URI of the subgroup IVb bHLH transcription factors (Kim et al., 2019; Gao et al., 2020; Lei et al., 2020), together with subgroup IVc bHLH transcription factors, bHLH34, bHLH104, bHLH115, and bHLH105/ILR3, induces expression of most of the known Fe-regulated genes, including the subgroup Ib bHLH transcription factors (Zhang et al., 2015; Li et al., 2016; Liang et al., 2017; Wang et al., 2017; Tissot et al., 2019).
Unlike the subgroup Ib bHLH transcription factors, consensus on the transcriptional regulation of the subgroup IVc bHLH transcription factors by Fe is yet to be reached (Zhang et al., 2015; Li et al., 2016; Liang et al., 2017; Wang et al., 2017; Samira et al., 2018). Therefore, more research needs to be done to better understand the transcriptional regulation of subgroup IVc bHLH transcription factors. The subgroup IVc bHLH transcription factors seem not to be functionally redundant but rather appear to be additive (Zhang et al., 2015; Li et al., 2016; Liang et al., 2017). Single mutants of all subgroup IVc members display defective induction of subgroup Ib bHLH transcription factors, with higher order mutants displaying increased Fe deficiency symptoms (Rampey et al., 2006; Li et al., 2016; Liang et al., 2017).
In a cell type-specific microarray study of Fe-deficient Arabidopsis roots, another bHLH transcription factor (subgroup IVb) named POPEYE (PYE) was identified, that regulates the Fe deficiency response independently of the FIT network (Long et al., 2010). PYE is induced by Fe deficiency, and promoter activity revealed that it has the highest expression in the pericycle of the root, although PYE protein may function in multiple cell types (Long et al., 2010). PYE has been shown to act as a transcriptional repressor that inhibits expression of Fe deficiency-induced genes in roots, such as NAS4, FRO3, and ZIF1, which encode a key gene involved in transport of Fe from the phloem, a plasma membrane-localized Fe chelate reductase, and a vacuolar transporter crucial for zinc regulation, respectively (Haydon and Cobbett, 2007; Jeong and Connolly, 2009; Klatte et al., 2009; Long et al., 2010). Interestingly, members of the subgroup IVc bHLH transcription factors, bHLH104, ILR3, and bHLH115, can interact in vivo forming heterodimers with PYE (Long et al., 2010). However, the functional consequences of these heterodimers and whether these heterodimers regulate the Fe deficiency response in plants is still unknown. Recently, ILR3 was shown to negatively regulate the expression of ferritin genes, which are induced when plants experience Fe excess (Tissot et al., 2019). Expression studies and ChIP assays revealed that ILR3 represses the expression of several genes involved in Fe homeostasis via dimerization with PYE and direct binding to their promoters. ILR3 appears then to be able to act as both a transcriptional activator and repressor, in regulating Fe homeostasis in Arabidopsis.
Transcriptional and post-translational regulation of FIT
In Arabidopsis, FIT acts a key factor in the transcriptional regulation of Fe uptake and homeostasis. It is a regulatory hub for integrating signals from multiple signaling pathways (Fig. 2). The constant turnover of FIT is proposed to facilitate rapid down-regulation of the Fe deficiency response when Fe becomes available in order to prevent Fe toxicity.
In this section, we will discuss the recent studies conducted on deciphering transcriptional and post-translational mechanisms which control FIT activity. Interaction of FIT with various binding partners can lead to different outcomes, including phosphorylation, proteasomal degradation, or stabilization of FIT–subgroup Ib bHLH heterodimers (Fig. 2) (Gratz et al., 2019; Wu and Ling, 2019; Schwarz and Bauer, 2020).
Subgroup Ib bHLH transcription factors
As described above, FIT is induced by Fe deficiency and interacts with subgroup Ib bHLH transcription factors (Yuan et al., 2008; Wang et al., 2013). Overexpression of FIT alone does not induce the Fe deficiency response (Colangelo and Guerinot, 2004) as heterodimerization with bHLH38, bHLH39, or bHLH101 is obligatory to activate Fe uptake genes (Wang et al., 2013). We still lack knowledge on how FIT transcription itself is induced under Fe deficiency, but it seems that its expression is probably controlled by a feedforward loop involving bHLH39 (Naranjo-Arcos et al., 2017). Overexpression of bHLH39 (39Ox) causes constitutive Fe uptake, leading to increased Fe content in leaves and seeds. FIT was shown to be essential for the 39Ox phenotype. The authors suggested that FIT is not only regulated at the protein level during Fe deficiency, but that FIT gene expression is also induced downstream of the bHLH transcriptional cascade, as part of a feedforward loop to amplify gene expression (Naranjo-Arcos et al., 2017). Recently, it was reported that subgroup Ib bHLH proteins might have a role in stabilizing FIT (Cui et al., 2018). Immunoblot analysis revealed that roots of plants overexpressing FIT (FITOx) alone did not accumulate FIT protein under Fe-sufficient conditions but abundant FIT protein could be detected in the roots of plants that overexpress both FIT and bHLH38 under Fe-sufficient conditions (Cui et al., 2018).
Jasmonic acid
Jasmonic acid (JA) is a plant hormone and a known negative regulator of the Fe deficiency response (Maurer et al., 2011; Kobayashi et al., 2016; Cui et al., 2018). The mechanism of inhibition of the Fe deficiency response due to JA treatment was recently elucidated (Cui et al., 2018). MYC2 (bHLH6 from bHLH transcription factor subgroup IIIe) is a known master regulator of the JA signaling pathway (Kazan and Manners, 2013). In the presence of JA, MYC2 leads to an increase in expression of subgroup IVa bHLH transcription factors (bHLH18, bHLH19, bHLH20, and bHLH25) (Cui et al., 2018). These four redundant transcription factors are mainly expressed in the roots and can interact with FIT to modulate FIT protein accumulation during Fe deficiency (Cui et al., 2018). Dimerization of FIT with bHLH subgroup IVa transcription factors is crucial to JA-induced FIT protein degradation (Cui et al., 2018). Plants overexpressing subgroup Ib bHLH transcription factor genes were able to reduce the JA-induced FIT protein degradation, which means that interaction with subgroup IVa bHLH leads to 26S proteasomal degradation of FIT; meanwhile, interaction with subgroup Ib bHLH promotes FIT stability (Cui et al., 2018). This suggests that the subgroup IVa bHLH proteins compete with the subgroup Ib bHLH proteins to bind FIT and promote its degradation.
