Abstract
Diatoms are one of the most successful group of photosynthetic eukaryotes in the contemporary ocean. They are ubiquitously distributed and are the most abundant primary producers in polar waters. Equally remarkable is their ability to tolerate iron deprivation and respond to periodic iron fertilization. Despite their relatively large cell sizes, diatoms tolerate iron limitation and frequently dominate iron-stimulated phytoplankton blooms, both natural and artificial. Here, we review the main iron use strategies of diatoms, including their ability to assimilate and store a range of iron sources, and the adaptations of their photosynthetic machinery and architecture to iron deprivation. Our synthesis relies on published literature and is complemented by a search of 82 diatom transcriptomes, including information collected from seven representatives of the most abundant diatom genera in the world’s oceans.
Keywords: Diatoms, ferritin, iron physiology, iron quotas, iron starvation-induced proteins (ISIPs), iron storage, iron uptake
Diatoms are responsible for an estimated fifth of the total carbon fixation on Earth. Here we review their relationship with a key micronutrient in a changing ocean: iron.
Introduction
Unicellular life began to evolve more than 3 billion years ago in an ocean that was devoid of molecular oxygen and rich in soluble ferrous iron (Fe2+). The metabolic processes that became core to all cellular life relied on the abundance of this transition element and its ability to stably occupy multiple valence states, as cofactor to enzymes catalysing reactions involving the transfer of electrons. Most notably, these processes include photosynthesis and respiration, as well as the synthesis of essential organic molecules such as amino acids, lipids, deoxyribonucleotides, and sterols. The minimum concentration of iron required by an individual cell to sustain its metabolic functions is referred to as its metabolic ‘iron quota’. Within cells, iron homeostasis is carefully controlled, since overabundance of Fe2+ can catalyse the formation of damaging reactive oxygen species. While the preference of cells for reduced iron has remained unchanged, that is, cells readily take up Fe2+ and carefully regulate its concentration intracellularly, the abiotic environment of unicellular species living in today’s ocean is vastly different from that existing at the start of these organisms’ evolution. The contemporary surface ocean is oxygenated and well mixed, with a mildly alkaline pH (global averages are around 8.2; Lauvset et al., 2015). This is an oxidizing environment that chemically shifts iron into its ferric state (Fe3+), so that the most abundant form of iron is as Fe3+ bound by organic material or colloid particles of oxyhydroxides (Gledhill and Buck, 2012). Organisms have therefore had to evolve a variety of molecular mechanisms to make such recalcitrant forms of iron bioavailable.
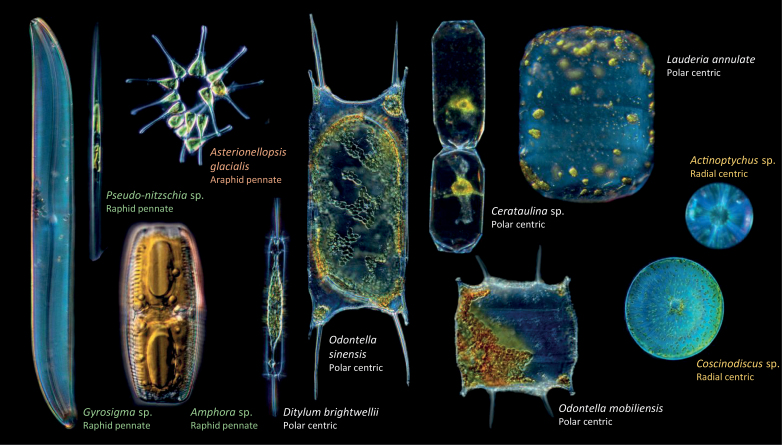
One of the dominant, ubiquitous groups of photosynthetic producers in the contemporary ocean is diatoms, which are estimated to account for 20% of total global primary production (~20×1015 g carbon fixed per year) (Field et al., 1998; Mann, 1999). Diatoms are unicellular stramenopiles (heterokont protists within the chromalveolates) and are, in an evolutionary context, a relatively young form of life. Molecular phylogenies date the origin of diatoms towards the beginning of the Mesozoic Era (Sims et al., 2006), while current estimates of the richness of diatom species vary between 40 000 and 200 000; counterintuitively, the higher end of this range is based on morphology-based estimates while the lower estimates are based on molecular markers (reviewed in Benoiston et al., 2017). The evolutionary history of diatoms is shaped by endosymbiotic events, of which it has been proposed that there were at least two. The first occurred an estimated 1.5 billion years ago, when a eukaryotic heterotroph assimilated or was invaded by a cyanobacterium (Yoon et al., 2004). Some 500 million years later, a secondary endosymbiosis occurred, in which a eukaryotic heterotroph captured or was invaded by a red alga, establishing the photosynthetic stramenopile lineage to which diatoms belong (Cavalier-Smith, 1999). Recent evidence suggests that the chimeric nature of diatoms is even more complicated, as a range of nucleus-encoded proteins are of green algal origin (Dorrell et al., 2017 and references therein). Diatoms are subclassified into ‘centric’ species, which include two suborders (radial and polar), and ‘pennate’ species, which are further divided into two suborders (raphid and araphid) (Fig. 1). These groups are not monophyletic, as centric diatoms grade into araphid pennates, and araphid pennates grade into the raphe-bearing pennate diatoms, which are a natural group (Alverson and Theriot, 2005).
Fig. 1.
Light microscopy images of representative species in the raphid and araphid pennate, and polar and radial centric groups of diatoms. Sizes are not to scale. Diatoms were collected and photographed on board Tara, in Roscoff, Villefranche-sur-Mer Marine Stations (France), and Bigelow Laboratory, Boothbay (USA). Photography by Christian and Noé Sardet, rearranged from C. Sardet, 2015. Plankton: Wonders of the Drifting World. Republished with permission of University of Chicago Press; permission conveyed through Copyright Clearance Center, Inc.
Diatom cells have a unique structure and features (Fig. 1). Cell sizes are large compared with other oceanic unicellular eukaryotes and are generally in excess of 20 μm in diameter (with maximum cell sizes of 2 mm recorded). Some diatoms are known to form chains. Unlike other organisms, diatom cell membranes are silicified and are referred to as frustules. Frustules are porous, with their nanopores ranging in size from 250 nm to 600 nm; they are generally assumed not to interfere with diatom nutrient uptake (Bhatta et al., 2009). The majority of pennate diatoms contain one central vacuole, while centric species can have many vacuoles. Diatom plastids are surrounded by four membranes, a vestige of the endosymbiosis events that generated the organelle. As with vacuoles, the majority of pennate species have only one plastid, but the number of plastids is variable in the centric species. Nine species of diatoms have had their genomes sequenced: Thalassiosira pseudonana (Armbrust et al., 2004), Phaeodactylum tricornutum (Bowler et al., 2008), Thalassiosira oceanica (Lommer et al., 2012), Pseudo-nitzschia multistriata strain B856 (NCBI assembly ASM90066040v1), Pseudo-nitzschia multiseries CLN47 (JGI Project ID: 16870), Synedra acus (Galachyants et al., 2015), Fistulifera solaris (Tanaka et al., 2015), Fragilariopsis cylindrus (Mock et al., 2017), and Plagiostriata sp. CCMP470 (Sato et al., 2020), while 92 species have available transcriptomes collected through the Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP) (Keeling et al., 2014). A further seven transcriptomes have been generated by Genoscope and represent seven of the most globally abundant diatom genera not covered by the MMETSP project (Dorrell et al., 2021). The pennate diatom P. tricornutum and the centric diatom T. pseudonana are laboratory models that can be readily cultured and transformed (Apt et al., 1996; Zaslavskaia et al., 2000; Poulsen et al., 2006; Hopes et al., 2016). Although neither is abundant in the environment, they often harbour genes that have been proposed to contribute to the success of diatoms in the wild.
The ecological success of diatoms is remarkable. They are present in all ocean provinces and are the most abundant photosynthetic producers in the Arctic and the Southern oceans (Malviya et al., 2016). Importantly, they are ruthless competitors for iron, often dominating iron-stimulated blooms (Boyd et al., 2007). This indicates that diatoms are adapted to survive conditions of low iron and are equipped with the molecular machinery necessary to take up and assimilate iron efficiently, without causing damage to their cells through oxidative stress. Rates of iron uptake and use are also linked to nitrogen assimilation and growth: on average, diatoms with higher iron uptake show proportionately greater nitrate uptake and productivity (e.g. Falkowski, 1997). Surprisingly, this is not reflected in their silica uptake, which remains constant under iron supplementation despite faster growth, leading to weakly silicified cells (Boyle, 1998 and references therein). Diatom iron nutrition therefore affects not only metabolism but also the resulting morphology and physical characteristics of diatom cells.
Comparing the cell metal concentrations of diatoms with those of autotrophic flagellates, heterotrophic flagellates, and autotrophic picoplankton in the equatorial Pacific Ocean, Twining et al. (2011) reported that phosphorus-normalized Mn, Fe, Ni, and Zn ratios were significantly higher in diatoms than in other cell types. Furthermore, total diatom Fe concentrations per cell exceeded minimum subsistence levels.
Here, we review what is known about iron physiology in diatoms. As far as is possible, we draw comparisons between different diatom species (Sunda and Huntsman, 1995; Strzepek and Harrison, 2004; Kustka et al., 2007). We take a broad perspective that considers how diatoms take up, store, and use iron, in order to shed light on the question of what makes diatoms competitive in handling this micronutrient.
A portfolio of iron uptake mechanisms shows diatoms are adapted to acquire iron from a variety of sources
The vast majority of iron present in surface ocean seawater is complexed by organic ligands (Gledhill and Buck, 2012). These are classified based on their affinity for iron. The strongest chelates are siderophore molecules, thought to be synthesized largely by bacteria, although fungi, which are of lower abundance in the ocean, are also capable of siderophore synthesis (Renshaw et al., 2002). There are three chemical classes of siderophores—hydroxamate, catecholate, and carboxylate—and their relative contribution to the iron pool of seawater is unknown. Hydroxamate siderophores, which have been detected both in coastal and open ocean waters (Mawji et al., 2008; Boiteau et al., 2016), are hydrophilic and are released by synthesizing species into the water column. In these microorganisms, siderophores are recaptured by specific recognition machinery. It has been proposed that it is these molecules in particular that are available to non-producing species, such as diatoms (Hopkinson and Morel, 2009). Amphiphilic siderophores, which are attached by lipid tails to the cell membranes of producing bacterial species, have received particular attention in the literature on account of their chimeric chemical structures (Martinez et al., 2000; Vraspir et al., 2011). However, they are an unlikely source of iron for non-producers. Other chelates include extracellular polysaccharides and humic acids, which bind iron weakly. Notably, many marine viruses are also capable of chelating iron on their tails and thus may represent a large sink of marine iron (Bonnain et al., 2016). Inorganic mineral iron is periodically available to surface communities, either through aeolian input (i.e. arising from the action of wind) or via upwelling currents that bring remineralized iron from the deep ocean (Mahowald et al., 2008; Boyd and Ellwood, 2010). The most bioavailable and scarcest source of iron is dissolved, uncomplexed iron (Fe′) (Liu and Millero, 2002; Lis et al., 2015).