Gibberellins
Gibberellins (GAs) are another class of plant growth hormones that have been shown to play a role in the Fe deficiency response (Matsuoka et al., 2014). Exogenous GA application was shown to induce the expression of Fe deficiency-responsive genes including IRT1, FRO2, bHLH38, and bHLH39 in the GA-deficient double mutant ga3ox1ga3ox2 (GA30X1 and GA3OX2 are GA biosynthetic genes) (Matsuoka et al., 2014). Recently, another group showed that GA signaling was involved in the Fe deficiency response through the DELLA proteins (Wild et al., 2016). Yeast two-hybrid, fluorescence resonance energy transfer imaging (FRET-FLIM), and co-immunoprecipitation (Co-IP) analyses revealed that the DELLA repressor protein of GA signaling can form heterodimers with FIT, bHLH38, and bHLH39 proteins. DELLA proteins interact with the DNA-binding domain of FIT (Wild et al., 2016). FRET-FLIM and EMSA revealed that the DELLA–FIT heterodimer does not prevent the formation of the FIT–subgroup Ib bHLH heterodimer, but rather it inhibits transcriptional activity of FIT via preventing the binding of the FIT–subgroup Ib bHLH heterodimer to their target gene promoters. ChIP assays using p35S:FIT-GFP (green fluorescent protein) seedlings demonstrated that interaction of FIT with its target gene promoters was reduced during Fe-deficient conditions in the presence of DELLA proteins (Wild et al., 2016). The authors reported that tissue-specific accumulation of DELLA proteins is required for activation of the Fe uptake pathway. During Fe deficiency, DELLA proteins accumulate in the root meristem and not in the epidermal cells in the root differentiation zone which ultimately leads to FIT–subgroup Ib bHLH heterodimer formation and activation of Fe uptake genes (Wild et al., 2016)
Ethylene
A lot of research has been conducted showing the role of ethylene as an important phytohormone in Fe deficiency signaling (Lingam et al., 2011; Hindt and Guerinot, 2012; Lucena et al., 2015). EIN3 (Ethylene Insensitive 3) and EIL1 (EIN3-Like 1) are two transcription factors that are activated through the ethylene signaling pathway (Chao et al., 1997). EIN3/EIL1 regulates ethylene responses through its post-transcription regulation (Guo and Ecker, 2003; Potuschak et al., 2003; Gagne et al., 2004). EIN3/EIL1 activity is regulated via 26S proteasomal degradation when EIN3/EIL1 is recognized by Skp, Cullin, F-box-containing complexes with EIN3 BINDING F-BOX PROTEINS1 and 2 (SCFEBF1/EBF2) complexes. During ethylene signaling, the F-box proteins EBF1 and EBF2 cannot bind to EIN3/EIL1, thus EIN3/EIL1 are not degraded but are stabilized to bind to downstream target genes to induce ethylene responses (Guo and Ecker, 2003; Potuschak et al., 2003; Gagne et al., 2004). The molecular link between ethylene signaling and Fe deficiency and the mechanism that regulates FIT protein abundance was recently elucidated (Lingam et al., 2011). The authors showed that EIN3 and EIL1 physically interact with FIT and promote its stability, consequently increasing the expression of Fe deficiency-induced genes. When FIT interacts with EIN3/EIL1, the proteasomal degradation of FIT is reduced, which in turn results in induction of Fe uptake genes. In the ein3 eil1 mutants, the protein levels of FIT were lower than in the wild type. The authors posed some interesting questions about the role of EIN3/EIL1 during Fe deficiency and the purpose of FIT interaction with EIN3/EIL1. Through transcriptomic analysis, the authors identified the targets of EIN3/EIL1 and proposed that EIN3/EIL1 might have a role in protection against photooxidative damage during Fe deficiency caused by increased Fe uptake.
ZAT12
ZAT12 is a C2H2-type zinc finger nuclear protein whose expression is induced by ROS (Davletova et al., 2005). Prolonged Fe deficiency causes oxidative stress which then leads to the induction of ZAT12 in roots (Le et al., 2016). Yeast two-hybrid and bimolecular fluorescence complementation (BiFC) assays revealed that ZAT12 interacts with the C-terminus of FIT via its ethylene-responsive element-binding factor-associated amphiphilic repression (EAR) motif. The C-terminus of the EAR motif of ZAT proteins has been shown to have repressive activity during abiotic stress conditions (Kagale et al., 2010; Le et al., 2016). Moreover, because during Fe deficiency-induced oxidative stress, FIT expression is down-regulated and ZAT12 expression is up-regulated, the authors proposed that ZAT12 might be a repressor. FIT–ZAT12 complexes would eventually inactivate FIT during prolonged Fe-deficient conditions. The zat12-3 mutant plants had higher Fe chelate reductase activity and shoot Fe content under Fe-sufficient conditions, suggesting that ZAT12 inhibits Fe uptake when there is a sufficient Fe supply, which is crucial for preventing harmful effects of Fe accumulation. However, the authors also suggested that ZAT12 could affect the uptake of Fe through other mechanisms which still need to be identified. The results demonstrate that ZAT12 is a mediator of oxidative stress during Fe deficiency and FIT integrates inputs from other signaling pathways (in this case ROS) to regulate the Fe uptake process.