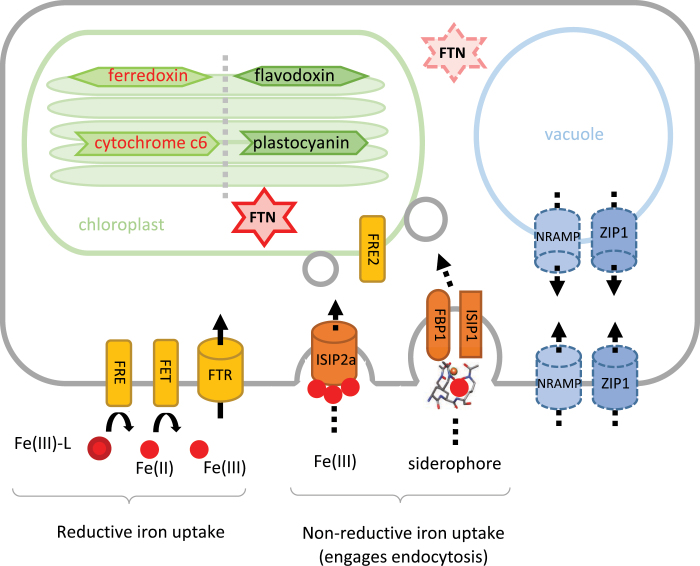
Diatoms have a diverse portfolio of iron uptake mechanisms adapted to the sources of iron available in seawater (summarized in Fig. 2). A mechanism whose function has been experimentally verified in the model pennate diatom P. tricornutum is the assimilation of uncomplexed iron bound by a transferrin-like protein. The iron starvation-induced protein 2A (ISIP2A), first identified in P. tricornutum (Allen et al., 2008), was shown to function as a ‘phytotransferrin’, a protein directed to the cell membrane of cells with carboxylate iron-binding domains, whose iron-chelating properties were pH sensitive (Morrissey et al., 2015; McQuaid et al., 2018). P. tricornutum cells deficient in ISIP2A showed reduced Fe′ uptake capabilities, suggesting that this was the primary but not the only mechanism of Fe′ uptake in this species (Kazamia et al., 2018). ISIP2A contains the functional domain PF07692, which was first characterized in the iron-assimilation proteins FEA1 and FEA2 of the green alga Chlamydomonas reinhardtii (Allen et al., 2007), and additionally was functionally described in Dunaliella salina (Paz et al., 2007). In C. reinhardtii, FEA1 and FEA2 were highly expressed under low-iron conditions and facilitated high-affinity iron uptake (Allen et al., 2007), likely in its ferrous form (Narayanan et al., 2011). By contrast, in diatoms, the iron-binding domains of ISIP2A coordinate ferric iron (McQuaid et al., 2018). We searched the available diatom genomes and transcriptomes for genes with a profile that matched a constructed Hidden Markov Model (HMM) for ISIP2A. Our results are presented as a heatmap in Fig. 3, where we show the detection of ISIP2A-like genes in 82 diatom species. We retrieved matches for ISIP2A-like genes in most diatoms, with some notable exceptions in some species of Chaetoceros.
Fig. 2.
Schematic diagram of iron uptake mechanisms and metabolism in diatoms. A summary schematic of the proteins discussed in this review and of their cellular location. In the chloroplast, we review the replacement of ferredoxin with its iron-free equivalent, flavodoxin, and the substitution of cytochrome c6 with plastocyanin. Reductive and non-reductive iron uptake mechanisms are considered, the latter employing the cell’s vesicular network to direct iron to the plastid. Two alternative iron storage mechanisms are reviewed: the accumulation of iron in the mineral core of ferritin (FTN), and vacuolar storage. For the storage proteins, the cellular localization of the putative proteins has not been confirmed through microscopy analyses (indicated with dashed lines).
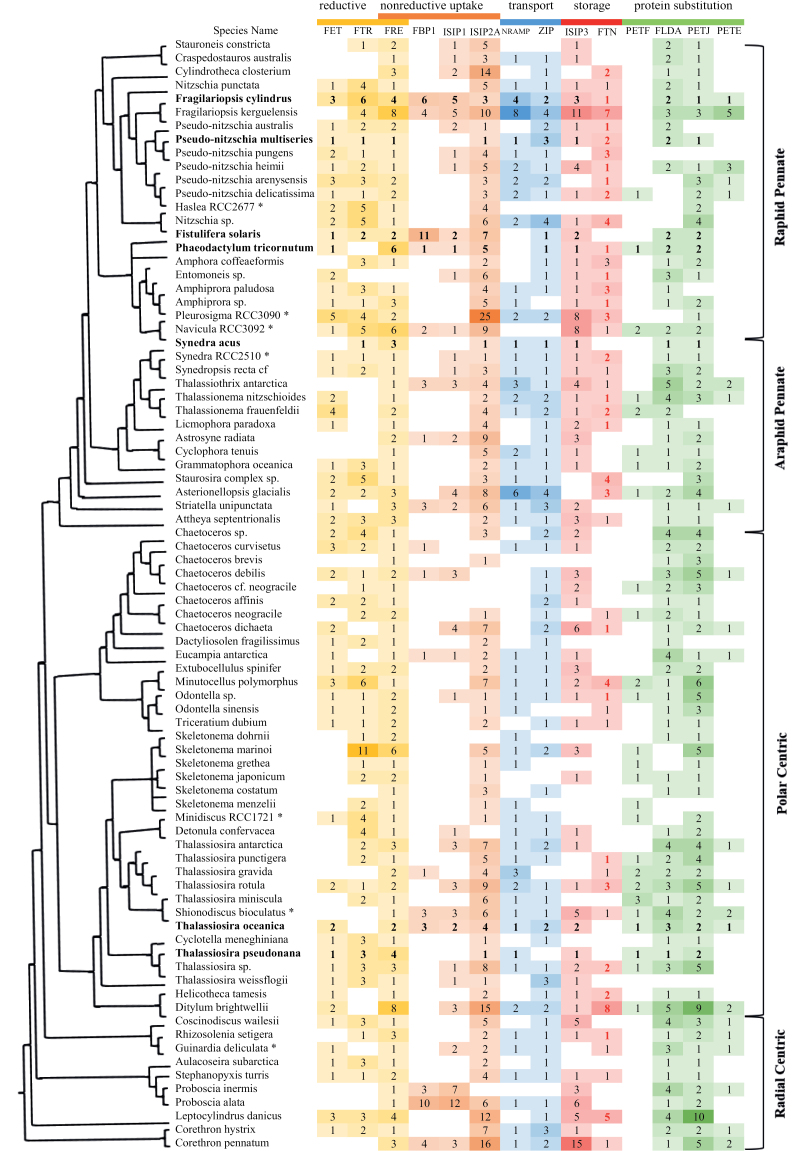
Fig. 3.
Iron-related genes in diatom transcriptomes. To investigate the evolutionary distribution of iron-related genes, first, representative protein sequences that are well annotated were downloaded from online databases (GenBank, Uniprot, and Ensembl). Further sequences were retrieved from NCBI by BLASTp with a threshold E value of 1×10–5 for each gene of interest. Together, these comprised the reference dataset; they were aligned by MUSCLE and transformed into a Hidden Markov Model using HMMER 3.0. The profiles were used for HMMER searches against the diatom transcriptomes from MMETSP alongside seven transcriptomes generated by Genoscope (indicated with asterisks) and diatom genomes (indicated in bold text). In total, our searchable database comprised 82 species, which met decontamination standards described in Johnson et al. (2019). The seven species sequenced by Genoscope are among the 10 most abundant diatom genera in the Tara Oceans dataset. Where no hits were detected, the grid is blank, otherwise the shade of the colour and the number within indicates the number of distinct transcripts encoding each gene (but not the expression levels). The subcellular localization of FTN was predicted using ASAFind, HECTAR, MitoFates, TargetP, and WolfPSort (Bendtsen et al., 2004; Emanuelsson et al., 2007; Horton et al., 2007; Gschloessl et al., 2008; Fukasawa et al., 2015; Gruber et al., 2015). Sequences predicted to target to the chloroplast are shown in red. It is important to note that this dataset likely misses genes that are under strong transcriptional control, which was not met by the culture conditions during RNA harvesting (e.g. low-Fe-induced genes in diatom cultures grown under Fe-replete conditions), as well as chloroplast-encoded genes. The sequencing depth between the transcriptomes and between genomes varied, and this may have influenced our results. The results presented here may be compared to similar published reviews, notably those by Blaby-Haas and Merchant (2013), Groussman et al. (2015), and Behnke and LaRoche (2020). Differences in the numbers of hits retrieved between analyses are likely due to differences in chosen methodologies. FET, ferroxidase; FTR, iron (III) permease; FRE, ferric reductase; FBP, ferrichrome-binding protein; ISIP, iron starvation-induced protein; FTN, ferritin; NRAMP, natural resistance-associated macrophage protein; ZIP, zinc transporter; PETF, ferredoxin; FLDA, flavodoxin; PETJ, cytochrome c6; PETE, plastocyanin.
Experimental evidence suggests that colloidal iron, as particulate Fe2+, is bioavailable to P. tricornutum (Shoenfelt et al., 2017). While aeolian inputs of iron are known to stimulate algal growth, and growth of diatoms in particular, both in short-term ecological experiments (De Baar et al., 2005) and over geological timeframes (Hain et al., 2014; Martínez-García et al., 2014), it has been a long-standing assumption that the iron stimulation is through the increase in Fe′ (Rich and Morel, 1990). Shoenfelt et al. (2017) challenged this assumption by demonstrating that the growth rates of P. tricornutum cultures improved by glacial dust stimulation beyond what would be expected by the increase in Fe′ alone (the assimilation rates did not match Monod dynamics, and a fit to Michaelis–Menten kinetics resulted in unrealistic half-saturation concentrations). In the same experiments, glaciogenic sediments were more bioavailable to P. tricornutum than non-glaciogenic sediments rich in iron, provided cells were in physical contact with the abiotic material. Together, these results implicate an evolved molecular mechanism for the ‘mining’ of mineral iron by some diatoms, which remains to be uncovered. A similar property was observed in the filamentous cyanobacterium Trichodesmium, although there too the molecular underpinnings are currently unresolved (Rubin et al., 2011).