Nitric oxide
Nitric oxide (NO) has also been shown to post-translationally modify FIT, and NO inhibitors cause loss of FIT protein and its activity (Meiser et al., 2011). The mechanism, however, is still unclear; therefore, it will be interesting to understand exactly how NO stabilizes and activates FIT.
Mediator
Mediator is a known positive regulator of the Fe uptake mechanism and stabilizes FIT protein (Yang et al., 2014; Zhang et al., 2014). The Mediator complex promotes transcription by increasing the rate of RNA polymerase II pre-initiation complex formation (Cantin et al., 2003). Because of Mediator interactions with specific transcriptional activators, Mediator subunits are involved in coordinating developmental and environmental cues to regulate the transcriptional machinery and expression of genes (Bäckström et al., 2007; Mathur et al., 2011; Zhang et al., 2012). MED16 (Mediator subunit 16) was shown to interact with FIT (Zhang et al., 2014), whereas MED25 (Mediator subunit 25) was able to interact with MED16 and EIN3/EIL1 (Yang et al., 2014). med16 and med25 mutants have lower expression of Fe deficiency-responsive genes IRT1, FRO2, and AHA2 during Fe deficiency (Yang et al., 2014; Zhang et al., 2014). The authors suggested that MED16 and MED25 might be regulating Fe deficiency via their interaction with FIT and EIN3/EIL1, respectively, all of these acting together to stabilize FIT protein and to recruit the FIT–subgroup Ib bHLH complex to their target gene promoters (Yang et al., 2014; Zhang et al., 2014). Many questions remain, such as if and how the Mediator tail senses Fe deficiency. Answering these questions will help us understand the environmental cues necessary for sensing and regulating Fe uptake.
Phosphorylation controls FIT activity
It has been proposed that low levels of FIT protein represent a small pool of active FIT that is important to sense any increase of Fe in the environment. Present models suggest that this small active FIT pool is degraded and replaced by new pool of inactive FIT which is crucial for regulated Fe uptake (Lingam et al., 2011; Meiser et al., 2011; Sivitz et al., 2012). Recently, it was shown that this small pool of FIT protein is actually present in the phosphorylated form, whereas the large non-phosphorylated FIT protein represents its inactive pool form (Gratz et al., 2019). The authors showed that during Fe deficiency, calcium- (Ca2+) induced serine protein kinase CIPK11 is activated, which phosphorylates FIT at Ser272, leading to an active pool of FIT. CIPK11 is activated due to an increase in concentration of cytosolic Ca2+ during Fe deficiency which is sensed through plasma membrane CBL1/CBL9. CBL–CIPK complexes play a crucial role in regulating Ca2+ signaling in response to abiotic stress conditions such as drought, abscisic acid (ABA), and alkaline soil conditions which all affect Fe uptake (Fuglsang et al., 2007; Lumba et al., 2014; Gratz et al., 2019; Ma et al., 2019). CIPK11 acts a positive regulator in ABA responses but causes down-regulation of some drought responses (Zhou et al., 2015; Ma et al., 2019). Under alkaline conditions, CIPK11 is known to negatively regulate AHA2, which is crucial for acidification of the rhizosphere for Fe uptake during Fe deficiency (Fuglsang et al., 2007). The involvement of the CBL–CIPK pathway in response to Fe deficiency in Arabidopsis raises the question of why CIPK11 has such contrasting roles in different stress responses. Thus, it would be interesting to look at how CIPK11 coordinates Fe deficiency with different abiotic stresses and the possible role of cytosolic Ca2+ concentration in Fe sensing or mediating the Fe deficiency response.
The formation of the FIT–bHLH39 heterodimer involves the C-terminal domain of FIT, and this dimerization preferentially occurs when FIT-C is phosphorylated at its Ser site (Gratz et al., 2019, 2020). FIT is activated via phosphorylation of Ser272 and deactivated via phosphorylation of Tyr237 and Tyr238, therefore suggesting that there are alternative phosphorylation pathways for FIT regulation (Gratz et al., 2019, 2020). Phosphorylation of FIT at its Tyr sites negatively affects FIT activity, causing a decrease in FIT mobility towards the nucleus and less interaction with bHLH39 (Gratz et al., 2020). The identification of two phosphorylated Tyr residues is very intriguing because, to date, Tyr phosphorylation has rarely been described in plants. This leads to the speculation that an unknown regulatory pathway is involved to controlling FIT activity via a Tyr kinase. The authors suggest that potential Tyr kinases might belong to Raf-like subfamilies of MAPKKKs (mitogen-activated protein kinase kinase kinase) in plants (Jouannic et al., 1999). Potential candidates could be MAPK3 or MAPK6 that are induced during Fe deficiency (Ye et al., 2015). Therefore, it would be interesting to see future research on identifying Tyr kinases that might be regulating Fe uptake. Another open question is where FIT phosphorylation occurs. It could be that Tyr phosphorylation occurs in the nucleus to eliminate used FIT and to keep a small pool of active/fresh FIT (Sivitz et al., 2011), or maybe it occurs in the cytoplasm where Tyr kinases might be present. However, all these are speculations which need to be confirmed, starting with the identification of a potential Tyr kinase.
Post-translational regulation of the bHLH transcription factor URI/bHLH121
Very recently, a bHLH transcription factor, upstream regulator of IRT1 (URI/bHLH121), was identified that plays a key role in the Fe deficiency signaling cascade in A. thaliana (Kim et al., 2019; Gao et al., 2020; Lei et al., 2020). Although all three papers report that URI is an important regulator of the Fe deficiency response, there are some results which appear contradictory and, at this point, remain unresolved. Lei et al. (2020) report that URI expression increases under Fe deficiency, but Gao et al. (2020) and Kim et al. (2019) reported that URI is expressed constitutively. Lei et al. (2020) employed yeast one-hybrid assay and ChIP, followed by quantitative PCR (qPCR) and EMSA to provide evidence that URI binds the promoter of FIT, whereas using ChIP-seq and ChIP-qPCR, Kim et al. (2019) did not see URI binding to the FIT promoter. Gao et al. (2020) also used ChIP-qPCR and reached a similar conclusion—FIT is not a direct target of URI. Gao et al. (2020) showed that when URI is overexpressed, there is an induction of Fe deficiency-responsive genes under Fe-deficient conditions, whereas Lei et al. (2020) did not see much change in expression of those genes. Lei et al. (2020) state that this difference could be attributed to the different isoforms of URI used for overexpression. Lei et al. (2020) used At3g19860.1, while At3g19860.2 was used by Gao et al. (2020). At3g19860.1 encodes a protein that lacks 53 amino acids at the N-terminus compared with the At3g19860.2-encoded protein. Thus, further research needs to be conducted to see if the different transcripts of URI play different roles in regulating the Fe deficiency response.
The uri mutant is defective in inducing the expression of many genes including IRT1, FRO2, and those encoding the subgroup Ib bHLH transcription factors (Kim et al., 2019). The authors used ChIP-seq to determine which of these genes were URI direct targets. URI was shown to bind to the promoters of various Fe-regulated genes, including the subgroup Ib bHLH transcription factors. Surprisingly, URI transcript and protein levels are not affected by Fe availability, but rather URI is post-translationally modified via phosphorylation when plants are Fe deficient (Kim et al., 2019). The current model suggests that during Fe deficiency, a phosphorylated form of URI is accumulated that forms heterodimers with subgroup IVc bHLH transcription factors. These heterodimers then bind to promoters of the subgroup Ib bHLH genes. Subgroup Ib transcription factors and FIT heterodimers then induce expression of Fe uptake genes IRT1 and FRO2 (Kim et al., 2019). During Fe re-supply, phosphorylated URI is degraded by proteasome-dependent degradation, and this turnover is dependent upon the E3 ligase BTS (discussed in more detail in the next section). Up until now, there were two Fe-regulated pathways established in the literature for Arabidopsis, an FIT-dependent and an FIT-independent pathway (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005; Sivitz et al., 2012; Mai et al., 2016; Schwarz and Bauer, 2020). With the discovery of URI regulating genes from both signaling pathways, it appears that URI plays a central role in the Fe regulatory network. FIT-independent genes including those encoding subgroup Ib bHLH transcription factors are direct targets of URI, which regulate the expression of FIT and other FIT-independent genes (Kim et al., 2019). The authors summarize that URI controls ~50% of the Fe-regulated genes via direct promoter binding, including PYE, BTS, BTSL1, IMA1, and IMA2. This also places the direct targets of these genes under the indirect control of URI. The authors proposed that URI works as a heterodimer with the subgroup IVc bHLH proteins because they do share direct targets. Previous studies (Zhang et al., 2015; Kim et al., 2019; Tissot et al., 2019) have shown that targets of bHLH104, ILR3, and bHLH115 are also the targets of URI. However, more work needs to be done to prove whether URI and subgroup IVc bHLH proteins bind as a heterodimer to their target gene promoters.
Post-translational regulation of bHLH transcription factors by members of the E3 ligase family
Recent studies have proposed that a family of three hemerythrin (HHE) E3 ligases, BTS, BTSL1, and BTSL2, sense Fe and act as negative regulators of the Fe deficiency response in Arabidopsis (Hindt et al., 2017; Rodríguez-Celma et al., 2019). BTS has several conserved domains, including three HHE domains, a cation-binding domain, a CHY zinc finger domain, and a RING domain near the C-terminus (Kobayashi et al., 2013; Li et al., 2016). As mentioned previously, FIT protein levels were thought to be controlled by 26S–proteosome-dependent turnover (Sivitz et al., 2012) but, until recently, no E3 ligase(s) had been identified that facilitated the turnover of FIT. In addition to BTS, E3 ubiquitin ligases BTSL1 and BTSL2 mediate FIT degradation under Fe-deficient conditions (Hindt et al., 2017; Rodríguez-Celma et al., 2019). Unlike BTS, the BTSL1 and BTSL2 proteins contain only two hemerythrin domains (Hindt et al., 2017). BTSL1 and BTSL2 were highly expressed in the root epidermis and cortex, whereas BTS was expressed in the root steele and in the shoot (Rodríguez-Celma et al., 2019). BTSL1 and BTSL2 are closely related in sequence, and mutant analysis showed that both the genes have common functions, but BTSL2 seems to be the dominant paralog (Rodríguez-Celma et al., 2019). The authors showed that BTSL1 or BTSL2 can interact with FIT. They also showed that BTSL2 was able to polyubiquitinate FIT in vitro and promote FIT degradation in vivo. BTSL2 is under the transcriptional control of FIT, which means that through a negative feedback loop FIT protein levels are constantly regulated. The role of BTSL1 still needs to be elucidated because the authors concluded that it also targets FIT based on its high similarity and partial redundancy with BTSL2. It would be interesting to see if BTSL1 is taking on a different function or has different degradation targets. More research is also needed to determine whether BTSL proteins are responsible for the turnover of other proteins besides FIT.