There is significant evidence to show that diatoms are able to access the organic pool of ligand-bound iron. Pioneering work by Soria-Dengg and Horstmann (1995) showed that P. tricornutum cells were able to access iron from the trihydroxamate siderophores ferrioxamine E (FOE) and ferrioxamine B (FOB), albeit with different kinetics. Working on the same model species, Kazamia et al. (2018) demonstrated that this uptake involved endocytosis of the siderophore complex into the cell, with a reduction step to disassociate the bound iron in the vicinity of the chloroplast. Through reverse genetics, the iron starvation-induced protein 1 (ISIP1) was identified as being necessary for the process of endocytosis and siderophore assimilation. Intriguingly, ISIP1 was shown to be largely a diatom-specific protein, implying an evolutionary innovation in this group (Kazamia et al., 2018). The presence of ISIP1 in diatom genomes and transcriptomes is shown in Fig. 3, although it is important to note that since this gene is highly sensitive to iron status in the cell, it may have been missed from transcriptomes collected from diatoms grown in iron-replete conditions.
Additionally, Coale et al. (2019) verified that a previously identified putative siderophore-binding protein, FBP1, which in bacteria binds the hydroxamate siderophore ferrichrome, was indeed required for siderophore assimilation. In our in silico searches, 21% of diatom genomes and transcriptomes displayed evidence of a FBP1 gene (Fig. 3). In physiological experiments on three diatom species reported in Kazamia et al. (2018), it was shown that siderophore recognition varied: P. tricornutum was unable to use the catecholate siderophore enterobactin as a source of iron, whereas T. oceanica assimilated this molecule, but did not respond to ferrioxamine supplementation despite having copies of FBP1, FRE2, and ISIP1 in its genome; T. pseudonana, which lacks FBP1 and ISIP1 genes, was unable to use either siderophore as a source of iron. These observations suggest that while the presence of FBP1 could be a marker for siderophore uptake in diatoms, further experiments are required to understand what determines substrate specificity in these species.
Laboratory studies on model diatom species, including P. tricornutum and T. oceanica, report that diatom cells exhibit ferric reductase activity (Maldonado and Price, 2001; Kazamia et al., 2018; Coale et al., 2019). Six genes in P. tricornutum have been putatively annotated as ferric reductases, but only two (FRE1 and FRE2) encode two domains indicative of ferric reductase function: the ferric reductase transmembrane component PF01794 and the NAD-binding domain PF08030 (Zhang et al., 2013). The in silico prediction for FRE2 is that it is targeted to the chloroplast, albeit with low confidence, whereas the localization of FRE1 is unassigned (Rastogi et al., 2018). We built a HMM for putative diatom ferric reductases based on reference genes from P. tricornutum, and searched the available diatom genomes and transcriptomes. Our results show that all species appear to harbour genes encoding ferric reductases, with around two per species on average (Fig. 3). Fragilariopsis kerguelensis and F. cylindrus, abundant diatoms in the Southern Ocean, stand out because their genomes encode eight and four distinct peptides annotated as ferric reductases, respectively. The coastal species Ditylum brightwellii has eight FRE genes.
The question arises whether these ferric reductases act on the surface of diatom cells, thus enabling the dissociation of iron from its seawater chelates before assimilation, or act intracellularly. On the one hand, extracellular reduction is unlikely in the highly oxidizing and diffusing environment of seawater. On the other hand, if it were to be coupled to efficient (i.e. rapid and proximal) iron-uptake systems, extracellular reduction could allow cells to dispense with the need to discriminate between iron sources (Shaked et al., 2005). The latter is a system well described in fungi, where ferric iron is displaced from weak ferric ligands by a ferrireductase, then re-oxidized by a multicopper oxidase and finally channelled across the plasma membrane through a Fe3+ permease (reviewed by Philpott, 2006). Transcriptional up-regulation of a ferrireductase and two iron permeases was recorded under limiting iron conditions in T. pseudonana cells (Kustka et al., 2007). Furthermore, bioinformatic analysis of transcriptome datasets confirm the existence of functional analogues of the yeast-like iron acquisition machinery [the ‘ferrireductase (FRE), multicopper oxidase (FET), iron permease (FTR)’ system] in the model green alga C. reinhardtii and across a range of diatom species, including all members of the Pseudo-nitzschia and Fragilariopsis genera (Fig. 3; also reviewed in Groussman et al., 2015). However, the question of whether this reductive system acts in consort on the surfaces of diatom cells requires experimental verification. Therefore, in Fig. 3 we have indicated the FRE genes as acting in both ‘reductive’ and ‘non-reductive’ uptake systems.
Low iron quota diatoms regulate the amount of iron required for photosynthesis
The metabolic iron requirement (or metabolic iron quota) of diatom cells differs from species to species. This is measured as the Fe:C ratio (μmol Fe mol C–1) and is estimated from co-measurements of iron‐limited growth rates and cellular iron content (Sunda and Huntsman, 1995). For example, in Pseudo-nitzschia diatoms grown under low-iron conditions, the Fe:C ratio ranged from 2.8 to 3.7 μmol Fe mol C–1 (Marchetti et al., 2006). Similar metabolic iron quotas were estimated for T. oceanica (at ~2 μmol Fe mol C–1), whereas other members of the Thalassiosira genus, such as T. pseudonana and T. weissflogii, had 10-fold higher requirements (Cohen et al., 2018b). In the same study, the metabolic Fe:C ratio for Corethron hystrix was estimated at ~40 μmol Fe mol C–1 (Cohen et al., 2018b). Diatoms with low metabolic iron quotas are remarkable for their ability to maintain uncompromised photosynthetic function, which is attributed to two main adaptations: (i) preferred use or complete replacement of iron-containing proteins with equivalents that are not dependent on iron, and (ii) re-arrangements of the photosynthetic architecture.
Diatoms replace iron-requiring proteins with iron-free functional equivalents
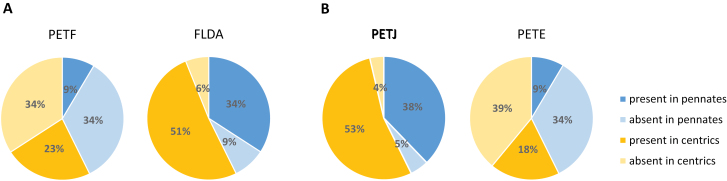
Quantitatively, iron is the most important trace metal in the photosynthetic apparatus because it is involved in the Photosystem II complex (PSII) (which requires two iron atoms per subunit), the cytochrome b6f complex (five iron atoms per monomer), cytochrome c6 (one iron atom per monomer), the Photosystem I complex (PSI) (12 iron atoms), and ferredoxin (two iron atoms) (Raven et al., 1999; Strzepek et al., 2012). Ferredoxin (PETF) is an iron–sulfur protein that is a key component of the chloroplast electron transport chain. Replacement of ferredoxin by its iso-functional carrier, the flavin-containing protein flavodoxin (FLDA), allows flavin rather than iron to be used for electron transport. The two proteins appear to have similar electrostatic potential profiles, although flavodoxin undergoes two successive single electron reductions, with only the second step matching ferredoxin potential (Sétif, 2001). Among the diatoms with sequence information available, 20 out of 82 species harbour both PETF and FLDA genes (Fig. 3). There appears to be no phylogenetic relationship between species that encode PETF, implying that it was present in the diatom ancestor and that its absence from some species is likely due to gene loss (Figs 3, 4A). The ability to synthesize flavodoxin is widespread among diatoms; we found FLDA in 70 out of 82 species (Fig. 3). Since no photosynthetic species are known to survive without either PETF or FLDA, in transcriptomes where neither transcript was detected, such as that of Pseudo-nitzschia pungens, we assume that this was due either to incomplete sequencing or to the stringency of the cut-off parameters chosen for detection in our analysis.
Fig. 4.
Genes encoding iron-switching proteins in diatom transcriptomes. Both ferredoxin (PETF) and cytochrome c6 (PETJ) are iron-containing enzymes. As some diatoms are capable of replacing them with the iron-free equivalents flavodoxin (FLDA) and plastocyanin (PETE), respectively, the relative proportions of transcripts encoding these proteins may indicate the dependence of diatoms on these proteins. The pie charts indicate the presence and absence of PETF versus FLDA (A), and PETJ versus PETE (B) in pennate and centric diatoms in our database of 82 species. It is important to note that our comparisons here rely on the numbers of genes detected in the transcriptomes of species, and therefore may not be a true reflection of species’ genomes. It is possible that genomes contain genes that were not detected in the transcriptomes, either because the transcriptomes were generated from iron-replete cultures or because species were not sequenced to sufficient depth.
For species that encode both FLDA and PETF, the ratio of PETF:FLDA in the cell is controlled by the availability or scarcity of iron. For example, cultures of F. cylindrus grown at high iron concentrations produced predominantly ferredoxin, with a small amount of flavodoxin. Ferredoxin was sequentially replaced by flavodoxin in cultures grown with less iron (Pankowski and McMinn, 2009). Similarly, in P. tricornutum, T. oceanica, and Pseudo-nitzschia granii, expression of the FLDA gene and the abundance of FLDA protein was much higher under iron limitation (La Roche et al., 1995; Allen et al., 2008; Lommer et al., 2012; Cohen et al., 2018a; Zhao et al., 2018). Under iron-replete conditions, PETF completely replaces FLDA, as observed in natural diatom communities (La Roche et al., 1995; Erdner and Anderson, 1999; McKay et al., 1999; Allen et al., 2008). However, some diatoms contain multiple copies of genes encoding flavodoxin, and only certain isoforms are differentially expressed in relation to iron status. Both F. cylindrus and T. oceanica have two isoforms of FLDA, but transcription of only one isoform is regulated by iron levels (Pankowski and McMinn, 2009; Whitney et al., 2011). Whether possessing a number of flavodoxin isoforms confers an ecological advantage in environments chronically deprived of iron is an interesting question for further investigation.
The strategy of differential transcription of selected genes does not appear to be used by all diatoms. Individual diatom species may be permanently adapted to specific iron regimes in the ocean rather than maintaining transcriptional flexibility. This was supported by analysis of the Tara Oceans global dataset for transcriptional regulation of ferredoxin and flavodoxin across a range of algal groups. The dataset combined comprehensive bio-oceanographic genomic and transcriptomic data with iron distributions derived from two global-scale biogeochemical models (Bork et al., 2015; Carradec et al., 2018). Unlike chlorophytes, haptophytes, and pelagophytes, diatoms showed constitutively higher expression of flavodoxin genes than ferredoxin genes, although there was more heterogeneity in expression between species than across these four major groups (Carradec et al., 2018). In P. granii, the transcript abundance of PETF was appreciably lower than that of FLDA regardless of iron status (Cohen et al., 2018a). The diatom Fragilariopsis curta appears to have lost PETF entirely, and transcription of this species’ remaining FLDA gene is not sensitive to iron concentrations (Pankowski and McMinn, 2009). Interestingly, PETF is localized in the chloroplast genome in T. pseudonana and other diatoms, but has been transferred to the nuclear genome in T. oceanica. Compared with its coastal relative T. pseudonana, the oceanic diatom T. oceanica is highly tolerant to iron limitation. It has been proposed that the transfer of PETF from the chloroplast to the nuclear genome might have contributed to the ecological success of T. oceanica (Lommer et al., 2010). Whether or not the gene transfer described for T. oceanica confers a competitive advantage still needs to be assessed through experimental approaches. That PETF is often chloroplast-encoded suggests that analyses of transcriptomes such as the one presented in Fig. 3 may be methodologically biased towards a lack of detection, as RNA harvesting and processing select for nuclear-encoded transcripts.