The discrepancy as to whether or not subgroup IVc bHLH transcription factors are transcriptionally regulated by Fe deficiency leads to the idea that they might be regulated post-translationally (Zhang et al., 2015; Li et al., 2016; Liang et al., 2017; Wang et al., 2017; Samira et al., 2018). In vitro analysis revealed that BTS can mediate 26S proteasomal degradation of subgroup IVc bHLH transcription factors under Fe sufficiency (Long et al., 2010; Selote et al., 2015). Compared with wild-type plants, the bts mutant is more tolerant of Fe deficiency but prone to Fe toxicity under Fe-replete conditions (Zhang et al., 2015; Hindt et al., 2017). This is due to constitutive activation of Fe uptake genes regardless of Fe availability in the bts mutant. Yeast two-hybrid analysis revealed BTS interacting with subgroup IVc members, including ILR3, bHLH104, and bHLH115 (Long et al., 2010; Selote et al., 2015), suggesting these to be potential degradation targets of BTS. bHLH104 and bHLH115 mutants were able to suppress the bts mutant phenotype and prevent constitutive expression of Fe uptake genes. The double mutant ilr3 bts was also able to suppress the Fe toxicity phenotype of the bts mutant, suggesting ILR3 functioning downstream of BTS (Li et al., 2019). These data suggest that BTS acts upstream of the Fe deficiency transcriptional network and regulates subgroup IVc bHLH transcription factors via proteasomal degradation. When there is sufficient Fe available, Fe binds via the HHE domain and stabilizes the BTS protein, allowing the E3 ligase complex to assemble and degrade its targets including subgroup IVc bHLH transcription factors, thus inhibiting the Fe deficiency response (Hindt et al., 2017). During Fe deficiency, BTS is inactive, allowing accumulation of subgroup IVc transcription factors which dimerize with URI to induce expression of Ib subgroup bHLH transcription factors, thus increasing Fe uptake (Hindt et al., 2017; Kim et al., 2019).
As mentioned before, Kim et al. (2019) showed that the phosphorylated form of URI only accumulates during Fe deficiency and interacts with subgroup IVc bHLH transcription factors. The phosphorylated URI undergoes BTS-mediated proteasomal-dependent degradation during Fe resupply. Because a prior yeast two-hybrid assay failed to show direct interaction between BTS and URI (Long et al., 2010), the authors introduced pURI:URI into the bts-3 mutant plants and URI protein levels were monitored (Kim et al., 2019). Phosphorylated URI was present in the bts-3 mutant and not in the wild-type plants under Fe-sufficient conditions. Whether BTS interacts directly with URI or whether there is a scaffold protein that brings these two proteins together remains to be determined.
Conclusion
In the past decade, significant progress has been made in understanding the mechanisms involved in Fe homeostasis in plants. Recent advances in understanding regulation of the bHLH transcriptional networks during Fe deficiency have identified key players from this complex network. Plants tightly regulate Fe homeostasis to avoid both Fe deficiency and Fe toxicity through multiple levels of controls that involve transcriptional and post-translational modes, as summarized in this review. Major discoveries in the field of the Fe deficiency-regulated bHLH transcriptional cascade include (i) phosphorylation-based regulation of FIT; (ii) URI as an Fe-dependent switch controlling both FIT-dependent and FIT-independent pathways; and (iii) identification of the FIT ortholog OsbHLH156 as the master regulator of Fe uptake in rice, providing insight into evolution of Fe uptake mechanisms in plants. However, we still lack knowledge on how the expression of most of the transcription factors upstream in the cascade is regulated. There is evidence that FIT transcription is controlled by bHLH39, but we still do not know which transcription factor(s) induce FIT transcription under Fe deficiency. Overexpression of the subgroup Ib bHLH transcription factors is sufficient to induce IRT1, which suggests that they also might induce FIT, probably as a heterodimer. Thus, given the importance of FIT in the regulatory network, a ChIP-seq analysis to determine all the direct targets of FIT as well as the subgroup Ib bHLH transcription factors should be carried out. If FIT and any or all of the subgroup Ib bHLH proteins do bind to the FIT promoter, it will also be important to determine if they bind as a heterodimer. Most of our understanding of the Fe deficiency responses is from roots, and little is known about how Fe is regulated in shoots. Only one RNA-seq analysis has been performed to compare the Fe deficiency response in roots and shoots of A. thaliana, indicating that the roots and shoots respond differently to Fe deficiency (Rodríguez-Celma et al., 2013). We do not know the role of PYE, URI or the subgroup IVc bHLH transcription factors in regulating the Fe deficiency response in the shoots. Thus, deciphering the regulation of Fe homeostasis in shoots should be the next important research area. Understanding the molecular mechanism of Fe-dependent regulation of Fe deficiency signaling in plants will allow breeding of crops that grow robustly in Fe-limited soils and produce high yield and high Fe content, which will consequently improve human health.
Acknowledgements
Work in the Guerinot lab is supported by grants from the National Science Foundation (IOS-1456290; IOS 1257722) and the National Institutes of Health (P42ES007373). We thank members of the Guerinot lab for helpful discussions. All figures were created with BioRender.com.
Author contributions
NR wrote the original draft and MLG edited the manuscript.