Cytochrome c6 acts as an electron carrier between the cytochrome b6f complex and PSI. Cytochrome c6 (encoded by PETJ) may be replaced with the copper-coordinating protein plastocyanin (encoded by PETE). The replacement of cytochrome c6 with plastocyanin is rarer than the replacement of ferredoxin by flavodoxin, and is presumed to have occurred via horizontal gene transfer (Strzepek and Harrison, 2004; Peers and Price, 2006). Only fifteen centric species and eight pennate species show evidence for the presence of a PETE gene (Figs 3, 4B). For species that encode both PETE and PETJ, differences in regulation have been noted. P. tricornutum and F. cylindrus were found to induce plastocyanin under iron limitation, to temporarily replace cytochrome c6, and to highly express the gene encoding cytochrome c6 under iron-replete conditions (La Roche et al., 1996; Cohen et al., 2018a). F. kerguelensis is an interesting candidate for investigation because it contains multiple isoforms of plastocyanin (five genes detected using our methods; Fig. 3). In a recent study, Moreno et al. (2020) investigated the response of three isoforms of PETE to iron in this species, and found that only two were significantly over-represented under low-iron conditions. In T. oceanica, which encodes PETE together with two functional copies of PETJ, the relative expression of plastocyanin was sensitive to iron status, while two genes encoding cytochrome c6 were weakly but constitutively expressed (Lommer et al., 2012). Similarly, in P. granii, transcripts for PETJ were weakly abundant regardless of iron status, while PETE was highly abundant under iron-replete conditions (Cohen et al., 2018a).
Only a few studies have addressed the question of what the consequences of using plastocyanin are for the copper requirements of cells. For example, the green alga C. reinhardtii switches to using cytochrome c6 instead of plastocyanin only under copper-limiting conditions (Merchant et al., 1991). In T. oceanica, copper deficiency inhibited electron transport regardless of iron status, implying that plastocyanin expression was not controlled by iron concentrations (Peers and Price, 2006).
Low iron quota diatoms lower the ratio of PSI to PSII without compromising photosynthetic output
The PSI complex has the highest iron demand of the light-dependent reactions of photosynthesis. The optimized ratio of PSI: PSII in land plants is approximately 1:1. Comparing the coastal diatom T. weissflogii with the open ocean diatom T. oceanica, Strzepek and Harrison (2004) found that this ratio was significantly lower in the latter species. This led to the hypothesis that diatoms, in particular those adapted to chronically iron-starved conditions, have streamlined their photosynthetic architecture, minimizing the iron quota necessary for growth. T. weissflogii cells contained twice as much PSII than PSI, and T. oceanica had reduced PSI demands even further, with a ratio of PSI:PSII of 1:10 (Strzepek and Harrison, 2004). In iron-replete media, the two diatoms grew at comparable rates. However, T. oceanica maintained high growth rates (~80% of the rate in iron-replete conditions) in low-iron media that restricted the growth of T. weissflogii to ~20% of its iron-replete growth rate. Two studies confirmed similar streamlining in F. kerguelensis and P. granii, polar diatoms that are abundant in the Southern Ocean and the Arctic Ocean, respectively (Cohen et al., 2018a; Moreno et al., 2020). In P. granii, the gene encoding PSI subunit IV (PSAE) was almost 4-fold more highly expressed under iron-replete conditions, and protein levels were more abundant by 35-fold compared with their levels under conditions of iron limitation (Cohen et al., 2018a).
Low iron quota diatoms such as T. oceanica and P. granii are not unique among algae and cyanobacteria in having undergone substitutions and rearrangements in their photosynthetic architecture in order to conserve iron. These traits and plasticity in iron use are widespread (Blaby-Haas and Merchant, 2013; Raven, 2013; Scheiber et al., 2019). However, in an experimental comparison of 29 species of eukaryotic algae and cyanobacteria, Quigg et al. (2003, 2011) demonstrated that algae that contain plastids of a secondary endosymbiotic origin, the ‘red plastid’ lineage, which includes the dinoflagellates, haptophytes, and chrysophytes as well as diatoms, had lower stoichiometric quantities of iron per cell compared with the ‘green lineage’ of algae and cyanobacteria. The results from these analyses are difficult to square with measurements of metabolic iron quotas, since the former are carried out under iron-replete conditions. Nevertheless, they suggest that members of the red plastid lineage are capitalizing on more than rearrangements of their photosynthetic architecture for efficient iron use. Studies that look into the adaptations of cell structure, such as the metabolic coupling of plastids and mitochondria (e.g. see some pioneering work by Bailleul et al., 2015), may shed further light on this question.
Diatoms are able to store iron and regulate its intracellular concentration efficiently
It has been observed that in situ iron fertilization, whether artificial or natural, results in blooms dominated by large diatoms, which are often rare in the standing microalgal community (De Baar et al., 2005). These include the chain-forming members of the genera Fragilariopsis, Pseudo-nitzschia, and Chaetoceros, in particular. Comparing their transcriptomes for the analysed genes (Fig. 3), there is no pattern that distinguishes them readily from other diatoms, as traits appear to be species-specific. However, it is important to note that these transcriptomes capture the behaviour of species under laboratory conditions and may not be indicative of their behaviour during a diatom bloom in the wild. The F. kerguelensis transcriptome and the F. cylindrus genome encode a more comprehensive portfolio of iron-sensitive genes, with multiple isoforms of each gene compared with other species. This is intriguing because, as model polar species, Fragilariopsis representatives are considered to be particularly adapted to fluctuating environmental conditions and life in sea ice, which is not ‘iron-limited’ (Mock et al., 2017). Our data suggest that Fragilariopsis species are interesting candidates for iron adaptation studies as well.
It is likely that species that respond to iron stimulation by forming blooms are efficient not only at iron uptake, but also in iron homeostasis and long-term storage. One way to assess the capacity for long-term iron storage is to measure the number of divisions that a cell is capable of when ambient iron concentrations drop back to growth-limiting conditions. This can be distinguished from short-term iron homeostasis, which is associated with mechanisms that act on a diurnal scale, where the requirement for iron depends on photosynthetic activity. An alternative is to measure the cell’s metabolic iron quota and subtract this from the total cellular iron carried by a cell under iron-replete conditions. This has been termed the ‘luxury iron’ quota (Sunda and Huntsman, 1997; Marchetti et al., 2009). For example, within the centric Thalassiosira genus, the cellular Fe:C ratio of coastal species reached values that were 20–30 times higher than those needed for maximum growth; by contrast, ‘plateau’ iron concentrations in oceanic species were only two to three times higher than maximum needed amounts (Sunda and Huntsman, 1995), indicating that coastal variants had greater capacity to accumulate iron when iron replete.
Two mechanisms have been proposed for the storage of iron inside diatom cells: sequestration into the mineral core of ferritin (FTN) proteins and/or vacuolar storage. FTNs are found in all domains of life, including animals, plants, and microorganisms, and are designed to accommodate large amounts of iron (Theil, 1987). Structurally, FTNs are large protein cages formed by arrays of self-assembling helices with nanocavities (5–8 nm) that catalytically couple iron and oxygen at protein sites for precursors of the cavity mineral. The mineral core of a single protein can store up to 4500 iron atoms (Liu and Theil, 2005). Iron is reversibly released from the core by reduction.
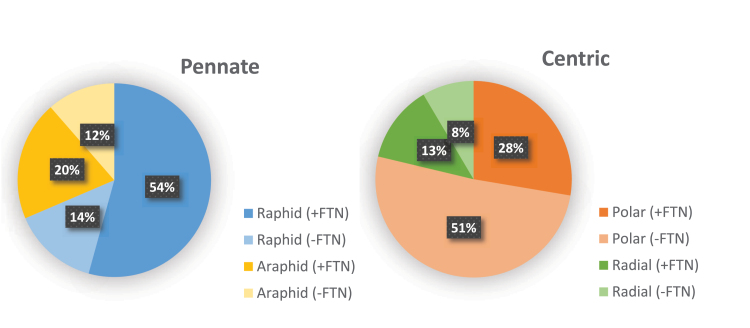
We retrieved putative transcripts for FTN in diatom transcriptomes and genomes by conducting a HMMER search using the methods described in Fig. 3. We found that ~74% of pennate diatoms contain at least one FTN homolog compared with 41% of centric diatoms. Among the centric diatoms the trend for lacking FTN skewed towards the polar centric species, of which approximately two thirds were missing the annotation (Fig. 5). The apparent loss of FTN in the polar centric diatoms is intriguing and requires further investigation. Looking at the transcriptomes of the three genera of frequently blooming diatoms, Fragilariopsis and Pseudo-nitzschia species appear to be rich in putative FTN genes (Fig. 3). In contrast, our search for FTN retrieved hits for only a quarter of the sampled Chaetoceros strains (two out of eight). There were notable absences among Thalassiosira species (among which five species out of nine do not have a FTN gene), including T. oceanica and T. pseudonana.
Fig. 5.
Genes encoding ferritins (FTNs) in diatom transcriptomes. Homologs of FTN were retrieved using a HMMER search within available diatom genomes and transcriptomes (see the caption of Fig. 3 for details). Pie charts summarize the presence and absence of FTN in raphid versus araphid pennate diatoms, and in polar versus radial centric species. It is important to note that the presence or absence of genes in a transcriptome may not be a true reflection of copy numbers in a genome, either because it was generated from iron-replete cultures or because it was not sequenced to sufficient depth.
In laboratory studies, it was shown that P. granii, which harbours a gene encoding FTN, was able to undergo several more cell divisions in the absence of iron than the comparably sized oceanic centric diatom T. oceanica, which lacks the FTN gene, supporting the hypothesis that FTN functions in the long-term storage of iron (Marchetti et al., 2009). A corollary of the hypothesis is that FTN transcript abundance should increase with iron concentration. However, this has not been universally upheld in laboratory studies. While P. granii and Thalassiosira sp. NH16 have been observed to increase FTN gene expression under high iron concentrations, Amphora coffeaeformis exhibited minimal iron-storage capacities and contained two distinct FTN genes, one of which increased in expression under iron limitation whereas the second showed no variation with cellular iron status (Cohen et al., 2018b). In fact, we detected three distinct FTN transcripts in this species using our methods (Fig. 3). Furthermore, a recent study investigated the community-level response of open ocean plankton ecosystems to iron availability and found that, with the exception of Pseudo-nitzschia species, no clear pattern in FTN gene abundance or expression and estimated iron levels could be observed, suggesting that iron storage may not be the main function of FTN in most eukaryotic marine phytoplankton (Caputi et al., 2019). Together, these results suggest that FTN in diatoms may have evolved to serve multiple functional roles.
A proposed alternative role for FTN in diatoms is as an iron oxidation enzyme rather than as a long-term iron-storage protein. Working with P. multiseries, Pfaffen et al. (2013) showed that FTN oxidizes Fe2+ at its ferroxidase centres rapidly but forms iron mineral only slowly. In a subsequent study, functional mutagenesis experiments showed that the protein is biochemically optimized for initial Fe2+ oxidation but not for mineralization. The authors argued that its primary function therefore might not be in long-term iron storage, but rather in iron homeostasis (Pfaffen et al., 2015). This is the situation in higher plants, where experiments on Arabidopsis thaliana demonstrated that FTN is regulated by the circadian clock cycle (Duc et al., 2009) and functions to buffer the iron released by degradation from PSI, a protective role in response to photo-oxidative stress (Rossel et al., 2002; Murgia et al., 2007). Similarly, in C. reinhardtii, FTN was shown to be required during high ambient iron availability or cellular degradation of iron-containing proteins and protein complexes such as ferredoxin during cellular acclimation to low-iron conditions, indicative of a function in iron homeostasis (La Fontaine et al., 2002; Busch et al., 2008; Long and Merchant, 2008). Finally, in the green picoalga Ostreococcus tauri, mutants lacking FTN were less tolerant of low-iron conditions, which induced greater recycling of iron within the cell than in the wild type, further underscoring the importance of this protein in homeostasis. In the same work, the authors proposed that nitrate reductase functioned as an iron-storage protein in this picoalga (Botebol et al., 2015).
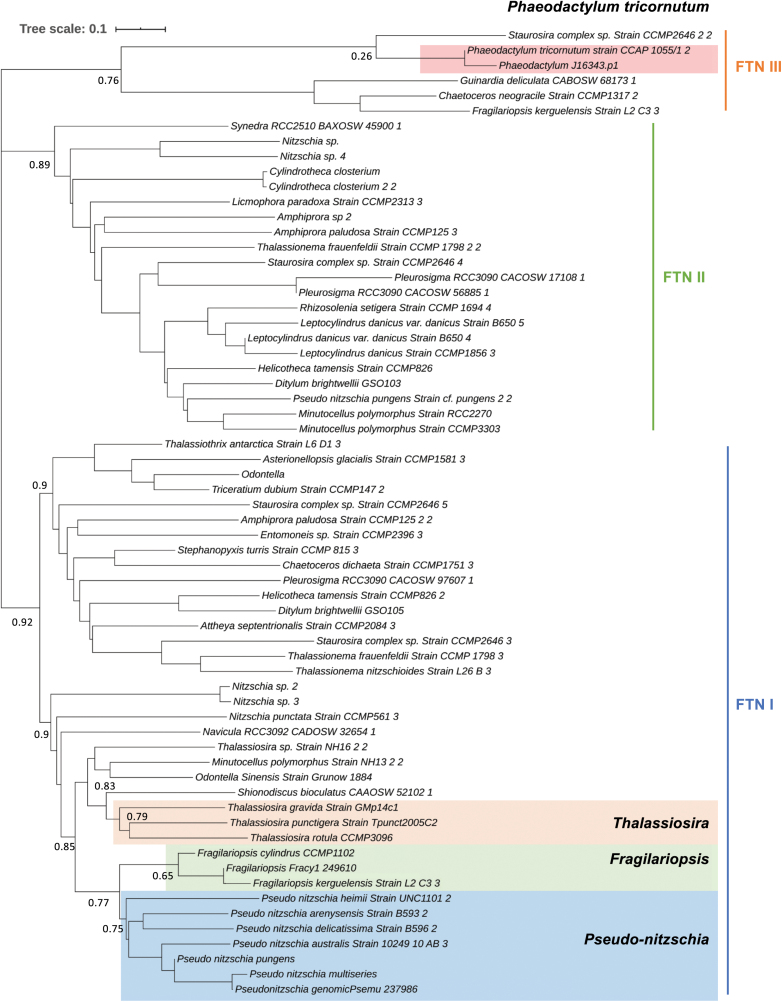
A closer look at diatom FTN phylogeny, which we updated using available diatom transcriptomes (Fig. 6), reveals that the FTNs within the diatoms resolve into several clades. Our phylogeny matches that previously reported by others (e.g. Marchetti et al., 2009; Groussman et al., 2015), but adds more species to the tree. There are two main FTN clades, one of which contains most members of the Thalassiosira, Pseudo-nitzschia, and Fragilariopsis genera, although phylogenetic relationships are not robustly upheld. The second clade contains members of 14 genera, including Cylindrotheca and Leptocylindrus. The species Helicotheca tamesis CCMP826, Amphiprora paludosa CCMP125, Thalassionema fraundfeldii CCMP1798, Staurosira complex sp. CCMP2646, and Pleurosigma sp. RCC3090 have annotated FTNs belonging to both clades. To our knowledge, there are no described functional differences between FTNs that belong to the two main groups. Additionally, we found a third group of putative FTNs (which we have annotated as FTN clade III in Fig. 6) with weaker bootstrap support, which includes genes from P. tricornutum, Staurosira complex sp., Guinardia deliculata, Chaetoceros neogracile, and F. kerguelensis.
Fig. 6.
Phylogenetic tree of ferritins (FTNs) in diatoms. The phylogenetic tree was constructed to clarify the evolutionary distance of FTN between species. A total of 244 sequences were first retrieved by a HMMER search using the FTN PFAM domain PF00210 as a query in the available 82 diatom transcriptomes/genomes with an E-value cut-off of 1×10–10. Retrieved sequences were analysed using the CD-HIT web server and sequences that met a similarity threshold >0.9 were presumed to be duplicates and eliminated. We generated a Hidden Markov Model for FTN using P. tricornutum and closely aligned sequences retrieved by BLASTp, and this was used as the basis for a second HMMER search of the remaining sequences, with an E-value cut-off of 1×10–10, to further reduce the redundancy. This left a total of 64 representative sequences, which captured the diversity of FTNs within the diatoms. Conserved sequences were aligned using the alignment builder in Geneious v.10.2 under default criteria. The tree was drawn with ITOL (https://itol.embl.de/). Numbers beside branches indicate RaxML bootstrap coefficients.
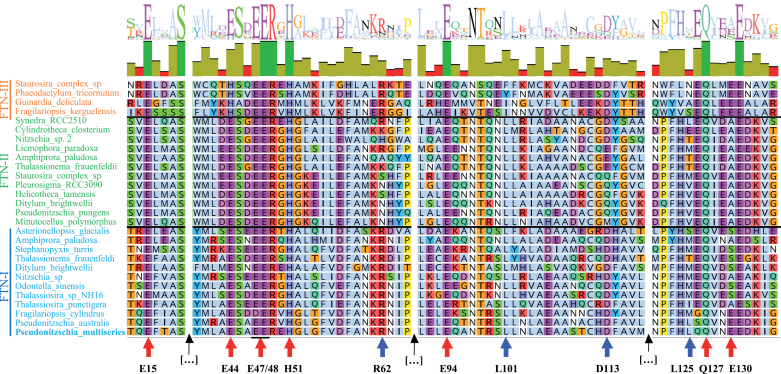
We compared the protein sequence of representative FTNs from the three groups by alignment (Fig. 7). We focused on a comparison of iron-binding sites identified by Pfaffen et al. (2013) working on P. multiseries and key residues identified by Jin et al. (2001) working on FTN isolated from the frog Rana catesbeiana. It would appear that essential residues involved in iron binding are more strictly conserved in FTN I and II (Fig. 7, red arrows), whereas residues involved in iron release are preserved more across FTN I and III. However, experimental verification is required to validate any implied functional differences.
Fig. 7.
Protein sequence alignment of representative diatom ferritins (FTNs). FTN protein sequence alignment (one-letter code) was carried out using MUSCLE in Geneious v.10.2. At the top is the sequence logo and identity. At the bottom, red arrows highlight the ferroxidase residue sites first identified in Pseudo-nitzschia multiseries (Pfaffen et al., 2013). The glutamic acid residues (E15, E47/48, and E94), one glutamine residue (Q127), and the histidine residue H51 are conserved across the three different clades (red arrows). However, the glutamic acid residue E44, which is conserved in FTN I/II, is replaced with histidine in FTN III. These residues are involved in iron binding. Blue arrows show conserved residue pairs essential for iron release in the frog species Rana catesbeiana (Jin et al., 2001). The amino acid pairs R62/D113 and L101/L125 are adjacent in three-dimensional space. R62/D113 is conserved in both FTN I and FTN III but not FTN II (which shows significant variation in these positions). Leucine is conserved in position 101 in FTN I and II but not FTN III, while leucine at position 125 is not conserved in any diatoms apart from two Pseudo-nitzschia species and Synedra sp. RCC2510. The most common replacement for leucine in this position is methionine, as observed in F. cylindrus and Thalassiosira sp. NH16. Black arrows indicate breaks in the sequence.
There is evidence from on-board experiments (which use environmental samples) to suggest that Pseudo-nitzschia ferritins serve a long-term storage role in cells, which is not the case in Thalassiosira and Corethron, in which a role in iron homeostasis is preferred as an explanation (Lampe et al., 2018). Broadly, this is also supported by Tara Oceans transcriptomic analyses, which detected up-regulated FTN expression in Pseudo-nitzschia cells in locations that are rich in iron (Caputi et al., 2019). One structural change that may explain this is the presence of glutamic acid at position 130 in P. multiseries FTN, which is absent from other FTN clade I and II species (Pfaffen et al., 2015). It should be noted that E130 is conserved in FTN III diatoms (Fig. 7).
It is likely that a range of proteins that have not yet been explored are involved in iron storage and homeostasis in diatom cells. One notable candidate is the iron starvation-induced protein ISIP3, the most abundant of the three ISIPs in global datasets (Carradec et al., 2018). ISIP3 is typified by a conserved domain of unknown function, DUF305, which belongs to the FTN superfamily (Behnke and LaRoche, 2020). Using our methods, we found transcripts for ISIP3 across most diatoms, with some notable absences in the coastal Skeletonema and Chaetocerus genera and an overabundance of putative hits in Fragilariopsis (Fig. 3).
An alternative mechanism for long-term iron storage that has been proposed is vacuolar. Iron loading into and release out of the vacuole was first identified in yeast, and was shown to be mediated by natural resistance-associated macrophage proteins (NRAMPs) in A. thaliana (Curie et al., 2000; Lanquar et al., 2005). Furthermore, some NRAMP proteins were shown to be functional equivalents of yeast FET proteins, and enabled the transport of iron into the cell (Curie et al., 2000). In T. pseudonana, a vacuolar storage mechanism was proposed when it was observed that NRAMP was dramatically regulated by intracellular iron concentrations (Kustka et al., 2007). However, it is important to note that vacuolar localization of NRAMP was proposed but not experimentally validated. In experiments using synchrotron X‐ray fluorescence element mapping, intracellular pockets of iron, indicative of storage features, were identified in both T. pseudonana and T. weissflogii (Nuester et al., 2012).