References
- Bäckström S, Elfving N, Nilsson R, Wingsle G, Björklund S. 2007. Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Molecular Cell 26, 717–729. [DOI] [PubMed] [Google Scholar]
- Bailey PC, Martin C, Toledo-Ortiz G, Quail PH, Huq E, Heim MA, Jakoby M, Werber M, Weisshaar B. 2003. Update on the basic helix–loop–helix transcription factor gene family in Arabidopsis thaliana. The Plant Cell 15, 2497–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir K, Rasheed S, Kobayashi T, Seki M, Nishizawa NK. 2016. Regulating subcellular metal homeostasis: the key to crop improvement. Frontiers in Plant Science 7, 1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat JF, Duc C, Ravet K, Gaymard F. 2010. Ferritins and iron storage in plants. Biochimica et Biophysica Acta 1800, 806–814. [DOI] [PubMed] [Google Scholar]
- Brumbarova T, Bauer P, Ivanov R. 2015. Molecular mechanisms governing Arabidopsis iron uptake. Trends in Plant Science 20, 124–133. [DOI] [PubMed] [Google Scholar]
- Brumbarova T, Ivanov R. 2019. The nutrient response transcriptional regulome of Arabidopsis. iScience 19, 358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantin GT, Stevens JL, Berk AJ. 2003. Activation domain–mediator interactions promote transcription preinitiation complex assembly on promoter DNA. Proceedings of the National Academy of Sciences, USA 100, 12003–12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretero-Paulet L, Galstyan A, Roig-Villanova I, Martínez-García JF, Bilbao-Castro JR, Robertson DL. 2010. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiology 153, 1398–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR. 1997. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89, 1133–1144. [DOI] [PubMed] [Google Scholar]
- Cheng L, Wang F, Shou H, et al. 2007. Mutation in nicotianamine aminotransferase stimulated the Fe(II) acquisition system and led to iron accumulation in rice. Plant Physiology 145, 1647–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML. 2004. The essential basic helix–loop–helix protein FIT1 is required for the iron deficiency response. The Plant Cell 16, 3400–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo C, Palumbo G, He J-Z, Pinton R, Cesco S. 2014. Review on iron availability in soil: interaction of Fe minerals, plants, and microbes. Journal of Soils and Sediments 14, 538–548. [Google Scholar]
- Connorton JM, Balk J, Rodríguez-Celma J. 2017. Iron homeostasis in plants—a brief overview. Metallomics 9, 813–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Chen CL, Cui M, Zhou WJ, Wu HL, Ling HQ. 2018. Four IVa bHLH transcription factors are novel interactors of FIT and mediate JA inhibition of iron uptake in Arabidopsis. Molecular Plant 11, 1166–1183. [DOI] [PubMed] [Google Scholar]
- Curie C, Panaviene Z, Loulergue C, Dellaporta SL, Briat JF, Walker EL. 2001. Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409, 346–349. [DOI] [PubMed] [Google Scholar]
- Davletova S, Schlauch K, Coutu J, Mittler R. 2005. The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiology 139, 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML. 1996. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proceedings of the National Academy of Sciences, USA 93, 5624–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglsang AT, Guo Y, Cuin TA, et al. 2007. Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. The Plant Cell 19, 1617–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JM, Smalle J, Gingerich DJ, Walker JM, Yoo SD, Yanagisawa S, Vierstra RD. 2004. Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proceedings of the National Academy of Sciences, USA 101, 6803–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Robe K, Bettembourg M, et al. 2020. The transcription factor bHLH121 interacts with bHLH105 (ILR3) and its closest homologs to regulate iron homeostasis in Arabidopsis. The Plant Cell 32, 508–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Robe K, Gaymard F, Izquierdo E, Dubos C. 2019. The transcriptional control of iron homeostasis in plants: a tale of bHLH transcription factors? Frontiers in Plant Science 10, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayomba SR, Zhai Z, Jung HI, Vatamaniuk OK. 2015. Local and systemic signaling of iron status and its interactions with homeostasis of other essential elements. Frontiers in Plant Science 6, 716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz R, Brumbarova T, Ivanov R, et al. 2020. Phospho-mutant activity assays provide evidence for alternative phospho-regulation pathways of the transcription factor FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR. New Phytologist 225, 250–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz R, Manishankar P, Ivanov R, et al. 2019. CIPK11-dependent phosphorylation modulates FIT activity to promote arabidopsis iron acquisition in response to calcium signaling. Developmental Cell 48, 726–740.e10. [DOI] [PubMed] [Google Scholar]
- Guerinot ML, Yi Y. 1994. Iron: nutritious, noxious, and not readily available. Plant Physiology 104, 815–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ecker JR. 2003. Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115, 667–677. [DOI] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JM. 1992. Biologically relevant metal ion-dependent hydroxyl radical generation. An update. FEBS Letters 307, 108–112. [DOI] [PubMed] [Google Scholar]
- Haydon MJ, Cobbett CS. 2007. A novel major facilitator superfamily protein at the tonoplast influences zinc tolerance and accumulation in Arabidopsis. Plant Physiology 143, 1705–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC. 2003. The basic helix–loop–helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Molecular Biology and Evolution 20, 735–747. [DOI] [PubMed] [Google Scholar]
- Hindt MN, Akmakjian GZ, Pivarski KL, Punshon T, Baxter I, Salt DE, Guerinot ML. 2017. BRUTUS and its paralogs, BTS LIKE1 and BTS LIKE2, encode important negative regulators of the iron deficiency response in Arabidopsis thaliana. Metallomics 9, 876–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindt MN, Guerinot ML. 2012. Getting a sense for signals: regulation of the plant iron deficiency response. Biochimica et Biophysica Acta 1823, 1521–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru Y, Suzuki M, Tsukamoto T, et al. 2006. Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. The Plant Journal 45, 335–346. [DOI] [PubMed] [Google Scholar]