A recent study investigated the responses of natural communities, sampled across a gradient of iron concentrations, to on-deck iron stimulation. Metatranscriptome data from incubations was used to assign relative expression of FTN and NRAMP genes on to three main diatom genera, Chaetoceros, Pseudo-nitzschia, and Thalassiosira (Lampe et al., 2018). The study found that Pseudo-nitzschia diatoms were unique utilizers of FTN (up-regulating its expression when iron was supplied), whereas the response from Thalassiosira species suggested vacuolar storage. In Chaetoceros, transcripts of neither FTN nor NRAMP were abundant, although iron quotas in Chaetoceros were often similar to those in Pseudo-nitzschia (Lampe et al., 2018). The authors proposed that Chaetoceros may have an alternative divalent metal transporter, a protein belonging to the ZIP family, that takes on the function of vacuolar iron transport in these species (Allen et al., 2008). ZIP proteins have been shown to facilitate passive metal transport, including that of ferrous iron, in a range of species (Eide, 2005).
We compared the portfolios of ZIP, NRAMP, and FTN genes across diatoms using all published diatom transcriptomes and genomes, by constructing HMMs for representative genes and scanning the dataset using HMMER (summarized in Fig. 3). This is an amalgamated view, since we did not refine our search for subclasses of NRAMP or ZIP proteins (i.e. the specificity of these genes for iron transport is not confirmed). We found that all diatoms have genes belonging to the ZIP family, and that the majority also have NRAMP genes (modelled on F. cylindrus NRAMP protein). Notable exceptions are most members of the Chaetoceros genus, which lack a NRAMP annotation. NRAMP genes appear to be less abundant among centric species than pennate species, but no robust phylogenetic relationship was observed.
Synthesis and future directions
There are multiple strands of evidence suggesting that diatoms as a group are successful competitors for iron. Significant research attention has focused on iron uptake and storage, and it is clear that diatoms use a portfolio of strategies to access various forms of iron in the ocean. Remarkably, they have adapted what appear to be ancestrally bacterial uptake proteins to eukaryotic mechanisms. The genes FBP1 and ISIP2A have bacterial origins yet rely on endocytosis to function. Diatom-specific genes have also been observed (e.g. ISIP1), implying further innovation in this group to facilitate access to iron. However, experimental verification of the molecular basis of proposed traits remains a major challenge. For example, validation of the in silico prediction that FTN is targeted to the chloroplast in diatoms, where iron demands are highest, should be an experimental priority. Additionally, multiple studies have indicated that ZIP and NRAMP proteins, as well as ISIP3, are important for iron physiology, making them clear targets for forward and reverse genetics.
One important avenue for future work is gauging which of these mechanisms is most strongly related to the success of diatoms in the wild. There is evidence that the replacement of ferredoxin with flavodoxin in the photosynthetic machinery of diatom cells is one of the more important adaptations. A recent global survey of gene expression in marine phytoplankton showed that oceanic members of the diatom lineage strongly express flavodoxin over ferredoxin, whereas certain coastal diatoms, which are likely adapted to environments experiencing more frequent and larger fluctuations in iron supply, expressed ferredoxin more highly under iron-replete conditions (Caputi et al., 2019). Of the seven most abundant genera in the ocean (indicated by asterisks in Fig. 3), four contain genes encoding flavodoxin.
Similarly, the expression of ISIPs showed the highest correlation in communities that were sampled from the lowest-iron environments (Kazamia et al., 2018; Caputi et al., 2019). ISIP1 is notable for being the most sensitive to low-iron status, and likely represents a diatom-specific innovation, although its function requires further elucidation. Its role in mediating siderophore uptake is intriguing, because it suggests a community link between diatoms and siderophore producers. FBP1 is the only siderophore-binding protein that has been described to date, and its distribution appears to correlate with that of ISIP1 in diatom transcriptomes, since most species that encode FBP1 also encode ISIP1, with the exception of Thalassiosira gravida and Chaetoceros curvisetus (Fig. 3). However, many more species encode ISIP1 than FBP1, so it is possible that ISIP1 has an additional role in cell iron homeostasis or that there are multiple siderophore-binding proteins interacting with ISIP1.
As we garner information on how diatoms put their metabolic portfolios to use in the wild, we will shed a refining light on diatom niches. This will be an important breakthrough in moving away from studies that either focus on individual model species or attempt to draw distinctions between ‘pennates’ and ‘centrics’, or ‘open ocean’ and ‘coastal’ diatoms, while comparing only a few representatives. For the iron uptake, homeostasis, or storage mechanisms that we have reviewed here, there do not appear to be any remarkable patterns that distinguish pennates from centrics. To meaningfully compare open ocean and coastal diatoms, we require more information on the distribution of individual species in the global ocean. The Tara Oceans global circumnavigation effort, which catalogued the community composition, metagenomes, and metatranscriptomes of aquatic microorganisms, can offer initial insight. For example, T. oceanica, the species most often referred to in the literature as an open ocean species, in fact has a ubiquitous distribution and is frequently found in coastal areas (based on 18S data from the Tara Oceans gene catalogue; data not shown). Of the 82 species reviewed here, only H. tamesis was not detected within 250 km from the coast during Tara Oceans sampling (J. Pierella Karlusich, Institut de Biologie de l’École Normale Supérieure, personal communication), and this species may therefore be considered a true open ocean representative. By contrast, there is a considerable number of coastal species represented in the MMETSP sequence set. The species that were not detected further offshore than 250 km are Proboscia inermis (found exclusively in coastal zones at high latitudes), D. brightwellii, Skeletonema marinoi, Ticeratium dubium, Odontella aurita, Navicula sp., P. tricornutum, A. coffeaeformis, and Cylindrotheca closterium.
Using Tara Oceans data, Caputi et al. (2019) found that diatom species thrived across a gradient of total iron concentrations and showed remarkable plasticity in their responses to iron availability. The authors concluded that it was not possible to correlate species assemblages to iron levels or transcriptional responses in iron-uptake systems. We believe that further progress will be made when in situ studies of species-specific diatom gene expression and analysis of community structure will be coupled to careful characterization of iron sources available in seawater, since it is possible that it is the chemical nature of iron sources that complicates diatom niche separation.
Acknowledgements
CB acknowledges funding from the European Research Council Advanced Awards Diatomite and Diatomic under the European Union’s Horizon 2020 research and innovation programme (grant agreement nos. 294823 and 835067), the ANR project PhytoMet (grant ANR-16-CE01-0008-02), the HFSP project Green Life in the Dark (grant RGP0003/2016), and the French Government ‘Investissements d’Avenir’ Programmes MEMO LIFE (grant ANR-10-LABX-54), Université de Recherche Paris Sciences et Lettres (Université PSL) (grant ANR-1253 11-IDEX-0001-02), and OCEANOMICS (grant ANR-11-BTBR-0008). XG acknowledges funding from the Chinese Scholarship Council (CSC-201706910081). We thank Dr Juan Pierella Karlusich and Dr Richard G. Dorrell for their advice on the bioinformatic analysis.
Author contributions
XG and EK wrote the original draft of the manuscript. EK and CB were responsible for reviewing and editing the writing. XG curated and analysed the data and was responsible for the visualization of the work. EK and CB conceptualized and supervised the project. CB and XG secured the funding support.
Conflict of interest
The authors declare no conflict of interest.