- Jakoby M, Wang HY, Reidt W, Weisshaar B, Bauer P. 2004. FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Letters 577, 528–534. [DOI] [PubMed] [Google Scholar]
- Jeong J, Connolly E. 2009. Iron uptake mechanisms in plants: functions of the FRO family of ferric reductases. Plant science 176, 709–714. [Google Scholar]
- Jouannic S, Hamal A, Leprince AS, Tregear JW, Kreis M, Henry Y. 1999. Plant MAP kinase kinase kinases structure, classification and evolution. Gene 233, 1–11. [DOI] [PubMed] [Google Scholar]
- Kagale S, Links MG, Rozwadowski K. 2010. Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiology 152, 1109–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM. 2013. MYC2: the master in action. Molecular Plant 6, 686–703. [DOI] [PubMed] [Google Scholar]
- Kim SA, LaCroix IS, Gerber SA, Guerinot ML. 2019. The iron deficiency response in Arabidopsis thaliana requires the phosphorylated transcription factor URI. Proceedings of the National Academy of Sciences, USA 116, 24933–24942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatte M, Schuler M, Wirtz M, Fink-Straube C, Hell R, Bauer P. 2009. The analysis of Arabidopsis nicotianamine synthase mutants reveals functions for nicotianamine in seed iron loading and iron deficiency responses. Plant Physiology 150, 257–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Itai RN, Senoura T, Oikawa T, Ishimaru Y, Ueda M, Nakanishi H, Nishizawa NK. 2016. Jasmonate signaling is activated in the very early stages of iron deficiency responses in rice roots. Plant Molecular Biology 91, 533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Nagasaka S, Senoura T, Itai RN, Nakanishi H, Nishizawa NK. 2013. Iron-binding haemerythrin RING ubiquitin ligases regulate plant iron responses and accumulation. Nature Communications 4, 2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Nishizawa NK. 2014. Iron sensors and signals in response to iron deficiency. Plant Science 224, 36–43. [DOI] [PubMed] [Google Scholar]
- Le CT, Brumbarova T, Ivanov R, Stoof C, Weber E, Mohrbacher J, Fink-Straube C, Bauer P. 2016. ZINC FINGER OF ARABIDOPSIS THALIANA12 (ZAT12) interacts with FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT) linking iron deficiency and oxidative stress responses. Plant Physiology 170, 540–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei R, Li Y, Cai Y, Li C, Pu M, Lu C, Yang Y, Liang G. 2020. bHLH121 functions as a direct link that facilitates the activation of FIT by bHLH IVc transcription factors for maintaining Fe homeostasis in Arabidopsis. Molecular Plant 13, 634–649. [DOI] [PubMed] [Google Scholar]
- Li M, Zhang J, Jin H, Feng D, Wang J, Wang H-B, Liu B. 2019. The iron deficiency response regulators IAA-LEUCINE RESISTANT3 and bHLH104 possess different targets and have distinct effects on photosynthesis in Arabidopsis. Journal of Plant Biology 62, 109–119. [Google Scholar]
- Li X, Zhang H, Ai Q, Liang G, Yu D. 2016. Two bHLH transcription factors, bHLH34 and bHLH104, regulate iron homeostasis in Arabidopsis thaliana. Plant Physiology 170, 2478–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Zhang H, Li X, Ai Q, Yu D. 2017. bHLH transcription factor bHLH115 regulates iron homeostasis in Arabidopsis thaliana. Journal of Experimental Botany 68, 1743–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HQ, Bauer P, Bereczky Z, Keller B, Ganal M. 2002. The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proceedings of the National Academy of Sciences, USA 99, 13938–13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingam S, Mohrbacher J, Brumbarova T, Potuschak T, Fink-Straube C, Blondet E, Genschik P, Bauer P. 2011. Interaction between the bHLH transcription factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis. The Plant Cell 23, 1815–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN. 2010. The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. The Plant Cell 22, 2219–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucena C, Romera FJ, García MJ, Alcántara E, Pérez-Vicente R. 2015. Ethylene participates in the regulation of Fe deficiency responses in strategy I plants and in rice. Frontiers in Plant Science 6, 1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumba S, Toh S, Handfield LF, et al. 2014. A mesoscale abscisic acid hormone interactome reveals a dynamic signaling landscape in Arabidopsis. Developmental Cell 29, 360–372. [DOI] [PubMed] [Google Scholar]
- Ma Y, Cao J, Chen Q, He J, Liu Z, Wang J, Li X, Yang Y. 2019. The kinase CIPK11 functions as a negative regulator in drought stress response in Arabidopsis. International Journal of Molecular Sciences 20, 2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai HJ, Pateyron S, Bauer P. 2016. Iron homeostasis in Arabidopsis thaliana: transcriptomic analyses reveal novel FIT-regulated genes, iron deficiency marker genes and functional gene networks. BMC Plant Biology 16, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Barranco A, Spielmann J, Dubeaux G, Vert G, Zelazny E. 2020. Dynamic control of the high-affinity iron uptake complex in root epidermal cells. Plant Physiology 184, 1236–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur S, Vyas S, Kapoor S, Tyagi AK. 2011. The Mediator complex in plants: structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (rice) during reproduction and abiotic stress. Plant Physiology 157, 1609–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K, Furukawa J, Bidadi H, Asahina M, Yamaguchi S, Satoh S. 2014. Gibberellin-induced expression of Fe uptake-related genes in Arabidopsis. Plant & Cell Physiology 55, 87–98. [DOI] [PubMed] [Google Scholar]
- Maurer F, Müller S, Bauer P. 2011. Suppression of Fe deficiency gene expression by jasmonate. Plant Physiology and Biochemistry 49, 530–536. [DOI] [PubMed] [Google Scholar]
- Meiser J, Lingam S, Bauer P. 2011. Posttranslational regulation of the iron deficiency basic helix–loop–helix transcription factor FIT is affected by iron and nitric oxide. Plant Physiology 157, 2154–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranjo-Arcos MA, Maurer F, Meiser J, Pateyron S, Fink-Straube C, Bauer P. 2017. Dissection of iron signaling and iron accumulation by overexpression of subgroup Ib bHLH039 protein. Scientific Reports 7, 10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozoye T, Nagasaka S, Kobayashi T, Takahashi M, Sato Y, Sato Y, Uozumi N, Nakanishi H, Nishizawa NK. 2011. Phytosiderophore efflux transporters are crucial for iron acquisition in graminaceous plants. Journal of Biological Chemistry 286, 5446–5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires N, Dolan L. 2010. Origin and diversification of basic-helix–loop–helix proteins in plants. Molecular Biology and Evolution 27, 862–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P. 2003. EIN3-dependent regulation of plant ethylene hormone signaling by two arabidopsis F box proteins: EBF1 and EBF2. Cell 115, 679–689. [DOI] [PubMed] [Google Scholar]