References
- Allen MD, del Campo JA, Kropat J, Merchant SS. 2007. FEA1, FEA2, and FRE1, encoding two homologous secreted proteins and a candidate ferrireductase, are expressed coordinately with FOX1 and FTR1 in iron-deficient Chlamydomonas reinhardtii. Eukaryotic Cell 6, 1841–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen AE, LaRoche J, Maheswari U, Lommer M, Schauer N, Lopez PJ, Finazzi G, Fernie AR, Bowler C. 2008. Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation. Proceedings of the National Academy of Sciences, USA 105, 10438–10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alverson AJ, Theriot EC. 2005. Comments on recent progress toward reconstructing the diatom phylogeny. Journal of Nanoscience and Nanotechnology 5, 57–62. [DOI] [PubMed] [Google Scholar]
- Apt KE, Grossman A, Kroth-Pancic P. 1996. Stable nuclear transformation of the diatom Phaeodactylum tricornutum. Molecular Genetics and Genomics 252, 572–579. [DOI] [PubMed] [Google Scholar]
- Armbrust EV, Berges JA, Bowler C, et al. 2004. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86. [DOI] [PubMed] [Google Scholar]
- Bailleul B, Berne N, Murik O, et al. 2015. Energetic coupling between plastids and mitochondria drives CO2 assimilation in diatoms. Nature 524, 366–369. [DOI] [PubMed] [Google Scholar]
- Behnke J, LaRoche J. 2020. Iron uptake proteins in algae and the role of Iron Starvation-Induced Proteins (ISIPs). European Journal of Phycology 55, 339–360. [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. Journal of Molecular Biology 340, 783–795. [DOI] [PubMed] [Google Scholar]
- Benoiston A-S, Ibarbalz FM, Bittner L, Guidi L, Jahn O, Dutkiewicz S, Bowler C. 2017. The evolution of diatoms and their biogeochemical functions. Philosophical Transactions of the Royal Society B: Biological Sciences 372, 20160397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatta H, Kong TK, Rosengarten G. 2009. Diffusion through diatom nanopores. Journal of Nano Research 7, 69–74. [DOI] [PubMed] [Google Scholar]
- Blaby-Haas CE, Merchant SS. 2013. Iron sparing and recycling in a compartmentalized cell. Current Opinion in Microbiology 16, 677–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteau RM, Mende DR, Hawco NJ, McIlvin MR, Fitzsimmons JN, Saito MA, Sedwick PN, DeLong EF, Repeta DJ. 2016. Siderophore-based microbial adaptations to iron scarcity across the eastern Pacific Ocean. Proceedings of the National Academy of Sciences, USA 113, 14237–14242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnain C, Breitbart M, Buck KN. 2016. The Ferrojan Horse hypothesis: iron-virus interactions in the ocean. Frontiers in Marine Science 3, 82. [Google Scholar]
- Bork P, Bowler C, de Vargas C, Gorsky G, Karsenti E, and Wincker P. 2015. Tara Oceans studies plankton at planetary scale. Science 348, 873. [DOI] [PubMed] [Google Scholar]
- Botebol H, Lesuisse E, Šuták R, et al. 2015. Central role for ferritin in the day/night regulation of iron homeostasis in marine phytoplankton. Proceedings of the National Academy of Sciences, USA 112, 14652–14657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Allen AE, Badger JH, et al. 2008. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239. [DOI] [PubMed] [Google Scholar]
- Boyd PW, Ellwood MJ. 2010. The biogeochemical cycle of iron in the ocean. Nature Geoscience 3, 675–682. [Google Scholar]
- Boyd PW, Jickells T, Law C, et al. 2007. Mesoscale iron enrichment experiments 1993–2005: synthesis and future directions. Science 315, 612–617. [DOI] [PubMed] [Google Scholar]
- Boyle E. 1998. Pumping iron makes thinner diatoms. Nature 393, 733–734. [Google Scholar]
- Busch A, Rimbauld B, Naumann B, Rensch S, Hippler M. 2008. Ferritin is required for rapid remodeling of the photosynthetic apparatus and minimizes photo‐oxidative stress in response to iron availability in Chlamydomonas reinhardtii. The Plant Journal 55, 201–211. [DOI] [PubMed] [Google Scholar]
- Caputi L, Carradec Q, Eveillard D, et al. 2019. Community‐level responses to iron availability in open ocean plankton ecosystems. Global Biogeochemical Cycles 33, 391–419. [Google Scholar]
- Carradec Q, Pelletier E, Da Silva C, et al. 2018. A global ocean atlas of eukaryotic genes. Nature Communications 9, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier‐Smith T. 1999. Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. Journal of Eukaryotic Microbiology 46, 347–366. [DOI] [PubMed] [Google Scholar]
- Coale TH, Moosburner M, Horák A, Oborník M, Barbeau KA, Allen AE. 2019. Reduction-dependent siderophore assimilation in a model pennate diatom. Proceedings of the National Academy of Sciences, USA 116, 23609–23617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen NR, Gong W, Moran DM, McIlvin MR, Saito MA, Marchetti A. 2018a. Transcriptomic and proteomic responses of the oceanic diatom Pseudo-nitzschia granii to iron limitation. Environmental Microbiology 20, 3109–3126. [DOI] [PubMed] [Google Scholar]
- Cohen NR, Mann E, Stemple B, Moreno CM, Rauschenberg S, Jacquot JE, Sunda WG, Twining BS, Marchetti A. 2018b. Iron storage capacities and associated ferritin gene expression among marine diatoms. Limnology and Oceanography 63, 1677–1691. [Google Scholar]
- Curie C, Alonso JM, Jean ML, Ecker JR, Briat J-F. 2000. Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochemical Journal 347, 749–755. [PMC free article] [PubMed] [Google Scholar]
- De Baar HJ, Boyd PW, Coale KH, et al. 2005. Synthesis of iron fertilization experiments: from the Iron Age in the Age of Enlightenment. Journal of Geophysical Research: Oceans 110, C09S16. [Google Scholar]
- Dorrell RG, Gile G, Mccallum G, et al. 2017. Chimeric origins of ochrophytes and haptophytes revealed through an ancient plastid proteome. eLife 6, e23717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrell RG, Villain A, Perez-Lamarque B, et al. 2021. Phylogenomic fingerprinting of tempo and functions of horizontal gene transfer within ochrophytes. Proceedings of the National Academy of Sciences 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duc C, Cellier F, Lobréaux S, Briat JF, Gaymard F. 2009. Regulation of iron homeostasis in Arabidopsis thaliana by the clock regulator time for coffee. Journal of Biological Chemistry 284, 36271–36281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide DJ. 2005. The ZIP family of zinc transporters. In Iuchi S, Kuldell N, eds. Zinc finger proteins: from atomic contact to cellular function. Boston: Springer, 261–264. [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nature Protocols 2, 953–971. [DOI] [PubMed] [Google Scholar]
- Erdner DL, Anderson DM. 1999. Ferredoxin and flavodoxin as biochemical indicators of iron limitation during open‐ocean iron enrichment. Limnology and Oceanography 44, 1609–1615. [Google Scholar]
- Falkowski PG. 1997. Evolution of the nitrogen cycle and its influence on the biological sequestration of CO2 in the ocean. Nature 387, 272–275. [Google Scholar]
- Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. 1998. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281, 237–240. [DOI] [PubMed] [Google Scholar]
- Fukasawa Y, Tsuji J, Fu SC, Tomii K, Horton P, Imai K. 2015. MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Molecular & Cellular Proteomics 14, 1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galachyants YP, Zakharova YR, Petrova D, et al. 2015. Sequencing of the complete genome of an araphid pennate diatom Synedra acus subsp. radians from Lake Baikal. Doklady Biochemistry and Biophysics 461, 84–88. [DOI] [PubMed] [Google Scholar]
- Gledhill M, Buck KN. 2012. The organic complexation of iron in the marine environment: a review. Frontiers in Microbiology 3, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groussman RD, Parker MS, Armbrust EV. 2015. Diversity and evolutionary history of iron metabolism genes in diatoms. PLoS One 10, e0129081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A, Rocap G, Kroth PG, Armbrust EV, Mock T. 2015. Plastid proteome prediction for diatoms and other algae with secondary plastids of the red lineage. The Plant Journal 81, 519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gschloessl B, Guermeur Y, Cock JM. 2008. HECTAR: a method to predict subcellular targeting in heterokonts. BMC Bioinformatics 9, 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hain M, Sigman D, Haug G. 2014. 8.18 The biological pump in the past. In Holland HD, Turekian KK, eds. Treatise on geochemistry (2nd edition), Vol. 8: The oceans and marine geochemistr y. Amsterdam: Elsevier, 485–517. [Google Scholar]
- Hopes A, Nekrasov V, Kamoun S, Mock T. 2016. Editing of the urease gene by CRISPR-Cas in the diatom Thalassiosira pseudonana. Plant Methods 12, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkinson BM, Morel FM. 2009. The role of siderophores in iron acquisition by photosynthetic marine microorganisms. Biometals 22, 659–669. [DOI] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. 2007. WoLF PSORT: protein localization predictor. Nucleic Acids Research 35, W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Takagi H, Pancorbo B, Theil EC. 2001. “Opening” the ferritin pore for iron release by mutation of conserved amino acids at interhelix and loop sites. Biochemistry 40, 7525–7532. [DOI] [PubMed] [Google Scholar]
- Johnson LK, Alexander H, Brown CT. 2019. Re-assembly, quality evaluation, and annotation of 678 microbial eukaryotic reference transcriptomes. GigaScience 8, giy158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazamia E, Sutak R, Paz-Yepes J, et al. 2018. Endocytosis-mediated siderophore uptake as a strategy for Fe acquisition in diatoms. Science Advances 4, eaar4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Burki F, Wilcox HM, et al. 2014. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biology 12, e1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kustka AB, Allen AE, Morel FM. 2007. Sequence analysis and transcriptional regulation of iron acquisition genes in two marine diatoms. Journal of Phycology 43, 715–729. [Google Scholar]
- La Fontaine S, Quinn JM, Nakamoto SS, Page MD, Göhre V, Moseley JL, Kropat J, Merchant S. 2002. Copper-dependent iron assimilation pathway in the model photosynthetic eukaryote Chlamydomonas reinhardtii. Eukaryotic Cell 1, 736–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe RH, Mann EL, Cohen NR, Till CP, Thamatrakoln K, Brzezinski MA, Bruland KW, Twining BS, Marchetti A. 2018. Different iron storage strategies among bloom-forming diatoms. Proceedings of the National Academy of Sciences, USA 115, E12275–E12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanquar V, Lelièvre F, Bolte S, et al. 2005. Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. The EMBO Journal 24, 4041–4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Roche J, Boyd PW, McKay RML, Geider RJ. 1996. Flavodoxin as an in situ marker for iron stress in phytoplankton. Nature 382, 802–805. [Google Scholar]
- La Roche JL, Murray H, Orellana M, Newton J. 1995. Flavodoxin expression as an indicator of iron limitation in marine diatoms. Journal of Phycology 31, 520–530. [Google Scholar]
- Lauvset SK, Gruber N, Landschützer P, Olsen A, Tjiputra JF. 2015. Trends and drivers in global surface ocean pH over the past 3 decades. Biogeosciences 12, 1285–1298. [Google Scholar]
- Lis H, Shaked Y, Kranzler C, Keren N, Morel FM. 2015. Iron bioavailability to phytoplankton: an empirical approach. The ISME Journal 9, 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Millero FJ. 2002. The solubility of iron in seawater. Marine Chemistry 77, 43–54. [Google Scholar]
- Liu X, Theil EC. 2005. Ferritins: dynamic management of biological iron and oxygen chemistry. Accounts of Chemical Research 38, 167–175. [DOI] [PubMed] [Google Scholar]
- Lommer M, Roy AS, Schilhabel M, Schreiber S, Rosenstiel P, LaRoche J. 2010. Recent transfer of an iron-regulated gene from the plastid to the nuclear genome in an oceanic diatom adapted to chronic iron limitation. BMC Genomics 11, 718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommer M, Specht M, Roy A-S, et al. 2012. Genome and low-iron response of an oceanic diatom adapted to chronic iron limitation. Genome Biology 13, R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Merchant SS. 2008. Photo-oxidative stress impacts the expression of genes encoding iron metabolism components in Chlamydomonas. Photochemistry and Photobiology 84, 1395–1403. [DOI] [PubMed] [Google Scholar]
- Mahowald NM, Engelstaedter S, Luo C, et al. 2008. Atmospheric iron deposition: global distribution, variability, and human perturbations. Annual Review of Marine Science 1, 245–278. [DOI] [PubMed] [Google Scholar]
- Maldonado MT, Price NM. 2001. Reduction and transport of organically bound iron by Thalassiosira oceanica (Bacillariophyceae). Journal of Phycology 37, 298–310. [Google Scholar]
- Malviya S, Scalco E, Audic S, et al. 2016. Insights into global diatom distribution and diversity in the world’s ocean. Proceedings of the National Academy of Sciences, USA 113, E1516–E1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann DG. 1999. The species concept in diatoms. Phycologia 38, 437–495. [Google Scholar]
- Marchetti A, Maldonado MT, Lane ES, Harrison PJ. 2006. Iron requirements of the pennate diatom Pseudo‐nitzschia: comparison of oceanic (high‐nitrate, low‐chlorophyll waters) and coastal species. Limnology and Oceanography 51, 2092–2101. [Google Scholar]
- Marchetti A, Parker MS, Moccia LP, Lin EO, Arrieta AL, Ribalet F, Murphy ME, Maldonado MT, Armbrust EV. 2009. Ferritin is used for iron storage in bloom-forming marine pennate diatoms. Nature 457, 467–470. [DOI] [PubMed] [Google Scholar]
- Martinez J, Zhang G, Holt P, Jung H-T, Carrano C, Haygood M, Butler A. 2000. Self-assembling amphiphilic siderophores from marine bacteria. Science 287, 1245–1247. [DOI] [PubMed] [Google Scholar]
- Martínez-García A, Sigman DM, Ren H, Anderson RF, Straub M, Hodell DA, Jaccard SL, Eglinton TI, Haug GH. 2014. Iron fertilization of the Subantarctic Ocean during the last ice age. Science 343, 1347–1350. [DOI] [PubMed] [Google Scholar]
- Mawji E, Gledhill M, Milton JA, Tarran GA, Ussher S, Thompson A, Wolff GA, Worsfold PJ, Achterberg EP. 2008. Hydroxamate siderophores: occurrence and importance in the Atlantic Ocean. Environmental Science & Technology 42, 8675–8680. [DOI] [PubMed] [Google Scholar]
- McKay RML, La Roche J, Yakunin AF, Durnford DG, Geider RJ. 1999. Accumulation of ferredoxin and flavodoxin in a marine diatom in response to Fe. Journal of Phycology 35, 510–519. [Google Scholar]
- McQuaid JB, Kustka AB, Oborník M, et al. 2018. Carbonate-sensitive phytotransferrin controls high-affinity iron uptake in diatoms. Nature 555, 534–537. [DOI] [PubMed] [Google Scholar]
- Merchant S, Hill K, Howe G. 1991. Dynamic interplay between two copper-titrating components in the transcriptional regulation of cyt c6. The EMBO journal 10, 1383–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock T, Otillar RP, Strauss J, et al. 2017. Evolutionary genomics of the cold-adapted diatom Fragilariopsis cylindrus. Nature 541, 536–540. [DOI] [PubMed] [Google Scholar]
- Moreno CM, Gong W, Cohen NR, DeLong K, Marchetti A. 2020. Interactive effects of iron and light limitation on the molecular physiology of the Southern Ocean diatom Fragilariopsis kerguelensis. Limnology and Oceanography 65, 1511–1531. [Google Scholar]
- Morrissey J, Sutak R, Paz-Yepes J, et al. 2015. A novel protein, ubiquitous in marine phytoplankton, concentrates iron at the cell surface and facilitates uptake. Current Biology 25, 364–371. [DOI] [PubMed] [Google Scholar]
- Murgia I, Vazzola V, Tarantino D, Cellier F, Ravet K, Briat JF, Soave C. 2007. Knock-out of ferritin AtFer1 causes earlier onset of age-dependent leaf senescence in Arabidopsis. Plant Physiology and Biochemistry 45, 898–907. [DOI] [PubMed] [Google Scholar]
- Narayanan NN, Ihemere U, Chiu WT, Siritunga D, Rajamani S, Singh S, Oda S, Sayre RT. 2011. The iron assimilatory protein, FEA1, from Chlamydomonas reinhardtii facilitates iron-specific metal uptake in yeast and plants. Frontiers in Plant Science 2, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuester J, Vogt S, Twining BS. 2012. Localization of iron within centric diatoms of the genus Thalassiosira. Journal of Phycology 48, 626–634. [DOI] [PubMed] [Google Scholar]
- Pankowski A, McMinn A. 2009. Iron availability regulates growth, photosynthesis, and production of ferredoxin and flavodoxin in Antarctic sea ice diatoms. Aquatic Biology 4, 273–288. [Google Scholar]
- Paz Y, Katz A, Pick U. 2007. A multicopper ferroxidase involved in iron binding to transferrins in Dunaliella salina plasma membranes. Journal of Biological Chemistry 282, 8658–8666. [DOI] [PubMed] [Google Scholar]
- Peers G, Price NM. 2006. Copper-containing plastocyanin used for electron transport by an oceanic diatom. Nature 441, 341–344. [DOI] [PubMed] [Google Scholar]
- Pfaffen S, Abdulqadir R, Le Brun NE, Murphy ME. 2013. Mechanism of ferrous iron binding and oxidation by ferritin from a pennate diatom. Journal of Biological Chemistry 288, 14917–14925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffen S, Bradley JM, Abdulqadir R, Firme MR, Moore GR, Le Brun NE, Murphy ME. 2015. A diatom ferritin optimized for iron oxidation but not iron storage. Journal of Biological Chemistry 290, 28416–28427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott CC. 2006. Iron uptake in fungi: a system for every source. Biochimica et Biophysica Acta - Molecular Cell Research 1763, 636–645. [DOI] [PubMed] [Google Scholar]
- Poulsen N, Chesley PM, Kröger N. 2006. Molecular genetic manipulation of the diatom Thalassiosira pseudonana (Bacillariophyceae). Journal of Phycology 42, 1059–1065. [Google Scholar]
- Quigg A, Finkel ZV, Irwin AJ, Rosenthal Y, Ho TY, Reinfelder JR, Schofield O, Morel FM, Falkowski PG. 2003. The evolutionary inheritance of elemental stoichiometry in marine phytoplankton. Nature 425, 291–294. [DOI] [PubMed] [Google Scholar]
- Quigg A, Irwin AJ, Finkel ZV. 2011. Evolutionary inheritance of elemental stoichiometry in phytoplankton. Proceedings of the Royal Society B: Biological Sciences 278, 526–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi A, Maheswari U, Dorrell RG, et al. 2018. Integrative analysis of large scale transcriptome data draws a comprehensive landscape of Phaeodactylum tricornutum genome and evolutionary origin of diatoms. Scientific Reports 8, 4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA. 2013. Iron acquisition and allocation in stramenopile algae. Journal of Experimental Botany 64, 2119–2127. [DOI] [PubMed] [Google Scholar]
- Raven JA, Evans MC, Korb RE. 1999. The role of trace metals in photosynthetic electron transport in O2-evolving organisms. Photosynthesis Research 60, 111–150. [Google Scholar]
- Renshaw JC, Robson GD, Trinci AP, Wiebe MG, Livens FR, Collison D, Taylor RJ. 2002. Fungal siderophores: structures, functions and applications. Mycological Research 106, 1123–1142. [Google Scholar]
- Rich HW, Morel FM. 1990. Availability of well‐defined iron colloids to the marine diatom Thalassiosira weissflogii. Limnology and Oceanography 35, 652–662. [Google Scholar]
- Rossel JB, Wilson IW, Pogson BJ. 2002. Global changes in gene expression in response to high light in Arabidopsis. Plant Physiology 130, 1109–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin M, Berman-Frank I, Shaked Y. 2011. Dust-and mineral-iron utilization by the marine dinitrogen-fixer Trichodesmium. Nature Geoscience 4, 529–534. [Google Scholar]
- Sardet, C. 2015. Plankton : wonders of the drifting world. University of Chicago Press – Books. [Google Scholar]
- Sato S, Nanjappa D, Dorrell RG, et al. 2020. Genome enabled phylogenetic and functional reconstruction of an araphid pennate diatom Plagiostriata sp. CCMP470, previously assigned as a radial centric diatom, and its bacterial commensal. Scientific Reports 10, 9449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiber IF, Pilátová J, Malych R, et al. 2019. Copper and iron metabolism in Ostreococcus tauri – the role of phytotransferrin, plastocyanin and a chloroplast copper-transporting ATPase. Metallomics 11, 1657–1666. [DOI] [PubMed] [Google Scholar]
- Sétif P. 2001. Ferredoxin and flavodoxin reduction by photosystem I. Biochimica et Biophysica Acta - Bioenergetics 1507, 161–179. [DOI] [PubMed] [Google Scholar]
- Shaked Y, Kustka AB, Morel FM. 2005. A general kinetic model for iron acquisition by eukaryotic phytoplankton. Limnology and Oceanography 50, 872–882. [Google Scholar]
- Shoenfelt EM, Sun J, Winckler G, et al. 2017. High particulate iron (II) content in glacially sourced dusts enhances productivity of a model diatom. Science Advances 3, e1700314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims PA, Mann DG, Medlin LK. 2006. Evolution of the diatoms: insights from fossil, biological and molecular data. Phycologia 45, 361–402. [Google Scholar]
- Soria-Dengg S, Horstmann U. 1995. Ferrioxamines B and E as iron sources for the marine diatom Phaeodactylum tricornutum. Marine Ecology Progress Series 127, 269–177. [Google Scholar]
- Strzepek RF, Harrison PJ. 2004. Photosynthetic architecture differs in coastal and oceanic diatoms. Nature 431, 689–692. [DOI] [PubMed] [Google Scholar]
- Strzepek RF, Hunter KA, Frew RD, Harrison PJ, Boyd PW. 2012. Iron–light interactions differ in Southern Ocean phytoplankton. Limnology and Oceanography 57, 1182–1200. [Google Scholar]
- Sunda WG, Huntsman SA. 1995. Iron uptake and growth limitation in oceanic and coastal phytoplankton. Marine Chemistry 50, 189–206. [Google Scholar]
- Sunda WG, Huntsman SA. 1997. Interrelated influence of iron, light and cell size on marine phytoplankton growth. Nature 390, 389–392. [Google Scholar]
- Tanaka T, Maeda Y, Veluchamy A, et al. 2015. Oil accumulation by the oleaginous diatom Fistulifera solaris as revealed by the genome and transcriptome. The Plant Cell 27, 162–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twining BS, Baines SB, Bozard JB, Vogt S, Walker EA, Nelson DM. 2011. Metal quotas of plankton in the equatorial Pacific Ocean. Deep Sea Research Part II: Topical Studies in Oceanography 58, 325–341. [Google Scholar]
- Theil EC. 1987. Ferritin: structure, gene regulation, and cellular function in animals, plants, and microorganisms. Annual Review of Biochemistry 56, 289–315. [DOI] [PubMed] [Google Scholar]
- Vraspir JM, Holt PD, Butler A. 2011. Identification of new members within suites of amphiphilic marine siderophores. Biometals 24, 85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney LP, Lins JJ, Hughes MP, Wells ML, Chappell PD, Jenkins BD. 2011. Characterization of putative iron responsive genes as species-specific indicators of iron stress in thalassiosiroid diatoms. Frontiers in Microbiology 2, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D. 2004. A molecular timeline for the origin of photosynthetic eukaryotes. Molecular Biology and Evolution 21, 809–818. [DOI] [PubMed] [Google Scholar]
- Zaslavskaia LA, Lippmeier JC, Kroth PG, Grossman AR, Apt KE. 2000. Transformation of the diatom Phaeodactylum tricornutum (Bacillariophyceae) with a variety of selectable marker and reporter genes. Journal of Phycology 36, 379–386. [Google Scholar]
- Zhang X, Krause K-H, Xenarios I, Soldati T, Boeckmann B. 2013. Evolution of the ferric reductase domain (FRD) superfamily: modularity, functional diversification, and signature motifs. PLoS One 8, e58126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Gu W, Huang A, Wu S, Liu C, Huan L, Gao S, Xie X, Wang G. 2018. Effect of iron on the growth of Phaeodactylum tricornutum via photosynthesis. Journal of Phycology 54, 34–43. [DOI] [PubMed] [Google Scholar]