- Rampey RA, Woodward AW, Hobbs BN, Tierney MP, Lahner B, Salt DE, Bartel B. 2006. An Arabidopsis basic helix–loop–helix leucine zipper protein modulates metal homeostasis and auxin conjugate responsiveness. Genetics 174, 1841–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML. 1999. A ferric-chelate reductase for iron uptake from soils. Nature 397, 694–697. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Celma J, Chou H, Kobayashi T, Long TA, Balk J. 2019. Hemerythrin E3 ubiquitin ligases as negative regulators of iron homeostasis in plants. Frontiers in Plant Science 10, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Celma J, Pan IC, Li W, Lan P, Buckhout TJ, Schmidt W. 2013. The transcriptional response of Arabidopsis leaves to Fe deficiency. Frontiers in Plant Science 4, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samira R, Li B, Kliebenstein D, Li C, Davis E, Gillikin JW, Long TA. 2018. The bHLH transcription factor ILR3 modulates multiple stress responses in Arabidopsis. Plant Molecular Biology 97, 297–309. [DOI] [PubMed] [Google Scholar]
- Santi S, Schmidt W. 2009. Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytologist 183, 1072–1084. [DOI] [PubMed] [Google Scholar]
- Schwarz B, Bauer P. 2020. FIT, a regulatory hub for iron deficiency and stress signaling in roots, and FIT-dependent and -independent gene signatures. Journal of Experimental Botany 71, 1694–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selote D, Samira R, Matthiadis A, Gillikin JW, Long TA. 2015. Iron-binding E3 ligase mediates iron response in plants by targeting basic helix–loop–helix transcription factors. Plant Physiology 167, 273–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivitz A, Grinvalds C, Barberon M, Curie C, Vert G. 2011. Proteasome-mediated turnover of the transcriptional activator FIT is required for plant iron-deficiency responses. The Plant Journal 66, 1044–1052. [DOI] [PubMed] [Google Scholar]
- Sivitz AB, Hermand V, Curie C, Vert G. 2012. Arabidopsis bHLH100 and bHLH101 control iron homeostasis via a FIT-independent pathway. PLoS One 7, e44843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringlis IA, de Jonge R, Pieterse CMJ. 2019. The age of coumarins in plant–microbe interactions. Plant & Cell Physiology 60, 1405–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi SI. 1976. Naturally occurring iron-chelating compounds in oat- and rice-root washings. Soil Science and Plant Nutrition 22, 423–433. [Google Scholar]
- Tissot N, Robe K, Gao F, et al. 2019. Transcriptional integration of the responses to iron availability in Arabidopsis by the bHLH factor ILR3. New Phytologist 223, 1433–1446. [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH. 2003. The Arabidopsis basic/helix–loop–helix transcription factor family. The Plant Cell 15, 1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trofimov K, Ivanov R, Eutebach M, Acaroglu B, Mohr I, Bauer P, Brumbarova T. 2019. Mobility and localization of the iron deficiency-induced transcription factor bHLH039 change in the presence of FIT. Plant Direct 3, e00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Yao X, Yu D, Liang G. 2017. Fe-deficiency-induced expression of bHLH104 enhances Fe-deficiency tolerance of Arabidopsis thaliana. Planta 246, 421–431. [DOI] [PubMed] [Google Scholar]
- Wang HY, Klatte M, Jakoby M, Bäumlein H, Weisshaar B, Bauer P. 2007. Iron deficiency-mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana. Planta 226, 897–908. [DOI] [PubMed] [Google Scholar]
- Wang N, Cui Y, Liu Y, Fan H, Du J, Huang Z, Yuan Y, Wu H, Ling HQ. 2013. Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Molecular Plant 6, 503–513. [DOI] [PubMed] [Google Scholar]
- Wang S, Li L, Ying Y, Wang J, Shao JF, Yamaji N, Whelan J, Ma JF, Shou H. 2020. A transcription factor OsbHLH156 regulates Strategy II iron acquisition through localising IRO2 to the nucleus in rice. New Phytologist 225, 1247–1260. [DOI] [PubMed] [Google Scholar]
- Wild M, Davière JM, Regnault T, Sakvarelidze-Achard L, Carrera E, Lopez Diaz I, Cayrel A, Dubeaux G, Vert G, Achard P. 2016. Tissue-specific regulation of gibberellin signaling fine-tunes Arabidopsis iron-deficiency responses. Developmental Cell 37, 190–200. [DOI] [PubMed] [Google Scholar]
- Wu H, Ling HQ. 2019. FIT-binding proteins and their functions in the regulation of Fe homeostasis. Frontiers in Plant Science 10, 844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ou B, Zhang J, Si W, Gu H, Qin G, Qu LJ. 2014. The Arabidopsis Mediator subunit MED16 regulates iron homeostasis by associating with EIN3/EIL1 through subunit MED25. The Plant Journal 77, 838–851. [DOI] [PubMed] [Google Scholar]
- Ye L, Li L, Wang L, Wang S, Li S, Du J, Zhang S, Shou H. 2015. MPK3/MPK6 are involved in iron deficiency-induced ethylene production in Arabidopsis. Frontiers in Plant Science 6, 953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ. 2008. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Research 18, 385–397. [DOI] [PubMed] [Google Scholar]
- Yuan YX, Zhang J, Wang DW, Ling HQ. 2005. AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Research 15, 613–621. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liu B, Li M, Feng D, Jin H, Wang P, Liu J, Xiong F, Wang J, Wang HB. 2015. The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. The Plant Cell 27, 787–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang C, Zhang Y, Sun Y, Mou Z. 2012. The Arabidopsis mediator complex subunit16 positively regulates salicylate-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. The Plant Cell 24, 4294–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wu H, Wang N, Fan H, Chen C, Cui Y, Liu H, Ling HQ. 2014. Mediator subunit 16 functions in the regulation of iron uptake gene expression in Arabidopsis. New Phytologist 203, 770–783. [DOI] [PubMed] [Google Scholar]
- Zhou X, Hao H, Zhang Y, et al. 2015. SOS2-LIKE PROTEIN KINASE5, an SNF1-RELATED PROTEIN KINASE3-type protein kinase, is important for abscisic acid responses in Arabidopsis through phosphorylation of ABSCISIC ACID-INSENSITIVE5. Plant Physiology 168, 659–676. [DOI] [PMC free article] [PubMed] [Google Scholar]